Abstract
We have modified RNase inhibitor (RI) protein so that it no longer detectably binds pancreatic RNases but retains near-native affinity for human angiogenin (ANG). The Ki value for RNase A is increased by a factor of >108, from 36 fM to >4 μM, and the selectivity factor for ANG is now >109. This dramatic change was achieved by remodeling the human RI loop segment Cys-408 -Leu-409 -Gly-410, which makes minor interactions with pancreatic RNase but does not contact ANG. The modifications selected were designed to sterically hinder docking of the undesired ligand. Three of the variants tested (C408W, G410W, and C408W/G410W) bind RNase A with almost the same avidity as WT RI. However, combination of the 408/410 double Trp replacement with deletion of the intervening residue, Leu-409, was sufficient to abolish inhibition of RNase A and human pancreatic RNase. The Ki value for ANG with the deletion variant is 1.1 fM, only 2-fold higher than with WT RI. This variant may have potential utility both as an anticancer drug targeting ANG and as a tool for the investigation of the biological function of ANG. More generally, these findings demonstrate that a protein-protein interaction can be effectively and specifically disrupted by redesigning an interface region that makes no major energetic contribution to complex stability. This finding, in turn, may have implications for the development of small molecules that modulate protein-protein interactions.
Most biological processes critically depend on accurate recognition of proteins by other proteins. Recognition is often highly selective, involving the binding of a single predominant partner by a particular protein surface or domain. However, in many cases, an individual domain must have the capacity to recognize multiple structurally related ligands to fulfill its function. This type of promiscuity can be based on recognition of a common short motif, as for the phosphorylated substrates bound by F-box proteins (1, 2) and Cbl (3). Alternatively, binding can involve extended regions of homologous proteins, as for the interactions of various hormones with different receptors (4), Cdk2 with cyclins (5), and cytosolic RNase inhibitor (RI) with members of the mammalian pancreatic RNase superfamily (see refs. 6 and 7 for reviews).
The RI-RNase system represents a particularly remarkable example of natural protein engineering. The affinity of human RI (hRI), a 50-kDa leucine-rich repeat protein, for its most avid ligand, the 14.1-kDa blood vessel-inducing RNase angiogenin (ANG), is among the highest on record for any protein-protein interaction [Ki = 0.5-0.7 fM (8, 9)]. Although nonorthologous ligands of RI share no more than ≈35% sequence identify with ANG, they all bind with Ki values that are still in the femtomolar range (7, 8, 10-12). This broad specificity may reflect the role of RI as a sentry to protect cells from all of the pancreatic RNase superfamily enzymes that inadvertently enter into the cytosol (13, 14).
In principle, RI might have achieved its relaxed selectivity during evolution by recognizing primarily those ligand elements that are highly conserved. However, the crystal structures of the complexes of porcine RI (pRI) with bovine pancreatic RNase A (15, 16) [Ki = 59 fM (10)] and hRI with human ANG (17) reveal that only seven contact residues on ANG and RNase A (of 24 in each case) are structurally analogous. Thus, although the two ligands dock similarly onto the RI ”horseshoe” (18) (Fig. 1A), occupying the central cavity and contacting the upper face of the horseshoe in the C-terminal region, the number of interactions that correspond is small. Mutational studies have shown that both interfaces do contain a common ”hot spot” (19) that provides a substantial portion of the binding energy, comprised of residues 434-438 and 460 on RI (hRI numbering) and the enzymatic active site of the ligand (20, 21). However, even here many of the specific contacts in the two complexes differ and, most strikingly, the hot spots exhibit opposite types of cooperativity (21). A recent mutational analysis of the complex of hRI with human eosinophil-derived neurotoxin (EDN, also known as RNase 2) shows even greater divergence in the structural basis for tight binding of this ligand (22).
Fig. 1.
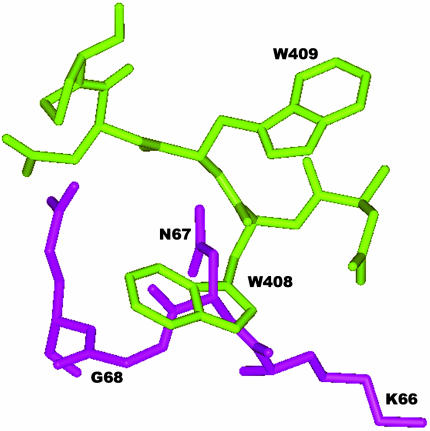
(A) The crystal structure of pRI in its complex with RNase A (16), showing the α-carbon trace, the side chains of the 28 residues that contact RNase A, and all main-chain atoms of contact residues 408-411. Hot spot residues whose replacement decreased the binding free energy by >2.5 kcal/mol (20) are shown in red, the 408-411 segment is in blue, and other contact residues are in green. (B) Region of the pRI-RNase A crystal structure containing the 408-411 segment of pRI (standard colors) and residues 66-69 of RNase A (red), with hydrogen bonds indicated by broken lines. ANG residues 66-69 (blue) from the hRI-ANG complex (17) are superimposed.
