Abstract
Recent evidence indicates that phosphatidylinositol 3-kinase (PI3K) is a central regulator of mitosis, apoptosis and oncogenesis. Nevertheless, the mechanisms by which PI3K regulates proliferation are not well characterized. Mitogens stimulate entry into the cell cycle by inducing the expression of immediate early genes (IEGs) that in turn trigger the expression of G1 cyclins. Here we describe a novel PI3K- regulated transcriptional cascade that is critical for mitogen regulation of the IEG, c-fos. We show that PI3K activates gene expression by transactivating SRF-dependent transcription independently of the previously described Rho and ETS TCF pathways. PI3K-stimulated cell cycle progression requires transactivation of SRF and expression of dominant- negative PI3K blocks mitogen-stimulated cell cycle progression. Furthermore, dominant-interfering SRF mutants attenuate mitogen-stimulated cell cycle progression, but are without effect on MEK-stimulated cell cycle entry. Moreover, expression of constitutively active SRF is sufficient for cell cycle entry. Thus, we delineate a novel SRF-dependent mitogenic cascade that is critical for PI3K- and growth factor-mediated cell cycle progression.
Keywords: cell cycle/PI3-kinase/serum response element/transcription
Introduction
Mitogens and growth factors activate phosphatidylinositol 3-kinase (PI3K) and promote the phosphorylation of phosphoinositides at the D3 position. These phospholipids play a critical role in the regulation of mitogenesis, apoptosis and cytoskeletal rearrangement (for a review see Rameh and Cantley, 1999). Type 1 PI3K is comprised of regulatory (p85) and catalytic (p110) subunits and was identified by virtue of its role in Src-mediated transformation (Sugimoto et al., 1984). Expression of constitutively active PI3K can trigger DNA synthesis (Frevert and Kahn, 1997; Klippel et al., 1998) and PI3K activity is required for mitosis in some cells (Jhun et al., 1994; Roche et al., 1994). Mice deficient for p110α catalytic subunits (Bi et al., 1999) have a proliferative defect that results in embryonic lethality. The identification of oncogenic forms of p110 and p85 PI3K further illustrates the critical role of PI3K in proliferation (Chang et al., 1997; Jimenez et al., 1998).
The identification of the tumor suppressor, PTEN, as a D3 phosphoinositide phosphatase underscores the critical role of PI3K in cell growth and oncogenesis (Myers et al., 1998). A significant fraction of human cancers contain deletions or mutations of PTEN (for review see Cantley and Neel, 1999). However, the mechanisms by which PTEN regulates cell cycle progression are not clear and the targets of PI3K signaling during mitosis remain obscure.
The ability of mitogens to trigger cell cycle progression depends on the rapid transcription of immediate early genes (IEGs). In particular, c-fos and related transcription factors play a critical role in mitogenesis (Riabowol et al., 1988; Kovary and Bravo, 1991) by inducing the expression of genes necessary for the activation of G1 cyclins (Brown et al., 1998). For example, cyclin D1 mRNA is increased by overexpression of c-fos (Miao and Curran, 1994) and mitosis and mitogen-stimulated cyclin D1 transcription are impaired in cells deficient for c-fos and FosB (Brown et al., 1998). Interestingly, mitogen-stimulated c-fos transcription requires PI3K activity (Yamauchi et al., 1993; Jhun et al., 1994) and activated PI3K stimulates c-fos transcription (Wang et al., 1998).
The cis-acting serum response element (SRE) is crucial for mitogen-induced c-fos transcription and is important for the mitogen-stimulated transcription of many IEGs (Treisman, 1995). The MADS box-containing protein, serum response factor (SRF), binds to the SRE as a dimer and is necessary for the activation of SRE-mediated gene expression by mitogens (Norman et al., 1988).
ETS family transcription factors (Elk1, Sap1a) bind a site adjacent to the SRE in the c-fos promoter and in some cells are required for growth factor-stimulated transcription (reviewed in Treisman, 1994). These ETS ternary complex factors (TCFs) are phosphorylated and transactivated by mitogen-activated protein kinases (MAPKs) (Treisman, 1995). Nevertheless, in many systems mitogens stimulate SRF-dependent transcription via an ETS-independent transcriptional pathway that targets the SRF DNA binding domain (Johansen and Prywes, 1994; Hill and Treisman, 1995). For example, in NIH 3T3 cells a Rho-dependent signaling pathway (Hill et al., 1995), a JNK-dependent histone acetylation cascade (Alberts et al., 1998) and the LIMK-1-dependent regulation of actin dynamics (Sotiropoulos et al., 1999) are required for ETS-independent transcription.
The role of PI3K signaling in cell cycle progression and the ability of activated PI3K to promote entry into S phase led us to examine whether PI3K signaling regulates mitogen-stimulated c-fos expression. Although PI3K may induce c-fos transcription indirectly via crosstalk with Ras and Rho signaling cascades (Yamauchi et al., 1993; Urich et al., 1997; Wang et al., 1998), other studies suggest a novel mechanism (Jhun et al., 1994; Bruning et al., 1997; Montmayeur et al., 1997). Here we demonstrate that SRF mediates PI3K-regulated c-fos transcription and that activation of SRF-dependent transcription by PI3K occurs via a novel ETS TCF and Rho-independent pathway. We also show that PI3K-dependent cell cycle progression depends on activation of SRF-dependent gene expression.
Results
Growth factor-stimulated c-fos transcription requires PI3K signaling
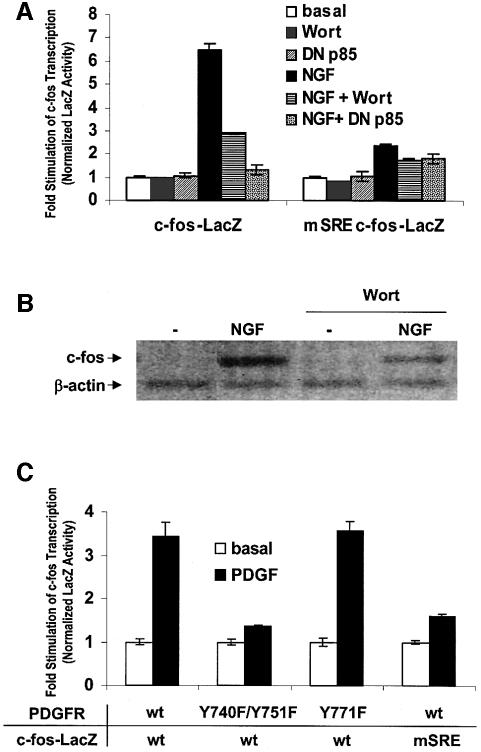
Because PI3K signaling regulates cell growth, we examined whether PI3K is required for growth factor-stimulated c-fos transcription. Treatment with nerve growth factor (NGF) increased c-fos transcription ∼6-fold in PC12 cells transfected with a c-fos promoter-regulated reporter construct (c-fos–LacZ) (Figure 1A). Treatment of cells with the PI3K inhibitor, wortmannin, or coexpression of a dominant-negative PI3K regulatory subunit (DN p85) significantly reduced NGF-mediated c-fos transcription. Mutations in the SRE (mSRE) that prevent SRF binding markedly attenuated NGF-stimulated c-fos transcription (Figure 1A). NGF-induced transcription of the endogenous c-fos gene also depended on PI3K signaling (Figure 1B).
Fig. 1. PI3K signaling is required for growth factor-stimulated c-fos transcription. (A) NGF stimulation of c-fos transcription required PI3K signaling through the SRE. PC12 cells were transfected with wild-type c-fos promoter–LacZ reporter construct (c-fos–LacZ) or a mutant SRE c-fos reporter construct (mSRE c-fos–LacZ). PI3K activity was blocked by cotransfection with a 3-fold excess (125 ng/well) of the dominant-negative regulatory subunit of PI3K (DN p85) DNA or pretreatment with 100 nM wortmannin (Wort). Cells were treated with 100 ng/ml NGF and assayed for LacZ activity. (B) Inhibition of PI3K attenuated NGF-induced c-fos transcription. PC12 cells were pretreated with 10 nM Wortmannin (Wort) or vehicle for 1 h and stimulated with 100 ng/ml NGF for 45 min. RT–PCR for c-fos was performed on total cellular RNA. β-actin transcription was used to normalize the level of c-fos transcription. (C) The PI3K signaling pathway was required for PDGF-mediated stimulation of c-fos expression. PC12 cells were transfected with the wild-type or mSRE c-fos LacZ reporter plasmids and either wild-type PDGFβ receptor (PDGFR wt), or mutants deficient in coupling to PI3K activation (Y741F/Y750F) or Shc/Ras GAP activation (Y771F) (375 ng/well). Cells were treated with 30 ng/ml PDGF and assayed for LacZ activity. The data are means ± standard deviation (SD) of triplicate assays normalized to luciferase activity driven from an SV40-luciferase plasmid.
