Abstract
DNA methylation is a common epigenetic signaling mechanism associated with silencing of repeated DNA and transcriptional regulation in eukaryotes. Here we report that DNA methylation in the human fungal pathogen Candida albicans is primarily localized within structural genes and modulates transcriptional activity. Major repeat sequences and multigene families are largely free of DNA methylation. Among the genes subject to DNA methylation are those associated with dimorphic transition between yeast and hyphal forms, switching between white and opaque cells, and iron metabolism. Transcriptionally repressed methylated loci showed increased frequency of C-to-T transitions during asexual growth, an evolutionarily stable pattern of repression associated mutation that could bring about genetic alterations under changing environmental or host conditions. Dynamic differential DNA methylation of structural genes may be one factor contributing to morphological plasticity that is cued by nutrition and host interaction.
Keywords: methylcytosine, dimorphic fungi, gene expression, genome evolution, bisulfite sequencing
The pathogenic yeast Candida albicans is the most prevalent cause of severe fungal infections in humans, especially in immunocompromised patients (1). The host environment cues complex responses from C. albicans such as dimorphic transition and phenotypic switching, which are important for virulence, survival, and mating of this fungus (2–4). In the laboratory, these unique morphological and phenotypic forms can be induced by changes in growth medium or environmental conditions (3). A twofold difference in DNA methylation, with 5-methylcytosine (meC) constituting 0.05–0.1% of the genome, has been observed between the hyphal and yeast morphologies, respectively (5). This finding suggests that transition between these forms could be regulated by epigenetic modifications.
Methylation of cytosine bases is the best characterized epigenetic DNA modification observed in the genome of eukaryotic organisms. This modification plays an important role in maintenance of genome integrity, genomic imprinting, transcriptional regulation, and developmental processes (6–8). Transposable elements, heterochromatic regions, and DNA repeats of many organisms are densely methylated, resulting in a repressive chromatin state and silencing of these genomic regions (7, 9). In mammals and plants, DNA methylation also occurs at the promoter region of genes, where it can cause gene repression by inhibition of transcriptional initiation (6, 8, 10). Recent studies have uncovered DNA methylation within the transcribed regions, or bodies, of genes in flowering plants and mammalian stem cells, where it is primarily involved in the inhibition of transcriptional elongation and antisense transcription (8, 10–12). In contrast to gene body methylation, DNA methylation in fungi such as Neurospora has been viewed as a genome defense mechanism because it silences transposable elements and repetitive DNA sequences that have been mutated by repeat-induced point mutation (7, 9). Compared with the levels of meC reported in other fungi (Neurospora, 1.5% meC; Agaricus bisporus ∼4% meC; Ustilago maydis, 2.3% meC; and Armillaria bulbosa, 4.3% meC), only 0.5% of cytosines are expected to be methylated in the yeast form of C. albicans (13–15). However, the distribution and biological significance of DNA methylation in this organism's genome are unknown.
Here we show that DNA methylation in C. albicans occurs largely within structural genes and regulates transcription dependent on reversible phenotypic pathways and iron metabolism. Our results further reveal that repeated DNA sequences in C. albicans are mostly devoid of DNA methylation in contrast to other eukaryotes where methylation occurs primarily in transposable elements and DNA repeats.
Results
Identification of Methylated Genomic Regions.
A His-tagged methyl-CpG binding domain (MBD) of the mouse MeCP2 protein (16) was prepared by expression in Escherichia coli and then coupled to Ni-nitrilotriacetic acid (NTA) agarose for purification of methylated DNA from bulk genomic DNA by affinity chromatography. We first validated the method using unique control DNAs that were methylated in vitro and mixed with unique unmethylated DNAs. Methylated DNAs eluted at high salt concentrations (0.6–1 M NaCl), whereas unmethylated DNAs eluted at low salt concentrations (Fig. S1A). We performed a screen for methylated DNAs from C. albicans strain SC5314 with Sau3AI-digested total genomic DNA. Putative methylated DNA fragments that eluted with high salt (≥0.6 M NaCl) were inserted into BamHI-digested pBluescript and propagated in E. coli. Plasmid DNA from each clone was used in slot blot analyses to identify clones enriched by the MBD column in comparison with unfractionated total genomic DNA (Fig. S1B). Enrichment was normalized to a known unmethylated DNA from the centromere of chromosome 1 (CEN1; ref. 17). Enrichment values twofold higher than CEN1 were considered significant. Slot blot analysis revealed 203 clones significantly enriched in the high salt eluted fractions.
The enriched candidate methylated clones were sequenced and then identified by using BLAST analysis against the C. albicans genome assembly 21 (www.candidagenome.org). The enriched sequences represented ∼0.2% of the C. albicans genome. Of 203 enriched clones, we chose 55 candidate methylated regions representing a range of low (2-fold) to high (46-fold) enrichment values (Dataset S1) for the validation of DNA methylation in the C. albicans genome (7.0-fold average and 4.7-fold median enrichments for the candidate group compared with 7.2- and 4.4-fold, respectively, for the total group). All 55 regions showed evidence of meC by bisulfite sequencing (18) and/or by methylation-specific restriction analysis using methylation-sensitive (Sau3AI) and methylation-insensitive (DpnII) restriction enzyme isoschizomers (Fig. 1 and Figs. S2 and S3) with genomic DNA that was not affinity-purified. These results confirmed that the DNA fragments enriched by MBD column chromatography correspond to methylated DNA regions in C. albicans. Sequence analysis revealed that the majority of C. albicans methylated DNAs identified in this study are categorized as genes (82%; n = 165 clones representing 150 coding regions) and nonrepetitive intergenic regions (13%; n = 27 clones), whereas only 5% of the methylated clones (n = 11 clones representing 8 repeated regions) are categorized as repetitive sequences (Dataset S1). DNA methylation in C. albicans occurs in both symmetric (meCpG) and asymmetric (meCpN) sequence contexts. There was a slight sequence preference for methylation in that two-thirds occurred in meCpA (43.3%) and meCpT (24.6%) dinucleotides (Fig. S4A) among 40 regions examined by bisulfite sequencing (34 coding, 1 intergenic, and 5 repeated regions; Dataset S1). In addition, the sequence analysis revealed a preference for a thymine upstream of meC in all sequence contexts (Fig. S4B).
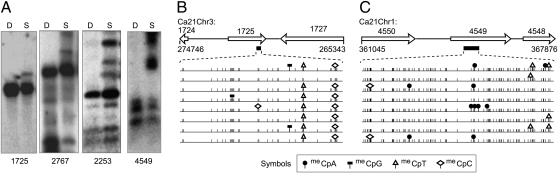
Fig. 1.
DNA methylation occurs in protein coding gene regions. (A) Genomic DNA from C. albicans strain SC5314 (yeast morphology) was digested with isoschizomers DpnII (D) and Sau3AI (S), fractionated on agarose gels, blotted to a nylon membrane, and hybridized with radiolabeled DNAs (probe lengths are specified parenthetically) derived from the coding regions indicated: orf19.1725 (2.0 kb), a predicted gene; orf19.2767 (1.9 kb), PGA59 gene, regulated according to cell morphology and encoding a cell wall GPI-anchored protein; orf19.2253 (1.5 kb), regulated by white–opaque switching; and orf19.4549 (0.9 kb), FGR38 gene, encoding a regulator of filamentous growth. (B and C) Bisulfite-converted genomic DNA corresponding to the MBD-affinity enriched DNA fragment, depicted as a filled black bar below the indicated gene, was analyzed by DNA sequencing. Sequences from independent clones are aligned. Cytosine, short vertical lines; meCpN, symbols as indicated. Ca21Chr1 and Ca21Chr3 represent C. albicans genome sequence assembly 21 chromosomes 1 and 3, respectively. All schematics are drawn to scale and are derived from C. albicans genome sequence assembly 21 (www.candidagenome.org).
Methylated Repeated DNAs.
Repeated regions found to be methylated include rRNA genes (nuclear and mitochondrial), subtelomeric DNA, and a few long terminal repeat (LTR) sequences. The methylation status of these DNA elements was independently verified by methylation-specific restriction analysis or bisulfite sequencing. Southern hybridizations of Sau3AI- and DpnII-digested genomic DNA with radiolabeled 25S ribosomal DNA (rDNA), CaRE1, or LTR theta showed variation in restriction patterns between isoschizomers that is consistent with the presence of meC in these DNA regions (Fig. S3A). Based on Southern hybridization, the occurrence of meC in 25S rDNA is conserved among three strains of C. albicans (Fig. S3B) that are separated by a divergence time of 1–3 million years (19). Bisulfite sequencing confirmed the presence of meC in 25S nuclear rDNA and mitochondrial rDNA of the large ribosomal subunit (Fig. S3 C and D), an observation consistent with the occurrence of methylation in these types of repeat sequences in other eukaryotes (7, 9).
C. albicans also harbors large blocks of repeated DNA known as major repeat sequences (MRS), which occur on seven of the eight chromosomes (20), as well as transposon derived sequences (21) and multigene families such as telomere length organization (TLO), secreted aspartyl proteinase (SAP), and agglutinin-like sequences (ALS) (22). None of these repeat families were enriched in high salt fractions from the MBD column, suggesting that these regions in C. albicans are not methylated. Nonetheless, we examined the status of DNA methylation in these repeated sequences. We were unable to detect DNA methylation using methylation-specific restriction analysis of Sau3AI-, DpnII-, HpaII-, and MspI-digested genomic DNA hybridized with a radiolabeled RPS1 region of MRS (Fig. S5A). Moreover, Southern hybridization of DNA fractions eluted from the MBD column with radiolabeled RPS1 revealed that MRS DNA eluted in 0.5 M salt, below the cutoff value for our collection of methylated sequences (Fig. S5B). This result suggests that an extremely low-level DNA methylation in this DNA region is possible, but it is below the detection limit of our assays. Although a single symmetrical meCpG pair is bound by MBD-affinity chromatography, low affinity interactions with non-CpG methylated residues are possible (23). Likewise, ALS3 and ALS4, encoding cell-surface glycoproteins required for adhesion of the organism to host surfaces, showed no evidence of DNA methylation as judged by methylation-specific restriction analysis (Fig. S5 C and D). Together these results indicate that the MRS and other repeated DNA sequences we examined in C. albicans are largely free of DNA methylation, unlike other fungi where DNA repeats and transposon-derived sequences are heavily methylated (7, 9, 12).
Methylated Gene Regions.
Classification according to predicted protein function revealed that many of the 150 methylated genes identified by our screen encode proteins that are involved in environmentally cued pathways, such as morphogenesis and hyphal growth (25 of 150 genes, or 16.7%), phenotypic switching (3.3%), iron utilization (6.7%), drug resistance and signaling (12%), and stress response (7.3%). Other methylated genes have predicted involvement in chromatin organization (3.3%), cell cycle or division (7.3%), protein biogenesis or transport (12.7%), DNA/RNA processing (5.3%), pathogenesis or virulence (2%), carbohydrate metabolism (1.3%), or unknown (22%) functions (Dataset S1). Southern analysis using methylation-sensitive and -insensitive restriction enzyme isoschizomers revealed the presence of methylation in all 13 genes examined (Fig. 1A and Dataset S1). Bisulfite sequencing was performed on 34 randomly selected methylated genes, and data were deposited with GenBank. Only data from representative genes (those shown in bold in Dataset S1) will be presented here and in Figs. S2–S7. Independent clones obtained from a population of cells growing in the yeast cell form displayed heterogeneous distribution of meC (Fig. 1 B and C and Fig. S2). Because we typically recovered small methylated Sau3AI fragments (<500 bp) from our screen, we randomly selected several methylated genes (orf19.5749, orf19.2593, and SEC20) and performed bisulfite sequencing of the entire gene including 100 bp upstream and downstream of the coding region. We found methylation only within the body of genes, whereas no meC was detected in the flanking region of any of the genes sequenced (Fig. S2C and SI Appendix). Only 6 (orf19.2639.1, orf19.4575, orf19.5788, orf19.6187, orf19.6286.2, and orf19.6718) of the 150 genes identified here contain introns (24), suggesting that meC does not mark exon–intron boundaries or regulate alternative splicing in C. albicans. Because DNA methylation is largely found within coding regions, it suggests that the major function of DNA methylation in C. albicans could be gene regulation.
Methylation Represses Transcription Dependent upon Reversible Phenotypic Pathways.
The prevalence of gene methylation in C. albicans suggests that DNA methylation may play an important regulatory role. We therefore examined the relationship between methylation and transcription among selected methylated genes in pathways for morphological transition between unicellular yeast and multicellular hyphal forms, or phenotypic switching between white and opaque cell forms. We examined the distribution of DNA methylation by bisulfite sequencing of a number of genes involved in morphogenesis and cell proliferation (orf19.2769/PBI2) and phenotypic switching (orf19.2834/RPD3, orf19.1288/FOX2, and orf19.2253) in C. albicans strain SC5314 and white–opaque switching strain WO1 (4), respectively. We found that methylation of these gene sets varied between yeast and hyphal cells and between white and opaque cells, respectively. Our results show a direct relationship between presence of methylation and transcriptional inhibition, as judged by Northern blots, correlated with phenotype or cell form (Fig. 2 and Fig. S6). We conducted a detailed analysis of meC and its impact on transcription of the PBI2 gene (orf19.2769), which is required for proteolysis during sporulation (25). Within a population of yeast-form cells, meC was found with variable positioning in the body of PBI2, whereas hyphal cells were largely free of methylation (Fig. 2A). Northern blot analysis revealed transcriptional repression of PBI2 in yeast cells; conversely, PBI2 was transcribed in hyphal cells (Fig. 2B). We used DNA methylation inhibitor 5-azacytidine (AzaC) to examine whether cytosine methylation is the cause of transcriptional repression of PBI2 in yeast-form cells. As observed in other organisms (10), AzaC treatment resulted in the loss of cytosine methylation in the methylated DNA regions examined (SI Appendix; orf19.7270 and orf19.5526). Growth of strain SC5314 in low-nitrogen medium containing AzaC efficiently prevented DNA methylation at the PBI2 locus and concomitantly activated transcription of this gene (Fig. 2 A and C), suggesting that DNA methylation directs repression of PBI2 in yeast-form cells.
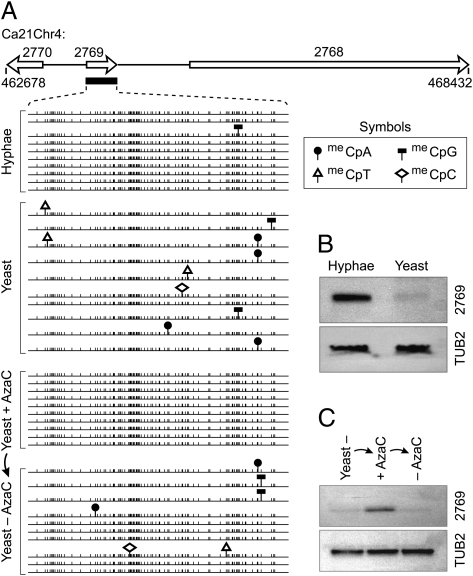
Fig. 2.
Transcriptional repression is correlated with DNA methylation and morphological phenotype. (A) DNA methylation in orf19.2769 (PBI2) determined by bisulfite DNA sequencing from hyphal or yeast-form cells grown with or without AzaC treatment. Schematic is drawn to scale and is derived from C. albicans genome sequence assembly 21. The DNA fragment corresponding to the filled black bar below PBI2 was analyzed by bisulfite sequencing. Sequences from independent clones are aligned. Cytosine, vertical lines; meCpN, symbols indicated. (B) Northern blot of total RNAs from cells with yeast or hyphal morphology were hybridized with radiolabeled PBI2 or TUB2 (loading control). (C) Northern blot of total RNA from cells with yeast morphology, yeast-form cells passaged in medium with AzaC, and yeast-form cells passaged from AzaC back to medium without AzaC. The Northern blot was probed with PBI2 or TUB2 (loading control). TUB2 was used to show that AzaC treatment does not affect gene expression.
In many organisms, if erased or lost, DNA methylation can be reestablished by de novo methylation (6, 7, 26). Thus, we next examined whether DNA methylation can reestablish in C. albicans once it is erased by AzaC treatment. We transferred cells to low-nitrogen medium after passage through AzaC medium, and cultures were allowed to grow for ∼20 generations. Our results show reestablishment of DNA methylation that is paralleled by repression of gene expression at the PBI2 locus (Fig. 2 A and C). Together, these results suggest that DNA methylation detected in C. albicans is associated with transcriptional repression of the linked gene.
DNA Methylation Is Associated with Iron-Regulated Gene Repression but Is Not Required for the Repression.
Iron is an essential component of the C. albicans pathogenesis pathway and plays an important role in signaling and progression of systemic infection (2, 27). Our screen uncovered methylated genes previously shown to exhibit iron dosage-dependent gene expression (2). We examined the distribution of meC and its effect on transcription of RBT5 and GEA2 in strain SC5314 grown in medium containing a low or high concentration of iron. RBT5, which encodes a glycosylphosphatidylinositol (GPI)-anchored protein required for the maintenance of C. albicans cell wall integrity, host adhesion, and hemoglobin utilization (27), was heterogeneously but extensively methylated in high iron conditions, but not in low iron conditions. Northern blotting showed that transcriptional repression was correlated with the presence of meC (Fig. 3 A and B). Similar results were observed for GEA2, which encodes an iron-regulated guanine nucleotide exchange factor involved in vesicular transport and cytoskeleton organization, although repression of GEA2 was not absolute under high iron conditions (Fig. S6D).
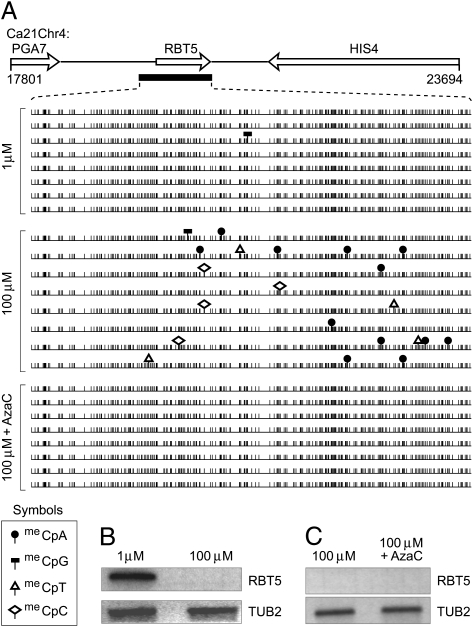
Fig. 3.
Iron-mediated gene silencing is independent of DNA methylation. (A) DNA methylation in the RBT5 gene was determined by bisulfite DNA sequencing in strain SC5314 grown in low (1 μM) and high (100 μM) iron conditions. Schematic is drawn to scale and is derived from C. albicans genome sequence assembly 21. The DNA fragment corresponding to the filled black bar below RBT5 was analyzed by bisulfite sequencing. Sequences from independent clones are aligned. Cytosine, vertical lines; meCpN, symbols as indicated. (B) Northern blot of total RNA from low- and high-iron cultures was probed with radiolabeled RBT5 or TUB2 (loading control). (C) Northern blot of total RNA isolated from cultures grown with or without AzaC treatment was probed with radiolabeled RBT5 or TUB2. TUB2 was used to show that AzaC treatment does not affect gene expression.
In the presence of high iron concentrations, RBT5 is regulated by an iron responsive GATA-like factor, Sfu1p, which has been proposed to act by blocking transcription initiation at genes it represses (2). Although deletion of the SFU1 gene would be expected to relieve RBT5 repression under high iron conditions, it has been previously shown that it is not significantly relieved (2). Our results reveal that under these conditions RBT5 would still be methylated. Conversely, we examined RBT5 in a wild-type SFU1 strain (SC5314) grown in high-iron medium containing AzaC. No DNA methylation was detected in RBT5 after AzaC treatment; however, RBT5 transcripts were not evident on a Northern blot (Fig. 3 A and C). This result suggests that Sfu1p directed silencing of RBT5 is independent of DNA methylation and that removal of DNA methylation is not sufficient to relieve iron-regulated gene repression.
Transcriptionally Repressed Methylated Gene Regions Show Increased Frequency of C-to-T Transitions.
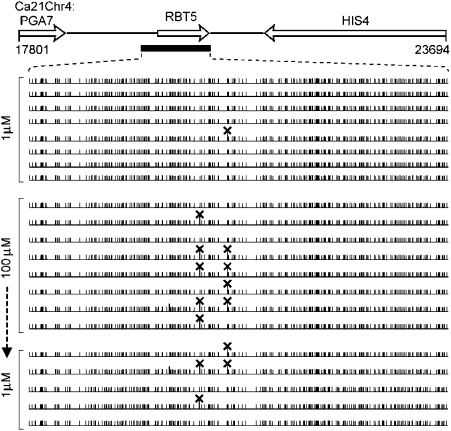
Genome sequencing of C. albicans revealed an unusually high overall AT content of ∼66.5% (15). Mechanisms that lead to these skewed genomic nucleotide ratios are largely unknown. In this study, we observed an increased rate of C-to-T mutations in methylated gene regions of C. albicans. Strain SC5314 was grown for ∼50 generations under conditions that result in the presence or near absence of methylation in iron-regulated RBT5 and GEA2 genes. These regions were PCR amplified, cloned into E. coli, and sequenced. DNA sequences of RBT5 and GEA2 gene clones from cells that had been transcriptionally repressed by growth under high exogenous iron conditions exhibited a higher frequency of C-to-T transitions compared with gene clones from cells grown for the same number of generations under low exogenous iron conditions (Fig. 4 and Fig. S7B, Rep-I). We conducted additional experiments with duplicate cultures to examine the reproducibility of the observed mutational patterns. We consistently observed a higher frequency of C-to-T transitions when genes were methylated and repressed (Fig. S7 A and B, Rep-II). In addition, sequence analysis of independent clones of RBT5 revealed that the mutation pattern established after growth in high-iron medium remains stably fixed in the C. albicans population reexposed to low iron conditions (Fig. 4 Lower).
Fig. 4.
Transcriptionally repressed methylated genes show an increased frequency of cytosine-to-thymine transitions. Schematic is drawn to scale and is derived from C. albicans genome sequence assembly 21. The RBT5 gene was PCR-amplified with proofreading Taq DNA polymerase using template DNA derived from strain SC5314 grown for ∼50 generations in low (1 μM) iron, high (100 μM) iron, or high iron followed by ∼50 generations in low iron conditions. Independent PCR products were cloned into E. coli and sequenced. Sequences from independent clones are aligned. Cytosine, vertical lines; C-to-T mutation, X.
To determine whether the elevated frequency of C-to-T transitions observed in genes under repression is influenced by the genetic background of strain SC5314 or the high-iron dosage used in the growth medium, we selected white–opaque switching strain WO1 to further examine the mutational spectrum of genes under repressive conditions. Notably, strains SC5314 and WO1 are estimated to be separated from each other by a divergence time of 1 million years (19). Strain WO1 was grown for ∼50 generations under conditions that result in the presence or near absence of methylation in the white–opaque regulated FOX2 gene (Fig. S6C). FOX2 genes cloned from white cells, in which the gene is transcriptionally repressed by DNA methylation, also showed a higher number of C-to-T transitions than did FOX2 genes cloned from opaque cells grown for a similar number of generations (Fig. S7C). The higher rate of mutation in FOX2 under repression was observed among independently grown cultures (Fig. S7C). We also examined orf19.2253, a white–opaque regulated gene, which exhibited C-to-T transitions under conditions when the gene was repressed and methylated (Fig. S7D). Sequence analysis revealed that all C-to-T transitions occurred at third-codon positions and were selectively neutral (synonymous mutations). Thus, C. albicans gene regions undergoing cytosine methylation appear to accumulate C-to-T transitions. This characteristic likely is one factor contributing to the evolution of an AT-rich genome in this organism.
We estimated the frequency of the C-to-T mutations by calculating the base substitutions per nucleotide per generation (28). Previous reports on C. albicans using reporter constructs estimated an average mutation frequency of 10−6 per cell per generation based on the formation of l-sorbose-utilizing mutants (29). We found that the frequency of mutations is about fivefold to ninefold higher and significantly different (Fisher's exact test, P = 0.0012) when target genes were methylated and transcriptionally silenced compared with the same genes transcribed in the absence of methylation (Fig. S7E). The mutation frequency for RBT5 in cells grown under high exogenous iron conditions was 2.88 (±0.94) × 10−5 base substitutions per nucleotide per generation, whereas it was 3.3 (±0.77) × 10−6 in cells grown in low-iron medium when RBT5 was transcribed (Fig. S7E). Similar mutational frequencies were also observed for other genes regulated by iron or white–opaque phenotypes (Fig. S7E). Because the frequency of mutation was significantly higher when the genes were under repression, we denote this process as repression associated mutation (RAM). As RAM is occurring on such a short timescale (∼50 generations) in coding regions that alternate between methylated and unmethylated states, it has the potential to evolve genes rapidly under conditions of fluctuating environmental changes or stress.
Discussion
DNA methylation in fungi is mainly regarded as a genome defense mechanism, where it is thought to play a role in preventing the duplication of transposable sequences and silencing of DNA repeats (7, 9, 12). Unlike other fungi examined, DNA methylation in C. albicans occurs mainly in gene bodies. The repeated DNA sequences, such as MRS and multigene families (for example, ALS genes) are largely free of DNA methylation. In higher eukaryotes, removal of DNA methylation from repeated DNA and transposable sequences results in the activation of these DNA elements leading to genomic instability (30). Apparently, C. albicans exhibits extensive chromosomal instability mediated by translocation between MRS regions of different chromosomes (20). Perhaps the absence of DNA methylation in MRS may contribute to instability in this repeated DNA locus. Overall, these observations indicate that the role of DNA methylation in repression of DNA repeats and transposon sequences has been lost or reduced in C. albicans.
DNA methylation in C. albicans is predominantly localized within intragenic regions and is propagated by de novo mechanisms. In other organisms, such as Neurospora, induction and propagation of nonsymmetric DNA methylation can also be achieved by maintenance activities (31); whether such mechanisms also exist in C. albicans is an open question. In Neurospora, symmetric and asymmetric methylation are propagated by a single DNA methyltransferase, dim-2 (32). In Arabidopsis, non-CpG methylation is propagated through RNA-directed DNA methylation and DRM2, a de novo DNA methyltransferase, whereas meCpNpG and meCpG are maintained by CMT3 and MET1, respectively (6). However, the enzyme and mechanism responsible for preferential methylation of CpA and CpT sequences in C. albicans has not been determined. We observed a direct link between the loss of DNA methylation and activation of gene transcription. Recent studies in plants, animals, and insects have uncovered gene body DNA methylation, resulting in transcription regulation (down or up depending on the organism examined), alternative splicing, chromatin alteration, and inhibition of transcription elongation (8, 10–12, 33). Our results in C. albicans further establish that a role for intragenic methylation in modulation of gene transcription is evolutionarily conserved.
This study has revealed that genes, whose expression is modulated in concert with complex transitions in cell morphology or maintenance of iron homeostasis in response to adverse growth conditions, exhibit heterogeneous methylation that is correlated with transcriptional repression. Notably, neurobiological studies have identified gene repression due to intragenic methylation that appears to be correlated with stressful early life experiences (34, 35). In these studies, heterogeneous and relatively low levels of methylation were detected for the brain-derived neurotrophic factor and glucocorticoid receptor genes within tissue-specific populations of mammalian brain cells (34, 35). Low-level DNA methylation is also sufficient to modulate genetic switches in bacteria (36). Together, these results suggest that only a few methylated residues are required and/or sufficient to establish a repressed chromatin state for the examined genes.
DNA methylation in C. albicans is associated with transcriptional repression of genes linked with dimorphic transition or phenotypic switching. Removal of methylation, however, is not sufficient for activation of gene pathways that are controlled by redundant negative regulation such as the iron-regulated gene RBT5. The latter result is consistent with observations from mammalian studies, where DNA methylation has a secondary role in transcriptional repression, such as mammalian X-chromosome inactivation (37). The mechanism by which the methylated state is reversed is not known. A demethylase activity has been postulated, but an enzyme that can accomplish this result in vitro has not been identified (38). It is significant that even among the small population of sequences analyzed for some genes in this study, a few unmethylated copies were present among the predominantly methylated population. This result could imply that the methylase is rate limiting, resulting in incomplete methylation, or that transition from the transcription OFF to the ON state proceeds by passive dilution of the methylated copies during DNA replication. Alternatively, DNA demethylation may actively occur by a multistep pathway using a meC DNA glycosylase and base excision repair as invoked for plants or a meC deaminase with subsequent removal of the T/G mismatch by thymine glycosylase as modeled for mammals (6). Unless deamination is tightly regulated, the latter mechanism has the potential to introduce transition mutations.
Methylated gene regions of C. albicans showed increased frequency of C-to-T mutations (RAM). RAM events occur during asexual growth and were elevated by a repressed transcriptional state of the genomic loci. Apparently, RAM is capable of introducing genetic diversity that would be exposed when previously repressed genes are induced upon changing growth conditions and may affect the rate of evolution in C. albicans, which lacks a complete meiotic cycle (22). Although the intrinsic rate of meC deamination is only 2- to 4-fold higher than C, in mammals the observed rate of mutation at meCpG is ∼10- to 50-fold higher than at CpG (39). A high density of single-nucleotide polymorphisms in regions with repressed chromatin has been observed in the human genome (40). Comparative analysis of the mutational patterns among organisms with divergent genomic complexity will be useful to determine whether the observed patterns of RAMs hold across the evolutionary scale and may provide insights into mechanisms that lead to increased frequency of mutation under repression.
The present study reports that DNA methylation in C. albicans occurs in structural genes and is involved in gene regulation, a previously undescribed finding. DNA methylation has an important role in maintaining and establishing transcriptional repression dependent upon the phenotype or metabolic state of the cell. These findings have broad implications because DNA methylation in C. albicans can be triggered by environmental stressors. Thus, this system offers an excellent model to dissect the mechanistic role of DNA methylation on gene regulation, genomic instability, and chromatin modification, as well as the reversal of this process. It would also be of general interest to use this fungus to address the unexplored evolutionary aspects associated with DNA methylation and genome evolution.
Methods
C. albicans strains SC5314, WO1, 1006, and NUM114 were used (19). Methylated DNA was isolated from strain SC5314 grown in Lee's medium at 37 °C containing either low or high nitrogen concentrations favoring yeast or hyphal growth, respectively (5). Phenotypic switching strain WO1 was used to examine the DNA methylation in white and opaque cells by using medium and growth conditions as described (4). Strain SC5314 was grown in a defined medium containing either low (1 μM) or high (100 μM) ferrous ammonium sulfate as described (2) to study the effect of iron dosage on DNA methylation and transcription. To examine the effect of loss of methylation, liquid medium were supplemented with 100 μM AzaC (Sigma). Total DNA and RNA were prepared from fungal tissues by using DNeasy and RNeasy mini kits, respectively (Qiagen).
A methyl DNA binding (MBD) column was prepared essentially as described (16) with some modifications. Sau3AI-digested C. albicans genomic DNA was fractionated by using the MBD column as outlined (Fig. S1B). The detailed procedure for MBD column preparation and use are described in SI Methods. Bisulfite sequencing (18) and Southern hybridization using methylation-sensitive and -insensitive restriction enzymes was used to determine the methylation status of C. albicans genomic regions. To examine the efficiency of bisulfite conversion, each bisulfite reaction included unique unmethylated pBluescript KS(−) vector DNA. Sequencing of bisulfite-treated vector DNA showed that bisulfite conversions were efficient and converted all cytosine residues to uracil (SI Appendix). Northern hybridization was used to measure gene expression following the procedure described in SI Methods. PCR primers used in this study are listed in SI Appendix.
Supplementary Material
Acknowledgments
We thank P. T. Magee and B. B. Magee for strains; A. Bird and S. Cross for plasmids; S. Poole for help with sequence analysis; and D. J. McLaren for artwork. This work was supported by National Institutes of Health (National Cancer Institute) Research Grant CA-11034.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. ET671383–ET671921, HQ690089, HR235325–HR235465, and JJ725187–JJ725299).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1109631108/-/DCSupplemental.
References
- 1.Lenz P, et al. Prevalence, associations, and trends of biliary-tract candidiasis: A prospective observational study. Gastrointest Endosc. 2009;70:480–487. doi: 10.1016/j.gie.2009.01.038. [DOI] [PubMed] [Google Scholar]
- 2.Lan CY, et al. Regulatory networks affected by iron availability in Candida albicans. Mol Microbiol. 2004;53:1451–1469. doi: 10.1111/j.1365-2958.2004.04214.x. [DOI] [PubMed] [Google Scholar]
- 3.Biswas S, Van Dijck P, Datta A. Environmental sensing and signal transduction pathways regulating morphopathogenic determinants of Candida albicans. Microbiol Mol Biol Rev. 2007;71:348–376. doi: 10.1128/MMBR.00009-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zordan RE, Galgoczy DJ, Johnson AD. Epigenetic properties of white-opaque switching in Candida albicans are based on a self-sustaining transcriptional feedback loop. Proc Natl Acad Sci USA. 2006;103:12807–12812. doi: 10.1073/pnas.0605138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Russell PJ, Welsch JA, Rachlin EM, McCloskey JA. Different levels of DNA methylation in yeast and mycelial forms of Candida albicans. J Bacteriol. 1987;169:4393–4395. doi: 10.1128/jb.169.9.4393-4395.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martienssen RA, Colot V. DNA methylation and epigenetic inheritance in plants and filamentous fungi. Science. 2001;293:1070–1074. doi: 10.1126/science.293.5532.1070. [DOI] [PubMed] [Google Scholar]
- 8.Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat Genet. 2007;39:61–69. doi: 10.1038/ng1929. [DOI] [PubMed] [Google Scholar]
- 9.Selker EU, et al. The methylated component of the Neurospora crassa genome. Nature. 2003;422:893–897. doi: 10.1038/nature01564. [DOI] [PubMed] [Google Scholar]
- 10.Rountree MR, Selker EU. DNA methylation inhibits elongation but not initiation of transcription in Neurospora crassa. Genes Dev. 1997;11:2383–2395. doi: 10.1101/gad.11.18.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maunakea AK, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466:253–257. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zemach A, McDaniel IE, Silva P, Zilberman D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science. 2010;328:916–919. doi: 10.1126/science.1186366. [DOI] [PubMed] [Google Scholar]
- 13.Foss HM, Roberts CJ, Claeys KM, Selker EU. Abnormal chromosome behavior in Neurospora mutants defective in DNA methylation. Science. 1993;262:1737–1741. doi: 10.1126/science.7505062. [DOI] [PubMed] [Google Scholar]
- 14.Storck R, Nobles MK, Alexopou CJ. The nucleotide composition of deoxyribonucleic acid of some species of hymenochaetaceae and polyporaceae. Mycologia. 1971;63:38–49. [Google Scholar]
- 15.Butler G, et al. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature. 2009;459:657–662. doi: 10.1038/nature08064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cross SH, Charlton JA, Nan X, Bird AP. Purification of CpG islands using a methylated DNA binding column. Nat Genet. 1994;6:236–244. doi: 10.1038/ng0394-236. [DOI] [PubMed] [Google Scholar]
- 17.Baum M, Sanyal K, Mishra PK, Thaler N, Carbon J. Formation of functional centromeric chromatin is specified epigenetically in Candida albicans. Proc Natl Acad Sci USA. 2006;103:14877–14882. doi: 10.1073/pnas.0606958103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clark SJ, Harrison J, Paul CL, Frommer M. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 1994;22:2990–2997. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mishra PK, Baum M, Carbon J. Centromere size and position in Candida albicans are evolutionarily conserved independent of DNA sequence heterogeneity. Mol Genet Genomics. 2007;278:455–465. doi: 10.1007/s00438-007-0263-8. [DOI] [PubMed] [Google Scholar]
- 20.Chibana H, et al. Diversity of tandemly repetitive sequences due to short periodic repetitions in the chromosomes of Candida albicans. J Bacteriol. 1994;176:3851–3858. doi: 10.1128/jb.176.13.3851-3858.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goodwin TJ, Poulter RT. Multiple LTR-retrotransposon families in the asexual yeast Candida albicans. Genome Res. 2000;10:174–191. doi: 10.1101/gr.10.2.174. [DOI] [PubMed] [Google Scholar]
- 22.van het Hoog M, et al. Assembly of the Candida albicans genome into sixteen supercontigs aligned on the eight chromosomes. Genome Biol. 2007;8:R52. doi: 10.1186/gb-2007-8-4-r52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meehan RR, Lewis JD, Bird AP. Characterization of MeCP2, a vertebrate DNA binding protein with affinity for methylated DNA. Nucleic Acids Res. 1992;20:5085–5092. doi: 10.1093/nar/20.19.5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mitrovich QM, Tuch BB, De La Vega FM, Guthrie C, Johnson AD. Evolution of yeast noncoding RNAs reveals an alternative mechanism for widespread intron loss. Science. 2010;330:838–841. doi: 10.1126/science.1194554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harcus D, Nantel A, Marcil A, Rigby T, Whiteway M. Transcription profiling of cyclic AMP signaling in Candida albicans. Mol Biol Cell. 2004;15:4490–4499. doi: 10.1091/mbc.E04-02-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lewis ZA, et al. Relics of repeat-induced point mutation direct heterochromatin formation in Neurospora crassa. Genome Res. 2009;19:427–437. doi: 10.1101/gr.086231.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weissman Z, Shemer R, Conibear E, Kornitzer D. An endocytic mechanism for haemoglobin-iron acquisition in Candida albicans. Mol Microbiol. 2008;69:201–217. doi: 10.1111/j.1365-2958.2008.06277.x. [DOI] [PubMed] [Google Scholar]
- 28.Lynch M, Crease TJ. The analysis of population survey data on DNA sequence variation. Mol Biol Evol. 1990;7:377–394. doi: 10.1093/oxfordjournals.molbev.a040607. [DOI] [PubMed] [Google Scholar]
- 29.Janbon G, Sherman F, Rustchenko E. Appearance and properties of L-sorbose-utilizing mutants of Candida albicans obtained on a selective plate. Genetics. 1999;153:653–664. doi: 10.1093/genetics/153.2.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bourc'his D, Bestor TH. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431:96–99. doi: 10.1038/nature02886. [DOI] [PubMed] [Google Scholar]
- 31.Selker EU, et al. Induction and maintenance of nonsymmetrical DNA methylation in Neurospora. Proc Natl Acad Sci USA. 2002;99(Suppl 4):16485–16490. doi: 10.1073/pnas.182427299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kouzminova E, Selker EU. dim-2 encodes a DNA methyltransferase responsible for all known cytosine methylation in Neurospora. EMBO J. 2001;20:4309–4323. doi: 10.1093/emboj/20.15.4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lyko F, et al. The honey bee epigenomes: Differential methylation of brain DNA in queens and workers. PLoS Biol. 2010;8:e1000506. doi: 10.1371/journal.pbio.1000506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roth TL, Lubin FD, Funk AJ, Sweatt JD. Lasting epigenetic influence of early-life adversity on the BDNF gene. Biol Psychiatry. 2009;65:760–769. doi: 10.1016/j.biopsych.2008.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roth TL, Sweatt JD. Epigenetic marking of the BDNF gene by early-life adverse experiences. Horm Behav. 2011;59:315–320. doi: 10.1016/j.yhbeh.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Low DA, Weyand NJ, Mahan MJ. Roles of DNA adenine methylation in regulating bacterial gene expression and virulence. Infect Immun. 2001;69:7197–7204. doi: 10.1128/IAI.69.12.7197-7204.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sado T, Okano M, Li E, Sasaki H. De novo DNA methylation is dispensable for the initiation and propagation of X chromosome inactivation. Development. 2004;131:975–982. doi: 10.1242/dev.00995. [DOI] [PubMed] [Google Scholar]
- 38.Buchen L. Neuroscience: In their nurture. Nature. 2010;467:146–148. doi: 10.1038/467146a. [DOI] [PubMed] [Google Scholar]
- 39.Pfeifer GP. Mutagenesis at methylated CpG sequences. Curr Top Microbiol Immunol. 2006;301:259–281. doi: 10.1007/3-540-31390-7_10. [DOI] [PubMed] [Google Scholar]
- 40.Prendergast JG, et al. Chromatin structure and evolution in the human genome. BMC Evol Biol. 2007;7:72. doi: 10.1186/1471-2148-7-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.