Abstract
Localized translation in mammalian dendrites may play a role in synaptic plasticity and contribute to the molecular basis for learning and memory. The regulatory mechanisms that control localized translation in neurons are not well understood. We propose a role for microRNAs (miRNAs), a class of noncoding RNAs, as mediators of neuronal translational regulation. We have identified 86 miRNAs expressed in mammalian neurons, of which 40 have not previously been reported. A subset of these miRNAs exhibits temporally regulated expression in cortical cultures. Moreover, all of the miRNAs that were tested cofractionate with polyribosomes, the sites of active translation. These findings indicate that a large, diverse population of miRNAs may function to regulate translation in mammalian neurons.
Keywords: translational control, noncoding RNA, messenger ribonucleoprotein complex (mRNP)
The role of microRNAs (miRNAs) in translational repression stems from studies of the first two miRNAs, lin-4 and let-7, identified genetically in Caenorhabditis elegans (1-6). lin-4 attenuates the translation, but not the mRNA abundance, of two target genes, lin-14 and lin-28, by imperfect base-pairing to complementary sequences in the 3′ UTR of the target mRNAs (2, 7). Similarly, let-7 regulates lin-41 and hbl-1/lin-57 through partially complementary sequences in the target mRNA 3′ UTR (3, 4, 8, 9). In Drosophila melanogaster, the bantam miRNA regulates the expression of its target, the proapoptotic hid gene (10). In these cases, the targets of miRNAs in animals are regulated at the posttranscriptional level through miRNA binding to imperfect antisense sites in the target mRNA 3′ UTR.
Over 200 miRNAs have been identified from animals, plants, and yeast (11-25). miRNA cloning from a panel of mouse organs revealed that many were expressed in a tissue-specific manner and supports roles for miRNAs in development (14, 15). In addition, computational analyses have revealed in genomes many more miRNAs that have eluded cloning strategies, often because of their low abundance and/or their brief temporal window of expression, and have suggested that as much as 1% to 1.5% of a genome may be represented by miRNAs (23-26).
The function of both lin-4 and let-7 in the control of developmental timing in C. elegans, and the temporal and spatial control of bantam miRNA expression during fly development, may indicate that many of the miRNAs with unknown functions regulate the temporal sequence of gene expression during development or other regulatory pathways (5, 10). The aggregate function of the miRNAs also can be inferred from mutations in the genes encoding components of the general machinery required for all miRNAs. lin-4 and let-7 are detected as ≈70-nt precursors with predicted hairpin secondary structures, which are cleaved by the Dicer RNase (DCR-1) to release the 22-nt mature miRNAs (1, 3, 27-29). Mutations in Dicer and Argonaute proteins, two components required in processing miRNAs, cause profound developmental defects, including meristem patterning in plants and embryonic development in C. elegans (6, 30, 31).
Regulatory pathways rich in evidence for translational control are expected to use miRNAs. Translational control has been implicated in neural development as well as the maintenance and plasticity of neural connections (32, 33). Select mRNAs and translational machinery, including ribosomes and other noncoding RNAs, are localized to dendritic regions of neurons (32-34). In addition, axons have been demonstrated to be capable of localized translation (35-37). Some of these mRNAs encode proteins such as kinases and translational control factors that are attractive candidates to mediate synaptic changes (33, 34, 38). Elements in the 3′ UTR of some mRNAs have been implicated in their localization (39-42). There is evidence that synaptic activity activates the translation of these localized mRNAs (32, 43). In addition, various neurotrophic signals regulate translation locally (37, 44-46). One attractive feature of localized translational control is that it can occur just in the region of synaptic activity, neatly solving the problem of how changes in gene expression in a neuron can modify only active synapses, when neurons can have thousands of synapses not destined for plastic changes (47). miRNAs would fit well into this model if their maturation, localization, or expression were modulated by synaptic activity.
We therefore sought to identify miRNAs from mammalian brain preparations and explored their regulation and possible function in translational control. We found many previously uncharacterized miRNAs and show that they are localized to polyribosomal fractions where translational control is likely to occur. We also found that the expression of particular brain miRNAs changes over time as neurons differentiate in culture. These data suggest that, as in the case of the lin-4 and let-7 miRNAs, some mammalian neural miRNAs may regulate the temporal sequence of developmental events by their control of translation of target mRNAs.
Methods
Primary Cortical Cultures and Sucrose Density Gradients. Procedures for culturing E18 rat cerebral cortices, cellular fractionation of cortical primary neurons, and linear sucrose gradients [15-45% (wt/wt)] have been described (48).
miRNA Cloning and RNA Analysis. The miRNA cloning procedure has been described (11). The starting material for the miRNA cloning combined primary cultures of dissociated E18 rat cortex that had been grown in vitro for 1.5, 7, and 14 days. Total RNA isolation from rat tissues, primary cell cultures, and sucrose gradients has been described (48). Antisense probes were designed to the cloned miRNA sequences and used for the Northern blot analysis as described (3, 49).
Annotation of miRNAs. Sequences from cloned miRNAs were grouped by sequence identity, and the longest representative from each group was searched against mouse genome sequence and rat genome trace sequences. Flanking sequence then was used to predict secondary structure by means of MFOLD 3.1 (50), and we propose as precursors the best sequence matches that, together with flanking sequence, are predicted to form hairpin secondary structure. All of the new miRNAs were submitted to the miRNA Registry web site at www.sanger.ac.uk/Software/Rfam/mirna for official annotation. See Table 2, which is published as supporting information on the PNAS web site, for the full set of cloned sequences and predicted structures. The clustering of miRNAs into sequence families has been described recently (24-26).