PDGFβ receptors (PDGFRs) couple to both the Ras/Raf/Erk pathway through Shc and Src, and PI3K lipid signaling through p85 (Valius and Kazlauskas, 1993). Cotransfection of the wild-type PDGFR and the c-fos reporter led to a 4-fold increase in c-fos–LacZ expression when cells were treated with PDGF (Figure 1C). PDGF stimulation was absent in cells transfected with the Y740F/Y751F receptor mutant, which does not couple to PI3K. In contrast, the Y771F mutant, which does not couple to Shc, showed activation of c-fos–LacZ expression comparable to the wild-type PDGFR-transfected cells. Taken together, these data suggest that the PI3K pathway is required for activation of c-fos transcription by NGF or PDGF.
PI3K signaling is necessary for full activation of SRE-mediated gene expression by growth factors
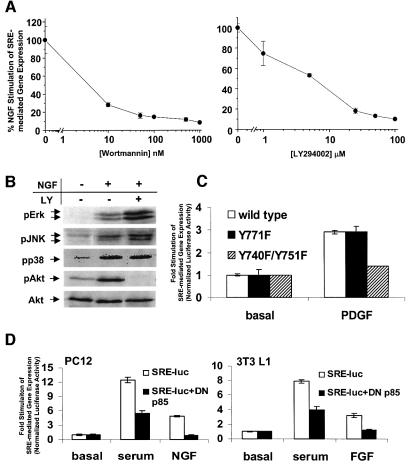
Because PI3K-regulated c-fos transcription targets the SRE, we explored the regulation of SRE-mediated transcription further by transfecting PC12 or 3T3-L1 cells with SRE–luciferase reporter constructs. NGF markedly activated PI3K as measured by Ser473 phosphorylation of the PI3K-regulated kinase, Akt (Figure 2B). Wortmannin and LY294002 inhibited NGF-stimulated SRE-mediated transcription in PC12 cells at concentrations consistent with specific inhibition of PI3K (Figure 2A). This inhibition was not due to non-specific effects on other SRE transactivating kinases since NGF-stimulated Erk, JNK and p38 phosphorylation were not attenuated by LY294002 treatment (Figure 2B). We also confirmed that inhibition of PI3K in PC12 cells and 3T3-L1 cells does not result in toxicity or downregulation of SRF expression (unpublished observations). As was the case for c-fos transcription, PC12 cells transfected with a PI3K-deficient PDGF receptor did not show PDGF-stimulated SRE-mediated transcription (Figure 2C). Cotransfection of DN p85 also attenuated mitogen-stimulated SRE-regulated transcription in PC12 and 3T3-L1 cells (Figure 2D). These data establish a role for PI3K in growth factor-stimulated SRE-mediated gene expression.
Fig. 2. NGF stimulation of SRE-mediated gene expression in PC12 is attenuated by inhibition of PI3K. (A) PI3K inhibitors decreased NGF-stimulated SRE-mediated gene expression. PC12 cells were transfected with a SRE–luciferase reporter plasmid, pretreated with the indicated concentrations of wortmannin or LY294002, treated with 100 ng/ml NGF and assayed for luciferase activity. NGF-stimulated, SRE-mediated transcription in the absence of inhibitor was set at 100%. (B) LY294002 inhibited PI3K signaling selectively. PC12 cells were serum starved for 18 h, pretreated with vehicle or 25 µM LY294002 (LY), treated with 100 ng/ml NGF for 10 min, lysed and analysed by western blot analysis for phosphorylated Erk, JNK, p38 and Akt. (C) PDGF receptor activation of SRE-mediated gene expression depended upon coupling to PI3K. PC12 cells were transfected with a SRE–luciferase reporter plasmid and PDGFβ receptor (PDGFR) (as described in Figure 1C). Cells were treated with 30 ng/ml PDGF and assayed for luciferase activity. (D) Dominant-negative PI3K inhibited SRE-mediated gene expression. PC12 and 3T3-L1 cells were transfected with a SRE–luciferase reporter plasmid along with a 3-fold excess of either empty vector or dominant-negative PI3K regulatory subunit (DN p85) expression vectors (125 ng/well). Cells were treated with 10% FBS, 100 ng/ml NGF or 50 ng/ml FGF, and assayed for luciferase activity. Where applicable, the data are means ± SD of triplicate assays normalized to EF1α promoter-driven LacZ expression.
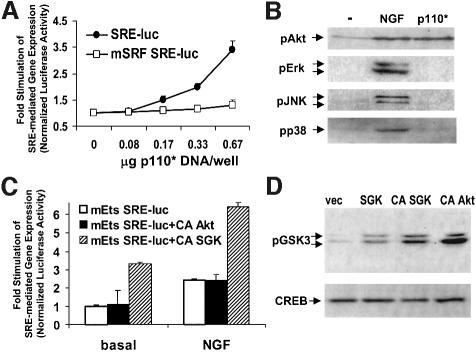
Expression of constitutively active PI3K (p110*) increased SRE-mediated transcription, indicating that PI3K activity is sufficient for SRE-dependent transcription (Figure 3A). Activated PI3K targeted SRF because mutation of the core SRF binding site (mSRF) blocked p110*-mediated transcription (Figure 3A). The ability of PI3K to couple to SRE-dependent transcription was not the result of crosstalk with MAPK cascades because p110* increased Akt phosphorylation but not phosphorylation of Erk, JNK or p38 MAPK (Figure 3B). Consistent with this, p110*-stimulated transcription was not inhibited by expression of dominant-negative (DN) MEK1 (MEK S222A), JBD (Jun kinase binding domain of JIP) or DN Rho N19 (unpublished observations). This led us to examine whether downstream targets of PI3K regulate SRE-mediated gene expression. Through the use of selective inhibitors or dominant negatives we ruled out a major role for atypical PKCs, conventional PKCs, p70 S6 kinase and Akt in mitogen-induced SRE-mediated transcription (unpublished observations). PI3K signaling regulates activation and nuclear translocation of serum- and glucocorticoid-inducible kinase (SGK) (Kobayashi and Cohen, 1999; Park et al., 1999), making it an attractive candidate for a downstream SRF effector. Expression of constitutively active SGK, but not Akt, markedly increased mEts SRE–luciferase reporter gene expression (Figure 3C). Both mutants were active because expression of activated Akt and SGK in PC12 cells increased GSK3 phosphorylation (Figure 3D). This suggests that SGK is a major PI3K-regulated effector of SRF-regulated transcription. Collectively these data indicate that PI3K is both necessary and sufficient for growth factor-stimulated SRE-mediated gene expression in PC12 cells and 3T3-L1 cells.
Fig. 3. Activation of PI3K and SGK induces SRE-mediated transcription. (A) Constitutively active PI3K increased SRE-mediated gene expression. PC12 cells were transfected with either a wild-type SRE luciferase reporter (SRE–luc) or a mutant SRE reporter (mSRF SRE–luc) that cannot bind SRF. Cells were cotransfected with increasing amounts of a constitutively active PI3K (p110*) expression vector. The data are means ± SEM of triplicate assays normalized to EF1α-driven LacZ expression. (B) Expression of p110* selectively activated PI3K-dependent signaling. PC12 cells were transfected with control or p110* expression vectors. Cells were treated with 100 ng/ml NGF for 10 min. Cells transfected with p110* were lysed 36 h post-transfection. Lysates were western blotted for phosphorylated Akt, Erk, JNK or p38 MAPK. (C) Expression of activated SGK increased SRF-dependent gene expression. PC12 cells were transfected with an SRF-regulated luciferase reporter that contains a mutation in the TCF binding Ets site (mEts SRE–luc) and a 3-fold excess of empty, constitutively active Akt (CA Akt) or constitutively active SGK (CA SGK) expression vectors. The data are means ± SEM of triplicate assays normalized to SV40-driven SEAP expression. (D) Expression of activated SGK and activated Akt increased GSK3 phosphorylation. PC12 cells were transfected with EF1α–LacZ and vector, activated Akt (CA Akt) or activated SGK (CA SGK), and extracts were blotted for phospho-GSK3. The extracts were then blotted for LacZ to control for transfection efficiency.
Since SRE-mediated gene expression can be activated by several MAPK cascades, including the ERK, p38 MAPK and JNK pathways (for a review see Treisman, 1995), we determined to what extent these pathways contribute to mitogen-stimulated transcription. We inhibited ERK, JNK and p38 MAPK signaling by expressing dominant-negative inhibitors or by treating cells with specific inhibitors (Table I). Inhibition of MAPK signaling either had no effect or only modestly inhibited SRE-mediated transcription (Table I). In every case, positive controls were performed to confirm that each inhibitor or dominant negative potently blocked their respective targets. For example, DN MEK and PD98059 blocked Gal4–Elk1-mediated transcription 87 and 98%, respectively (Figure 4G). These observations indicate that the PI3K pathway plays a major role in mitogen-stimulated SRE-mediated transcription.
