Abstract
Microorganisms have become an increasingly important platform for the production of drugs, chemicals, and biofuels from renewable resources. Advances in protein engineering, metabolic engineering, and synthetic biology enable redesigning microbial cellular networks and fine-tuning physiological capabilities, thus generating industrially viable strains for the production of natural and unnatural value-added compounds. In this review, we describe the recent progress on engineering microbial factories for synthesis of valued-added products including alkaloids, terpenoids, flavonoids, polyketides, non-ribosomal peptides, biofuels, and chemicals. Related topics on lignocellulose degradation, sugar utilization, and microbial tolerance improvement will also be discussed.
Keywords: Synthetic biology, Metabolic engineering, Microbial synthesis, Value-added products, Natural products, Fuels and chemicals
Introduction
Microorganisms have been increasingly used to produce value-added compounds with numerous applications in the food, agriculture, chemical, and pharmaceutical industries. Examples of these value-added compounds include many antibacterial and anticancer drugs, amino acids, organic acids, vitamins, industrial chemicals, and biofuels. Compared to synthetic chemistry methodologies, microbial bio-synthesis has several advantages. First, it avoids the use of heavy metals, organic solvents, and strong acids and bases, thus allowing the synthetic process to take place through an environmentally benign route. Second, enzymes usually have a relatively high substrate specificity, which helps reduce the formation of byproducts. Third, some compounds with complex structures already have natural synthetic pathways while establishing chemical synthetic routes for these complex compounds is very difficult. Finally, metabolic engineering offers ways to further improve the yield and productivity of a target compound while combinatorial biosynthesis allows the creation of novel derivatives.
It is straightforward to think about directly extracting natural products from their native producers. However, most of these native producers are not cultivable in the laboratory and many microorganisms grow very slowly and produce minute amounts of the target compounds. It has been estimated that only 1% of bacteria and 5% of fungi have been cultivated in the laboratory [16, 26, 27, 66]. Even when it is possible to cultivate the native producers, their growth conditions have to be extensively optimized. In addition, due to the lack of genetic tools to manipulate these hosts, it is very difficult to improve the product yield and productivity. Therefore, well-characterized microorganisms that can be used as universal platform organisms are highly desired. Escherichia coli and Saccharomyces cerevisiae are two of the most widely used platform organisms due to their well-characterized physiology and genetics, fast cell-growth rates, and the availability of abundant genetic tools. Other platform microorganisms include Bacillus subtilis, Pseudomonas putida, and Streptomyces species.
Recent advances in protein engineering, metabolic engineering, and synthetic biology have revolutionized our ability to discover and construct new biosynthetic pathways and engineer platform organisms or so-called microbial factories to produce a wide variety of value-added products such as alkaloids, terpenoids, flavonoids, polyketides, non-ribosomal peptides, biofuels, and chemicals in a cost-effective manner. This review will highlight a few representative examples from the past 5 years. Related topics on lignocellulose degradation, sugar utilization, and microbial tolerance improvement will also be discussed.
Natural products
Alkaloids
Alkaloids are nitrogen-containing compounds of low molecular weight produced by a large variety of organisms, including bacteria, fungi, plants, and animals. Most alkaloids are derived through decarboxylation of amino acids such as tryptophan, tyrosine, ornithine, histidine, and lysine, and possess important pharmacological activities [84]. For example, the antimicrobial agent berberine has cholesterol-lowering activity [62], sanguinarine has shown potential as an anticancer therapeutic [60], bisbenzyliso-quinoline alkaloid tetrandrine has been used to treat autoimmune disorders and hypertension [64, 65], and a number of indolocarbazole alkaloids have entered clinical trials for diabetic retinopathy, cancer treatment or Parkinson's disease [18]. They have a very high diversity and molecular complexity in structure and can be classified into a number of groups, such as morphinane-, protoberberine-, ergot-, pyrrolizidine-, quinolizidine- and furanoquinoline-alkaloids according to the amino acids from which they originate [117]. Even for the plant alkaloids alone, there are over 10,000 structurally characterized members. Due to their high structural diversity and molecular complexity, chemical synthesis of alkaloids has not been very effective. On the other hand, although metabolic engineering strategies have been tried in plants to increase the amount of alkaloid products [3, 36, 116], success was limited and difficult to generalize due to the lack of convenient tools for engineering biosynthetic pathways in plants [115], the complexity of alkaloid biosynthetic pathways and their regulation [150] and the unavoidable transport of synthetic intermediates in and out among multiple intracellular organelles in plants [73].
Reconstitution of alkaloid biosynthesis in an engineered microbe has several advantages including rapid growth and biomass accumulation, abundant availability of genetic tools for pathway expression and optimization, and ease of characterizing and isolating final product and key intermediates due to a relatively cleaner background and the lack of interference from other metabolites in plants [48]. To this end, E. coli and S. cerevisiae were recently explored as production hosts. Sato and coworkers combined microbial and plant enzymes to synthesize benzylisoquinoline alkaloids including magnoflorine and scoulerine from dopamine via reticuline by co-culturing E. coli and S. cerevisiae [84] (Fig. 1a). The key intermediate (S)-reticuline was synthesized from dopamine by crude enzymes from recombinant E. coli, and was subsequently channeled into S. cerevisiae, generating magnoflorine and scoulerine with final yields of 7.2 and 8.3 mg/l, respectively. Smolke and coworkers engineered yeast alone expressing combinations of enzymes from different sources to produce the key intermediate reticuline and downstream metabolites along with two of the major branches from reticuline: the sanguinarine/berberine branch and the morphinan branch (Fig. 1b). In this system, a galactose-inducible enzyme tuning strategy was designed to balance enzyme expression and product yield, conserving cellular resources without compromising pathway flux [48].
Fig. 1.
Production of benzylisoquinoline alkaloids either by a co-culturing system of E. coli and S. cerevisiae (a) or by S. cerevisiae alone (b)
In addition to alkaloids produced naturally, combinatorial strategies were applied to further diversify existing alkaloids with the goal of improving their potency as therapeutic molecules. For example, indolocarbazole biosynthetic enzymes possess useful degrees of substrate flexibility, thus they are able to accept different intermediates to yield novel derivatives. More specifically, partial clusters of rebeccamycin and staurosporine biosynthesis were combined and expressed together with additional sugar biosynthetic genes in Streptomyces albus. This resulted in generation of a series of novel indolocarbazole derivatives bearing different deoxysugars, some of which showed potent, subnanomolar, yet selective inhibition against kinases, one of the major targets in current drug discovery and development processes [113, 114]. It is believed that the sugar moieties play an important role in the selectivity of protein kinase inhibition.
Terpenoids
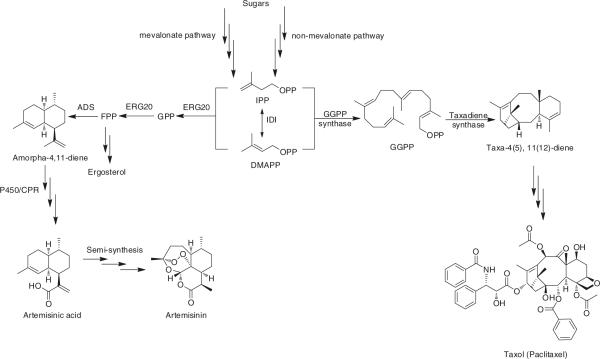
Terpenoids (also called isoprenoids), derived from five-carbon isoprene units assembled and modified in thousands of ways, compose the largest class of naturally occurring molecules with important medicinal and industrial properties. They are found in all classes of living organisms and approximately 25,000 structures have already been elucidated [40]. Despite the enormous structural diversity, terpenoids are synthesized from two basic isoprene building blocks, isopentenyl pyrophosphate (IPP) and dimethylallyl pyrophosphate (DMAPP), originated through either the mevalonate pathway or the non-mevalonate pathway depending on the species [22]. They serve a wide variety of functions such as respiration and electron transport (quinones), membrane fluidity and hormone signaling (steroids), and photosynthesis and antioxidant agents (carotenoids). Many terpenoids, especially the ones isolated from plants and marine invertebrates, are bioactive and are used in the pharmaceutical, cosmetic, and food industries [88]. The transformation of IPP and its derivatives to terpenoids of high complexity has been an active area in synthetic biology and metabolic engineering [22]. For example, S. cerevisiae has been engineered to produce artemisinic acid, a key precursor for the anti-malarial drug artemisinin [105] (Fig. 2a). The farnesyl pyrophosphate (FPP) biosynthetic pathway was engineered to increase FPP production and its use for sterols was decreased. The amorphadiene synthase (ADS) gene, a cytochrome P450 monooxygenase and its redox partner from Artemisia annua were introduced to convert FPP to amorphadiene, which was further converted to artemisinic acid through a three-step oxidation, with a titer of 115 mg/l. This production was further improved to reach 250 mg/l in shake-flask conditions and 1 g/l in bioreactors by modulating the selection markers and the culture composition [104].
Fig. 2.