Results and Discussion
To investigate the role of miRNAs in translational regulatory mechanisms in mammalian neurons, we first isolated miRNAs from rat cerebro-cortical dissociated cultures of E18 rat embryos. RNAs of 20 to 25 nt in length were purified, cloned, and sequenced. Eighty-six distinct miRNAs were identified (Tables 1 and 2); 40 were distinct from previously reported mammalian miRNAs, and the remaining 46 matched those previously identified (13-16, 21, 23, 51). Of the 40 new miRNAs, 32 miRNAs (mir-322 to mir-352) were entirely new and, in the case of mir-324, a mature miRNA was identified from both the 5′ and 3′ arms of the precursor hairpin (annotated as mir-324-5p and mir-324-3p, respectively). Five other new miRNAs were cloned also from the opposite strand of known precursors and have been tagged with an * to indicate the origins of these miRNAs (e.g., the miRNA cloned from the opposite strand of mir-20 was annotated as mir-20*; the others are mir-7-1*, mir-129-2*, mir-140*, and mir-151*; see Tables 1 and 2). Finally, three new miRNAs were close enough to existing miRNAs, but derived from distinct loci, to receive the same mir number followed by a new letter suffix (mir-135b, mir-148b, and mir-101b).
Table 1. Rat miRNA sequences identified by cloning from E18 primary cortical neurons.
| Precursors
|
||||
|---|---|---|---|---|
| miRNA names | miRNA clone sequence | Northern | Rat | Mouse |
| mir-322 | AAACATGAAGCGCTGCAACA | — | + | + |
| mir-323 | GCACATTACACGGTCGACCTCT (8×) | + | + | + |
| mir-324-3p | CCACTGCCCCAGGTGCTGCTGG (6×) | + | + | + |
| mir-324-5p | CGCATCCCCTAGGGCATTGGTGT (7×) | + | + | + |
| mir-325 | CCTAGTAGGTGCTCAGTAAGTGT (4×) | — | + | + |
| mir-326 | CCTCTGGGCCCTTCCTCCAGT | + | + | + |
| mir-327 | CCTTGAGGGGCATGAGGGT | — | + | |
| mir-328 | CTGGCCCTCTCTGCCCTTCCGT (2×) | + | + | + |
| mir-329 | AACACACCCAGCTAACCTTTTT (2×) | + | + | + |
| mir-330 | GCAAAGCACAGGGCCTGCAGAGAA (2×) | — | + | + |
| mir-331 | GCCCCTGGGCCTATCCTAGAA (4×) | + | + | + |
| mir-332 | GGTCCCACCAGAGTCGCCA | — | + | |
| mir-333 | GTGGTGTGCTAGTTACTTTT | — | + | |
| mir-334 | TAAACGGTGCAGAGATGTG | — | + | |
| mir-335 | TCAAGAGCAATAACGAAAAATGT | + | + | + |
| mir-336 | TCACCCTTCCATATCTAGTCT | — | + | + |
| mir-337 | TTCAGCTCCTATATGATGCCTTT (2×) | — | + | + |
| mir-338 | TCCAGCATCAGTGATTTTGTTGA | + | + | + |
| mir-339 | TCCCTGTCCTCCAGGAGCTCATT | — | + | + |
| mir-340 | TCCGTCTCAGTTACTTTATAGCC (2×) | — | + | + |
| mir-341 | TCGATCGGTCGGTCGGTCAGT | — | + | + |
| mir-342 | TCTCACACAGAAATCGCACCCGTC | + | + | + |
| mir-343 | TCTCCCTCCGTGTGCCCAGTT | — | + | + |
| mir-344 | TGATCTAGCCAAAGCCTGACCGT | + | + | + |
| mir-345 | TGCTGACCCCTAGTCCAGTGC (2×) | + | + | + |
| mir-346 | TGTCTGCCTGAGTGCCTGCCTCTAA | — | + | + |
| mir-347 | TGTCCCTCTGGGTCGCCA | — | + | |
| mir-348 | TGCCCACCCTTTACCCCACTCCAGT | — | + | |
| mir-349 | CAGCCCTGCTGTCTTAACCTCT | + | + | |
| mir-350 | ATTCACAAAGCCCATACACTTTCAC | — | + | + |
| mir-351 | TCCCTGAGGAGCCCTTTGAGCCTGT (7×) | + | + | + |
| mir-352 | AGAGTAGTAGGTTGCATAGTA (2×) | — | + | |
| let-7a | TGAGGTAGTAGGTTGTATAGTT (5×) | NA | + | + |
| let-7b | TGAGGTAGTAGGTTGTGTGGTTT (11×) | NA | + | + |
| let-7c | TGAGGTAGTAGGTTGTATGGTTT (28×) | NA | + | + |
| let-7d | AGAGGTAGTAGGTTGCATAGTT (13×) | NA | + | |
| let-7d* | CTATACGACCTGCTGCCTTTCTA (2×) | + | + | + |
| let-7e | TGAGGTAGGAGGTTGTATAGTTA (2×) | NA | + | + |
| let-7f | TGAGGTAGTAGATTGTATAGTT (10×) | NA | + | + |
| let-7gL | TGAGGTAGTAGTTTGTACAGTT (5×) | + | + | |
| let-7i | TGAGGTAGTAGTTTGTGCTGTT (4×) | NA | + | + |
| mir-7-1* | CAACAAATCACAGTCTGCCATA (2×) | + | + | + |
| mir-9 | TCTTTGGTTATCTAGCTGTATGA (9×) | NA | + | + |
| mir-15 | TAGCAGCACATAATGGTTTGT (2×) | NA | + | |
| mir-15b | TAGCAGCACATCATGGTTTACA (2×) | NA | + | + |
| mir-16 | TAGCAGCACGTAAATATTGGCGT (20×) | + | + | + |
| mir-18 | TAAGGTGCATCTAGTGCAGATAG (3×) | NA | + | + |
| mir-19b | TGTGCAAATCCATGCAAAACTGA | NA | + | + |
| mir-20 | TAAAGTGCTTATAGTGCAGGTAG (3×) | NA | + | + |
| mir-20* | ACTGCATTACGAGCACTTACA | — | + | + |
| mir-21 | TAGCTTATCAGACTGATGTTGA (3×) | NA | + | + |
| mir-22 | AAGCTGCCAGTTGAAGAACTGT | NA | + | + |
| mir-29a | TAGCACCATCTGAAATCGGTTA | NA | + | + |
| mir-30b | TGTAAACATCCTACACTCAGCT (6×) | + | + | + |
| mir-30c | TGTAAACATCCTACACTCTCAGCT (7×) | NA | + | + |
| mir-30d | TGTAAACATCCCCGACTGGAAGCT (3×) | NA | + | + |
| mir-34 | TGGCAGTGTCTTAGCTGGTTGT (3×) | — | + | + |
| mir-91 | CAAAGTGCTTACAGTGCAGGTAG (21×) | NA | + | + |
| mir-93 | CAAAGTGCTGTTCGTGCAGGTAG (9×) | NA | + | + |
| mir-98 | TGAGGTAGTAAGTTGTATTGTT | NA | + | |
| mir-99a/miR-99 | AACCCGTAGATCCGATCTTGTG (18×) | NA | + | + |
| mir-99b | CACCCGTAGAACCGACCTTGCG (3×) | + | + | + |
| mir-100 | AACCCGTAGATCCGAACTTGTG (6×) | + | + | + |
| mir-101b | TACAGTACTGTGATAGCTGAAG | — | + | + |
| mir-103 | AGCAGCATTGTACAGGGCTATGA (10×) | + | +(2) | +(2) |
| mir-118-b, -c | ACCATCGACCGTTGATTGTACC (3×) | + | + | + |
| mir-124a | TTAAGGCACGCGGTGAATGCCAA (59×) | + | +(2) | +(2) |
| mir-125b | ATCCCTGAGACCCTAACTTGTGA (71×) | + | + | + |
| mir-127 | TCGGATCCGTCTGAGCTTGG (2×) | NA | + | + |
| mir-128 | TCACAGTGAACCGGTCTCTTTC (17×) | + | + | + |
| mir-129-2* | AAGCCCTTACCCCAAAAAGCAT (3×) | + | + | + |
| mir-130 | CAGTGCAATGTTAAAAGGGCAT (23×) | + | + | + |
| mir-130b | CAGTGCAATGATGAAAGGGCAT (2×) | + | + | + |
| mir-131 | TAAAGCTAGATAACCGAAAGTA (6×) | NA | +(2) | +(2) |
| mir-132 | TAACAGTCTACAGCCATGGTCG (16×) | NA | + | + |
| mir-135b | TATGGCTTTTCATTCCTATGTG (2×) | — | + | + |
| mir-138 | AGCTGGTGTTGTGAATCAGGCCG | + | +(2) | + |
| mir-140* | TACCACAGGGTAGAACCACGGACA | + | + | + |
| mir-148b | TCAGTGCATCACAGAACTTTGT | — | + | + |
| mir-149 | TCTGGCTCCGTGTCTTCACTCCCT | NA | + | + |
| mir-151* | TCGAGGAGCTCACAGTCTAGTA (2×) | — | + | + |
| mir-181 | AACATTCAACGCTGTCGGTGAG | + | +(2) | +(2) |
| mir-191 | CAACGGAATCCCAAAAGCAGCTGT (6×) | + | + | + |
| mir-218-1, -2 | TTGTGCTTGATCTAACCATGTG | + | +(3) | +(2) |
| mir-221 | AGCTACATTGTCTGCTGGGTTTC | + | + | + |
| mir-301 | CAGTGCAATAGTATTGTCAAAGCAT (2×) | — | + | + |
The new miRNAs identified from this study have been assigned mir annotations from mir-322 to -352. In addition, new miRNAs cloned from the opposite strand of a known precursor had an asterisk appended to the known mir annotation. Finally, new miRNAs deemed close enough to existing miRNAs, but derived from distinct loci, received the same mir annotation as the known miRNA but with a ”b” suffix. Sequences represent the longest of each miRNA sequence identified by cloning and that matches to the greatest extent with available rat or mouse genome sequence. Frequency of cloning is indicated in parentheses after the miRNA sequences, and variations between clones and genomic sequences are denoted in bold. All of the previously uncharacterized miRNAs were examined by Northern blotting with total RNA isolated from adult rat cortex, those detected by Northern blot are indicated (+). The presence of predicted precursor hairpin structures in mouse and rat genome sequences is noted for each miRNA. The number of times a particular miRNA appears in the genome is noted in parentheses in the mouse and rat ”precursor” columns when there is more than one observed occurrence. miRNAs found only in the rat genome may be rat-specific, although those found only in the mouse genome may not yet be represented in available rat genome sequence traces. Clone sequence information and precursor hairpin sequences and structures are presented in Table 2; NA, not assayed.
The number of times a miRNA was cloned varied greatly, ranging from 71 clones for mir-125 to a single clone for 32 of the 86 miRNAs; in all, 29 of the 86 miRNAs were isolated four or more times (Tables 1 and 2). The corresponding rat and/or mouse genomic sequences and their locations were identified for each miRNA. The 5′ or 3′ flanking genomic sequences then were tested for the ability to fold into canonical ≈70-nt miRNA precursor hairpin structures by using the MFOLD program (www.bioinfo.rpi.edu/~zukerm/rna; refs. 50 and 52). Predicted precursors for 32 of the 40 newly identified rat miRNAs were found in both rat genome traces and the assembled mouse genome sequence (ftp://ftp.ncbi.nih.gov/genomes/M_musculus and ftp://ftp.ncbi.nih.gov/pub/TraceDB/rattus_norvegicus), and, in nearly all of those cases, sequence conservation included the predicted precursor (Table 2). The six new rat miRNAs that were not identified in the mouse genome may be rat-specific, and two rat miRNAs identified in the mouse genome but not in rat genome traces may reflect incompleteness of the rat genome assembly (see Tables 1 and 2).
