Abstract
Cytochrome P450 oxidoreductase (POR) is required for drug metabolism by all microsomal cytochrome P450 enzymes. The aim of this study was to investigate whether any of the common single nucleotide polymorphisms (SNPs) in the POR gene and its flanking intergenic sequences correlate with interindividual variations in the warfarin maintenance dose (which is determined partly by rates of warfarin metabolism) in patients undergoing anticoagulation therapy. Warfarin dose and patients' demographic and clinical information were collected from 124 patients, who had attained a stable warfarin dose while receiving treatment at the Stratton VA Medical Center. Genomic DNAs were isolated from blood samples and were genotyped for 15 SNPs (including 10 SNPs on the POR gene). Association analysis was performed on 122 male patients by linear regression. Simple regression analysis revealed that vitamin K epoxide reductase complex subunit 1 (VKORC1) −1639A>G, CYP2C9*2, CYP2C9*3, age, and chronic aspirin therapy were significantly associated with warfarin dose. In contrast, multiple regression analysis revealed that, in addition to several known factors contributing to the variations in warfarin maintenance dose (VKORC1 −1639A>G, CYP2C9*2, CYP2C9*3, CYP4F2 rs2108622, and chronic aspirin therapy), three common POR SNPs (−173C>A, −208C>T, and rs2868177) were also significantly associated with variations in warfarin maintenance dose. These results indicate, for the first time, that three common SNPs in the POR gene may contribute to the interindividual variability in warfarin maintenance dose. Further studies on functional characterization of the POR SNPs identified, including their impact on the in vivo metabolism of additional drugs, are needed.
Introduction
Cytochrome P450 oxidoreductase (POR) is the obligate electron donor to all microsomal cytochrome P450 (P450) enzymes, which play major roles in the metabolism of most drugs (Porter and Coon, 1991; Guengerich, 2008). Interindividual variations in POR activity (at least 4- to 5-fold differences) have been found in human liver microsomes (Kaminsky et al., 1984; Hart et al., 2008). The notion that variations in POR expression or activity influence the rates of P450-mediated drug metabolism in patients is supported by several lines of data, including the impact of Por gene knockout on drug clearance in mouse models (Gu et al., 2003; Henderson et al., 2003; Zhang et al., 2009), positive correlations between POR activity and P450-mediated drug metabolism activities in human liver microsomes (Kaminsky et al., 1984; Hart et al., 2008), and the impact of many POR single nucleotide polymorphisms (SNPs) on P450 activities toward drugs and other xenobiotics in reconstituted enzyme systems (Agrawal et al., 2010; Flück et al., 2010; Marohnic et al., 2010; Nicolo et al., 2010; Sandee et al., 2010; Miller et al., 2011).
At present, more than 800 POR SNPs (Hart et al., 2008; Huang et al., 2008; Gomes et al., 2008; NCBI dbSNP database, http://www.ncbi.nlm.nih.gov/snp) and 41 POR alleles (Sim et al., 2009; http://www.cypalleles.ki.se/por.htm) have been identified. Rare POR coding region mutations, identified in patients with Antley-Bixler syndrome and congenital adrenal hyperplasia, cause dramatic decreases in POR activity and the activities of microsomal steroidogenic P450 enzymes (Arlt et al., 2004; Flück et al., 2004; Huang et al., 2005). The impact of the rare POR mutant A287P on drug metabolism in a patient (Tomalik-Scharte et al., 2010), the effects of various POR SNPs on human liver microsomal drug metabolism activity (Hart et al., 2008; Gomes et al., 2009), and the potential role of the common A503V variation (POR*28) in interindividual differences in midazolam metabolism (Oneda et al., 2009) have been reported. However, few studies have examined the impact of common POR SNPs on drug metabolism in a clinical setting.
In this study, we aimed to investigate whether any of the common SNPs in the POR gene and its flanking intergenic sequences correlate with interindividual variations in the warfarin maintenance dose (which is determined partly by rates of warfarin metabolism) in patients undergoing anticoagulation therapy. Our hypothesis was that POR SNPs influence POR activity or expression, thereby influencing warfarin metabolism, through P450 pathways, and contributing to the interindividual variations in warfarin dose requirement. Warfarin is a widely prescribed and effective oral anticoagulant for the treatment and prevention of thromboembolic diseases. However, warfarin has a very narrow therapeutic range and a large potential to produce hemorrhage and thrombotic complications. Interindividual variability in the warfarin maintenance dose is very high (estimated to be >10 fold) (Rettie and Tai, 2006). The safe achievement of a maintenance dose requires intensive monitoring of patients to ensure their safety.
Recent pharmacogenomics studies have demonstrated that the polymorphisms in genes that are involved in warfarin metabolism or action, such as CYP2C9, CYP4F2, and vitamin K epoxide reductase complex subunit 1 (VKORC1), contribute to the interindividual variability in response to warfarin (Marsh and McLeod, 2006; Wadelius and Pirmohamed, 2007; Yin and Miyata, 2007; Caldwell et al., 2008; Limdi and Veenstra, 2008; Takeuchi et al., 2009). These findings indicated that personalized dosing, based on genotyping data, may be a solution for safe warfarin therapy. However, the currently identified genetic factors, combined with nongenetic factors, can only explain approximately 33 to 60% of the interindividual variations in warfarin dose required to achieve therapeutic effects, leaving the factors influencing the remaining 40 to 67% unknown.
In the current study, DNA samples were genotyped for the common SNPs (with ≥5% variant allele frequency) in the POR gene, using newly developed rapid, high-throughput genotyping assays based on Luminex xTAG technology. The genotypes were then correlated with levels of the warfarin maintenance dose for the individual donor patients. Statistically significant correlations indicate association of the genotypes with the warfarin dose requirements of the individual patients and further imply that the relevant POR SNPs can lead to changes in POR expression or activity. The identification of “relevant” genetic polymorphisms in the POR gene and of other genetic or nongenetic interacting factors will not only contribute to the efforts to develop accurate warfarin dosing algorithms for predicting the required warfarin maintenance dose for each patient but also provide the basis for further determination of the impact of the relevant POR SNPs on the in vivo metabolism of numerous other clinical drugs.
Materials and Methods
Participating Patients.
The study was approved by the institutional review boards of the Stratton VA Medical Center and the New York State Department of Health. The study subjects were patients who were receiving warfarin anticoagulation therapy in the Stratton VA Medical Center during 2008 and 2009 and who gave written informed consent. The inclusion criteria were 1) target therapeutic international normalized ratio (INR) range of 2 to 3 (INR is standard unit used to report the result of a prothrombin time test) and 2) attainment of a stable warfarin maintenance dose as reflected in the medical records. The most recent three or more consecutive INR measurements that are in the target range (2–3) were identified, for calculation of the average weekly dose (median dose), which was taken to represent the stable warfarin maintenance dose. In a few cases, the INR range was extended to 1.8 or 3.2, because a difference of 0.2 in INR values would not cause dose modification (Aquilante et al., 2006). A total of 124 patients (including both inpatients and outpatients) met the inclusion criteria, of which only 2 were female. Thus, only males were included in the study. The demographic and clinical information for these patients is summarized in Table 1. The age of patients ranged from 42 to 93 years at the time their medical records were reviewed. Of the patients, 115 patients were white, six were black, and one was Hispanic. The indications for treatment included atrial fibrillation or flutter, deep venous thrombosis, cerebrovascular accident, pulmonary embolism, and other diseases.
TABLE 1.
Summary of demographics and clinical variables
A total of 122 male patients were studied. Data are n (%) unless indicated otherwise.
| Values | |
|---|---|
| Demographic variables | |
| Age | |
| Mean ± S.D. | 72 ± 10 |
| 40–49 yr | 2 (2) |
| 50–59 yr | 11 (9) |
| 60–69 yr | 37 (30) |
| 70–79 yr | 41 (34) |
| 80–89 yr | 29 (24) |
| >89 yr | 2 (2) |
| Weight (lb) | |
| Mean ± S.D. | 209 ± 48 |
| Race | |
| White | 115 (94) |
| African American | 6 (5) |
| Other | 1 (1) |
| Clinical variables | |
| Warfarin dose (mg/wk), median (minimum, maximum) | 33 (8, 71) |
| Target INR (range) | 2–3 |
| Clinical indications for warfarin treatment | |
| Atrial fibrillation or flutter | 79 (65) |
| Deep venous thrombosis | 22 (18) |
| Cerebrovascular accident | 3 (2) |
| Pulmonary embolism | 2 (2) |
| Other diseases | 16 (13) |
| Other medication | |
| Aspirin | 26 (21) |
| Amlodipine | 14 (11) |
| Medicines containing statin | 86 (70) |
| Multivitamin | 22 (18) |
| Insulin | 11 (9) |
Data Collection.
Weekly warfarin dose (milligrams per week); INRs; aspirin (dose range: 0, 81, or 325 mg/day); use of other concomitant medications (yes or no), including statins (all statin-containing medicines), multivitamins, insulin, and amlodipine; and patients' demographic information including age (years), gender, race (white or nonwhite), and weight (pounds) were obtained by reviewing the patient's medical records. All of the extracted information was deidentified and linked to the corresponding blood samples by coding. Peripheral blood samples (∼5 ml) were taken during the patient's blood draw for INR measurement and stored at −30°C until isolation of genomic DNA.
Genotyping.
Genomic DNA was isolated from whole blood using a QIAamp DNA Blood Mini Kit (QIAGEN, Valencia, CA). Genotyping assays were performed for the following four groups of SNPs:
Five SNPs, four of which are known to be associated with varied warfarin dose requirement: VKORC1 −1639A>G (rs9923231), CYP2C9*2 (rs1799853), CYP2C9*3 (rs1057910), and CYP4F2 V33M; the SNP rs4889606 in STX1B intron 4 (located at ∼90 kilobases downstream of VKORC1 and significantly associated with varying levels of VKORC1 mRNA expression) (Schadt et al., 2008) was also included (to our knowledge, there has been no report on the relationship between SNP rs4889606 in STXIB with varied warfarin dose requirement).
Two POR SNPs known to be associated with POR functional consequences: POR A503V (rs1057868) (causes ∼30% decrease in POR activity) (Huang et al., 2008) and POR rs41301472 (within intron 12, G>A change, correlated with decreased POR activity in liver microsomes) (Hart et al., 2008).
Six tag SNPs in POR and its flanking intergenic sequences (chr7:75356180-75454513), which were identified in the CEU population from HapMap (Data Rel 27, PhaseII+III, Feb09, on NCBI B36 assembly, dbSNP b126), using the Annotate tag SNP Picker (Tagger Pairwise, R2 cutoff: 0.8, MAF cutoff: 0.05): rs10280802, rs28737229, rs1057870, rs17148944, rs2868177, and rs17685 (http://www.hapmap.org/).
Two SNPs located in the POR proximal promoter region: POR −173C>A (a newly identified SNP at the time of the study, now also included in dbSNP; rs72553971) and POR −208C>T (rs12537282) (Huang et al., 2008); the transcription start site (first nucleotide of POR mRNA sequence) was designated as +1.
Genotyping assays for the 15 SNPs above were developed on the basis of Luminex xTAG technology, which uses a proprietary universal tag system. Multiplex PCR primers and allele-specific primer extension (ASPE) primers were designed according to the NCBI GenBank sequences [CYP2C9: NT_030059.12 (GI: 51467897); VKORC1: NT_010393.15 (GI: 51472974); POR: NT_007933.14 (GI: 51493052); CYP4F2: NT_011295.10 (GI: 29801560); and STX1B: NT_010393.15 (GI: 51472974)]. All the primers were synthesized at Integrated DNA Technologies, Inc. (Coralville, IA), and the ASPE primers were high-performance liquid chromatography-purified. Details of PCR and ASPE primers are listed in Supplemental Tables S1 and S2, respectively.
The 15 SNPs above were detected in four multiplex genotyping assays: assay 1 for 6 SNPs (CYP2C9*2, CYP2C9*3, VKORC1 −1639G>A, STX1B rs4889606, CYP4F2 rs2108622, and POR A503V); assay 2 for 3 SNPs (POR rs10280802, POR rs28737229, and POR rs41301427); assay 3 for 3 SNPs (POR rs1057870, POR rs17148944, and POR rs2868177); and assay 4 for 3 SNPs (POR −173C>A, POR −208C>T, and POR rs17685). The experimental procedures of the four assays differed only at the multiplex PCR step. The genotyping assays consisted of the following five steps.
Step 1. Multiplex PCR.
DNA fragments covering the SNPs studied for each genotyping assay were amplified in one reaction on a model 9800 Fast Thermal Cycler (Applied Biosystems, Foster City, CA). For assays 1 to 3, after an initial denaturation at 95°C for 15 min, the amplification reaction was performed for 35 cycles, with each cycle consisting of a denaturation at 95°C for 30 s, an annealing at 55°C for 30 s, and an extension at 72°C for 40 s, followed by a final extension at 72°C for 10 min. The reaction mixtures, in a total volume of 25 μl, contained 1× PCR buffer (20 mM Tris-HCl, pH 8.4, and 50 mM KCl), 0.2 mM concentrations each of dNTPs, 0.2 μM concentrations each of the PCR primers, 0.625 unit of HotStarTaq DNA Polymerase (QIAGEN, Valencia, CA), and 40 ng of genomic DNA. For assay 4, the TaKaRa LA Taq system (TaKaRa BIO, Inc., Madison, WI) was used, and after initial denaturation at 94°C for 2 min, 35 PCR cycles were performed, with each consisting of a denaturation at 94°C for 30 s, and an annealing and extension at 64°C for 100 s, followed by a final extension at 68°C for 10 min; the reaction mixture contained, in a total volume of 25 μl, 1× LA PCR buffer II (Mg2+ PLUS), 1.25 mM concentrations each of dNTPs, 0.2 μM concentrations each of the 12 PCR primers, 1.25 units of TaKaRa LA Taq Polymerase, and 40 ng of genomic DNA. An H2O blank (no template) control was routinely used for detecting potential contamination of reagents; the results were also used later for fluorescence background correction at step 5. Identity of each PCR product was confirmed by direct DNA sequence analysis.
Step 2. Shrimp alkaline phosphatase and exonuclease I treatment.
To remove unincorporated deoxynucleotide triphosphates and primers from the last PCR amplification step, the PCR products were treated with shrimp alkaline phosphatase and exonuclease I (ExoSAP-IT; USB, Cleveland, OH). A mixture of ExoSAP-IT (2 μl) and the PCR product (5 μl) was incubated at 37°C for 30 min, followed by an incubation at 80°C for 15 min (to inactivate the enzymes), in the model 9800 Fast Thermal Cycler. The treated PCR products were kept at 4°C until use.
Step 3. Multiplex ASPE.
Multiplex ASPE reactions were performed in the model 9800 Fast Thermal Cycler, with 5 μl of ExoSAP-IT-treated PCR product, in a total volume of 20 μl. Each reaction mixture consisted of 1× PCR buffer, 1.25 mM MgCl2, 25 nM concentrations each of Tag-ASPE primers, 5 μM concentrations each of dGTP, dATP, and dTTP (Invitrogen, Carlsbad, CA), 5 μM biotin-14-dCTP (Invitrogen), and 0.75 unit of Platinum Genotype TspDNA Polymerase (Invitrogen). The reaction was started with preincubation at 96°C for 2 min (to denature DNA) and then followed by 30 cycles of PCR, with each consisting of 94°C for 30 s, 55°C for 1 min, and 74°C for 2 min. The ASPE products were kept at 4°C until use.
Step 4. Hybridization to Luminex MicroPlex-xTAG microspheres and acquisition of data.
The hybridization of ASPE products to the MicroPlex-xTAG microspheres was performed in a PerkinElmer thermal cycler 9600 instrument (Applied Biosystems). Data collection was performed on a Luminex 100 IS System (Luminex Corporation, Austin, TX). All procedures of this step were performed in the dark. Microsphere suspension was prepared by gentle vortexing, followed by sonication, in an Ultrasonic cleaner (model 08849-00; Cole-Parmer Instrument Co., Vernon Hills, IL), for ∼20 s. Each hybridization reaction was performed with ∼2500 each of the populations of carboxylated fluorescent microspheres that are precoupled with the xTAG oligonucleotide sequences (anti-tags) (Luminex Corporation).
Equal amounts of the microsphere sets were combined, concentrated, and then resuspended to 100 beads each/μl, in 2× Tm hybridization buffer (0.2 M Tris-HCl, pH 8.0, 0.4 M NaCl, and 0.16% Triton X-100). The resuspended beads (25 μl) were combined with 5 μl of the ASPE products from step 3 and H2O to make a total volume of 50 μl; the mixture was heated at 96°C for 90 s to denature DNA, followed immediately by incubation at 37°C for 60 min. After this hybridization step, the reaction mixtures were transferred to filter plates (Millipore Corporation, Billerica, MA) and prewetted in 1× Tm hybridization buffer, and the supernatant was removed under vacuum, as described in the manufacturer's instruction manual. After two washes with 100 μl of 1× Tm hybridization buffer, the beads were resuspended in 100 μl of 1× Tm hybridization buffer containing 2 μg/μl streptavidin-R-phycoerythrin (Invitrogen) and incubated at 37°C for 15 min. A 80-μl portion of the mixture was then analyzed at 35°C on a Luminex 100 analyzer, according to the system manual. For each sample, the instrument was set to read a minimum of 100 events for each type of beads, with the sample timeout setting being 90 s and the doublet discriminator gate setting being 8000 to 13,500. The bead sets were sorted, and the intensity of R-phycoerythrin fluorescence on each of the bead sets, corresponding to the alleles analyzed in the assay, was measured.
Step 5. Genotype determination.
Genotypes of a given DNA sample for a specific SNP were determined on the basis of the calculated median fluorescence intensity ratios between variant allele and the total (variant + reference), after subtraction of background fluorescence intensity. The threshold ratio values for each genotype were empirically determined: a ratio of <0.25 was found to indicate a homozygous reference call, a ratio between 0.25 and0.75 was found to indicate a heterozygous call, and a ratio of >0.75 was found to indicate a homozygous variant call, with exceptions for CYP2C9*3 (<0.13, 0.13–0.35, and >0.35), VKORC1 (<0.25, 0.25–0.77, and >0.77), and POR −173C>A (<0.19, 0.19–0.36, and >0.36). The relative fluorescence intensities of the specific beads used to label each pair of primers (reference versus variant) differed for each SNP, which led to the occasional SNP-specific differences in threshold values. The accuracy of the genotyping assays was validated for each SNP during method development using >30 samples with known genotypes and by additional genotype analysis of two to three randomly selected DNA samples using direct DNA sequencing; 100% concordance was obtained between the genotype calls assessed by direct sequencing and the results of Luminex assays. In addition, in rare cases when a DNA sample had a median fluorescence intensity ratio close to the threshold value, the sample was also resequenced.
Data Analysis.
The χ2 test for goodness of fit was used first to examine whether the genotype distribution of the SNPs studied in the patient population deviated from Hardy-Weinberg equilibrium. Spearman correlation analysis between every two SNPs was performed to see whether they were linked with each other. If the correlation coefficient (r) between the two SNPs was greater than 0.85 (arbitrary), we would choose only one of the two SNPs in the multivariate analysis.
Before the analysis of potential associations between the genotype (SNPs) and the phenotype (varying warfarin dose), the SNP genotype data were converted into quantitative codes: code = 2 if a SNP was a homozygous variant genotype, code = 1 if a SNP was a heterozygous variant genotype, and code = 0 if a SNP was a homozygous reference genotype. The stable warfarin maintenance dose (milligrams per week), which was treated as the dependent variable for linear regression analysis, was square root-transformed to meet the normal distribution requirement. Simple and multiple linear regression analyses were performed to determine whether the SNPs and the nongenetic factors were significantly associated with variations in warfarin dose requirement (using P < 0.05 as the criterion for significance). The square of the correlation coefficient (R2) was calculated, which shows the proportion of variations of the dependent variable (square root of the warfarin dose) that can be accounted for by variations of the independent variable(s), such as the SNPs studied. All analyses were performed using SAS 9.2 (SAS Institute, Cary, NC).
Results
The genotype data and the variant allele frequency data for the 15 selected SNPs are show in Table 2. Among the 122 male patients, frequencies of variant alleles ranged from 4.5 to 46%. Genotype distribution for none of the 15 SNPs deviated from Hardy-Weinberg equilibrium, on the basis of the results from the χ2 test for goodness of fit, at the P < 0.05 criterion. Correlation analysis performed on each SNP pair among the 15 SNPs identified three pairs that are highly correlated with each other: POR rs41301427 and POR rs17148944 (r = 0.858, P < 0.0001); POR rs17685 and POR A503V (r = 0.969, P < 0.0001); and STX1B rs4889606 and VKORC1 −1639G>A (r = 0.935, P < 0.0001). Therefore, rs41301427, rs17685, and rs4889606 were not included in subsequent multiple linear regression analysis.
TABLE 2.
Genotype and allele frequency data for the 15 studied SNPs
| SNPs Examined | Homozygous Variant Allele | Heterozygous Variant Allele | Homozygous Reference Allele | No. Samples Examined | Variant Allele Frequency |
|---|---|---|---|---|---|
| CYP2C9*2 | 1 | 25 | 96 | 122 | 0.111 |
| CYP2C9*3 | 0 | 11 | 111 | 122 | 0.045 |
| VKORC1 −1639G>A | 23 | 53 | 46 | 122 | 0.406 |
| STX1B rs4889606 | 23 | 54 | 45 | 122 | 0.410 |
| CYP4F2 rs2108622 | 10 | 55 | 57 | 122 | 0.307 |
| POR A503V | 8 | 42 | 72 | 122 | 0.238 |
| POR rs10280802 | 8 | 42 | 72 | 122 | 0.238 |
| POR rs28737229 | 25 | 61 | 36 | 122 | 0.455 |
| POR rs41301427 | 0 | 15 | 107 | 122 | 0.061 |
| POR rs1057870 | 24 | 58 | 40 | 122 | 0.434 |
| POR rs17148944 | 0 | 17 | 105 | 122 | 0.070 |
| POR rs2868177 | 12 | 57 | 53 | 122 | 0.332 |
| POR −173C>A | 0 | 17 | 105 | 122 | 0.070 |
| POR −208C>T | 3 | 20 | 99 | 122 | 0.107 |
| POR rs17685 | 7 | 41 | 74 | 122 | 0.225 |
Association analysis of genetic and nongenetic factors with stable warfarin maintenance dose (square root-transformed) was performed through linear regression. Simple regression analysis of all the variables [15 SNPs, age, race, weight, and concomitant medications (aspirin, statins, multivitamin, insulin, and amlodipine)] revealed that age, CYP2C9*2, CYP2C9*3, VKORC1 −1639G>A, STX1B rs4889606, and aspirin are significantly associated with the stable warfarin dose. The contribution (R2) to dose variation was ∼3.2% by age, ∼12.2% by CYP2C9*2, ∼5.2% by CYP2C9*3, ∼21.4% by VKORC1 −1639G>A, ∼18.6% by STX1B rs4889606, and ∼5.0% by aspirin (Table 3). However, note that the apparent contribution by STX1B probably reflects the contribution by VKORC1, given the close linkage between the two genes. None of the POR SNPs or the CYP4F2 SNP showed a significant correlation with warfarin dose in simple regression analysis.
TABLE 3.
Significant factors in the simple linear regression analysis (P < 0.05)
| Variables | Parameter Estimate | Variable Significance in Model | R2 | Adjusted R2 |
|---|---|---|---|---|
| Age (yr) | −0.02065 | P = 0.0489 | 0.0319 | 0.0239 |
| CYP2C9*2 | −0.97160 | P < 0.0001 | 0.1224 | 0.1151 |
| CYP2C9*3 | −0.96394 | P = 0.0112 | 0.0524 | 0.0445 |
| VKORC1 −1639G>A | −0.76681 | P < 0.0001 | 0.2141 | 0.2076 |
| STX1B rs4889606 | −0.71861 | P < 0.0001 | 0.1862 | 0.1794 |
| Aspirin | −0.00426 | P = 0.0095 | 0.0499 | 0.0420 |
Multiple regression analysis was performed on the variables listed above with rs41301427, rs17685, and rs4889606 excluded from the analysis. Table 4 shows the analysis results from multiple linear regression (backward: P value must be <0.05 for a variable to stay in the model). The SNPs CYP2C9*2, CYP2C9*3, VKORC1 −1639G>A, CYP4F2 rs2108622, POR rs2868177, POR −173C>A, and POR −208C>T, weight, and aspirin were significantly associated with stable warfarin dose. The direction of association was negative for CYP2C9*2, CYP2C9*3, VKORC1 −1639G>A, POR −173C>A, POR −208C>T, and aspirin, meaning that a patient with the indicated variant allele(s) or with a higher aspirin dose required lower doses of warfarin to achieve effective anticoagulation compared with a patient with the reference alleles or with a lower aspirin dose. In contrast, the direction of association was positive for CYP4F2 rs2108622, POR rs2868177, and weight, meaning that a patient with the indicated variant allele(s) or higher body weight range required a higher dose of warfarin to achieve effective anticoagulation compared with a patient with the reference alleles or with lower body weight.
TABLE 4.
Significant factors in the multiple linear regression analysis
Backward: P < 0.05 for variables to stay in the model.
| Variable | Parameter Estimate | S.E. | F Value | P > F |
|---|---|---|---|---|
| Intercept | 5.55931 | 0.40179 | 191.44 | <0.0001 |
| Weight | 0.00431 | 0.00176 | 5.99 | 0.0159 |
| CYP2C9*2 | −0.87096 | 0.19831 | 19.29 | <0.0001 |
| CYP2C9*3 | −1.07031 | 0.29295 | 13.35 | 0.0004 |
| VKORC1 −1639G>A | −0.81933 | 0.11751 | 48.62 | <0.0001 |
| CYP4F2 rs2108622 | 0.37928 | 0.14053 | 7.28 | 0.0080 |
| POR rs2868177 | 0.35491 | 0.15440 | 5.28 | 0.0234 |
| POR −173C>A | −0.70359 | 0.26143 | 7.24 | 0.0082 |
| POR −208C>T | −0.50989 | 0.21827 | 5.46 | 0.0213 |
| Aspirin | −0.00287 | 0.00139 | 4.27 | 0.0411 |
Various regression models were generated for estimation of the relative contributions of the variables studied to the variations in warfarin maintenance dose in the study population. The variables were categorized into four groups: 1) patient's demographic information, including age, weight, and race; 2) SNPs that are known to affect warfarin dose, including CYP2C9*2, CYP2C9*3, VKORC1 −1639A>G, and CYP4F2 rs2108622; 3) SNPs in the POR gene, including A503V, rs10280802, rs28737229, rs1057870, rs17148944, rs2868177, −173C>A, and −208C>T; and 4) other medications, including aspirin, amlodipine, multivitamin, insulin, and medicines containing statin. As shown in Table 5, when all of the variables studied were included at the first step of regression (model 1), nine significant variables (as listed in Table 4) remained in the regression model, and the nine combined can explain 47.7% (R2) of warfarin dose variation. After removal of concurrent medications (i.e., aspirin) from the analysis (model 2), eight significant variables remained in the model, and they can explain 45.7% (R2) of warfarin dose variation. The difference in R2 values (2%) between the two regression models represents the contribution of aspirin to warfarin dose variation.
TABLE 5.
Linear regression models for estimation of relative contributions of the variables studied to the variations in warfarin maintenance dose
Backward, P < 0.05 for variables to stay in the model.
| Regression Models | Variables Entered at the Beginning of Regression | Final Regression Equation | Model P Value | R2 |
|---|---|---|---|---|
| 1. All variables | Patient's demographic information,a SNPs known to affect warfarin dose,b SNPs in POR gene,c other concomitant medicationsd | SQRT(D) = 5.559 + 0.004 (weight) − 0.871 (CYP2C9*2) − 1.070 (CYP2C9*3) − 0.819 (VKORC1 −1639 A>G) + 0.379 (CYP4F2 rs2108622) + 0.355 (POR rs2868177) − 0.704 (POR −173C>A) − 0.510 (POR −208C>T) − 0.003 (aspirin) | <0.0001 | 0.477 |
| 2. Without other medications | Patient's demographic information,a SNPs known to affect warfarin dose,b SNPs in POR genec | SQRT(D) = 5.567 + 0.004 (weight) − 0.941 (CYP2C9*2) − 1.087 (CYP2C9*3) − 0.831 (VKORC1 −1639 A>G) + 0.312 (CYP4F2 rs2108622) + 0.357 (POR rs2868177) − 0.680 (POR −173C>A) − 0.556 (POR −208C>T) | <0.0001 | 0.457 |
| 3. Without other medications and POR SNPs | Patient's demographic information,a SNPs known to affect warfarin doseb | SQRT(D) = 5.650 + 0.005 (weight) − 0.869 (CYP2C9*2) − 1.091 (CYP2C9*3) − 0.751 (VKORC1 −1639 A>G) | <0.0001 | 0.395 |
Age, weight, and race.
CYP2C9*2, CYP2C9*3, VKORC1 −1639A>G, and CYP4F2 rs2108622.
POR A503V, rs10280802, rs28737229, rs1057870, rs17148944, rs2868177, −173C>A, and −208C>T.
Aspirin, amlodipine, multivitamin, insulin, and medicines containing statin.
After further removal of the three significant POR SNPs (rs2868177, −173C>A, and −208C>T) from the analysis (model 3), only four variables (CYP2C9*2, CYP2C9*3, VKORC1 −1639G>A, and weight) remained significant in the model, and they can explain only 39.5% (R2) of the warfarin dose variation. The difference in R2 values between model 2 and model 3 was 6.2%, which represents the combined contributions of the three POR SNPs and the CYP4F2 SNP to warfarin dose variation. Of note, because of the small sample size, we could not determine the specific contributions by the POR SNPs; however, given the previous reports (Caldwell et al., 2008; Takeuchi et al., 2009) indicating contributions by CYP4F2 to be 1 to 2%, we estimate the actual contributions by the POR SNPs to be at least 4%. Further studies on a larger population are warranted to confirm this estimation.
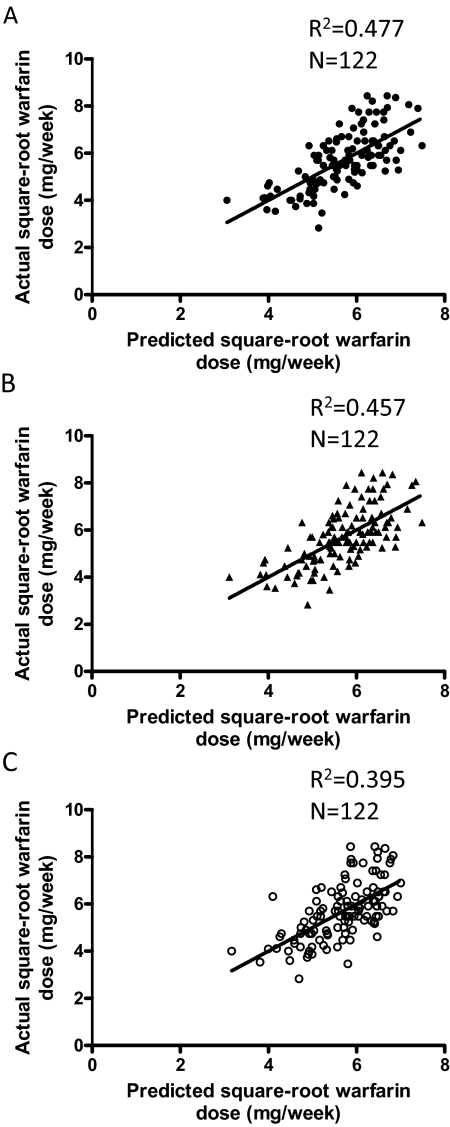
The assumption of linearity in each of the three regression models was confirmed by residual analysis, as visualized on scatter plots of standardized residuals versus predicted values of the square root of warfarin dose (data not shown). In addition, as shown in Fig. 1, in plots of the square root of the actual warfarin dose that maintained the target INR within 2 and 3 versus the square root of the warfarin dose predicted by each of the three linear regression models, the scatter above and below the straight regression line was roughly the same.
Fig. 1.
Correlation between actual and predicted warfarin doses. The square root of the actual warfarin dose that maintained the target INR within 2 and 3 was compared with the square root of the warfarin dose predicted by the model, for different groups of variables used in the linear regression models (as in Table 5). A, all variables (model 1); B, without other medications (model 2); C, without other medications and POR SNPs (model 3).