Utilizing S. cerevisiae to synthesize artemisinic acid (a) and taxol (b)
One of the most important classes of enzymes involved in plant-derived natural products is the membrane-bound cytochrome P450 superfamily, which is ubiquitously involved in terpenoid biosynthesis and takes part in a wide variety of reactions. For example, eight out of the approximately 20 steps in paclitaxel biosynthesis are catalyzed by P450 enzymes [23]. Despite their essential nature, functional expression of plant P450 s in bacteria is extremely challenging and hinders the biosynthesis of many functional molecules by recombinant bacteria. This is mainly due to the absence of cytochrome P450 reductase (CPR) redox partners in bacteria for electron transfer [119] and the absence of an endoplasmic reticulum, which results in the translational incompatibility of the membrane signal modules [141]. In order to enable E. coli for production of functionalized terpenoids using plant P450 s, Keasling and coworkers cloned the codon optimized 8-cadinene hydroxylase (CAH) along with a CPR from Candida tropicalis in E. coli and obtained production of 8-hydroxycadinene at approximately 25 ± 2 mg/l. The N-terminal membrane anchor was subsequently replaced with various N-terminal sequences from three heterologous P450 s, two secretion/ solubilization sequences, or a self-assembling membrane protein, among which the N-terminal sequence of the bovine CAH [11] yielded an additional fiefold improvement in productivity to 105 ± 7 mg/l [21]. Such a strategy of generating a chimeric P450 with a fie-tuned N-terminal domain has also been successfully applied in isoflavone production in E. coli [70].
Another successful example of combining protein engineering, metabolic engineering, and synthetic biology strategies to design microbes for production of value-added compounds was demonstrated in the biosynthesis of taxadiene, a taxol precursor (Fig. 2b). Taxol and its structural analogs are potent and commercially successful anticancer drugs [61], originally isolated from the bark of the Pacific yew tree [136]. The traditional direct extraction method [100], the later-developed method of total chemical synthesis [96], and the currently used semisynthetic route [100] all suffered from low productivity and the accompanying constraints of being generalized to other derivatives in the search for more efficacious drugs [53, 106]. As the first step towards the production of Taxol in S. cerevisiae, co-expression of Taxus chinensis taxadiene synthase (TStc) and geranylgeranyl pyrophosphate synthase (GGPPStc) only resulted in a production of 204 μg/l taxadiene [33]. In yeast, the isoprenoid building blocks are mostly used for steroid biosynthesis, and the mevalonate pathway is subject to complex feedback regulation, with HMG-CoA reductase as the major regulatory target [29]. Expression of a truncated version of yeast HMG-CoA reductase (tHMG1) in combination with Sulfolobus acidocaldarius GGPPS, which does not compete with steroid synthesis and the codon-optimized TStc led to a 40-fold increase in taxadiene to 8.7 mg/l [33]. To further improve it, Stephanopoulos and coworkers designed a multivariate modular approach and succeeded in increasing the titer of taxadiene to approximately 1 g/l [1]. In this approach, the pathway was partitioned into two modules at IPP: the upstream native methylerythritol phosphate (MEP) pathway and the downstream heterologous taxadiene pathway. Systematically varying promoters of different strengths and plasmid copy-numbers resulted in identifying conditions that optimally balance the two pathway modules, such that the taxadiene production was maximized with minimal accumulation of any toxic intermediate. Such a modular pathway engineering strategy has the potential for engineering other terpenoid biosynthesis.
Flavonoids
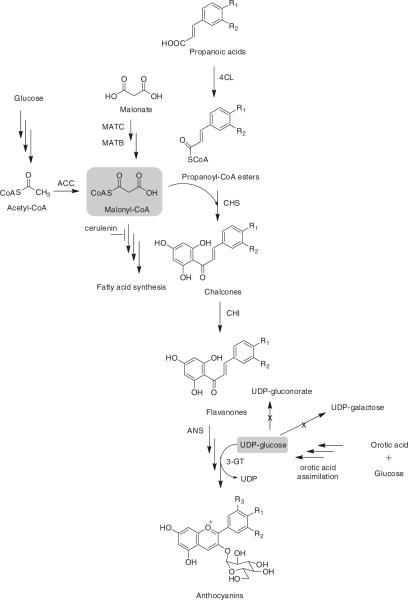
Flavonoids are an important group of plant secondary metabolites, which in general have linear C6-C3-C6 skeletons derived from a phenylpropanoid (C6-C3) starter and three C2 elongation units [31]. Such a 15-carbon phenylpropanoid core is extensively modified by rearrangement, methylations, methoxylations, alkylation, oxidation, C- and O-glycosylation, and hydroxylation [74, 132], forming a fascinating group of over 9,000 members exhibiting antioxidant, antibacterial, antiviral, and anti-cancer activities [35]. Through the phenylpropanoid pathway, phenylalanine ammonia lyase (PAL) deaminates phenylalanine to cinnamic acid, which is hydroxylated by cinnamic-4-hydroxylases (C4H), activated by 4-coumarate/cinnamate coenzymes, and condensed with three malonyl-CoA units to form a chalcone catalyzed by chalcone synthase (CHS). Chalcones are converted by chalcone isomerases (CHI) in a ring closing step to form the heterocyclic C ring [55]. The variability in molecular structure further divides flavonoids into flavones, flavanones, flavonols, isoflavones, anthocyanins, and catechines [74]. Similar to the production of alkaloids and terpenoids, E. coli and S. cerevisiae are widely used as model systems to produce flavonoids. However, in many cases, the biosynthetic efficacy is greatly limited by precursor and cofactor availability in the host. Therefore, alternative carbon assimilation pathways such as the malonate utilization pathway were introduced and competitive reactions including fatty acid synthesis and UDP-glucose consumption were inhibited and deleted, respectively, in order to improve the availability of malonyl-CoA and UDP-glucose. Such a strain expressing plant 4-coumarate:CoA ligase (4CL), CHS, CHI, anthocyanin synthase (ANS) and 3-O-glycosyltransferase (3-GT) produced flavanones up to 700 mg/l and anthocyanins up to 113 mg/l [74] (Fig. 3).
Fig. 3.
Engineered pathways for increased flavanone and anthocyanin biosynthesis in E. coli
In another study, overexpression of the four acetyl-CoA carboxylase (ACC) subunits under a constitutive promoter resulted in an over fivefold increase in flavanone production. Acetate assimilation pathways were also amplified to convert acetate to malonyl-CoA via acetyl-CoA. Auxiliary expression of ACC with a chimeric biotin ligase (BirA) consisting of the N-terminus of E. coli BirA and the C terminus of Photorhabdus luminescens BirA further increased the production of pinocembrin, naringenin, and eriodictyol to 429, 119, and 52 mg/l, respectively [71].
Due to the relatively broad substrate specificity of the flavonoid biosynthetic genes, providing the pathway with unusual precursors could result in generation of new flavonoids. For example, UDP-glucose dehydrogenase (CalS8) and UDP-glucuronic acid decarboxylase (CalS9) from Micromonospora echinospora spp. calichensis and 7-O-glycosyltransferase (ArGt-4) from Arabidopsis thaliana were expressed together with an integrated copy of E. coli K12 UDP-glucose pyrophosphorylase (GalU) in background strain E. coli BL21 (DE3) with the glucose-phosphate isomerase (Pgi) gene deleted. When the resulting strain was fed with naringenin and naringenin 7-O-xyloside, a glycosylated naringenin product, was detected [124]. Due to the importance of flavonoids for human health, further derivatization might offer a chance to create new members with improved or novel properties.
Polyketides and non-ribosomal peptides
Polyketides, mostly derived from bacteria, Wlamentous fungi, and plants, are among the most important metabolites in human medicine, used clinically as antibiotics (erythromycin A, rifamycin S), antifungals (amphotericin B), anticancer drugs (doxorubicin, epothilone), antiparasitics (avermectin), cholesterol-lowering agents (lovastatin), and immunosuppressants (rapamycin) [138]. They are synthesized by a family of multifunctional enzymes known as polyketide synthases (PKSs). The core structures of polyketides are assembled through sequential Claisen-like condensations of extender units derived from carboxylated acyl-CoA precursors mainly including malonyl-CoA, methyl-malonyl-CoA, methoxy-malonyl-CoA, and ethyl-malonyl-CoA [126]. PKSs are classified into types I, II and III based on their biochemical features. The products synthesized by different types of PKSs can undergo different sets of tailoring modifications such as oxygenation, hydroxylation, cyclization, methylation, acylation, and glycosylation to form products with a great level of structural diversity [34, 98].