The broad specificity of RI limits its potential use as an anticancer drug targeting ANG and as a tool for the investigation of ANG function. Monoclonal antibodies and an antisense oligonucleotide against ANG have proved to be highly effective against several types of cancer in athymic mice (23, 24), and clinical studies have shown an association between ANG and cancer (e.g., refs. 25 and 26). RI itself has been used successfully against tumors in mice (27, 28), but is largely impractical as a drug for humans because of the relatively high concentrations of endogenous nontarget ligands. [The sensitivity of hRI to oxidation is another limitation, but can likely be surmounted by replacing free cysteine residues (29, 30).]
Here, we have engineered hRI to selectively reduce its affinity for pancreatic RNase, the major RNase present in human plasma and many organs (31). Our approach was to introduce modifications that would impede RNase A docking. The hRI region selected for mutation was a four-residue loop segment (408-411) that contacts RNase A but not ANG and lies adjacent to the energetic hot spot (Fig. 1 A). Remarkably, remodeling of this segment eliminated all detectable binding by pancreatic RNase but had little effect on affinity for ANG.
Experimental Procedures
Materials. WT hRI, human ANG, and ANG variants were produced in Escherichia coli as described (20, 32, 33). Recombinant EDN and human pancreatic RNase (both produced in E. coli) were provided by K. R. Acharya (University of Bath, Bath, U.K.) and A. Russo (University of Naples, Caserta, Italy), respectively. Oligonucleotides and fluorogenic substrates were from Integrated DNA Technologies (Coralville, IA). Sources of other materials and procedures for quantification of RNase A, ANG, and WT hRI are described in refs. 21 and 22. Concentrations of hRI variants were determined by titration with RNase A or EDN. EDN was quantified by titration with WT hRI (22).
Site-Directed Mutagenesis. Mutated forms of the expression plasmid pTRP-PRI (32) were generated by overlap extension PCR as described (21). Sequencing confirmed the presence of the intended mutations and the absence of any spurious changes.
Expression Plasmids for hRI Fusion Proteins. Expression plasmids for WT and variant hRI were digested with BstXI, which cleaves at a single site near the start codon. The cleaved plasmids were treated with calf intestinal phosphatase (New England Biolabs), purified with a PCR Purification Kit from Qiagen (Chatsworth, CA), and then ligated with a synthetic oligonucleotide duplex (5′-pGGCCATCATCATCATCATCATAGCAGCGGCATCGAAGGTCGTACGAGC-3′ and 5′-pGTACGACCTTCGATGCCGCTGCTATGATGATGATGATGATGGCCGCTC-3′); the resultant plasmid encodes hRI with the sequence GHHHHHHSSGIEGRT [(His)6-pep] inserted between Met-(-1) and Ser-1 and contains a new BsiWI cleavage site for screening.
Production of Variant Proteins. C408W, G410W, and C408W/G410W hRI were produced in E. coli strain W3110, purified to homogeneity, and stored as described (34). (His)6-pep-hRI fusion proteins were produced in E. coli Rosetta strain (Novagen) cells grown at 20-22°C. The bacteria were harvested 6-7 h after induction, frozen at –20°C, thawed, and resuspended at 4°C in BugBuster reagent (Novagen; 20-25 ml per liter of culture) containing 15 mM 2-mercaptoethanol. The mixture was then sonicated and ultracentrifuged as in the earlier procedure. The supernatant was applied to a column of Ni-nitrilotriacetic acid agarose (Novagen; 2-ml per liter of culture) that had been equilibrated with 50 mM sodium phosphate, 10 mM imidazole, 300 mM NaCl, pH 8.0, containing 15% (vol/vol) glycerol, and 15 mM 2-mercaptoethanol, at 4°C. The column was washed with 25 vol of the same buffer supplemented with an additional 10 mM imidazole, and elution was achieved by increasing the imidazole concentration to 250 mM. Fractions that contained inhibitory activity were supplemented with 5 mM DTT, diluted 5-fold with 10 mM Mes, 1 mM EDTA, 0.1% PEG 8000, 5 mM DTT, pH 6.0, and loaded onto a Mono Q column (HR 5/5; Amersham Pharmacia) that had been equilibrated in the same buffer. A 30-min linear gradient of 0-500 mM NaCl was then applied at a flow rate of 0.8 ml/min at ambient temperature. Partially purified inhibitor eluted at 23 min and was rechromatographed by the same method. This procedure yielded preparations that were >95% pure (as judged by SDS/PAGE), which were then dialyzed, concentrated, and stored as in the earlier procedure. The fusion proteins retained full activity after storage for at least 6 weeks.
Kinetics. Inhibition assays and measurements of kinetic parameters for RI complexes were performed in 0.1 M Mes-NaOH, 0.1 M NaCl, pH 6.0, with 10 μg/ml BSA at 25°C, unless stated otherwise. The activity of (His)6-pep-C408W/ΔL409/G410W hRI during purification was monitored in assays measuring RNase A-, T80A/Q117A-ANG-, and recombinant EDN-catalyzed cleavage of FAM-(mA)2rU(mA)2-Dabcyl (for RNase A and EDN) or FAM-(mA)2rC(mA)2-Dabcyl (for T80A/Q117A-ANG), where FAM is 6-carboxyfluorescein, mA is 2′-O-methyl-riboadenosine, rC is ribocytidine, rU is ribouridine, and Dabcyl is 4-(4-dimethylaminophenylazo)benzoic acid. The increase in fluorescence emission upon cleavage of substrate was measured as described (22, 35). The concentrations of RNase A, T80A/Q117A-ANG, and EDN were 150 pM, 2.5 nM, and 440 pM, respectively, and the corresponding substrate concentrations were 25, 100, and 25 nM. For (His)6-pep-WT hRI and the remaining hRI variants, inhibitory activity during purification was monitored only with RNase A (1.5 nM) in 20 mM Hepes-NaOH, 120 mM KCl, pH 7.2. In all cases, enzyme and inhibitor were preincubated for 5-10 min before addition of substrate.
