Abstract
In Saccharomyces cerevisiae the rate of DNA replication is slowed down in response to DNA damage as a result of checkpoint activation, which is mediated by the Mec1 and Rad53 protein kinases. We found that the Srs2 DNA helicase, which is involved in DNA repair and recombination, is phosphorylated in response to intra-S DNA damage in a checkpoint-dependent manner. DNA damage-induced Srs2 phosphorylation also requires the activity of the cyclin-dependent kinase Cdk1, suggesting that the checkpoint pathway might modulate Cdk1 activity in response to DNA damage. Moreover, srs2 mutants fail to activate Rad53 properly and to slow down DNA replication in response to intra-S DNA damage. The residual Rad53 activity observed in srs2 cells depends upon the checkpoint proteins Rad17 and Rad24. Moreover, DNA damage-induced lethality in rad17 mutants depends partially upon Srs2, suggesting that a functional Srs2 helicase causes accumulation of lethal events in a checkpoint-defective context. Altogether, our data implicate Srs2 in the Mec1 and Rad53 pathway and connect the checkpoint response to DNA repair and recombination.
Keywords: checkpoint/DNA recombination/DNA repair/DNA replication/Srs2
Introduction
All living cells respond to DNA damage by promoting the transcription of several DNA metabolism genes and by coordinating chromosome replication and segregation with DNA repair and recombination (for reviews see Carr and Hoekstra, 1995; Paulovich et al., 1997; Weinert, 1998; Lowndes and Murguia, 2000). Many of the genes involved in the DNA damage checkpoint pathway, which is specifically responsible for delaying cell cycle progression in response to DNA damage in eukaryotic cells, have been identified in budding and in fission yeast (Foiani et al., 2000). Most of these genes have been highly conserved throughout evolution and their function is important for preventing genome instability and cancer in mammalian cells (Hartwell and Kastan, 1994; Weinert, 1997).
It is generally believed that the DNA damage checkpoint pathway is necessary to delay the cell cycle in response to DNA damage in order to provide the cell with enough time to repair the lesions prior to chromosome replication or segregation. Once the DNA damage has been removed, the cell can re-establish a normal cell cycle through a process known as recovery (Sandell and Zakian, 1993). Recent evidence indicates that the recovery process is associated with checkpoint inactivation (Pellicioli et al., 1999), although the nature of this mechanism is still unclear. Moreover, in the presence of an irreparable DNA lesion, the cells do not remain permanently arrested and they resume cell cycle progression through a process known as adaptation (Sandell and Zakian, 1993; Toczyski et al., 1997; Lee et al., 1998).
In the yeast Saccharomyces cerevisiae the DNA damage checkpoint pathway is controlled by a cascade of phosphorylation events mediated principally by the MEC1, RAD53 and DUN1 gene products (Foiani et al., 2000). Mec1 is a member of the evolutionarily conserved subfamily of phosphatidylinositol 3-kinase (PI3-kinase) that includes budding yeast Tel1, fission yeast Rad3, mammalian ATM and ATR and DNA-dependent protein kinase (DNA-PK) (Elledge, 1996). It is generally assumed that Mec1 is a protein kinase, but in the absence of direct biochemical evidence the physiological targets of Mec1 remain speculative. The Rad53 protein kinase is highly homologous to human Chk2 and Schizosaccharomyces pombe Cds1 (Lowndes and Murguia, 2000), and it is phosphorylated and activated in response to DNA damage through a process that requires a functional Mec1 (Sanchez et al., 1996; Sun et al., 1996). The C-terminal of Rad53 contains a forkhead-associated domain that mediates the interaction with Rad9, another checkpoint protein (Sun et al., 1998). Rad53 is also required for phosphorylation of Dun1, another protein kinase involved in the checkpoint response (Zhou and Elledge, 1993; Gardner et al., 1999). Dun1 plays a major role in the transcriptional induction of several DNA metabolism genes in response to genotoxic treatments (Zhou and Elledge, 1993) and in channelling DNA repair into a non-recombinational pathway (Fasullo et al., 1999).
Other factors involved in the DNA damage response include Mec3, Ddc1, Rad17 and Rad24. These proteins are absolutely required for checkpoint activation in G1, while they are only partially needed in response to DNA damage during S phase (Pellicioli et al., 1999). Even though the role of these checkpoint proteins is still unknown, it has been suggested that they might participate in DNA damage recognition and/or processing (Lydall and Weinert, 1997; Foiani et al., 2000; Lowndes and Murguia, 2000). DNA polymerase ε, replication factor C (RF-C) and the DNA helicase Sgs1 have also been implicated in the checkpoint response and in Rad53 activation (Navas et al., 1995; Sugimoto et al., 1997; Frei and Gasser, 2000; Pellicioli et al., 1999). DNA polymerase ε and RF-C are required for DNA replication (Jonsson and Hubscher, 1997), although the catalytic domain of DNA polymerase ε appears to be dispensable (Kesti et al., 1999). The role of Sgs1 seems to be more complex since its function has been invoked not only in checkpoint activation (Frei and Gasser, 2000), but also in DNA recombination, replication and transcription (Chakraverty and Hickson, 1999; Lee et al., 1999; Gangloff et al., 2000).
While significant progress has been made in identifying some of the factors acting in the DNA damage checkpoint pathway, very little is known about the physiological targets of this signal transduction cascade. The DNA replication machinery seems to be one of the final targets of DNA damage checkpoint, which expands the length of S phase in the presence of genotoxic agents (Paulovich and Hartwell, 1995). In fact, two essential replication factors, replication protein A (RP-A) and the DNA polymerase α–primase complex (pol–prim), are modulated by the checkpoint response (Brush et al., 1996; Pellicioli et al., 1999). RP-A is involved in replication, recombination and repair (Wold, 1997) and, in response to DNA damage, it is phosphorylated through a mechanism dependent upon Mec1, but not on Rad53 (Brush et al., 1996). The pol–prim complex, which is required for initiation of DNA synthesis at origins of replication and for lagging strand DNA synthesis (Foiani et al., 1997), is also involved in the checkpoint pathway (Marini et al., 1997) and is kept unphosphorylated under damaging conditions (Pellicioli et al., 1999). Moreover, it has been shown that the cyclin-dependent kinases Cdc7/Dbf4 and Cdc28/Cyclin B (Cdk1), which control DNA synthesis under normal conditions, are also regulated in response to DNA damage checkpoint activation (Cheng et al., 1999; Dohrmann et al., 1999; Pellicioli et al., 1999). However, it is not yet clear whether the delay in S phase progression caused by checkpoint activation in response to genotoxic treatments is simply due to negative regulation of the replication process or, instead, results from intrinsically slow replication mechanisms that couple replication to recombination and repair (Foiani et al., 2000). In fact, several pieces of data indicate that prokaryotic and eukaryotic cells use specialized replication mechanisms such as template switching or break-induced replication (BIR) to replicate a damaged template (Higgins et al., 1976; Malkova et al., 1996; Kogoma, 1997; Holmes and Haber, 1999). These two processes need the function of proteins involved in leading and lagging strand synthesis and, therefore, require a reprogramming of the replication machinery since most of the factors involved are the same as those used under normal conditions. Strand switching models, leading to the formation of Holliday junctions by annealing of the two newly synthesized strands upon encounter of replication forks with DNA lesions, have been proposed to explain translesion synthesis in Escherichia coli and mammalian cells (Higgins et al., 1976; Seigneur et al., 1998) and to account for the accumulation of recombination intermediates in certain yeast replication mutants (Zou and Rothstein, 1997). BIR has been demonstrated in prokaryotes (Kogoma, 1997) and yeast cells (Malkova et al., 1996). Although these replication-coupled recombination processes may be responsible for the increase in the length of S phase as a consequence of genotoxic treatments, so far there are no indications that they are regulated by the checkpoint response.
In this paper we show that the SRS2 gene product is a regulatory target of the checkpoint response. Srs2 is a DNA helicase with 3′–5′ polarity (Rong and Klein, 1993) and mutations in the SRS2 gene result in an increased rate of gene conversion (Rong et al., 1991). Srs2 has been implicated in DNA repair (Aboussekhra et al., 1989) and recombination (Pâques and Haber, 1997). Here we show that Srs2 is phosphorylated in response to DNA damage and that this modification is dependent upon a functional checkpoint pathway and on Cdk1 activity. Moreover, we provide evidence that srs2 mutants are unable to activate Rad53 properly in response to intra-S DNA damage and consequently are defective in slowing down the DNA replication process. Furthermore, our findings suggest that a functional Srs2 helicase causes lethal events in a rad17 mutant background in response to intra-S DNA damage. Altogether our data implicate Srs2 helicase in the DNA damage checkpoint response.