Research has been directed towards reconstitution of PKS biosynthetic pathways in more technically amenable microbes including E. coli, S. cerevisiae, B. subtilis, P. putida, and various Streptomyces species [13]. Despite several obvious advantages, E. coli and S. cerevisiae have their own drawbacks as heterologous hosts, including unavailability of some biosynthetic building blocks and lack of post-translational enzymes to modify PKSs [39, 89]. In addition, difficulties in efficient translation and functional folding of key enzymes including megasynthases and the P450 family of enzymes were encountered in E. coli. These limitations have to be addressed before synthesis of a wider range of molecules is attempted in these hosts. In fact, some of them were already addressed to a certain degree through metabolic engineering and protein engineering efforts. Nowadays, examples of polyketides synthesized by all three types of PKSs have been reported in E. coli, although the titers do vary case by case [39]. On the other hand, P. putida has a high GC content in its genome, suggesting it could better support the heterologous expression of genes with a high GC content, especially for the clusters isolated from actinomycetes [13]. Several Streptomyces species [10] and B. subtilis possess a native ability to provide needed substrates, which is an advantage as a heterologous host. However, the genetic manipulations of these three organisms are not as easy and convenient as the ones developed for E. coli and S. cerevisiae.
The most successful example of in vivo PKS reconstitution is the biosynthesis of 6-deoxyerythronolide B (6-dEB), the 14-membered macrocyclic core of erythromycin, in E. coli. The native and heterologous metabolism were engineered, and the resulting strain BAP1 could supply the required starter unit propionyl-CoA and the extender unit (2S)-methylmalonyl-CoA. The three 6-deoxyerythronolide B synthase (DEBS) proteins catalyze six chain extension cycles, converting exogenous propionate into 6-dEB with a specific productivity that compares well with a high-producing mutant of the original host [99]. The titer was further improved by overexpressing the S-adenosylmethionine synthetase MetK from Streptomyces spectabilis to increase the synthesis of signaling molecules [135] or by deleting the propionyl-CoA: succinate CoA transferase [147]. The DEBS system could serve as a paradigm model to study and engineer modular type I PKSs. Numerous 6-dEB and erythromycin derivatives were synthesized through domain or module insertions, deletions, and replacements [20, 83, 138].
The epothilone PKS currently represents the largest modular type I PKS reconstituted in E. coli. The epothilone family is synthesized by a hybrid non-ribosomal peptide synthetase (NRPS)/polyketide synthase in the myxobacterium Sorangium cellulosum. The cluster is composed of one NRPS module and eight PKS modules, and the largest gene, the 21.8 kb epoD encodes a protein larger than 760 kDa. Through codon optimization, lowered growth temperature, chaperone coexpression, and replacement of the T7 promoter by the arabinose-inducible PBAD promoter, all pathway proteins except EpoD were solubly expressed. The expression of EpoD was finally achieved by dividing the large enzyme into two polypeptides, each consisting of two modules and the compatible linker pairs from related polyketide synthases. The entire cluster was expressed in the strain BAP1 mentioned above, resulting in the production of epothilones C and D. The success of the epothilone example provides an ideal platform for generation of novel epothilone derivatives and shows the importance of using protein engineering strategies to redesign a large gene cluster [90].
Although examples of nearly all major classes of natural products have been synthesized and engineered in E. coli, a noticeable exception is type II PKSs from actinomycete, which produce pharmaceutically important aromatic polyketides such as tetracyclines and anthracyclines [50]. The main obstacle is the inability to express the ketosynthase (KS)-chain length factor (CLF) heterodimer in soluble form. To bypass this obstacle using bacterial type II PKSs, Tang and coworkers targeted fungal iterative nonreducing PKSs, dissected and extracted the minimal PKS components of Gibberella fujikuroi PKS4, and reassembled it into a synthetic PKS, which synthesized a spectrum of aromatic polyketides in E. coli with cyclization regioselectivity not observed among fungal polyketides [148].
The products of the type III PKSs can be categorized into three groups based on their specific activities [39]. Chalcone synthase cyclizes the intermediates into flavanones, the synthetic precursors of a variety of flavonoids [137]. Stilbene synthase (STS) catalyzes the cyclization of the intermediate polyketide to form a stilbene backbone, which is further modified to stilbenoids [8]. Curcuminoid synthase (CUS) only catalyzes condensation reactions without cyclization, producing curcuminoids [58, 59]. Coexpression of phenylalanine ammonia lyase (PAL) and 4CL with different type III PKSs produced various compounds such as resveratrol, pinosylvin [57], bisdemethoxycurcumin, and dicinnamoylmethane [59]. Precursor-directed biosynthesis [57, 58] and coexpression of post-PKS modification enzymes [85, 144] further led to the generation of a variety of unnatural compounds.
As mentioned earlier, Streptomyces are naturally better host microorganisms for the production of polyketides [69]. About half of the bioactive microbial metabolites discovered to date are produced by actinomycetes, with the genus Streptomyces being the primary producer [98]. The recent progress on synthesis of bioacitve compounds or their precursors by metabolic engineering of Streptomyces was summarized recently in other reviews [69, 97, 98].
Similar to polyketides, non-ribosomal peptides (NRPs) are synthesized by large modular non-ribosomal peptide synthetases and represent a diverse family of secondary metabolites exhibiting a broad range of biological activities. This family includes antibiotics such as vancomycin and bacitracin, antibiotic precursors like ACV, immunosuppressive agents such as cyclosporine, and siderophores [122]. Baltz and coworkers expanded the modification of the daptomycin amino acid core by replacing single or multiple modules in the DptBC subunit with modules from daptomycin and A54145 NRPSs. A combination of module exchanges, NRPS subunit exchanges, and inactivation of tailoring enzymes enabled Streptomyces roseosporus and Streptomyces fradiae to generate libraries of novel lipopeptide antibiotics related to daptomycin and A54145 [2, 94, 95]. Nielsen and coworkers initiated the development of a yeast platform for heterologous production of NRPs using ACV as a model NRP. ACV synthetase was expressed in S. cerevisiae in a high-copy plasmid together with phosphopantetheinyl transferases (PPTase) from three sources, all leading to production of ACV. The synthesis was improved by lowering the cultivation temperature and integrating the cluster into the genome. This work represents the first success in production of an NRP in yeast [123].
Biofuels
Due to environmental, economic, and energy security considerations, there is an increasing interest in the development of bio-derived fuel alternatives [78]. Dominant biofuel alternatives, such as corn-derived ethanol, however, proved to be only marginally profitable even with the application of governmental subsidies [51]. Thus, the current biofuel production scheme must be modified through the use of cheaper, non-edible lignocellulosic biomass as feedstock or via the production of advanced biofuels (Fig. 4).
Fig. 4.
Production of fuels and chemicals from lignocelluosic biomass
Utilization of lignocellulosic feedstocks
Lignocelluloses, the non-edible portion of plant-derived biomass, are considered preferable feedstock for biofuel production due to their low requirement for energy, fertilizer, and pesticide input [77]. Unfortunately, the recalcitrant structure of plant cell wall presents a great challenge for the efficient deconstruction and complete utilization of lignocellulosic feedstocks. Lignocellulosic biomass has a very distinctive structure, with its most abundant component, cellulose, tightly surrounded by a hemicellulose and lignin complex that protects the inner cellulose from hydrolytic enzymes [139].
Considerable research effort has been dedicated to breaking down the crystalline structure of cellulose to release the fermentable monosaccharide glucose for use in biofuel production. The most recent breakthrough in this area is the development of a microorganism capable of consolidated bioprocessing (CBP). CBP is a process where enzyme production, cellulose hydrolysis, and monosaccharide fermentation for fuel production are combined into a single step. CBP has been proposed to significantly lower biofuel production cost as it eliminates the large-scale production of cellulases [133]. One common strategy to develop a CBP microorganism is to introduce cellulolytic ability into ethanol producing, non-cellulolytic organisms [79].
Gram-negative bacterium Zymomonas mobilis is one attractive candidate for CBP microorganism development due to its high ethanol productivity and tolerance. Darzins and coworkers successfully expressed two cellulolytic enzymes, E1 and GH12, from Acidothermus cellulolyticus, in Z. mobilis. Furthermore, both cellulolytic enzymes can be secreted extracellularly due to the inclusion of native Z. mobilis secretion signals. Functional expression and secretion of cellulolytic enzymes in Z. mobilis indicate its high potential for serving as a CBP platform microorganism [76].
Another very attractive CBP organism candidate is S. cerevisiae, which has served as the major producer of ethanol for thousands of years [134]. van Zyl and coworkers first demonstrated that introducing the endoglucanase of Trichoderma reesei and the β-glucosidase of Saccharomycopsis fibuligera into S. cerevisiae can result in a recombinant strain with the ability to grow on phosphoric acid-swollen cellulose (PASC) as its sole carbon source and produce up to 1.0 g/l of ethanol [28]. Inspired by the structure of cellulosomes, Zhao and coworkers developed a recombinant yeast strain in which endoglucanases, cellobiohydrolases, and β-glucosidases were assembled into a trifunctional minicellulosome through cohesin and dockerin. In the recombinant yeast strain, a miniscaffoldin composed of a cellulose-binding domain and three distinct cohesin modules were expressed using yeast surface display, while three cellulolytic enzymes, each fused with a C-terminal dockerin corresponding to a different cohesin, were co-expressed in the same strain. The cellulolytic enzymes were assembled into minicellulosomes through cohesin–dockerin interaction onto miniscaffoldings anchored onto the yeast's surface. The recombinant yeast exhibited a higher cellulolytic activity due to enzyme–enzyme and enzyme–substrate synergistic effects. As a result, recombinant strains can simultaneously break down and ferment PASC to ethanol with a titer of 1.8 g/l [140]. Around the same time, Chen and coworkers reported their work involving another CBP process that utilized a minicellulosome. In their study, instead of creating a single recombinant strain, a synthetic yeast consortium was used for the expression and assembly of its minicellulosome. The synthetic consortium was composed of four different recombinant yeast strains: a strain displaying a trifunctional scaffoldin and three strains each expressing a dockerin-tagged cellulolytic enzyme. After optimization of the ratio of different populations in the synthetic consortium, a 1.87 g/l ethanol titer was achieved, which is 93% of the theoretical yield [131].
