Abstract
Type III secretion systems enable plant and animal bacterial pathogens to deliver virulence proteins into the cytosol of eukaryotic host cells, causing a broad spectrum of diseases including bacteremia, septicemia, typhoid fever, and bubonic plague in mammals, and localized lesions, systemic wilting, and blights in plants. In addition, type III secretion systems are also required for biogenesis of the bacterial flagellum. The HrcQB protein, a component of the secretion apparatus of Pseudomonas syringae with homologues in all type III systems, has a variable N-terminal and a conserved C-terminal domain (HrcQB-C). Here, we report the crystal structure of HrcQB-C and show that this domain retains the ability of the full-length protein to interact with other type III components. A 3D analysis of sequence conservation patterns reveals two clusters of residues potentially involved in protein–protein interactions. Based on the analogies between HrcQB and its flagellum homologues, we propose that HrcQB-C participates in the formation of a C-ring-like assembly.
In plant pathogens, the type III secretion system, termed Hrp, is required for pathogenicity on susceptible plants and is also essential for induction of a defense reaction called the hypersensitive response, on resistant plants (1–4). The Hrp system consists of a secretion apparatus encoded by ≈25 hrp genes and its main function is to deliver virulence proteins (effectors) into plant cells (5–7). A broadly conserved subset of the hrp genes (designated hrc) appears to encode the core components of the delivery apparatus (2). It has been proposed that these core components are involved in the recognition of the secretion signals carried by the effectors and are thus responsible for the promiscuous character of type III systems for heterologous secreted proteins (8–10).
The HrcQB protein from Pseudomonas syringae belongs to the conserved core of the secretion apparatus of the type III system, with homologues in Erwinia, Xanthomonas, Ralstonia, and also in Yersinia, Shigella, Salmonella, and the flagellum (2). Notably, sequence similarity among the HrcQB homologues is restricted to their C-terminal regions (1). This conservation together with the variability of the N termini led to the proposal that these proteins may mediate interactions between the conserved and the species-specific components of type III systems (1).
The conserved C-terminal domain of HrcQB (HrcQB-C; residues 50–128) from P. syringae pv. phaseolicola was cloned in Escherichia coli and its structure was determined by x-ray crystallography. To our knowledge, this is the first structure reported for a conserved protein of the type III secretion apparatus.
Methods
Protein Expression and Data Collection. HrcQB-C comprises 84 amino acids, of which the first five have been introduced by the construct, and the remaining 79 correspond to residues 50–128 of the full-length protein (HrcQ2, SwissProt-ALL accession no. O85094). The expression, purification, and crystallization of HrcQB-C has been reported (11). For structure determination, Leu-72 was replaced by a methionine and the protein was expressed by the Met-auxotrophic E. coli strain B834 in the presence of selenomethionine. The anomalous dispersion of the incorporated selenium was exploited in a multiwavelength anomalous dispersion experiment. Diffraction data were collected at three wavelengths (Table 1) from a single, frozen crystal at the EMBL Hamburg Outstation (Deutsches Elektronen Synchrotron) and processed with the hkl package (12). A native data set collected at 2.3-Å resolution at the European Synchrotron Radiation Facility was used for the refinement.
Table 1. Summary of crystallographic analysis.