Results
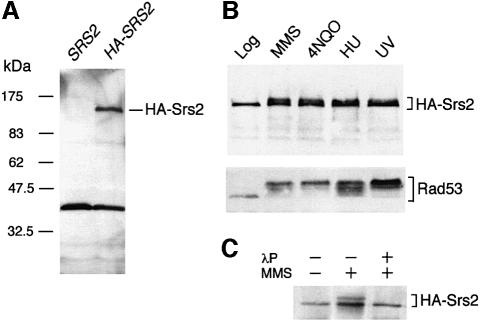
We have produced a HA-tagged version of the S.cerevisiae SRS2 gene to analyse the level and modifications of the corresponding gene product under normal growth conditions and in response to DNA damage. The HA-tagged SRS2 gene behaves like wild type both under normal growing conditions and in response to DNA damage (data not shown). Western blot analysis performed on a crude extract, prepared from logarithmically growing cells carrying the HA-SRS2 gene, revealed a major polypeptide with an apparent mol. wt of 140 kDa (Figure 1A). This immunoreactive polypeptide was not present in extracts prepared from untagged cells and its size is that predicted for a fusion protein carrying three copies of the HA epitope.
Fig. 1. HA-SRS2 is phosphorylated in response to DNA damage. (A) Aliquots of total protein extracts prepared from strains K699 (SRS2) and CY2715 (HA-SRS2), as described in Materials and methods, were separated by SDS–PAGE and analysed by western blotting with the 12CA5 monoclonal antibody (mAb) directed against the HA epitope tag. (B) Western blot analysis with 12CA5 mAb and anti-Rad53 polyclonal antibodies (Ab) was performed on aliquots of total protein extracts prepared from CY2715 cells after treatment with 0.02% MMS for 3 h, 2 µg/ml 4-NQO for 1 h, 0.2 M HU for 3 h or 100 J/m2 UV. (C) HA-Srs2 immunoprecipitates, obtained from CY2715 cells grown in the presence or absence of MMS, were analysed by western blotting with the 12CA5 mAb. Where indicated, the HA-Srs2 immunoprecipitate was treated with λ-phosphatase (λP) prior to gel loading.
Since Srs2 is involved in DNA repair, we tested whether the level of the protein increased in cells treated with a variety of DNA damaging agents, and/or whether the protein was post-translationally modified under these conditions. As shown in Figure 1B, the amount of HA-Srs2 increased slightly (2- to 4-fold) in response to DNA damaging treatments, similar to that found by measuring SRS2-lacZ expression after UV irradiation (Heude et al., 1995). Moreover, an additional immunoreactive band migrating more slowly than the HA-Srs2 polypeptide found in extracts from untreated cells was clearly visible after genotoxic treatments. This modified polypeptide was no longer detectable after phosphatase treatment (Figure 1C), indicating that it represents a HA-Srs2 phosphorylated isoform.
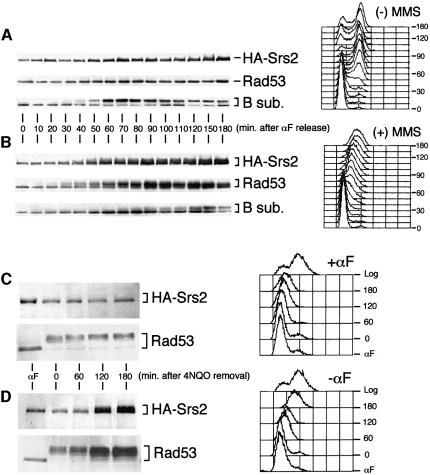
We then analysed the HA-Srs2 phosphorylation state in synchronized cells during an unperturbed cell cycle and in response to DNA damage. As shown in Figure 2A, under normal conditions, both HA-Srs2 and the checkpoint protein kinase Rad53 were present in their unphosphorylated forms at any stage of the cell cycle, whereas the B subunit of the pol–prim complex was clearly modified during S phase (Foiani et al., 1995). The level of the three proteins approximately doubled when cells were entering S phase, a finding that is likely to be related to the presence of an MCB box in the promoter of the corresponding genes (Johnston and Lowndes, 1992). When wild-type cells were released from the G1 block in the presence of sublethal concentrations of methyl methane sulfonate (MMS) the intra-S DNA damage checkpoint was induced, leading to phosphorylation and activation of the Rad53 protein kinase (Figure 2B; Pellicioli et al., 1999) and to a delay in pol–prim B subunit phosphorylation (compare western blots in Figure 2A and B and Pellicioli et al., 1999). As a consequence of checkpoint activation, the kinetics of S phase progression was dramatically slowed (Figure 2A and B). In response to MMS treatment, HA-Srs2 became phosphorylated and this modification occurred slightly later than Rad53 activation, raising the possibility that HA-Srs2 phosphorylation might be mediated by the checkpoint response. However, when treatment with the DNA damaging agent 4-nitroquinoline-N-oxide (4NQO) was carried out on α-factor-arrested cells that were kept in G1 by maintaining the α-factor block, cells were able to activate Rad53, but failed to phosphorylate HA-Srs2 (Figure 2C). Conversely, when α-factor-arrested cells were treated with 4NQO and then allowed to proceed through the cell cycle after removing the G1 block and the DNA damaging agent, Rad53 remained active throughout the experiment, while HA-Srs2 became phosphorylated at the approximate time of S phase entry and remained phosphorylated for at least 3 h (Figure 2D). This finding indicates that HA-Srs2 cannot be modified in response to DNA damage in G1, but phosphorylation can occur only when cells are allowed to proceed further in the cell cycle. It is possible that in G1 cells either the kinase responsible for HA-Srs2 phosphorylation is limiting, or HA-Srs2 is not available for modification.
Fig. 2. HA-Srs2 phosphorylation requires checkpoint activation and entry into S phase. (A and B) Aliquots of protein extracts, prepared from CY2715 cells taken at the indicated time after α-factor (αF) release in the absence (A) or presence (B) of 0.02% MMS, were analysed by western blotting with 12CA5 mAb, anti-Rad53 Ab and the 6D2 mAb recognizing the pol–prim B subunit. Aliquots of cells taken at the indicated time were also processed for FACS analysis as described in Materials and methods. (C and D) CY2715 cells were blocked in G1 by αF treatment, as described in Materials and methods. G1-arrested cells were treated for 15 min with 0.25 µg/ml 4-NQO; the drug was then removed by washing the cells with YPD containing (C) or not containing (D) αF to maintain the G1 block or to allow cell cycle progression. At the indicated times after 4-NQO removal, aliquots of protein extracts were analysed by western blotting with 12CA5 mAb and anti-Rad53 Ab. At the same time points, cell samples were also processed for FACS analysis.
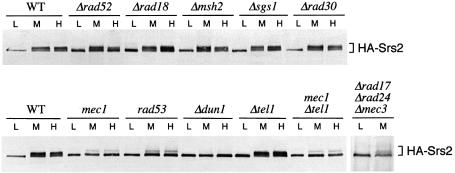
To identify genetically the pathway required for HA-Srs2 phosphorylation, we tested whether this modification was impaired in certain genetic backgrounds defective in DNA repair or in the DNA damage checkpoint. As shown in Figure 3, Δrad52, Δmsh2, Δrad18, Δsgs1 and Δrad30 mutant cells, which are defective in different repair pathways (Friedberg et al., 1995), were still able to phosphorylate HA-Srs2 in response to hydroxyurea (HU) and MMS treatments. Conversely, DNA damage-induced HA-Srs2 phosphorylation was prevented or strongly reduced in mec1, rad53 and Δdun1 mutant cells, which are defective in three protein kinases required for a proper DNA damage response (Figure 3). This finding indicates that a functional checkpoint is required for HA-Srs2 phosphorylation in response to MMS and HU treatments. The observation that HA-Srs2 was still modified in Δtel1 cells and that mec1 and Δtel1 mec1 mutants exhibited the same level of residual phosphorylation of Srs2 suggests that Tel1 is not required for Srs2 phosphorylation. Several factors including Rad17, Mec3, Ddc1, Rad24 and Rad9 are required for proper Rad53 activation in response to MMS treatment, while they appear to be dispensable in the presence of HU (Pellicioli et al., 1999). The finding that in a triple Δrad17 Δrad24 Δmec3 mutant strain HA-Srs2 phosphorylation was reduced in response to MMS treatment (Figure 3) further confirms that HA-Srs2 phosphorylation is dependent upon a functional checkpoint pathway.