A similar effort was carried out to tackle the problem of hemicellulose utilization. Hemicellulose is the second-most abundant component of lignocellulosic biomass and can make up to 20–30% of the total feedstock. Unlike cellulose, which is composed of glucose, hemicellulose is primarily composed of five-carbon sugars (pentoses) such as D-xylose and L-arabinose [111]. Unfortunately, the industrial microorganism currently used for large-scale production of ethanol, S. cerevisiae, cannot utilize pentoses contained within the hydrolysates of the hemicellulose component of biomass feedstocks [49]. The incomplete utilization of sugar substrates present in lignocellulosic biomass hydrolysates is one of the major causes of elevated bioethanol production cost, making this environmentally friendly energy alternative typically far less economically competitive compared to fossil fuels [38, 46]. To be utilized by ethanol producing S. cerevisiae, pentose sugars need to be first transported into cells and then converted into D-xylulose-5-phosphate, which can then be further metabolized through the pentose phosphate pathway. In order to enable the effcient conversion of D-xylose and L-arabinose into D-xylulose-5-phosphate, heterologous pathways need to be introduced into S. cerevisiae and further optimized [46, 63]. For example, to improve the fermentative ability of the fungal D-xylose utilization pathway, much effort was devoted to identify heterologous enzymes with better catalytic efficiency and cofactor specificity [109], balance the cofactor usage of the pathway [75], and optimize ethanol production using more robust industrial ethanol-producing yeast strains [80].
Another factor hampering effcient production of lignocellulosic ethanol in S. cerevisiae is the “glucose repression” that occurs during mixed sugar fermentation. Because S. cerevisiae preferably utilizes glucose over other carbon sources, utilization of pentose sugars is severely repressed before glucose is depleted [107]. A novel approach was recently developed to overcome glucose repression by introducing a cellobiose transporter and a β-glucosidase into recombinant xylose-utilizing S. cerevisiae strains [45, 75] (Fig. 5). In these strains, cellobiose, an incomplete hydrolysis product of cellulose, is fermented instead of glucose in the presence of xylose. Since cellobiose enters S. cerevisiae cells through a dedicated cellobiose transporter, the competition of glucose and xylose at the sugar uptake step is eliminated. Cellobiose is then hydrolyzed into glucose intracellularly and continuously consumed for the production of ethanol. The simultaneous hydrolysis and utilization of cellobiose avoids intracellular accumulation of glucose, thus alleviating glucose repression. In the engineered cellobiose and xylose co-utilization strains, cellobiose and xylose are consumed simultaneously and synergistically with an ethanol productivity of 0.65 g/l h and overall ethanol yield of 0.39 g/g glucose.
Fig. 5.
Co-fermentation of cellobiose and xylose using recombinant S. cerevisiae co-expressing a cellobiose transporter and an intracellular β-glucosidase. This novel approach eliminates the use of exogenously added β-glucosidase and alleviates glucose repression on xylose uptake and utilization (shown in blue)
Production of advanced biofuels
The development of biologically derived ethanol has achieved significant success in the past few decades [4, 9]. However, ethanol exhibits some intrinsic limitations, such as low energy content and corrosiveness, which hampers its large-scale application as a fuel alternative. In contrast, advanced biofuels, such as higher alcohols, fatty acid derived fuels, and hydrocarbons, are considered to be better fuel alternatives as their physiochemical properties are more compatible with the current gasoline-based infrastructure [145].
Isopropanol and n-butanol are both better fuel alternatives compared to ethanol due to their higher energy content, higher octane number, and lower water solubility. Fortunately, unlike other long-chain alcohols, both isopropanol and n-butanol can be produced by Clostridium species in nature. However, since Clostridium species are Gram-positive anaerobes with a relatively slow growth rate and spore-forming life cycles, it is hard to control the yield of desired long-chain alcohols in industrial fermentation [145]. To address this issue, long-chain alcohols were produced in non-native hosts such as E. coli and S. cerevisiae [6, 7, 127, 145]. For heterologous production of isopropanol, various combinations of genes from different Clostridium and E. coli species have been introduced into E. coli for production through the coenzyme-A-dependent fermentative pathway. The resulting optimized recombinant strain can achieve an isopropanol production titer of 5 g/l with a yield of 43.5% mol/mol glucose [47]. Similarly, the n-butanol production pathway from a Clostridium species has also been introduced into E. coli and extensive metabolic engineering efforts have been dedicated to increasing the titer. However, the highest titer for n-butanol production only achieved 552 mg/l, most likely due to limitations imposed by low heterologous enzyme activity, insuffcient carbon precursors and inadequate reducing power [6]. At the same time, Keasling and coworkers introduced a similar pathway into S. cerevisiae and achieved an n-butanol production titer of 2.5 mg/l through the optimization of isozymes used in the pathway [127]. One reason for the low efficiency in the heterologous production of long-chain alcohols may be the cytotoxicity caused by the accumulation of intermediate metabolites as well as redox imbalance due to the introduction of heterologous pathways [6]. To address this issue, Liao and coworkers investigated the production of long-chain alcohols through existing non-fermentative keto acid pathways. Using this strategy, 2-keto acids, which are intermediates in amino acid biosynthesis pathways, are converted into aldehydes by broad range 2-keto-acid decarboxylases (KDC) and then reduced to alcohols by alcohol dehydrogenases (ADH). Compared to fermentative pathways, only two heterologous steps need to be introduced for alcohol production through 2-keto-acid pathways. Through the choice of different KDCs, 2-keto acids from various amino acid synthesis pathways can be used to produce long-chain alcohols including isobutanol, 1-butanol, 2-methyl-1-butanol, and 2-phenylethanol [5, 7, 25].
In addition to alcohols, fatty acid-derived fuel alternatives such as fatty acid esters and fatty alcohols are also potential fuel alternatives. In a recent study by Keasling and coworkers [128], recombinant E. coli strains were engineered to overproduce free fatty acid through cytosolic expression of a native E. coli thioesterase and the deletion of fatty acid degradation genes. Furthermore, the chain length and saturation of the fatty acid chain can be controlled by simply altering the thioesterase used in the pathway. The recombinant strain was further modified to directly produce fatty acid ethyl esters (FAEEs) via the introduction of ethanol production genes from Z. mobilis and overexpression of endogenous wax-ester synthase. Finally, hemicellulases from various species were expressed in recombinant fatty acid derivative producers and secreted into the medium to realize consolidated bioprocessing of hemicellulose biomass directly into biodiesels.
Aliphatic hydrocarbons, such as alkenes and alkanes, are highly attractive targets for advanced biofuel production as they are currently the major constituents of jet fuel, gasoline, and diesel. Though alkenes are naturally produced by many species, the genetic and biochemical mechanism for alkene synthesis remains unclear. Keasling and coworkers achieved long-chain alkene production through the expression of a three-gene cluster from M. luteus in a fatty acid-overproducing E. coli strain. After a series of biochemical characterizations of the strain, a metabolic pathway for alkene biosynthesis was proposed involving acyl-CoA thioester and decarboxylative Claisen condensation catalyzed by OleA [12]. In another report, del Cardayre and coworkers elucidated an alkane biosynthesis pathway from cyanobacteria [118]. In this pathway, intermediates of fatty acid metabolism are converted to alkanes and alkenes by an acyl-acyl carrier protein reductase and an aldehyde decarbonylase. Heterologous production of C13–C17 mixtures of alkanes and alkenes was achieved in E. coli by the expression of this pathway.
Chemicals
While issues such as the recurring oil crisis and global climate change have spurred the development of new fuel alternatives, they have also led to an increased awareness of the world's traditional petroleum-based chemical production processes. Consequently, clean, mild, and safe chemical synthesis processes utilizing renewable resources are highly desirable.
Organic acids
The wide application of organic acids as platform chemicals, along with the few catalytic steps required for their production, has led to intensive investigation into the microbial synthesis of organic acids. One prominent example is the production of lactic acid and its derivatives. Lactic acid is commercially produced by glucose fermentation using Lactobacillus species [15]. Additionally, recent studies have reported the production of lactic acid from other renewable substrates such as glycerol, cellobiose, and even cellulose [81, 125].