| MAD data sets
|
||||
|---|---|---|---|---|
| Native data | Peak | Remote | Inflection | |
| Space group | P21 | |||
| Unit cell dimensions | a = 52.9 b = 27.7 c = 99.0, Å | |||
| α = 90.0 β = 99.8 γ = 90.0, ° | ||||
| Content of the asymmetric unit | four molecules | |||
| Data collection and processing | ||||
| Beamlines | BM14 (ESRF) | BW7A (DESY) | BW7A (DESY) | BW7A (DESY) |
| Wavelength, Å | 0.9800 | 0.9760 | 0.9221 | 0.9770 |
| Resolution, Å | 39.0-2.30 (2.42-2.30) | 26.0-2.50 (2.54-2.50) | 31.0-2.67 (2.72-2.67) | 35.0-2.69 (2.47-2.69) |
| Mosaicity, ° | 0.8 | 1.5 | 1.5 | 1.5 |
| Rsym, % | 5.8 (15.6) | 8.1 (19.9) | 7.6 (27.9) | 7.6 (31.0) |
| I/σ(I) | 9.3 (4.6) | 17.1 (9.5) | 21.0 (9.6) | 16.3 (7.3) |
| Unique reflections | 12,352 | 10,064 | 8,578 | 8,673 |
| Multiplicity | 3.4 (1.8) | 4.9 (3.9) | 5.4 (5.0) | 4.2 (4.1) |
| Completeness, % | 98.2 (88.6) | 91.7 (51.3) | 96.5 (88.2) | 94.5 (84.0) |
| Phasing | ||||
| No. of sites | 4 | |||
| Mean figure of merit from experimental (multiwavelength anomalous dispersion) phasing | 0.33 | |||
| Refinement and analysis of molecular model | ||||
| Resolution, Å | 2.30 | |||
| R/Rfree, % | 18.2/22.8 | |||
| Atoms modelled (protein/water) | 2,056/153 | |||
| rmsd bond lengths, Å | 0.009 | |||
| rmsd angles, ° | 1.261 | |||
Values in parentheses are for the highest—resolution shell. Rsym = ΣhΣj∥Ih,j — Îh∥/Σh ΣjIh,j, for the intensity I of j observations of reflection h. R = Σh∥Fo∥ — ∥Fo∥/Σh ∥Fo∥ and Rfree is calculated for a randomly chosen subset of reflections that are omitted from refinement. rmsd, rms deviation.
Structure Determination and Refinement. The Se sites were identified by the program solve (13). The experimental phases were improved by density modification techniques, including iterative noncrystallographic symmetry averaging and solvent flattening, and most of the model was autobuild and rebuild by resolve (14). Several rounds of refinement using cns (15) and refmac5 (16) and manual rebuilding resulted in the final model (Table 1), which includes residues 55–126 of chain A, residues 55–125 of chains B and C, and residues 54–125 of chain D.
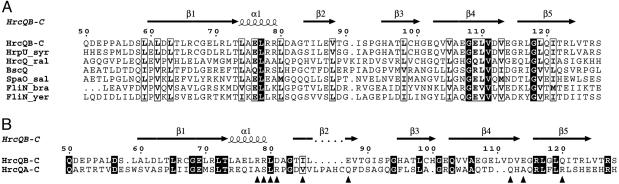
Multiple Sequence Alignment. Proteins used in the multiple sequence alignment shown in Fig. 3A are P. syringae pv. syringae HrpU (HrpU_syr, SwissProt-ALL accession no. Q60235, 89% identity), Ralstonia solanacearum HrcQ (HrcQ_ral, Q52489, 33% identity), Yersinia pestis FliN (FliN_yer, Q8ZF94, 29% identity), Bradyrhizobium japonicum FliN (FliN_bra, BAC52267, 33% identity), Salmonella typhimurium SpaO (SpaO_sal, Q56022, 26% identity), and Bordetella bronchiseptica BscQ (BscQ, Q9L9E0, 32% identity).
Fig. 3.
Sequence alignments. The numbering scheme corresponds to the sequence of the full-length HrcQB, and the secondary structural elements shown at Upper are those of the HrcQB-C structure. Alignment was performed with clustal w (18) and plotted with the espript program (19). Strictly conserved residues are highlighted, and similar residues are boxed. (A) Sequence alignment of the HrcQB-C and the C termini of homologous proteins identified by a blasta search with an identity of 26% or higher. Three pairs of sequences from plant pathogens, animal pathogens, and flagella are included. The pairs are chosen to be as different from each other as possible. The proteins aligned are listed in Methods. (B) Sequence alignment between the HrcQB-C and the C-terminal region (residues 154–235) of the HrcQA. Triangles indicate positions corresponding to HrcQB-C residues that participate in the dimer–dimer interface.
Yeast Two-Hybrid System. The hrcQA gene was inserted into pAS2-1 plasmid to create an in-frame fusion with the GAL4 DNA-binding domain. The hrcQB gene was cloned in the pACT2 plasmid and fused with the transcription activation domain of GAL4. Plasmids were transformed into Saccharomyces cerevisiae strain PJ69-4A carrying three different GAL4-responsive reporter genes (ade, his, and lacZ), each driven by a different GAL4-responsive promoter, to reduce the incidence of false positives. The interaction of hybrid proteins was examined by growing the transformed cells on complete minimal medium lacking Trp, Leu, Ade, and His, for 3 days at 30°C. Plasmids pVA3-1 and pTD1-1 are DNA-binding domain and activation domain fusion plasmids that provide a positive control for interacting proteins.