Ki values for ANG-RI complexes cannot be determined from inhibition plots owing to the extreme tightness of binding and are calculated instead from the rate constants for dissociation and association (kd and ka, respectively). Although Ki values for some RNase A and EDN complexes can be measured directly, we used kd and ka values whenever feasible because they provide additional mechanistic information. Values of kd for ANG, RNase A, and EDN complexes were determined by following the appearance of free ligand after addition of a large molar excess of a scavenger for free inhibitor (8). Free ligand was quantified chromatographically [for ANG (20)] or by activity [RNase A (8) and EDN (22)]. Values of ka for RNase A and EDN complexes were determined by monitoring the onset of inhibition (22). The concentrations of enzyme, inhibitor, and substrate [FAM-(mA)2rU(mA)2-Dabcyl] were 2-6 pM, 50-100 pM, and 100 nM, respectively. Values of ka for ANG complexes were derived from those for EDN by adding inhibitor (final concentration 4.4 nM) to a mixture of ANG (4-6 nM) and EDN (5 nM), and measuring the partitioning of inhibitor between the two ligands in an initial rate assay with UpA (110 μM) as substrate; see refs. 11 and 36 for additional methodological details.
Ki values for all inhibitor complexes except for those of the pancreatic RNases with (His)6-pep-C408W/ΔL409/G410W hRI were calculated from kd and ka (Ki = kd/ka). Inhibition of RNase A by (His)6-pep-C408W/ΔL409/G410W hRI was assessed in assays monitoring complete cleavage of FAM-(mA)2rU(mA)2-Dabcyl (25 nM) or CpG (60 μM) (21) by 150 pM and 20 nM enzyme, respectively; the concentrations of variant used were 15 nM [with FAM-(mA)2rU(mA)2-Dabcyl] and 186-700 nM (with CpG). Inhibition of human pancreatic RNase (50 nM) was assessed with the CpG assay. Standard errors were calculated as described (21).
Irreversible Thermal Inactivation. Aliquots of inhibitor (28 nM) were incubated in assay buffer supplemented with 8 mM DTTred and 100 μg/ml BSA for 15 min at a series of temperatures from 40°C to 65°C (every 2°-3°). After 10 min of cooling on ice, inhibitor activity was measured in an assay for EDN-catalyzed cleavage of FAM-(mA)2rU(mA)2-Dabcyl (1 nM natural EDN, 0.5 nM total inhibitor, 25 nM substrate).
Modeling of RI Variants. 3D structures of hRI variants were modeled from the crystal structures of free pRI, pRI in its complex with RNase A, and hRI bound to human ANG [Protein Data Bank ID codes 1BNH (16), 1DFJ (16), and 1A4Y (17), respectively] at the swiss-model server [http://swissmodel.expasy.org (37)]. The pRI complex was used for predicting the effects of the hRI mutations on RNase A binding because the RI horseshoe opens to different extents to accommodate the two ligands, and RNase A does not dock properly to the structure of hRI from the ANG complex. Apart from the difference in cavity size, the backbone structures of RI in the two complexes are nearly indistinguishable. The sequences of pRI and hRI are 77% identical, and ≈50% of the replacements are conservative.
Results
Design of RI Variants with Increased Selectivity for ANG over RNase A. In the crystal structures of the pRI-RNase A (16) and hRI-ANG (17) complexes, the 404-411 β/α loop of RI forms multiple contacts with RNase A (Table 1), but none with ANG (Fig. 1B). The RNase A residues involved are part of a disulfide loop that is not present in ANG. The portion of the RI-RNase A interface that contains these loops lies adjacent to the energetic hot spot identified by mutagenesis (Fig. 1A) (20).
Table 1. Contacts between RI segment 408-411 and RNase A in the crystal structure of the complex.
| RI residue | Contacts on RNase A |
|---|---|
| C408 | K66: HB (Sγ-O, 2.8 Å), VDW (Sγ-C, 3.7 Å) |
| N67: VDW (O-Cβ, 3.9 Å) | |
| V409 | N67: HB (O-Nδ2, 3.1 Å), VDW (O-Cβ, 3.6 Å; O-Cγ, 3.8 Å) |
| G410 | N67: VDW (Cα-Nδ2, 3.8 Å) |
| D407 | Q69: VDW (N-Oε1, 3.6 Å; Cγ-Oε1, 3.9 Å) |
Hydrogen bond (HB) and van der Waals (VDW) contacts in the pRI-RNase A complex (16), listed with the RI atom first and RNase A atom second. Residue numbering is that for hRI. Residue 409 is a leucine in hRI.
We sought to impede binding of RNase A to hRI by altering the 408-410 segment of the 404-411 loop to generate an obstruction. Two different approaches were applied. In one, residues 408 and 410 were replaced by Trp either singly or together. The β-carbons of both Trps were expected to be directed toward the ligand, and in energy-minimized models of the C408W, G410W, and C408W/G410W variants the indole groups occupy positions that would clash with RNase A. At the same time, we considered it possible that the backbone of the RI loop and the side chains of RI and RNase A might have sufficient flexibility so that a conflict could be averted. Therefore, in our second approach, we designed an additional variant in which the loop would be more constrained: residues 408 and 410 were again substituted by Trp, but in addition, the intervening residue (Leu-409) was deleted. The rationale was that the deletion might diminish any slack in the loop and thereby force the Trp-Trp segment farther out toward the space occupied by RNase A in the WT complex. Modeling of this variant (designated C408W/ΔL409/G410W) supports this hypothesis, as discussed in detail below.
Kinetic Characterization of C408W, G410W, and C408W/G410W hRI.The C408W, G410W, and C408W/G410W variants were produced in E. coli and purified by standard methods, which use RNase A affinity chromatography as the final step. Yields were similar to that for WT, and only minor increases in Ki were observed (Table 2).
Table 2. Kinetic constants for the complexes of RNase A and hRI variants C408W, G410W, and C408W/G410W.
| hRI | kd, 10-5 s-1 | ka, 108 M-1·s-1 | Ki, fM | Ki,var/Ki,WT |
|---|---|---|---|---|
| WT* | 1.2 ± 0.1 | 2.8 ± 0.1 | 43 ± 7 | |
| C408W | 0.59 ± 0.03 | 1.07 ± 0.03 | 55 ± 3 | 1.3 |
| G410W | 0.57 ± 0.02 | 0.96 ± 0.02 | 59 ± 2 | 1.4 |
| C408W/G410W | 0.63 ± 0.02 | 0.73 ± 0.02 | 86 ± 4 | 2.0 |
Values for WT hRI are from ref. 9.
Production of C408W/ΔL409/G410W hRI with the Standard Recombinant System. RNase A affinity chromatography of extracts from bacteria expressing C408W/ΔL409/G410W hRI yielded no RNase A-inhibitory activity, suggesting that binding of the variant was drastically weakened or that an insufficient amount of inhibitor had been produced. To distinguish between these possibilities, we loaded the material that had not been retained on the RNase A column onto a column of K40G-ANG-Sepharose. [The ANG variant was used because binding of WT hRI to WT ANG is too tight to allow elution under nondenaturing conditions; the affinities of K40G-ANG and WT RNase A for WT hRI are similar (21).] No protein was obtained under the standard conditions used for inhibitor elution. To determine whether any inhibitor had remained bound to either column, the resins were boiled in SDS/PAGE sample buffer, and the eluates were analyzed by Western blotting with polyclonal anti-hRI. The K40G-ANG resin yielded a band at the same molecular weight as hRI (corresponding to <1 μg per liter of culture), whereas the RNase A resin did not. These observations suggested that C408W/ΔL409/G410W hRI has the intended functional properties (low affinity for RNase A, high affinity for ANG), but that a different production and purification system would be required to obtain adequate amounts of protein for characterization.
Development of an Expression System for (His)6-pep-hRI Fusion Proteins. To provide a simple method for inhibitor purification that is not based on activity, we inserted into the hRI cDNA a sequence encoding (His)6-pep, a 15-aa peptide that contains a hexahistidine segment for isolation by nickel-chelate chromatography. The peptide is attached to the N terminus of the protein, which extends away from the ”bottom” face of the horseshoe (Fig. 1 A), i.e., the face opposite from that where ligands bind; the extension also contains a factor Xa recognition site so that the peptide could be removed if it were found to hinder ligand binding. An additional change was the use of a bacterial strain that has supplemental tRNAs for improved efficiency of translation. When this system was used for the WT fusion protein, the yield of inhibitory activity from the Ninitrilotriacetic acid column was equivalent to ≈500 μg of hRI per liter, i.e., ≈2-fold higher than with the standard system. The protein was only ≈10% pure at this stage, but became homogeneous after Mono Q chromatography. The Ki values for the complexes of the fusion protein with the three ligands tested were similar or identical to those determined previously for hRI, i.e., 36 vs. 43 fM with RNase A, 0.5 vs. 0.5 fM with ANG, and 4.4 vs. 2.7 fM with EDN (9, 22).
Production of (His)6-pep-C408W/ΔL409/G410W hRI. When bacteria expressing (His)6-pep-C408W/ΔL409/G410W were grown at the standard temperature of 37°C, the yields were only 2.5-5 μg/liter of culture, as judged by Western blotting. However, yields increased to 50 μg/liter when the temperature was lowered to 20-22°C after induction. Addition of up to 20 μl of the Ni-nitrilotriacetic acid eluate had no effect on the activity of RNase A (at 150 pM in a volume of 2 ml), whereas 10 μl was sufficient to abolish the activity of the ANG variant T80A/Q117A (at 2.5 nM) (Fig. 2). [WT ANG was not used in these assays because its activity is extremely weak; T80A/Q117A is ≈100-fold more active (33) and its affinity for hRI is similar to that of WT.] The inhibitor preparation obtained after Mono Q chromatography was again >95% pure.
Fig. 2.
Effects of (His)6-pep-C408W/ΔL409/G410W hRI preparation from Ni-nitrilotriacetic acid column on the enzymatic activities of T80A/Q117A-ANG (▴) and RNase A (•). The volumes of eluate indicated were taken from a total of 1.2 ml that contained the majority of the variant from 1 liter of bacterial culture.