Succinic acid, a compound commonly used as a surfactant, is another widely investigated platform chemical. Succinic esters are precursors to many known petrochemical products (e.g., 1,4-butanediol). Succinate acid can be biochemically produced using bacteria such as Actinobacillus succinogenes, Mannheimia succiniproducens, as well as recombinant E. coli [68, 112]. Lang and coworkers reported the production of succinic acid in recombinant S. cerevisiae with an interrupted TCA cycle that resulted from a quadruple gene deletion. This engineered strain can produce succinic acid at a titer of 3.62 g/l with a yield of 0.11 mol/mol glucose [101].
3-Hydroxypropionic acid (3-HPA) has received significant attention mainly due to its applications in the polymer industry. 3-HPA can be produced from glucose or glycerol through various pathways [54]. Park and coworkers achieved production of 3-HPA from glycerol in a recombinant E. coli strain that expresses heterologous glycerol dehydratase and aldehyde dehydrogenase with a titer of 0.58 g/l. However, the imbalanced enzyme activity and instability of glycerol dehydratase hampers the efficient production of 3-HPA in the recombinant strains. Further study has shown that the use of an α-ketoglutaric semialdehyde dehydrogenase instead of aldehyde dehydrogenase, along with proper fermentation condition optimization, could improve the 3-HPA production to a titer of 38.7 g/l [103].
D-Glucaric acid, a compound found in fruits, vegetables and mammals, has been investigated for a variety of therapeutic purposes. D-Glucaric acid can be synthesized by the mammalian D-glucuronic acid pathway initiated with D-galactose or D-glucose. Prather and coworkers constructed a recombinant D-glucaric acid producing E. coli strain by heterologous expression of the myo-inositol-1-phosphate synthase (Ino1) from S. cerevisiae and myoinositol oxygenase (MIOX) from mice with a titer of 0.3 g/l [87]. The activity of MIOX was identified as rate limiting in the pathway, resulting in the accumulation of both myo-inositol and D-glucuronic acid. Co-expressing the urinate dehydrogenase from Pseudomonas syringae that facilitates the conversion of D-glucuronic acid into D-glucaric acid improved the production titer to more than 1 g/l. In a follow-up study, synthetic scaffolds were introduced into the recombinant system to help improve the effective concentration of myo-inositol [86]. Specifically, polypeptide scaffolds built from protein–protein interaction domains were used to co-localize three heterologous pathway enzymes involved in D-glucaric acid synthesis in a complex. The synthetic scaffolds significantly increased the specific activity of MIOX and resulted in a recombinant strain with a Wvefold improved D-glucaric acid production titer.
Rare sugars and sugar alcohols
Xylitol is a favorable sugar substitute with low caloric content and anticariogenic properties. The traditional xylitol production method involves chemical or enzymatic hydrogenation of hemicellulosic hydrolysate and extensive puri-Wcation of non-specific reduction products. However, one of the major impurities, L-arabinitol, is an epimer of xylitol, and the subsequent complex impurity separation process ultimately culminates in a prohibitively high cost for xylitol production. To overcome this obstacle, Zhao and coworkers developed a recombinant E. coli strain to produce pure xylitol from hemicellulose hydrolysate. First, an engineered aldose reductase was constructed to specifically reduce D-xylose. This engineered enzyme was obtained through rounds of directed evolution on a promiscuous aldose reductase using an in vivo selection method. The resulting enzyme exhibits a 50-fold lower catalytic efficiency toward L-arabinose while maintaining most of its D-xylose activity [91]. Furthermore, the recombinant strain was subjected to extensive metabolic engineering to further improve the selective reduction of D-xylose to xylitol. The resultant engineered E. coli strain was capable of production of nearly 100% pure xylitol from an equiweight mixture of D-xylose, L-arabinose, and D-glucose [92].
L-Ribose, a rare sugar in nature, is a very important intermediate for the preparation of pharmaceutical, food, and agrochemical products. Woodyer and coworkers developed a new synthetic platform for production of L-ribose involving the use of a unique NAD-dependent mannitol-1-dehydrogenase (MDH). The recombinant E. coli strain expressing this enzyme can be used as a whole-cell catalyst for the production of L-ribose from ribitol. As a result, L-ribose productivity of 17.4 g/l day was achieved using this system at 25°C in one-liter fermentation [142]. In a follow-up study, a directed evolution strategy was applied to improve the activity and thermal stability of MDH. The recombinant strain harboring the improved MDH achieved a conversion rate of 46.6% and a productivity of 3.88 g/l day in shake Xasks at 34°C with a overall 19.2-fold improvement. Since MDH can catalyze the interconversion of several polyols and their L-sugar counterparts, the L-ribose production system can be potentially applied in the production of other rare sugars as well [24].
1,3-Propanediol
1,3-Propanediol (1,3-PD) is a platform chemical with applications in the production of plastics, cosmetics, lubricants, and drugs. Production of 1,3-PD was achieved in E. coli using either glucose or glycerol as a substrate with a high production titer of 135 and 104 g/l, respectively [93, 129]. yqhD from E. coli and dhaB from Citrobacter freundii were cloned into a temperature-inducible vector and introduced into a recombinant E. coli strain to enable the heterologous production of 1,3-PD. Through optimization of the cultivation and supplementation of vitamin B12 (a coenzyme required for 1,3-PD production), heterologous production of 1,3-PD with a titer of 43.1 g/l could be achieved in E. coli [149]. In a more recent study, introduction of dhaB1 and dhaB2 genes from C. butyrium and yqhD from E. coli, along with a novel two-stage fed-batch fermentation strategy, achieved 1,3-PD production with a titer of 104 g/l in E. coli using glycerol as a substrate [129]. In addition to E. coli, S. cerevisiae, a well-known glycerol producer, was also engineered to produce 1,3-PD from glucose. In a recent study, both dhaB from K. pneumonia and yqhD from E. coli were introduced into S. cerevisiae via the Agrobacterium tumefaciens genetic transfer system [102]. It was shown that stable expression of the 1,3-PD production genes can be achieved in S. cerevisiae. However, the production titer of 1,3-PD only reached 0.4 g/l due to low glycerol availability.
Vitamins and amino acids
Vitamins and amino acids are important food supplements and microbial production of these compounds has been extensively investigated [17, 30]. Recently, new metabolic engineering tools have been applied to improve the production of vitamins and amino acids. For example, the carbon storage regulator (Csr), a global regulatory system of E. coli, was engineered to improve phenylalanine biosynthesis [130]. In a follow-up study, Madhyastha and coworkers explored the effects of csrA and csrD mutations and csrB overexpression on phenylalanine production in E. coli NST37 (NST) and discovered that the overexpression of csrB led to a significantly greater phenylalanine production than csrA and csrD mutations. Together with tktA overexpression, a phenylalanine production titer of 2.39 g/l was achieved [143].
Improving cellular properties
To achieve a high-level of value-added compound production, a microorganism must exhibit a high tolerance towards any substrate inhibitors and toxic products as well as the currently utilized strict industrial fermentation conditions. One good example for engineering microbial tolerance towards substrate inhibitors is the improvement of lignocellulosic hydrolysate tolerance in biofuel producing microorganisms. Lignocellulosic hydrolysate contains a variety of cell-growth inhibitors due to the harsh chemical/enzymatic hydrolysis process. These inhibitors mainly consist of acetic acid, furan derivatives, and phenolic compounds. Different engineering approaches, such as long-course adaptation, genomic library selection, and rational design, have all been applied to improve microbial tolerance and have each yielded different results [14, 44]. In a recent study, Brown and coworkers investigated the hydrolysate inhibitor tolerance of both Z. mobilis and S. cerevisiae in regards to the expression level of a conserved bacterial member of the Sm-like family of RNA-binding proteins Hfq and its homologue Lsm. Their results indicate that these regulator proteins are very important for pretreatment inhibitor tolerance [146].
Extensive investigation has been carried out to explore the tolerance of Wnal products. The understanding and engineering of microbial ethanol and butanol tolerance is a particularly active and interesting example. For example, Keasling and coworkers examined the transcript, protein, and metabolite levels in E. coli to construct a cell-wide view of the n-butanol stress response. Their results indicate that butanol stress includes the perturbation of respiratory, oxidative, heat shock, and cell envelope stresses, as well as disrupting general metabolite transport and biosynthesis [110]. Jin and coworkers studied alcohol tolerance in S. cerevisiae using transformation of a genomic DNA library and serial subculture into media containing isobutanol. Sequence analysis revealed overexpression of INO1, DOG1, HAL1, or a truncated form of MSN2 in the enriched population and provided a potential target for the understanding and engineering of an alcohol-tolerant phenotype [52].