Fig. 3. The checkpoint kinases Mec1, Rad53 and Dun1 are required for HA-Srs2 phosphorylation in response to DNA damage. Cultures of strains CY2715 (WT), CY2823 (Δrad52), CY2822 (Δrad18), CY2884 (Δmsh2), CY3137 (Δsgs1), CY3135 (Δrad30), CY2835 (mec1), CY2837 (rad53), CY3138 (Δdun1), CY2885 (Δtel1), CY2888 (mec1Δtel1) and CY2827 (Δrad17Δrad24Δmec3) were collected in log phase (L) or treated for 3 h with 0.02% MMS (M) or 0.2 M HU (H). Aliquots of protein extracts were analysed by western blotting with the 12CA5 mAb.
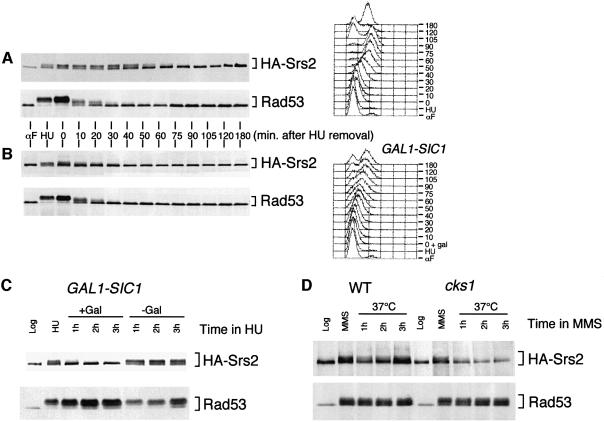
We then analysed the Srs2 phosphorylation state during the recovery process, which allows cells to restore cell cycle progression once the genotoxic stress has been removed. Wild-type cells were treated with HU to activate the checkpoint fully and then released from the HU block to allow recovery. Under these conditions, Rad53 is rapidly inactivated (Pellicioli et al., 1999). As shown in Figure 4A, HA-Srs2 was still phosphorylated at 40–50 min after HU removal, while, at the same time, Rad53 was completely dephosphorylated and its kinase activity strongly reduced (data not shown).
Fig. 4. HA-Srs2 is phosphorylated during recovery from HU and its phosphorylation requires a functional Cdk1. (A and B) A log-phase culture of strain CY2735 (GAL1-SIC1) was grown in raffinose, presynchronized by αF treatment and released from the G1 block in 0.2 M HU-containing media. Half of the culture was maintained in raffinose (A), while galactose was added to the other half of the culture to overexpress Sic1 (B). The HU block was then removed by washing the cells with YPD containing either raffinose (A) or galactose (B). Samples were taken at the time points indicated and processed for FACS or for protein extraction. Aliquots of protein extracts were analysed by western blotting performed with the 12CA5 mAb and with anti-Rad53 Ab. (C) Log-phase cultures (Log) of strain CY2735 (GAL1-SIC1) were grown in raffinose and treated with 0.2 M HU for 3 h. The culture was then divided into two parts, which were maintained in HU with (+Gal) or without (–Gal) galactose addition to induce Sic1 expression. Aliquots of protein extracts were analysed by western blotting with the 12CA5 mAb and anti-Rad53 Ab. (D) Log-phase cultures of strains CY2829 (WT) and CY2830 (cks1) were grown at 25°C and treated for 3 h with 0.02% MMS. The cultures were then shifted to 37°C in the presence of the drug. Aliquots of protein extracts were analysed by western blot analysis performed with the 12CA5 mAb and anti-Rad53 Ab.
We have recently found that Rad53 probably modulates the activity of the Cdc28 protein kinase in response to DNA damage (Pellicioli et al., 1999). We thus tested whether Cdk1 was playing a role in HA-Srs2 phosphorylation by overexpressing the Cdk1 inhibitor Sic1 when the cells were recovering from the HU block. Cdk1 inhibition caused a faster dephosphorylation of HA-Srs2, while the kinetics of Rad53 dephosphorylation was unaffected (Figure 4B). Analogous results on the timing of HA-Srs2 dephosphorylation were obtained by overexpressing the Cdc14 phosphatase, another Cdk1 inhibitor (data not shown). Cdk1 inhibition by Sic1 overexpression also caused a slower progression through S phase (Figure 4B), probably because the timing of late origin firing was altered under these conditions. To test directly the effect of Cdk1 inhibition on HA-Srs2 dephosphorylation in the absence of any cell cycle effect, Sic1 was overexpressed in HU-arrested cells. As shown in Figure 4C, HA-Srs2 became dephosphorylated in HU-blocked cells, while the Rad53 kinase remained fully phosphorylated and active (Figure 4C and data not shown). Again, identical results were obtained by overexpressing the Cdc14 phosphatase (data not shown). To further confirm the involvement of Cdk1 in Srs2 modification, we tested whether Srs2 phosphorylation was dependent upon the Cdk1-associated protein Cks1, which is required for kinase function of Cdk1 (Hadwiger et al., 1989). cks1 temperature-sensitive mutant cells were treated with MMS at the permissive temperature to allow checkpoint activation, and then the culture was shifted to the restrictive temperature to inactivate Cks1. As shown in Figure 4D, we found that Cks1 inactivation causes Srs2 dephosphorylation without interfering with the Rad53 phosphorylation state. This result suggests that a functional Cks1 is required to maintain Srs2 phosphorylation in response to DNA damage.
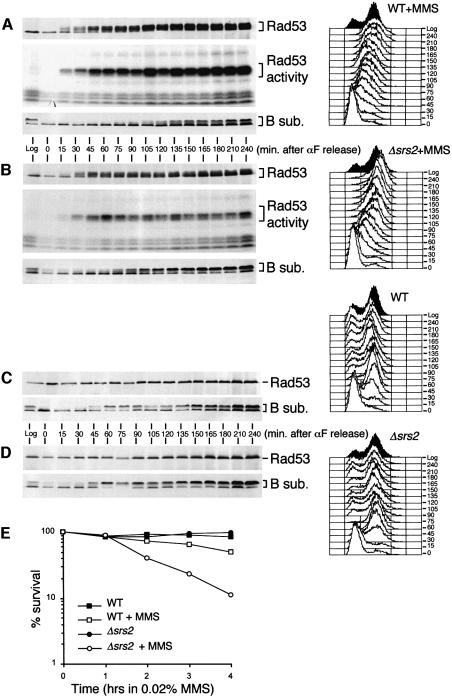
All the data presented until now indicate that HA-Srs2 modification depends upon a functional checkpoint and is mediated by Cdk1, whose activity is in turn modulated by the checkpoint response. To test more directly a possible role of SRS2 in the checkpoint response, Δsrs2 cells were arrested in G1 by α-factor treatment and released from the G1 block in the presence of MMS. As shown in Figure 5A and B, the timing of Rad53 activation is not significantly altered in Δsrs2 cells compared with wild-type cells. However, the level of Rad53 kinase activity was reduced in MMS-treated Δsrs2 cells, although the level of the protein was the same in wild-type and Δsrs2 cells (Figure 5A and B). We have previously shown that the defective intra-S DNA damage checkpoint response of a rad53 mutant allele correlates with premature phosphorylation of the pol–prim B subunit (Pellicioli et al., 1999). We found that the inability to activate Rad53 properly in Δsrs2 cells also correlates with premature phosphorylation of the B subunit, faster cell cycle progression and increased cell lethality (Figure 5A, B and E). However, Rad53 was properly activated in Δsrs2 cells treated with 4NQO or UV radiation in G1-blocked cells (data not shown). Further more, under unperturbed conditions, progression through the cell cycle was indistinguishable in wild-type and Δsrs2 cells, and Rad53 remained unphosphorylated and maintained the same basal level of kinase activity (Figure 5C and D and data not shown).
Fig. 5. Srs2 is required for proper Rad53 activation in response to DNA damage and it is dispensable for cell cycle progression under unperturbed conditions. (A–E) Log-phase cultures (Log) of strain K699 (WT) and CY2643 (Δsrs2) were synchronized by αF treatment and released from the G1 block in the presence (A and B) or absence (C and D) of 0.02% MMS. At the indicate time points, aliquots of cells were taken for FACS analysis and protein extraction. Aliquots of the protein exctracts were analysed by western blotting using the 12CA5 and 6D2 mAbs and anti-Rad53 Ab and tested using the ISA assay (A and B). Cell survival (E) was determined as plating efficiency on YPD plates.