All 40 new miRNAs were examined by Northern blotting to total RNA from adult rat cortex, and 17 of the miRNAs were detected (Table 1). The remaining 23 either may be expressed at levels below the detection threshold for Northern blotting or may exhibit temporally restricted expression patterns. In general, the number of times a particular miRNA was cloned correlated with the intensity of the signal by Northern blotting. In fact, 22 of the 23 new miRNAs that escaped detection by Northern blotting were cloned only once or twice (Table 1). Two recent studies demonstrate that many bona fide miRNAs evade detection by Northern blotting; it is only when an amplified library of small RNAs is queried by a second PCR-based amplification method with primers complementary to the miRNA under study that it can be detected (24, 26). Furthermore, although the miRNAs were cloned from primary E18 cortical cultures, which are enriched for neurons, testing expression by Northern blotting for all of the cloned miRNAs necessitated a large quantity of total RNA, which we procured from the abundant RNA source of adult rat cortex. Therefore, the developmental difference in the cortical material used for miRNA cloning and Northern blotting also may explain the lack of detection of 23 of the 40 new miRNAs identified in this study.
Sequence analysis indicates that clusters of miRNAs share common sequence elements, especially within 6- to 8-nt regions in the 5′ half of the mature miRNA sequences. Based on sequence motif conservation, the mature miRNAs were grouped into sequence “families” that include previously cloned miRNAs from other organisms (e.g., Fig. 1). Examples of such miRNA sequence families have been reported recently (24-26). A dendrogram representation of the miRNA sequence relatedness is shown in Fig. 5, which is published as supporting information on the PNAS web site.
Fig. 1.
miRNAs from rat cortical neurons cluster in miRNA “families.” Previously identified miRNA sequences and miRNAs cloned in this study from combined E18 rat cortical neurons cultured for 1.5, 7, and 14 days were compiled and each sequence was aligned against all others by using a Smith-Waterman algorithm (EMBOSS 2.3.1; ref. 70). Complete hierarchical clustering was performed on the dissimilarity matrix generated from the scores of the pairwise sequence alignments. A dendrogram was generated from the clustering analysis (see Fig. 5) and was cut to yield a set of clusters, which were hand-adjusted to improve groupings of miRNAs that share common subsequences. Two examples of such clusters are presented here. Conserved sequences are highlighted in yellow. miRNAs identified in rat cortical neurons are labeled in red. R.n., Rattus norvegicus; M.m., Mus musculus; H.s., Homo sapiens; C.e., Caenorhabditis elegans. H/M/R designates clones found in human, mouse, and rat.
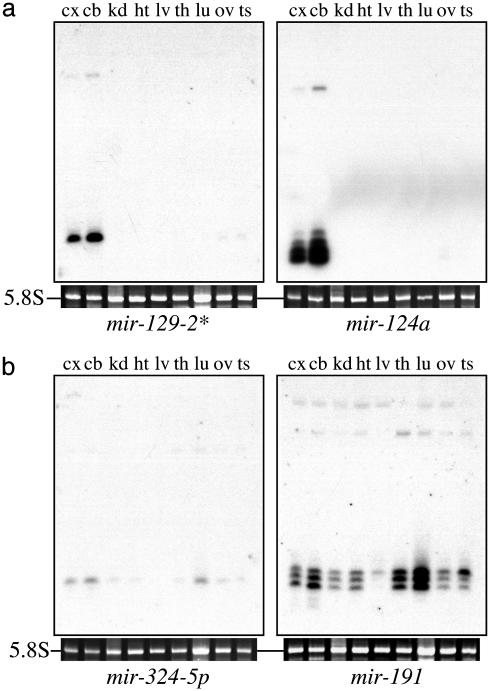
To determine whether the expression of the present set of miRNAs cloned from rat neurons was restricted to neurons or more broadly expressed, we analyzed the subset of miRNAs that was easily detectable by Northern blotting with total RNA from a panel of rat tissues (Fig. 2). Of the 12 miRNAs examined, eight (mir-103, -124a, -128, -323, -326, -329, -344, and -129-2*) were expressed largely in the cortex and cerebellum (Fig. 2a), suggesting a neural-specific function; the remaining four (mir-191, -324-5p, -335, and -151*) exhibited a broader, but distinct, tissue distribution pattern (Fig. 2b).
Fig. 2.
Tissue-specific expression patterns of miRNAs cloned from rat cortex. Northern blots of total RNA isolated from adult rat tissues were assayed with probes against 12 miRNAs. Eight miRNAs were expressed predominantly in the cortex and cerebellum. Two miRNAs are shown in a: mir-129-2* and mir-124a, previously cloned from mouse (14); the others (not shown) with brain-restricted expression patterns include the rat homologue of mir-103 cloned from the Gemin3-Gemin4 - eIF2C2 complex in human HeLa cells (16), mir-128 from mouse (14), and four previously uncharacterized miRNAs (mir-323,-326, -329, and -344). Four miRNAs exhibit a broader expression pattern. Two miRNAs are shown in b: mir-191 and mir-324-5p; the others with broad expression patterns were mir-335 and mir-151*. The tissues analyzed were cortex (cx), cerebellum (cb), kidney (kd), heart (ht), liver (lv), thymus (th), lung (lu), ovary (ov), and testis (ts).
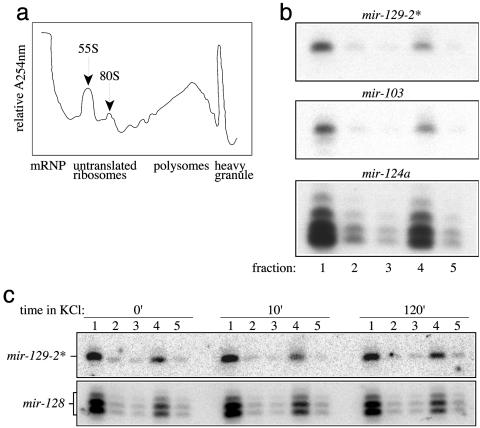
We investigated a possible role of the new miRNAs in neuronal translation by asking whether miRNAs were localized to sites of active translation on polyribosomes. Dissociated cultured neurons from the E18 rat forebrain were fractionated and resolved on sucrose density gradients (see Methods). Measurements of the total RNA in the gradient fractions reveal signature patterns of peaks corresponding to messenger ribonucleoprotein complexes (mRNP), followed by the 55S and 80S ribosomal fractions, the polyribosomal fraction, and a dense fraction containing RNA granules (Fig. 3a). Northern blot analysis of the eight neuronally expressed miRNAs (mir-103, -124a, -128, -323, -326, -329, -344, and -129-2*) indicates that, in all cases, the miRNAs displayed a bimodal distribution with peak miRNA abundance in the light fraction containing mRNPs and, in the polyribosome fraction, the site of active translation (Fig. 3b). Interestingly, miRNAs were not detected in the fraction containing RNA granules, which are macromolecular structures that have been proposed to transport untranslated mRNA cargo to dendritic sites before activity-dependent translation (48, 53-55). The steadystate localization of miRNAs with polyribosomes suggests that miRNA-mediated regulation occurs at active sites of translation in neurons. Whether the fraction of miRNAs localized to the mRNP fraction represents an intermediate population in the transport pathway leading to the translational machinery remains to be determined.
Fig. 3.
Localization of miRNAs to the mRNP and polyribosomes in cortical neurons. (a) Cytoplasmic extracts of primary cortical neurons were lysed and resolved on 15-45% sucrose gradients after sedimentation of nuclei and the majority of mitochondria. Continuous RNA measurements (A254) of the gradient fractions were measured. (b) Fraction 1 contains mRNPs, fractions 2 and 3 contain 55S and 80S ribosomal subunits, respectively, fraction 4 contains polyribosomes, and fraction 5 contains RNA granules. Total RNA isolated from each gradient fraction was analyzed by Northern blots with complementary probes for 8 miRNAs [mir-129-2*, -103, and -124a (shown) and mir-128,-323,-326,-329, and 344 (not shown)]. All of the miRNAs tested are predominantly localized to the mRNP and polyribosome fractions. (c) Four miRNAs [mir-128 and -129-2* (shown) and mir-326 and -344 (not shown)] were analyzed on cortical neuron cultures that were untreated (0′) or incubated in 50 mM KCl for 10 min (10′) or 2 h (120′) and resolved by sucrose density gradients as in b. A change in miRNA expression or distribution in the mRNP and polyribosome fractions was not detected after KCl treatment for these miRNAs.
Depolarization of neurons results in the translation of somatodendritic, plasticity-related mRNAs (33, 38, 46, 48, 56). However, depolarization of primary neuronal cultures with KCl did not alter the localization or expression of the miRNAs tested (mir-128, -326, -344, and -129-2*) in the mRNP and polyribosome fractions (Fig. 3c), suggesting that if miRNAs regulate such translational transitions, they occur without the release of these miRNAs from active translational machinery. However, to conclude that all miRNAs behave in the same manner as the four miRNAs examined here will require a survey of many more miRNAs and the analysis of neurons under a greater variety of stimulatory and quiescent conditions.
The localization of miRNAs to polyribosomes is suggestive that all of the miRNAs tested, and perhaps all miRNAs in mammalian neurons, mediate the translational control of target mRNAs on polyribosomes. These findings are consistent with the observation in C. elegans: lin-4 and its target lin-14 mRNA both associate with polyribosomes, even after lin-4 down-regulates the translation of the lin-14 mRNA and indicates that repression occurs after the initiation of translation (57).
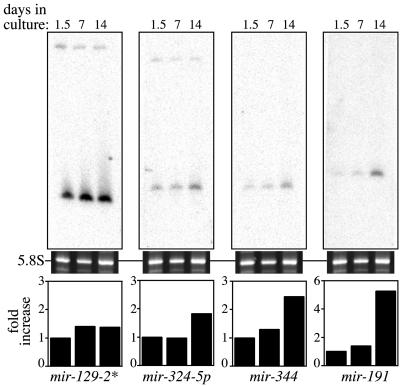
Recent studies demonstrate that translational control in dendrites and axons is essential for developmental processes such as axon growth, guidance, and collapse, as well as dendritic maturation (36). We examined whether the expression of miRNAs identified in neurons might be temporally regulated and correspond to the stages of neuronal differentiation. Primary neurons in culture follow a well established set of morphological events that lead to mature, differentiated neurons (58-61). Cultures of rat E18 cortical neurons were plated and harvested at three stages of growth: 1.5 days in vitro (DIV) cultures, when cortical neurons adhere to the plates, begin growing minor neurites and extend one of these minor neurites as the incipient axon; 7 DIV, when axonal projections have emerged, dendritic elaboration is well underway, and the number of synapses increases dramatically; and 14 DIV, when dendritic processes have formed and additional synapses are established and stabilized. We examined the expression of 12 miRNAs by Northern blot analysis and discovered that 7 (mir-128, -191, -323, -324-5p, -326, -329, and -344) increased in expression ≈2- to 5-fold from day 1.5 to day 14 in cortical cultures (Fig. 4). The expression level of the other miRNAs tested remained relatively constant over the time course. The temporal regulation of a subset of miRNAs correlated with these neural developmental events, and the precedence for miRNA function in regulation of timing during animal development suggests a potential role for miRNA-mediated gene regulation during neuronal differentiation and development.
Fig. 4.
Temporal expression of miRNAs in primary rat cortical cultures. Dissociated neurons from E18 rat forebrain were plated and harvested after 1.5, 7, and 14 days in culture. Northern blots of equal amounts of total RNA isolated from these primary cultures were assayed with probes against 12 miRNAs. Three of seven miRNAs (mir-191, -324-5p, and -344) that increase in expression level from day 1.5 to day 14 cultures are shown (the remaining miRNAs, mir-128, -323, -326, and -329, are not shown). mir-129-2* (shown) is among five miRNAs tested (mir-103,-124a,-335,-129-2*, and -151*) that display a relatively constant level of expression. The quantitation of the miRNA expression levels was normalized to day 1.5 cultures.
Our finding that all miRNAs tested are localized to polyribosomes indicates that neuronal miRNAs, as a class, may act as translational regulators of neuronal gene expression. miRNAs are attractive candidates for this translational control. Those that are not expressed in early cortical cultures but are expressed later may regulate only their target mRNAs at later stages of neural development, for example, to mediate a temporal switch in neural type or pathfinding. Temporal sequences in neural type have been noted in the development of the vertebrate retina (62). In Drosophila, neuroblasts of the central nervous system produce specific lineages of neurons and glia according to the position and timing of division (63-65). This temporal sequence of neuroblast divisions is controlled by a temporal progression of transcription factor expression (hunchback → Kruppel → pdm → castor) in neuroblasts (66, 67). Given the precedent for plant miRNAs to target transcription factors, miRNAs are good candidates to mediate the temporal regulation of these transcription factors in D. melanogaster. The mammalian miRNAs that we have shown to be temporally regulated during cortical development may act on analogous or even homologous target mRNAs.
Given that the miRNA localization on polysomes did not change in KCl-activated neural preparations (Fig. 3c), it is unlikely that the activity of these miRNAs is regulated by their localization to polyribosomes. In addition, we did not see changes in the precursor to mature miRNA product ratio in these models of synaptic activity (data not shown), suggesting that it also is unlikely that the activity of these miRNAs is regulated at the level of processing. However, because we have not yet assigned any targets or functions to these miRNAs, it is possible that the particular miRNAs tested are regulated by neural events other than KCl exposure. Alternatively, only a subset of miRNAs may be regulated through localization to the polyribosome or by conversion of the precursor miRNA to the mature form.
The identification of translational machinery within dendritic spines suggests that the polyribosome-associated miRNAs also may be localized to dendrites to regulate local translation. Although in situ hybridization of the 21- to 25-nt miRNAs to subcellular locations remains a challenging technical problem, a recent study in D. melanogaster indicates that the fragile X mental retardation protein (FMRP in mammals; dFMR1 in fly) colocalizes with ribosomal translational apparatus, as well as with miRNAs, in vivo (68). FMRP also localizes to dendritic sites and interacts with polyribosomes in mammalian neurons (69). Given the requirement of FMRP for normal human cortical function, miRNAs also may be present in translationally active zones in dendrites, functioning in a complex with FMRP.
Despite the identification of >200 miRNAs from plants and animals, very few miRNA targets have been experimentally confirmed. A major challenge in elucidating the function of miRNAs will be to identify the specific target genes that they regulate. Unlike the near-perfect complementarity between plant miRNAs and the targets, computational approaches for target discovery have been complicated by the imperfect complementarity between an miRNA and its target transcript in animals. However, as dendritically localized mRNAs are isolated, it is possible that this more limited sequence space of potential complementarity to the neuronally expressed miRNAs will allow the computational mapping of miRNAs to target mRNAs. Assigning mRNA targets to these miRNAs will prove an important step in the delineation of their roles in the control of the temporal sequence of neural development and in the regulation of synaptic remodeling.
Supplementary Material
Acknowledgments
We thank Dr. Brenda Reinhart for extensive advice and assistance in the miRNA cloning procedures. We also thank Drs. Juergen Brosius, Alison Frand, Maria Grunwald, Patrick Hu, Amy Pasquinelli, and Paul Worley for helpful discussions and comments on the manuscript. This research was supported by a Department of Energy Human Genome Project grant and the Lipper Foundation (to G.M.C.), National Institutes of Health and March of Dimes grants (to K.S.K.), and National Institutes of Health Grant GM44619 (to G.R.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: miRNA, microRNAs; mRNP, messenger ribonucleoprotein complex.
Data deposition: The miRNAs reported in this paper have been deposited in the miRNA Registry database, www.sanger.ac.uk/Software/Rfam/mirna (miRNA nos. mir-322-mir352).
References
- 1.Lee, R. C., Feinbaum, R. L. & Ambros, V. (1993) Cell 75, 843-854. [DOI] [PubMed] [Google Scholar]
- 2.Wightman, B., Ha, I. & Ruvkun, G. (1993) Cell 75, 855-862. [DOI] [PubMed] [Google Scholar]
- 3.Reinhart, B. J., Slack, F. J., Basson, M., Pasquinelli, A. E., Bettinger, J. C., Rougvie, A. E., Horvitz, H. R. & Ruvkun, G. (2000) Nature 403, 901-906. [DOI] [PubMed] [Google Scholar]
- 4.Slack, F. J., Basson, M., Liu, Z., Ambros, V., Horvitz, H. R. & Ruvkun, G. (2000) Mol. Cell 5, 659-669. [DOI] [PubMed] [Google Scholar]
- 5.Rougvie, A. E. (2001) Nat. Rev. Genet. 2, 690-701. [DOI] [PubMed] [Google Scholar]
- 6.Pasquinelli, A. E. & Ruvkun, G. (2002) Annu. Rev. Cell. Dev. Biol. 18, 495-513. [DOI] [PubMed] [Google Scholar]
- 7.Moss, E. G., Lee, R. C. & Ambros, V. (1997) Cell 88, 637-646. [DOI] [PubMed] [Google Scholar]
- 8.Lin, S. Y., Johnson, S. M., Abraham, M., Vella, M. C., Pasquinelli, A., Gamberi, C., Gottlieb, E. & Slack, F. J. (2003) Dev. Cell 4, 639-650. [DOI] [PubMed] [Google Scholar]
- 9.Abrahante, J. E., Daul, A. L., Li, M., Volk, M. L., Tennessen, J. M., Miller, E. A. & Rougvie, A. E. (2003) Dev. Cell 4, 625-637. [DOI] [PubMed] [Google Scholar]
- 10.Brennecke, J., Hipfner, D. R., Stark, A., Russell, R. B. & Cohen, S. M. (2003) Cell 113, 25-36. [DOI] [PubMed] [Google Scholar]
- 11.Lau, N. C., Lim, L. P., Weinstein, E. G. & Bartel, D. P. (2001) Science 294, 858-862. [DOI] [PubMed] [Google Scholar]
- 12.Lee, R. C. & Ambros, V. (2001) Science 294, 862-864. [DOI] [PubMed] [Google Scholar]
- 13.Lagos-Quintana, M., Rauhut, R., Lendeckel, W. & Tuschl, T. (2001) Science 294, 853-858. [DOI] [PubMed] [Google Scholar]
- 14.Lagos-Quintana, M., Rauhut, R., Yalcin, A., Meyer, J., Lendeckel, W. & Tuschl, T. (2002) Curr. Biol. 12, 735-739. [DOI] [PubMed] [Google Scholar]
- 15.Lagos-Quintana, M., Rauhut, R., Meyer, J., Borkhardt, A. & Tuschl, T. (2003) RNA 9, 175-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mourelatos, Z., Dostie, J., Paushkin, S., Sharma, A., Charroux, B., Abel, L., Rappsilber, J., Mann, M. & Dreyfuss, G. (2002) Genes Dev. 16, 720-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rhoades, M. W., Reinhart, B. J., Lim, L. P., Burge, C. B., Bartel, B. & Bartel, D. P. (2002) Cell 110, 513-520. [DOI] [PubMed] [Google Scholar]
- 18.Llave, C., Kasschau, K. D., Rector, M. A. & Carrington, J. C. (2002) Plant Cell 14, 1605-1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reinhart, B. J. & Bartel, D. P. (2002) Science 297, 1831. [DOI] [PubMed] [Google Scholar]
- 20.Reinhart, B. J., Weinstein, E. G., Rhoades, M. W., Bartel, B. & Bartel, D. P. (2002) Genes Dev. 16, 1616-1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dostie, J., Mourelatos, Z., Yang, M., Sharma, A. & Dreyfuss, G. (2003) RNA 9, 180-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hall, I. M., Noma, K. & Grewal, S. I. (2003) Proc. Natl. Acad. Sci. USA 100, 193-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lim, L. P., Glasner, M. E., Yekta, S., Burge, C. B. & Bartel, D. P. (2003) Science 299, 1540. [DOI] [PubMed] [Google Scholar]
- 24.Lim, L. P., Lau, N. C., Weinstein, E. G., Abdelhakim, A., Yekta, S., Rhoades, M. W., Burge, C. B. & Bartel, D. P. (2003) Genes Dev. 17, 991-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ambros, V., Lee, R. C., Lavanway, A., Williams, P. T. & Jewell, D. (2003) Curr. Biol. 13, 807-818. [DOI] [PubMed] [Google Scholar]
- 26.Grad, Y., Aach, J., Hayes, G. D., Reinhart, B. J., Church, G. M., Ruvkun, G. & Kim, J. (2003) Mol. Cell 11, 1253-1263. [DOI] [PubMed] [Google Scholar]
- 27.Grishok, A., Pasquinelli, A. E., Conte, D., Li, N., Parrish, S., Ha, I., Baillie, D. L., Fire, A., Ruvkun, G. & Mello, C. C. (2001) Cell 106, 23-34. [DOI] [PubMed] [Google Scholar]
- 28.Hutvagner, G., McLachlan, J., Pasquinelli, A. E., Balint, E., Tuschl, T. & Zamore, P. D. (2001) Science 293, 834-838. [DOI] [PubMed] [Google Scholar]
- 29.Ketting, R. F., Fischer, S. E., Bernstein, E., Sijen, T., Hannon, G. J. & Plasterk, R. H. (2001) Genes Dev. 15, 2654-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carmell, M. A., Xuan, Z., Zhang, M. Q. & Hannon, G. J. (2002) Genes Dev. 16, 2733-2742. [DOI] [PubMed] [Google Scholar]
- 31.Schauer, S. E., Jacobsen, S. E., Meinke, D. W. & Ray, A. (2002) Trends Plant Sci 7, 487-491. [DOI] [PubMed] [Google Scholar]
- 32.Jiang, C. & Schuman, E. M. (2002) Trends Biochem. Sci. 27, 506-513. [DOI] [PubMed] [Google Scholar]
- 33.Steward, O. & Schuman, E. M. (2001) Annu. Rev. Neurosci. 24, 299-325. [DOI] [PubMed] [Google Scholar]
- 34.Job, C. & Eberwine, J. (2001) Nat. Rev. Neurosci. 2, 889-898. [DOI] [PubMed] [Google Scholar]
- 35.Brittis, P. A., Lu, Q. & Flanagan, J. G. (2002) Cell 110, 223-235. [DOI] [PubMed] [Google Scholar]
- 36.Steward, O. (2002) Cell 110, 537-540. [DOI] [PubMed] [Google Scholar]
- 37.Zhang, X. & Poo, M. M. (2002) Neuron 36, 675-688. [DOI] [PubMed] [Google Scholar]
- 38.Eberwine, J., Miyashiro, K., Kacharmina, J. E. & Job, C. (2001) Proc. Natl. Acad. Sci. USA 98, 7080-7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mayford, M., Baranes, D., Podsypanina, K. & Kandel, E. R. (1996) Proc. Natl. Acad. Sci. USA 93, 13250-13255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blichenberg, A., Schwanke, B., Rehbein, M., Garner, C. C., Richter, D. & Kindler, S. (1999) J. Neurosci. 19, 8818-8829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rook, M. S., Lu, M. & Kosik, K. S. (2000) J. Neurosci. 20, 6385-6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mori, Y., Imaizumi, K., Katayama, T., Yoneda, T. & Tohyama, M. (2000) Nat. Neurosci. 3, 1079-1084. [DOI] [PubMed] [Google Scholar]
- 43.Angenstein, F., Evans, A. M., Settlage, R. E., Moran, S. T., Ling, S. C., Klintsova, A. Y., Shabanowitz, J., Hunt, D. F. & Greenough, W. T. (2002) J. Neurosci. 22, 8827-8837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kang, H. & Schuman, E. M. (1996) Science 273, 1402-1406. [DOI] [PubMed] [Google Scholar]
- 45.Campbell, D. S. & Holt, C. E. (2001) Neuron 32, 1013-1026. [DOI] [PubMed] [Google Scholar]
- 46.Aakalu, G., Smith, W. B., Nguyen, N., Jiang, C. & Schuman, E. M. (2001) Neuron 30, 489-502. [DOI] [PubMed] [Google Scholar]
- 47.Martin, K. C. & Kosik, K. S. (2002) Nat. Rev. Neurosci. 3, 813-820. [DOI] [PubMed] [Google Scholar]
- 48.Krichevsky, A. M. & Kosik, K. S. (2001) Neuron 32, 683-696. [DOI] [PubMed] [Google Scholar]
- 49.Ausubel, F. M., Brent, R., Kingston, R., Moore, D., Seidman, J., Smith, J. & Struhl, K., eds. (1995) Current Protocols in Molecular Biology (Wiley, New York).
- 50.Mathews, D. H., Sabina, J., Zuker, M. & Turner, D. H. (1999) J. Mol. Biol. 288, 911-940. [DOI] [PubMed] [Google Scholar]
- 51.Houbaviy, H. B., Murray, M. F. & Sharp, P. A. (2003) Dev. Cell 5, 351-358. [DOI] [PubMed] [Google Scholar]
- 52.Zuker, M., Mathews, D. H. & Turner, D. H. (1999) in RNA Biochemistry and Biotechnology, ed. Clark, B. F. C. (Kluwer, Dordrecht, The Netherlands), pp. 11-43.
- 53.Knowles, R. B., Sabry, J. H., Martone, M. E., Deerinck, T. J., Ellisman, M. H., Bassell, G. J. & Kosik, K. S. (1996) J. Neurosci. 16, 7812-7820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.De Diego Otero, Y., Severijnen, L. A., van Cappellen, G., Schrier, M., Oostra, B. & Willemsen, R. (2002) Mol. Cell. Biol. 22, 8332-8341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kosik, K. S. & Krichevsky, A. M. (2002) Sci. STKE 2002, PE16. [DOI] [PubMed] [Google Scholar]
- 56.Steward, O. & Worley, P. F. (2001) Proc. Natl. Acad. Sci. USA 98, 7062-7068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Olsen, P. H. & Ambros, V. (1999) Dev. Biol. 216, 671-680. [DOI] [PubMed] [Google Scholar]
- 58.Dotti, C. G., Sullivan, C. A. & Banker, G. A. (1988) J. Neurosci. 8, 1454-1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Craig, A. M. & Banker, G. (1994) Annu. Rev. Neurosci. 17, 267-310. [DOI] [PubMed] [Google Scholar]
- 60.Fletcher, T. L., Cameron, P., De Camilli, P. & Banker, G. (1991) J. Neurosci. 11, 1617-1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fletcher, T. L., De Camilli, P. & Banker, G. (1994) J. Neurosci. 14, 6695-6706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cepko, C. L., Austin, C. P., Yang, X., Alexiades, M. & Ezzeddine, D. (1996) Proc. Natl. Acad. Sci. USA 93, 589-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bossing, T., Udolph, G., Doe, C. Q. & Technau, G. M. (1996) Dev. Biol. 179, 41-64. [DOI] [PubMed] [Google Scholar]
- 64.Schmidt, H., Rickert, C., Bossing, T., Vef, O., Urban, J. & Technau, G. M. (1997) Dev. Biol. 189, 186-204. [DOI] [PubMed] [Google Scholar]
- 65.Schmid, A., Chiba, A. & Doe, C. Q. (1999) Development (Cambridge, U.K.) 126, 4653-4689. [DOI] [PubMed] [Google Scholar]
- 66.Isshiki, T., Pearson, B., Holbrook, S. & Doe, C. Q. (2001) Cell 106, 511-521. [DOI] [PubMed] [Google Scholar]
- 67.Harris, W. A. (2001) Dev. Cell 1, 313-314. [DOI] [PubMed] [Google Scholar]
- 68.Caudy, A. A., Myers, M., Hannon, G. J. & Hammond, S. M. (2002) Genes Dev. 16, 2491-2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ohashi, S., Koike, K., Omori, A., Ichinose, S., Ohara, S., Kobayashi, S., Sato, T. A. & Anzai, K. (2002) J. Biol. Chem. 277, 37804-37810. [DOI] [PubMed] [Google Scholar]
- 70.Rice, P., Longden, I. & Bleasby, A. (2000) Trends Genet. 16, 276-277. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.