For industrial microorganisms, resistance to stress is highly desirable due to the simultaneous or sequential combinations of different environmental stressors present in biotechnological processes. The molecular basis of stress resistances is very complicated, making it difficult to engineer stress resistances by rational approaches. However, using evolutionary engineering approaches, engineered strains with multiple-stress resistances are possible. Sauer and coworkers tested various selection procedures in chemostats and batch cultures systematically for a multiple-stress resistant S. cerevisiae phenotype. Mutant populations harvested at different time points as well as clones were randomly chosen and grown in batch cultures to be exposed to high-temperature, oxidative, freeze–thaw, and ethanol stresses. A unique high-throughput procedure utilizing 96-well plates combined with a most-probable-number assay was developed for the selection of multi-stress resistant strains. In this research, the best selection strategy to obtain highly improved multiple-stress-resistant yeast was found to be batch selection for freeze–thaw stress. Mutants not only significantly improved in freeze–thaw stress resistance but also in the other stress resistances identified by this strategy. The best isolated clone exhibited a 102-, 89-, 62-, and 1,429-fold increased resistance to freeze–thaw, high-temperature, ethanol, and oxidative stress, respectively [19].
Transport issues
Efficient uptake of a substrate into microbial cells and export of a product outside microbial cells are critical to value-added compound production. Recently, some progress has been made to improve substrate uptake, especially lignocellulose hydrolysate product uptake in biofuel producing microorganisms.
Pentose uptake through sugar transporters is the Wrst step of pentose utilization in S. cerevisiae. However, pentose sugars only enter S. cerevisiae cells through the hexose uptake system, a system that has two orders of magnitude lower affinity for pentose sugars than hexoses [63]. As a result, pentose uptake in pentose-assimilating yeast strains is very slow and inhibited by the presence of D-glucose in the growth media. To improve D-xylose uptake, heterologous D-xylose transporters were introduced into recombinant S. cerevisiae strains. Spencer-Martins and coworkers discovered one high affinity D-xylose/D-glucose symporter (GXS1) and one low affinity D-xylose/D-glucose facilitator (GXF1) from Candida intermedia and characterized them at the molecular level in S. cerevisiae [67]. It was observed that overexpression of the Gxf1 transporter improved fermentation performance in a recombinant D-xylose-utilizing S. cerevisiae strain [108]. Similarly, S. cerevisiae strains overexpressing heterologous D-xylose transporters from Arabidopsis thaliana showed up to 2.5-fold increased D-xylose consumption and 70% increased ethanol production [49]. In addition, overexpression of the D-glucose transporter Sut1 from Pichia stipitis was also shown to improve ethanol productivity during D-xylose and D-glucose co-fermentation by a D-xylose-assimilating S. cerevisiae strain [56]. However, all of the transporters mentioned above still have a lower affinity for D-xylose when compared to glucose. Recently, two D-xylose-specific transporters from pentose assimilating fungal species N. crassa and P. stipitis were discovered, heterologously expressed, and characterized in S. cerevisiae. Although the overexpression of these two D-xylose-specific transporters failed to improve D-xylose utilization in recombinant S. cerevisiae strains, the sequencing of these types of transporters may provide some insight that could eventually lead to the discovery and engineering of highly active pentose-specific sugar transporters [32].
Cellodextrins are glucose polymers with various lengths that cannot be metabolized by ethanol-producing S. cerevisiae. Cate and coworkers discovered a group of cellodextrin transporters from hemicellulose-assimilating species N. crassa through a microarray study. By introducing the newly discovered cellodextrin transporters along with β-glucosidase into S. cerevisiae, cellodextrin-assimilating recombinant strains could be constructed [37]. Follow-up studies have shown that by enabling intracellular cellodextrin utilization, the long-lasting glucose repression that occurs during mixed sugar utilization can be circumvented through the use of cellobiose instead of glucose as the carbon source [75]. Simultaneous and synergistic utilization of cellobiose and xylose could significantly reduce the cost of biomass-based fuel alternatives [45].
Conclusions and future prospects
Numerous impressive accomplishments have been made in the engineering of microbial factories for synthesis of value-added products in the past few years. However, continuous efforts towards exploring new production hosts, creating novel enzymes that catalyze unnatural reactions, and developing more powerful tools for functional genomics and proteomics will be necessary to expand the range of products that can be synthesized by microbial factories. Of special note, innovative synthetic biology approaches for pathway and genome engineering are expected to play an increasingly important role in this effort.
For example, Zhao and coworkers developed a DNA assembler approach for rapid construction and engineering of a biochemical pathway either on a plasmid or on a chromosome in S. cerevisiae in a single-step fashion [121]. This approach was further extended for discovery, characterization, and engineering of natural product biosynthetic pathways [120]. In this method, the entire expression vector containing the target biosynthetic pathway and the genetic elements required for DNA maintenance and replication in various hosts is synthesized. Because the DNA fragments to be assembled are completely mobile and amenable to all sorts of sophisticated genetic manipulations accessible to PCR, or can be chemically synthesized de novo with optimized codons, this strategy offers the ultimate versatility and Xexibility in characterizing and engineering a biochemical pathway. More importantly, the recent success in chemical synthesis of entire bacterial genomes implies the possibility of constructing artificial organisms [41–43].
In the future, synthetic biology could become as powerful as synthetic chemistry, and could greatly expand the range of products that can be produced from renewable sources. In particular, a combination of synthetic biology platforms with current protein and metabolic engineering tools is expected to give rise to a new generation of organisms that function as highly robust and programmable biological machines [72, 82].
Acknowledgments
We thank the National Institutes of Health (GM077596), the National Academies Keck Futures Initiative on Synthetic Biology, the Biotechnology Research and Development Consortium (BRDC) (Project 2-4-121), the British Petroleum Energy Biosciences Institute, and the National Science Foundation as part of the Center for Enabling New Technologies through Catalysis (CENTC), CHE-0650456, and the National Research Foundation of Korea (NRF) (220-2009-1-D00033) for Wnancial support. J. Du also acknowledges both the Chia-chen Chu graduate fellowship from the School of Chemical Sciences and the Henry Drickamer Fellowship support from the Department of Chemical and Biomolecular Engineering at the University of Illinois.
Footnotes
Jing Du and Zengyi Shao contributed equally to this work.
References
- 1.Ajikumar PK, Xiao WH, Tyo KE, Wang Y, Simeon F, Leonard E, Mucha O, Phon TH, Pfeifer B, Stephanopoulos G. Isoprenoid pathway optimization for taxol precursor overproduction in Escherichia coli. Science. 2010;330:70–74. doi: 10.1126/science.1191652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexander DC, Rock J, Gu JQ, Mascio C, Chu M, Brian P, Baltz RH. Production of novel lipopeptide antibiotics related to A54145 by Streptomyces fradiae mutants blocked in biosynthesis of modified amino acids and assignment of lptJ, lptK and lptL gene functions. J Antibiot (Tokyo) 2011;64:79–87. doi: 10.1038/ja.2010.138. [DOI] [PubMed] [Google Scholar]
- 3.Allen RS, Millgate AG, Chitty JA, Thisleton J, Miller JA, Fist AJ, Gerlach WL, Larkin PJ. RNAi-mediated replacement of morphine with the nonnarcotic alkaloid reticuline in opium poppy. Nat Biotechnol. 2004;22:1559–1566. doi: 10.1038/nbt1033. [DOI] [PubMed] [Google Scholar]
