Abstract
A down-regulation of reelin and glutamic acid decarboxylase (GAD) 67 mRNAs was detected in γ-aminobutyric acid (GABA)ergic cortical interneurons of schizophrenia (SZ) postmortem brains (10), suggesting that the availability of GABA and reelin may be decreased in SZ cortex. In situ hybridization of the mRNA encoding for DNA-methyltransferase 1, which catalyzes the methylation of promoter CpG islands, shows that the expression of this mRNA is increased in cortical GABAergic interneurons but not in pyramidal neurons of SZ brains. Counts of reelin mRNA-positive neurons in Brodmann's area 10 of either nonpsychiatric subjects or SZ patients show that the expression of reelin mRNA is decreased in layer-I, -II, and -IV GABAergic interneurons of SZ patients. These findings are consistent with the hypothesis that the increase of DNA-methyltransferase 1 expression in telencephalic GABAergic interneurons of SZ patients causes a promoter hypermethylation of reelin and GAD67 and perhaps of other genes expressed in these interneurons. It is difficult to decide whether this dysfunction of GABAergic neurons detected in SZ is responsible for this disease or is a consequence of this disorder. Although at present we cannot differentiate between these two alternatives, it is important to consider that so far a molecular pathology of cortical GABAergic neurons appears to be the most consistent finding associated with SZ morbidity.
In vertebrate neurons, the 5-methylation of the cytosine pyrimidine ring within CpG dinucleotides is catalyzed by DNA-methyltransferases (DNMTs) (1, 2). When the methylation of promoter CpG islands is compared to that of CpGs scattered throughout the genome, promoter CpGs appear to be hypomethylated (3–5). During replication, this difference in DNA methylation pattern can be inherited by daughter DNA strands. In fact, DNMT1 is considered a maintenance enzyme that preferentially catalyzes the methylation of those CpG sequences that are already hemimethylated (for review, see ref. 2).
DNA methylation is frequent in genomic regions that are usually transcriptionally silent, suggesting that DNMT1 and the de novo DNMT3a and DNMT3b activities contribute to gene silencing and to the repression of transposable element activities (1, 2). These elements comprise nearly half the genome, whereas the protein-coding sequences make up ≈5% of the human genome (6). DNA methylation also can be a marker of the gene parental origin, thereby enabling somatic cells to “distinguish” the parental source of each of the two gene copies (7). Because the DNMT1 catalytic activity plays a role in gene silencing and because a decrease of gene expression in cortical γ-aminobutyric acid (GABA)ergic neurons may be operative in schizophrenia (SZ) (8–15), we decided to investigate whether in SZ, the expression of mRNA encoding for DNMT1 is selectively increased in cortical GABAergic neurons. These neurons, by secreting GABA and reelin, may contribute to the orchestration of the intermittent event-related patterns of pyramidal neuron columnary firing. It is pertinent to mention that reelin, which in adult cortex is selectively expressed in GABAergic neurons (16, 17), is secreted from these neurons in the extracellular matrix, presumably by a constitutive mechanism (18, 19). In the extracellular matrix, reelin interacts with integrin receptors expressed in pyramidal neuron dendritic spine postsynaptic densities (19–22) and thereby can modulate the translation of extrasomatic mRNAs expressed in the proximity of pyramidal neuron dendritic spines (22). For instance, reelin, by regulating extrasomatic Arc (activity-regulated cytoskeleton-associated protein) biosynthesis in dendrites, modulates plasticity of excitatory synapses (21–23). Hence, the down-regulation of reelin expression in SZ (21, 22) may contribute to the cortical neuropil hypoplasia and to the low expression density of dendritic spines found in SZ cortex (24–26). In turn, this reduces stimulation by glutamatergic afferents impinging on pyramidal cortical neurons.
The present report investigates whether DNMT1 expression is changed in telencephalic GABAergic interneurons of SZ patients (SZP) compared to nonpsychiatric subjects (NPS) and evaluates whether such a change in DNMT1 expression contributes to a down-regulation of cortical GABAergic inhibition that may be a sign of this disease. The results obtained are consistent with the view that a selective increase of DNMT1 expression in telencephalic GABAergic interneurons of SZP may cause hypermethylation of the gene promoters expressed in these interneurons. This hypermethylation may compromise the GABAergic function in a manner that may lead to a specific molecular neuropathology and associated functional changes characteristic of SZ morbidity.
Methods
Tissue Collection. Specimens of prefrontal cortex (PFC), Brodmann's area 10 (BA10), primary visual cortex BA17, caudate nucleus (CN), and cerebellar hemisphere (cerebellum) from 15 subjects (Table 1) were obtained from the Harvard Brain Tissue Resource Center (Belmont, MA). All specimens were fixed in 4% formaldehyde at the time of autopsy occurring after a postmortem interval (PMI) ranging from 4.75 to 27.5 h. The samples were kept in formaldehyde for 612 ± 265 (SD) days for NPS and 728 ± 105 days for SZP. Each sample studied was transferred into 30% sucrose in 0.1 M PBS 72 h before histological preparation. The psychiatric diagnosis was established by two senior psychiatrists based on clinical and family histories and according to Diagnostic and Statistical Manual of Mental Disorders IV criteria. The demographic data are summarized in Table 1. The mean age ± SD was 56.7 ± 11.2 yr for SZP and 60.9 ± 13.4 yr for NPS (difference was nonsignificant). The PMI was 20.1 ± 5.6 h for SZP and 17.3 ± 6.7 h for NPS (nonsignificant). The male-to-female ratio was 4/3 for SZP and 4/4 for NPS. The left-to-right hemisphere ratio was 4/3 for SZP and 4/4 for NPS. In the SZP group, the age of illness onset was 23 ± 2.5 yr and the duration of illness was 33 ± 8 yr.
Table 1. Demographic characteristics of brains used in this study.
| Diagnosis | Gender | Hemisphere | Age, years | PMI, hours | Age of onset, years | Cause of death* | Antipsychotic medication† | Abuse or dependency history |
|---|---|---|---|---|---|---|---|---|
| NPS | M | R | 60 | 23.27 | — | 1 | — | No |
| NPS | M | R | 68 | 16.58 | — | 1 | — | No |
| NPS | F | R | 41 | 14 | — | 1 | — | No |
| NPS | M | L | 51 | 4.75 | — | 1 | — | No |
| NPS | M | L | 52 | 22.36 | — | 3 | — | No |
| NPS | F | L | 74 | 23 | — | 1 | — | No |
| NPS | F | L | 78 | 23.91 | — | 2 | — | No |
| NPS | F | R | 62 | 16.4 | — | 2 | — | No |
| SZP-U‡ | M | L | 64 | 15.43 | 28 | 1 | Fl, Di, Lo | No |
| SZP-D | M | R | 48 | 27.5 | 19 | 1 | Clz | No |
| SZP-U | F | R | 40 | 18.4 | 18 | 4 | § | No |
| SZP-U | M | R | 49 | 24.5 | — | 3 | Ha, Am | No |
| SZP-U | F | L | 61 | 11 | 32 | 3 | Clz, Par, Clo | Yes¶ |
| SZP-P | F | L | 72 | 21.75 | 24 | 2 | Ri, Par | No |
| SZP-U | M | L | 63 | 22.35 | 21 | 3 | Clz, Clo, Lo | No |
—, Not applicable or not known.
1, Unknown; 2, cancer; 3, myocardial infarction; 4, overdose
Fl, fluphenazine; Ha, haloperidol; Clz, clozapine; Ri, risperidone; Par, paroxetine; Di, valproate; Lo, lorazepam; Am, amitriptyline; Clo, clonazepam
U, undifferentiated; P, paranoia; D, dementia
Did not receive antipsychotic medication in the last 6 months; death by overdose of aspirin and olanzapine
Drug of abuse, alcohol
In Situ Hybridization. To visualize DNMT1 mRNA, free-floating 40-μm sections were incubated for 40–48 h with a mixture of 50 pmol/ml of two antisense oligonucleotide probes complementary to bases 1627–1650 (P1) and 4801–4824 (P2) of the human DNMT1 cDNA (GenBank accession no. NM_001379). These nucleotides did not match either DNMT3a or DNMT3b or any other known mRNA sequences as determined by multiple genomewide BLAST comparisons. However, a BLAT search of P1 and P2 against the UCSC Genome Bioinformatics group (http://genome.ucsc.edu) detected some sequence identity with other genomic sequences. Because oligonucleotide arrays have shown that genomic sequences that have not previously been reported as part of the intron/exon structures of transcribed genes can be transcribed (27), we performed in situ hybridization studies with the two DNMT1 antisense probes separately. In coronal PFC 20-μm slices, the distribution of neurons positively stained with P1 is virtually identical to the distribution of neurons independently detected with P2. We counted the number of neurons positively stained with P1 [layer (L) I = 3.2 ± 0.9 (SD) per mm2 × 102; LII = 8.0 ± 1.3; LIII = 1.7 ± 0.2; LIV = 8.6 ± 1.9; LV = 1.8 ± 0.5; LVI = 3.1 ± 0.5] in the six cortical layers of one NPS (four boxes for each cortical layer) and found this staining to be almost identical to that obtained by using P2 [LI = 3.2 ± 0.9 (SD) per mm2 × 102; LII = 8.3 ± 0.4; LIII = 2.1 ± 0.4; LIV = 9.5 ± 1.2; LV = 2.1 ± 0.5; LVI = 3.4 ± 0.5]. Because these two probes, with the exception of the region encoding DNMT1 mRNA, show identity with different genomic sequences, the results argue against this being due to aberrant or spurious mRNA transcription.
The oligonucleotide 3′ terminals were labeled with a digoxigenin kit (Roche Diagnostics). In situ hybridization was performed according to Rodriguez et al. (28). After the hybridization reactions were completed, tissue sections were incubated for 72 h at 4°C in 0.1 M PBS containing 1% normal goat serum and 0.1 μg/ml of an antibody against digoxigenin (mouse antidigoxigenin monoclonal antiserum from Roche Diagnostics). This procedure was followed by an incubation with biotinylated goat anti-mouse antiserum (Vector Laboratories, 1:250) for 1 h and reacted with the avidin-biotin-peroxidase complex (Vector Laboratories) followed by 3–3′-diaminobenzidine tetrahydrochloride (DAB; Sigma) with 0.1% nickel ammonium sulfate (2–3 min) to obtain a gray-black shade in the precipitate. These immunohistochemical conditions were sufficient to label all neurons expressed in a 20–40 μm section (28).
To detect reelin mRNA, we used a hybridization with antisense probes 1729–1752, 5505–5528, and 10102–10125 of the human reelin cDNA (GenBank accession no. NM_005045). The glutamic acid decarboxylase (GAD) 65 mRNA was also detected with antisense probes 187–231 and 334–378 of the human GAD65 cDNA (accession no. NM_00818).
In pilot experiments with 40-μm floating sections counterstained with neutral red, we established that incubation with 20- to 25-base oligonucleotide probes for 20–24 h was sufficient to label the highest percent of neurons; in fact, the percentage of labeled neurons failed to change significantly when the incubation was prolonged to 72 h. Using the in situ hybridization and immunochemistry conditions described above, there were no differences in the percentage level of staining across a 40-μm section.
To test the specificity of the immunological detection of digoxigenin, oligonucleotides were omitted from the hybridization buffer, and no staining was observed. The oligoprobe specificity was tested by using the digoxigenin-labeled scrambled oligonucleotide sequences. As expected, the specific neuronal staining was not detected.
Immunohistochemistry. To obtain neuron-specific nuclear protein (NeuN) immunolabeling, 40-μm floating sections were incubated for 3 days with a mouse anti-NeuN monoclonal antiserum (Chemicon) diluted 1:500 according to the procedure described by Rodriguez et al. (28).
Double in Situ Hybridization and Immunohistochemistry. Using 16-μm sections, after the DNMT1 mRNA in situ hybridization was completed, these sections were processed for immunohistochemistry with antibodies directed against (i) GAD65/67 (Chemicon, 1:1,000), (ii) glial fibrillary acidic protein (GFAP) (Chemicon, 1:500), and (iii) vesicular glutamate transporter 2 (VGLUT2) (Sysy, Gottingen, Germany, 1:500). The sections were incubated with the antibody for 48 h at 4°C and 2 h at room temperature. After the addition of the secondary antibody, the sections were stained following the protocol described by Rodriguez et al. (28), except that in this case, the sections were reacted with DAB without the addition of nickel to obtain a brown reaction product.
Confocal Fluorescence Microscopy. After the double immunohistochemistry and in situ hybridization procedures, the slices were incubated with Cy5-conjugated goat anti-rabbit IgG (Amersham Biosciences, 1:1,000) to label the antibodies reacted with GAD65/67 (see above) or DNMT1 proteins (New England Biolabs, 1:500). Cy2-conjugated streptavidin (1:500) was used to label DNMT1 or reelin mRNAs. The reactions were carried out in 1% normal goat serum, 1% BSA, and 1% normal rabbit serum in PBS for 1 h. After washing, the sections were incubated in 10 mM CuSO4 and 50 mM CH3COONH4 for 1 h to eliminate the lipofuscin-mediated autofluorescence (29).
Stereological Neuronal Counts. From each brain area of each subject, three sections were taken (one every fourth successive slices), and by using the modifications of Rodriguez et al. (28) and Liu et al. (23) of the three-dimensional cell-counting procedure described by Williams and Rakic (30), the cells were stereologically counted. This method takes advantage of confocal microscopy at a magnification of ×40 to obtain serial optical sections focused through the z axis. The number of DAB-positive cells coming into view (or alternatively disappearing from view) is counted in a box (100 × 100 × 20 (z) μm) entirely embedded within the tissue section. The neurons were counted in three randomly selected columns in each of three brain sections from each subject (SZP or NPS); thus, a total of nine columns per area per subject were counted.
For the CN, three cell-containing fields were randomly selected from each of three sections per subject, and the above-described counting criteria were followed.
The number of cells counted in each box was corrected for tissue shrinkage, which, in our experimental conditions, was estimated at ≈30–35%. The thickness of the sections after histological preparation was measured with confocal microscopy focusing, from the upper to the lower surface of the slice, on sections with a division spacing of 2 μm according to the specification of the manufacturer (Leica).
OD Measurements. Relative OD measurements were performed with SCION IMAGE (www.scioncorp.com) in WINDOWS. OD was measured in randomly selected visual fields (200 × 270 μm) from cortical area BA10 layers II and V and from the CN of seven SZP and seven NPS. After the background was subtracted from each group, the number of positive cells was plotted against the OD.
Digital Photomicrography. Images were captured by AXIOVISION 3.1 (Zeiss). The final composites were processed by using PHOTOSHOP (Adobe Systems, Mountain View, CA) and POWERPOINT (Microsoft) and were printed on photographic paper.
Statistical Analyses. Differences between the SZP and NPS groups in PMI (in hours) between death and autopsy or in clinical variables; age and gender were evaluated with one-way ANOVA or χ2 analyses. To further test whether gender, age, and PMI affect differences between SZP and NPS on DNMT1, reelin mRNAs, or NeuN protein-positive neurons in different cortical layers, we used analyses of covariance (ANCOVA). The differences between SZP and NPS in the number of DNMT1 mRNA, reelin mRNA, NeuN positive neurons, or Nissl-stained cells in the different layers of coronal sections from areas BA10 and BA17 were statistically evaluated by repeated measures ANOVA for the six layers, followed by unpaired t tests of each layer. The differences between SZP and NPS by ANCOVA were compared with those obtained with the unpaired t tests.
Results
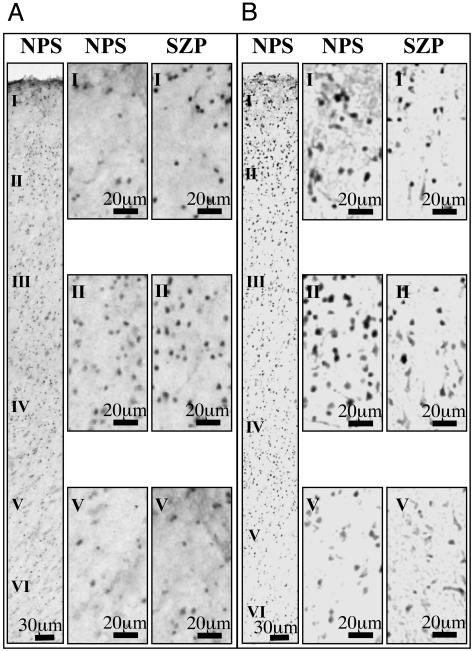
Cortical Overexpression of DNMT1 mRNA in GABAergic Interneurons of SZ Brains. In situ hybridization signals show that in BA10, DNMT1, and reelin mRNAs are detectable in some neurons of every layer of SZP and NPS. In addition, in layers I–IV, DNMT1 mRNA is more abundantly expressed in interneurons of SZP than in those of NPS (Fig. 1A). In contrast, in the same layers, the reelin mRNA hybridization signal was less abundantly expressed in SZP than in NPS (Fig. 1B).
Fig. 1.
DNMT1 mRNA and reelin mRNA in BA10 of NPS and SZP. To the left of each panel are low-magnification micrographs of the six cortical layers of one NPS. Adjacent to the low-magnification images are high-magnification micrographs of selected regions of layers I, II, and V. Note the increase in DNMT1 mRNA labeling in the SZP (A) and the decrease in reelin mRNA labeling in samples from the same SZP (B).
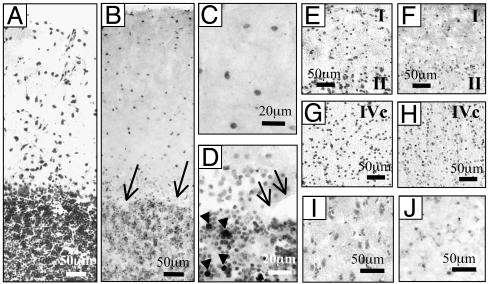
A similar pattern of DNMTI mRNA expression extended to BA17, where most of the interneurons that express DNMT1 were located in layers I, II, and IV (Fig. 2 F and H). In the cerebellum (Fig. 2 B–D), DNMT1 mRNA was expressed in the granular and molecular layers but not in Purkinje neurons. Arrowheads in Fig. 2D indicate heavy DNMT1 expression in a neuronal subgroup of the granular layer. A study of the morphology of these neurons suggests that they may be classified as Golgi type II neurons. Many CN neurons (Fig. 2 I and J) express a moderate hybridization signal for DNMT1 mRNA.
Fig. 2.
DNMT1 mRNA in situ hybridization in several NPS brain areas. (A–D) Cerebellum. (A) Nissl staining. (B–D) DNMT1 in situ hybridization. Positive neurons are found in the granular and molecular layer, whereas Purkinje cells (arrows) fail to show the hybridization signal (B and D). At a larger magnifi-cation, note the presence of heavily stained neurons in the molecular layer (C) and the granular layer (D) that possibly correspond to Golgi type II cells (arrowheads). (E–H) BA17. (E and G) Nissl-stained images of layers I and II (E) and IVc (G). (F and H) DNMT1 in situ hybridization images in layers I and II (F) and layer IVc (H). (I and J) CN. (I) Nissl staining. (J) Moderately stained DNMT1 mRNA-positive neurons.
Colocalization of DNMT1 with GAD65/67 and Reelin. By combining in situ hybridization and immunochemistry, we studied the colocalization of DNMT1 mRNA with a number of other cellular markers in BA10 interneurons (Figs. 3, 4, 5).
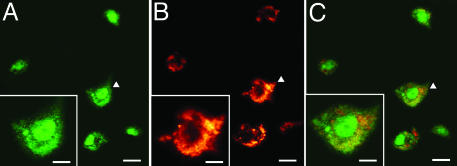
Fig. 3.
Colocalization of DNMT1mRNA and GAD65/67 in BA10 (layer I/II) of one SZP. (A) DNMT1 mRNA-positive neurons, color-coded in green. (Scale bar, 10 μm.) (B) GAD65/67 immunoreactivity, color-coded in red. (Scale bar, 10 μm.) (C) Overlay of A and B. (Scale bar, 10 μm.) At the bottom left of each panel is a large magnification of the cells indicated by arrowheads. (Scale bars, 5 μm.)
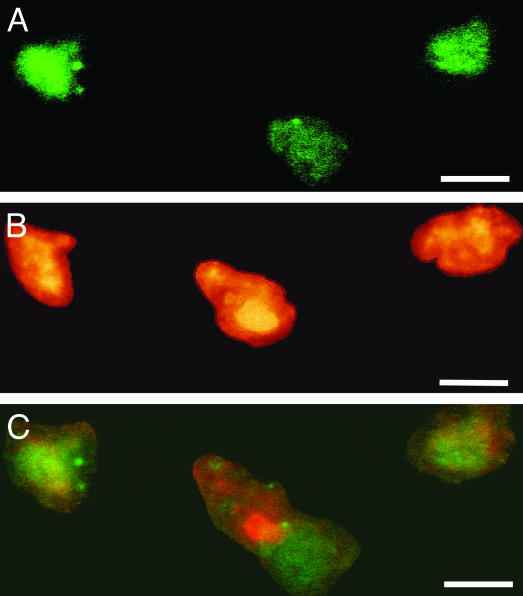
Fig. 4.
Colocalization of reelin mRNA and DNMT1protein in BA10 (layer I/II) of one SZP. (A) Reelin mRNA-positive neurons color-coded in green. (Scale bar, 10 μm.) (B) DNMT1-positive neurons color-coded in red. (Scale bar, 10 μm.) (C) Overlay of A and B. (Scale bar, 10 μm.)
Fig. 5.
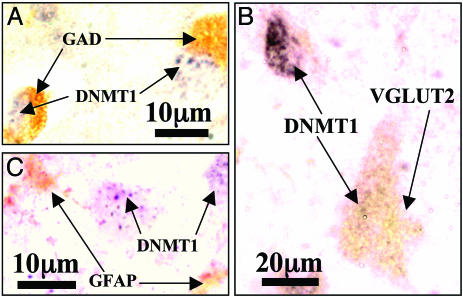
Colocalization of DNMT1 mRNA with various histochemical markers in BA 10 of NPS. (A) DNMT1 mRNA (blue-gray, nucleus) in situ hybridization and GAD65/67 immunolabeling (brown, cytosol) in layer II. (B) DNMT1 mRNA (blue-gray nucleus) and vesicular glutamate transporter 2 (VGLUT2) (brown, cytoplasm) immunolabeling in layer V. (C) DNMT1 mRNA and GFAP immunolabeling (brown, cytosol) in layer II.
Fig. 3 shows that DNMT1 mRNA (A, green fluorescence) and GAD65/67 proteins (B, red fluorescence) are colocalized (overlay of DNMT1 and GAD65/67, C) in the same GABAergic neurons of layer I/II. Fig. 4 shows colocalization (C) of the reelin mRNA in situ hybridization signal (A, green fluorescence) with DNMT1 protein immunostaining (B, red fluorescence) in GABAergic neurons of layer I/II. Fig. 5A shows a double-DAB staining of DNMT1 mRNAs (blue-gray, nucleus) and GAD65/67 proteins (brown, cytosol) in the GABAergic neurons of layer II; Fig. 5B illustrates that the labeling of DNMT1 mRNA in GABAergic neurons and the immunostaining of vesicular glutamate transporter (a marker of glutamatergic neurons) are not colocalized. Fig. 5C shows that DNMT1 mRNA and GFAP staining also is not colocalized.
Taken together, these findings suggest that DNMT1 and reelin are colocalized in a cortical interneuron population that appears to be GABAergic (GAD65/67-positive). Both DNMT1 and reelin mRNA expression are virtually absent in glutamatergic pyramidal neurons (identified morphologically for their typical somata profiles and histochemically for the presence of the vesicular glutamate transporter) or in glial cells (identified by the GFAP expression). Only a few GFAP-positive cells in layer I at the border with pia mater appeared to be DNMT1 mRNA-positive.
The Number of DNMT1-Positive Neurons Is Increased in BA10, BA17, and CN of SZP. When neurons expressing DNMT1 and reelin mRNA were individually labeled by in situ hybridization (Fig. 1 A and B) and counted stereologically, an apparent increase in DNMT1 mRNA-positive neurons and a decrease in reelin mRNA-positive neurons was detected in SZP compared to NPS (Figs. 1 and 6).
Fig. 6.
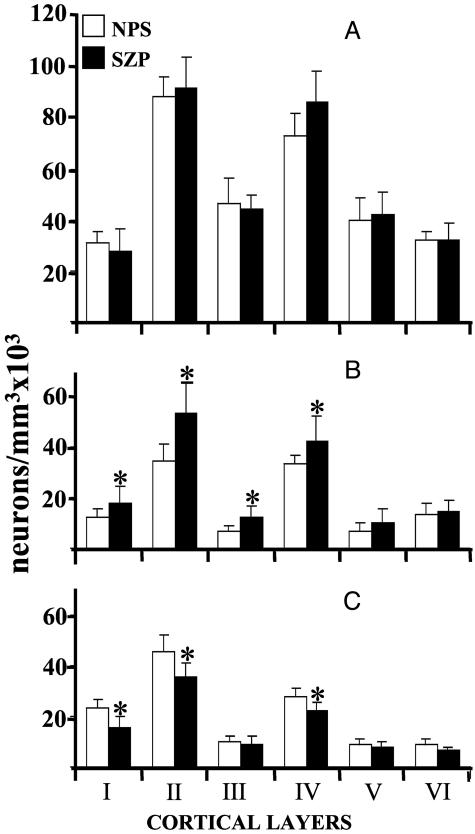
Stereological counts of NeuN (A), DNMT1 mRNA (B), or reelin mRNA (C) in various layers of BA10 from NPS and SZP. Each value is the mean ± SD of seven NPS (open bars) or seven SZP (filled bars). *, Statistically significant difference when SZP are compared with NPS (for statistical analyses, see ‡).
Very likely, a change in the total number of GABAergic neurons cannot explain this difference. In fact, the number of GAD65 mRNA-positive neurons was similar in BA10 coronal sections of NPS and SZP. For example, in layer II of BA10, the number of GAD65 mRNA-positive neurons was 60 ± 8.0 (SD) in NPS and 62 ± 4.2 in SZP (df = 12, t = 0.19, P = 0.85, nonsignificant).
In BA10 layers II and V, the intensity of DNMT1 mRNA-positive neurons in NPS and SZP was measured in terms of relative OD. In keeping with the data of Fig. 1, in layer II, the number of neurons with a high expression of DNMT1 mRNA is greater in SZP than in NPS. However, a similar difference fails to occur in layer V. In contrast, the number of interneurons expressing a relevant signal for reelin mRNA is smaller in SZP than in NPS (data not shown).
Fig. 6 reports the stereological counts of NeuN-immunopositive neurons (A), neurons expressing a high intensity DNMT1 labeling (B), and reelin mRNA-positive neurons (C) in various cortical layers of BA10 from NPS and SZP. There is no difference in the counts of NeuN-immunopositive neurons among the cortical layers of NPS and SZP.
In contrast, the number of DNMT1 (Fig. 6B) mRNA-positive neurons in layers I–IV is more abundant (30–40%) in SZP than in NPS, whereas the number of reelin mRNA-positive neurons in layers I, II, and IV of SZP is lower than that in NPS (Fig. 6C) (for a statistical evaluation of the differences, see ‡).
The differences in reelin and DNMT1 mRNA signal intensities (Fig. 6), which are coexpressed in PFC GABAergic neurons (Figs. 3, 4, and 5A), prompted us to perform stereological counts of neurons expressing DNMT1 mRNA in BA17 and in the CN. In BA17, the counts of neurons positive for DNMT1 mRNA were found to be significantly higher only in cortical layer II of SZP (56 ± 16 SD positive neurons per mm3 × 103, n = 6) compared with NPS (40 ± 5.0, n = 8, P = 0.025). In the CN, the number of DNMT1 mRNA-positive neurons was significantly higher in SZP (41 ± 6 SD positive neurons per mm3 × 103, n = 6) than in NPS (30 ± 6 SD, n = 8; P = 0.023).
Because the accuracy of stereological counts conducted in 40-μm sections may be limited by differences in reagent permeability and because of difficulties in calculating tissue shrinkage during labeling procedures (31), in a few instances we have repeated the counts by using a 2D method and 20-μm sections, obtaining results comparable to the stereological counts in 40-μm sections.
All SZP cases but one (see Table 1) were receiving antipsychotic medications at the time of death. In BA10 of SZP, the number of DNMT1 mRNA-positive neurons in layers I–IV was increased but the number of reelin mRNA-positive neurons was decreased by a comparable extent, regardless of the type of neuroleptic used. Moreover, in one SZP with no history of antipsychotic medication, the expression intensity of DNMT1 and reelin in GABAergic neurons of BA10 was well within the range of the other SZP treated with antipsychotic drugs. In the SZP cohort, only one patient had a history of alcohol and drug abuse. In addition, in this case, in BA10 the expression of DNMT1-positive neurons was higher, whereas that of reelin was lower than the corresponding averages calculated from NPS neuronal counts.
Discussion
The results obtained demonstrate that the overexpression of DNMT1 mRNA and the down-regulation of reelin and GAD67 in cortical GABAergic neurons are specific features of SZ neuropathology. Because DNMT1 methylates the five position of the cytosine pyrimidine ring, we infer that the overexpression of DNMT1 mRNA and protein may be a mechanism causing hypermethylation of reelin and GAD67 promoters leading to the down-regulation of cognate mRNAs and proteins.
In proliferating somatic cells of vertebrates, the regulation of cytosine methylation by DNMT1 is associated with specific sites of DNA synthesis (replication foci) throughout the S phase of the cell cycle and it is expected to correlate with cellular proliferation (32, 33). However, our in situ hybridization studies show that DNMT1 is highly expressed in nonproliferating cortical interneurons, medium spiny CN neurons, and cerebellar granule cells.
These data suggest that, in addition to maintenance of methylation at “replication foci” in proliferating cells, DNMT1 very likely contributes to cell-specific promoter methylation patterns at genomic loci that may control specific gene expression in nonreplicating cell phenotypes such as cortical neurons.
Considering the abundant expression of DNMT1 mRNA in cortical GABAergic interneurons and the much lower expression of this gene in cortical pyramidal neurons, one may suggest that an interneuron-specific epigenetic mechanism may be selective for GABAergic interneurons but may not include glutamatergic pyramidal neurons.
Our studies also show that the presence of high levels of DNMT1 mRNA is not a characteristic of every GABAergic neuronal population; in fact, cerebellar Purkinje cells, which are bona fide GABAergic neurons, do not express high levels of DNMT1 or reelin mRNAs. Hence, the coexpression of reelin and DNMT1 is a property of cortical GABAergic interneurons that cannot be extrapolated to GABAergic Purkinje cells, which in fact are not interneurons but the principal neurons of the cerebellar cortex innervating the deep cerebellar nuclei.
It is of interest to mention that cortical GABAergic interneurons of layers I, II, and IV, which are among the fastest spiking neurons expressed in the cortex (34), are those that also contain the highest amounts of DNMT1 (Figs. 3, 4, and 6), suggesting that the expression intensity of this enzyme may regulate some characteristics of neuronal function and/or activity. The present demonstration that the DNMT1 mRNA expression is increased in GABAergic cortical neurons of SZP (Figs. 1, 3, and 6), taken together with the increase in the cytosine methylation of the reelin promoter (35) and the decrease of reelin and GAD67 mRNA expression (Fig. 6C), supports the hypothesis that an increase of the epigenetically induced methylation in GABAergic neurons may be a neuropathological event that characterizes SZ morbidity (36).
Reelin mRNA-positive interneurons are particularly abundant in layers I, II, and IV of BA10. This finding is in keeping with the results of a reelin study in monkeys, in which the brain specimens were fixed with paraformaldehyde shortly after death (28), and is also supported by independent evidence (14) showing that GABAergic interneurons present in BA22 cortical layers and in the interstitial white matter also express reelin mRNA.
Our quantitative RT-PCR measurements of reelin mRNA in PFC of NPS and SZP (10) suggest that the number of interneurons expressing reelin in layers I, II, and IV of BA10 is decreased in SZP in the absence of changes in the number of NeuN or GAD65-positive neurons. Thus, the decrease in the number of interneurons expressing reelin observed in BA10 of SZP most likely is not due to neuronal loss but probably relates to changes in the extent of reelin down-regulation. However, the magnitude of the reelin expression deficit measured histochemically (20–30%) in SZP is lower than that evaluated with quantitative RT-PCR measures of reelin mRNA expression in PFC samples of SZP, which showed a decrease by ≈40% (10, 11). This discrepancy is explainable, because a histochemical evaluation of mRNA expression is devoid of an internal standard correction and has a low discrimination capacity. Hence, the histochemical method may not be appropriate to conduct quantitative comparative studies of reelin or DNMT1 mRNA expression intensity.
Except for one patient, the SZP group was receiving antipsychotic medications at the time of death. To investigate whether increased DNMT1 expression rather than the antipsychotic treatment is at the root of the above-mentioned GABAergic molecular pathology, we studied mice treated with haloperidol (1 mg/kg s.c. twice a day for 21 days) or clozapine (5 mg/kg s.c. twice a day for 21 days). This treatment failed to change DNMT1 mRNA expression, which was quantified with competitive RTPCR in cortex and cerebellum.
The transcriptional mechanism(s) that regulate DNMT1 mRNA expression in cortical GABAergic neurons is (are) presently unknown. It is possible that the increase in DNMT1 expression we detected in cortical GABAergic neurons of SZP may be secondary to the formation of a chromatin remodeling complex including DNMT1 and various corepressors, a molecular species of histone deacetylase and/or methyl transferase, and/or other factors attending chromatin remodeling that regulate the DNA access of transcription modulators (37–39).
A fraction of DNMT1 that we measured in cortical GABAergic neurons is most likely associated with chromatin remodeling complexes that surround nucleosome domains expressed in GABAergic neuronal nuclei. Presumably, the DNMT1 overexpression detected in cortical GABAergic interneurons of SZP is targeted to specific nucleosomal chromatin domains.
Taken together, these considerations reinforce our view that SZ is characterized by signs of epigenetic molecular neuropathology in GABAergic neurons that decrease transcription of a number of genes including reelin and GAD67 (10, 21, 36). This change in gene regulation very likely contributes to a GABAergic neuronal dysfunction raising the following question: Does SZ specifically or predominantly consist in an alteration of cortical GABAergic function? The data presented in this paper not only are consistent with this hypothesis but also suggest that the increase of DNMT1 expression in cortical and NC GABAergic neurons of SZP may reflect an alteration of some higher-order chromatin remodeling processes. If this were the case, future study of the molecular mechanisms regulating chromatin remodeling could unveil new targets for the treatment of SZ that may directly address a specific pathological process of chromatin remodeling operative in GABAergic neurons of SZP.
In closing, it is interesting to consider that chromatin remodeling may be the next target for pharmacological intervention aimed at the treatment of psychotic symptoms.
Acknowledgments
We thank Dr. Francine M. Benes (Laboratories for Structural Neuroscience, McLean Hospital, Belmont, MA, and the Program in Neuroscience and the Department of Psychiatry, Harvard Medical School, Boston) and Dr. Arturas Petronis (Center for Addiction and Mental Health and the University of Toronto) for constructive criticism and suggestions in the preparation of the manuscript. This work was supported in part by National Institute of Mental Health Grants MH062188 (to A.G.), MH062090 (to E.C.), and MH62682-01 (to D.R.G.).
Abbreviations: BA, Brodmann's area; CN, caudate nucleus; DNMT, DNA methyltransferase; GAD, glutamic acid decarboxylase; GFAP, glial fibrillary acidic protein; NPS, nonpsychiatric subjects; PFC, prefrontal cortex; PMI, postmortem interval; SZ, schizophrenia; SZP, SZ patients; GABA, γ-aminobutyric acid.
Footnotes
Statistical analyses of the data in Fig. 6 with repeated measures ANOVA indicates that the number of DNMT1 mRNA-positive neurons in SZP is significantly higher overall (all layers included) compared to NPS: F = 21.4, df = 1, 12, P = 0.0006. In contrast, the overall number of reelin mRNA-positive neurons is statistically decreased in SZP: F = 14.3 df = 12, P = 0.03. The number of DNMT1-positive neurons in SZP is statistically higher in layers I (t12 = 2.7, P = 0.018), II (t12 = 3.5, P = 0.004), III (t12 = 2.3, P = 0.039), and IV (t12 = 2.4, P = 0.032) but not in layers V (t12 = 1.7, P = 0.11) and VI (t12 = 0.6, P = 0.58). The number of reelin mRNA-positive neurons in SZP is statistically lower in layers I (t12 = 3.6, P = 0.0041), II (t12 = 2.8, P = 0.016), and IV (t12 = 2.7, P = 0.02) but not in layers III (t12 = 0.41, P = 0.69), V (t12 = 0.74, P = 0.47), and VI (t12 = 2.1, P = 0.06). The evaluation of the results with ANCOVA is virtually identical to the evaluation by Student's t test. It is highly unlikely that the statistical differences are false positives given the consistency of these differences across the layers. There are no significant effects due to age or PMI. There is no significant interaction between gender and diagnosis in any layer except for layer I, where there is an interaction with P < 0.04. In layer I, the mean value ± SD of DNMT1-positive neurons is higher in male SZP (29 ± 6.5 per mm3 × 103, n = 4) than in male NPS (14 ± 0.29, n = 3). No differences were observed in females: 18 ± 2.0 (n = 3) in SZP and 16 ± 5.0 (n = 4) in NPS. Because the number of samples is small, the SD is large, and layer I is often damaged, it is difficult at present to interpret the significance of this difference.
References
- 1.Burgers, W. A., Fuks, F. & Kouzarides, T. (2002) Trends Genet. 18, 275-277. [DOI] [PubMed] [Google Scholar]
- 2.Robertson, K. D. (2002) Oncogene 21, 5361-5379. [DOI] [PubMed] [Google Scholar]
- 3.Gardiner-Garden, M. & Frommer, M. (1987) J. Mol. Biol. 196, 261-282. [DOI] [PubMed] [Google Scholar]
- 4.Yoder, J. A. & Bestor, T. H. (1998) Hum. Mol. Genet. 7, 279-284. [DOI] [PubMed] [Google Scholar]
- 5.Bird, A. P. (1986) Nature 321, 209-213. [DOI] [PubMed] [Google Scholar]
- 6.Lander, E. S., Linton, L. M., Birren, B., Nusbaum, C., Zody, M. C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. (2001) Nature 409, 860-921. [DOI] [PubMed] [Google Scholar]
- 7.Li, E., Beard, C. & Jaenish, R. (1993) Nature 366, 362-365. [DOI] [PubMed] [Google Scholar]
- 8.Akbarian, S., Kim, J. J., Potkin, S. G., Hagman, J. O., Tafazzoli, A., Bunney, W. E., Jr., & Jones, E. G. (1995) Arch. Gen. Psychiatry 52, 258-266. [DOI] [PubMed] [Google Scholar]
- 9.Heckers, S., Stone, D., Walsh, J., Shick, J., Koul, P. & Benes, F. M. (2002) Arch. Gen. Psychiatry 59, 521-529. [DOI] [PubMed] [Google Scholar]
- 10.Guidotti, A., Auta, J., Davis, J. M., Di-Giorgi Gerevini, V., Dwivedi, Y., Grayson, D. R., Impagnatiello, F., Pandey, G., Pesold, C., Sharma, R., et al. (2000) Arch. Gen. Psychiatry 57, 1061-1069. [DOI] [PubMed] [Google Scholar]
- 11.Impagnatiello, F., Guidotti, A., Pesold, C., Dwivedi, Y., Caruncho, H., Pisu, M. G., Uzunov, D. P., Smalheiser, N. R., Davis, J. M., Pandey, G., et al. (1998) Proc. Natl. Acad. Sci. USA 95, 15718-15723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Woo, T. U., Whitehead, R. E., Melchitzky, D. S. & Lewis, D. A. (1998) Proc. Natl. Acad. Sci. USA 95, 5341-5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Volk, D. W. & Lewis, D. A. (2002) Physiol. Behav. 77, 501-505. [DOI] [PubMed] [Google Scholar]
- 14.Eastwood, S. L. & Harrison, P. J. (2003) Mol. Psychiatry 8, 821-831. [DOI] [PubMed] [Google Scholar]
- 15.Hashimoto, T., Volk, D. W., Eggan, S. M., Mirnics, K., Pierri, J. N., Sun, Z., Sampson, A. R. & Lewis, D. A. (2003) J. Neurosci. 23, 6315-6326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pesold, C., Impagnatiello, F., Pisu, M. G., Uzunov, D. P., Costa, E., Guidotti, A. & Caruncho, H. J. (1998) Proc. Natl. Acad. Sci. USA 95, 3221-3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alcantera, S., Ruiz, M., D'Arcangelo, G., Ezan, F., deLecea, L., Curran, T., Sotelo, C. & Soriano, E. (1998) J. Neurosci. 18, 7779-7799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lacor, P. N., Grayson, D. R., Auta, J., Sugaya, I., Costa, E. & Guidotti, A. (2000) Proc. Natl. Acad. Sci. USA 97, 3556-3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rodriguez, M. A., Pesold, C., Liu, W. S., Kriho, V., Guidotti, A., Pappas, G. D. & Costa, E. (2000) Proc. Natl. Acad. Sci. USA 97, 3550-3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Costa, E., Davis, J., Grayson, D. R., Guidotti, A., Pappas, G. D. & Pesold, C. (2001) Neurobiol. Dis. 8, 723-742. [DOI] [PubMed] [Google Scholar]
- 21.Costa, E., Chen, Y., Davis, J., Dong, E., Noh, J. S., Tremolizzo, L., Veldic, M., Grayson, D. R. & Guidotti, A. (2002) Mol. Interv. 2, 47-57. [DOI] [PubMed] [Google Scholar]
- 22.Dong, E., Caruncho, H., Liu, W. S., Smalheiser, N. R., Grayson, D. R., Costa, E. & Guidotti, A. (2003) Proc. Natl. Acad. Sci. USA 100, 5479-5484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu, W. S., Pesold, C., Rodriguez, M. A., Carboni, G., Auta, J., Lacor, P., Larson, J., Condie, B., Guidotti, A. & Costa, E. (2001) Proc. Natl. Acad. Sci. USA 98, 3477-3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garey, L. J., Ong, W. Y., Patel, T. S., Kanani, M., Davis, A., Mortimer, A. M., Barnes, T. R. & Hirsch, S. R. (1998) J. Neurol. Neurosurg. Psychiatry 65, 446-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Glantz, L. A. & Lewis, D. A. (2000) Arch. Gen. Psychiatry 57, 65-73. [DOI] [PubMed] [Google Scholar]
- 26.Selemon, L. D. & Goldman-Rakic, P. S. (1999) Biol. Psychiatry 45, 17-25. [DOI] [PubMed] [Google Scholar]
- 27.Kapranov, P., Cawley, S. E., Drenkow, J., Bekiranov, S., Strausberg, R. L., Fodor, S. P. A. & Gingeras, T. R. (2002) Science 296, 916-919. [DOI] [PubMed] [Google Scholar]
- 28.Rodriguez, M. A., Caruncho, H. J., Costa, E., Pesold, C., Liu, W. S. & Guidotti, A. (2002) J. Comp. Neurol. 451, 279-288. [DOI] [PubMed] [Google Scholar]
- 29.Schnell, S. A., Staines, W. A. & Wessendorf, M. W. (1999) J. Histochem. Cytochem. 47, 719-730. [DOI] [PubMed] [Google Scholar]
- 30.Williams, R. W. & Rakic, P. (1988) J. Comp. Neurol. 278, 344-352. [DOI] [PubMed] [Google Scholar]
- 31.Benes, F. M. & Lange, N. (2001) Trends Neurosci. 24, 11-17. [DOI] [PubMed] [Google Scholar]
- 32.Leonhardt, H., Page, A. V., Weier, H. & Bestor, T. H. (1992) Cell 71, 865-873. [DOI] [PubMed] [Google Scholar]
- 33.Liu, Y., Oakeley, E. J., Sun, L. & Jost, J. P. (1998) Nucleic Acids Res. 26, 1038-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mountcastle, V. B. (1998) Perceptual Neuroscience: The Cerebral Cortex (Harvard Univ. Press, Cambridge, MA).
- 35.Chen, Y., Sharma, R., Costa, R. H., Costa, E. & Grayson, D. R. (2002) Nucleic Acids Res. 30, 2930-2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Costa, E., Grayson, D. & Guidotti, A. (2003) Mol. Interv. 3, 220-229. [DOI] [PubMed] [Google Scholar]
- 37.Fuks, F., Hurd, P. J., Deplus, R. & Kouzarides, T. (2003) Nucleic Acids Res. 31, 2305-2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jenuwein, T. (2002) Science 297, 2215-2218. [DOI] [PubMed] [Google Scholar]
- 39.Fischle, W., Wang, Y. & Allis, C. D. (2003) Curr. Opin. Cell Biol. 15, 172-183. [DOI] [PubMed] [Google Scholar]