Table I. Effects of dominant negative and chemical inhibitors on growth factor stimulation of SRE-mediated gene expression.
| Dominant negative/inhibitor | % Inhibition of stimulationa by |
||
|---|---|---|---|
| Serum PC12 cells | NGF PC12 cells | Serum 3T3-L1 cells | |
| MEK1b | 12.4 ± 0.6 | 3.8 ± 1.0 | 0 ± 0.6 |
| PD98059c | 0 ± 0.2 | 10.5 ± 1.0 | 8.4 ± 0.3 |
| JBDb | 0 ± 4.6 | 10.9 ± 1.0 | 0 ± 7.0 |
| SB203580c | 5.3 ± 1.0 | 15.1 ± 1.0 | 5.6 ± 1.7 |
| LY294002c | 78.2 ± 27.4 | 82.7 ± 2.4 | 49.7 ± 7.8 |
aPC12 and 3T3-L1 cells were transfected with a SRE–luciferase reporter, serum starved for 18 h, then treated with either 10% fetal bovine serum (Serum) or 100 ng/ml NGF for 5 h.
bDominant-negative MEK1 and JBD were cotransfected at a 3-fold excess (133 ng/well) of dominant negative to reporter constructs. Constructs are referenced in Materials and methods.
cCells were pretreated with 25 µM PD98059, 5 µM SB203580, 25 µM LY294002 for 1 h. Values are the mean of triplicate assays ± SD normalized to EF1α-driven LacZ expression.
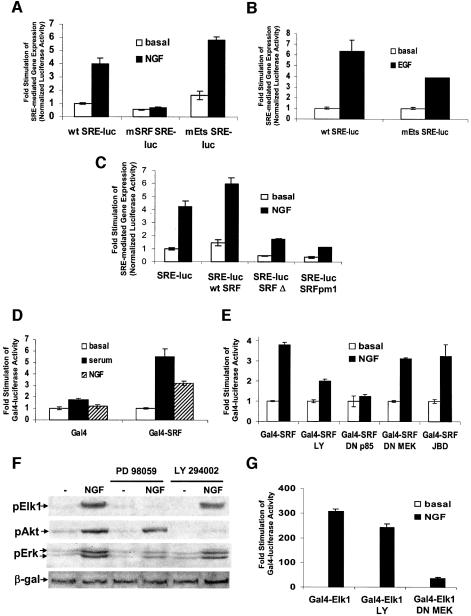
Fig. 4. PI3K and mitogens regulate SRE-mediated gene expression by transactivating SRF but not TCF. (A) Mutations in the DNA binding sequences for SRF, but not Ets, abolished NGF-stimulated SRE-mediated transcription. SRE reporter plasmids were constructed containing mutations in the SRF (mSRF SRE–luc) or Ets (mEts SRE–luc) binding sites (see Materials and methods for sequences) and transfected into PC12 cells. Cells were treated with 100 ng/ml NGF, then assayed for luciferase activity. (B) Ets DNA binding mutations reduced growth factor-induced SRE-mediated gene expression in HeLa cells. Cells were transfected as in (A) with the wild-type and mEts SRE–luciferase reporter plasmids, treated with 50 ng/ml EGF and assayed. (C) SRF was required for NGF-stimulated SRE-mediated transcription. PC12 cells were transfected with equal amounts (75 ng/well) of a SRE–luciferase reporter plasmid and expression vectors for wild-type SRF (wt SRF), a dominant-negative SRF with a C-terminal truncation (SRF Δ) or a dominant-negative SRF deficient for DNA binding (SRFpm1). Cells were treated with 100 ng/ml NGF and assayed for luciferase activity. (D) Mitogens transactivated Gal4–SRF. PC12 cells were transfected with a Gal4–luciferase reporter plasmid and a 2-fold excess (67 ng/well) of either Gal4 DNA binding domain (Gal4) or Gal4-SRF expression vectors. Cells were stimulated with 10% fetal bovine serum (serum) or 100 ng/ml NGF, and assayed for luciferase activity. (E) Blocking PI3K, but not MEK or JNK, inhibited NGF stimulation of Gal4–SRF-mediated transcription. PC12 cells were transfected with a Gal4–luciferase reporter, Gal4–SRF and a 4-fold excess of DN p85 (200 ng/well), a 3-fold excess of DN MEK (133 ng/well) or JBD (133 ng/well) expression constructs. Cells were stimulated with 100 ng/ml NGF and assayed for luciferase activity. (F) MEK inhibitors, but not PI3K inhibitors, blocked phosphorylation of Elk1 at Ser383. PC12 cells transfected with Elk1 (pCMV Elk1) DNA, were pretreated with dimethylsulfoxide (DMSO), 10 µM PD98059 or 25 µM LY294002, then blotted for Elk1, Akt and Erk phosphorylation following treatment with 100 ng/ml NGF for 10 min. An SV40-LacZ expression vector was cotransfected and lysates were blotted for β-galactosidase as a control for transfection efficiency. (G) Inhibition of MEK, but not PI3K, attenuated transactivation of Gal4–Elk1. PC12 cells were transfected with a Gal4–luciferase reporter construct, a 2-fold excess of Gal4–Elk1 expression vector (89 ng/well) and a 3-fold excess of control or DN MEK expression plasmids (167 ng/well). Cells were pretreated with 25 µM LY294002, then treated with 100 ng/ml NGF and assayed for luciferase activity. Where applicable, the data are means ± SD of triplicate assays normalized to EF1α promoter-driven LacZ expression.
Growth factors stimulate SRE-mediated transcription independently of Rho signaling
The Rho family of small G-proteins regulate c-fos expression and SRF-dependent transcription via LIMK-1-regulated actin treadmilling (Sotiropoulos et al., 1999). To determine whether Rho family G-proteins contribute to PI3K-regulated gene expression, PC12 cells were transfected with a SRE–luciferase reporter construct and inhibitors of Ras (Ras N17), Rho (C3 exoenzyme, Rho GDI and Rho N19), Rac (Rac N17) or Cdc42 (Cdc42 N17). Surprisingly, expression of these dominant negatives was either without effect or only slightly decreased mitogen-induced SRE-regulated transcription (Table II). Interestingly, the modest inhibition of SRE-mediated transcription by these dominant negatives (5–13%; Table II) was far less than that seen following inhibition of PI3K (57–82%; Figure 2D and Table I), indicating that in PC12 cells the Rho transcriptional pathway is dispensable for mitogen-stimulated SRE-regulated transcription. The potent efficacy of each inhibitory construct was confirmed in PC12 and NIH 3T3 cells (unpublished observations). Therefore, we propose a novel PI3K-dependent pathway for the activation of SRF-mediated transcription that does not involve Rho family G-proteins.
Table II. Inhibition of Rho family small G proteins does not affect mitogen stimulation of SRE-mediated gene expression.
| Expression constructsb | % Inhibition of stimulationa by |
|
|---|---|---|
| Serum | NGF | |
| Ras N17 | 0 ± 1.0 | 0 ± 6.1 |
| C3 | 12.5 ± 0.7 | 13.0 ± 0.5 |
| Rho GDI | 0 ± 0.1 | 4.5 ± 0.1 |
| Rho N19 | 0 ± 1.1 | 7.7 ± 0.9 |
| Rac N17 | 0 ± 2.2 | 10.6 ± 1.2 |
| Cdc42 N17 | 0 ± 1.5 | 10.4 ± 0.4 |
aPC12 cells were transfected with a SRE–luciferase reporter plasmid, serum starved for 18 h, then treated with either 10% FBS (Serum) or 100 ng/ml NGF for 5 h, then assayed for luciferase activity.
bA 3-fold excess (133 ng/well) of either control vector, C3 botulinum toxin expression vector, a Rho guanine nucleotide dissociation inhibitor (Rho GDI) dominant-negative Rho (Rho N19), Cdc42 (Cdc42 N17) or Rac (Rac N17) to reporter was cotransfected.
Values are the mean of triplicate assays ± SD. Values are normalized for transfection efficiency by cotransfection of an EF1α–LacZ expression vector and measurement of β-galactosidase activity from cell lysates.
PI3K activation of SRE-mediated gene expression occurs via SRF
Several transcription factors bind to the c-fos SRE and regulate transcription. To determine the target of PI3K-dependent signaling, we generated mutant reporter constructs that are deficient for binding of SRF (mSRF) or TCF (mEts) to the SRE. PC12 cells transfected with the mSRF luciferase reporter construct did not show NGF-induced transcription (Figure 4A). Disruption of the TCF site 5′ of the CArG had little effect on NGF stimulation of the SRE. Although the ETS TCF pathway plays an important role in SRE-regulated transcription in fibroblasts (Hill et al., 1993), in other cell types different pathways predominate (Johansen and Prywes, 1994; Hill and Treisman, 1995). For example, disruption of the TCF binding site reduced EGF-induced SRE-mediated transcription (Figure 4B). Nevertheless, significant stimulation remained, indicating a TCF-independent target for polypeptide growth factors at the SRE.
We also confirmed that SRE-mediated gene expression required SRF via the use of two specific SRF dominant negatives. One is a mutant with a deleted C-terminal activation domain, which forms transcriptionally inactive heterodimers with wild-type SRF (SRF Δ). The other mutant is a DNA binding domain mutant that forms heterodimers with wild-type SRF, reducing the pool of DNA binding competent SRF (SRFpm1). Both dominant-negative SRF mutants inhibited NGF-stimulated SRE-mediated transcription (Figure 4C), but not forskolin-stimulated CRE-mediated transcription (unpublished observations).
To confirm that SRF is transactivated by PI3K signaling, we examined the ability of fusions of full-length SRF to the yeast Gal4 DNA binding domain (Gal4–SRF) to drive Gal4–luciferase transcription. Serum and NGF transactivated Gal4–SRF (Figure 4D). Transactivation of SRF required PI3K, as LY294002 and DN p85 both inhibited NGF stimulation of transcription (Figure 4E). Similarly, expression of DN MEK and JBD had no effect on activation of Gal4–SRF by NGF (Figure 4E).
These data argue that SRF or a novel accessory protein is a direct target of PI3K-dependent signaling. Because the ETS family TCFs, Elk1 and Sap1, play a crucial role in mitogen-stimulated SRE-dependent transcription (Treisman, 1995), we examined whether they are also a target of PI3K signaling. Treatment of PC12 cells with LY294002 did not reduce NGF-stimulated phosphorylation of Ser383 of Elk1, a residue critical for Elk1 transactivation (Figure 4E). While LY294002 treatment did not affect transactivation of Gal4–Elk1 (Figure 4G), it markedly attenuated the transactivation of Gal4–SRF. Conversely, PD98059, which had no effect on NGF stimulation of SRE-mediated transcription (Table I), strongly inhibited Elk1 phosphorylation (Figure 4F). Similarly, expression of DN MEK attenuated transactivation of Gal4–Elk1 (Figure 4G) but was without effect on Gal4–SRF. These data strongly suggest that SRF or a novel accessory factor is the primary target of a PI3K-dependent and polypeptide growth factor-mediated transcriptional cascade.
PI3K and SRF are required for mitogen-induced cell cycle progression
Several lines of evidence suggest that PI3K plays a critical role in cellular proliferation. Although several studies report that PI3K may promote cell cycle progression by activating p70 S6 kinase (Klippel et al., 1998), other studies suggest an unknown mechanism (Gille and Downward, 1999; Takuwa et al., 1999). Thus, the mechanism(s) by which PI3K signaling regulates cell cycle progression is not clear.
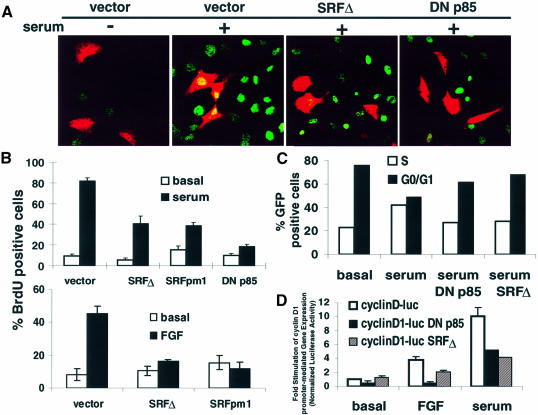
Because PI3K can potently activate SRF-dependent gene expression, we explored whether SRF is a potential target of PI3K during cell cycle progression. Cotransfection of regulatory constructs with a chloramphenicol acetyl transferase (CAT) expression vector enabled us to identify transfected cells and assess whether DNA synthesis occurred, using immunocytochemistry (ICC) for CAT detection and BrdU incorporation, respectively. Treatment of quiescent 3T3-L1 cells with serum or fibroblast growth factor (FGF) resulted in a marked increase in DNA synthesis measured by ICC for BrdU incorporation (Figure 5A and B). Expression of DN p85 attenuated serum-induced BrdU labelling, confirming that PI3K signaling is critical for the G1–S transition (Figure 5A and B). If SRF is an important target of PI3K during the G1–S transition, then inhibition of SRF-dependent transcription should also attenuate entry into the cell cycle. Expression of SRF Δ or SRFpm1 attenuated serum- or FGF-induced cell cycle progression relative to vector control (Figure 5A and B). FACS analysis confirmed that expression of SRF Δ and DN p85 inhibits serum- stimulated cell cycle progression in G0/G1 (Figure 5C).
Fig. 5. SRF-regulated transcription and PI3K activity are obligatory for mitogen-stimulated cell cycle progression. (A) Inhibition of SRF-dependent gene expression or PI3K activity blocked mitogen-stimulated cell cycle entry. Representative images depict BrdU staining (green) and CAT staining (red). 3T3-L1 cells were transfected with a vector control (vector), a C-terminally truncated dominant-negative SRF (SRF Δ) or a dominant-negative PI3K regulatory subunit (DN p85) expression vector (150 ng/well). Cotransfection with a CAT expression vector facilitated identification of transfected cells. Cells were treated with 5% FBS (serum) for 20 h. DNA synthesis was measured by ICC for BrdU. (B) Bar graph shows cumulative data from (A). Cells were transfected as described in (A) except that SRFpm1 was also transfected and cells were also treated with 50 ng/ml FGF. Quantitation of BrdU staining was conducted as described in Materials and methods (n = 5–10; mean ± SEM). (C) Blocking SRF-mediated gene expression or PI3K activity caused cell cycle arrest in G1. 3T3-L1 cells were transfected with EF1α–green fluorescent protein (GFP) and a 2-fold excess of vector control, SRF Δ or DN p85 expression vector. Transfected cells were isolated by sorting for GFP fluorescence and FACS analysis of DNA content was performed as described in Materials and methods. (D) Blocking SRF-dependent gene expression attenuated mitogen-induced cyclin D1 transcription. 3T3-L1 cells were transfected with a cyclin D1 promoter-regulated luciferase reporter construct and a 2-fold excess of a vector control, SRF Δ or DN p85 expression vector. Cells were treated with 50 ng/ml FGF or 10% FBS (serum) for 10 h and assayed for luciferase activity. The data are means ± SEM of triplicate assays normalized to EF1α-driven LacZ expression.
Mitogen-induced cyclin D1 expression is believed to be an essential event in the G1–S transition (Jiang et al., 1993; Musgrove et al., 1994). Therefore, we explored the role of SRF in mitogen-stimulated cyclin D1 gene expression and found that SRF Δ and DN p85 both inhibited mitogen-stimulated cyclin D1 transcription (Figure 5D). Thus, cyclin D1 is a potential downstream target of PI3K-stimulated SRF-dependent gene expression and increased expression of cyclin D1 is likely to contribute to PI3K-stimulated cell cycle progression. Because the cyclin D1 promoter does not contain SREs, the requirement for SRF for cyclin D1 transcription is likely to be the result of the induction of SRF-regulated IEG transcription factors, such as c-fos and FosB, which in turn induce cyclin D1 transcription (Brown et al., 1998). Since PI3K is critical for the G1–S transition and is a potent activator of SRF-dependent gene expression, these data led us to explore whether PI3K-regulated SRF-dependent transcription plays an important role in mitosis.
Activation of SRF-mediated gene expression is sufficient for cell cycle entry
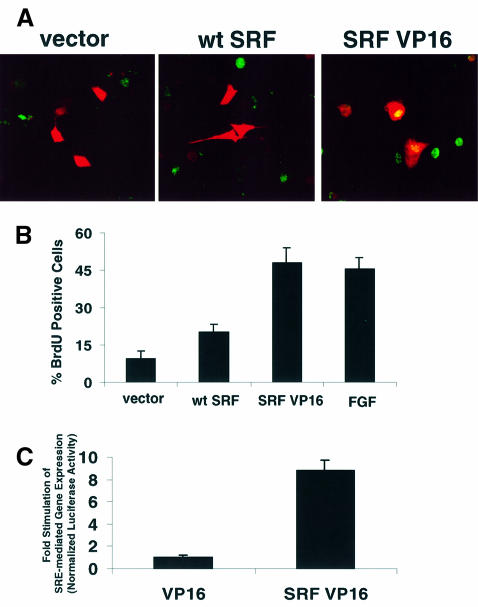
Our data suggest that both PI3K- and SRE-mediated gene expression are required for mitogen-induced cell cycle progression. Therefore, we examined whether constitutive activation of SRF-dependent gene expression is sufficient to trigger entry into S phase. We made a constitutively active SRF by fusing SRF to the VP16 transactivation domain. Expression of SRF VP16 robustly activated SRF-regulated transcription (Figure 6C). Interestingly, coexpresion of SRF VP16 triggered DNA synthesis (Figure 6A and B). Coexpression of SRF VP16 promoted levels of DNA synthesis comparable to those in FGF-treated cells. These results indicate that genes induced by SRF are sufficient for cell cycle entry.
Fig. 6. Activation of SRF-regulated gene expression is sufficient to induce cell cycle entry. (A) Activation of SRF-dependent transcription induced DNA synthesis. Representative images depict BrdU staining (green) and CAT staining (red). 3T3-L1 cells were transfected with control, wild-type SRF (wt SRF) or constitutively active SRF (SRF VP16) expression vectors (150 ng/well). (B) Bar graph shows cumulative data from (A). Cells were transfected as described in (A). Quantitation of BrdU staining was conducted as described in Materials and methods (n = 4–6; mean ± SEM). Cells were treated with 50 ng/ml FGF for 20 h where indicated. (C) SRF VP16 induced SRE-mediated gene expression in 3T3-L1 cells. Cells were transfected with a SRE–luciferase reporter construct and a 2-fold excess (75 ng/well) of control (VP16) or SRF VP16 expression vector. The data are means ± SEM of triplicate assays normalized to EF1α-driven LacZ expression.
Activation of PI3K triggers cell cycle progression
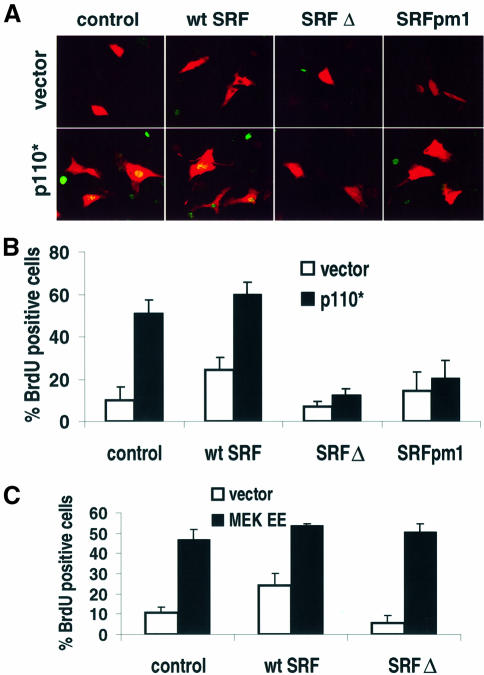
Because PI3K is a critical regulator of mitogen-stimulated cell cycle entry we examined whether activation of PI3K was sufficient for cell cycle progression. Expression of constitutively active PI3K (p110*) in quiescent 3T3-L1 cells was sufficient to promote entry into the cell cycle (Figure 7A and B). PI3K-stimulated DNA synthesis was robust and was equivalent to that induced by an oncogenic activated Ras mutant (unpublished observations). Importantly, coexpression of SRF dominant negatives (SRF Δ and SRFpm1) largely attenuated p110*-mediated cell cycle entry (Figure 7A and B). Thus, we provide direct evidence that SRF is a critical target of PI3K-stimulated cell cycle progression.
Fig. 7. PI3K-induced cell cycle entry requires SRF-dependent gene expression. (A) Expression of dominant-negative SRF attenuated PI3K-stimulated cell cycle progression. Representative images depict BrdU staining (green) and CAT staining (red). 3T3-L1 cells were transfected with control, wild-type SRF (wt SRF), a dominant-negative SRF with a DNA binding domain mutation (SRFpm1) or a C-terminal truncated dominant-negative SRF (SRF Δ) expression vector in the presence or absence of a 2-fold excess (300 ng/well) of p110* expression construct. A CAT expression vector was cotransfected to allow for detection of transfected cells by ICC. (B) Bar graph shows cumulative data from (A). Cells were transfected as described in (A). Quantitation of BrdU staining was conducted blind as described in Materials and methods (n = 4–6; error is SEM). (C) MEK-induced cell cycle progression did not require PI3K signaling. 3T3-L1 cells were transfected as in (A) except that a 4-fold excess (600 ng/well) constitutively active MEK (MEK EE) replaced p110*. Quantitation of BrdU staining was conducted as described in Materials and methods (n = 4–6; error is SEM).
Although SRF-dependent transcription was critical for growth factor and PI3K-mediated cell cycle entry, we also examined whether SRF-dependent transcription was universally required for cell cycle entry. Expression of activated MEK induces cell cycle progression in a number of cell lines (Wright et al., 1999). We also found that expression of constitutively active MEK (MEK S218E/S222E) induced S-phase entry in 3T3-L1 cells (Figure 7C). Contrary to what we observed for p110* expression, coexpression of SRF Δ did not significantly decrease MEK-induced S-phase entry. Thus, PI3K signaling regulates G1–S progression by distinct transcriptional mechanisms from the ERK MAPK cascade.
Discussion
Although PI3K plays a pivotal role in mitosis, cell transformation and oncogenesis, the mechanisms by which PI3K regulates cell growth remain unclear. Here, we characterized a novel mitogen-induced PI3K signaling cascade that potently increases SRF-dependent transcription and regulates the transcription of some growth-associated IEGs and cyclins. We also provide evidence that SRF-dependent gene expression is required for PI3K-mediated cell cycle entry. Therefore, we describe a novel mechanism for the coupling of mitogenic signals to increases in gene expression and cell proliferation.
PI3K regulates IEG expression by transactivating SRF
The IEG, c-fos and related transcription factors are believed to play a critical role in mitogenesis by inducing rapid increases in the expression of G1 cyclins that promote entry into S phase (Riabowol et al., 1988; Kovary and Bravo, 1991; Brown et al., 1998). Because PI3K has recently been implicated as a crucial regulator of cell cycle entry (Carpenter and Cantley, 1996), we investigated the mechanisms by which PI3K signaling activates c-fos transcription. Consistent with earlier work implicating PI3K in mitogen-stimulated c-fos transcription (Yamauchi et al., 1993; Jhun et al., 1994), we found that PI3K activity is required for growth factor- and serum-induced increases in c-fos transcription in PC12 cells and 3T3-L1 cells. We also found that PI3K activity is necessary and sufficient for mitogen-stimulated SRE-regulated transcription.
In fibroblasts, growth factor induction of SRE-mediated transcription can occur via activation of MAPK cascades and phosphorylation of the TCFs, Elk1 and Sap1a (Hill et al., 1993; Miranti et al., 1995; Treisman, 1995). Interestingly, in PC12 cells we found that PI3K was neither necessary nor sufficient for the transactivation of Gal4–Elk1 and Gal4–Sap1. Furthermore, inhibition of JNK, p38 and ERK MAPKs, the known activators of ETS TCFs, only slightly reduced growth factor-stimulated transcription in PC12 and 3T3-L1 cells. Because PI3K signaling rather than MAPK signaling was required for the transactivation of Gal4–SRF by growth factors, we conclude that PI3K activates SRF via an ETS TCF-independent pathway. Confirming this idea, mutation of the ETS TCF binding site adjacent to the core c-fos SRE had little effect on PI3K or mitogen-induced SRF-dependent transcription.
Although in certain fibroblast cell lines, such as HeLa, the ETS TCFs play a role in growth factor-induced transcription (Hill et al., 1993; Miranti et al., 1995), we found that a significant component of growth factor-induced transcription is TCF independent. Interestingly, in these cells potent activators of MAPK signaling have been shown to regulate transcription via an ETS TCF-independent pathway (Johansen and Prywes, 1994; Hill et al., 1995). Clearly, whether ETS-dependent or -independent pathways are recruited depends not only on cell type but also on poorly understood combinatorial signaling. The relative contribution of ETS TCFs may also have been overstated because several studies used DNA binding domain SRF mutants that block ETS-independent transcription while leaving ETS-dependent transcription intact (see Hill et al., 1994 for a discussion of this issue).
This work highlights the potential for growth factors to increase IEG transcription via an SRF- and PI3K-dependent pathway. PI3K-mediated gene expression is likely to be of particular importance in the regulation of promoters that contain core SREs but lack adjacent ETS binding sites, such as those found in Egr-1, Egr-2, SRF, JunB and atrial natriuretic factor.
PI3K activates a novel SRF-dependent transcriptional pathway
The Rho family of small G-proteins are potent regulators of SRF-dependent transcription (Hill et al., 1995). Thus, we investigated whether Rho family members contribute to PI3K-stimulated gene expression. Dominant-negative inhibitors of Rho, Rac and Cdc42 had little effect on NGF- or FGF-stimulated SRF-dependent gene expression in PC12 cells or 3T3-L1 cells. Conversely, the ability of activated Rho family G-proteins to increase transcription also did not depend on PI3K activity (unpublished observations). Taken together, these results show that the ability of PI3K to transactivate SRF does not depend on crosstalk with Rho-dependent transcriptional cascades.
How does PI3K activate SRF-dependent transcription?
PI3K can activate the ERK and JNK MAP kinase cascades (Klippel et al., 1996); however, we show that selective inhibition of these pathways does not block SRF-dependent transcription. Activation of PI3K also promotes the activation of the AGC family of PI3K-regulated kinases, including Akt, p70 S6 kinases, PKC isoforms and SGK. Through the use of selective dominant-negative constructs we ruled out a major role for Akt, p70 S6 kinase and the atypical and conventional PKCs (unpublished observations). The mitogen-induced activation and nuclear trans location of SGK depends on PI3K signaling (Kobayashi and Cohen, 1999; Park et al., 1999), making SGK a possible candidate for an SRF effector. Interestingly, expression of a constitutively active SGK mutant markedly stimulated SRF-dependent transcription. This suggests that mitogens and growth factors can activate SRF-dependent transcription via a PI3K-PDK1-SGK signaling cassette.
Potential transcriptional targets of PI3K signaling
Although mitogen- and Rho-dependent SRF-dependent transcription are described in several studies, there is at present little insight into the mechanisms by which SRF is transactivated (Hill et al., 1994, 1995; Johansen and Prywes, 1994; Hill and Treisman, 1995). Mitogen stimulation induces the phosphorylation of the N-terminus of SRF by multiple kinases (for review see Treisman, 1995). However, it is not known if or how these phosphorylation events influence SRF transactivation (Johansen and Prywes, 1993; Hill et al., 1994). Multiple transcription factors interact with SRF, including, phox1/MHox, C/EBPβ, NF-κB and ATF6, and could potentially mediate SRF-dependent transcription. Recent work suggests that the SRF DNA binding domain interacts with an unknown factor required for SRF-dependent transcription (Hill et al., 1993, 1994; Johansen and Prywes, 1994). Further work is clearly required to identify the composition of the PI3K-stimulated SRF-dependent transcriptional complex and to identify the targets of PI3K signaling.
PI3K-regulated SRF-dependent transcription and cellular proliferation
A substantial body of evidence demonstrates a critical role for PI3K in cell cycle progression. For example, oncogenic mutations have been isolated in both the regulatory and catalytic subunits of type I PI3Ks (Chang et al., 1997; Jimenez et al., 1998). Moreover, activation of PI3K is sufficient for cell cycle entry (Frevert and Kahn, 1997; Klippel et al., 1998) and PI3K activity is necessary for cell cycle progression (Jhun et al., 1994; Roche et al., 1994; Gille and Downward, 1999). Interestingly, expression of constitutively active PI3K is not sufficient for completion of the cell cycle (Klippel et al., 1998; unpublished observations). Because expression of oncogenic Ras is also not sufficient for completion of the cell cycle in 3T3-L1 cells (Benito et al., 1991), this result is consistent with a requirement for additional deregulation of cell cycle checkpoints for chronic activation of mitogenic cascades to promote proliferation. Nevertheless, the finding that mice deficient for the p110α catalytic subunit of PI3K have a profound proliferative defect points to a critical role for PI3K signaling in cell growth (Bi et al., 1999). Although Ras/MAPK signaling clearly plays an important role in mitogenesis in some cells, increasing evidence indicates that PI3K signaling represents an independent pathway essential for mitogen-induced cell cycle progression (Roche et al., 1994; Frevert and Kahn, 1997; Gille and Downward, 1999).
SRF-dependent gene expression is regulated by the cell cycle and has been proposed to play a role in cell cycle progression (Gauthier-Rouviere et al., 1991; Liu et al., 1994). Because SRF is also a transcriptional target of PI3K, we explored whether it is a target of PI3K during cell cycle progression. Interestingly, expression of dominant-negative SRF and PI3K constructs inhibited FGF-, serum- and PI3K-induced cell cycle entry, suggesting that PI3K can couple mitogenic signals to SRF. Moreover, expression of a constitutively active SRF was sufficient to trigger entry into the cell cycle. These results indicate that PI3K can stimulate cell cycle entry by inducing SRF-dependent gene expression. Nevertheless, the incomplete inhibition of serum-induced mitosis by SRF dominant negatives suggests that PI3K can also target other mitogenic signaling cascades. We also demonstrate that PI3K and ERK recruit distinct transcriptional pathways for cell cycle progression because MEK-mediated cell cycle entry does not require SRF-dependent gene expression.
What are the mitogenic targets of PI3K-stimulated gene expression?
Cyclin D1 transcription is induced by mitogens (Winston and Pledger, 1993) and cyclin D1 is thought to play a key role in mitogen-stimulated G1 progression (Musgrove et al., 1994; Aktas et al., 1997). Recent evidence indicates that mitogens induce the expression and transcription of cyclin D1 in some cells via PI3K signaling (Gille and Downward, 1999; Takuwa et al., 1999). Interestingly, we found that growth factor-induced cyclin D1 transcription was blocked by expression of dominant-negative PI3K or SRF. Because the cyclin D1 promoter does not contain an SRE, the requirement for SRF for cyclin D1 transcription is the result of the indirect expression of SRF-regulated IEG transcription factors. For example, fibroblasts deficient for the SRF-regulated IEGs, c-fos and FosB, have a defect in mitogen-induced proliferation and cyclin D1 transcription (Brown et al., 1998). Taken together, these results suggest that a mitotic target of PI3K-stimulated SRF-mediated transcription is cyclin D1, which in turn mediates repression of Rb and activation of E2F.
In conclusion, we delineate a novel SRF-dependent transcriptional pathway critical for mitogen- and PI3K-stimulated mitogenesis in 3T3-L1 cells. Moreover, this SRF-dependent transcriptional pathway is also critical for PI3K- and growth factor-stimulated c-fos and cyclin D1 transcription. While the identity of SRF-regulated genes required for cell proliferation is not known, the proliferative defect in fibroblasts lacking the SRF-regulated IEGs, c-fos and FosB, is suggestive (Brown et al., 1998). The coupling of PI3K to the SRE might be critically important for the early development of organisms as well as for proliferation.
Materials and methods
Cell culture
Early passage PC12 cells were cultured as described in Mark et al. (1995). 3T3-L1 and HeLa cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 5% bovine calf serum (BCS). For reporter gene assays, PC12 and 3T3-L1 cells were plated on 24-well plates (Costar) coated with 100 µg/ml poly-d-lysine (Sigma). For proliferation assays, 3T3-L1 cells were plated on glass coverslips (Fisher) coated with 200 µg/ml poly-d-lysine.
Plasmids
Gal4–Elk1 (Whitmarsh et al., 1995), c-fos–LacZ reporter plasmids (Smeyne et al., 1993), JBD (Dickens et al., 1997), DN p85 (Dhand et al., 1994), p110* (Klippel et al., 1996), Gal4–SRF (Hanlon and Sealy, 1999) and PDGFR (Valius and Kazlauskas, 1993) expression plasmids are described elsewhere. The Rho N19, Rac N17 and Cdc42 N17 expression constructs were described in Coso et al. (1995). The wt SRF (CGN SRF) and SRF Δ (CGN SRF 1–338) plasmids were described in Johansen and Prywes (1993). DN MEK (MEKS222A), MEK EE (MEK S218E/S222E), Ras N17 and Gal4–E1B–luc are described in Impey et al. (1998). The SGK and Rho GDI cDNAs were isolated by low-cycle PCR from human cDNA (Clontech) and cloned into pCDNA3.1 (Invitrogen). CA SGK-pCDNA3.1 contained an S422D mutation as described in Kobayashi and Cohen (1999) and was made using the Quickchange mutagenesis system (Stratagene). SRFpm1 was created by making R143L, K145A and I146G mutations in SRF (CGN SRF) using the Quickchange mutagenesis system (Stratagene). SRF VP16 was made by subcloning human SRF into pVP16 (Clontech). The following double-stranded oligos were inserted into the SRE–luc vector (Johansen and Prywes, 1994) to make mEts SRE–luc and mSRF SRE–luc, respectively: AAGCTTGCTCGAAAATGTCCATAT TAGGTAATTAAGATCT, AAGCTTGCTCGAGGATGTCCCTATTG GGACAAGTAGATCT. The mEts and mSRF mutations were described previously in Johansen and Prywes (1994).
Transient transfections and reporter assays
Cells were transfected with Transfast (Promega) as described by the manufacturer. Reporter assays were conducted 48 h post-transfection unless otherwise noted. Cells were serum starved in DMEM containing 25 mM HEPES for 16–24 h, pretreated with inhibitors for 1 h, then treated with or without agonists for 5 h. Luciferase, β-gal and secreted alkaline phosphatase (SEAP) activity were measured using Luciferase Assay, Galacoto-Light, and Phospha-Light assay kits as described by the manufacturer (Tropix, Bedford, MA).
RT–PCR analysis of c-fos expression
PC12 cells were plated onto 35 mm plates coated with 100 µg/ml poly-d-lysine. Total cellular RNA was isolated using Trizol (Life Technologies) and RT–PCR was conducted with SuperScript II (Gibco-BRL). Primers for c-fos and β-actin expression are described in Gardier et al. (2000).
Western blot analysis
Agonist- and inhibitor-treated PC12 cells were lysed in 3 × boiling SDS–PAGE sample buffer and boiled for 10 min at 95°C. The samples were subjected to SDS–PAGE and blotted as described in Impey et al. (1998). Antibodies were used at the following dilutions: 1:1000 [rabbit anti-phospho Akt, rabbit anti-Akt, rabbit anti-phospho Gsk3 (NEB), mouse anti-phospho MAP kinase, mouse anti-CREB, goat anti-Erk (Santa Cruz)]; 1:2000 [rabbit anti-phospho p38 (NEB), AP-conjugated anti-IgG (Cappel) and HRP-conjugated anti-IgG (Jackson Immunoresearch)]; 1:5000 rabbit anti-active JNK (Promega); 1:500 rabbit anti-phospho Elk (NEB). Blots were developed using alkaline phosphatase (Tropix) or horseradish peroxidase (Amersham) chemiluminescence as described by the manufacturer.
Immunochemistry
3T3-L1 cells were serum starved for 24 h and treated with 10 µg/ml BrdU and growth factors for 20 h. Thirty-six hours post-transfection, cells were washed twice with phosphate-buffered saline (PBS) pH 7.4 and fixed with 4% formaldehyde in PBS. Coverslips were blocked in PBS–5% BSA (USB, Fraction V) and incubated at 23°C for 2 h with the following primary antibodies: 1:500 goat anti-mouse BrdU (Calbiochem) and 1:250 goat anti-rabbit CAT (5′–3′) in PBST containing 5% BSA. Coverslips were then incubated at 23°C for 1 h with the following secondary antibodies at 1:500: Alexa594 anti-rabbit and Alexa488 anti-mouse (Molecular Probes). Following antibody incubations coverslips were washed 3 × 5 min in PBST.
Image analysis
Images were captured on a Bio-Rad MRC-600 laser scanning confocal microscope. Quantification of BrdU staining was conducted blind using Metamorph software (Universal Imaging). Briefly, integrated pixel intensity was measured in a 10 pixel diameter circle and a measurement in excess of an integrated pixel value of 2000 was counted as positive. For each experiment, 15–30 CAT transfected cells were measured.
Acknowledgments
Acknowledgements
We thank E.Krebs, R.Prywes, T.Curran, A.Hall, S.Gutkind, R.Treisman, L.Sealy, E.Wang, L.Williams and M.Kasuga for plasmids and cDNAs. Imaging and analysis were conducted at the Keck Center, University of Washington. S.P. was supported in part by PHS NRSA T32 from NIGMS. This research was supported by National Institutes of Health grants NS 20498 and HL 44948.
References
- Aktas H., Cai,H. and Cooper,G.M. (1997) Ras links growth factor signaling to the cell cycle machinery via regulation of cyclin D1 and the Cdk inhibitor p27KIP1. Mol. Cell. Biol., 17, 3850–3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberts A.S., Geneste,O. and Treisman,R. (1998) Activation of SRF-regulated chromosomal templates by Rho-family GTPases requires a signal that also induces H4 hyperacetylation. Cell, 92, 475–487. [DOI] [PubMed] [Google Scholar]
- Benito M., Porras,A., Nebreda,A.R. and Santos,E. (1991) Differentiation of 3T3-L1 fibroblasts to adipocytes induced by transfection of ras oncogenes. Science, 253, 565–568. [DOI] [PubMed] [Google Scholar]
- Bi L., Okabe,I., Bernard,D.J., Wynshaw-Boris,A. and Nussbaum,R.L. (1999) Proliferative defect and embryonic lethality in mice homozygous for a deletion in the p110α subunit of phosphoinositide 3-kinase. J. Biol. Chem., 274, 10963–10968. [DOI] [PubMed] [Google Scholar]
- Brown J.R., Nigh,E., Lee,R.J., Ye,H., Thompson,M.A., Saudou,F., Pestell,R.G. and Greenberg,M.E. (1998) Fos family members induce cell cycle entry by activating cyclin D1. Mol. Cell. Biol., 18, 5609–5619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruning J.C., Winnay,J., Cheatham,B. and Kahn,C.R. (1997) Differential signaling by insulin receptor substrate 1 (IRS-1) and IRS-2 in IRS-1-deficient cells. Mol. Cell. Biol., 17, 1513–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantley L.C. and Neel,B.G. (1999) New insights into tumor suppression: PTEN suppresses tumor formation by restraining the phosphoinositide 3-kinase/AKT pathway. Proc. Natl Acad. Sci. USA, 96, 4240–4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter C.L. and Cantley,L.C. (1996) Phosphoinositide 3 kinase and the regulation of cell growth. Biochim. Biophys. Acta, 1288, m11–m16. [DOI] [PubMed] [Google Scholar]
- Chang H.W., Aoki,M., Fruman,D., Auger,K.R., Bellacosa,A., Tsichlis,P.N., Cantley,L.C., Roberts,T.M. and Vogt,P.K. (1997) Transform ation of chicken cells by the gene encoding the catalytic subunit of PI 3-kinase. Science, 276, 1848–1850. [DOI] [PubMed] [Google Scholar]
- Coso O.A., Chiariello,M., Yu,J.C., Teramoto,H., Crespo,P., Xu,N., Miki,T. and Gutkind,J.S. (1995) The small GTP-binding proteins Rac1 and Cdc42 regulate the activity of the JNK/SAPK signaling pathway. Cell, 81, 1137–1146. [DOI] [PubMed] [Google Scholar]
- Dhand R., Hara,K., Hiles,I., Bax,B., Gout,I., Panayotou,G., Fry,M.J., Yonezawa,K., Kasuga,M. and Waterfield,M.D. (1994) PI 3-kinase: structural and functional analysis of intersubunit interactions. EMBO J., 13, 511–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickens M., Rogers,J.S., Cavanagh,J., Raitano,A., Xia,Z., Halpern,J.R., Greenberg,M.E., Sawyers,C.L. and Davis,R.J. (1997) A cytoplasmic inhibitor of the JNK signal transduction pathway. Science, 277, 693–696. [DOI] [PubMed] [Google Scholar]
- Frevert E.U. and Kahn,B.B. (1997) Differential effects of constitutively active phosphatidylinositol 3-kinase on glucose transport, glycogen synthase activity and DNA synthesis in 3T3-L1 adipocytes. Mol. Cell. Biol., 17, 190–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardier A.M., Moratalla,R., Cuellar,B., Sacerdote,M., Guibert,B., Lebrec,H. and Graybiel,A.M. (2000) Interaction between the serotoninergic and dopaminergic systems in d-fenfluramine-induced activation of c-fos and jun B genes in rat striatal neurons. J. Neurochem., 74, 1363–1373. [DOI] [PubMed] [Google Scholar]
- Gauthier-Rouviere C., Cavadore,J.-C., Blanchard,J.-M., Lamb,N.J.C. and Fernandez,A. (1991) p67SRF is a constitutive nuclear protein implicated in the modulation of genes required throughout the G1 period. Cell Regul., 2, 575–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gille H. and Downward,J. (1999) Multiple ras effector pathways contribute to G(1) cell cycle progression. J. Biol. Chem., 274, 22033–22040. [DOI] [PubMed] [Google Scholar]
- Hanlon M. and Sealy,L. (1999) Ras regulates the association of serum response factor and CCAAT/enhancer-binding protein beta. J. Biol. Chem., 274, 14224–14228. [DOI] [PubMed] [Google Scholar]
- Hill C.S. and Treisman,R. (1995) Differential activation of c-fos promoter elements by serum, lysophosphatidic acid, G proteins and polypeptide growth factors. EMBO J., 14, 5037–5047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C.S., Marais,R., John,S., Wynne,J., Dalton,S. and Treisman,R. (1993) Functional analysis of a growth factor-responsive transcription factor complex. Cell, 73, 395–406. [DOI] [PubMed] [Google Scholar]
- Hill C.S., Wynne,J. and Treisman,R. (1994) Serum-regulated transcription by serum response factor (SRF): a novel role for the DNA binding domain. EMBO J., 13, 5421–5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C.S., Wynne,J. and Treisman,R. (1995) The Rho family GTPases RhoA, Rac1 and CDC42Hs regulate transcriptional activation by SRF. Cell, 81, 1159–1170. [DOI] [PubMed] [Google Scholar]
- Impey S., Obrietan,K., Wong,S.T., Poser,S., Yano,S., Wayman,G., Deloulme,J.C., Chan,G. and Storm,D.R. (1998) Cross talk between ERK and PKA is required for Ca2+ stimulation of CREB-dependent transcription and ERK nuclear translocation. Neuron, 21, 869–883. [DOI] [PubMed] [Google Scholar]
- Jhun B.H., Rose,D.W., Seely,B.L., Rameh,L., Cantley,L., Saltiel,A.R. and Olefsky,J.M. (1994) Microinjection of the SH2 domain of the 85-kilodalton subunit of phosphatidylinositol 3-kinase inhibits insulin-induced DNA synthesis and c-fos expression. Mol. Cell. Biol., 14, 7466–7475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W., Kahn,S.M., Zhou,P., Zhang,Y.J., Cacace,A.M., Infante,A.S., Doi,S., Santella,R.M. and Weinstein,I.B. (1993) Overexpression of cyclin D1 in rat fibroblasts causes abnormalities in growth control, cell cycle progression and gene expression. Oncogene, 8, 3447–3457. [PubMed] [Google Scholar]
- Jimenez C. et al. (1998) Identification and characterization of a new oncogene derived from the regulatory subunit of phosphoinositide 3-kinase. EMBO J., 17, 743–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen F.E. and Prywes,R. (1993) Identification of transcriptional activation and inhibitory domains in serum response factor (SRF) by using GAL4-SRF constructs. Mol. Cell. Biol., 13, 4640–4647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen F.E. and Prywes,R. (1994) Two pathways for serum regulation of the c-fos serum response element require specific sequence elements and a minimal domain of serum response factor. Mol. Cell. Biol., 14, 5920–5928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klippel A., Reinhard,C., Kavanaugh,W.M., Apell,G., Escobedo,M.A. and Williams,L.T. (1996) Membrane localization of phosphatidyl inositol 3-kinase is sufficient to activate multiple signal-transducing kinase pathways. Mol. Cell. Biol., 16, 4117–4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klippel A., Escobedo,M.A., Wachowicz,M.S., Apell,G., Brown,T.W., Giedlin,M.A., Kavanaugh,W.M. and Williams,L.T. (1998) Activation of phosphatidylinositol 3-kinase is sufficient for cell cycle entry and promotes cellular changes characteristic of oncogenic transformation. Mol. Cell. Biol., 18, 5699–5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T. and Cohen,P. (1999) Activation of serum- and glucocorticoid-regulated protein kinase by agonists that activate phosphatidylinositide 3-kinase is mediated by 3- phosphoinositide-dependent protein kinase-1 (PDK1) and PDK2. Biochem. J., 339, 319–328. [PMC free article] [PubMed] [Google Scholar]
- Kovary K. and Bravo,R. (1991) The jun and fos protein families are both required for cell cycle progression in fibroblasts. Mol. Cell. Biol., 11, 4466–4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S.H., Lee,H.H., Chen,J.J., Chuang,C.F. and Ng,S.Y. (1994) Serum response element-regulated transcription in the cell cycle: possible correlation with microtubule reorganization. Cell Growth Differ., 5, 447–455. [PubMed] [Google Scholar]
- Mark M.D., Liu,Y., Wong,S.T., Hinds,T.R. and Storm,D.R. (1995) Stimulation of neurite outgrowth in PC12 cells by EGF and KCl depolarization: a Ca(2+)-independent phenomenon. J. Cell Biol., 130, 701–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao G.G. and Curran,T. (1994) Cell transformation by c-fos requires an extended period of expression and is independent of the cell cycle. Mol. Cell. Biol., 14, 4295–4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranti C.K., Ginty,D.D., Huang,G., Chatila,T. and Greenberg,M.E. (1995) Calcium activates serum response factor-dependent transcription by a Ras- and Elk-1-independent mechanism that involves a Ca2+/calmodulin-dependent kinase. Mol. Cell. Biol., 15, 3672–3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montmayeur J.P., Valius,M., Vandenheede,J. and Kazlauskas,A. (1997) The platelet-derived growth factor beta receptor triggers multiple cytoplasmic signaling cascades that arrive at the nucleus as distinguishable inputs. J. Biol. Chem., 272, 32670–32678. [DOI] [PubMed] [Google Scholar]
- Musgrove E.A., Lee,C.S., Buckley,M.F. and Sutherland,R.L. (1994) Cyclin D1 induction in breast cancer cells shortens G1 and is sufficient for cells arrested in G1 to complete the cell cycle. Proc. Natl Acad. Sci. USA, 91, 8022–8026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers M.P., Pass,I., Batty,I.H., Van der Kaay,J., Stolarov,J.P., Hemmings,B.A., Wigler,M.H., Downes,C.P. and Tonks,N.K. (1998) The lipid phosphatase activity of PTEN is critical for its tumor supressor function. Proc. Natl Acad. Sci. USA, 95, 13513–13518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman C., Runswick,M., Pollock,R. and Treisman,R. (1988) Isolation and properties of cDNA clones encoding SRF, a transcription factor that binds to the c-fos serum response element. Cell, 55, 989–1003. [DOI] [PubMed] [Google Scholar]
- Park J., Leong,M.L., Buse,P., Maiyar,A.C., Firestone,G.L. and Hemmings,B.A. (1999) Serum and glucocorticoid-inducible kinase (SGK) is a target of the PI 3-kinase-stimulated signaling pathway. EMBO J., 18, 3024–3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rameh L.E. and Cantley,L.C. (1999) The role of phosphoinositide 3-kinase lipid products in cell function. J. Biol. Chem., 274, 8347–8350. [DOI] [PubMed] [Google Scholar]
- Riabowol K.T., Vosatka,R.J., Ziff,E.B., Lamb,N.J. and Feramisco,J.R. (1988) Microinjection of fos-specific antibodies blocks DNA synthesis in fibroblast cells. Mol. Cell. Biol., 8, 1670–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche S., Koegl,M. and Courtneidge,S.A. (1994) The phosphatidyl inositol 3-kinase alpha is required for DNA synthesis induced by some, but not all, growth factors. Proc. Natl Acad. Sci. USA, 91, 9185–9189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeyne R.J., Schilling,K., Oberdick,J., Robertson,L., Luk,D., Curran,T. and Morgan,J.I. (1993) A fos–lacZ transgenic mouse that can be used for neuroanatomic mapping. Adv. Neurol., 59, 285–291. [PubMed] [Google Scholar]
- Sotiropoulos A., Gineitis,D., Copeland,J. and Treisman,R. (1999) Signal-regulated activation of serum response factor is mediated by changes in actin dynamics. Cell, 98, 159–169. [DOI] [PubMed] [Google Scholar]
- Sugimoto Y., Whitman,M., Cantley,L.C. and Erikson,R.L. (1984) Evidence that the Rous sarcoma virus transforming gene product phosphorylates phosphatidylinositol and diacylglycerol. Proc. Natl Acad. Sci. USA, 81, 2117–2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takuwa N., Fukui,Y. and Takuwa,Y. (1999) Cyclin D1 expression mediated by phosphatidylinositol 3-kinase through mTOR-p70(S6K)-independent signaling in growth factor-stimulated NIH 3T3 fibroblasts. Mol. Cell. Biol., 19, 1346–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treisman R. (1994) Ternary complex factors: growth factor regulated transcriptional activators. Curr. Opin. Genet. Dev., 4, 96–101. [DOI] [PubMed] [Google Scholar]
- Treisman R. (1995) Journey to the surface of the cell: Fos regulation and the SRE. EMBO J., 14, 4905–4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urich M., Senften,M., Shaw,P.E. and Ballmer-Hofer,K. (1997) A role for the small GTPase Rac in polyomavirus middle-T antigen-mediated activation of the serum response element and in cell transformation. Oncogene, 14, 1235–1241. [DOI] [PubMed] [Google Scholar]
- Valius M. and Kazlauskas,A. (1993) Phospholipase C-γ1 and phosphatidylinositol 3 kinase are the downstream mediators of the PDGF receptor’s mitogenic signal. Cell, 73, 321–334. [DOI] [PubMed] [Google Scholar]
- Wang Y., Falasca,M., Schlessinger,J., Malstrom,S., Tsichlis,P., Settleman,J., Hu,W., Lim,B. and Prywes,R. (1998) Activation of the c-fos serum response element by phosphatidyl inositol 3-kinase and rho pathways in HeLa cells. Cell Growth Differ., 9, 513–522. [PubMed] [Google Scholar]
- Whitmarsh A.J., Shore,P., Sharrocks,A.D. and Davis,R.J. (1995) Integration of MAP kinase signal transduction pathways at the serum response element. Science, 269, 403–407. [DOI] [PubMed] [Google Scholar]
- Winston J.T. and Pledger,W.J. (1993) Growth factor regulation of cyclin D1 mRNA expression through protein synthesis-dependent and -independent mechanisms. Mol. Biol. Cell, 4, 1133–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright J.H., Munar,E., Jameson,D.R., Andreassen,P.R., Margolis,R.L., Seger,R. and Krebs,E.G. (1999) Mitogen-activated protein kinase kinase activity is required for the G(2)/M transition of the cell cycle in mammalian fibroblasts. Proc. Natl Acad. Sci. USA, 96, 11335–11340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi K., Holt,K. and Pessin,J.E. (1993) Phosphatidylinositol 3-kinase functions upstream of Ras and Raf in mediating insulin stimulation of c-fos transcription. J. Biol. Chem., 268, 14597–14600. [PubMed] [Google Scholar]