Kinetic Characterization of (His)6-pep-C408W/ΔL409/G410W hRI.Concentrations of deletion variant as high as 0.7 μM failed to inhibit RNase A detectably. Because replicate determinations of reaction velocity in the presence and absence of inhibitor showed <5% variation, we estimate that an amount of inhibition as low as 20% could have been measured reliably. Therefore, the lower limit in the Ki value for (His)6-pep-C408W/ΔL409/G410W with RNase A is 4 μM, representing an increase of at least 8 orders of magnitude with respect to the WT fusion protein (Table 3). The variant (again at up to 0.7 μM) also did not affect the activity of human pancreatic RNase. In contrast, its affinity for ANG is nearly the same as that of WT: kd values are indistinguishable (Fig. 3), whereas the ka value for the variant is ≈2-fold lower (Table 3). Binding to EDN also remains extremely tight, with Ki in this case increased by a factor of 3.3, reflecting changes in both kd and ka (Table 3 and Fig. 3).
Table 3. Kinetic constants for the complexes of (His)6-pep-hRI fusion proteins with RNase A, ANG, and EDN.
| Enzyme | hRI | kd, S-1 | ka, M-1·S-1 | Ki, M | Ki,var/Ki,WT | ΔΔG, kcal/mol |
|---|---|---|---|---|---|---|
| RNase A | WT* | (8.0 ± 0.3) × 10-6 | (2.2 ± 0.1) × 108 | (3.6 ± 0.2) × 10-14 | ||
| RNase A | C408W/ΔL409/G410W | — | — | >4 × 10-6 | >100,000,000 | >10.9 |
| ANG | WT | (8.1 ± 1.7) × 10-8 | (1.5 ± 0.1) × 108 | (5.4 ± 1.2) × 10-16 | ||
| ANG | C408W/ΔL409/G410W | (8.7 ± 1.1) × 10-8 | (7.6 ± 0.2) × 107 | (1.1 ± 0.2) × 10-15 | 2.0 | 0.3 ± 0.1 |
| EDN | WT | (6.9 ± 0.3) × 10-7 | (1.5 ± 0.1) × 108 | (4.6 ± 0.4) × 10-15 | ||
| EDN | C408W/ΔL409/G410W | (1.1 ± 0.1) × 10-6 | (7.2 ± 0.1) × 107 | (1.5 ± 0.1) × 10-14 | 3.3 | 0.7 ± 0.1 |
Natural EDN was used.
The preparation of WT hRI fusion protein used for these measurements was obtained from bacteria grown at the same temperature as the variant.
Fig. 3.
Dissociation of the complexes of WT and C408W/ΔL409/G410W hRI fusion proteins with ANG (• and ▴, respectively) and EDN (▾ and ▪, respectively). Data were normalized with respect to the amount of dissociation at the first time point (3-10% of the total). Lines are fits of the data to a single-exponential decay.
Irreversible Thermal Denaturation of WT and C408W/ΔL409/G410W hRI Fusion Proteins. Profiles for the irreversible loss of inhibitory activity at elevated temperatures were determined to assess the effect of the loop remodeling on structural integrity and stability. The WT fusion protein retained full potency up to 52°C and then lost essentially all activity as the temperature was raised to 59°C, with the midpoint at 55°C. For the deletion variant, the transition was lowered only slightly (midpoint 52°C).
Discussion
In principle, three general approaches can be used to modulate protein-protein interactions so as to improve ligand selectivity: (i) introduction of new interactions that are specific for the target ligand; (ii) elimination of interactions that are specific for the undesired ligand(s); and (iii) introduction of mismatches (steric, charge, or hydrophilicity) with respect to the nontarget ligand. For conversion of RI from a broadly specific RNase inhibitor to one that is highly selective for ANG, the first of these strategies is not feasible. The ka values of the hRI complexes, a major determinant of RI distribution among its ligands in vivo, are all close to the theoretical upper limit for a protein-protein complex (see ref. 38). Moreover, the kd value for the ANG complex is already so small (t1/2 = 2-3 months) that any appreciable decrease would be extremely challenging to measure experimentally.
The second strategy has been used to design variants of human growth hormone (hGH) that bind specifically to either the hGH or the prolactin receptor (39). Each of the shifts in binding preference (factors of 34,000 and 150 toward the hGH and prolactin receptors, respectively) was achieved by truncation of two side chains that only participate in recognition by the nontarget receptor. Although this approach was successful for hGH, mutational studies on hRI complexes (9, 20, 21) have revealed no promising avenues for applying it to this system. The only single-site mutations in hRI shown to increase the Ki for RNase A by more than a factor of 20 are those involving the 434-437/460 hot spot (where increases range from 84- to 23,000-fold). The impacts on ANG binding are for the most part less dramatic, but none are negligible. Moreover, the combined effects of mutations on affinity for RNase A are subadditive, whereas with ANG they are superadditive. Thus, for example, the losses in affinity for RNase A and ANG with Y434A/Y437A are similar although the effects of the individual 434 and 437 replacements on ANG binding are 93-and 20-fold smaller than on that of RNase A. These opposite types of cooperativities are widely operative in the two complexes (9) and would probably undermine any attempts to develop substantial differential effects by accumulating large sets of mutations.
Examination of the 3D structures of the pRI-RNase A and hRI-ANG complexes reveals some ways in which the third strategy, the introduction of ligand-specific clashes, might be implemented. Three RI residues (Lys-320, Glu-401, and Arg-457) form salt links with RNase A residues (Glu-86, Arg-39, and Asp-38, respectively) that are not conserved in ANG, and we initially considered the possibility that the creation of charge mismatches at these positions would hinder RNase A binding. However, testing of the E401R hRI showed there was no significant loss in affinity for RNase A (unpublished results). In addition, D38R-RNase A was reported to retain high sensitivity to RI (40). As an alternative, we sought to identify interface regions where enlargement of RI might produce a steric clash with RNase A, but not with ANG. Raines and coworkers (40) have shown that substitution of an Arg for Gly-88 of RNase A, which forms close contacts with Trp-261 and Trp-263 of RI, increases Ki by nearly 4 orders of magnitude. Enlargement of the RI side of this interface region is problematic because most of the RI residues that approach Gly-88 or its neighbors on the RNase A already bear large side chains (Trp, Glu, Lys) and participate in fixed secondary structures. Moreover, ANG forms even more extensive contacts with this region of RI than does RNase A, and any modifications that interfere with RNase A binding would probably diminish affinity for ANG as well.
Our analysis identified only one RI region, the 404-411 β/α loop, that seems to possess the requisite characteristics to serve as a target for successful steric restriction of ligand selectivity (Fig. 1B). Residues 408-411 pack against the 66-69 segment of RNase A (Table 1), whereas the closest point of approach with ANG is 7.8 Å. Three of the loop modifications that we designed were simple replacements of small residues (Cys-408 and Gly-410, both singly and together) by bulky tryptophans. In all cases, the Ki for RNase A was increased by <2-fold. Interestingly, association of these variants was 2- to 3-fold slower than for WT, suggesting that the Trps might present an initial impediment that is resolvable through minor conformational adjustments.
In designing the fourth RI loop variant, we aimed to introduce a more constrained obstruction. Addition of residues to the loop was rejected because it seemed likely that this would increase the number of conformational options available. Instead, we retained the two tryptophans, but deleted the intervening residue to tighten up the loop. Homology modeling and energy minimization of the C408W/ΔL409/G410W variant predicted that the deletion would not perturb the structural elements that anchor the ends of the loop, and that the modified loop would clash severely with RNase A (Fig. 4). In the superposition with the pRI-RNase A crystal structure, the 406-408 main chain extends 4-5 Å farther out toward the ligand than does 406-409 of WT hRI. The indole group of Trp-408 overlaps all four backbone atoms of RNase A Asn-67 and lies too close to main-chain atoms of Lys-66 and Gly-68 and the side chain of Asn-67; the Cβ, Cα, and O atoms of Trp-408 and NH of Trp-409 also bump against the Asn-67 side chain. Trp-409 occupies a position similar to that of Val/Leu-409 (pRI/hRI), which forms part of the hydrophobic core of the inhibitor. If the backbone of this loop is now relatively well fixed, then RNase A can only dock onto the inhibitor if the conformation of the RNase A segment 66-68 changes dramatically. Although these RNase A residues are on a loop, their backbone structure is highly constrained by hydrogen bonds between β2 (61-64) and β3 (70-74) and between Cys-65 O and Asn-67 NH.
Fig. 4.
Model of C408W/ΔV409/G410W pRI (green) superimposed onto the crystal structure of RNase A (purple) in the pRI-RNase A complex (16).
Consistent with the modeling predictions, RNase A was not inhibited to any measurable extent by C408W/ΔL409/G410W hRI, whereas ANG (which is >4 Å from the variant in the modeled complex) binds with affinity similar to that of WT. The increase in Ki for the RNase A complex is, minimally, 8 orders of magnitude, corresponding to a loss of >60% of the 18.3 kcal/mol binding energy of the WT complex. The resultant selectivity factor for ANG vs. RNase A is >3 × 109 (compared to ≈60 for WT hRI). We are not aware of any precedent for such an enormous shift in specificity achieved through protein engineering, whether by rational approaches or random selection methods. Stoop and Craik (41) have recently increased the selectivity of the serine protease inhibitor ecotin for plasma kallikrein by factors as high as 4 × 105 through partial randomization of residues on the four surface loops that contact the protease.
A detailed understanding of the inability of C408W/ΔL409/G410W hRI to bind RNase A must await the determination of the 3D structure of this variant. One theoretical alternative to the steric obstruction hypothesis presented here would be that the RI-RNase A interactions (Table 1) that are disrupted by the loop remodeling contribute a large fraction of the binding energy. Several pieces of direct and indirect evidence, taken together, rule this out: (i) neither of the hydrogen bonds is energetically important, as demonstrated here [and previously (20)] for the Cys-408-Lys-66 interaction and inferred for the Val(Leu)-409-Asn-67 interaction from the high B factor of the Asn-67 group involved (see ref. 17); (ii) this part of the interface accounts for <10% of the total solvent-accessible surface area buried in the complex (and most of the atoms buried are polar); and (iii) the interactions of other interface regions adequately account for the total binding energy of the complex (9). Although the present model of the variant structure provides a satisfactory explanation for the experimental findings, other plausible models can be envisioned. Indeed, when the free pRI structure, rather than pRI from the RNase A complex, is used as the starting point for modeling, Trp-408 again clashes badly with RNase A but Trp-409 extends toward the ligand rather than into the interior of RI.
Like RNase A, human pancreatic RNase [Ki = 200 fM for WT hRI (12)] is not inhibited detectably by C408W/ΔL409/G410W hRI. Thus, all physiologically relevant binding to the most prevalent human RNase has been eliminated. The only other human RNase that might compete effectively with ANG for WT RI in blood and various tissues is EDN, but here the effect of remodeling was small, and the preference factor for ANG increased only from 8.5 to 14 (Table 3). The region of (free) EDN analogous to the RNase A loop that approaches RI residues 408-411 adopts a similar shape (42), despite a low degree of sequence conservation. Thus, the high avidity of EDN for the hRI deletion variant suggests that the precise ways in which EDN and RNase A dock to RI may differ. This suggestion is consistent with mutagenesis results showing that the 434-437/460 region of hRI is less important for binding EDN than the other two ligands (22). Additional mutational and structural studies may reveal ways in which EDN binding can be curtailed without weakening that of ANG, either by eliminating unique contacts or creating another steric hindrance.
The findings reported here may have more general implications for the modulation of protein-protein interactions by protein engineering or the design or selection of small molecule effectors. We have demonstrated that an extremely large change in affinity and selectivity can be achieved by modifying an interface region that contributes little to complex stability. This strategy may be effective for diminishing avidity in other cases where attention has hitherto focused primarily on energetic hot spots. For proteins that have broad ligand specificity based on recognition of short common motifs such as phosphodegrons (2, 3), it may also provide a means to restrict specificity by remodeling outlying regions that have distinct spatial relationships with various ligands.‡ Finally, our results raise the possibility that steric interference with protein-protein interactions can also be achieved by targeting small molecules to nonhotspot regions.
Acknowledgments
We thank Drs. K. R. Acharya and A. Russo for generously supplying recombinant EDN and human pancreatic RNase, respectively, and Drs. B. L. Vallee and J. F. Riordan for helpful discussions. This work was supported by National Institutes of Health Grant CA88738 (to R.S.).
Abbreviations: RI, RNase inhibitor; hRI, human RI; pRI, porcine RI; ANG, angiogenin; EDN, eosinophil-derived neurotoxin; FAM, 6-carboxyfluorescein; mA, 2′-O-methyl-riboadenosine; rU, ribouridine; Dabcyl, 4-(4-dimethylaminophenylazo)benzoic acid.
Footnotes
In a recent report (43), the selectivity of the protease inhibitor eglin for furin versus PC7 was enhanced by a factor of ≈80 through optimization of ”adventitious” contacts outside the core interface region that involves similar active site residues on the two proteases. The mutations of potential contact residues on the inhibitor were random, and it is unclear to what extent, if any, the improvement reflects steric hindrance of nontarget ligand binding.
References
- 1.Skowyra, D., Craig, K. L., Tyers, M., Elledge, S. J. & Harper, J. W. (1997) Cell 91, 209-219. [DOI] [PubMed] [Google Scholar]
- 2.Orlicky, S., Tang, X. J., Willems, A., Tyers, M. & Sicheri, F. (2003) Cell 112, 243-256. [DOI] [PubMed] [Google Scholar]
- 3.Lupher, M. L., Rao, N., Eck, M. J. & Band, H. (1999) Immunol. Today 20, 375-382. [DOI] [PubMed] [Google Scholar]
- 4.Wells, J. A. & de Vos, A. M. (1996) Annu. Rev. Biochem. 65, 609-634. [DOI] [PubMed] [Google Scholar]
- 5.Jeffrey, P. D., Russo, A. A., Polyak, K., Gibbs, E., Hurwitz, J., Massague, J. & Pavletich, N. P. (1995) Nature 376, 313-320. [DOI] [PubMed] [Google Scholar]
- 6.Lee, F. S. & Vallee, B. L. (1993) Prog. Nucleic Acid Res. Mol. Biol. 44, 1-30. [DOI] [PubMed] [Google Scholar]
- 7.Hofsteenge, J. (1997) in Ribonucleases: Structures and Functions, eds. D'Alessio, G. & Riordan, J. F. (Academic, New York), pp. 621-658.
- 8.Lee, F. S., Shapiro, R. & Vallee, B. L. (1989) Biochemistry 28, 225-230. [DOI] [PubMed] [Google Scholar]
- 9.Shapiro, R., Ruiz-Gutierrez, M. & Chen, C. Z. (2000) J. Mol. Biol. 302, 497-519. [DOI] [PubMed] [Google Scholar]
- 10.Vicentini, A. M., Kieffer, B., Matthies, R., Meyhack, B., Hemmings, B. A., Stone, S. R. & Hofsteenge, J. (1990) Biochemistry 29, 8827-8834. [DOI] [PubMed] [Google Scholar]
- 11.Shapiro, R. & Vallee, B. L. (1991) Biochemistry 30, 2246-2255. [DOI] [PubMed] [Google Scholar]
- 12.Boix, E., Wu, Y., Vasandani, V. M., Saxena, S. K., Ardelt, W., Ladner, J. & Youle, R. J. (1996) J. Mol. Biol. 257, 992-1007. [DOI] [PubMed] [Google Scholar]
- 13.Beintema, J. J., Schuller, C., Irie, M. & Carsana, A. (1988) Prog. Biophys. Mol. Biol. 51, 165-192. [DOI] [PubMed] [Google Scholar]
- 14.Haigis, M. C., Kurten, E. L. & Raines, R. T. (2003) Nucleic Acids Res. 31, 1024-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kobe, B. & Deisenhofer, J. (1995) Nature 374, 183-186. [DOI] [PubMed] [Google Scholar]
- 16.Kobe, B. & Deisenhofer, J. (1996) J. Mol. Biol. 264, 1028-1043. [DOI] [PubMed] [Google Scholar]
- 17.Papageorgiou, A. C., Shapiro, R. & Acharya, K. R. (1997) EMBO J. 16, 5162-5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kobe, B. & Deisenhofer, J. (1993) Nature 366, 751-756. [DOI] [PubMed] [Google Scholar]
- 19.Clackson, T. & Wells, J. A. (1995) Science 267, 383-386. [DOI] [PubMed] [Google Scholar]
- 20.Chen, C. Z. & Shapiro, R. (1997) Proc. Natl. Acad. Sci. USA 94, 1761-1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen, C. Z. & Shapiro, R. (1999) Biochemistry 38, 9273-9285. [DOI] [PubMed] [Google Scholar]
- 22.Teufel, D. P., Kao, R. Y. T., Acharya, K. R. & Shapiro, R. (2003) Biochemistry 42, 1451-1459. [DOI] [PubMed] [Google Scholar]
- 23.Olson, K. A., Fett, J. W., French, T. C., Key, M. E. & Vallee, B. L. (1995) Proc. Natl. Acad. Sci. USA 92, 442-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Olson, K. A., Byers, H. R., Key, M. E. & Fett, J. W. (2001) Clin. Cancer Res. 7, 3598-3605. [PubMed] [Google Scholar]
- 25.Shimoyama, S., Gansauge, F., Gansauge, S., Negri, G., Oohara, T. & Beger, H. G. (1996) Cancer Res. 56, 2703-2706. [PubMed] [Google Scholar]
- 26.Etoh, T., Shibuta, K., Barnard, G. F., Kitano, S. & Mori, M. (2000) Clin. Cancer Res. 6, 3545-3551. [PubMed] [Google Scholar]
- 27.Polakowski, I. J., Lewis, M. K., Muthukkaruppan, V. R., Erdman, B., Kubai, L. & Auerbach, R. (1993) Am. J. Pathol. 143, 507-517. [PMC free article] [PubMed] [Google Scholar]
- 28.Botella-Estrada, R., Malet, G., Revert, F., Dasi, F., Crespo, A., Sanmartin, O., Guillen, C. & Alino, S. F. (2001) Cancer Gene Ther. 8, 278-284. [DOI] [PubMed] [Google Scholar]
- 29.Kim, B. M., Schultz, L. W. & Raines, R. T. (1999) Protein Sci. 8, 430-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stumpp, M. T., Forrer, P., Binz, H. K. & Pluckthun, A. (2003) J. Mol. Biol. 332, 471-487. [DOI] [PubMed] [Google Scholar]
- 31.Weickmann, J. L. & Glitz, D. G. (1982) J. Biol. Chem. 257, 8705-8710. [PubMed] [Google Scholar]
- 32.Lee, F. S. & Vallee, B. L. (1989) Biochem. Biophys. Res. Commun. 160, 115-120. [DOI] [PubMed] [Google Scholar]
- 33.Shapiro, R. (1998) Biochemistry 37, 6847-6856. [DOI] [PubMed] [Google Scholar]
- 34.Lee, F. S. & Vallee, B. L. (1990) Biochemistry 29, 6633-6638. [DOI] [PubMed] [Google Scholar]
- 35.Kelemen, B. R., Klink, T. A., Behlke, M. A., Eubanks, S. R., Leland, P. A. & Raines, R. T. (1999) Nucleic Acids Res. 27, 3696-3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee, F. S., Auld, D. S. & Vallee, B. L. (1989) Biochemistry 28, 219-224. [DOI] [PubMed] [Google Scholar]
- 37.Schwede, T., Kopp, J., Guex, N. & Peitsch, M. C. (2003) Nucleic Acids Res. 31, 3381-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schreiber, G. & Fersht, A. R. (1996) Nat. Struct. Biol. 3, 427-431. [DOI] [PubMed] [Google Scholar]
- 39.Cunningham, B. C. & Wells, J. A. (1991) Proc. Natl. Acad. Sci. USA 88, 3407-3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leland, P. A., Schultz, L. W., Kim, B. M. & Raines, R. T. (1998) Proc. Natl. Acad. Sci. USA 95, 10407-10412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stoop, A. A. & Craik, C. S. (2003) Nat. Biotechnol. 21, 1063-1068. [DOI] [PubMed] [Google Scholar]
- 42.Swaminathan, G. J., Holloway, D. E., Veluraja, K. & Acharya, K. R. (2002) Biochemistry 41, 3341-3352. [DOI] [PubMed] [Google Scholar]
- 43.Komiyama, T., VenderLugt, B., Fugere, M., Day, R., Kaufman, R. J. & Fuller, R. S. (2003) Proc. Natl. Acad. Sci. USA 100, 8205-8210. [DOI] [PMC free article] [PubMed] [Google Scholar]