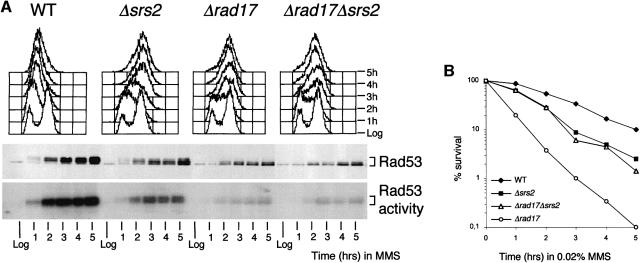
We then tested whether the residual Rad53 activity observed in MMS-treated Δsrs2 cells was dependent upon the checkpoint activating factor Rad17 or Rad24. We found that, in response to MMS treatment, the decrease in the level of Rad53 activity and the rate of S phase progression were indistinguishable in Δrad17 and Δrad17Δsrs2 cells, which were both more defective than Δsrs2 cells (Figure 6A). Therefore, at least for the checkpoint function, RAD17 seems to be epistatic to SRS2. However, cell viability in Δsrs2 and Δsrs2Δrad17 mutant cells was higher than in Δrad17 mutants (Figure 6B), suggesting that cell lethality caused by intra-S DNA damage in a Δrad17 mutant background is due partially to a functional Srs2 protein.
Fig. 6. SRS2 deletion partially rescues the MMS sensitivity but not the checkpoint defect of Δrad17 mutant cells. (A and B) Log-phase cultures (Log) of strains K699 (WT), CY2643 (Δsrs2), DMP1913/11C (Δrad17), CY3221(Δrad17Δsrs2) were treated with 0.02% MMS. Samples were taken at the times indicated for FACS analysis. Aliquots of protein extracts were analysed by western blotting performed using anti-Rad53 Ab and were used for the ISA assay (A). Cell survival (B) was determined as plating efficiency on YPD plates.
Analogous results were obtained when we tested the epistasis relationship between RAD24 and SRS2 (data not shown).
Discussion
The cellular response to DNA damage, which is mediated by the Mec1, Rad53 and Dun1 protein kinases, allows cell survival by delaying cell cycle progression and by promoting transcription of several genes involved in DNA metabolism. Thus, the repair machinery is able to remove the DNA lesions and the cell can recover by restoring a normal cell cycle progression.
Although an intimate relationship between DNA repair and DNA damage checkpoint is somehow expected, it is still unclear whether the DNA damage checkpoint is directly involved in promoting specific repair pathways. Similarly, it is presently unknown whether the recovery process is dependent upon DNA repair or, instead, the checkpoint is turned off in a time-dependent manner.
We have recently suggested a role for Mec1 and Rad53 in triggering a replication-coupled repair pathway requiring the lagging strand DNA replication apparatus (Foiani et al., 2000), but direct experimental evidence connecting the checkpoint to the repair machinery is still lacking.
The Srs2 DNA helicase is a likely candidate to connect several DNA transactions. In fact, Srs2 seems to be involved in channelling damaged DNA towards specific repair pathways (Aboussekhra et al., 1989; Schiestl et al., 1990; Rong et al., 1991) and in stabilizing some recombination intermediates (Pâques and Haber, 1997). srs2 mutants are also defective in DNA replication and transcription (Lee et al., 1999), although this phenotype is probably caused by an indirect effect due to unrestrained recombination (Gangloff et al., 2000). Here we have shown that the Srs2 DNA helicase is phosphorylated in response to DNA damage and that this modification requires a functional checkpoint pathway, thus suggesting that Srs2 may directly connect the checkpoint pathway to the repair process.
Our data indicate that damage-induced Srs2 phosphorylation is mediated by the Mec1, Rad53 and Dun1 protein kinases, but it does not occur in G1. Several observations suggest that Srs2 is not a Rad53 substrate: (i) Rad53 is activated in G1-arrested cells in response to DNA damage, but Srs2 remains unphosphorylated; (ii) Rad53 is fully functional in Δdun1 cells treated with genotoxic agents (data not shown), but Srs2 is not phosphorylated; (iii) Rad53 is rapidly inactivated during recovery from HU, while Srs2 remains phosphorylated; (iv) Cdk1 inactivation drives Srs2 dephosphorylation even though Rad53 remains active. Dun1 has been placed downstream of Rad53 (Zhou and Elledge, 1993) and our data indicate that Srs2 phosphorylation is abolished in Δdun1 mutant cells. Dun1-dependent Srs2 phosphorylation might be explained in view of recent results that have pointed out a role for Dun1 in channelling DNA repair into a non-recombinogenic pathway (Fasullo et al., 1999). Since Srs2 plays a similar role (Aboussekhra et al., 1989), it has been suggested that the higher rate of mitotic recombination observed in Δdun1 mutants might result from the inability to phosphorylate specific repair proteins, such as the Srs2 helicase (Fasullo et al., 1999). Although we cannot exclude the possibility that Srs2 is a direct substrate of Dun1, we would like to suggest that the pathway dependent upon Mec1, Rad53 and possibly Dun1 acts to modulate Cdk1, which, either directly or indirectly, would cause Srs2 phosphorylation. Srs2 contains seven putative consensus sites for Cdk1 and their mutagenesis is under way to test directly the role of Cdk1 in Srs2 phosphorylation.
Interestingly, it has been shown recently that the Rad55 protein, which is required for double strand break repair, is also phosphorylated in response to checkpoint activation through a process that requires a functional checkpoint. However, differently from Srs2, Rad55 protein can also be phosphorylated in G1 and does not seem to play any role in Rad53 activation in response to DNA damage (Bashkirov et al., 2000).
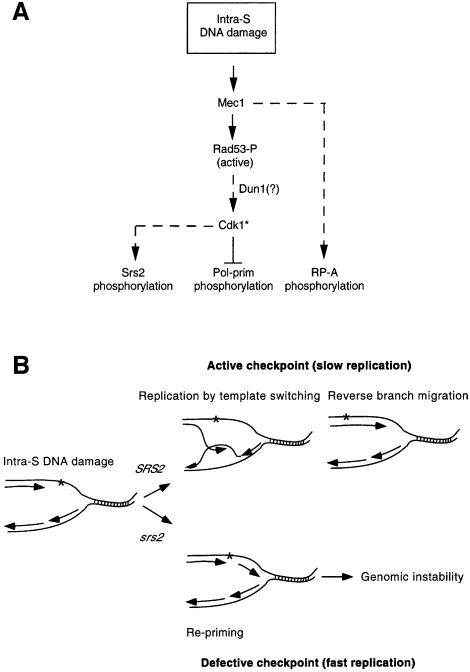
There is an apparent paradox between the observation that the phosphorylation of the pol–prim B subunit, which requires Cdk1 activity under normal conditions, is negatively regulated by the checkpoint response (Pellicioli et al., 1999), while the checkpoint-dependent Srs2 phosphorylation requires a functional Cdk1. A possible explanation could be that activation of the DNA damage checkpoint changes the substrate specificity of Cdk1 towards targets required for cell survival under damaging conditions (Figure 7A). Hence this checkpoint-dependent Cdk1 (Cdk1*) will not be able to phosphorylate the usual targets (i.e. pol–prim), but would promote the phosphorylation of specific repair proteins (i.e. Srs2). This hypothesis is in agreement with previous observations indicating that Cdc28 activity is high in HU-treated cells (Amon et al., 1992; Sorger and Murray, 1992) and might account for the finding that any attempt to override the checkpoint by ectopic activation of Cdk1 has so far been unsuccessful (Pellicioli et al., 1999).
Fig. 7. Model for the intra-S DNA damage checkpoint. (A) DNA damage during S phase leads to phosphorylation of RP-A and Rad53 mediated by Mec1. Active Rad53 modulates the target specificity of Cdk1*, causing inhibition of pol–prim phosphorylation and modification of Srs2. (B) Replication forks encountering DNA damage have the option, in the presence of a functional SRS2 gene product, to switch the template copying the newly synthesized lagging strand, thus bypassing the DNA lesion. The re-establishment of a conventional replication fork can occur by reverse branch migration. This mechanism allows duplication of a damaged template without leaving gaps opposite the lesions. In the absence of Srs2, a faster replication mechanism, involving repriming downstream of the damage, is favoured. This generates gapped DNA molecules that will be highly recombinogenic, likely to lead to increased genomic instability (see text for details).