Discussion
The current study investigated whether the common SNPs in the POR gene are associated with warfarin dose variation in patients undergoing anticoagulation therapy. Simple regression analysis confirmed that VKORC1 −1639A>G, CYP2C9*2, CYP2C9*3, age, and chronic aspirin therapy were significantly associated with warfarin dose variation in our study population, whereas multiple regression analysis confirmed CYP4F2 rs2108622 as an additional contributing factor and revealed that three common POR SNPs (−173C>A, −208C>T, and rs2868177) were also significantly associated with variations in warfarin maintenance dose. Our findings regarding the contributions of CYP2C9*2, CYP2C9*3, VKORC1 −1639A>G, and CYP4F2 rs2108622 to warfarin dose variation are consistent with results reported previously by others (Marsh and McLeod, 2006; Wadelius and Pirmohamed, 2007; Yin and Miyata, 2007; Caldwell et al., 2008; Limdi and Veenstra, 2008; Takeuchi et al., 2009). Our simple regression analysis also showed an association between the STX1B SNP and warfarin dose variation, which has not been reported previously; however, the result was not unexpected, given the known association of the STX1B SNP with VKORC1 mRNA expression.
Both simple and multiple regression analyses were used here and in previous studies (see Takeuchi et al., 2009) to examine the relationships between various predictors and warfarin maintenance dose. Simple regression analysis, which estimates the impact of each individual predictor on the response variable without considering the impact of any other predictors, may fail to identify predictors that have a weak impact, as was demonstrated in previous work. Multiple regression analysis considers multiple predictors simultaneously; it yields an overall estimate of the impact of multiple predictors on a response variable, as well as estimates of the impact of individual predictors after controlling for the effects of other predictors. Multiple regression analysis may provide enhanced detection of predictors with weak impact on a response variable, as well as potential interactions among the variables tested; however, given the inclusion of multiple variables, it also requires larger sample sizes.
In our simple regression analysis, none of the POR SNPs was identified as being significantly associated with warfarin dose variation. Similar negative results were also found in the forward multiple regression analysis, in which the variables were sequentially added to the model in order of greatest significance to the model, rather than sequentially removed from the model in order of least significance to the model (data not shown). The fact that three POR SNPs were identified as being significantly associated with warfarin dose variation in the backward multiple regression analysis reflects the known dependence of P450 function on POR function and the reality that neither enzyme alone can metabolize warfarin. Furthermore, the impact of POR SNPs on warfarin dose requirement would be maximal in the presence of the P450 SNPs, given the theoretical possibility that a deficiency in P450 function can be either exacerbated or compensated for by variations in POR function.
It should be noted that, given the relatively small sample size and the large number of variables tested in the multiple regression models, our results should be viewed as exploratory, and the estimates of regression will need to be replicated in additional, independent studies. Nonetheless, the positive identification of CYP4F2, a previously known minor contributor, in our multiple regression analysis seems to further support the validity of our findings with POR. In that connection, the specific contribution by CYP4F2, which could not be determined in our study because of the limited sample size, was estimated to be 1 to 2% on the basis of the results from two previous studies on a U.S. population and a northern European (Swedish) population (Caldwell et al., 2008; Takeuchi et al., 2009). The majority of the study subjects in these two latter studies, as in our study, were white. Differing results have also been reported in studies of other populations, with predicted CYP4F2 contributions ranging from 0 to 11%. We did not consider the results from these other studies, given that they were conducted on populations from regions other than the United States or northern Europe, such as Italy (Borgiani et al., 2009), Singapore (Singh et al., 2011), and Japan (Harada et al., 2010), and that there are known differences in the extent of genetic polymorphisms in genes relevant to warfarin dose requirement among various populations, even between northern (e.g., Swedish) and southern (e.g., Italian) European populations (Schelleman et al., 2008; Borgiani et al., 2009; Singh et al., 2011).
Our results indicate, for the first time, that three common SNPs in the POR gene may contribute to the interindividual variability in warfarin maintenance dose. Several reasons may explain why POR did not emerge as a significant contributor to warfarin dose in previous genome-wide association studies (GWASs) (Caldwell et al., 2008; Takeuchi et al., 2009). Takeuchi et al. (2009), who determined that CYP4F2 accounted for ∼1.5% of warfarin dose variance by multiple regression analysis, suggested that, although their study probably detected the most common SNPs that contribute >1.5% to warfarin dose variation, it may have failed to detect other common SNPs that have lower effects or rare variants with greater effects than that of CYP4F2. For the three POR SNPs found to be significant in our study, it is possible that their individual contributions were each <1.5%, thus making it difficult to detect in a GWAS. Furthermore, of the three POR SNPs, −173C>A was newly identified and hence was not included in dbSNP or in any previous GWAS. As for the other two POR SNPs (POR −208C>T and POR rs2868177), they were not included on the DMET chip (Caldwell et al., 2008), and it is not clear whether they were included in the published GWASs, given the reported call rates of <95% (Takeuchi et al., 2009). In addition, it is possible that the opposing directions of the influence of the CYP2C9 and CYP4F2 variant alleles might have prevented detection of the POR effects in the GWASs.
Given the small sample size, we were not able to estimate the relative contributions by each of the three significant POR SNPs. However, the fact that the three POR SNPs (which are all noncoding) had opposing effects (one positive and two negative) on warfarin dose suggests that the SNPs have a differential impact on POR expression, and this is currently being tested experimentally. POR −173C>A (genomic position: 75382183) and POR −208C>T (genomic position: 75382148) are located just upstream of the POR transcription start site and were identified here to be negatively associated with warfarin dose. Database analysis for potential transcription factor binding sites, using TFsearch (http://www.cbrc.jp/research/db/TFSEARCH.html) (Heinemeyer et al., 1998), showed that POR −173C>A and POR −208C>T are both located within potential binding sites for multiple transcription factors, such as the homeo domain factor Nkx-2.5 (Chen and Schwartz, 1995) (identity score 81.4 for POR −173C>A) and alcohol dehydrogenase gene regulator 1 (Cheng et al., 1994) (identity score 81.5 for POR −208C>T). There are two possible mechanisms to explain how these two POR SNPs might affect warfarin metabolism: one is that these two upstream SNPs may decrease POR expression and thus lower metabolism of warfarin through P450 pathways. Another possible mechanism is that the two SNPs are in linkage disequilibrium with other unknown SNPs that affect POR expression. In that connection, Tee et al. (2011) reported that neither of the two promoter region SNPs had an impact on the transcription of a 325-base pair POR basal promoter in reporter gene assays. Thus, it appears that the second mechanism is more likely; i.e., other unknown SNPs that are in linkage disequilibrium with the SNP at −178 or −208 are critical for POR expression. Further studies are needed to clarify these issues. POR rs2868177, which showed positive correlation with warfarin dose, is in linkage disequilibrium with rs2868180, rs7804806, and rs2868178; all four SNPs are located centrally in intron 2 of POR. The underlying mechanisms for the involvement of POR rs2868177 are unclear.
The common POR coding region polymorphism A503V, with an allele frequency of 19 to 36%, causes 30 to 40% reduction in POR activity (Huang et al., 2005, 2008). However, the impact of A503V on P450 activity has been found to be variable, depending on the P450 enzyme involved and the substrate studied (Arlt et al., 2004; Gomes et al., 2008, 2009; Miller et al., 2009, 2011; Oneda et al., 2009; Sandee et al., 2010). The ability of this variant to support P450-mediated warfarin metabolism has not been examined in vitro. In the present study, we did not find a significant correlation between POR A503V and warfarin dose in patients. It appears that this SNP may not affect warfarin metabolism through either the CYP2C9 or the CYP4F2 pathway.
In conclusion, three SNPs in the POR gene are significantly associated with variable warfarin dose. Further examination of a complete list of common POR SNPs, in a larger patient population and molecular studies on the functional impact of the significant SNPs on POR expression/function are needed to better understand the nature and extent of the impact of POR SNPs on warfarin dose requirement. In addition, studies on the potential functional impact of the POR SNPs on the in vivo metabolism of numerous additional drugs are warranted.
Supplementary Material
Acknowledgments
We gratefully acknowledge the use of the Molecular Genetics Core of the Wadsworth Center and the role of Joann Finn of the Stratton VA Medical Center in the collection and deidentification of medical records. We also thank Dr. Ken Pass of the Wadsworth Center for the use of his Luminex instrument.
This work was supported in part by the National Institutes of Health National Cancer Institute [Grant CA092596]; and the New York State Attorney General's Office [Grant for Drug Safety Studies].
Article, publication date, and citation information can be found at http://dmd.aspetjournals.org.
doi:10.1124/dmd.111.038836.
 The online version of this article (available at http://dmd.aspetjournals.org) contains supplemental material.
The online version of this article (available at http://dmd.aspetjournals.org) contains supplemental material.
- POR
- cytochrome P450 oxidoreductase
- P450
- cytochrome P450
- SNP
- single nucleotide polymorphism
- NCBI
- National Center for Biotechnology Information
- VKORC1
- vitamin K epoxide reductase complex subunit 1
- INR
- international normalized ratio
- PCR
- polymerase chain reaction
- ASPE
- allele-specific primer extension
- GWAS
- genome-wide association study.
Authorship Contributions
Participated in research design: Zhang, Ding, and Kaminsky.
Conducted experiments: Zhang and Li.
Performed data analysis: Zhang and Li.
Wrote or contributed to the writing of the manuscript: Zhang, Li, Ding, and Kaminsky.
References
- Agrawal V, Choi JH, Giacomini KM, Miller WL. (2010) Substrate-specific modulation of CYP3A4 activity by genetic variants of cytochrome P450 oxidoreductase. Pharmacogenet Genomics 20:611–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aquilante CL, Langaee TY, Lopez LM, Yarandi HN, Tromberg JS, Mohuczy D, Gaston KL, Waddell CD, Chirico MJ, Johnson JA. (2006) Influence of coagulation factor, vitamin K epoxide reductase complex subunit 1, and cytochrome P450 2C9 gene polymorphisms on warfarin dose requirements. Clin Pharmacol Ther 79:291–302 [DOI] [PubMed] [Google Scholar]
- Arlt W, Walker EA, Draper N, Ivison HE, Ride JP, Hammer F, Chalder SM, Borucka-Mankiewicz M, Hauffa BP, Malunowicz EM, et al. (2004) Congenital adrenal hyperplasia caused by mutant P450 oxidoreductase and human androgen synthesis: analytical study. Lancet 363:2128–2135 [DOI] [PubMed] [Google Scholar]
- Borgiani P, Ciccacci C, Forte V, Sirianni E, Novelli L, Bramanti P, Novelli G. (2009) CYP4F2 genetic variant (rs2108622) significantly contributes to warfarin dosing variability in the Italian population. Pharmacogenomics 10:261–266 [DOI] [PubMed] [Google Scholar]
- Caldwell MD, Awad T, Johnson JA, Gage BF, Falkowski M, Gardina P, Hubbard J, Turpaz Y, Langaee TY, Eby C, et al. (2008) CYP4F2 genetic variant alters required warfarin dose. Blood 111:4106–4112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Schwartz RJ. (1995) Identification of novel DNA binding targets and regulatory domains of a murine tinman homeodomain factor, nkx-2.5. J Biol Chem 270:15628–15633 [DOI] [PubMed] [Google Scholar]
- Cheng C, Kacherovsky N, Dombek KM, Camier S, Thukral SK, Rhim E, Young ET. (1994) Identification of potential target genes for Adr1p through characterization of essential nucleotides in UAS1. Mol Cell Biol 14:3842–3852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flück CE, Mullis PE, Pandey AV. (2010) Reduction in hepatic drug metabolizing CYP3A4 activities caused by P450 oxidoreductase mutations identified in patients with disordered steroid metabolism. Biochem Biophys Res Commun 401:149–153 [DOI] [PubMed] [Google Scholar]
- Flück CE, Tajima T, Pandey AV, Arlt W, Okuhara K, Verge CF, Jabs EW, Mendonça BB, Fujieda K, Miller WL. (2004) Mutant P450 oxidoreductase causes disordered steroidogenesis with and without Antley-Bixler syndrome. Nat Genet 36:228–230 [DOI] [PubMed] [Google Scholar]
- Gomes AM, Winter S, Klein K, Turpeinen M, Schaeffeler E, Schwab M, Zanger UM. (2009) Pharmacogenomics of human liver cytochrome P450 oxidoreductase: multifactorial analysis and impact on microsomal drug oxidation. Pharmacogenomics 10:579–599 [DOI] [PubMed] [Google Scholar]
- Gomes LG, Huang N, Agrawal V, Mendonça BB, Bachega TA, Miller WL. (2008) The common P450 oxidoreductase variant A503V is not a modifier gene for 21-hydroxylase deficiency. J Clin Endocrinol Metab 93:2913–2916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J, Weng Y, Zhang QY, Cui H, Behr M, Wu L, Yang W, Zhang L, Ding X. (2003) Liver-specific deletion of the NADPH-cytochrome P450 reductase gene: impact on plasma cholesterol homeostasis and the function and regulation of microsomal cytochrome P450 and heme oxygenase. J Biol Chem 278:25895–25901 [DOI] [PubMed] [Google Scholar]
- Guengerich FP. (2008) Cytochrome p450 and chemical toxicology. Chem Res Toxicol 21:70–83 [DOI] [PubMed] [Google Scholar]
- Harada T, Ariyoshi N, Shimura H, Sato Y, Yokoyama I, Takahashi K, Yamagata S, Imamaki M, Kobayashi Y, Ishii I, et al. (2010) Application of Akaike information criterion to evaluate warfarin dosing algorithm. Thromb Res 126:183–190 [DOI] [PubMed] [Google Scholar]
- Hart SN, Wang S, Nakamoto K, Wesselman C, Li Y, Zhong XB. (2008) Genetic polymorphisms in cytochrome P450 oxidoreductase influence microsomal P450-catalyzed drug metabolism. Pharmacogenet Genomics 18:11–24 [DOI] [PubMed] [Google Scholar]
- Heinemeyer T, Wingender E, Reuter I, Hermjakob H, Kel AE, Kel OV, Ignatieva EV, Ananko EA, Podkolodnaya OA, Kolpakov FA, et al. (1998) Databases on transcriptional regulation: TRANSFAC, TRRD and COMPEL. Nucleic Acids Res 26:362–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson CJ, Otto DM, Carrie D, Magnuson MA, McLaren AW, Rosewell I, Wolf CR. (2003) Inactivation of the hepatic cytochrome P450 system by conditional deletion of hepatic cytochrome P450 reductase. J Biol Chem 278:13480–13486 [DOI] [PubMed] [Google Scholar]
- Huang N, Agrawal V, Giacomini KM, Miller WL. (2008) Genetics of P450 oxidoreductase: sequence variation in 842 individuals of four ethnicities and activities of 15 missense mutations. Proc Natl Acad Sci USA 105:1733–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang N, Pandey AV, Agrawal V, Reardon W, Lapunzina PD, Mowat D, Jabs EW, Van Vliet G, Sack J, Flück CE, et al. (2005) Diversity and function of mutations in p450 oxidoreductase in patients with Antley-Bixler syndrome and disordered steroidogenesis. Am J Hum Genet 76:729–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminsky LS, Dunbar DA, Wang PP, Beaune P, Larrey D, Guengerich FP, Schnellmann RG, Sipes IG. (1984) Human hepatic cytochrome P-450 composition as probed by in vitro microsomal metabolism of warfarin. Drug Metab Dispos 12:470–477 [PubMed] [Google Scholar]
- Limdi NA, Veenstra DL. (2008) Warfarin pharmacogenetics. Pharmacotherapy 28:1084–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marohnic CC, Panda SP, McCammon K, Rueff J, Masters BS, Kranendonk M. (2010) Human cytochrome P450 oxidoreductase deficiency caused by the Y181D mutation: molecular consequences and rescue of defect. Drug Metab Dispos 38:332–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh S, McLeod HL. (2006) Pharmacogenomics: from bedside to clinical practice. Hum Mol Genet 15 (Spec No 1):R89–R93 [DOI] [PubMed] [Google Scholar]
- Miller WL, Agrawal V, Sandee D, Tee MK, Huang N, Choi JH, Morrissey K, Giacomini KM. (2011) Consequences of POR mutations and polymorphisms. Mol Cell Endocrinol 336:174–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller WL, Huang N, Agrawal V, Giacomini KM. (2009) Genetic variation in human P450 oxidoreductase. Mol Cell Endocrinol 300:180–184 [DOI] [PubMed] [Google Scholar]
- Nicolo C, Flück CE, Mullis PE, Pandey AV. (2010) Restoration of mutant cytochrome P450 reductase activity by external flavin. Mol Cell Endocrinol 321:245–252 [DOI] [PubMed] [Google Scholar]
- Oneda B, Crettol S, Jaquenoud Sirot E, Bochud M, Ansermot N, Eap CB. (2009) The P450 oxidoreductase genotype is associated with CYP3A activity in vivo as measured by the midazolam phenotyping test. Pharmacogenet Genomics 19:877–883 [DOI] [PubMed] [Google Scholar]
- Porter TD, Coon MJ. (1991) Cytochrome P-450. Multiplicity of isoforms, substrates, and catalytic and regulatory mechanisms. J Biol Chem 266:13469–13472 [PubMed] [Google Scholar]
- Rettie AE, Tai G. (2006) The pharmacogenomics of warfarin: closing in on personalized medicine. Mol Interv 6:223–227 [DOI] [PubMed] [Google Scholar]
- Sandee D, Morrissey K, Agrawal V, Tam HK, Kramer MA, Tracy TS, Giacomini KM, Miller WL. (2010) Effects of genetic variants of human P450 oxidoreductase on catalysis by CYP2D6 in vitro. Pharmacogenet Genomics 20:677–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt EE, Molony C, Chudin E, Hao K, Yang X, Lum PY, Kasarskis A, Zhang B, Wang S, Suver C, et al. (2008) Mapping the genetic architecture of gene expression in human liver. PLoS Biol 6:e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schelleman H, Limdi NA, Kimmel SE. (2008) Ethnic differences in warfarin maintenance dose requirement and its relationship with genetics. Pharmacogenomics 9:1331–1346 [DOI] [PubMed] [Google Scholar]
- Sim SC, Miller WL, Zhong XB, Arlt W, Ogata T, Ding X, Wolf CR, Flück CE, Pandey AV, Henderson CJ, et al. (2009) Nomenclature for alleles of the cytochrome P450 oxidoreductase gene. Pharmacogenet Genomics 19:565–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh O, Sandanaraj E, Subramanian K, Lee LH, Chowbay B. (2011) Influence of CYP4F2 rs2108622 (V433M) on warfarin dose requirement in Asian patients. Drug Metab Pharmacokinet 26:130–136 [DOI] [PubMed] [Google Scholar]
- Takeuchi F, McGinnis R, Bourgeois S, Barnes C, Eriksson N, Soranzo N, Whittaker P, Ranganath V, Kumanduri V, McLaren W, et al. (2009) A genome-wide association study confirms VKORC1, CYP2C9, and CYP4F2 as principal genetic determinants of warfarin dose. PLoS Genet 5:e1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tee MK, Huang N, Damm I, Miller WL. (2011) Transcriptional regulation of the human P450 oxidoreductase gene: hormonal regulation and influence of promoter polymorphisms. Mol Endocrinol 25:715–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomalik-Scharte D, Maiter D, Kirchheiner J, Ivison HE, Fuhr U, Arlt W. (2010) Impaired hepatic drug and steroid metabolism in congenital adrenal hyperplasia due to P450 oxidoreductase deficiency. Eur J Endocrinol 163:919–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadelius M, Pirmohamed M. (2007) Pharmacogenetics of warfarin: current status and future challenges. Pharmacogenomics J 7:99–111 [DOI] [PubMed] [Google Scholar]
- Yin T, Miyata T. (2007) Warfarin dose and the pharmacogenomics of CYP2C9 and VKORC1—rationale and perspectives. Thromb Res 120:1–10 [DOI] [PubMed] [Google Scholar]
- Zhang QY, Fang C, Zhang J, Dunbar D, Kaminsky L, Ding X. (2009) An intestinal epithelium-specific cytochrome P450 (P450) reductase-knockout mouse model: direct evidence for a role of intestinal P450s in first-pass clearance of oral nifedipine. Drug Metab Dispos 37:651–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.