- 4.Alper H, Stephanopoulos G. Engineering for biofuels: exploiting innate microbial capacity or importing biosynthetic potential? Nat Rev Microbiol. 2009;7:715–723. doi: 10.1038/nrmicro2186. [DOI] [PubMed] [Google Scholar]
- 5.Atsumi S, Cann AF, Connor MR, Shen CR, Smith KM, Brynildsen MP, Chou KJY, Hanai T, Liao JC. Metabolic engineering of Escherichia coli for 1-butanol production. Metab Eng. 2008;10:305–311. doi: 10.1016/j.ymben.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 6.Atsumi S, Hanai T, Liao JC. Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature. 2008;451:86–89. doi: 10.1038/nature06450. [DOI] [PubMed] [Google Scholar]
- 7.Atsumi S, Wu TY, Eckl EM, Hawkins SD, Buelter T, Liao JC. Engineering the isobutanol biosynthetic pathway in Escherichia coli by comparison of three aldehyde reductase/alcohol dehydrogenase genes. Appl Microbiol Biotechnol. 2010;85:651–657. doi: 10.1007/s00253-009-2085-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Austin MB, Bowman ME, Ferrer JL, Schroder J, Noel JP. An aldol switch discovered in stilbene synthases mediates cyclization specificity of type III polyketide synthases. Chem Biol. 2004;11:1179–1194. doi: 10.1016/j.chembiol.2004.05.024. [DOI] [PubMed] [Google Scholar]
- 9.Bajwa PK, Pinel D, Martin VJJ, Trevors JT, Lee H. Strain improvement of the pentose-fermenting yeast Pichia stipitis by genome shuffling. J Microbiol Methods. 2010;81:179–186. doi: 10.1016/j.mimet.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 10.Baltz RH. Streptomyces and Saccharopolyspora hosts for heterologous expression of secondary metabolite gene clusters. J Ind Microbiol Biotechnol. 2010;37:759–772. doi: 10.1007/s10295-010-0730-9. [DOI] [PubMed] [Google Scholar]
- 11.Barnes HJ, Arlotto MP, Waterman MR. Expression and enzymatic activity of recombinant cytochrome P450 17 alpha-hydroxylase in Escherichia coli. Proc Natl Acad Sci USA. 1991;88:5597–5601. doi: 10.1073/pnas.88.13.5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beller HR, Goh EB, Keasling JD. Genes involved in long-chain alkene biosynthesis in Micrococcus luteus. Appl Environ Microbiol. 2010;76:1212–1223. doi: 10.1128/AEM.02312-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boghigian BA, Pfeifer BA. Current status, strategies, and potential for the metabolic engineering of heterologous polyketides in Escherichia coli. Biotechnol Lett. 2008;30:1323–1330. doi: 10.1007/s10529-008-9689-2. [DOI] [PubMed] [Google Scholar]
- 14.Bonomo J, Warnecke T, Hume P, Marizcurrena A, Gill RT. A comparative study of metabolic engineering anti-metabolite tolerance in Escherichia coli. Metab Eng. 2006;8:227–239. doi: 10.1016/j.ymben.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 15.Bozell JJ, Petersen GR. Technology development for the production of biobased products from biorefinery carbohydratesthe US Department of Energy's “Top 10” revisited. Green Chem. 2010;12:539–554. [Google Scholar]
- 16.Bull AT, Goodfellow M, Slater JH. Biodiversity as a source of innovation in biotechnology. Annu Rev Microbiol. 1992;46:219–252. doi: 10.1146/annurev.mi.46.100192.001251. [DOI] [PubMed] [Google Scholar]
- 17.Burgess CM, Smid EJ, Dv Sinderen Bacterial vitamin B2, B11 and B12 overproduction: an overview. Int J Food Microbiol. 2009;133:1–7. doi: 10.1016/j.ijfoodmicro.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 18.Butler MS. Natural products to drugs: natural product derived compounds in clinical trials. Nat Prod Rep. 2005;22:162–195. doi: 10.1039/b402985m. [DOI] [PubMed] [Google Scholar]
- 19.Cakar ZP, Seker UOS, Tamerler C, Sonderegger M, Sauer U. Evolutionary engineering of multiple-stress resistant Saccharomyces cerevisiae. FEMS Yeast Res. 2005;5:569–578. doi: 10.1016/j.femsyr.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 20.Cane DE. Programming of erythromycin biosynthesis by a modular polyketide synthase. J Biol Chem. 2010;285:27517–27523. doi: 10.1074/jbc.R110.144618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang MC, Eachus RA, Trieu W, Ro DK, Keasling JD. Engineering Escherichia coli for production of functionalized terpenoids using plant P450 s. Nat Chem Biol. 2007;3:274–277. doi: 10.1038/nchembio875. [DOI] [PubMed] [Google Scholar]
- 22.Chang MC, Keasling JD. Production of isoprenoid pharmaceuticals by engineered microbes. Nat Chem Biol. 2006;2:674–681. doi: 10.1038/nchembio836. [DOI] [PubMed] [Google Scholar]
- 23.Chau M, Jennewein S, Walker K, Croteau R. Taxol biosynthesis: molecular cloning and characterization of a cytochrome P450 taxoid 7 beta-hydroxylase. Chem Biol. 2004;11:663–672. doi: 10.1016/j.chembiol.2004.02.025. [DOI] [PubMed] [Google Scholar]
- 24.Christ TN, Deweese KA, Woodyer RD. Directed evolution toward improved production of L-ribose from ribitol. Comb Chem High Throughput Screen. 2010;13:302–308. doi: 10.2174/138620710791054277. [DOI] [PubMed] [Google Scholar]
- 25.Connor MR, Liao JC. Engineering of an Escherichia coli strain for the production of 3-methyl-1-butanol. Appl Environ Microbiol. 2008;74:5769–5775. doi: 10.1128/AEM.00468-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davies J. Millennium bugs. Trends Cell Biol. 1999;9:M2–M5. [PubMed] [Google Scholar]
- 27.Demain AL. From natural products discovery to commercialization: a success story. J Ind Microbiol Biotechnol. 2006;33:486–495. doi: 10.1007/s10295-005-0076-x. [DOI] [PubMed] [Google Scholar]
- 28.Den Haan R, Rose SH, Lynd LR, van Zyl WH. Hydrolysis and fermentation of amorphous cellulose by recombinant Saccharomyces cerevisiae. Metab Eng. 2007;9:87–94. doi: 10.1016/j.ymben.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 29.Dimster-Denk D, Thorsness MK, Rine J. Feedback regulation of 3-hydroxy-3-methylglutaryl coenzyme A reductase in Saccharomyces cerevisiae. Mol Biol Cell. 1994;5:655–665. doi: 10.1091/mbc.5.6.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dong X, Quinn PJ, Wang X. Metabolic engineering of Escherichia coli and Corynebacterium glutamicum for the production of L-threonine. Biotechnol Adv. 2011;29:11–23. doi: 10.1016/j.biotechadv.2010.07.009. [DOI] [PubMed] [Google Scholar]
- 31.Du H, Huang Y, Tang Y. Genetic and metabolic engineering of isoflavonoid biosynthesis. Appl Microbiol Biotechnol. 2010;86:1293–1312. doi: 10.1007/s00253-010-2512-8. [DOI] [PubMed] [Google Scholar]
- 32.Du J, Li SJ, Zhao H. Discovery and characterization of novel D-xylose-specific transporters from Neurospora crassa and Pichia stipitis. Mol Biosyst. 2010;6:2150–2156. doi: 10.1039/c0mb00007h. [DOI] [PubMed] [Google Scholar]
- 33.Engels B, Dahm P, Jennewein S. Metabolic engineering of taxadiene biosynthesis in yeast as a Wrst step towards taxol (paclitaxel) production. Metab Eng. 2008;10:201–206. doi: 10.1016/j.ymben.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 34.Fischbach MA, Walsh CT. Assembly-line enzymology for polyketide and nonribosomal peptide antibiotics: logic, machinery, and mechanisms. Chem Rev. 2006;106:3468–3496. doi: 10.1021/cr0503097. [DOI] [PubMed] [Google Scholar]
- 35.Forkmann G, Martens S. Metabolic engineering and applications of Xavonoids. Curr Opin Biotechnol. 2001;12:155–160. doi: 10.1016/s0958-1669(00)00192-0. [DOI] [PubMed] [Google Scholar]
- 36.Fujii N, Inui T, Iwasa K, Morishige T, Sato F. Knockdown of berberine bridge enzyme by RNAi accumulates (S)-reticuline and activates a silent pathway in cultured California poppy cells. Transgenic Res. 2007;16:363–375. doi: 10.1007/s11248-006-9040-4. [DOI] [PubMed] [Google Scholar]
- 37.Galazka JM, Tian CG, Beeson WT, Martinez B, Glass NL, Cate JHD. Cellodextrin transport in yeast for improved biofuel production. Science. 2010;330:84–86. doi: 10.1126/science.1192838. [DOI] [PubMed] [Google Scholar]
- 38.Galbe M, Zacchi G. A review of the production of ethanol from softwood. Appl Microbiol Biotechnol. 2002;59:618–628. doi: 10.1007/s00253-002-1058-9. [DOI] [PubMed] [Google Scholar]
- 39.Gao X, Wang P, Tang Y. Engineered polyketide biosynthesis and biocatalysis in Escherichia coli. Appl Microbiol Biotechnol. 2010;88:1233–1242. doi: 10.1007/s00253-010-2860-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gershenzon J, Dudareva N. The function of terpene natural products in the natural world. Nat Chem Biol. 2007;3:408–414. doi: 10.1038/nchembio.2007.5. [DOI] [PubMed] [Google Scholar]
- 41.Gibson DG, Benders GA, Andrews-Pfannkoch C, Denisova EA, Baden-Tillson H, Zaveri J, Stockwell TB, Brownley A, Thomas DW, Algire MA, Merryman C, Young L, Noskov VN, Glass JI, Venter JC, Hutchison CA, III, Smith HO. Complete chemical synthesis, assembly, and cloning of a Mycoplasma genitalium genome. Science. 2008;319:1215–1220. doi: 10.1126/science.1151721. [DOI] [PubMed] [Google Scholar]
- 42.Gibson DG, Benders GA, Axelrod KC, Zaveri J, Algire MA, Moodie M, Montague MG, Venter JC, Smith HO, Hutchison CA., III One-step assembly in yeast of 25 overlapping DNA fragments to form a complete synthetic Mycoplasma genitalium genome. Proc Natl Acad Sci USA. 2008;105:20404–20409. doi: 10.1073/pnas.0811011106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gibson DG, Glass JI, Lartigue C, Noskov VN, Chuang RY, Algire MA, Benders GA, Montague MG, Ma L, Moodie MM, Merryman C, Vashee S, Krishnakumar R, Assad-Garcia N, Andrews-Pfannkoch C, Denisova EA, Young L, Qi ZQ, Segall-Shapiro TH, Calvey CH, Parmar PP, Hutchison CA, III, Smith HO, Venter JC. Creation of a bacterial cell controlled by a chemically synthesized genome. Science. 2010;329:52–56. doi: 10.1126/science.1190719. [DOI] [PubMed] [Google Scholar]
- 44.Gonzalez R, Tao H, Purvis JE, York SW, Shanmugam KT, Ingram LO. Gene array-based identification of changes that contribute to ethanol tolerance in ethanologenic Escherichia coli: comparison of KO11 (parent) to LY01 (resistant mutant) Biotechnol Prog. 2003;19:612–623. doi: 10.1021/bp025658q. [DOI] [PubMed] [Google Scholar]
- 45.Ha SJ, Galazka JM, Rin Kim S, Choi JH, Yang X, Seo JH, Louise Glass N, Cate JH, Jin YS. Engineered Saccharomyces cerevisiae capable of simultaneous cellobiose and xylose fermentation. Proc Natl Acad Sci USA. 2010 doi: 10.1073/pnas.1010456108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hahn-Hagerdal B, Karhumaa K, Fonseca C, Spencer-Martins I, Gorwa-Grauslund MF. Towards industrial pentose-fermenting yeast strains. Appl Microbiol Biotechnol. 2007;74:937–953. doi: 10.1007/s00253-006-0827-2. [DOI] [PubMed] [Google Scholar]
- 47.Hanai T, Atsumi S, Liao JC. Engineered synthetic pathway for isopropanol production in Escherichia coli. Appl Environ Microbiol. 2007;73:7814–7818. doi: 10.1128/AEM.01140-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hawkins KM, Smolke CD. Production of benzylisoquinoline alkaloids in Saccharomyces cerevisiae. Nat Chem Biol. 2008;4:564–573. doi: 10.1038/nchembio.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hector RE, Qureshi N, Hughes SR, Cotta MA. Expression of a heterologous xylose transporter in a Saccharomyces cerevisiae strain engineered to utilize xylose improves aerobic xylose consumption. Appl Microbiol Biotechnol. 2008;80:675–684. doi: 10.1007/s00253-008-1583-2. [DOI] [PubMed] [Google Scholar]
- 50.Hertweck C, Luzhetskyy A, Rebets Y, Bechthold A. Type II polyketide synthases: gaining a deeper insight into enzymatic teamwork. Nat Prod Rep. 2007;24:162–190. doi: 10.1039/b507395m. [DOI] [PubMed] [Google Scholar]
- 51.Hill J, Nelson E, Tilman D, Polasky S, Tiffany D. Environmental, economic, and energetic costs and benefits of biodiesel and ethanol biofuels. Proc Natl Acad Sci USA. 2006;103:11206–11210. doi: 10.1073/pnas.0604600103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hong ME, Lee KS, Yu BJ, Sung YJ, Park SM, Koo HM, Kweon DH, Park JC, Jin YS. Identification of gene targets eliciting improved alcohol tolerance in Saccharomyces cerevisiae through inverse metabolic engineering. J Biotechnol. 2010;149:52–59. doi: 10.1016/j.jbiotec.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 53.Goodman J, Walsh V. The story of taxol: nature and politics in the pursuit of an anti-cancer drug. Cambridge University Press; Cambridge: 2001. [Google Scholar]
- 54.Jiang XL, Meng X, Xian M. Biosynthetic pathways for 3-hydroxypropionic acid production. Appl Microbiol Biotechnol. 2009;82:995–1003. doi: 10.1007/s00253-009-1898-7. [DOI] [PubMed] [Google Scholar]
- 55.Julsing MK, Koulman A, Woerdenbag HJ, Quax WJ, Kayser O. Combinatorial biosynthesis of medicinal plant secondary metabolites. Biomol Eng. 2006;23:265–279. doi: 10.1016/j.bioeng.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 56.Katahira S, Ito M, Takema H, Fujita Y, Tanino T, Tanaka T, Fukuda H, Kondo A. Improvement of ethanol productivity during xylose and glucose co-fermentation by xylose-assimilating S. cerevisiae via expression of glucose transporter Sut1. Enzyme Microb Technol. 2008;43:115–119. [Google Scholar]
- 57.Katsuyama Y, Funa N, Miyahisa I, Horinouchi S. Synthesis of unnatural flavonoids and stilbenes by exploiting the plant biosynthetic pathway in Escherichia coli. Chem Biol. 2007;14:613–621. doi: 10.1016/j.chembiol.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 58.Katsuyama Y, Hirose Y, Funa N, Ohnishi Y, Horinouchi S. Precursor-directed biosynthesis of curcumin analogs in Escherichia coli. Biosci Biotechnol Biochem. 2010;74:641–645. doi: 10.1271/bbb.90866. [DOI] [PubMed] [Google Scholar]
- 59.Katsuyama Y, Matsuzawa M, Funa N, Horinouchi S. Production of curcuminoids by Escherichia coli carrying an artificial biosynthesis pathway. Microbiology. 2008;154:2620–2628. doi: 10.1099/mic.0.2008/018721-0. [DOI] [PubMed] [Google Scholar]
- 60.Kemeny-Beke A, Aradi J, Damjanovich J, Beck Z, Facsko A, Berta A, Bodnar A. Apoptotic response of uveal melanoma cells upon treatment with chelidonine, sanguinarine and chelerythrine. Cancer Lett. 2006;237:67–75. doi: 10.1016/j.canlet.2005.05.037. [DOI] [PubMed] [Google Scholar]
- 61.Kingston DG. The shape of things to come: structural and synthetic studies of taxol and related compounds. Phytochemistry. 2007;68:1844–1854. doi: 10.1016/j.phytochem.2006.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kong WJ, Wei J, Abidi P, Lin MH, Inaba S, Li C, Wang YL, Wang ZZ, Si SY, Pan HN, Wang SK, Wu JD, Wang Y, Li ZR, Liu JW, Jiang JD. Berberine is a novel cholesterol-lowering drug working through a unique mechanism distinct from statins. Nat Med. 2004;10:1344–1351. doi: 10.1038/nm1135. [DOI] [PubMed] [Google Scholar]
- 63.Kotter P, Ciriacy M. Xylose fermentation by Saccharomyces cerevisiae. Appl Microbiol Biotechnol. 1993;38:776–783. [Google Scholar]
- 64.Kwan CY, Achike FI. Tetrandrine and related bis-benzylisoquinoline alkaloids from medicinal herbs: cardiovascular effects and mechanisms of action. Acta Pharmacol Sin. 2002;23:1057–1068. [PubMed] [Google Scholar]
- 65.Lai JH. Immunomodulatory effects and mechanisms of plant alkaloid tetrandrine in autoimmune diseases. Acta Pharmacol Sin. 2002;23:1093–1101. [PubMed] [Google Scholar]
- 66.Leadbetter JR. Cultivation of recalcitrant microbes: cells are alive, well and revealing their secrets in the 21st century laboratory. Curr Opin Microbiol. 2003;6:274–281. doi: 10.1016/s1369-5274(03)00041-9. [DOI] [PubMed] [Google Scholar]
- 67.Leandro MJ, Goncalves P, Spencer-Martins I. Two glucose/xylose transporter genes from the yeast Candida intermedia: first molecular characterization of a yeast xylose-H+ symporter. Biochem J. 2006;395:543–549. doi: 10.1042/BJ20051465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lee SJ, Song H, Lee SY. Genome-based metabolic engineering of Mannheimia succiniciproducens for succinic acid production. Biochem J. 2006;72:1939–1948. doi: 10.1128/AEM.72.3.1939-1948.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lee SY, Kim HU, Park JH, Park JM, Kim TY. Metabolic engineering of microorganisms: general strategies and drug production. Drug Discov Today. 2009;14:78–88. doi: 10.1016/j.drudis.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 70.Leonard E, Koffas MA. Engineering of artificial plant cytochrome P450 enzymes for synthesis of isoflavones by Escherichia coli. Appl Environ Microbiol. 2007;73:7246–7251. doi: 10.1128/AEM.01411-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Leonard E, Lim KH, Saw PN, Koffas MA. Engineering central metabolic pathways for high-level flavonoid production in Escherichia coli. Appl Environ Microbiol. 2007;73:3877–3886. doi: 10.1128/AEM.00200-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Leonard E, Nielsen D, Solomon K, Prather KJ. Engineering microbes with synthetic biology frameworks. Trends Biotechnol. 2008;26:674–681. doi: 10.1016/j.tibtech.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 73.Leonard E, Runguphan W, O'Connor S, Prather KJ. Opportunities in metabolic engineering to facilitate scalable alkaloid production. Nat Chem Biol. 2009;5:292–300. doi: 10.1038/nchembio.160. [DOI] [PubMed] [Google Scholar]
- 74.Leonard E, Yan Y, Fowler ZL, Li Z, