Abstract
Because G-protein coupled receptors (GPCRs) continue to represent excellent targets for the discovery and development of small-molecule therapeutics, it is posited that additional protein components of the signal transduction pathways emanating from activated GPCRs themselves are attractive as drug discovery targets. This review considers the drug discovery potential of two such components: members of the “regulators of G-protein signaling” (RGS protein) superfamily, as well as their substrates, the heterotrimeric G-protein α subunits. Highlighted are recent advances, stemming from mouse knockout studies and the use of “RGS-insensitivity” and fast-hydrolysis mutations to Gα, in our understanding of how RGS proteins selectively act in (patho)physiologic conditions controlled by GPCR signaling and how they act on the nucleotide cycling of heterotrimeric G-proteins in shaping the kinetics and sensitivity of GPCR signaling. Progress is documented regarding recent activities along the path to devising screening assays and chemical probes for the RGS protein target, not only in pursuits of inhibitors of RGS domain-mediated acceleration of Gα GTP hydrolysis but also to embrace the potential of finding allosteric activators of this RGS protein action. The review concludes in considering the Gα subunit itself as a drug target, as brought to focus by recent reports of activating mutations to GNAQ and GNA11 in ocular (uveal) melanoma. We consider the likelihood of several strategies for antagonizing the function of these oncogene alleles and their gene products, including the use of RGS proteins with Gαq selectivity.
I. Introduction
A. Biological and Pharmaceutical Importance of G-Protein Coupled Receptor Signaling
For a cell to adapt to its environment, it must be able to receive extracellular cues and then elicit an appropriate intracellular response to those cues. Although there are multiple receptor families (i.e., receptor tyrosine kinases, ion channels, nuclear receptors), G-protein-coupled receptors (GPCRs1) represent the largest and most pharmacologically important family. Approximately 1% of the human genome is dedicated to these receptors (Takeda et al., 2002; Fredriksson et al., 2003; Vassilatis et al., 2003), and nearly a third of the pharmaceuticals currently on the market target one or more of these receptors (Jacoby et al., 2006; Overington et al., 2006; Lagerström and Schiöth, 2008). In addition to being the largest component of the “druggable” proteome, GPCRs are also responsible for our ability to perceive the visual, olfactory, and gustatory cues in our environment. Missense or truncation mutations to individual codons in genes encoding GPCRs result in a myriad of pathological conditions, including color blindness, retinitis pigmentosa, pseudohermaphroditism, and Hirschsprung's disease (Spiegel and Weinstein, 2004). Given the importance of GPCRs in both pathologic conditions and treatment of disease, it is critical that we comprehensively understand these receptors and their downstream signaling components.
At the most basic level, GPCRs consist of seven α-helical transmembrane stretches with an extracellular N terminus and an intracellular C terminus. These diverse receptors can be further divided into subfamilies named by their hallmark member: glutamate-, rhodopsin-, adhesion-, frizzled-, and secretin-like (Fredriksson et al., 2003; Perez, 2003). Although the precise mechanism of activation of the heterotrimeric G-protein probably varies from family to family and remains elusive, in simplest terms upon binding of a hormone, neurotransmitter, ion, or other stimuli, the GPCR undergoes conformational changes that allow the activation of the Gα-GDP/Gβγ complex. Upon the binding of an activating ligand, the GPCR catalyzes the release of GDP and subsequent binding of GTP on the Gα subunit (Gilman, 1987; Johnston and Siderovski, 2007; Oldham and Hamm, 2008).
B. The Classic Guanine Nucleotide Cycle of Heterotrimeric G-Protein Subunits
Heterotrimeric G-proteins act as molecular switches that are considered in the off state when bound to GDP and in the on state (“activated”) when GTP-bound. In the basal state, the GDP-bound Gα subunit is in complex with the Gβγ dimer (Fig. 1). The Gα/Gβγ interaction serves to enhance localization to the membrane, to enhance coupling, and to slow the spontaneous dissociation of GDP (so-called “GDP dissociation inhibitory” function that reduces basal activity) (Brandt and Ross, 1985; Higashijima et al., 1987; Robillard et al., 2000; Evanko et al., 2001). Upon an agonist-induced conformational change, the receptor acts as a GEF resulting in the displacement of GDP and subsequent binding of GTP (which is in higher abundance). The nucleotide pocket of the heterotrimeric G-protein α subunit is surrounded by three flexible switch regions that undergo dramatic conformational changes depending on nucleotide state (Bohm et al., 1997; Wall et al., 1998). The binding of GTP and subsequent change in the switch regions results in the dissociation of the GTP-bound Gα from Gβγ. At this point, the activated Gα subunit and the Gβγ obligate heterodimer are able to interact with effectors such as adenylyl cyclase, phospholipase C isoforms, RhoGEFs, and ion channels (Clapham and Neer, 1997; Kozasa et al., 1998; Simonds, 1999; Kammermeier et al., 2000; Rhee, 2001; Lutz et al., 2007).
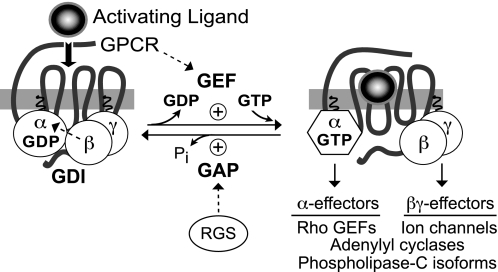
Fig. 1.
The standard model of guanine nucleotide cycle of G-protein coupled receptors. The heterotrimeric G-protein consists of a GDP-bound Gα subunit associated with the Gβγ heterodimer. The Gβγ serves not only to assist the coupling of Gα to the GPCR, but also as a guanine nucleotide dissociation inhibitor (GDI) for Gα, preventing the release of GDP. Upon binding of an activating ligand to the receptor, conformation changes result in the GPCR acting as a GEF, causing the release of GDP and subsequent binding of GTP. This exchange of bound nucleotide results in the dissociation of Gβγ and both Gα-GTP and Gβγ are free to signal to downstream effectors. Downstream effectors are activated until the GTP is hydrolyzed by the intrinsic GTP hydrolysis activity of the Gα subunit [which can be further accelerated by particular downstream effectors such as PLCβ1 and p115RhoGEF (Berstein et al., 1992; Kozasa et al., 1998)]. Upon hydrolysis of GTP, Gα-GDP rebinds Gβγ and the system returns to the inactive state. The rate of GTP hydrolysis can be dramatically enhanced by RGS proteins, which serve as GAPs for Gα subunits in vitro (Berman et al., 1996) and in vivo (Lambert et al., 2010).
C. Structural Determinants of G-Protein Subunit Function
1. Gα Subunit.
The Gα subunit, in its inactive state, binds GDP within a nucleotide-binding pocket circumscribed by residues derived from both of its constituent domains: a Ras-like domain (resembling the structural fold of “small” G-proteins) and an all α-helical domain unique to the “large” Gα family, comprising a structurally distinct six-helix bundle (Fig. 2A). An extended N-terminal α-helix is modified by covalent attachment of the fatty acids myristate and/or palmitate, which facilitates membrane targeting as well as assembly with Gβγ subunits (Wedegaertner et al., 1995). Exchange of GDP for GTP is catalyzed in a poorly understood process by an activated GPCR acting as a GEF for the Gα-GDP/Gβγ heterotrimer (Johnston and Siderovski, 2007; Oldham and Hamm, 2008). This receptor-catalyzed nucleotide exchange results in nucleotide-pocket residues interacting with the γ-phosphoryl group of the newly bound GTP (Lambright et al., 1994; Posner et al., 1998) and causes a structural rearrangement within three switch regions (I–III) of Gα (Fig. 2B). The particular conformations of these three switch regions are critical to the protein/protein interactions that Gα makes with its nucleotide-state-selective binding partners, such as Gβγ, effectors, RGS proteins, and GoLoco motifs (Bohm et al., 1997; Tesmer et al., 1997a; Wall et al., 1998; Willard et al., 2004b).
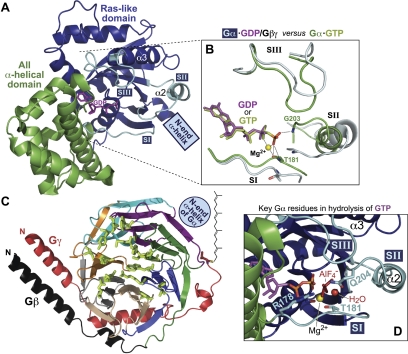
Fig. 2.
Structural features of the heterotrimeric G-protein subunits. A, overall structural fold of the heterotrimeric G-protein Gα subunit in its inactive, GDP-bound form. The Gα subunit (PDB number 1GP2) is composed of a Ras-like domain (blue) and an all α-helical domain (green), between which is found the guanine nucleotide binding pocket (GDP in magenta). The three flexible switch regions (SI, SII, and SIII) are highlighted in cyan. B, details of structural differences between GDP- and GTP-bound states. The additional (third) phosphoryl group (orange and red) of bound GTP establishes contacts with residues Thr-181 and Gly-203 of switches I and II, respectively, thus leading to changes in all three switch regions (green; PDB number 1GIA) versus their conformation in the GDP-bound state (cyan; PDB number 1GP2). Magnesium cation is highlighted in yellow. C, overall structural fold of the Gβγ heterodimer. The Gβγ subunit (PDB number 1OMW) is colored to highlight the seven WD40 repeats that comprise the β-propeller fold: WD1, green; WD2, purple; WD3, cyan; WD4, orange; WD5, gray; WD6, wheat; and WD7; blue. The cysteine residue within Gβγ (red) that receives post-translational geranylgeranylation is highlighted in sticks configuration. The relative positioning of the N-terminal α-helix of the Gα subunit (when in the Gα-GDP/Gβγ heterotrimeric complex) is also highlighted. D, structural basis of GTP hydrolysis by Gα. Residues within Gα that are critical to the GTP hydrolysis mechanism include Arg-178 and Thr-181 from switch I and Gln-204 from switch II (colored as in A and numbered as in Gαi1; coordinates are from PDB number 1GFI). Magnesium cation is highlighted in yellow. The planar anion AlF4−, which mimics the γ-phosphate leaving group of the GTP → GDP + Pi hydrolysis reaction, is depicted in metallic red.
2. Gβγ Dimer.
Gβ and Gγ subunits form tightly associated heterodimers (Fig. 2C). Gβ begins with an extended N-terminal α-helix and is composed mainly of a β-propeller fold formed by seven individual segments of a ∼40-amino acid sequence known as the WD-40 repeat. Gγ is an extended stretch of two α-helices joined by an intervening loop. Assuming no significant tertiary structure on its own, the N terminus of Gγ participates in a coiled-coil interaction with the N-terminal α-helix of Gβ (Fig. 3); much of the remainder of Gγ binds along the outer edge of the Gβ toroid (Wall et al., 1995; Sondek et al., 1996). Gγ is prenylated post-translationally on a cysteine residue found four amino acids from the C terminus. Most Gγ subunits receive a 20-carbon geranylgeranyl group at this position (as illustrated in Fig. 2C), whereas Gγ1, Gγ8, and Gγ11 receive a 15-carbon farnesyl group instead (Wedegaertner et al., 1995). Such lipid modification facilitates membrane localization of the Gβγ heterodimer that is important to receptor coupling. GDP-bound Gα and the Gβγ dimer form the G-protein heterotrimer via two principal sites of interaction: 1) extensive burial of the β3/α2 loop and α2 helix (switch II) of Gα within six of the seven WD repeats of Gβ, and 2) contact between the side of the first β-propeller blade of Gβ and the extended N-terminal helix of Gα (see Fig. 2C) (Bohm et al., 1997; Wall et al., 1998). These extensive interactions form the basis for competition for Gβγ binding between Gα-GDP and βγ-effectors. Structures of Gβγ bound to the regulatory protein phosducin, the receptor kinase (and “βγ-effector”) GRK2, and SIRK/SIGK [peptides capable of disrupting βγ-effector activation (Scott et al., 2001)] have shown that the effector-binding site on Gβγ overlaps significantly with the region responsible for binding switch II of Gα near the central pore of the Gβ torus (Gaudet et al., 1996; Lodowski et al., 2003; Bonacci et al., 2006).
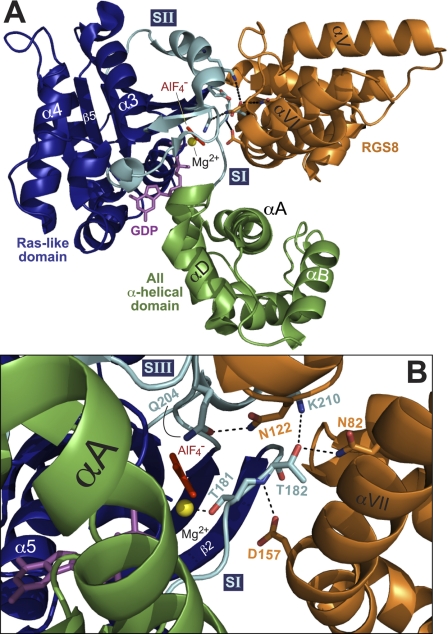
Fig. 3.
RGS proteins stabilize the transition state of Gα subunits and coordinate the positioning of the Gα catalytic glutamate residue that is critical to intrinsic GTPase activity. Diagram of the RGS8/Gαi3 structure [PDB number 2ODE (Soundararajan et al., 2008)], as rendered using PyMOL (Schrödinger, Inc., Portland, OR). The all-helical subdomain of Gαi3 is shown in green, whereas the Ras-like nucleotide binding domain in shown in dark blue. The three flexible switch regions (SI, SII, and SIII) are highlighted in cyan. The guanine nucleotide, AlF4−, and Mg2+ are highlighted in magenta, red, and yellow, respectively, whereas RGS8 is illustrated in orange. A, the RGS8 Gα-binding interface consists primarily of the SI and SII regions of Gαi3. B, the Asn-122 amide forms a hydrogen bond with Gln-204 of Gαi3, orienting it to help stabilize the planar leaving group, whereas the Asn-82 of RGS8 forms contacts with side-chain carbonyl of Thr-182, allowing the side-chain carbonyl to make a contact with the SII Lys-210 of Gαi3, stabilizing SI and SII in their transition state orientations. In addition, Asp-157 of RGS8 stabilizes the backbone amine of Thr-182, allowing the Thr-181 side-chain hydroxyl group to stabilize the Mg2+ ion (yellow).
3. Structural Basis for Intrinsic GTP Hydrolysis Activity by Gα Subunits.
The mechanism of GTP hydrolysis by Gα has been discerned from X-ray diffraction crystallographic structures, especially of the Gα transition state-mimetic form (i.e., Gα bound to GDP and AlF4−) (Coleman et al., 1994), as well as hydrolysis reaction intermediates, including Gα bound to guanosine 5′-(βγ-imido)triphosphate or GDP plus inorganic phosphate (Raw et al., 1997; Coleman and Sprang, 1999). The GTP hydrolysis reaction is mediated by three conserved Gα amino acids (Fig. 2D). Gln-204 in switch II (residues numbered as found in Gαi1) coordinates the critical nucleophilic water molecule responsible for hydrolysis of the γ-phosphate, whereas Arg-178 and Thr-181 (both from switch I) help to stabilize the leaving group (as mimicked by the planar anion AlF4−), the latter residue coordinating a bound Mg2+ ion (Coleman et al., 1994).
4. Structural Features of Regulators of G-Protein Signaling—the Gα GTPase-Accelerating Proteins.
The GTP hydrolysis activity intrinsic to the Gα subunit was initially thought to control the lifetime of G-protein α subunits in their GTP-bound state and the in vitro kinetics of GTP hydrolysis observed by Gαs supported this hypothesis (Cassel et al., 1979); however, intrinsic rates of GTP hydrolysis measured in vitro could not account for the fast deactivation kinetics seen with other G-proteins in the cellular context. For instance, purified transducin, which is the heterotrimeric G-protein that couples to the phototransduction GPCR rhodopsin, hydrolyzes GTP with a t1/2 of ∼15 s; however, the rate of retinal deactivation is <1 s (Vuong and Chabre, 1991). In addition, G-protein coupled inwardly rectifying potassium channels (GIRKs), which are activated by Gβγ freed from Gαi subunits, are deactivated 100 times faster than would be predicted based on the intrinsic GTP hydrolysis rate exhibited by Gαi subunits in vitro (Breitwieser and Szabo, 1988; Yatani et al., 1988). The first evidence that the cycle of nucleotide binding and hydrolysis by Gα subunits could be modulated by binding partners other than Gβγ came from the report of Berstein et al. (1992) demonstrating that the Gαq/11 effector phospholipase C (PLC) β1 could also increase the rate of GTP hydrolysis by Gαq/11. Although PLCβ1 seemed to have paradoxical roles, being both an effector and a GTPase-accelerating protein (GAP) for Gαq/11, this report provided an early demonstration of a GAP for heterotrimeric G-protein α subunits, although GAPs had been known for Ras-family GTPases for at least 5 years before this (Trahey and McCormick, 1987). The first evidence of noneffector GAPs for heterotrimeric G-proteins came from a yeast-based genetic screen for mutants that increased sensitivity of Saccharomyces cerevisiae to α-factor pheromone. These screens identified two primary factors, supersensitive-1 (Sst1) and supersensitive-2 (Sst2), that made yeast supersensitive to α-factor (Chan and Otte, 1982a,b). In these initial studies, Sst1 acted as a “barrier” inhibiting the diffusion of α-factor in solution (Chan and Otte, 1982b). Consistent with this initial description, Sst1 (also known as Bar1) is now known to encode an extracellular protease that degrades α-factor in the environment (MacKay et al., 1988). Although the molecular details of Sst2's function remained enigmatic for 7 more years (unlike Sst1's protease activity), Sst2 was speculated to inhibit the pheromone response in the intracellular compartment (Chan and Otte, 1982b). Once the components of the pheromone pathway had been rigorously elucidated (Hartwell, 1980; Dietzel and Kurjan, 1987; Miyajima et al., 1987; Nakayama et al., 1988), work by Dohlman et al. (1995) demonstrated that the overexpression of the yeast Gα subunit (GPA1) suppressed the pheromone supersensitivity of Sst2 mutant yeast, but overexpression of Gβ subunit was not able to suppress the phenotype. Although these experiments were not able to demonstrate conclusively a direct binary interaction between Sst2 and GPA1, they helped establish the groundwork for discovery of a then-novel family of negative regulators of GPCR signaling by multiple groups studying multiple different systems, from Caenorhabditis elegans body-bending and egg-laying behaviors to human T- and B-lymphocyte immediate-early gene activation programs (De Vries et al., 1995; Berman et al., 1996; Druey et al., 1996; Hunt et al., 1996; Koelle and Horvitz, 1996; Siderovski et al., 1996; Watson et al., 1996).
This large family of Gα-directed GAPs, the “regulators of G-protein signaling” (RGS proteins), is characterized by the presence of a nine-α-helix bundle that binds most avidly to the Gα transition state for GTP hydrolysis (Tesmer et al., 1997a; Slep et al., 2001; Soundararajan et al., 2008). The nine helices are subdivided into two subdomains, the first of which is composed of helices αI, II, III, VIII, and IX; the remaining subdomain comprises helices αIV, V, VI, and VII, with each subdomain arranged in antiparallel helical bundles (Tesmer et al., 1997a; Slep et al., 2001; Soundararajan et al., 2008). Unlike the GAPs for small G-proteins that contribute a critical arginine (or other residue) into the active site for nucleotide hydrolysis (Vetter and Wittinghofer, 2001), RGS proteins do not contribute any single residue to the nucleotide binding pocket that is directly necessary for the catalytic mechanism (and that can be eliminated yet still retain Gα-binding affinity). Their catalytic activity has been established by X-ray diffraction crystallography and NMR structures of isolated RGS proteins, as well as RGS protein/Gα protein complexes (Tesmer et al., 1997a; Moy et al., 2000; Slep et al., 2001; Slep et al., 2008; Soundararajan et al., 2008; Kimple et al., 2009). RGS proteins are selective for binding to the transition state of Gα(GTP → GDP + Pi), which can be mimicked by Gα-GDP bound with the planar ion aluminum tetrafluoride (AlF4−) (Sondek et al., 1994; Berman et al., 1996; Popov et al., 1997). Three critical contacts are formed between RGS proteins and their Gα partners (Tesmer et al., 1997a; Slep et al., 2001; Slep et al., 2008; Soundararajan et al., 2008). The highly conserved amide of Asn-122 (Fig. 3; residue numbered as in human RGS8) forms a hydrogen bond with the critical glutamine of Gα responsible for GTP hydrolysis (Gln-204 of Gαi3). This helps orient the glutamine residue to stabilize the terminal phosphate that is being hydrolyzed from GTP (as mimicked by the AlF4− ion). A second Asn (Asn-82 of RGS8) contacts the side-chain hydroxyl of a switch I threonine (Thr-182 in Gαi3), allowing the side-chain hydroxyl to contact the switch II Lys (Lys-210 in Gαi3). This locks switches I and II into their transition state conformations, promoting accelerated GTPase activity. In addition, an aspartate residue in the C terminus of the RGS domain (Asp-157 in RGS8), which is conserved in all RGS proteins except RGS2, serves to stabilize the backbone amine of the Gα switch I Thr-182 (allowing the neighboring Thr-181 side-chain hydroxyl group to stabilize the Mg2+ cation; Fig. 3).
II. Selected Functional Vignettes among the Complement of Regulators of G-Protein Signaling
A. The Utility of the “Regulators of G-Protein Signaling-Insensitivity” Point Mutation
The numerous contacts made by Thr-182 as described above (Fig. 3B) highlight the importance of this switch I region in Gα in stabilizing the RGS domain/Gα interaction and, in addition, explain the profound loss of binding and GAP activity that occurs when the neighboring glycine (Gly-183 in Gαi3) is subtly changed to serine. This “RGS-insensitivity” point mutation (glycine to serine) was originally identified by DiBello et al. (1998) in GPA1, the Gα subunit of the yeast S. cerevisiae, functions equivalently in mammalian Gα subunits such as Gαi1, Gαo, and Gαq (Lan et al., 1998; Clark and Traynor, 2004) and has also been shown to leave all other functions of Gα intact, including intrinsic nucleotide binding and hydrolysis activities, as well as coupling to Gβγ, receptor, and effectors (Lan et al., 1998; Chen et al., 2004; Day et al., 2004; Fu et al., 2004; Ikeda and Jeong, 2004). Separate mouse strains bearing this RGS-insensitivity point mutation (G184S) within their Gαo or Gαi2 gene loci, respectively, have been generated (Fu et al., 2004; Huang et al., 2006); these mice possess select changes in various organ system functions controlled by GPCR signaling, including central nervous system, cardiovascular, and endocrine functions (Fu et al., 2006, 2007; Huang et al., 2006, 2008; Goldenstein et al., 2009; Icaza et al., 2009; Signarvic et al., 2010; Talbot et al., 2010). The most recent findings of Ruiz de Azua et al. (2010), that RGS4 is a negative regulator of M3 mAChR signaling to insulin secretion in pancreatic β-cells, articulate well with the findings of Huang et al. (2008) that RGS-insensitive Gαi2(G184S) knock-in mice exhibit increased glucose tolerance when on a high-fat diet.
Collectively, the results obtained thus far with the RGS-insensitivity mutation have highlighted the importance of RGS protein action on Gα nucleotide cycling in the context of the whole organism but do not necessarily identify the specific member(s) of the RGS protein superfamily at play in these various organ systems that could be directly exploited as a drug discovery target. Thirty-seven RGS proteins are encoded by gene loci in the human genome; this collection of related proteins has been divided into 10 different subfamilies based on the relatedness of their RGS domain sequence and their multiple domain architectures (Fig. 4). Several excellent recent reviews have been published regarding the specialized physiological functions now known for many of these individual RGS proteins, including in the central nervous system (Hooks et al., 2008; Anderson et al., 2009; Sjögren and Neubig, 2010; Traynor, 2010) and in cancer (Hurst and Hooks, 2009; Sjögren et al., 2010). Thus, to complement these recent publications, we have chosen to highlight below several vignettes regarding appreciation of the roles of RGS proteins in cardiovascular and immune system functions. These highlights are in no way meant to be comprehensive but are intended to emphasize the potential for proteins from among this large family as targets for therapeutic exploitation.
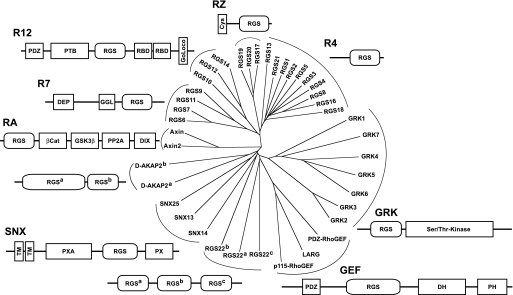
Fig. 4.
Subfamily categorizations of the 37 RGS domain-containing proteins identified in humans, based on sequence similarities and domain architectures. An unrooted dendrogram was generated using ClustalW (http://www.clustal.org; Thompson et al., 1994) and visualized using TreeView (Page, 1996). Domain boundaries were predicted using the SMART database (Letunic et al., 2009). PDZ, PSD-95/Dlg/ZO-1 domain; PTB, phosphotyrosine-binding domain; RBD, Ras-binding domain; DEP, Dishevelled/EGL-10/Pleckstrin domain; GGL, Gγ-like domain; βCat, β-catenin interaction region; GSK3β, glycogen synthase kinase-3β interaction region; PP2A, protein phosphatase 2A; DIX, Dishevelled interaction region; SNX, sorting nexin; TM, transmembrane domain; PXA, domain associated with a PX domain; PX, p40/p47-Phox homology domain; DH, Dbl-homology domain; PH, Pleckstrin-homology domain.
The largest RGS protein subfamily is known as the R4 family and contains 10 members: RGS1, -2, -3, -4, -5, -8, -13, -16, -18, and -21. R4 family members represent some of the smallest and simplest of the RGS proteins and, with the exception of RGS3 (Kehrl et al., 2002), consist of a single RGS domain with minimal additional amino acids at their N and C termini. With the exception of RGS2 (Heximer et al., 1997b; Kimple et al., 2009), members of the R4 family accelerate the hydrolysis of GTP by both Gi and Gq family Gα subunits (Arshavsky et al., 2002; Soundararajan et al., 2008). With little biochemical selectivity between Gαi1, Gαi2, Gαi3, Gαo, and Gαq substrates, and no additional regions containing obvious domain structures, members of this subfamily would be predicted to act promiscuously as negative regulators of Gi- and Gq-coupled GPCRs; however, early work by Zeng et al. (1998) and Xu et al. (1999) demonstrated that the N termini of R4 family members, outside of the canonical RGS domain, can provide specificity to the in vivo potency of R4 protein GAP activity on specific receptors. Although it is not entirely clear how these terminal extensions on R4 family RGS domains enhance specificity, it has been suggested that selectivity toward particular GPCR signaling pathways is mediated by the binding of adaptor proteins such as spinophilin (Wang et al., 2005) or through direct interactions with GPCRs (Xu et al., 1999; Bernstein et al., 2004; Wang et al., 2009). In addition to receptor specificity that is dependent on the N terminus, point mutations have been identified that affect the overall in vivo stability of RGS protein in overexpression studies (Bodenstein et al., 2007). The physiological relevance of the N terminus in regulating degradation of RGS proteins is supported by the identification of a hypertensive cohort that had a single nucleotide polymorphism in the gene loci of RGS2 that results in a Gln-2-Leu mutation resulting in destabilization of RGS2 and subsequent hypertension (Yang et al., 2005).
B. Regulators of G-Protein Signaling 1, 2, and 13 in Immune System Regulation
Although the biochemical role of RGS proteins as GTPase-accelerating proteins has been well characterized (Berman et al., 1996; Apanovitch et al., 1998; Snow et al., 1998a), and the cellular role of RGS proteins in attenuating GPCR-mediated signaling is also established (Doupnik et al., 1997; Saitoh et al., 1997; He et al., 1998; Lambert et al., 2010), it has remained a more arduous task to characterize the specific roles of RGS proteins in the context of whole-organism homeostasis and pathophysiology. Several mouse knockout strains of R4 family members have been published to date that have shed light on this issue. Using an RGS1-deficient mouse strain, Moratz et al. (2004) reported on the importance of RGS1 in negatively regulating CXCR4 and CXCR5 chemokine receptor signaling in B-lymphocytes and the necessity of RGS1 expression for the proper maturation of germinal centers. Bansal et al. (2008) identified a different immune system phenotype in RGS13-deficient mice. Their research demonstrated that loss of RGS13 resulted in increased mast-cell degranulation and anaphylaxis. Deregulation typically occurs when the antigen-bound IgE interacts with the IgE receptor (FcεRI), which is not a G-protein coupled receptor (Kinet, 1999; Rivera and Gilfillan, 2006; Metz and Maurer, 2007; Gilfillan and Rivera, 2009); however, Bansal et al. (2008) showed that RGS13 acts in a GAP-independent manner to negatively regulate IgE-mediated degranulation. They determined that the amino-terminal 51 amino acids (outside of the RGS domain) bind the p85α regulatory subunit of phosphatidylinositol-3-OH kinase, preventing the activation of antigen-induced phosphatidylinositol 3-kinase-mediated degranulation.
In addition to their role in B cells and mast cells, RGS proteins are also important in other immune system functions. RGS2 was originally identified as “G0-switch gene-8” (G0S8), a gene up-regulated upon activation of blood mononuclear cells by the plant lectin concanavalin A or treated with cycloheximide (Siderovski et al., 1990, 1994; Heximer et al., 1997a). In studies of RGS2-deficient mice, these authors were able to show that, unlike RGS1-deficient mice, RGS2-deficient mice have normal B cell quantities and differentiation; however, RGS2-deficient mice were unable to mount a robust T-cell mediated immune response. RGS2-deficient T cells, compared with wild-type T cells, were impaired in their ability to proliferate in response to T-cell receptor engagement, to treatment with phorbol 12-myristate 13-acetate and Ca2+ ionophores, or to anti-CD3ε cross-linking, with or without CD28 coreceptor engagement (Oliveira-Dos-Santos et al., 2000). In addition, T cells had an impaired ability to secrete interleukin-2 (IL-2) in response to an immune challenge. Although the diminished IL-2 secretion could account for the decreased proliferation (Cantrell and Smith, 1984), supplementation of IL-2 was unable to stimulate T-cell proliferation to levels seen in wild-type T-cells, suggesting that the observed phenotype was not the result of decreased IL-2 production (Oliveira-Dos-Santos et al., 2000).
C. Regulators of G-Protein Signaling 2, 4, 5, and 6 in Cardiovascular System Regulation
In addition to their roles in modulating immune responses, R4 family RGS proteins have been shown to regulate cardiovascular development and physiology (Manzur and Ganss, 2009). Maintenance of vascular perfusion of the entire body is a delicate balance. If arterial pressure is significantly decreased, regional hypoxia and coagulative necrosis will destroy tissue; however, if arterial pressure is elevated, the risk of heart failure, stroke, and kidney disease are dramatically increased (Harris et al., 2008). One crucial component to maintaining normotension is vascular resistance, which is dynamically modulated by the vascular smooth muscle that lines blood vessels. Given that vessel resistance (R) is inversely proportional to vessel radius (r) (R ∝ 1/r4), small changes in the lumen of a vessel can have dramatic change in the resistance and thus the vascular pressure (Levy et al., 2007). GPCRs are crucial mediators of vasodilation and vasoconstriction (Brinks and Eckhart, 2010); for example, angiotensin II, norepinephrine, vasopressin, and acetylcholine cause vasoconstriction by activating GPCRs coupled to Gαq/11, which subsequently activate PLC.
The unique in vitro specificity of RGS2 toward Gαq (Heximer et al., 1997b; Kimple et al., 2009) and the multitude of Gq-coupled GPCRs that control vasoconstriction suggest that RGS2 might be an important negative regulator in inhibiting vascular smooth muscle constriction. The clearest demonstration of the importance of RGS2 in regulating blood pressure came from studies of RGS2-deficient mice (Oliveira-Dos-Santos et al., 2000) that characterized RGS2-deficient animals as having constitutive hypertension (Heximer et al., 2003; Tang et al., 2003; Gu et al., 2008). Further evidence supporting the role of RGS2 in maintaining normostatic blood pressure has come from human population-based studies of hypertensive cohorts. These studies have identified single nucleotide polymorphisms within the coding region of RGS2 that result in a decrease of proper localization of RGS2 to the plasma membrane and a resultant decrease of its inhibitory influence on Gαq-mediated vasoconstrictive hormone signal transduction (Yang et al., 2005; Gu et al., 2008). Promoter polymorphisms in the RGS2 gene locus and renal actions of RGS2 have also been highlighted as contributing factors to the intrinsic homeostatic and extrinsic therapeutic control of blood pressure maintenance (Gurley et al., 2010; Semplicini et al., 2010; Sugimoto et al., 2010). In addition, the RGS2-deficient mouse was instrumental in demonstrating the role for RGS2 protein action in cardiac compensation to blood pressure overload and the antihypertrophic effects of PDE5 inhibition (Takimoto et al., 2009).
Although the loss of RGS2 results in constitutive hypertension, mice deficient in RGS5, which is highly expressed in pericytes, exhibit constitutive hypotension, suggesting that RGS5 might be a critical negative regulator of vasodilatory signaling or vascular development (Cho et al., 2008). Although the mechanism by which RGS5 assists in the maintenance of normal blood pressure remains to be established, RGS5 has been observed to be highly expressed in vascular smooth muscle and pericytes (Bondjers et al., 2003; Cho et al., 2003). The high expression of RGS5 in pericytes of angiogenic tumor vessels (Berger et al., 2005) led Hamzah et al. (2008) to cross rgs5-deficient mice with a tumorigenic mouse strain that rapidly develops insulinomas. The tortuosity and dilated nature of the vessels characteristic of insulinomas derived from wild-type mice were lost in the RGS5-deficient line. Instead, the blood supply in RGS5-deficient insulinomas had a regular appearance with normal branching reminiscent of normal developmental angiogenesis in organs (Ryschich et al., 2002; Hamzah et al., 2008). Although the precise role that RGS5 is playing in the neovascularization of tumors is unclear, it is apparent that RGS5 is a critical component of maintaining normal blood pressure and proper angiogenesis.
An additional RGS-deficient mouse model identified to have a cardiovascular phenotype is the RGS4-deficient mouse (Grillet et al., 2005). In this mouse, the RGS4 promoter was used to drive the expression of β-galactosidase, thereby allowing expression of the Rgs4 gene to be characterized by histochemical staining. The authors reported high levels of expression of the Rgs4 gene locus in the sinoatrial node (Cifelli et al., 2008), an anatomical region of the heart that serves to initiate and control the timing of cardiac contractions (Efimov et al., 2010). In the absence of RGS4 expression, basal heart rates were identical to those of wild type; however, upon activation of the parasympathetic system by the administration of carbachol, RGS4-deficient mice had an exaggerated decrease in heart rate compared with wild-type control mice (Cifelli et al., 2008). In examining isolated sinoatrial myocytes from RGS4-deficient and control animals, Cifelli et al. (2008) also observed a decreased frequency of action potential initiation in response to activation of the parasympathetic nervous system by carbachol administration.
More recently, RGS6-deficient mice have been generated by independent groups, highlighting a role for this particular R7 family RGS protein in modulating parasympathetic regulation of heart rate (Posokhova et al., 2010; Yang et al., 2010). Reduction in heart rate by parasympathetic innervation is known to involve the sequential activation of m2 muscarinic acetylcholine receptors (M2-mAChR), heterotrimeric G proteins of the Gi/o subfamily, and the atrial potassium channel IKACh, composed of GIRK1 and GIRK4 channel subunits. RGS6 deficiency was found to yield a profound delay in M2-mAChR/IKACh deactivation kinetics in both adult sinoatrial nodal cells and neonatal atrial myocytes; mice lacking RGS6 have a mild resting bradycardia and altered heart rate responses to pharmacological manipulations consistent with enhanced M2-mAChR/IKACh signaling. This function for RGS6 is consistent with its obligate association with the unique Gβ5 subunit (Posner et al., 1999; Snow et al., 1999; Zhang and Simonds, 2000) (an association shared with the other R7 family members RGS7, -9, and -11 (Cabrera et al., 1998; Snow et al., 1998b; Makino et al., 1999); it has recently been suggested that this Gβ5 association can recruit R7 family RGS proteins to GIRK channels and thereby regulate the kinetics of neuronal signaling through the receptor/G-protein/channel axis (Xie et al., 2010).
III. Pursuing Chemical Probes for Regulators of G-Protein Signaling GTPase-Accelerating Protein Activity
A. GTPase-Accelerating Protein Activity as the Sole Determinant of Regulator of G-Protein Signaling Action on G-Protein-Coupled Receptor Signaling Sensitivity and Kinetics
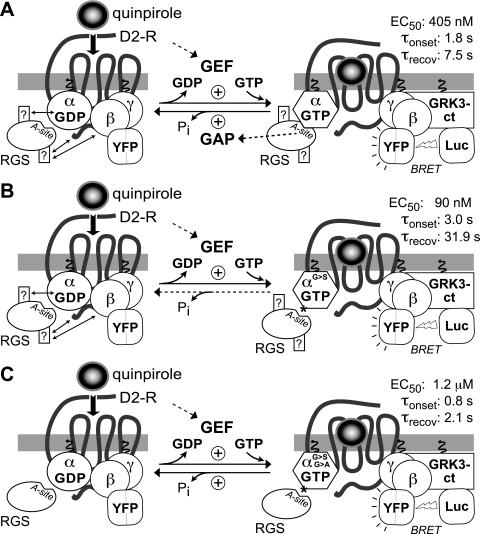
As the above passage regarding R7 family members illustrates, many RGS proteins bear additional protein-protein interaction domains beyond their signature RGS domain with Gα GAP activity (see Fig. 4). As another example, two of the R12 family members (RGS12 and RGS14) contain a second, Gα-GDP-interaction motif [alternatively known as the GoLoco motif or GPR motif (Ponting, 1999; Kimple et al., 2001; Lanier, 2004)] and additionally coordinate components of the Ras/Raf/mitogen-activated protein kinase signaling pathway (Traver et al., 2000; Mittal and Linder, 2006; Willard et al., 2007, 2009; Shu et al., 2010). These additional interaction points, and their logical implication of signaling protein “physical scaffolding” (e.g., Benians et al., 2005), have been thought to underlie a “paradoxical” function of RGS proteins observed in reconstituted cellular systems early after their initial discovery: namely, acceleration of the onset of GPCR signaling without a demonstrable change in activating ligand potency per se (Doupnik et al., 1997; Saitoh et al., 1997; Zerangue and Jan, 1998). To address this long-standing paradox, Lambert et al. (2010) recently revisited the cellular reconstitution studies of Doupnik et al. (1997) and Saitoh et al. (1997) but used a rapid, bioluminescence-based measure of Gβγ liberation from activated receptors (rather than whole-cell electrophysiological measurements of GIRK channel activity) and employed the classic Gα “RGS-insensitivity” point mutation (section II.A) in combination with a second, cis-acting “fast-hydrolysis” point mutation that accelerates the intrinsic GTPase activity of Gα subunits (Thomas et al., 2004) (schematized in Fig. 5). X-ray diffraction crystallography was performed to validate that the cis-acting fast-hydrolysis point mutation poises the Gα in a pretransition state for GTP hydrolysis and thereby accelerates what normally the trans-acting RGS domain performs for the Gα (Lambert et al., 2010). In both yeast pheromone response assays and measurements of reconstituted neurotransmitter GPCR signaling, observed increases in receptor sensitivity to respective activating ligands (i.e., leftward shifts in EC50) upon using “RGS-insensitive” Gα subunits (e.g., Fig. 5B) (Lambert et al., 2010) reaffirmed the prevailing notion that the critical function of endogenous RGS proteins is to regulate GPCR agonist sensitivity (Neubig and Siderovski, 2002; Ishii and Kurachi, 2003). Moreover, adding the fast-hydrolysis mutation on top of the “RGS-insensitive” mutation restored normal agonist sensitivity and kinetics of both signal onset and decay to that observed from wild-type Gα acted on by endogenous RGS proteins (Lambert et al., 2010); indeed, addition of faster-than-intrinsic GTPase activity by the cis-acting mutation produced slightly faster onset and recovery kinetics than the wild-type situation as well as a rightward shift in EC50 (e.g., Fig. 5C). This restoration of “normalcy” to GPCR signaling by the Gα fast-hydrolysis mutant strongly suggests that the critical function for RGS proteins in regulating kinetics and (ultimately) agonist sensitivity of GPCR signaling comes solely from GTPase acceleration (Lambert et al., 2010), suggesting that kinetic (and not physical) events are the likely mechanism of RGS protein action in this context (Zhong et al., 2003; Turcotte et al., 2008) and supporting the idea (Neubig and Siderovski, 2002; Chasse and Dohlman, 2003; Cho et al., 2004; Liebmann, 2004) that modulating the GAP activity of RGS domains by small molecules will be a valuable new approach in changing GPCR responsiveness in pathophysiological settings.
Fig. 5.
Use of the RGS-insensitivity (“G>S”) and fast-hydrolysis (“G>A”) point mutants of Gα in establishing the central role of GTPase acceleration by RGS proteins in their modulatory actions on GPCR signaling kinetics and sensitivity. A cell-based system employing Gα reconstitution, along with a temporally sensitive and -reversible measure of Gαβγ activation, was established to interrogate whether “non-GAP” activities of RGS proteins (illustrated as question marks in rectangles) exist that influence the kinetics of signal onset (τonset) and receptor sensitivity (EC50 for agonist) beyond the GAP activity embodied by the RGS domain A-site. In this experimental system set up by Lambert et al. (2010), G-protein heterotrimer activation, by the binding of the agonist quinpirole to the dopamine D2 receptor (“D2-R”), increases bioluminescence resonance energy transfer (BRET) between the Gβγ-binding reporter protein masGRK3ct-Rluc8 (GRK3-ct Luc) and Gβ1γ2-Venus (YFP). Human embryonic kidney 293 cells were pretreated with pertussis toxin to inactivate native Gαi/o subunits, and so quinpirole responses were mediated by heterotrimers composed of ectopically expressed, pertussis toxin-insensitive GαoA (wt; A), PTX- and RGS-insensitive GαoA (G184S; called “G>S” in B and denoted with an asterisk), or additionally containing the fast hydrolysis switch II point mutation (G203A; called “G>A” in C). The glycine-to-alanine switch II mutation (G203A, “fast-hydrolysis”) was seen to blunt agonist sensitivity [EC50 of 1.2 μM, nearer to that of wild-type GαoA (EC50 of 405 nM)] over the more sensitive responses mediated by the use of RGS-insensitive GαoA alone (G184S; EC50 of 90 nM). In addition, normalized BRET plotted against time during sequential addition of the agonist quinpirole (30 μM) and the antagonist haloperidol (10 μM) indicated that the fast-hydrolysis mutation (“G>A”) restored rapid onset and recovery kinetics (τonset and τrecov values nearer to that of wild-type GαoA use) over the more languid responses mediated by the use of RGS-insensitive (“G>S”) GαoA alone. These results served to negate the necessity of evoking non-GAP activities of RGS proteins to explain earlier observations of RGS proteins leading to accelerated GPCR signaling onset without demonstrable changes in activating ligand potency (Doupnik et al., 1997; Saitoh et al., 1997; Zerangue and Jan, 1998), hence the removal of the question marks in rectangles from the RGS protein in the final panel.
B. The Potential for Allosteric Control over Regulator of G-Protein Signaling GTPase-Accelerating Protein Activity
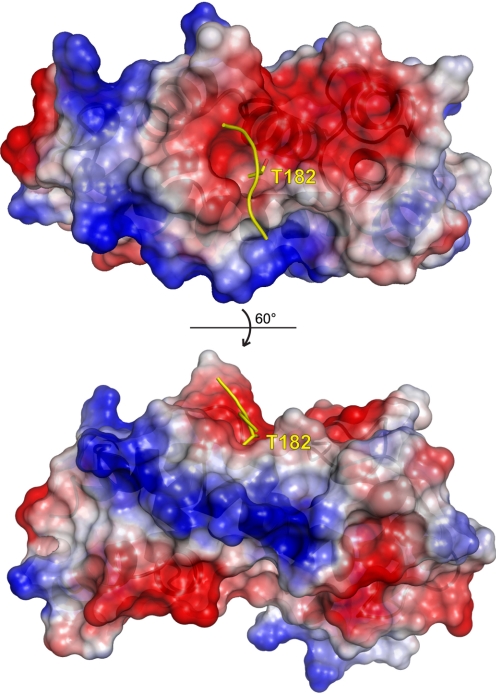
As previously stated, GPCRs are the single largest target for currently prescribed pharmaceuticals, and RGS proteins are potent negative regulators of GPCR-mediated signaling. RGS proteins thus provide an attractive target to either modulate the action of currently prescribed pharmaceuticals or modulate tonic signaling in a pathway-dependent manner (Neubig and Siderovski, 2002; Cho et al., 2004; Liebmann, 2004; Riddle et al., 2005). The specific targeting of RGS proteins by small molecules is still in its infancy (see discussion in section III.C), yet the “druggability” of RGS domains was suggested early on the basis of observations from the first crystal structure of the RGS4/Gαi1 complex (Tesmer et al., 1997a), coupled with discovery of the RGS-insensitivity point mutation on Gα (DiBello et al., 1998). As shown in Fig. 6, threonine-182 of switch I within Gαi1, among all switch region contacts, experiences the largest change in accessible surface area upon complex formation (Tesmer et al., 1997a) by becoming buried within a depression on the Gα-interacting “A-site” of the RGS4 RGS domain; the critical nature of this burial of threonine-182 is underscored by the profound loss of GAP activity upon subtle substitution of the neighboring glycine (Gly-183) within Gαi1 to serine. Thus, early speculations have been made (e.g., Neubig and Siderovski, 2002) that an inhibitor binding within this pocket of the RGS domain should block the Thr-182 interaction and thus abrogate the Gα/RGS domain binding event.
Fig. 6.
The Gαi1 interaction surface (“A-site”) of the RGS4 RGS domain contains a charged depression. Electrostatic surface rendering of RGS4 (PDB number 1AGR; potentials contoured at ±5 kT/e), highlighting the electronegative region (red) into which the threonine-182 residue of Gαi1 is buried. Electropositive potential is highlighted in blue.
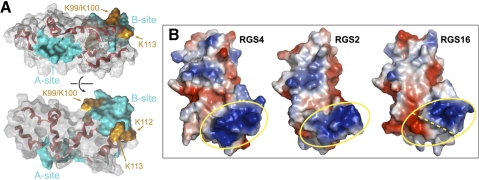
Although a small molecule that binds to the A-site depression on an RGS domain and blocks its interaction with Gα would be an invaluable proof of principle for the “druggability” of RGS proteins, it would be equally useful to have a small molecule that could allosterically enhance the GAP function of endogenous RGS proteins [i.e., in pathological states in which an enhancement in RGS protein GAP activity is desired; for example, as believed to be the case with RGS4 in schizophrenia (Mirnics et al., 2001; Volk et al., 2010)]. Bioinformatic methods (Sowa et al., 2000) and mutagenesis (Popov et al., 2000) have implicated a region between helices IV and V in RGS4 as an allosteric site on the RGS domain responsible for the influences of phosphatidylinositol-3,4,5-trisphosphate (PIP3) and Ca2+/calmodulin (CaM) on GAP activity. This allosteric site (“B-site”), flanked by key basic residues (Fig. 7), is distinct from the Gα-interacting A-site and, upon binding of PIP3, decreases GAP activity in vitro. In a Ca2+-dependent manner, CaM can competitively inhibit PIP3-mediated GAP inhibition (Popov et al., 2000; Ishii et al., 2001; Ishii et al., 2002). Modulation of GAP activity via PIP3 and Ca2+/CaM is also seen in cellular assays using cardiac myocytes in electrophysiological recordings of GPCR signaling to ion channel gating (Ishii et al., 2002, 2005; Ishii and Kurachi, 2004; Ishii et al.). Based on sequence conservation in the B-site, it is possible that the allosteric modulation of RGS1, -2, -10, and -19 also occurs (Tu and Wilkie, 2004); however, this remains to be experimentally validated. This B-site could potentially be exploited by small molecules to either mimic the effect of PIP3 in inhibiting GAP activity or the effect of Ca2+/CaM in preventing the allosteric inhibition of GAP activity by PIP3.
Fig. 7.
Predicted structural determinants within RGS4 that enable allosteric control over Gα-directed GAP activity by PIP3 and Ca2+/CaM. A, structural coordinates of the RGS domain of RGS4 were rendered in PyMOL using data from the RGS4/Gαi1-GDP-AlF4− complex [PDB number 1AGR (Tesmer et al., 1997a)]. The conserved RGS domain fold is composed of nine α-helices (displayed in red). Orange regions depict lysines thought to be required for PIP3 binding, whereas solid cyan areas depict the proposed A- and B-sites. Rotation about the horizontal axis by 90° is shown in the lower panel. B, electrostatic surface rendering of RGS4 (PDB number 1AGR), RGS2 (PDB number 2AF0), and RGS16 (PDB number 2BT2) using PyMol, highlighting the electropositive (blue) region considered the putative CaM-binding B-site within RGS2 and RGS4 (yellow oval). RGS16, shown to be insensitive to PIP3 and CaM modulation (Tu and Wilkie, 2004), has less electropositive potential in this region, as well as a patch of electronegative potential (red).
C. Current State of Identifying Chemical Probes that Modulate Regulator of G-Protein Signaling Protein Activity
The most direct means to disrupt the RGS domain/Gα interaction is via point mutations on either protein's interaction surface. Single amino acid substitutions on either side of the interface can completely abolish binding and the catalytic activity of RGS proteins (e.g., DiBello et al., 1998; Srinivasa et al., 1998; Wieland et al., 2000; Willard et al., 2005; Kimple et al., 2009). The ability to disrupt this large protein/protein interface (1290 Å2; Tesmer et al., 1997a) with single point mutations suggests that small perturbations in the RGS domain A-site surface, such as those caused by a bound small molecule, could have dramatic results in inhibiting RGS domain GAP activity.
Measuring RGS domain-mediated acceleration of GTP hydrolysis in vitro, as part of a compound library screening campaign, for example, is complicated by the fact that GDP release by Gα (not GTP hydrolysis per se) is the rate-limiting step in steady-state nucleotide cycling by Gα subunits (Higashijima et al., 1987; Ross, 2002). Thus, to quantify the effects of RGS domain GAP activity, one typically preloads the Gα subunit with radiolabeled GTP (in the absence of the critical cofactor for hydrolysis: Mg2+) and then measures one round of hydrolysis in a so-called “single-turnover” assay (e.g., Berman et al., 1996; Hunt et al., 1996; Watson et al., 1996; Snow et al., 1998a). This experimental design requires one to establish a pool of Gα-([γ-32P]GTP), initiate the assay at time 0 with the addition of Mg2+, sample aliquots from this reaction over time, precipitate all unhydrolyzed [γ-32P]GTP with charcoal, separate the charcoal, and then quantify the inorganic 32P-phosphate that was produced (and that resides in the supernatant) using liquid scintillation counting. This cumbersome assay design is not readily amenable to rapid automation, so alternative, fluorescence-based assays for RGS domain GAP activity have been developed that should be more suitable for high-throughput screening (HTS) of compound libraries (e.g., Willard and Siderovski, 2004; Willard et al., 2004a, 2005; Roman et al., 2007, 2009; Blazer et al., 2010).
1. Pharmaceutical Company Activities Published in the Literature.
Wyeth Laboratory has published a yeast two-hybrid assay-based screening method for identifying RGS4 and RGS20 inhibitors (Nieuwenhuijsen et al., 2003; Wang and Young, 2004). Although their screen was reported to have identified small molecule inhibitors, these compounds have not yet been made public. In a functional screen to identify novel treatments for urinary incontinence using ex vivo rat bladder smooth muscle cultures, a Bristol-Myers Squibb group identified two compounds (BMS-192364 and BMS-195270) that had no known molecular target yet resulted in relaxation of bladder tone (Fitzgerald et al., 2006). Using a nematode genetics approach to identifying the target of these two drugs, this group concluded that these two compounds targeted the Gα/RGS domain interaction and specifically locked the pair in an unproductive complex (Fitzgerald et al., 2006). There is precedence for such a proposed mechanism of action, given that brefeldin A, a naturally occurring antibiotic, can trap the Ras-family GTPase ARF1 in an unproductive complex with the ARF1 GEF, Sec7 (Mossessova et al., 2003). However, no confirmatory report has yet appeared in the literature indicating that these two BMS compounds directly target the Gα/RGS domain interaction.
2. Academic Laboratory Activities with Binding Assays.
In addition to the efforts that are ongoing by the pharmaceutical industry, academic laboratories have been developing novel high-throughput screening assays for the RGS domain/Gα interaction target and searching for small-molecule modulators of RGS protein GAP activity. A high-throughput flow cytometry method has been described for screening for small molecules that can disrupt the binding of RGS proteins to Gα subunits (Roman et al., 2007, 2009). This elegant assay design uses fluorescently labeled Gαo protein and a LumAvidin microsphere-coupled RGS protein to look for compounds that disrupted their interaction. The advantage of this assay is the ability to multiplex different biotinylated RGS proteins to different LumAvidin microspheres (Roman et al., 2007, 2009). Results of an initial “in house” screening of a small, ∼3000-compound collection from ChemBridge have been published (Roman et al., 2007); a single compound from this collection [methyl-N-[(4-chlorophenyl)sulfonyl]-4-nitrobenzenesulfinimidoate (CCG-4986)] was reported to inhibit the primary in vitro RGS4/Gαo binding assay as well as RGS4-mediated acceleration of Gαo GTPase activity in the in vitro single-turnover assay format. This compound was reported to lack activity in assays of intact, RGS4-transfected cells but was shown to work on permeabilized C6 glioma cells in potentiating [d-Ala2,N-Me-Phe4,Gly5-ol]-enkephalin-mediated inhibition of forskolin-stimulated cAMP production (i.e., negating the inhibitory influence of added recombinant RGS4 on activated, Gi/o-coupled μ-opioid receptor signaling to the inhibition of adenylyl cyclase).
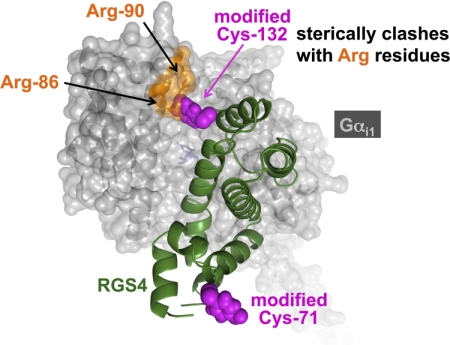
The lack of effect of CCG-4986 on intact cellular GPCR signaling was originally interpreted as reflecting a lack of cell permeability by the compound (Roman et al., 2007). This issue of cell permeability is a general concern for any future chemical probes identified as specific to the Gα/RGS domain interaction because, unlike the orthosteric and allosteric binding sites of GPCRs (May et al., 2007; Smith et al., 2011), the RGS protein target is obviously intracellular. However, subsequent work with this particular compound suggested that the lack of action of CCG-4986 on intact cells is more likely to be due to its sensitivity to a reducing environment (e.g., as is present inside the cytosol of intact, nonpermeabilized cells). CCG-4986 was thus categorized as a generic, thiol-reactive compound likely to be ill-suited for further development as a proof-of-principle chemical probe for modulating RGS domain GAP activity in a cellular context (Kimple et al., 2007); results from mutagenesis and mass spectrometry studies have suggested that CCG-4986 works in an indiscriminate fashion, covalently modifying more than one cysteine within recombinant RGS4 (including Cys-132, present in the Gα-binding A-site) and thereby blocking the binding of its Gα substrate by steric hindrance (Fig. 8).
Fig. 8.
CCG-4986 is a generic thiol-reactive compound and should not be considered an RGS4-selective GAP-inhibitory drug. Mass spectrometry data from recombinant RGS4 (green) incubated with CCG-4986 (Kimple et al., 2007) indicates that at least two, surface-exposed cysteine residues (Cys-71 and Cys-132) become covalently modified with a 153-Da moiety (magenta) derived from the 4-nitrobenzenethiol radical liberated from CCG-4986. One of these positions (Cys-132) is near two arginines (Arg-86, Arg-90; orange) within the all α-helical domain of the Gα subunit (rendered here in a gray, space-filling translucent cloud overlying the Cα ribbon trace). Figure was rendered in PyMol based on the structural coordinates of the RGS4/Gαi1-GDP-AlF4− complex (PDB id 1AGR).
More recently, a different binding assay (involving time-resolved FRET with terbium-labeled Gαo and Alexa Fluor 488-labeled RGS4) was employed to identify two additional candidate RGS4 inhibitory compounds: (2E)-2-(1,3-benzothiazol-2-yl)-3-[9-methyl-2-(3-methylphenoxy)-4-oxo-4H-pyrido[1,2-a]pyrimidin-3-yl]prop-2-enenitrile (CCG-63802) and (2E)-2-(1,3-benzothiazol-2-yl)-3-[9-methyl-2-(4-fluorolphenoxy)-4-oxo-4H-pyrido[1,2-a]pyrimidin-3-yl]prop-2-enenitrile (CCG-63808) (Blazer et al., 2010). These two compounds are highly related and share a soft electrophilic functionality (α,β-unsaturated nitrile) that probably explains their cysteine dependence for in vitro activity on RGS4 (akin to that of CCG-4986) (Blazer et al., 2010); no congeners of CCG-63802 and CCG-63808 have yet been reported to have activity in RGS protein inhibition without also having this reactive acrylonitrile (vinyl cyanide) group. The likely reactive nature of these two compounds complicates their further development as an RGS domain-selective probe in a cellular context. To quote a recent review by Frye on what constitutes a high-quality chemical probe worthy of further pursuit in cell-based assays: “[E]lectrophiles… must be credentialed with extensive mechanistic studies to demonstrate specificity” (Frye, 2010). However, further in vitro work with CCG-63802 and CCG-63808 could provide valuable insight into what one should consider “druggable” targets within the RGS domain, especially if structural studies such as X-ray crystallography or NMR spectroscopy could establish the disposition of these compounds within the RGS domain to high-resolution. This also holds true for the compound 4-[(4-fluorophenyl)methyl]-2-(4-methylphenyl)-1,2,4-thiadiazolidine-3,5-dione (CCG-50014) (Blazer et al., 2011), the most recent cysteine-reactive chemical probe identified as a covalent modifier and GAP-activity inhibitor of RGS4 and RGS8; a high-resolution structure of one of these RGS protein targets in complex with CCG-50014 would hopefully support existing computational docking studies (Blazer et al., 2011) that this compound inhibits GAP activity through direct actions on the allosteric B-site of the RGS domain (section III.B).
Both the Wyeth yeast two-hybrid screen and the fluorescence-based binding assays described previously are unable to measure the actual catalytic activity of RGS domains in vitro. Instead, it has been common practice in RGS protein assay design and high-throughput screens to use binding of Gα to the RGS domain as a surrogate for GAP activity. Based on the mechanism by which RGS proteins stabilize the switch regions in their transition state conformation, this is a valid assumption; however, using binding as a surrogate for GAP activity has some potential pitfalls. A compound such as brefeldin-A (which traps the G-protein in an unproductive complex with its regulatory partner) would be missed. It is possible that, for the RGS domain/Gα target, a small molecule might inhibit RGS domain-mediated stabilization of the switch regions in a conformation that facilitates hydrolysis or otherwise traps the RGS domain/Gα complex. In addition, it is possible that, relying on binding rather than enzymatic activity in a compound library screen, one may have false negatives given weak binding of an low-potency inhibitor that would be lost to the noise of the binding assay. Instead, if one were able to read out successive rounds of GTP hydrolysis by Gα, and acceleration of that steady-state hydrolysis by the RGS protein, the effects of such weak inhibitors may become apparent. Finally, small molecules might exist that can selectively bind and allosterically potentiate the GAP activity of RGS proteins. With this logic in mind, a true enzymatic assay for RGS domain GAP activity that is amenable to HTS automation has recently been developed.
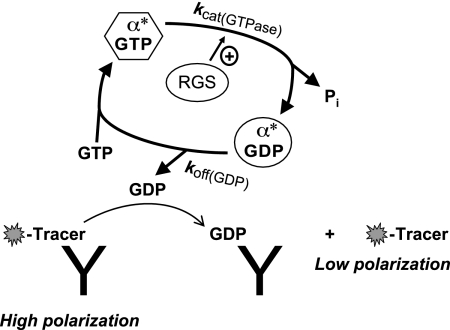
3. An Enzymatic Assay Using Rate-Altered Gα and Fluorescence Polarization Detection of GDP Generation.
To facilitate an enzymatic approach to HTS for RGS domain GAP activity modulators, the nucleotide cycle of Gαi1 was first changed so that GTP hydrolysis [kcat(GTPase)], not spontaneous GDP release [koff(GDP)], would become the rate-limiting step. Gαi1 and closely related Gα proteins have been the focus of extensive structure/function studies (Coleman et al., 1994; Sondek et al., 1994; Nishina et al., 1995; Posner et al., 1998), and point mutations have been identified previously (Coleman et al., 1994; Nishina et al., 1995; Posner et al., 1998; Marin et al., 2001) that affect both koff(GDP) and kcat(GTPase) without affecting functional interaction with the RGS domain. Two of the most striking Gα mutations have been made to the highly conserved active-site arginine, which greatly reduces intrinsic GTPase activity but does not eliminate RGS domain-mediated GTPase acceleration [Arg-178 in Gαi1, Arg-183 in Gαq; e.g., Fig. 2D (Coleman et al., 1994; Berman et al., 1996)], and to the alanine residue within the conserved TCAT loop known to contact the guanine ring of bound nucleotide (A326S; Posner et al., 1998), which results in a ∼25-fold increase in koff(GDP) relative to wild-type Gα. In testing these and other rate-altering mutations (Zielinski et al., 2009), a combination of R178M and A326S substitutions to Gαi1 was identified that produces sufficient alteration in nucleotide binding and cycling rates to allow the steady-state measurement of RGS domain-mediated acceleration of [γ-32P]GTP hydrolysis. It is noteworthy that multiple RGS proteins in binding and GTP hydrolysis assays were used to ascertain that these two substitutions within the guanine nucleotide binding pocket of Gαi1 had no effect on either the affinity or the specificity of the RGS domain interaction (Zielinski et al., 2009).
To detect RGS protein-accelerated GTPase activity without the use of radioactivity, a monoclonal antibody and fluorescent tracer system, previously developed by Bellbrook Labs for their Transcreener ADP assay (Kleman-Leyer et al., 2009), was adapted for selective immunodetection of GDP with a fluorescence polarization (FP) readout (Fig. 9). Measuring GTPase activity through the detection of generated GDP product using this “mix and measure” FP assay overcomes the low signal-to-noise ratio and other limitations of inorganic phosphate detection methods [e.g., malachite green and phosphate-binding protein assays (Van Veldhoven and Mannaerts, 1987; Geladopoulos et al., 1991; Willard and Siderovski, 2004; Shutes and Der, 2005)] and has previously been validated as a robust HTS method in the case of ADP detection for kinases and ATPases (Huss et al., 2007; Liu et al., 2007; Klink et al., 2008). Moreover, because this Transcreener GDP-based assay for the Gα/RGS domain interaction is one based on enzymatic activity, it should enable detection of all types of modulators of RGS domain GAP activity, including those that bind at allosteric sites and affect RGS domain catalytic activity without directly targeting the RGS domain Gα binding-site per se (see section III.B). In a pilot 960-compound library screen in the 384-well form factor with the rate-altered Gαi1(R178M/A326S) mutant (Zielinski et al., 2009), wild-type (i.e., surface cysteine-containing) RGS4 protein was employed so that the thiol-reactive compound CCG-4986 could serve as a positive control for RGS4 inhibition; subsequent screens of larger compound collections should necessarily use a cysteine-less mutant RGS4 [e.g., RGS4(C71N/C132E)] and/or addition of reductant, as has previously been considered (Kimple et al., 2007; Blazer et al., 2010; Roman et al., 2010) to avoid additional thiol-reactive hits. In addition, a counterscreen using rate-altered Gαi1(R178M/A326S) mutant without any input RGS protein, as has been demonstrated previously in a pilot screen (Zielinski et al., 2009), will be necessary to exclude hits that act directly on the intrinsic GTPase activity of the Gα subunit or otherwise interfere with assay components outside of the desired RGS protein target. However, a compound that has selective modulatory activity on the intrinsic GTPase activity of Gα subunit(s), especially one capable of enhancing weak or dormant intrinsic GTPase activity, may be valuable in its own right as a lead for future therapeutics (e.g., see next section below).
Fig. 9.
HTS-compatible enzymatic assay developed for the Gα/RGS domain target using a rate-altered Gα mutant and a fluorescence polarization immunoassay for the detection of generated GDP. The Gα subunit used in this assay bears two point mutations (denoted with asterisk) that dampen intrinsic GTPase and enhance spontaneous GDP release, respectively, thereby shifting the rate-limiting step in steady-state GTP hydrolysis away from product release [koff(GDP)] toward GTP hydrolysis [kcat(GTPase)] so that the influence of RGS domain GAP activity can be observed (section III.C.3). Fluorescent tracer is illustrated with a jagged oval; when bound to the GDP-selective monoclonal antibody, emitted light remains polarized, whereas there is low polarization of emitted light when tracer is displaced by free GDP as generated by the reaction.
IV. Activated Gαq/11 Point Mutants as Targets for Ocular Melanoma Therapeutic Strategies
As previously mentioned, recent reviews (Hurst and Hooks, 2009; Sjögren et al., 2010) have surveyed the burgeoning evidence that RGS proteins can be involved in aberrant survival, growth, and motility/invasiveness signal transduction pathways that underlie tumorigenesis and metastasis, including correlative data from gene microarray studies of cancerous versus normal tissue indicating differential RGS transcript expression patterns and from single-nucleotide polymorphism analyses of RGS gene loci indicating altered risks for cancer development. These findings are consistent with the idea that the predominant activity of RGS proteins is negative regulation of GPCR signaling, coupled with the ample existing evidence that GPCRs can be involved in dysregulated autocrine/paracrine signaling (or frankly hijacked by oncogenic viruses) to drive abnormal cell growth, survival, and motility (Sodhi et al., 2004; Dorsam and Gutkind, 2007; Spiegelberg and Hamm, 2007). Further support for the carcinogenic potential of dysregulated RGS protein GAP function comes from early evidence that GTPase-inactivating point mutations to their substrates (i.e., Gα subunits) are found in human tumors [e.g., pituitary and endocrine tumors (Landis et al., 1989; Lyons et al., 1990)] and can transform rodent fibroblasts cells upon ectopic overexpression (for review, see Dhanasekaran et al., 1995; Fukuhara et al., 2001). More recently, a somatic mutation to the GNAO1 gene locus (Gαo) was described in breast carcinoma (Kan et al., 2010) as leading to a missense mutation (R243H) of unknown functional consequence; Garcia-Marcos and colleagues have since identified that the R243H substitution renders Gαo constitutively active by enhancing spontaneous nucleotide exchange [rather than by affecting either intrinsic or GAP-accelerated GTPase activity per se (Garcia-Marcos et al., 2011)]. Here, in this final section, we consider another recent finding of Gα subunit involvement in cancer—that of Gαq and Gα11 in ocular (or “uveal”) melanoma—and how the field's existing knowledge base, including the structural determinants of Gαq/11 signal regulation, could be exploited for therapeutic development for this particular cancer.
Recent genetic analyses of ocular melanoma have revealed a high percentage (>80%) of activating point mutations in either the GNAQ (Gαq) or GNA11 (Gα11) gene loci (Onken et al., 2008; Van Raamsdonk et al., 2009, 2010), most predominantly a Q209L (“exon 5”) mutation that completely cripples the intrinsic GTPase activity of these Gα subunits and thereby creates a persistent GTP-bound, activated state (Kleuss et al., 1994). The Gln-209 of Gαq and Gα11 is analogous to the Gln-204 residue of Gαi depicted in Figs. 2D and 3B as critical to GTP hydrolysis, to the Gln-227 of Gαs found mutated in pituitary tumors (Landis et al., 1989), and to the Gln-61 residue of the small GTPase Ras found mutated in multiple cancers (Feig and Cooper, 1988; Cox and Der, 2002). Such constitutive activation of mutant Gαq or Gα11 in melanocytes is thought to drive inappropriate proliferative signaling through the extracellular signal-regulated kinase/mitogen-activated protein kinase signaling cascade in a manner similar to that of the activating mutations to B-Raf that are observed with high frequency in cutaneous melanoma [e.g., B-Raf(V600E) (Davies et al., 2002)].
With the recent success of targeting activated B-Raf in cutaneous melanoma using novel chemotherapeutic agents such as the kinase inhibitor PLX4032 (Bollag et al., 2010; Flaherty et al., 2010), developing similar therapeutic strategies toward the oncogenic alleles of Gαq and Gα11 might have equal success in uveal melanoma treatment. This is most germane for developing new treatments for the life-threatening metastatic dissemination of the melanoma to the liver (Vahrmeijer et al., 2008); although debilitating, eye removal (“enucleation”) because of the primary tumor is not life-threatening per se.
A. Regulator of G-Protein Signaling Domain-Based Gene Therapy?
In the current absence of small molecules able to reactivate the GTPase activity of Gα mutants (Ja et al., 2006; Siderovski et al., 2009), what other strategies could be employed to antagonize constitutively activated Gαq and Gα11 in uveal melanoma? Given the theme of this review, some targeted gene-therapy application of the RGS proteins might spring to mind. Indeed, one member of the R4 family, RGS2, exhibits a unique selectivity in its in vitro GAP activity for Gα subunits of the Gq family (Heximer et al., 1997b). X-ray diffraction crystallography was recently used to elucidate the particular structural determinants that engender this unique selectivity shown by RGS2 (Kimple et al., 2009). Unfortunately, RGS domain GAP activity has no effect on Gα subunits lacking the critical hydrolytic glutamine absent in the Q209L alleles of Gαq and Gα11; this lack of activity was first established in the original description of RGS proteins as Gα-directed GAPs (Berman et al., 1996). Fortunately, RGS domain GAP activity is able to accelerate GTP hydrolysis by other “GTPase-deficient” Gα point mutants (Berman et al., 1996) that bear a less-crippling mutation to the arginine residue that also contributes to the hydrolysis reaction, known as Arg-183 in Gαq and Gα11 and highlighted as Arg-178 of Gαi1 in Fig. 2D. This particular mutation (R183Q or R183C in exon 4) is seen in Gαq and Gα11 in uveal melanoma, but is considered less potent as an oncogenic allele and is less frequently observed (<10%) versus the Q209L mutation [>70% (Van Raamsdonk et al., 2010)].
RGS domains not only possess GAP activity but also bind the active (GTP-bound) species of Gα subunits and thus can curtail downstream signal transduction in cells by a process of “effector antagonism” (e.g., Hepler et al., 1997; Anger et al., 2004). Indeed, the N-terminal RGS domain identified in GRK2 exhibits little if any GAP activity yet binds readily to the activated state of Gαq/11 subunits, for which it demonstrates considerable selectivity over other Gα subfamilies (Siderovski et al., 1996; Carman et al., 1999; Sallese et al., 2000; Usui et al., 2000; Tesmer et al., 2005). Tesmer et al. (2005) have shown by X-ray crystallography that the GRK2 RGS domain binds Gαq in a mode unlike that of conventional (GAP-active) RGS proteins, not only explaining the lack of observable GAP activity but also affording a high-resolution structure of the key elements of Gαq/11 effector antagonism that should prove useful for any future gene-therapy or “deliverable protein” therapeutic designs derived from the GRK2 N terminus.
B. Gene- or Peptide-Based Strategies?
Antagonizing the oncogenic alleles of GNAQ and GNA11 at the mRNA transcript level, rather than at the protein level, may be an achievable pursuit given recent advances in RNA interference-based therapeutics (Tiemann and Rossi, 2009; Ashihara et al., 2010). There is precedence for a single siRNA oligonucleotide duplex affording effective knock-down of both Gαq and Gα11 protein levels in cells (Barnes et al., 2005), given the high degree of nucleotide sequence relatedness between GNAQ and GNA11. For example, in an RNA interference screen for RGS protein specificity in modulating endogenous GPCR signaling, a single siRNA duplex targeted to both GNAQ and GNA11 was shown to be quite effective in human cells in dampening GPCR signaling to intracellular calcium store mobilization (Laroche et al., 2010)—a well established signaling outcome downstream of activated Gαq/11-mediated by the stimulation of PLCβ and the resultant generation of the second messenger inositol-1,4,5-trisphosphate (Harden, 1992; Berridge, 2009). Sharpening the melanoma-selective nature of such siRNA-based therapy could possibly be achieved by exploiting the nucleotide-level changes in the oncogenic GNAQ/GNA11 alleles (versus their wild-type sequence) and/or using established physical approaches to liver-focused delivery such as transcatheter arterial chemoembolization (Sato, 2010; Schuster et al., 2010), given that hepatic metastatic disease is the critical, life-threatening feature of this disease.