Although at the moment we do not know the functional significance of Srs2 phosphorylation, the finding that cells overexpressing Srs2 exhibit enhanced DNA damage sensitivity (Kaytor et al., 1995), together with the observation that Srs2 plays a crucial role in channelling DNA damage to specific repair pathways (Aboussekhra et al., 1989), strongly suggest that the cell has to tightly regulate the Srs2 helicase and that its phosphorylation might be relevant to allow cell survival under damaging conditions. Since Srs2 phosphorylation is concomitant with the modulation of the replication machinery, i.e. the lagging strand DNA polymerase and RF-A (Brush et al., 1996; Pellicioli et al., 1999), it is tempting to speculate that, in response to intra-S DNA damage, the cell promotes a specialized replication-coupled repair process dependent upon phosphorylated Srs2 and the lagging strand replication apparatus.
However, it should be pointed out that Srs2 phosphorylation might be related to other cellular processes, such as recovery or adaptation. In fact, we have recently shown that, when the checkpoint is activated by HU treatment, the cell has already synthesized all the protein factors required to execute recovery (Pellicioli et al., 1999). Moreover, recent results implicate Srs2 in the adaptation response (S.E.Lee and J.E.Haber, personal communication) and, in this view, Srs2 phosphorylation might resemble Cdc5 phosphorylation, which is dependent upon Rad53 and is also required for adaptation (Toczyski et al., 1997; Sanchez et al., 1999).
The finding that Δsrs2 exhibits a reduced level of Rad53 activity under damaging conditions further enforces the connections between the checkpoint response and the Srs2 helicase, suggesting that Srs2 is indeed a relevant component of the pathway. In an attempt to elucidate the functional relationships between Srs2 and other components of the pathway, the phenotypes of a number of double mutant cells were tested. We found that in Δsrs2Δrad17 double mutant cells, the level of residual Rad53 activity and the checkpoint defect in response to MMS treatment were indistinguishable from those found in a Δrad17 single mutant. However, SRS2 deletion partially rescues the cell lethality of Δrad17 cells under damaging conditions, and the cell viability of the Δsrs2Δrad17 double mutant was comparable to that of Δsrs2 cells. This finding suggests that, in a Δrad17 background, a functional SRS2 gene product may cause lethal events. Hence, cell viability and cell cycle progression in Δrad17Δsrs2 cells seem to be uncoupled. The picture is further complicated by the observation that the DNA damage-induced cell lethality in Δsrs2 mutant cells is caused by unrestrained recombination (Milne et al., 1995; Schild, 1995; Chanet et al., 1996). Moreover, we have shown previously that Rad53 activation in response to MMS treatment depends on Rad17 completely in G1, but only partially in S phase (Pellicioli et al., 1999). Conversely, Srs2 seems to be relevant specifically in response to intra-S DNA damage.
In an attempt to reconcile our results with a role for Srs2 in preventing certain lethal recombination pathways (Aboussekhra et al., 1989) and, at the same time, in promoting the formation of other recombination intermediates (Pâques and Haber, 1997), we suggest the following model (Figure 7B).
Cells experiencing DNA damage while replicating DNA have different options (Friedberg et al., 1995). Among them, cells can restart replication downstream of the lesion through a process that requires repriming and therefore DNA primase. Such an option would inevitably lead to the formation of highly recombinogenic gaps and, therefore, massive intra-S recombination would jeopardize genome integrity. An alternative option would be to promote the formation of other recombination intermediates by annealing newly synthesized leading and lagging strands through template switching. Such intermediates (Holliday junctions) might occur spontaneously to relieve the superhelical tension that accumulates ahead of the replication fork due to the replication pausing caused by DNA damage (Doe et al., 2000). These DNA structures could then migrate through replication by allowing the leading strand to copy the newly synthesized lagging strand (Figure 7B). This mechanism of replication would prevent genotoxic recombination, allowing error-free DNA synthesis. Furthermore, once the damage has been bypassed, the cell could resolve the Holliday junctions, resetting a normal replication fork through a process that is better known as reverse branch migration (Figure 7B). Indeed, it is possible to envisage a role for a 3′–5′ DNA helicase either in promoting replication by template switching by stabilizing the Holliday junctions during branch migration, or in the subsequent reverse branch migration step. This mechanism has been proposed in mammalian cells to describe DNA replication in response to MMS treatment (Higgins et al., 1976) and to explain the role of Rqh1 helicase in S.pombe (Doe et al., 2000).
We would therefore like to suggest that the Srs2 helicase may channel intra-S DNA damage into a template switching mode of replication. In srs2 mutants, instead, replication would be achieved by extensive repriming causing accumulation of recombinogenic gaps, similar to what has already been suggested in the case of the checkpoint-defective DNA primase mutant pri1-M4 (Marini et al., 1997). This hypothesis is in accordance with a role of Srs2 during S phase and with the finding that Srs2 stabilizes certain recombination intermediates (Pâques and Haber, 1997). Moreover, this model might explain the observation that srs2 mutants exhibit unrestrained recombination in response to damaged DNA and that srs2 DNA damage sensitivity can be rescued by preventing certain recombination pathways dependent upon Rad52 and Rad51 (Milne et al., 1995; Chanet et al., 1996).
Δsrs2 cells are defective in maintaining Rad53 activity in response to intra-S DNA damage. This can be explained by assuming that Srs2 might generate checkpoint signals during DNA replication perhaps by processing the primary damage or by accumulating DNA structures that contribute to activate the pathway leading to Rad53 activation. Alternatively, since Δsrs2 cells are unable to prevent the recruitment of certain recombination pathways, some recombination factors might actively mask the checkpoint signals by binding to damaged DNA, thus causing a defect in Rad53 activation. On the other hand, it is also possible that unscheduled error-prone recombination might accelerate the processing of DNA lesions, causing premature inactivation of Rad53.
The functional relationship between Rad17 and Srs2 still remains unclear. It is possible that Rad17 plays an indirect role during S phase in restraining specific repair processes that might be dangerous in a replication context, and this function is relevant if Srs2 is functional. Indeed, it has been suggested recently that Rad17 and other checkpoint genes may play a role in accommodating DNA damage during replication by antagonizing certain recombination pathways (Paulovich et al., 1998). Hence, Rad17 and Srs2 might act in the same direction in channelling damage towards a replication-coupled repair process such as template switching. However, in a Δrad17 background the cell may not be able to prevent certain repair events, which, in the presence of a functional Srs2, might cause accumulation of abortive or abnormal repair intermediates. The finding that Srs2 triggers lethal events in a Δrad17 background could also be ascribed to an excessive amount of unphosphorylated Srs2 under damaging conditions, since in Δrad17 Srs2 does not get properly phosphorylated (Figure 3 and data not shown).
Another DNA helicase, Sgs1, has recently been implicated in the checkpoint response. Interestingly, Δsgs1 and Δsrs2 mutants exhibit intriguing analogies since they are both required to activate Rad53 properly. However, Sgs1 that has been placed upstream of Rad53 (Frei and Gasser, 2000) is not required for Srs2 phosphorylation. Moreover, we found that although Δsgs1, like Δsrs2 cells, are unable to activate Rad53 properly in response to intra-S DNA damage, Δsgs1 does not rescue the cell viability of Δrad17 mutants (our unpublished observations). It has been found recently that Δsrs2Δsgs1 double mutants show a synthetic slow growth phenotype associated with enhanced genomic instability, which can be rescued by RAD51 deletion (Gangloff et al., 2000), and it has been suggested that the S.pombe Sgs1 homologue might be involved in catalysing reverse branch migration of Holliday junctions arising from template switching (Doe et al., 2000). It will then be relevant to establish the functional relationship between Sgs1 and Srs2 within the DNA damage response pathway.
Materials and methods
Plasmids
The HA-tagged version of SRS2 gene was produced as follows. The N-terminal region of SRS2 (from –536 to +180) was amplified by PCR using the primers 5′-CCCAAGCTTCTCACGATCTACGAG ATGC-3′ (Srs-HindIII#3) and 5′-CGGGATCCGGGATGAATATGG TGGTGCT-3′ (Srs-BamHI#4), and it was cloned into the HindIII– BamHI sites of plasmid YIplac211 (Gietz and Sugino, 1998) to generate plasmid pG34. pG34 was then amplified by PCR using the primers 5′-GCGGCCGCCCATTTGCTATCCCTAAGTAC-3′ (Srs-NotI#1) and 5′-GGCGGCCGCTCGTCGAACAATGATCTTTG-3′ (Srs-NotI#2) to generate plasmid pG35, which contains a NotI site at the ATG codon of SRS2 ORF. Three tandem copies of the HA epitope were cloned into the NotI site of pG35 in-frame with SRS2 to generate plasmid pG36. All the PCRs were performed using the Pfu Turbo DNA polymerase (Stratagene) and the final products of amplification were controlled by sequence analysis.
Plasmids pMHT (GAL1-SIC1) and p100 (GAL1-CDC14) were provided by J.Diffley and A.Amon, respectively.
Yeast strains
The genotypes of the strains used are listed in Table I. Strains CY2715, CY2835, CY2827, CY2837, CY3138, CY2829 and CY2830 were originated by integrating a PstI-linearized pG36 plasmid at the SRS2 locus in strains K699, DMP2541/8A (Paciotti et al., 1998), DMP2161/25B (Paciotti et al., 1998), CY2034 (Pellicioli et al., 1999), YA145 (Fasullo et al., 1999), K5247 and K5248 (kindly provided by K.Nasmyth), respectively, and by selecting for 5-FOA-resistant cells. Correct HA-SRS2 integration was controlled by Southern analysis.
Table I. Strains used in this study.
| Strain | Genotype | Reference/source |
|---|---|---|
| K699 | MATa ade2-1 trp1-1 leu2-3,112 his3-11,15 ura3 can1-100 | K.Nasmyth |
| CY2715 | MATa ade2-1 trp1-1 leu2-3,112 his3-11,15 ura3 can1-100 HA3SRS2::srs2 | this study |
| CY2735 | MATa ade2-1 trp1-1 leu2-3,112 his3-11,15 ura3 can1-100 HA3SRS2::srs2 GAL1-SIC1::URA3 | this study |
| CY2904 | MATa ade2-1 trp1-1 leu2-3,112 his3-11,15 ura3 can1-100 HA3SRS2::srs2 GAL1-CDC14::URA3 | this study |
| CY2823 | MATa ade2-1 trp1-1 leu2-3,112 his3-11,15 ura3 can1-100 HA3SRS2::srs2 rad52Δ::HIS3MX6 | this study |
| CY2822 | MATa ade2-1 trp1-1 leu2-3,112 his3-11,15 ura3 can1-100 HA3SRS2::srs2 rad18Δ::HIS3MX6 | this study |
| CY2884 | MATa ade2-1 trp1-1 leu2-3,112 his3-11,15 ura3 can1-100 HA3SRS2::srs2 msh2Δ::HIS3MX6 | this study |
| CY3135 | MATa ade2-1 trp1-1 leu2-3,112 his3-11,15 ura3 can1-100 HA3SRS2::srs2 rad30Δ::KanMX4 | this study |
| CY3137 | MATa ade2-1 trp1-1 leu2-3,112 his3-11,15 ura3 can1-100 HA3SRS2::srs2 sgs1Δ::KanMX4 | this study |
| CY2835 | MATa ade2-1 trp1-1 leu2-3,112 his3-11,15 ura3 can1-100 HA3SRS2::srs2 HA2DDC1::LEU2::ddc1 mec1-1 sml1-1 | this study |
| CY2837 | MATa ade2-1 trp1-1 leu2-3,112 his3-11,15 ura3 can1-100 HA3SRS2::srs2 rad53-K227A::KanMX4::rad53 | this study |
| CY3138 | MATα ade2-1 trp1-1 leu2-3,112 his3-11,15 ura3-1 can1-100 HA3SRS2::srs2 dun1-Δ100::HIS3 | this study |
| CY2885 | MATa ade2-1 trp1-1 leu2-3,112 his3-11,15 ura3 can1-100 HA3SRS2::srs2 tel1Δ::HIS3MX6 | this study |
| CY2888 | MATa ade2-1 trp1-1 leu2-3,112 his3-11,15 ura3 can1-100 HA3SRS2::srs2 HA2DDC1::LEU2::ddc1 tel1Δ::HIS3MX6 mec1-1 sml1-1 | this study |
| CY2827 | MATa ade2-1 trp1-1 leu2-3,112 his3-11,15 ura3 can1-100 HA3SRS2::srs2 HA2DDC1::LEU2::ddc1 rad17Δ::LEU2 rad24Δ::TRP1 mec3Δ::TRP1 | this study |
| CY2643 | MATa ade2-1 trp1-1 leu2-3,112 his3-11,15 ura3 can1-100 srs2Δ::KanMX4 | this study |
| CY2829 | MATa ade2-1 trp1-1 leu2-3,112 his3-11,15 ura3 can1-100 HA3SRS2::srs2 cks1::LEU2 [CKS1 TRP1 ARS1 CEN4] | this study |
| CY2830 | MATa ade2-1 trp1-1 leu2-3,112 his3-11,15 ura3 can1-100 HA3SRS2::srs2 cks1::LEU2 [cks1-ts38 TRP1 ARS1 CEN4] | this study |
| DMP1913/11C | MATa ade2-1 trp1-1 leu2-3,112 his3-11,15 ura3 can1-100 rad17Δ::LEU2 | M.P. Longhese |
| CY3221 | MATa ade2-1 trp1-1 leu2-3,112 his3-11,15 ura3 can1-100 rad17Δ::LEU2 srs2Δ::KanMX4 | this study |
| DMP1913/20B | MATa ade2-1 trp1-1 leu2-3,112 his3-11,15 ura3 can1-100 rad24Δ::TRP1 | M.P. Longhese |
| CY3223 | MATa ade2-1 trp1-1 leu2-3,112 his3-11,15 ura3 can1-100 rad24Δ::TRP1 srs2Δ::KanMX4 | this study |
Plasmids are indicated by brackets.
Since strains CY2715 and K699 exhibited the same DNA damage sensitivity in response to UV and MMS treatment, we concluded that the HA-SRS2-tagged gene is fully functional. Gene disruptions were produced according to Wach et al. (1994, 1997) using either the KanMX4 or HIS3MX6 cassettes. The strains originated were controlled by genomic PCR. Strains CY2823, CY2822, CY2884, CY3135 and CY3137 were obtained by deleting the RAD52, RAD18, MSH2, RAD30 and SGS1 genes, respectively, in strain CY2715. Strains CY2888 and CY2885 were obtained by deleting the TEL1 gene in strains CY2835 and CY2715, respectively. Strains CY2643, CY3221 and CY3223 were obtained by deleting the SRS2 gene in strains K699, DMP1913/11C (kindly provided by M.P.Longhese) and DMP1913/20B (kindly provided by M.P.Longhese), respectively. Strains CY2735 and CY2904 were obtained by integrating plasmids pMHT or p100, respectively, at the URA3 locus.
Media and growth conditions
Unless otherwise indicated, strains were grown at 28°C in YP (1% yeast extract, 2% Bacto-peptone; Oxoid) containing glucose, galactose or raffinose at 2% w/v.
G1 cells synchronization was achieved by adding 2 µg/ml α-factor to the cultures, except for the experiment described in Figure 2C in which we used 20 µg/ml α-factor.
Western blot analysis and immunological reagents
Crude extract was prepared as described in Foiani et al. (1999). Total protein extract (25 µg) was used for western blotting or in situ autophosphorylation assay (ISA) (see below). The western blot procedure has already been described (Foiani et al., 1995), except that the secondary antibodies were peroxidase labelled (Amersham). The monoclonal antibody against the pol–prim B subunit has been described previously (Foiani et al., 1995) and the polyclonal antibodies against Rad53 were a generous gift from C.Santocanale and J.Diffley.
Immunoprecipitation and phosphatase treatment
Cells (2 ×∼109) grown under normal conditions or in the presence of 0.02% MMS, were resuspended in 1 ml of lysis buffer (0.4 M sorbitol, 150 mM potassium acetate, 20 mM PIPES–KOH pH 6.8, 2 mM magnesium acetate, 1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride, supplemented with protease inhibitor cocktail; Roche). Cells were broken in the presence of glass beads in a mini beadbeater (Biospec) and the extract was clarified in a microfuge. Lysis buffer (800 µl) was added to 200 µl of crude extract and incubated for 2 h at 4°C in the presence of 1 ×∼107 magnetic beads (Dynal), which were pre-incubated with 12CA5 monoclonal antibody. The beads were then washed three times with 1 ml of lysis buffer and treated with 4000 U of λ-phosphatase (Biolabs). An identical aliquot of beads was treated under the same conditions without λ-phosphatase addition. Samples were incubated for 10 min at 30°C. The beads were then washed with 1 ml of lysis buffer and resuspended in 20 µl of Laemmli buffer.
ISA
The procedure to measure Rad53 activity in situ has already been described (Pellicioli et al., 1999).
FACS analysis
FACS analysis was performed as described in Foiani et al. (1999).
Acknowledgments
Acknowledgements
We wish to thank J.E.Haber and S.E.Lee (Brandeis University, Waltham, MA), J.X.Diffley (ICRF, Clare Hall, UK), M.Kupiec (Tel Aviv University, Tel Aviv, Israel) and the members of our laboratory for enthusiastic discussions and suggestions. We also thank J.E.Haber and S.E.Lee for communicating unpublished results, and J.X.Diffley, C.Santocanale (PNU, Milan), K.Nasmyth (IMP, Vienna), M.Fasullo (AMC, Albany, NY) and A.Amon (MIT, Boston, MA) who kindly provided plasmids, strains and reagents. This work was supported by Associazione Italiana per la Ricerca sul Cancro and partially by grants from Telethon-Italy (Grant No. E1108), Programma per la Ricerca Finalizzata 1998—Ministero della Sanità, Cofinanziamento 1999 MURST—Università di Milano, MURST (5%) Biomolecole per la Salute Umana and CNR Target Project on Biotechnology.
References
- Aboussekhra A., Chanet,R., Zgaga,Z., Cassier-Chauvat,C., Heude,M. and Fabre,F. (1989) RADH, a gene of Saccharomyces cerevisiae encoding a putative DNA helicase involved in DNA repair. Characteristics of radH mutants and sequence of the gene. Nucleic Acids Res., 17, 7211–7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amon A., Surana,U., Muroff,I. and Nasmyth,K. (1992) Regulation of p34CDC28 tyrosine phosphorylation is not required for entry into mitosis in S.cerevisiae. Nature, 355, 368–371. [DOI] [PubMed] [Google Scholar]
- Bashkirov V.I., King,J.S., Bashkirova,E.V., Schmukli-Maurer,J. and Heyer,W.-D. (2000) DNA repair protein Rad55 is a terminal substrate of the DNA damage checkpoints. Mol. Cell. Biol., 20, 4393–4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brush G., Morrow,D.M., Heiter,P. and Kelly,T.J. (1996) The ATM homolog MEC1 is required for phosphorylation of replication protein A in yeast. Proc. Natl Acad. Sci. USA, 93, 15075–15080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr A. and Hoekstra,M.F. (1995) The cellular response to DNA damage. Trends Cell Biol., 5, 32–40. [DOI] [PubMed] [Google Scholar]
- Chakraverty R.K. and Hickson,I.D. (1999) Defending genome integrity during replication: a proposed role for RecQ family helicases. BioEssays, 21, 286–294. [DOI] [PubMed] [Google Scholar]
- Chanet R., Heude,M., Adjiri,A., Maloisel,L. and Fabre,F. (1996) Semidominant mutations in the yeast Rad51 protein and their relationships with the Srs2 helicase. Mol. Cell. Biol., 16, 4782–4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L., Collyer,T. and Hardy,C.F.J. (1999) Cell cycle regulation of DNA replication initiator factor Dbf4p. Mol. Cell. Biol., 19, 4270–4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doe C.L., Dixon,J., Osman,F. and Whitby,M.C. (2000) Partial suppression of the fission yeast rqh1– phenotype by expression of a bacterial Holliday junction resolvase. EMBO J., 19, 2751–2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohrmann P.R., Oshiro,G., Tecklenburg,M. and Sclafani,R.A. (1999) RAD53 regulates DBF4 independently of checkpoint function in Saccharomyces cerevisiae. Genetics, 151, 965–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elledge S.J. (1996) Cell cycle checkpoints: preventing an identity crisis. Science, 274, 1664–1672. [DOI] [PubMed] [Google Scholar]
- Fasullo M., Koudelik,J., AhChing,P., Giallanza,P. and Cera,C. (1999) Radiosensitive and mitotic recombination phenotypes of the Saccharomyces cerevisiae dun1 mutant defective in DNA damage-inducible gene expression. Genetics, 152, 909–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foiani M., Liberi,G., Lucchini,G. and Plevani,P. (1995) Cell cycle-dependent phosphorylation and dephosphorylation of the yeast DNA polymerase α–primase B subunit. Mol. Cell. Biol., 15, 883–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foiani M., Lucchini,G. and Plevani,P. (1997) The DNA polymerase α–primase complex couples DNA replication, cell cycle progression and DNA-damage response. Trends Biochem. Sci., 22, 424–427. [DOI] [PubMed] [Google Scholar]
- Foiani M., Liberi,G., Piatti,S. and Plevani,P. (1999) Saccharomyces cerevisiae as a model system to study DNA replication. In Cotterill,S. (ed.), Eukaryotic DNA Replication. A Practical Approach. Oxford University Press, Oxford, UK, pp. 185–200. [Google Scholar]
- Foiani M., Pellicioli,A., Lopes,M., Lucca,C., Ferrari,M., Liberi,G., Muzi-Falconi,M. and Plevani,P. (2000) DNA damage checkpoints and DNA replication controls in Saccharomyces cerevisiae. Mutat. Res., 451, 187–196. [DOI] [PubMed] [Google Scholar]
- Frei C. and Gasser,S.M. (2000) The yeast Sgs1p helicase acts upstream of Rad53p in the DNA replication checkpoint and colocalizes with Rad53p in S-phase-specific foci. Genes Dev., 14, 81–96. [PMC free article] [PubMed] [Google Scholar]
- Friedberg E.C., Walker,G.C. and Siede,W.D. (1995) DNA Repair and Mutagenesis. ASM Press, Washington, DC. [Google Scholar]
- Gangloff S., Soustelle,C. and Fabre,F. (2000) Homologous recombination is responsible for cell death in the absence of the Sgs1 and Srs2 helicases. Nature Genet., 25, 192–194. [DOI] [PubMed] [Google Scholar]
- Gardner C.W., Putnam,T. and Weinert,T. (1999) RAD53, DUN1 and PDS1 define two parallel G2/M checkpoint pathways in budding yeast. EMBO J., 18, 3173–3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz R.D. and Sugino,A. (1988) New yeast–Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six base-pair restriction sites. Gene, 74, 527–534. [DOI] [PubMed] [Google Scholar]
- Hadwiger J.A., Wittenberg,C., Mendenhall,M.D. and Reed,S.I. (1989) The Saccharomyces cerevisiae CKS1 gene, a homolog of the Schizosaccharomyces pombe suc1+ gene, encodes a subunit of the Cdc28 protein kinase complex. Mol. Cell. Biol., 9, 2034–2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L.H. and Kastan,M.B. (1994) Cell cycle controls and cancer. Science, 266, 1821–1828. [DOI] [PubMed] [Google Scholar]
- Heude M., Chanet,R. and Fabre,F. (1995) Regulation of the Saccharomyces cerevisiae Srs2 helicase during the mitotic cell cycle, meiosis and after irradiation. Mol. Gen. Genet., 248, 59–68. [DOI] [PubMed] [Google Scholar]
- Higgins N.P., Kato,K. and Strauss,B. (1976) A model for replication repair in mammalian cells. J. Mol. Biol., 101, 417–425. [DOI] [PubMed] [Google Scholar]
- Holmes A.M. and Haber,J.E. (1999) Double-strand break repair in yeast requires both leading and lagging strand DNA polymerases. Cell, 96, 415–424. [DOI] [PubMed] [Google Scholar]
- Johnston L.H. and Lowndes,N.F. (1992) Cell cycle control of DNA synthesis in budding yeast. Nucleic Acids Res., 20, 2403–2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson Z.O. and Hubscher,U. (1997) Proliferating cell nuclear antigen: more than a clamp for DNA polymerases. BioEssays, 19, 967–975. [DOI] [PubMed] [Google Scholar]
- Kaytor M.D., Nguyen,M. and Livingston,D.M. (1995) The complexity of the interaction between RAD52 and SRS2. Genetics, 140, 1441–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesti T., Flick,K., Keranen,S., Syvaoja,J.E. and Wittenberg,C. (1999) DNA polymerase ε catalytic domains are dispensable for DNA replication, DNA repair and cell viability. Mol. Cell, 3, 679–685. [DOI] [PubMed] [Google Scholar]
- Kogoma T. (1997) Stable DNA replication: interplay between DNA replication, homologous recombination and transcription. Microbiol. Mol. Biol. Rev., 61, 212–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.E., Moore,J.K., Holmes,A., Umezu,K., Kolodner,R.D. and Haber,J.E. (1998) Ku70, Mre11/Rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell, 94, 399–409. [DOI] [PubMed] [Google Scholar]
- Lee S.K., Johnson,R.E., Yu,S.L., Prakash,L. and Prakash,S. (1999) Requirement of yeast SGS1 and SRS2 genes for replication and transcription. Science, 286, 2339–2343. [DOI] [PubMed] [Google Scholar]
- Lowndes N.F. and Murguia,J.R. (2000) Sensing and responding to DNA damage. Curr. Opin. Genet. Dev., 10, 17–25. [DOI] [PubMed] [Google Scholar]
- Lydall D. and Weinert,T. (1997) G2/M checkpoint genes of Saccharomyces cerevisiae: further evidence for roles in DNA replication and/or repair. Mol. Gen. Genet., 256, 638–651. [DOI] [PubMed] [Google Scholar]
- Malkova A., Ivanov,E.L. and Haber,J.E. (1996) Double-strand break repair in the absence of RAD51 in yeast: a possible role for break-induced DNA replication. Proc. Natl Acad. Sci. USA, 93, 7131–7136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marini F., Pellicioli,A., Paciotti,V., Lucchini,G., Plevani,P., Stern,D.F. and Foiani,M. (1997) A role for DNA primase in coupling DNA replication to DNA damage response. EMBO J., 16, 639–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne G.T., Ho,T. and Weaver,D.T. (1995) Modulation of Saccharomyces cerevisiae DNA double-strand break repair by SRS2 and RAD51. Genetics, 139, 1189–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navas T.A., Zhou,Z. and Elledge,S. (1995) DNA polymerase ε links the DNA replication machinery to the S-phase checkpoint. Cell, 80, 29–39. [DOI] [PubMed] [Google Scholar]
- Paciotti V., Lucchini,G., Plevani,P. and Longhese,M.P. (1998) Mec1p is essential for phosphorylation of the yeast DNA damage checkpoint protein Ddc1p, which physically interacts with Mec3p. EMBO J., 17, 4199–4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pâques F. and Haber,J.E. (1997) Two pathways for removal of nonhomologous DNA ends during double-strand break repair in Saccharomyces cerevisiae. Mol. Cell. Biol., 17, 6765–6771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulovich A.G. and Hartwell,L.H. (1995) A checkpoint regulates the rate of progression through S-phase in S.cerevisiae in response to DNA damage. Cell, 82, 841–847. [DOI] [PubMed] [Google Scholar]
- Paulovich A.G., Toczyski,D.P. and Hartwell,L.H. (1997) When checkpoints fail. Cell, 88, 315–321. [DOI] [PubMed] [Google Scholar]
- Paulovich A.G., Armour,C.D. and Hartwell,L.H. (1998) The Saccharomyces cerevisiae RAD9, RAD17, RAD24 and MEC3 genes are required for tolerating irreparable, ultraviolet-induced DNA damage. Genetics, 150, 75–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellicioli A., Lucca,C., Liberi,G., Marini,F., Lopes,M., Plevani,P., Romano,A., Di Fiore,P.P. and Foiani,M. (1999) Activation of Rad53 kinase in response to DNA damage and its effect in modulating phosphorylation of the lagging strand DNA polymerase. EMBO J., 18, 6561–6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong L. and Klein,H.L. (1993) Purification and characterization of the SRS2 DNA helicase of the yeast Saccharomyces cerevisiae. J. Biol. Chem., 268, 1252–1259. [PubMed] [Google Scholar]
- Rong L., Palladino,F., Aguilera,A. and Klein,H.L. (1991) The hyper-gene conversion hpr5-1 mutation of Saccharomyces cerevisiae is an allele of the SRS2/RADH gene. Genetics, 127, 75–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez Y., Desany,B.A., Jones,W.J., Liu,Q., Wang,B. and Elledge,S.J. (1996) Regulation of RAD53 by the ATM-like kinases MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science, 271, 357–360. [DOI] [PubMed] [Google Scholar]
- Sanchez Y., Bachant,J., Wang,H., Hu,F., Liu,D., Tetzlaff,M. and Elledge,S.J. (1999) Control of the DNA damage checkpont by Chk1 and Rad53 protein kinases through distinct mechanisms. Science, 286, 1166–1171. [DOI] [PubMed] [Google Scholar]
- Sandell L.L. and Zakian,V. (1993) Loss of a yeast telomere: arrest, recovery, and chromosome loss. Cell, 75, 729–739. [DOI] [PubMed] [Google Scholar]
- Schiestl R., Prakash,S. and Prakash,L. (1990) The SRS2 suppressor of rad6 mutations of Saccharomycs cerevisiae acts by channeling DNA lesions into the RAD52 DNA repair pathway. Genetics, 124, 817–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schild D. (1995) Suppression of a new allele of the yeast RAD52 gene by overexpression of RAD51, mutations in srs2 and ccr4, or mating type heterozygosity. Genetics, 140, 115–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seigneur M., Bidnenko,V., Ehrlich,S.D. and Michel,B. (1998) RuvAB acts at the arrested replication forks. Cell, 95, 419–430. [DOI] [PubMed] [Google Scholar]
- Sorger P.K. and Murray,A.W. (1992) S-phase feedback control in budding yeast is independent of tyrosine phosphorylation of p34CDC28. Nature, 355, 365–368. [DOI] [PubMed] [Google Scholar]
- Sugimoto K., Ando,S., Shimura,T. and Matsumoto,K. (1997) Rfc5, a replication factor C component, is required for regulation of Rad53 protein kinase in the yeast checkpoint pathway. Mol. Cell. Biol., 17, 5905–5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z., Fay,D.S., Marini,F., Foiani,M. and Stern,D.F. (1996) Spk1/Rad53 is regulated by Mec1-dependent protein phosphorylation in DNA replication and DNA damage checkpoint pathway. Genes Dev., 10, 395–406. [DOI] [PubMed] [Google Scholar]
- Sun Z., Hsiao,J., Fay,D.S. and Stern,D.F. (1998) Rad53 FHA domain is associated with phosphorylated Rad9 in the DNA damage checkpoint. Science, 281, 272–274. [DOI] [PubMed] [Google Scholar]
- Toczyski D.P., Galgoczy,D.J. and Hartwell,L.H. (1997) CDC5 and CKII control adaptation to the yeast DNA damage checkpoint. Cell, 90, 1097–1106. [DOI] [PubMed] [Google Scholar]
- Wach A., Brachat,A., Pohlmann,R. and Philippsen,P. (1994) New heterologous modules for classical or PCR-based gene disruption in Saccharomyces cerevisiae. Yeast, 10, 1793–1808. [DOI] [PubMed] [Google Scholar]
- Wach A., Brachat,A., Alberti-Segui,C., Rebischung,C. and Philippsen,P. (1997) Heterologous HIS3 marker and GFP reporter modules for PCR-targeting in Saccharomyces cerevisiae. Yeast, 13, 1065–1075. [DOI] [PubMed] [Google Scholar]
- Weinert T. (1997) Yeast checkpoints controls and relevance to cancer. Cancer Surv., 29, 109–132. [PubMed] [Google Scholar]
- Weinert T. (1998) DNA damage checkpoints update: getting molecular. Curr. Opin. Genet. Dev., 8, 185–193. [DOI] [PubMed] [Google Scholar]
- Wold M.S. (1997) Replication protein A: a heterotrimeric, single-stranded DNA-binding potein required for eukaryotic DNA metabolism. Annu. Rev. Biochem., 66, 61–92. [DOI] [PubMed] [Google Scholar]
- Zhou Z. and Elledge,S.J. (1993) DUN1 encodes a protein kinase that controls the DNA damage response in yeast. Cell, 75, 1119–1127. [DOI] [PubMed] [Google Scholar]
- Zou H. and Rothstein,R. (1997) Holliday junctions accumulate in replication mutants via RecA homolog-independent mechanism. Cell, 90, 87–96. [DOI] [PubMed] [Google Scholar]