Immunoblotting and Far Western Blot Analysis. Purified proteins were electrophoresed through SDS-polyacrylamide gel and transferred to poly(vinylidene difluoride) (PVDF) membrane. For immunoblotting, the membrane was blocked in a solution of Tris-buffered saline/0.1% Tween 20/5% nonfat milk, incubated with the appropriate antibodies and the signal was detected by nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate substrates of AP-conjugate. For Far Western blot analysis, the membrane was immersed with 8 M urea in 20 mM Tris·HCl, pH 7.5/60 mM NaCl/10 mM MgCl2/0.1 mM EDTA/5% glycerol/0.02% Nonidet P-40. Proteins absorbed to the membrane were renatured and the membrane was blocked as described above and incubated overnight at 4°C with 35S-labeled protein. The signal was detected by autoradiography. Rabbit polyclonal antisera specific for HrcQA and HrcQB were used for the immunoblotting assays.
Subcellular Fractionation Experiment. P. syringae pv. phaseolicola NPS3121 cells were grown for 14 h in hrp-inducing medium and separated into culture supernatant and cell-bound fraction by centrifugation at 10,000 × g. The supernatant was passed through a 0.22-μm filter and concentrated by centricon-10 (Amicon). Both fractions were analyzed by SDS/PAGE and immunoblotting. The cell-bound fraction was further fractionated into a cytoplasmic and a total membrane fraction by ultracentrifugation at 65,000 × g for 40 min. The total membrane fraction was mixed with urea or NaCl or Triton X-114 and incubated on ice for 30 min before ultracentrifugation at 65,000 × g for 40 min. Both the supernatant and the pellet were analyzed by SDS/PAGE, followed by immunoblot analysis.
Results and Discussion
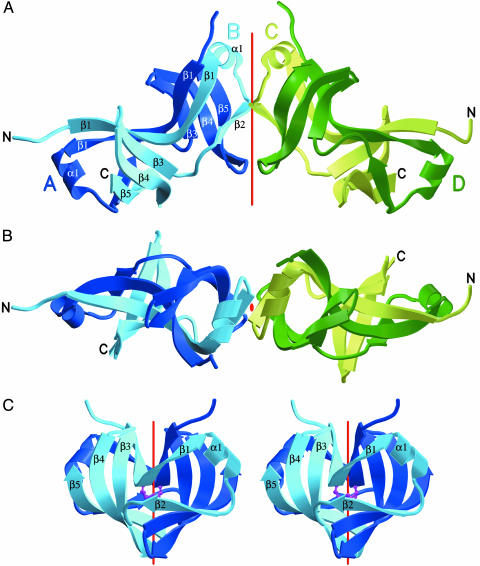
Overview of the Structure. The crystal structure was determined by multiwavelength anomalous dispersion and refined to 2.3-Å resolution. Table 1 shows details of the data collection, processing, and refinement statistics. As shown in Fig. 1 A and B, HrcQB-C is an elongated, gently curved homotetramer with approximate dimensions of 25 × 30 × 90 Å3. The four monomers assemble into two tightly bound homodimers that are packed together to form a dimer of dimers. The dimers associate with each other in such a way that the angle between their major axes of inertia is ≈120°. The secondary structure of each monomer consists of five β-strands (labeled β1-β5 in Fig. 1) and one short helix (labeled α1). Two monomers (A and B or C and D chains in Fig. 1C) fold together in a symmetrical manner to form a compact and intertwined dimeric structure that comprises two six-stranded antiparallel β-sheets bridged through a long β-ribbon (β1-β1 in Fig. 1 A) that spans the length of the whole dimer and is shared by the β-sheets (Fig. 2). The β-ribbon connects the monomers through an extended array of hydrogen bonds and is further stabilized by a disulfide bond between the symmetry related cysteines 67 (Fig. 1C). This covalent bond (being completely buried in the interior of the molecule) remains intact in the crystals despite the presence of reducing agent (10 mM 1,4 dithiothreitol). Strands β3, β4, and β5 of monomer A and strand β2 of B (Fig. 1 A) are successively hydrogen bonded and together with the first half of the β-ribbon form the complete six-stranded β-sheet.
Fig. 1.
A schematic representation of the HrcQB-C structure. Each chain is individually colored and labeled. (A) A view of the complete tetramer. The dimers are related by a two-fold axis that is indicated by a red line. (B) A view of the tetramer along the two-fold axis. The molecule is rotated 90° relative to A. (C) A stereoview of the HrcQB-C dimer. The red line indicates the local two-fold axis. Symmetry related cysteines that form the disulfide bond are shown as a ball-and-stick model.
Fig. 2.
A stereoview of the 2Fo – Fc electron density map contoured at 1.5 σ with the final model overlaid on it. The region shown corresponds to part of the sheet formed by the strands β1 (chain A), β1 (chain B), and β3 (chain B).
The dimer–dimer interface consists of a six-stranded antiparallel β-barrel formed by the β2 strands of chains B and C and the β4, β5 strands of chains A and D (Fig. 1 A and B). A total surface area of 1,228 Å2 is buried on tetramer formation through dimer association. In addition, the tetramer is stabilized by hydrogen bonds formed between carbonyl and amino groups of the protein backbone at the positions of Ile-85 of B and C chains and Asp-112 and Glu-114 of A and D. The Stokes' radius of the protein in solution, which was estimated by size exclusion chromatography to be ≈28.5 Å, is in excellent agreement with the value of 29.1 Å as calculated (17) from the crystal structure of the whole tetramer.
Sequence Conservations and Identification of Functional Residues. HrcQB-C shares a sequence identity ranging between 33% and 89% with the C termini of homologues from other phytopathogenic bacteria. It also has an identity between 29% to 33% with the flagellar FliN proteins and between 28% to 32% with homologues from animal pathogens. A multiple sequence alignment of these homologues reveals well conserved individual amino acids and extended regions of high similarity (Fig. 3A). Four residues, Leu-77, Gly-108, Val-111, and Gly-118, are strictly conserved among all aligned sequences. Glycines, in particular, located on strands β4 and β5, respectively, are completely buried into the hydrophobic core of the HrcQB-C, in a tightly packed region where there is no space for side chains to be accommodated. The great majority of the other conserved residues are hydrophobic; mainly leucines. The conservation pattern extensively coincides with the secondary structure elements implying fold similarities among the aligned sequences.
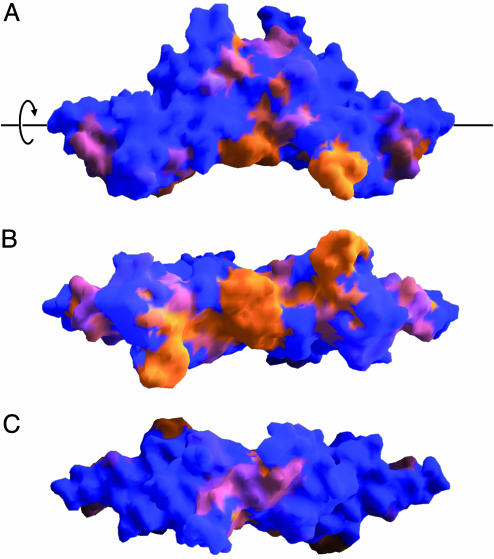
The structure of HrcQB-C together with a multiple sequence alignment was used in a 3D cluster analysis procedure (20), in an attempt to identify putatively functional residues. For this alignment, only proteins from plant-associated bacteria with an identity of 47% or better were used. Two clusters of conserved, mainly surface-exposed residues (Fig. 4) are found. The first of them (residues 71–79) comprises amino acids from the end of strand β1 and the whole helix α1, whereas the second one (residues 94, 95, and 108–121) is more extended. As shown in Fig. 4, the distribution of these conserved residues on the molecular surface is markedly asymmetric, with the majority of them being concentrated on the concave side of the tetramer. Some of the residues (78, 79, 111, 112, 114, and 120) of these clusters participate in the formation of the tetramer interface. The remaining residues of the clusters could have functional roles, e.g., interactions with other proteins.
Fig. 4.
Molecular surface of the HrcQB-C that is colored according to the degree of amino acid conservation as calculated by the 3D cluster analysis program 3dca (20). A multiple sequence alignment (identity of 47% or better) among the proteins P. syringae pv. syringae HrpU, P. syringae pv. tomato HrcQb, Pseudomonas fluorescens RscQB, E. amylovora HrcQ, and Pantoea agglomerans HrcQB was performed. The coloring code ranges from blue for nonconserved to orange for highly conserved residues. Three views of the molecule, related by rotations about a horizontal axis (indicated as a black line in A) are presented. (A) Side view. (B) Concave surface (rotated 90° clockwise relative to A). (C) Convex surface (rotated 90° counterclockwise relative to A).
Interactions of HrcQB-C with Other Type III Components. It has been previously shown by electron microscopy that the type III secretion apparatus is a supramolecular structure (21). In this context, and considering the indications for the existence of protein–protein interfaces, we investigated potential interactions between the HrcQB and other type III components, through genetic (yeast two-hybrid system) and biochemical (Far Western and immunoprecipitation) experiments.
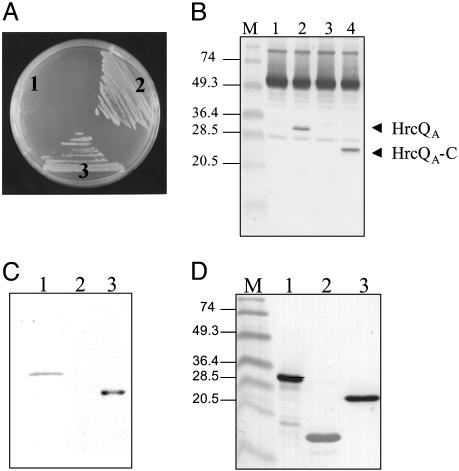
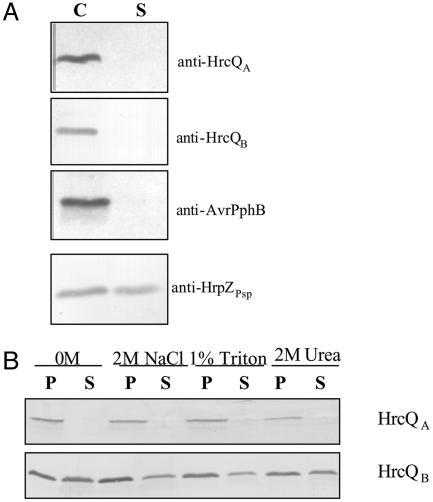
Our studies demonstrate that in the P. syringae secretion apparatus: (i) HrcQB strongly binds to HrcQA, a protein encoded by a gene immediately upstream of hrcQB, (Fig. 5A), (ii) HrcQB-C retains the ability of the full-length protein to interact with HrcQA (Fig. 5B), and (iii) only the C terminus (residues 66–238) of HrcQA is involved in the association with HrcQB (Fig. 5 C and D). The HrcQB–HrcQA interaction is consistent with the observation that these proteins are expressed as a single product, named HrcQ (22), in the genera Erwinia, Xanthomonas, and Ralstonia. We have also performed subcellular localization experiments (Fig. 6) that show that neither HrcQB nor HrcQA are found in the culture supernatant of P. syringae cells, indicating that they are not secreted components. The same experiments showed that HrcQB fractionates in both cytoplasmic and membrane extracts suggesting that is cytoplasmic or loosely associated with a membrane component. In addition, HrcQA is exclusively found in the membrane fraction; this finding is consistent with the presence of a transmembrane domain that is predicted on the basis of sequence data (data not shown). Our results are fully consistent with earlier findings for flagellar system: (i) the FliN/FliM proteins, which are the flagellum counterparts of HrcQB/HrcQA, interact with each other (23–26), (ii) the C terminus of the FliN protein (which is homologous to HrcQB-C) is sufficient for both flagellar assembly and function (24), (iii) the C terminus of FliM is sufficient for binding FliN (25), and (iv) the FliN–FliM complex is also localized in the cytoplasm. Its attachment to the inner membrane-embedded MS ring is established through FliM (23–27); this result could indicate analogous roles between the FliM and HrcQA, which fractionates from the membrane. In addition, it has been shown that FliN/FliM association results in the formation of a multiprotein, ring-shaped assembly, known as C-ring (23–26), in which, most likely, the C-terminal parts of both proteins participate.
Fig. 5.
HrcQB–HrcQA protein interactions. (A) Yeast two-hybrid analysis of HrcQB--HrcQA protein interaction (for details see Methods). Yeast cells expressing the hybrid proteins DBD-HrcQA and activation domain HrcQB (area 3) were grown in the absence of Ade and His and formed red colonies, as did the positive control cells, indicating that the hybrid proteins interact with each other. Plasmids pVA3-1 and pTD1-1 were used as positive controls, (area 2) and the vectors pAS2-1 and pACT2 without inserts served as negative controls (area 1). (B) HrcQB-C is involved in the association with the HrcQA. Wild-type and mutant HrcQA proteins were mixed in equimolar amounts with HrcQB-C and were incubated for 6 h at 4°C, then antibody specific for HrcQB (1:5,000) was added and was further incubated overnight at 4°C. Immunocomplexes were precipitated with protein A-agarose at 4°C overnight. The immunoprecipitates were separated by SDS/PAGE and visualized by immunoblotting with HrcQA-specific antibody. Lane 1, HrcQB; lane 2, HrcQA; lane 3, HrcQA-N (residues 1–65); lane 4, HrcQA-C (residues 66–238). Prestained molecular mass markers (M) in kDa are indicated on the left. The arrows indicate the HrcQA and HrcQA-C proteins that coimmunoprecipitated with HrcQB-C. (C) An autoradiogram that shows the binding of HrcQB to His-10-HrcQA protein and to the C-terminal truncated version. Lane 1, HrcQA; lane 2, HrcQA-N; lane 3, HrcQA-C. Far Western blot analysis was performed as described in Methods. The membrane was probed with 35S-labeled HrcQB and subjected to autoradiography. (D) The presence of proteins on the membrane shown in C was confirmed by Western blot analysis using anti-His-AP conjugate (1:1,000). Prestained molecular mass markers (M) in kDa are indicated on the left.
Fig. 6.
Subcellular localization of HrcQB and HrcQA proteins in P. syringae pv. Phaseolicola. (A) Cell-bound (C) and cell-free supernatant (S) fractions (see Methods) were subjected to SDS/PAGE and immunoblot analysis with antisera specific for HrcQA, HrcQB, HrpZ, and AvrPphB. The proteins HrpZ and AvrPphB are used as positive and negative controls (secreted and nonsecreted in culture), respectively. (B) The effects of washing with NaCl, urea, or Triton X-114 on the membrane association of HrcQA and HrcQB. P and S indicate the supernatants pellets and from ultracentrifugation, respectively (see Methods). Molar concentrations of NaCl, urea, and Triton X-114 are indicated above the respective panels.
Based on the striking analogies of HrcQB/HrcQA with FliN/FliM, we propose that in the P. syringae Hrp secretion apparatus, HrcQB-C and the C terminus of the HrcQA, may associate to form a cytoplasmic assembly analogous to the flagellar C-ring. This hypothesis is consistent with electron microscopy studies (21, 28) that show significant morphological similarities between type III systems of animal pathogens and the flagellum, implying the presence of a common architectural theme present in type III systems. Based on the sequence similarities observed between FliN and FliM, it has been suggested that the two proteins might share some common structural features that participate in the formation of the C-ring, possibly by means of the occupation of quasiequivalent positions (25). Consistent with this hypothesis, we also observe a significant homology (20.5% identity and 54.5% similarity; Fig. 3B) between the HrcQB-C and the C-terminal segment of HrcQA with a concomitant agreement on the predicted (29) secondary structure (data not shown). A complete coincidence is found between the predicted and experimentally determined secondary structural elements for residues 170–185 of HrcQA and the homologous HrcQB-C segment (residues 65–80), respectively; this HrcQB-C segment contains a cluster of residues possibly involved in protein–protein interfaces (Fig. 4).
This structure determination of a component of the bacterial type III secretion apparatus, highly conserved among plant, animal, and human pathogens, has broad implications in the understanding at an atomic level of bacterial pathogenesis and in drug design.
Acknowledgments
We thank Dr. P. Tucker for advice on data collection at the Deutsches Elektronen Synchrotron and Dr. C. Trinh for assistance with data collection at the European Synchrotron Radiation Facility. This work was partially supported by grants in the framework of the Access to Research Infrastructure/Improving Human Potential and the Marie Curie Fellowship programs of the European Union.
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1O9Y).
References
- 1.Hueck, C. J. (1998) Microbiol. Mol. Biol. Rev. 62, 379–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alfano, J. R. & Collmer, A. (1997) J. Bacteriol. 179, 5655–5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Galán, J. E. & Collmer, A. (1999) Science 284, 1322–1328. [DOI] [PubMed] [Google Scholar]
- 4.Büttner, D. & Bonas, U. (2000) EMBO J. 21, 5313–5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jin, Q. & He, S.-Y. (2001) Science 294, 2556–2558. [DOI] [PubMed] [Google Scholar]
- 6.Stebbins, C. E. & Galán, J. E. (2001) Nature 414, 77–81. [DOI] [PubMed] [Google Scholar]
- 7.Luo, Y., Bertero, M. G., Frey, E. A., Pfuetzner, R. A., Wenk, M. R., Creagh, L., Marcus, S. L., Lim, D., Sicheri, F., Kay, C., et al. (2001) Nat. Struct. Biol. 8, 1031–1036. [DOI] [PubMed] [Google Scholar]
- 8.Rossier, O., Wengelnik, K., Hahn, K. & Bonas, U. (1999) Proc. Natl. Acad. Sci. USA 96, 9368–9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collmer, A., Badel, J. L., Charkowski, A. O., Deng, W. L., Fouts, D. E., Ramos, A. R., Rehm, A. H., Anderson, D. M., Schneewind, O., van Dijk, K., et al. (2000) Proc. Natl. Acad. Sci. USA 97, 8770–8777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anderson, D. M., Fouts, D. E., Collmer, A. & Schneewind, O. (1999) Proc. Natl. Acad. Sci. USA 96, 12839–12834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fadouloglou, V. E., Tampakaki, A. P., Panopoulos, N. J. & Kokkinidis M. (2001) Acta Crystallogr. D 57, 1689–1691. [DOI] [PubMed] [Google Scholar]
- 12.Otwinowski, Z. & Minor, W. (1997) Methods Enzymol. 276, 307–326. [DOI] [PubMed] [Google Scholar]
- 13.Terwilliger, T. C. & Berendzen, J. (1999) Acta Crystallogr. D 55, 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Terwillinger, T. C. (2000) Acta Crystallogr. D 56, 965–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905–921. [DOI] [PubMed] [Google Scholar]
- 16.Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997) Acta Crystallogr. D 53 240–255. [DOI] [PubMed] [Google Scholar]
- 17.García de la Torre, J., Huertas, M. L. & Carrasco, B. (2000) Biophys. J. 78, 719–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thomson, J. D., Higgins, D. G. & Gibson, T. J. (1997) Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gouet, P., Courcelle, E., Stuart, D. I. & Metoz, F. (1999) Bioinformatics 15, 305–308. [DOI] [PubMed] [Google Scholar]
- 20.Landgraf, R., Xenarios, I. & Eisenberg, D. (2001) J. Mol. Biol. 307, 1487–1502. [DOI] [PubMed] [Google Scholar]
- 21.Kubori, T., Matsushima, Y., Nakamura, D., Uralil, J., Lara-Tejero, M., Sukhan, A., Galan, J. E. & Aizawa, S. I. (1998) Science 280, 602–605. [DOI] [PubMed] [Google Scholar]
- 22.Charkowski, A. O., Huang, H. C. & Collmer, A. (1997) J. Bacteriol. 179, 3866–3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Francis, N. R., Sosinsky, G. E., Thomas, D. & DeRosier, D. J. (1994) J. Mol. Biol. 235, 1261–1270. [DOI] [PubMed] [Google Scholar]
- 24.Tang, H., Billings, S., Wang, X., Sharp, L. & Blair, D. F. (1995) J. Bacteriol. 177, 3496–3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mathews, M. A. A., Tang, H. L. & Blair, D. F. (1998) J. Bacteriol. 180, 5580–5590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao, R., Pathak, N., Jaffe, H., Reese, T. S. & Khan, S. (1996) J. Mol. Biol. 261, 195–208. [DOI] [PubMed] [Google Scholar]
- 27.Lloyd, S. A., Whitby, F. G., Blair, D. F. & Hill, C. P. (1999) Nature 400, 472–475. [DOI] [PubMed] [Google Scholar]
- 28.Tamano, K., Aizawa, S., Katayama, E., Nonaka, T., Imajoh-Ohmi, S., Kuwae, A., Nagai, S. & Sasakawa, C. (2000) EMBO. J. 19, 3876–3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McGuffin, L. J., Bryson, K. & Jones, D. T. (2000) Bioinformatics 16, 404–405. [DOI] [PubMed] [Google Scholar]