Considering peptide-based strategies, the cyclic depsipeptide (1R)-1-{(3S,6S,9S,12S,18R,21S,22R)-21-acetamido-18-benzyl-3-[(1R)-1-methoxyethyl]-4,9,10,12,16,22-hexamethyl-15-methylene-2,5,8,11,14,17,–20-heptaoxo-1,19-dioxa-4,7,10,13,16-pentaazacyclodocosan-6-yl}-2-methylpropyl rel-(2S,3R)-2-acetamido-3-hydroxy-4-methylpentanoate (YM-254890) might be useful but likely only for the lower frequency Arg-183 mutants of Gαq and Gα11 seen in uveal melanoma. YM-254890 was originally identified from Chromobacterium spp. QS3666 culture broth (Taniguchi et al., 2003) as a secreted agent capable of blocking ADP-induced platelet aggregation [a physiological response to ADP dependent on signaling from purinergic GPCRs and Gαq-containing heterotrimers (Kunapuli et al., 2003)]. The structural determinants by which this cyclic depsipeptide selectively inhibited Gαq-mediated signaling were resolved via X-ray crystallography of a Gαq-GDP/Gβγ heterotrimer bound to YM-254890 (Nishimura et al., 2010). YM-254890 was found to bind a hydrophobic cleft between two interdomain linkers that connect the Ras-like and α-helical domains of the Gαq subunit, thereby restricting interdomain movement presumed to be required for GDP/GTP exchange (Remmers et al., 1999). This mechanism of action as a GDP dissociation inhibitor explains earlier findings (Takasaki et al., 2004) that YM-254890 is unable to inhibit cellular signaling by Gαq(Q209L), yet can inhibit signaling by Gαq(R183C). As mentioned previously, the Q209L mutation creates a persistent GTP-bound, activated state and thus does not require on-going GDP/GTP exchange for persistent cellular signaling; in contrast, the fractional occupancy of the R183C mutant has been reported as only 40% GTP-loaded (Kleuss et al., 1994), presumably reflecting a remaining degree of GTP hydrolysis that, in the cellular context, requires either spontaneous nucleotide exchange or a return to Gβγ complexation and receptor-catalyzed exchange for continued function in driving downstream signaling. These exchange events by Gαq(R183C) can be blocked by YM-254890.
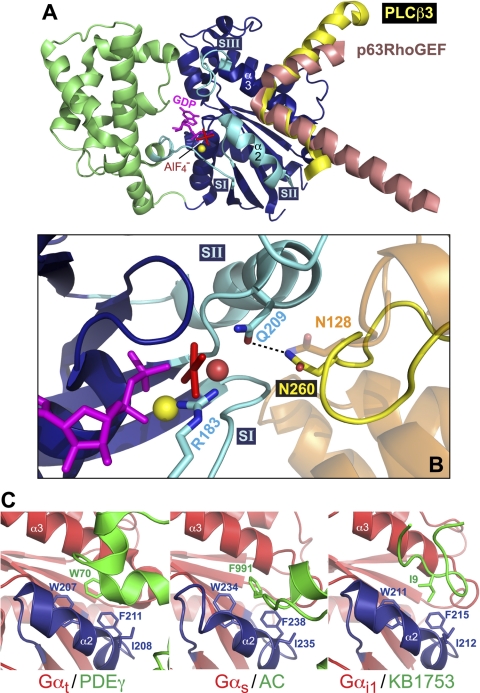
In considering peptide-based therapeutics inspired from Gαq-binding proteins, in addition to the GRK2/Gαq complex mentioned above, two more recent high-resolution structures have been reported of active-state Gαq in complex with key binding partners/effector proteins: namely p63RhoGEF (Lutz et al., 2007) and, more recently, PLCβ3 (Waldo et al., 2010). Both effector proteins bind selectively to activated Gαq through the use of a helix-turn-helix configuration of their respective secondary structures that engages a hydrophobic patch between switch II (α2-helix) and α3-helix of the Gα subunit (Fig. 10); this engagement is a common structural theme seen in multiple other Gα subunit/Gα-effector pairings (e.g., Tesmer et al., 1997b; Slep et al., 2001; Chen et al., 2005; Johnston et al., 2006), including Gαi1-GDP-AlF4− bound to KB-1753 (Fig. 10C), the effector-mimetic peptide originally identified in a phage-display screen for nucleotide-state-selective binding peptides specific for the Gαi/o family (for review, see Johnston et al., 2008). Strategies to identify analogous peptides that could serve as effector antagonists of activated Gαq and Gα11 could therefore include phage or mRNA display with either a semirandom peptide library as starting material (Ja et al., 2006; Johnston et al., 2008) or a peptide library that preserves core contact residues from the helix-turn-helix region of p63RhoGEF or PLCβ3 (Lutz et al., 2007; Waldo et al., 2010). Gαq/11-binding peptides thus identified might be able to serve directly as leads for therapeutic development or else provide useful affinity probes for the creation of HTS assays of compound libraries for small molecules able to displace their binding, thereby identifying “effector antagonist” chemicals for further development as therapeutics. A similar strategy of HTS assay development was recently pursued with a short peptide from the RGS12 GoLoco motif that binds selectively to inactive-state Gαi subunits (Kimple et al., 2001); fluorescent dye-labeling of this GoLoco-peptide enabled a fluorescence polarization assay to be developed for the 1536-well form factor (Kimple et al., 2008) with excellent signal-to-noise and reproducibility characteristics (Z factor >0.8). It is hoped that one of these outlined strategies will prove fruitful in developing a Gαq/11-targeted therapy to this particular cancer.
Fig. 10.
Shared structural determinants of binding and GAP activity on activated Gαq, as elucidated via recent high-resolution structures of Gαq/PLCβ3 and Gαq/p63RhoGEF complexes. A, the two main effectors of Gαq-mediated signaling, PLCβ (yellow) and p63RhoGEF (salmon), share a similar helix-turn-helix configuration in their binding sites for activated Gαq and engage the same hydrophobic cleft circumscribed by the switch II (SII) (α2) and α3 helices of Gα. Structural coordinates were rendered in PyMol using PDB numbers 2RGN and 3OHM. Elements of Gαq are colored similarly to the Gα illustrations of Fig. 2. SI, switch I. B, PLCβ isoforms are not only effectors but also GAPs for Gαq/11 subunits (section I.C.4). The crystal structure of the Gαq/PLCβ3 complex revealed a mechanism for accelerating Gα GTPase activity similar to that of the RGS proteins such as RGS4 (light orange; PDB number 1AGR): namely, helping to orient the critical hydrolytic glutamine residue (Gln-209 in Gαq) via contact with an asparagine residue (Asn-260 in PLCβ3, Asn-128 in RGS4). This Gln-209 residue is mutated in Gαq and Gα11 with high frequency in uveal melanoma, so it is unlikely that the GAP activity of PLCβ or a Gαq/11-selective RGS protein would be beneficial in a gene-therapy strategy for uveal melanoma (section IV.A). Magnesium cation and water are illustrated as yellow and red spheres, respectively. C, consistent with the Gαq/PLCβ3 and Gαq/p63RhoGEF structures, the α2-α3 groove of activated Gα subunits is recognized by other effector molecules. In the GTP-bound state, switch II of Gα (α2 helix in blue) is oriented alongside the α3 helix (red), providing a hydrophobic groove that is used by diverse effector molecules (green). Left, transducin-α engages the inhibitory γ subunit of retinal phosphodiesterase (PDB number 1FQJ). Middle, Gαs recognizes adenylyl cyclase (PDB number 1AZS). Right, the phage display peptide KB1753 binds to activated Gαi1 (PDB number 2G83). These three effector molecules each insert a hydrophobic side chain into the α2-α3 groove (Trp-70, Phe-991, and Ile-9, respectively), suggesting this α2-α3 groove position as a potential site for small molecule manipulation of the Gα subunit/effector interaction.
Acknowledgments
This work was supported by the National Institutes of Health National Institute on Drug Abuse [Grant R03-DA030555] (to D.P.S.); the National Institutes of Health National Institute of General Medical Sciences [Grants R01-GM062338, R01-GM074268, R01-GM082892] (to D.P.S.), [Grant T32-GM008719] (to A.J.K., D.E.B.), [Grant T32-GM007040 (to D.E.B.)]; the National Institutes of Health National Institute of Mental Health [F30-MH074266] (to A.J.K.); and a postdoctoral fellowship from the Heart and Stroke Foundation of Canada (to P.M.G.).
Authorship Contributions
Wrote or contributed to the writing of the manuscript: Kimple, Bosch, Giguère, and Siderovski.
This article is available online at http://pharmrev.aspetjournals.org.
doi:10.1124/pr.110.003038.
- CaM
- calmodulin
- CCG-4986
- methyl-N-[(4-chlorophenyl)sulfonyl]-4-nitrobenzenesulfinimidoate
- CCG-50014
- 4-[(4-fluorophenyl)methyl]-2-(4-methylphenyl)-1,2,4-thiadiazolidine-3,5-dione
- CCG-63802
- (2E)-2-(1,3-benzothiazol-2-yl)-3-[9-methyl-2-(3-methylphenoxy)-4-oxo-4H-pyrido[1,2-a]pyrimidin-3-yl]prop-2-enenitrile
- CCG-63808
- (2E)-2-(1,3-benzothiazol-2-yl)-3-[9-methyl-2-(4-fluorolphenoxy)-4-oxo-4H-pyrido[1,2-a]pyrimidin-3-yl]prop-2-enenitrile
- FP
- fluorescence polarization
- GAP
- GTPase-accelerating protein
- GEF
- guanine nucleotide exchange factor
- GIRK
- G-protein coupled inwardly rectifying potassium channel
- GoLoco
- Gαi/o-Loco interaction motif
- GPA1
- yeast Gα subunit
- GPCR
- G-protein-coupled receptor
- HTS
- high-throughput screen
- IL
- interleukin
- mAChR
- muscarinic acetylcholine receptor
- PDB
- Protein Data Bank
- PIP3
- phosphatidylinositol-3,4,5-trisphosphate
- PLC
- phospholipase C
- RGS
- regulator of G-protein signaling
- siRNA
- small interfering RNA
- Sst
- supersensitive
- YM-254890
- (1R)-1-{(3S,6S,9S,12S,18R,21S,22R)-21-acetamido-18-benzyl-3-[(1R)-1-methoxyethyl]-4,9,10,12,16,22-hexamethyl-15-methylene-2,5,8,11,14,17,–20-heptaoxo-1,19-dioxa-4,7,10,13,16-pentaazacyclodocosan-6-yl}-2-methylpropyl rel-(2S,3R)-2-acetamido-3-hydroxy-4-methylpentanoate.
References
- Anderson GR, Posokhova E, Martemyanov KA. (2009) The R7 RGS protein family: multi-subunit regulators of neuronal G protein signaling. Cell Biochem Biophys 54:33–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anger T, Zhang W, Mende U. (2004) Differential contribution of GTPase activation and effector antagonism to the inhibitory effect of RGS proteins on Gq-mediated signaling in vivo. J Biol Chem 279:3906–3915 [DOI] [PubMed] [Google Scholar]
- Apanovitch DM, Slep KC, Sigler PB, Dohlman HG. (1998) Sst2 is a GTPase-activating protein for Gpa1: purification and characterization of a cognate RGS-Galpha protein pair in yeast. Biochemistry 37:4815–4822 [DOI] [PubMed] [Google Scholar]
- Arshavsky VY, Lamb TD, Pugh EN., Jr (2002) G proteins and phototransduction. Annu Rev Physiol 64:153–187 [DOI] [PubMed] [Google Scholar]
- Ashihara E, Kawata E, Maekawa T. (2010) Future prospect of RNA interference for cancer therapies. Curr Drug Targets 11:345–360 [DOI] [PubMed] [Google Scholar]
- Bansal G, Xie Z, Rao S, Nocka KH, Druey KM. (2008) Suppression of immunoglobulin E-mediated allergic responses by regulator of G protein signaling 13. Nat Immunol 9:73–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes WG, Reiter E, Violin JD, Ren XR, Milligan G, Lefkowitz RJ. (2005) beta-Arrestin 1 and Galphaq/11 coordinately activate RhoA and stress fiber formation following receptor stimulation. J Biol Chem 280:8041–8050 [DOI] [PubMed] [Google Scholar]
- Benians A, Nobles M, Hosny S, Tinker A. (2005) Regulators of G-protein signaling form a quaternary complex with the agonist, receptor, and G-protein. A novel explanation for the acceleration of signaling activation kinetics. J Biol Chem 280:13383–13394 [DOI] [PubMed] [Google Scholar]
- Berger M, Bergers G, Arnold B, Hämmerling GJ, Ganss R. (2005) Regulator of G-protein signaling-5 induction in pericytes coincides with active vessel remodeling during neovascularization. Blood 105:1094–1101 [DOI] [PubMed] [Google Scholar]
- Berman DM, Wilkie TM, Gilman AG. (1996) GAIP and RGS4 are GTPase-activating proteins for the Gi subfamily of G protein alpha subunits. Cell 86:445–452 [DOI] [PubMed] [Google Scholar]
- Bernstein LS, Ramineni S, Hague C, Cladman W, Chidiac P, Levey AI, Hepler JR. (2004) RGS2 binds directly and selectively to the M1 muscarinic acetylcholine receptor third intracellular loop to modulate Gq/11alpha signaling. J Biol Chem 279:21248–21256 [DOI] [PubMed] [Google Scholar]
- Berridge MJ. (2009) Inositol trisphosphate and calcium signalling mechanisms. Biochim Biophys Acta 1793:933–940 [DOI] [PubMed] [Google Scholar]
- Berstein G, Blank JL, Jhon DY, Exton JH, Rhee SG, Ross EM. (1992) Phospholipase C-beta 1 is a GTPase-activating protein for Gq/11, its physiologic regulator. Cell 70:411–418 [DOI] [PubMed] [Google Scholar]
- Blazer LL, Roman DL, Chung A, Larsen MJ, Greedy BM, Husbands SM, Neubig RR. (2010) Reversible, allosteric small-molecule inhibitors of regulator of G protein signaling proteins. Mol Pharmacol 78:524–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazer LL, Zhang H, Casey EM, Husbands SM, Neubig RR. (2011) A nanomolar-potency small molecule inhibitor of regulator of G protein signaling proteins. Biochemistry 50:3181–3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodenstein J, Sunahara RK, Neubig RR. (2007) N-terminal residues control proteasomal degradation of RGS2, RGS4, and RGS5 in human embryonic kidney 293 cells. Mol Pharmacol 71:1040–1050 [DOI] [PubMed] [Google Scholar]
- Bohm A, Gaudet R, Sigler PB. (1997) Structural aspects of heterotrimeric G-protein signaling. Curr Opin Biotechnol 8:480–487 [DOI] [PubMed] [Google Scholar]
- Bollag G, Hirth P, Tsai J, Zhang J, Ibrahim PN, Cho H, Spevak W, Zhang C, Zhang Y, Habets G, Burton EA, Wong B, Tsang G, West BL, Powell B, Shellooe R, Marimuthu A, Nguyen H, Zhang KY, Artis DR, Schlessinger J, Su F, Higgins B, Iyer R, D'Andrea K, Koehler A, Stumm M, Lin PS, Lee RJ, Grippo J, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, Chapman PB, Flaherty KT, Xu X, Nathanson KL, Nolop K. (2010) Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature 467:596–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonacci TM, Mathews JL, Yuan C, Lehmann DM, Malik S, Wu D, Font JL, Bidlack JM, Smrcka AV. (2006) Differential targeting of Gbetagamma-subunit signaling with small molecules. Science 312:443–446 [DOI] [PubMed] [Google Scholar]
- Bondjers C, Kalén M, Hellström M, Scheidl SJ, Abramsson A, Renner O, Lindahl P, Cho H, Kehrl J, Betsholtz C. (2003) Transcription profiling of platelet-derived growth factor-B-deficient mouse embryos identifies RGS5 as a novel marker for pericytes and vascular smooth muscle cells. Am J Pathol 162:721–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt DR, Ross EM. (1985) GTPase activity of the stimulatory GTP-binding regulatory protein of adenylate cyclase, Gs. Accumulation and turnover of enzyme-nucleotide intermediates. J Biol Chem 260:266–272 [PubMed] [Google Scholar]
- Breitwieser GE, Szabo G. (1988) Mechanism of muscarinic receptor-induced K+ channel activation as revealed by hydrolysis-resistant GTP analogues. J Gen Physiol 91:469–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinks HL, Eckhart AD. (2010) Regulation of GPCR signaling in hypertension. Biochim Biophys Acta 1802:1268–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera JL, de Freitas F, Satpaev DK, Slepak VZ. (1998) Identification of the Gbeta5-RGS7 protein complex in the retina. Biochem Biophys Res Commun 249:898–902 [DOI] [PubMed] [Google Scholar]
- Cantrell DA, Smith KA. (1984) The interleukin-2 T-cell system: a new cell growth model. Science 224:1312–1316 [DOI] [PubMed] [Google Scholar]
- Carman CV, Parent JL, Day PW, Pronin AN, Sternweis PM, Wedegaertner PB, Gilman AG, Benovic JL, Kozasa T. (1999) Selective regulation of Galpha(q/11) by an RGS domain in the G protein-coupled receptor kinase, GRK2. J Biol Chem 274:34483–34492 [DOI] [PubMed] [Google Scholar]
- Cassel D, Eckstein F, Lowe M, Selinger Z. (1979) Determination of the turn-off reaction for the hormone-activated adenylate cyclase. J Biol Chem 254:9835–9838 [PubMed] [Google Scholar]
- Chan RK, Otte CA. (1982a) Isolation and genetic analysis of Saccharomyces cerevisiae mutants supersensitive to G1 arrest by a factor and alpha factor pheromones. Mol Cell Biol 2:11–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan RK, Otte CA. (1982b) Physiological characterization of Saccharomyces cerevisiae mutants supersensitive to G1 arrest by a factor and alpha factor pheromones. Mol Cell Biol 2:21–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chasse SA, Dohlman HG. (2003) RGS proteins: G protein-coupled receptors meet their match. Assay Drug Dev Technol 1:357–364 [DOI] [PubMed] [Google Scholar]
- Chen H, Clark MA, Lambert NA. (2004) Endogenous RGS proteins regulate presynaptic and postsynaptic function: functional expression of RGS-insensitive Galpha subunits in central nervous system neurons. Methods Enzymol 389:190–204 [DOI] [PubMed] [Google Scholar]
- Chen Z, Singer WD, Sternweis PC, Sprang SR. (2005) Structure of the p115RhoGEF rgRGS domain-Galpha13/i1 chimera complex suggests convergent evolution of a GTPase activator. Nat Struct Mol Biol 12:191–197 [DOI] [PubMed] [Google Scholar]
- Cho H, Harrison K, Kehrl JH. (2004) Regulators of G protein signaling: potential drug targets for controlling cardiovascular and immune function. Curr Drug Targets Immune Endocr Metabol Disord 4:107–118 [DOI] [PubMed] [Google Scholar]
- Cho H, Kozasa T, Bondjers C, Betsholtz C, Kehrl JH. (2003) Pericyte-specific expression of Rgs5: implications for PDGF and EDG receptor signaling during vascular maturation. FASEB J 17:440–442 [DOI] [PubMed] [Google Scholar]
- Cho H, Park C, Hwang IY, Han SB, Schimel D, Despres D, Kehrl JH. (2008) Rgs5 targeting leads to chronic low blood pressure and a lean body habitus. Mol Cell Biol 28:2590–2597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cifelli C, Rose RA, Zhang H, Voigtlaender-Bolz J, Bolz SS, Backx PH, Heximer SP. (2008) RGS4 regulates parasympathetic signaling and heart rate control in the sinoatrial node. Circ Res 103:527–535 [DOI] [PubMed] [Google Scholar]
- Clapham DE, Neer EJ. (1997) G protein beta gamma subunits. Annu Rev Pharmacol Toxicol 37:167–203 [DOI] [PubMed] [Google Scholar]
- Clark MJ, Traynor JR. (2004) Assays for G-protein-coupled receptor signaling using RGS-insensitive Galpha subunits. Methods Enzymol 389:155–169 [DOI] [PubMed] [Google Scholar]
- Coleman DE, Berghuis AM, Lee E, Linder ME, Gilman AG, Sprang SR. (1994) Structures of active conformations of Gi alpha 1 and the mechanism of GTP hydrolysis. Science 265:1405–1412 [DOI] [PubMed] [Google Scholar]
- Coleman DE, Sprang SR. (1999) Structure of Gialpha1.GppNHp, autoinhibition in a galpha protein-substrate complex. J Biol Chem 274:16669–16672 [DOI] [PubMed] [Google Scholar]
- Cox AD, Der CJ. (2002) Ras family signaling: therapeutic targeting. Cancer Biol Ther 1:599–606 [DOI] [PubMed] [Google Scholar]
- Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, Davis N, Dicks E, Ewing R, Floyd Y, Gray K, Hall S, Hawes R, Hughes J, Kosmidou V, Menzies A, Mould C, Parker A, Stevens C, Watt S, Hooper S, Wilson R, Jayatilake H, Gusterson BA, Cooper C, Shipley J, Hargrave D, Pritchard-Jones K, Maitland N, Chenevix-Trench G, Riggins GJ, Bigner DD, Palmieri G, Cossu A, Flanagan A, Nicholson A, Ho JW, Leung SY, Yuen ST, Weber BL, Seigler HF, Darrow TL, Paterson H, Marais R, Marshall CJ, Wooster R, Stratton MR, Futreal PA. (2002) Mutations of the BRAF gene in human cancer. Nature 417:949–954 [DOI] [PubMed] [Google Scholar]
- Day PW, Tesmer JJ, Sterne-Marr R, Freeman LC, Benovic JL, Wedegaertner PB. (2004) Characterization of the GRK2 binding site of Galphaq. J Biol Chem 279:53643–53652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vries L, Mousli M, Wurmser A, Farquhar MG. (1995) GAIP, a protein that specifically interacts with the trimeric G protein G alpha i3, is a member of a protein family with a highly conserved core domain. Proc Natl Acad Sci USA 92:11916–11920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhanasekaran N, Heasley LE, Johnson GL. (1995) G protein-coupled receptor systems involved in cell growth and oncogenesis. Endocr Rev 16:259–270 [DOI] [PubMed] [Google Scholar]
- DiBello PR, Garrison TR, Apanovitch DM, Hoffman G, Shuey DJ, Mason K, Cockett MI, Dohlman HG. (1998) Selective uncoupling of RGS action by a single point mutation in the G protein alpha-subunit. J Biol Chem 273:5780–5784 [DOI] [PubMed] [Google Scholar]
- Dietzel C, Kurjan J. (1987) The yeast SCG1 gene: a G alpha-like protein implicated in the a- and alpha-factor response pathway. Cell 50:1001–1010 [DOI] [PubMed] [Google Scholar]
- Dohlman HG, Apaniesk D, Chen Y, Song J, Nusskern D. (1995) Inhibition of G-protein signaling by dominant gain-of-function mutations in Sst2p, a pheromone desensitization factor in Saccharomyces cerevisiae. Mol Cell Biol 15:3635–3643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsam RT, Gutkind JS. (2007) G-protein-coupled receptors and cancer. Nat Rev Cancer 7:79–94 [DOI] [PubMed] [Google Scholar]
- Doupnik CA, Davidson N, Lester HA, Kofuji P. (1997) RGS proteins reconstitute the rapid gating kinetics of gbetagamma-activated inwardly rectifying K+ channels. Proc Natl Acad Sci USA 94:10461–10466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druey KM, Blumer KJ, Kang VH, Kehrl JH. (1996) Inhibition of G-protein-mediated MAP kinase activation by a new mammalian gene family. Nature 379:742–746 [DOI] [PubMed] [Google Scholar]
- Efimov IR, Fedorov VV, Joung B, Lin SF. (2010) Mapping cardiac pacemaker circuits: methodological puzzles of the sinoatrial node optical mapping. Circ Res 106:255–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evanko DS, Thiyagarajan MM, Siderovski DP, Wedegaertner PB. (2001) Gbeta gamma isoforms selectively rescue plasma membrane localization and palmitoylation of mutant Galphas and Galphaq. J Biol Chem 276:23945–23953 [DOI] [PubMed] [Google Scholar]
- Feig LA, Cooper GM. (1988) Relationship among guanine nucleotide exchange, GTP hydrolysis, and transforming potential of mutated ras proteins. Mol Cell Biol 8:2472–2478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald K, Tertyshnikova S, Moore L, Bjerke L, Burley B, Cao J, Carroll P, Choy R, Doberstein S, Dubaquie Y, Franke Y, Kopczynski J, Korswagen H, Krystek SR, Lodge NJ, Plasterk R, Starrett J, Stouch T, Thalody G, Wayne H, van der Linden A, Zhang Y, Walker SG, Cockett M, Wardwell-Swanson J, Ross-Macdonald P, Kindt RM. (2006) Chemical genetics reveals an RGS/G-protein role in the action of a compound. PLoS Genet 2:e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaherty KT, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, O'Dwyer PJ, Lee RJ, Grippo JF, Nolop K, Chapman PB. (2010) Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med 363:809–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredriksson R, Lagerström MC, Lundin LG, Schiöth HB. (2003) The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol Pharmacol 63:1256–1272 [DOI] [PubMed] [Google Scholar]
- Frye SV. (2010) The art of the chemical probe. Nat Chem Biol 6:159–161 [DOI] [PubMed] [Google Scholar]
- Fu Y, Huang X, Piao L, Lopatin AN, Neubig RR. (2007) Endogenous RGS proteins modulate SA and AV nodal functions in isolated heart: implications for sick sinus syndrome and AV block. Am J Physiol Heart Circ Physiol 292:H2532–H2539 [DOI] [PubMed] [Google Scholar]
- Fu Y, Huang X, Zhong H, Mortensen RM, D'Alecy LG, Neubig RR. (2006) Endogenous RGS proteins and Galpha subtypes differentially control muscarinic and adenosine-mediated chronotropic effects. Circ Res 98:659–666 [DOI] [PubMed] [Google Scholar]
- Fu Y, Zhong H, Nanamori M, Mortensen RM, Huang X, Lan K, Neubig RR. (2004) RGS-insensitive G-protein mutations to study the role of endogenous RGS proteins. Methods Enzymol 389:229–243 [DOI] [PubMed] [Google Scholar]
- Fukuhara S, Chikumi H, Gutkind JS. (2001) RGS-containing RhoGEFs: the missing link between transforming G proteins and Rho? Oncogene 20:1661–1668 [DOI] [PubMed] [Google Scholar]
- Garcia-Marcos M, Ghosh P, Farquhar MG. (2011) Molecular basis of a novel oncogenic mutation in GNAO1. Oncogene doi:10.1038/onc.2010.645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudet R, Bohm A, Sigler PB. (1996) Crystal structure at 2.4 angstroms resolution of the complex of transducin betagamma and its regulator, phosducin. Cell 87:577–588 [DOI] [PubMed] [Google Scholar]
- Geladopoulos TP, Sotiroudis TG, Evangelopoulos AE. (1991) A malachite green colorimetric assay for protein phosphatase activity. Anal Biochem 192:112–116 [DOI] [PubMed] [Google Scholar]
- Gilfillan AM, Rivera J. (2009) The tyrosine kinase network regulating mast cell activation. Immunol Rev 228:149–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilman AG. (1987) G proteins: transducers of receptor-generated signals. Annu Rev Biochem 56:615–649 [DOI] [PubMed] [Google Scholar]
- Goldenstein BL, Nelson BW, Xu K, Luger EJ, Pribula JA, Wald JM, O'Shea LA, Weinshenker D, Charbeneau RA, Huang X, Neubig RR, Doze VA. (2009) Regulator of G protein signaling protein suppression of Galphao protein-mediated alpha2A adrenergic receptor inhibition of mouse hippocampal CA3 epileptiform activity. Mol Pharmacol 75:1222–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grillet N, Pattyn A, Contet C, Kieffer BL, Goridis C, Brunet JF. (2005) Generation and characterization of Rgs4 mutant mice. Mol Cell Biol 25:4221–4228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu S, Tirgari S, Heximer SP. (2008) The RGS2 gene product from a candidate hypertension allele shows decreased plasma membrane association and inhibition of Gq. Mol Pharmacol 73:1037–1043 [DOI] [PubMed] [Google Scholar]
- Gurley SB, Griffiths RC, Mendelsohn ME, Karas RH, Coffman TM. (2010) Renal actions of RGS2 control blood pressure. J Am Soc Nephrol 21:1847–1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamzah J, Jugold M, Kiessling F, Rigby P, Manzur M, Marti HH, Rabie T, Kaden S, Gröne HJ, Hämmerling GJ, Arnold B, Ganss R. (2008) Vascular normalization in Rgs5-deficient tumours promotes immune destruction. Nature 453:410–414 [DOI] [PubMed] [Google Scholar]
- Harden TK. (1992) G-protein-regulated phospholipase C. Identification of component proteins. Adv Second Messenger Phosphoprotein Res 26:11–34 [PubMed] [Google Scholar]
- Harris DM, Cohn HI, Pesant S, Eckhart AD. (2008) GPCR signalling in hypertension: role of GRKs. Clin Sci (Lond) 115:79–89 [DOI] [PubMed] [Google Scholar]
- Hartwell LH. (1980) Mutants of Saccharomyces cerevisiae unresponsive to cell division control by polypeptide mating hormone. J Cell Biol 85:811–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W, Cowan CW, Wensel TG. (1998) RGS9, a GTPase accelerator for phototransduction. Neuron 20:95–102 [DOI] [PubMed] [Google Scholar]
- Hepler JR, Berman DM, Gilman AG, Kozasa T. (1997) RGS4 and GAIP are GTPase-activating proteins for Gq alpha and block activation of phospholipase C beta by gamma-thio-GTP-Gq alpha. Proc Natl Acad Sci USA 94:428–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heximer SP, Cristillo AD, Forsdyke DR. (1997a) Comparison of mRNA expression of two regulators of G-protein signaling, RGS1/BL34/1R20 and RGS2/G0S8, in cultured human blood mononuclear cells. DNA Cell Biol 16:589–598 [DOI] [PubMed] [Google Scholar]
- Heximer SP, Knutsen RH, Sun X, Kaltenbronn KM, Rhee MH, Peng N, Oliveira-dos-Santos A, Penninger JM, Muslin AJ, Steinberg TH, Wyss JM, Mecham RP, Blumer KJ. (2003) Hypertension and prolonged vasoconstrictor signaling in RGS2-deficient mice. J Clin Invest 111:445–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heximer SP, Watson N, Linder ME, Blumer KJ, Hepler JR. (1997b) RGS2/G0S8 is a selective inhibitor of Gqalpha function. Proc Natl Acad Sci USA 94:14389–14393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashijima T, Ferguson KM, Sternweis PC, Smigel MD, Gilman AG. (1987) Effects of Mg2+ and the beta gamma-subunit complex on the interactions of guanine nucleotides with G proteins. J Biol Chem 262:762–766 [PubMed] [Google Scholar]
- Hooks SB, Martemyanov K, Zachariou V. (2008) A role of RGS proteins in drug addiction. Biochem Pharmacol 75:76–84 [DOI] [PubMed] [Google Scholar]
- Huang X, Charbeneau RA, Fu Y, Kaur K, Gerin I, MacDougald OA, Neubig RR. (2008) Resistance to diet-induced obesity and improved insulin sensitivity in mice with a regulator of G protein signaling-insensitive G184S Gnai2 allele. Diabetes 57:77–85 [DOI] [PubMed] [Google Scholar]
- Huang X, Fu Y, Charbeneau RA, Saunders TL, Taylor DK, Hankenson KD, Russell MW, D'Alecy LG, Neubig RR. (2006) Pleiotropic phenotype of a genomic knock-in of an RGS-insensitive G184S Gnai2 allele. Mol Cell Biol 26:6870–6879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt TW, Fields TA, Casey PJ, Peralta EG. (1996) RGS10 is a selective activator of G alpha i GTPase activity. Nature 383:175–177 [DOI] [PubMed] [Google Scholar]
- Hurst JH, Hooks SB. (2009) Regulator of G-protein signaling (RGS) proteins in cancer biology. Biochem Pharmacol 78:1289–1297 [DOI] [PubMed] [Google Scholar]
- Huss KL, Blonigen PE, Campbell RM. (2007) Development of a Transcreener kinase assay for protein kinase A and demonstration of concordance of data with a filter-binding assay format. J Biomol Screen 12:578–584 [DOI] [PubMed] [Google Scholar]
- Icaza EE, Huang X, Fu Y, Neubig RR, Baghdoyan HA, Lydic R. (2009) Isoflurane-induced changes in righting response and breathing are modulated by RGS proteins. Anesth Analg 109:1500–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda SR, Jeong SW. (2004) Use of RGS-insensitive Galpha subunits to study endogenous RGS protein action on G-protein modulation of N-type calcium channels in sympathetic neurons. Methods Enzymol 389:170–189 [DOI] [PubMed] [Google Scholar]
- Ishii M, Fujita S, Yamada M, Hosaka Y, Kurachi Y. (2005) Phosphatidylinositol 3,4,5-trisphosphate and Ca2+/calmodulin competitively bind to the regulators of G-protein-signalling (RGS) domain of RGS4 and reciprocally regulate its action. Biochem J 385:65–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii M, Inanobe A, Fujita S, Makino Y, Hosoya Y, Kurachi Y. (2001) Ca(2+) elevation evoked by membrane depolarization regulates G protein cycle via RGS proteins in the heart. Circ Res 89:1045–1050 [DOI] [PubMed] [Google Scholar]
- Ishii M, Inanobe A, Kurachi Y. (2002) PIP3 inhibition of RGS protein and its reversal by Ca2+/calmodulin mediate voltage-dependent control of the G protein cycle in a cardiac K+ channel. Proc Natl Acad Sci USA 99:4325–4330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii M, Kurachi Y. (2003) Physiological actions of regulators of G-protein signaling (RGS) proteins. Life Sci 74:163–171 [DOI] [PubMed] [Google Scholar]
- Ishii M, Kurachi Y. (2004) Assays of RGS protein modulation by phosphatidylinositides and calmodulin. Methods Enzymol 389:105–118 [DOI] [PubMed] [Google Scholar]
- Ja WW, Wiser O, Austin RJ, Jan LY, Roberts RW. (2006) Turning G proteins on and off using peptide ligands. ACS Chem Biol 1:570–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby E, Bouhelal R, Gerspacher M, Seuwen K. (2006) The 7 TM G-protein-coupled receptor target family. ChemMedChem 1:761–782 [DOI] [PubMed] [Google Scholar]
- Johnston CA, Lobanova ES, Shavkunov AS, Low J, Ramer JK, Blaesius R, Fredericks Z, Willard FS, Kuhlman B, Arshavsky VY, Siderovski DP. (2006) Minimal determinants for binding activated G alpha from the structure of a G alpha(i1)-peptide dimer. Biochemistry 45:11390–11400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston CA, Siderovski DP. (2007) Receptor-mediated activation of heterotrimeric G-proteins: current structural insights. Mol Pharmacol 72:219–230 [DOI] [PubMed] [Google Scholar]
- Johnston CA, Willard FS, Ramer JK, Blaesius R, Roques CN, Siderovski DP. (2008) State-selective binding peptides for heterotrimeric G-protein subunits: novel tools for investigating G-protein signaling dynamics. Comb Chem High Throughput Screen 11:370–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammermeier PJ, Ruiz-Velasco V, Ikeda SR. (2000) A voltage-independent calcium current inhibitory pathway activated by muscarinic agonists in rat sympathetic neurons requires both Galpha q/11 and Gbeta gamma. J Neurosci 20:5623–5629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan Z, Jaiswal BS, Stinson J, Janakiraman V, Bhatt D, Stern HM, Yue P, Haverty PM, Bourgon R, Zheng J, Moorhead M, Chaudhuri S, Tomsho LP, Peters BA, Pujara K, Cordes S, Davis DP, Carlton VE, Yuan W, Li L, Wang W, Eigenbrot C, Kaminker JS, Eberhard DA, Waring P, Schuster SC, Modrusan Z, Zhang Z, Stokoe D, de Sauvage FJ, Faham M, Seshagiri S. (2010) Diverse somatic mutation patterns and pathway alterations in human cancers. Nature 466:869–873 [DOI] [PubMed] [Google Scholar]
- Kehrl JH, Srikumar D, Harrison K, Wilson GL, Shi CS. (2002) Additional 5′ exons in the RGS3 locus generate multiple mRNA transcripts, one of which accounts for the origin of human PDZ-RGS3. Genomics 79:860–868 [DOI] [PubMed] [Google Scholar]
- Kimple AJ, Soundararajan M, Hutsell SQ, Roos AK, Urban DJ, Setola V, Temple BR, Roth BL, Knapp S, Willard FS, Siderovski DP. (2009) Structural determinants of G-protein alpha subunit selectivity by regulator of G-protein signaling 2 (RGS2). J Biol Chem 284:19402–19411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimple AJ, Willard FS, Giguère PM, Johnston CA, Mocanu V, Siderovski DP. (2007) The RGS protein inhibitor CCG-4986 is a covalent modifier of the RGS4 Galpha-interaction face. Biochim Biophys Acta 1774:1213–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimple AJ, Yasgar A, Hughes M, Jadhav A, Willard FS, Muller RE, Austin CP, Inglese J, Ibeanu GC, Siderovski DP, Simeonov A. (2008) A high throughput fluorescence polarization assay for inhibitors of the GoLoco motif/G-alpha interaction. Comb Chem High Throughput Screen 11:396–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimple RJ, De Vries L, Tronchère H, Behe CI, Morris RA, Gist Farquhar M, Siderovski DP. (2001) RGS12 and RGS14 GoLoco motifs are G alpha(i) interaction sites with guanine nucleotide dissociation inhibitor Activity. J Biol Chem 276:29275–29281 [DOI] [PubMed] [Google Scholar]
- Kinet JP. (1999) The high-affinity IgE receptor (Fc epsilon RI): from physiology to pathology. Annu Rev Immunol 17:931–972 [DOI] [PubMed] [Google Scholar]
- Kleman-Leyer KM, Klink TA, Kopp AL, Westermeyer TA, Koeff MD, Larson BR, Worzella TJ, Pinchard CA, van de Kar SA, Zaman GJ, Hornberg JJ, Lowery RG. (2009) Characterization and optimization of a red-shifted fluorescence polarization ADP detection assay. Assay Drug Dev Technol 7:56–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleuss C, Raw AS, Lee E, Sprang SR, Gilman AG. (1994) Mechanism of GTP hydrolysis by G-protein alpha subunits. Proc Natl Acad Sci USA 91:9828–9831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klink TA, Kleman-Leyer KM, Kopp A, Westermeyer TA, Lowery RG. (2008) Evaluating PI3 kinase isoforms using Transcreener ADP assays. J Biomol Screen 13:476–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koelle MR, Horvitz HR. (1996) EGL-10 regulates G protein signaling in the C. elegans nervous system and shares a conserved domain with many mammalian proteins. Cell 84:115–125 [DOI] [PubMed] [Google Scholar]
- Kozasa T, Jiang X, Hart MJ, Sternweis PM, Singer WD, Gilman AG, Bollag G, Sternweis PC. (1998) p115 RhoGEF, a GTPase activating protein for Galpha12 and Galpha13. Science 280:2109–2111 [DOI] [PubMed] [Google Scholar]
- Kunapuli SP, Dorsam RT, Kim S, Quinton TM. (2003) Platelet purinergic receptors. Curr Opin Pharmacol 3:175–180 [DOI] [PubMed] [Google Scholar]
- Lagerström MC, Schiöth HB. (2008) Structural diversity of G protein-coupled receptors and significance for drug discovery. Nat Rev Drug Discov 7:339–357 [DOI] [PubMed] [Google Scholar]
- Lambert NA, Johnston CA, Cappell SD, Kuravi S, Kimple AJ, Willard FS, Siderovski DP. (2010) Regulators of G-protein signaling accelerate GPCR signaling kinetics and govern sensitivity solely by accelerating GTPase activity. Proc Natl Acad Sci USA 107:7066–7071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambright DG, Noel JP, Hamm HE, Sigler PB. (1994) Structural determinants for activation of the alpha-subunit of a heterotrimeric G protein. Nature 369:621–628 [DOI] [PubMed] [Google Scholar]
- Lan KL, Sarvazyan NA, Taussig R, Mackenzie RG, DiBello PR, Dohlman HG, Neubig RR. (1998) A point mutation in Galphao and Galphai1 blocks interaction with regulator of G protein signaling proteins. J Biol Chem 273:12794–12797 [DOI] [PubMed] [Google Scholar]
- Landis CA, Masters SB, Spada A, Pace AM, Bourne HR, Vallar L. (1989) GTPase inhibiting mutations activate the alpha chain of Gs and stimulate adenylyl cyclase in human pituitary tumours. Nature 340:692–696 [DOI] [PubMed] [Google Scholar]
- Lanier SM. (2004) AGS proteins, GPR motifs and the signals processed by heterotrimeric G proteins. Biol Cell 96:369–372 [DOI] [PubMed] [Google Scholar]
- Laroche G, Giguère PM, Roth BL, Trejo J, Siderovski DP. (2010) RNA interference screen for RGS protein specificity at muscarinic and protease-activated receptors reveals bidirectional modulation of signaling. Am J Physiol Cell Physiol 299:C654–C664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Doerks T, Bork P. (2009) SMART 6: recent updates and new developments. Nucleic Acids Res 37:D229–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy MN, Pappano AJ, Berne RM. (2007) Cardiovascular Physiology, Mosby Elsevier, Philadelphia [Google Scholar]
- Liebmann C. (2004) G protein-coupled receptors and their signaling pathways: classical therapeutical targets susceptible to novel therapeutic concepts. Curr Pharm Des 10:1937–1958 [DOI] [PubMed] [Google Scholar]
- Liu Y, Zalameda L, Kim KW, Wang M, McCarter JD. (2007) Discovery of acetyl-coenzyme A carboxylase 2 inhibitors: comparison of a fluorescence intensity-based phosphate assay and a fluorescence polarization-based ADP Assay for high-throughput screening. Assay Drug Dev Technol 5:225–235 [DOI] [PubMed] [Google Scholar]
- Lodowski DT, Pitcher JA, Capel WD, Lefkowitz RJ, Tesmer JJ. (2003) Keeping G proteins at bay: a complex between G protein-coupled receptor kinase 2 and Gbetagamma. Science 300:1256–1262 [DOI] [PubMed] [Google Scholar]
- Lutz S, Shankaranarayanan A, Coco C, Ridilla M, Nance MR, Vettel C, Baltus D, Evelyn CR, Neubig RR, Wieland T, Tesmer JJ. (2007) Structure of Galphaq-p63RhoGEF-RhoA complex reveals a pathway for the activation of RhoA by GPCRs. Science 318:1923–1927 [DOI] [PubMed] [Google Scholar]
- Lyons J, Landis CA, Harsh G, Vallar L, Grünewald K, Feichtinger H, Duh QY, Clark OH, Kawasaki E, Bourne HR. (1990) Two G protein oncogenes in human endocrine tumors. Science 249:655–659 [DOI] [PubMed] [Google Scholar]
- MacKay VL, Welch SK, Insley MY, Manney TR, Holly J, Saari GC, Parker ML. (1988) The Saccharomyces cerevisiae BAR1 gene encodes an exported protein with homology to pepsin. Proc Natl Acad Sci USA 85:55–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino ER, Handy JW, Li T, Arshavsky VY. (1999) The GTPase activating factor for transducin in rod photoreceptors is the complex between RGS9 and type 5 G protein beta subunit. Proc Natl Acad Sci USA 96:1947–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzur M, Ganss R. (2009) Regulator of G protein signaling 5: a new player in vascular remodeling. Trends Cardiovasc Med 19:26–30 [DOI] [PubMed] [Google Scholar]
- Marin EP, Krishna AG, Sakmar TP. (2001) Rapid activation of transducin by mutations distant from the nucleotide-binding site: evidence for a mechanistic model of receptor-catalyzed nucleotide exchange by G proteins. J Biol Chem 276:27400–27405 [DOI] [PubMed] [Google Scholar]
- May LT, Leach K, Sexton PM, Christopoulos A. (2007) Allosteric modulation of G protein-coupled receptors. Annu Rev Pharmacol Toxicol 47:1–51 [DOI] [PubMed] [Google Scholar]
- Metz M, Maurer M. (2007) Mast cells–key effector cells in immune responses. Trends Immunol 28:234–241 [DOI] [PubMed] [Google Scholar]
- Mirnics K, Middleton FA, Stanwood GD, Lewis DA, Levitt P. (2001) Disease-specific changes in regulator of G-protein signaling 4 (RGS4) expression in schizophrenia. Mol Psychiatry 6:293–301 [DOI] [PubMed] [Google Scholar]
- Mittal V, Linder ME. (2006) Biochemical characterization of RGS14: RGS14 activity towards G-protein alpha subunits is independent of its binding to Rap2A. Biochem J 394:309–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyajima I, Nakafuku M, Nakayama N, Brenner C, Miyajima A, Kaibuchi K, Arai K, Kaziro Y, Matsumoto K. (1987) GPA1, a haploid-specific essential gene, encodes a yeast homolog of mammalian G protein which may be involved in mating factor signal transduction. Cell 50:1011–1019 [DOI] [PubMed] [Google Scholar]
- Moratz C, Hayman JR, Gu H, Kehrl JH. (2004) Abnormal B-cell responses to chemokines, disturbed plasma cell localization, and distorted immune tissue architecture in Rgs1-/- mice. Mol Cell Biol 24:5767–5775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossessova E, Corpina RA, Goldberg J. (2003) Crystal structure of ARF1*Sec7 complexed with Brefeldin A and its implications for the guanine nucleotide exchange mechanism. Mol Cell 12:1403–1411 [DOI] [PubMed] [Google Scholar]
- Moy FJ, Chanda PK, Cockett MI, Edris W, Jones PG, Mason K, Semus S, Powers R. (2000) NMR structure of free RGS4 reveals an induced conformational change upon binding Galpha. Biochemistry 39:7063–7073 [DOI] [PubMed] [Google Scholar]
- Nakayama N, Kaziro Y, Arai K, Matsumoto K. (1988) Role of STE genes in the mating factor signaling pathway mediated by GPA1 in Saccharomyces cerevisiae. Mol Cell Biol 8:3777–3783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neubig RR, Siderovski DP. (2002) Regulators of G-protein signalling as new central nervous system drug targets. Nat Rev Drug Discov 1:187–197 [DOI] [PubMed] [Google Scholar]
- Nieuwenhuijsen BW, Huang Y, Wang Y, Ramirez F, Kalgaonkar G, Young KH. (2003) A dual luciferase multiplexed high-throughput screening platform for protein-protein interactions. J Biomol Screen 8:676–684 [DOI] [PubMed] [Google Scholar]
- Nishimura A, Kitano K, Takasaki J, Taniguchi M, Mizuno N, Tago K, Hakoshima T, Itoh H. (2010) Structural basis for the specific inhibition of heterotrimeric Gq protein by a small molecule. Proc Natl Acad Sci USA 107:13666–13671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishina H, Nimota K, Kukimoto I, Maehama T, Takahashi K, Hoshino S, Kanaho Y, Katada T. (1995) Significance of Thr182 in the nucleotide-exchange and GTP-hydrolysis reactions of the alpha subunit of GTP-binding protein Gi2. J Biochem 118:1083–1089 [DOI] [PubMed] [Google Scholar]
- Oldham WM, Hamm HE. (2008) Heterotrimeric G protein activation by G-protein-coupled receptors. Nat Rev Mol Cell Biol 9:60–71 [DOI] [PubMed] [Google Scholar]
- Oliveira-Dos-Santos AJ, Matsumoto G, Snow BE, Bai D, Houston FP, Whishaw IQ, Mariathasan S, Sasaki T, Wakeham A, Ohashi PS, Roder JC, Barnes CA, Siderovski DP, Penninger JM. (2000) Regulation of T cell activation, anxiety, and male aggression by RGS2. Proc Natl Acad Sci USA 97:12272–12277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onken MD, Worley LA, Long MD, Duan S, Council ML, Bowcock AM, Harbour JW. (2008) Oncogenic mutations in GNAQ occur early in uveal melanoma. Invest Ophthalmol Vis Sci 49:5230–5234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overington JP, Al-Lazikani B, Hopkins AL. (2006) How many drug targets are there? Nat Rev Drug Discov 5:993–996 [DOI] [PubMed] [Google Scholar]
- Page RD. (1996) TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12:357–358 [DOI] [PubMed] [Google Scholar]
- Perez DM. (2003) The evolutionarily triumphant G-protein-coupled receptor. Mol Pharmacol 63:1202–1205 [DOI] [PubMed] [Google Scholar]
- Ponting CP. (1999) Raf-like Ras/Rap-binding domains in RGS12- and still-life-like signalling proteins. J Mol Med 77:695–698 [DOI] [PubMed] [Google Scholar]
- Popov S, Yu K, Kozasa T, Wilkie TM. (1997) The regulators of G protein signaling (RGS) domains of RGS4, RGS10, and GAIP retain GTPase activating protein activity in vitro. Proc Natl Acad Sci USA 94:7216–7220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popov SG, Krishna UM, Falck JR, Wilkie TM. (2000) Ca2+/Calmodulin reverses phosphatidylinositol 3,4,5-trisphosphate-dependent inhibition of regulators of G protein-signaling GTPase-activating protein activity. J Biol Chem 275:18962–18968 [DOI] [PubMed] [Google Scholar]
- Posner BA, Gilman AG, Harris BA. (1999) Regulators of G protein signaling 6 and 7. Purification of complexes with gbeta5 and assessment of their effects on g protein-mediated signaling pathways. J Biol Chem 274:31087–31093 [DOI] [PubMed] [Google Scholar]
- Posner BA, Mixon MB, Wall MA, Sprang SR, Gilman AG. (1998) The A326S mutant of Gialpha1 as an approximation of the receptor-bound state. J Biol Chem 273:21752–21758 [DOI] [PubMed] [Google Scholar]
- Posokhova E, Wydeven N, Allen KL, Wickman K, Martemyanov KA. (2010) RGS6/Gβ5 complex accelerates IKACh gating kinetics in atrial myocytes and modulates parasympathetic regulation of heart rate. Circ Res 107:1350–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raw AS, Coleman DE, Gilman AG, Sprang SR. (1997) Structural and biochemical characterization of the GTPgammaS-, GDP.Pi-, and GDP-bound forms of a GTPase-deficient Gly42→Val mutant of Gialpha1. Biochemistry 36:15660–15669 [DOI] [PubMed] [Google Scholar]
- Remmers AE, Engel C, Liu M, Neubig RR. (1999) Interdomain interactions regulate GDP release from heterotrimeric G proteins. Biochemistry 38:13795–13800 [DOI] [PubMed] [Google Scholar]
- Rhee SG. (2001) Regulation of phosphoinositide-specific phospholipase C. Annu Rev Biochem 70:281–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle EL, Schwartzman RA, Bond M, Insel PA. (2005) Multi-tasking RGS proteins in the heart: the next therapeutic target? Circ Res 96:401–411 [DOI] [PubMed] [Google Scholar]
- Rivera J, Gilfillan AM. (2006) Molecular regulation of mast cell activation. J Allergy Clin Immunol 117:1214–1225; quiz 1226 [DOI] [PubMed] [Google Scholar]
- Robillard L, Ethier N, Lachance M, Hébert TE. (2000) Gbetagamma subunit combinations differentially modulate receptor and effector coupling in vivo. Cell Signal 12:673–682 [DOI] [PubMed] [Google Scholar]
- Roman DL, Blazer LL, Monroy CA, Neubig RR. (2010) Allosteric inhibition of the regulator of G protein signaling-Galpha protein-protein interaction by CCG-4986. Mol Pharmacol 78:360–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman DL, Ota S, Neubig RR. (2009) Polyplexed flow cytometry protein interaction assay: a novel high-throughput screening paradigm for RGS protein inhibitors. J Biomol Screen 14:610–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman DL, Talbot JN, Roof RA, Sunahara RK, Traynor JR, Neubig RR. (2007) Identification of small-molecule inhibitors of RGS4 using a high-throughput flow cytometry protein interaction assay. Mol Pharmacol 71:169–175 [DOI] [PubMed] [Google Scholar]
- Ross EM. (2002) Quantitative assays for GTPase-activating proteins. Methods Enzymol 344:601–617 [DOI] [PubMed] [Google Scholar]
- Ruiz de Azua I, Scarselli M, Rosemond E, Gautam D, Jou W, Gavrilova O, Ebert PJ, Levitt P, Wess J. (2010) RGS4 is a negative regulator of insulin release from pancreatic beta-cells in vitro and in vivo. Proc Natl Acad Sci USA 107:7999–8004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryschich E, Schmidt J, Hämmerling GJ, Klar E, Ganss R. (2002) Transformation of the microvascular system during multistage tumorigenesis. Int J Cancer 97:719–725 [DOI] [PubMed] [Google Scholar]
- Saitoh O, Kubo Y, Miyatani Y, Asano T, Nakata H. (1997) RGS8 accelerates G-protein-mediated modulation of K+ currents. Nature 390:525–529 [DOI] [PubMed] [Google Scholar]
- Sallese M, Mariggiò S, D'Urbano E, Iacovelli L, De Blasi A. (2000) Selective regulation of Gq signaling by G protein-coupled receptor kinase 2: direct interaction of kinase N terminus with activated Galphaq. Mol Pharmacol 57:826–831 [PubMed] [Google Scholar]
- Sato T. (2010) Locoregional management of hepatic metastasis from primary uveal melanoma. Semin Oncol 37:127–138 [DOI] [PubMed] [Google Scholar]
- Schuster R, Lindner M, Wacker F, Krössin M, Bechrakis N, Foerster MH, Thiel E, Keilholz U, Schmittel A. (2010) Transarterial chemoembolization of liver metastases from uveal melanoma after failure of systemic therapy: toxicity and outcome. Melanoma Res 20:191–196 [DOI] [PubMed] [Google Scholar]
- Scott JK, Huang SF, Gangadhar BP, Samoriski GM, Clapp P, Gross RA, Taussig R, Smrcka AV. (2001) Evidence that a protein-protein interaction ‘hot spot’ on heterotrimeric G protein betagamma subunits is used for recognition of a subclass of effectors. EMBO J 20:767–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semplicini A, Strapazzon G, Papparella I, Sartori M, Realdi A, Macchini L, Calò LA, Ceolotto G. (2010) RGS2 expression and aldosterone: renin ratio modulate response to drug therapy in hypertensive patients. J Hypertens 28:1104–1108 [DOI] [PubMed] [Google Scholar]
- Shu FJ, Ramineni S, Hepler JR. (2010) RGS14 is a multifunctional scaffold that integrates G protein and Ras/Raf MAPkinase signalling pathways. Cell Signal 22:366–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shutes A, Der CJ. (2005) Real-time in vitro measurement of GTP hydrolysis. Methods 37:183–189 [DOI] [PubMed] [Google Scholar]
- Siderovski DP, Blum S, Forsdyke RE, Forsdyke DR. (1990) A set of human putative lymphocyte G0/G1 switch genes includes genes homologous to rodent cytokine and zinc finger protein-encoding genes. DNA Cell Biol 9:579–587 [DOI] [PubMed] [Google Scholar]
- Siderovski DP, Hessel A, Chung S, Mak TW, Tyers M. (1996) A new family of regulators of G-protein-coupled receptors? Curr Biol 6:211–212 [DOI] [PubMed] [Google Scholar]
- Siderovski DP, Heximer SP, Forsdyke DR. (1994) A human gene encoding a putative basic helix-loop-helix phosphoprotein whose mRNA increases rapidly in cycloheximide-treated blood mononuclear cells. DNA Cell Biol 13:125–147 [DOI] [PubMed] [Google Scholar]
- Siderovski DP, Kimple AJ, Willard FS. (2009) Large G-proteins, in Wiley Encyclopedia of Chemical Biology (Begley TP. ed), John Wiley & Sons, Hoboken, NJ [Google Scholar]
- Signarvic RS, Cierniewska A, Stalker TJ, Fong KP, Chatterjee MS, Hess PR, Ma P, Diamond SL, Neubig RR, Brass LF. (2010) RGS/Gi2alpha interactions modulate platelet accumulation and thrombus formation at sites of vascular injury. Blood 116:6092–6100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonds WF. (1999) G protein regulation of adenylate cyclase. Trends Pharmacol Sci 20:66–73 [DOI] [PubMed] [Google Scholar]
- Sjögren B, Blazer LL, Neubig RR. (2010) Regulators of G protein signaling proteins as targets for drug discovery. Prog Mol Biol Transl Sci 91:81–119 [DOI] [PubMed] [Google Scholar]
- Sjögren B, Neubig RR. (2010) Thinking outside of the “RGS box”: new approaches to therapeutic targeting of regulators of G protein signaling. Mol Pharmacol 78:550–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slep KC, Kercher MA, He W, Cowan CW, Wensel TG, Sigler PB. (2001) Structural determinants for regulation of phosphodiesterase by a G protein at 2.0 A. Nature 409:1071–1077 [DOI] [PubMed] [Google Scholar]
- Slep KC, Kercher MA, Wieland T, Chen CK, Simon MI, Sigler PB. (2008) Molecular architecture of Galphao and the structural basis for RGS16-mediated deactivation. Proc Natl Acad Sci USA 105:6243–6248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith NJ, Bennett KA, Milligan G. (2011) When simple agonism is not enough: emerging modalities of GPCR ligands. Mol Cell Endocrinol 331:241–247 [DOI] [PubMed] [Google Scholar]
- Snow BE, Betts L, Mangion J, Sondek J, Siderovski DP. (1999) Fidelity of G protein beta-subunit association by the G protein gamma-subunit-like domains of RGS6, RGS7, and RGS11. Proc Natl Acad Sci USA 96:6489–6494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow BE, Hall RA, Krumins AM, Brothers GM, Bouchard D, Brothers CA, Chung S, Mangion J, Gilman AG, Lefkowitz RJ, Siderovski DP. (1998a) GTPase activating specificity of RGS12 and binding specificity of an alternatively spliced PDZ (PSD-95/Dlg/ZO-1) domain. J Biol Chem 273:17749–17755 [DOI] [PubMed] [Google Scholar]
- Snow BE, Krumins AM, Brothers GM, Lee SF, Wall MA, Chung S, Mangion J, Arya S, Gilman AG, Siderovski DP. (1998b) A G protein gamma subunit-like domain shared between RGS11 and other RGS proteins specifies binding to Gbeta5 subunits. Proc Natl Acad Sci USA 95:13307–13312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodhi A, Montaner S, Gutkind JS. (2004) Viral hijacking of G-protein-coupled-receptor signalling networks. Nat Rev Mol Cell Biol 5:998–1012 [DOI] [PubMed] [Google Scholar]
- Sondek J, Bohm A, Lambright DG, Hamm HE, Sigler PB. (1996) Crystal structure of a G-protein beta gamma dimer at 2.1A resolution. Nature 379:369–374 [DOI] [PubMed] [Google Scholar]
- Sondek J, Lambright DG, Noel JP, Hamm HE, Sigler PB. (1994) GTPase mechanism of Gproteins from the 1.7-A crystal structure of transducin alpha-GDP-AIF-4. Nature 372:276–279 [DOI] [PubMed] [Google Scholar]
- Soundararajan M, Willard FS, Kimple AJ, Turnbull AP, Ball LJ, Schoch GA, Gileadi C, Fedorov OY, Dowler EF, Higman VA, Hutsell SQ, Sundström M, Doyle DA, Siderovski DP. (2008) Structural diversity in the RGS domain and its interaction with heterotrimeric G protein alpha-subunits. Proc Natl Acad Sci USA 105:6457–6462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowa ME, He W, Wensel TG, Lichtarge O. (2000) A regulator of G protein signaling interaction surface linked to effector specificity. Proc Natl Acad Sci USA 97:1483–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegel AM, Weinstein LS. (2004) Inherited diseases involving g proteins and g protein-coupled receptors. Annu Rev Med 55:27–39 [DOI] [PubMed] [Google Scholar]
- Spiegelberg BD, Hamm HE. (2007) Roles of G-protein-coupled receptor signaling in cancer biology and gene transcription. Curr Opin Genet Dev 17:40–44 [DOI] [PubMed] [Google Scholar]
- Srinivasa SP, Watson N, Overton MC, Blumer KJ. (1998) Mechanism of RGS4, a GTPase-activating protein for G protein alpha subunits. J Biol Chem 273:1529–1533 [DOI] [PubMed] [Google Scholar]
- Sugimoto K, Katsuya T, Kamide K, Fujisawa T, Shimaoka I, Ohishi M, Morishita R, Ogihara T, Rakugi H. (2010) Promoter polymorphism of RGS2 gene is associated with change of blood pressure in subjects with antihypertensive treatment: the azelnidipine and temocapril in hypertensive patients with type 2 diabetes study. Int J Hypertens 2010:196307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takasaki J, Saito T, Taniguchi M, Kawasaki T, Moritani Y, Hayashi K, Kobori M. (2004) A novel Galphaq/11-selective inhibitor. J Biol Chem 279:47438–47445 [DOI] [PubMed] [Google Scholar]
- Takeda S, Kadowaki S, Haga T, Takaesu H, Mitaku S. (2002) Identification of G protein-coupled receptor genes from the human genome sequence. FEBS Lett 520:97–101 [DOI] [PubMed] [Google Scholar]
- Takimoto E, Koitabashi N, Hsu S, Ketner EA, Zhang M, Nagayama T, Bedja D, Gabrielson KL, Blanton R, Siderovski DP, Mendelsohn ME, Kass DA. (2009) Regulator of G protein signaling 2 mediates cardiac compensation to pressure overload and antihypertrophic effects of PDE5 inhibition in mice. J Clin Invest 119:408–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot JN, Jutkiewicz EM, Graves SM, Clemans CF, Nicol MR, Mortensen RM, Huang X, Neubig RR, Traynor JR. (2010) RGS inhibition at G(alpha)i2 selectively potentiates 5-HT1A-mediated antidepressant effects. Proc Natl Acad Sci USA 107:11086–11091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang KM, Wang GR, Lu P, Karas RH, Aronovitz M, Heximer SP, Kaltenbronn KM, Blumer KJ, Siderovski DP, Zhu Y, Mendelsohn ME, Tang M, Wang G. (2003) Regulator of G-protein signaling-2 mediates vascular smooth muscle relaxation and blood pressure. Nat Med 9:1506–1512 [DOI] [PubMed] [Google Scholar]
- Taniguchi M, Nagai K, Arao N, Kawasaki T, Saito T, Moritani Y, Takasaki J, Hayashi K, Fujita S, Suzuki K, Tsukamoto S. (2003) YM-254890, a novel platelet aggregation inhibitor produced by Chromobacterium sp. QS3666. J Antibiot (Tokyo) 56:358–363 [DOI] [PubMed] [Google Scholar]
- Tesmer JJ, Berman DM, Gilman AG, Sprang SR. (1997a) Structure of RGS4 bound to AlF4− activated G(i alpha1): stabilization of the transition state for GTP hydrolysis. Cell 89:251–261 [DOI] [PubMed] [Google Scholar]
- Tesmer JJ, Sunahara RK, Gilman AG, Sprang SR. (1997b) Crystal structure of the catalytic domains of adenylyl cyclase in a complex with Gsalpha.GTPgammaS. Science 278:1907–1916 [DOI] [PubMed] [Google Scholar]
- Tesmer VM, Kawano T, Shankaranarayanan A, Kozasa T, Tesmer JJ. (2005) Snapshot of activated G proteins at the membrane: the Galphaq-GRK2-Gbetagamma complex. Science 310:1686–1690 [DOI] [PubMed] [Google Scholar]
- Thomas CJ, Du X, Li P, Wang Y, Ross EM, Sprang SR. (2004) Uncoupling conformational change from GTP hydrolysis in a heterotrimeric G protein alpha-subunit. Proc Natl Acad Sci USA 101:7560–7565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiemann K, Rossi JJ. (2009) RNAi-based therapeutics-current status, challenges and prospects. EMBO Mol Med 1:142–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trahey M, McCormick F. (1987) A cytoplasmic protein stimulates normal N-ras p21 GTPase, but does not affect oncogenic mutants. Science 238:542–545 [DOI] [PubMed] [Google Scholar]
- Traver S, Bidot C, Spassky N, Baltauss T, De Tand MF, Thomas JL, Zalc B, Janoueix-Lerosey I, Gunzburg JD. (2000) RGS14 is a novel Rap effector that preferentially regulates the GTPase activity of galphao. Biochem J 350:19–29 [PMC free article] [PubMed] [Google Scholar]
- Traynor J. (2010) Regulator of G protein-signaling proteins and addictive drugs. Ann NY Acad Sci 1187:341–352 [DOI] [PubMed] [Google Scholar]
- Tu Y, Wilkie TM. (2004) Allosteric regulation of GAP activity by phospholipids in regulators of G-protein signaling. Methods Enzymol 389:89–105 [DOI] [PubMed] [Google Scholar]
- Turcotte M, Tang W, Ross EM. (2008) Coordinate regulation of G protein signaling via dynamic interactions of receptor and GAP. PLoS Comput Biol 4:e1000148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usui H, Nishiyama M, Moroi K, Shibasaki T, Zhou J, Ishida J, Fukamizu A, Haga T, Sekiya S, Kimura S. (2000) RGS domain in the amino-terminus of G protein-coupled receptor kinase 2 inhibits Gq-mediated signaling. Int J Mol Med 5:335–340 [DOI] [PubMed] [Google Scholar]
- Vahrmeijer AL, van de Velde CJ, Hartgrink HH, Tollenaar RA. (2008) Treatment of melanoma metastases confined to the liver and future perspectives. Dig Surg 25:467–472 [DOI] [PubMed] [Google Scholar]
- Van Raamsdonk CD, Bezrookove V, Green G, Bauer J, Gaugler L, O'Brien JM, Simpson EM, Barsh GS, Bastian BC. (2009) Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature 457:599–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Raamsdonk CD, Griewank KG, Crosby MB, Garrido MC, Vemula S, Wiesner T, Obenauf AC, Wackernagel W, Green G, Bouvier N, Sozen MM, Baimukanova G, Roy R, Heguy A, Dolgalev I, Khanin R, Busam K, Speicher MR, O'Brien J, Bastian BC. (2010) Mutations in GNA11 in uveal melanoma. N Engl J Med 363:2191–2199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Veldhoven PP, Mannaerts GP. (1987) Inorganic and organic phosphate measurements in the nanomolar range. Anal Biochem 161:45–48 [DOI] [PubMed] [Google Scholar]
- Vassilatis DK, Hohmann JG, Zeng H, Li F, Ranchalis JE, Mortrud MT, Brown A, Rodriguez SS, Weller JR, Wright AC, Bergmann JE, Gaitanaris GA. (2003) The G protein-coupled receptor repertoires of human and mouse. Proc Natl Acad Sci USA 100:4903–4908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetter IR, Wittinghofer A. (2001) The guanine nucleotide-binding switch in three dimensions. Science 294:1299–1304 [DOI] [PubMed] [Google Scholar]
- Volk DW, Eggan SM, Lewis DA. (2010) Alterations in metabotropic glutamate receptor 1alpha and regulator of G protein signaling 4 in the prefrontal cortex in schizophrenia. Am J Psychiatry 167:1489–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuong TM, Chabre M. (1991) Deactivation kinetics of the transduction cascade of vision. Proc Natl Acad Sci USA 88:9813–9817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldo GL, Ricks TK, Hicks SN, Cheever ML, Kawano T, Tsuboi K, Wang X, Montell C, Kozasa T, Sondek J, Harden TK. (2010) Kinetic scaffolding mediated by a phospholipase C-beta and Gq signaling complex. Science 330:974–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall MA, Coleman DE, Lee E, Iñiguez-Lluhi JA, Posner BA, Gilman AG, Sprang SR. (1995) The structure of the G protein heterotrimer Gi alpha 1 beta 1 gamma 2. Cell 83:1047–1058 [DOI] [PubMed] [Google Scholar]
- Wall MA, Posner BA, Sprang SR. (1998) Structural basis of activity and subunit recognition in G protein heterotrimers. Structure 6:1169–1183 [DOI] [PubMed] [Google Scholar]
- Wang Q, Liu-Chen LY, Traynor JR. (2009) Differential modulation of mu- and delta-opioid receptor agonists by endogenous RGS4 protein in SH-SY5Y cells. J Biol Chem 284:18357–18367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zeng W, Soyombo AA, Tang W, Ross EM, Barnes AP, Milgram SL, Penninger JM, Allen PB, Greengard P, Muallem S. (2005) Spinophilin regulates Ca2+ signalling by binding the N-terminal domain of RGS2 and the third intracellular loop of G-protein-coupled receptors. Nat Cell Biol 7:405–411 [DOI] [PubMed] [Google Scholar]
- Wang Y, Young KH. (2004) Analysis of RGSZ1 protein interaction with Galphai subunits. Methods Enzymol 390:31–52 [DOI] [PubMed] [Google Scholar]
- Watson N, Linder ME, Druey KM, Kehrl JH, Blumer KJ. (1996) RGS family members: GTPase-activating proteins for heterotrimeric G-protein alpha-subunits. Nature 383:172–175 [DOI] [PubMed] [Google Scholar]
- Wedegaertner PB, Wilson PT, Bourne HR. (1995) Lipid modifications of trimeric G proteins. J Biol Chem 270:503–506 [DOI] [PubMed] [Google Scholar]
- Wieland T, Bahtijari N, Zhou XB, Kleuss C, Simon MI. (2000) Polarity exchange at the interface of regulators of G protein signaling with G protein alpha-subunits. J Biol Chem 275:28500–28506 [DOI] [PubMed] [Google Scholar]
- Willard FS, Kimple AJ, Johnston CA, Siderovski DP. (2005) A direct fluorescence-based assay for RGS domain GTPase accelerating activity. Anal Biochem 340:341–351 [DOI] [PubMed] [Google Scholar]
- Willard FS, Kimple RJ, Kimple AJ, Johnston CA, Siderovski DP. (2004a) Fluorescence-based assays for RGS box function. Methods Enzymol 389:56–71 [DOI] [PubMed] [Google Scholar]
- Willard FS, Kimple RJ, Siderovski DP. (2004b) Return of the GDI: the GoLoco motif in cell division. Annu Rev Biochem 73:925–951 [DOI] [PubMed] [Google Scholar]
- Willard FS, Siderovski DP. (2004) Purification and in vitro functional analysis of the Arabidopsis thaliana regulator of G-protein signaling-1. Methods Enzymol 389:320–338 [DOI] [PubMed] [Google Scholar]
- Willard FS, Willard MD, Kimple AJ, Soundararajan M, Oestreich EA, Li X, Sowa NA, Kimple RJ, Doyle DA, Der CJ, Zylka MJ, Snider WD, Siderovski DP. (2009) Regulator of G-protein signaling 14 (RGS14) is a selective H-Ras effector. PLoS One 4:e4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willard MD, Willard FS, Li X, Cappell SD, Snider WD, Siderovski DP. (2007) Selective role for RGS12 as a Ras/Raf/MEK scaffold in nerve growth factor-mediated differentiation. EMBO J 26:2029–2040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie K, Allen KL, Kourrich S, Colón-Saez J, Thomas MJ, Wickman K, Martemyanov KA. (2010) Gbeta5 recruits R7 RGS proteins to GIRK channels to regulate the timing of neuronal inhibitory signaling. Nat Neurosci 13:661–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Zeng W, Popov S, Berman DM, Davignon I, Yu K, Yowe D, Offermanns S, Muallem S, Wilkie TM. (1999) RGS proteins determine signaling specificity of Gq-coupled receptors. J Biol Chem 274:3549–3556 [DOI] [PubMed] [Google Scholar]
- Yang J, Huang J, Maity B, Gao Z, Lorca RA, Gudmundsson H, Li J, Stewart A, Swaminathan PD, Ibeawuchi SR, Shepherd A, Chen CK, Kutschke W, Mohler PJ, Mohapatra DP, Anderson ME, Fisher RA. (2010) RGS6, a modulator of parasympathetic activation in heart. Circ Res 107:1345–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Kamide K, Kokubo Y, Takiuchi S, Tanaka C, Banno M, Miwa Y, Yoshii M, Horio T, Okayama A, Tomoike H, Kawano Y, Miyata T. (2005) Genetic variations of regulator of G-protein signaling 2 in hypertensive patients and in the general population. J Hypertens 23:1497–1505 [DOI] [PubMed] [Google Scholar]
- Yatani A, Mattera R, Codina J, Graf R, Okabe K, Padrell E, Iyengar R, Brown AM, Birnbaumer L. (1988) The G protein-gated atrial K+ channel is stimulated by three distinct Gi alpha-subunits. Nature 336:680–682 [DOI] [PubMed] [Google Scholar]
- Zeng W, Xu X, Popov S, Mukhopadhyay S, Chidiac P, Swistok J, Danho W, Yagaloff KA, Fisher SL, Ross EM, Muallem S, Wilkie TM. (1998) The N-terminal domain of RGS4 confers receptor-selective inhibition of G protein signaling. J Biol Chem 273:34687–34690 [DOI] [PubMed] [Google Scholar]
- Zerangue N, Jan LY. (1998) G-protein signaling: fine-tuning signaling kinetics. Curr Biol 8:R313–316 [DOI] [PubMed] [Google Scholar]
- Zhang JH, Simonds WF. (2000) Copurification of brain G-protein beta5 with RGS6 and RGS7. J Neurosci 20:RC59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong H, Wade SM, Woolf PJ, Linderman JJ, Traynor JR, Neubig RR. (2003) A spatial focusing model for G protein signals. Regulator of G protein signaling (RGS) protein-mediated kinetic scaffolding. J Biol Chem 278:7278–7284 [DOI] [PubMed] [Google Scholar]
- Zielinski T, Kimple AJ, Hutsell SQ, Koeff MD, Siderovski DP, Lowery RG. (2009) Two Galpha(i1) rate-modifying mutations act in concert to allow receptor-independent, steady-state measurements of RGS protein activity. J Biomol Screen 14:1195–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]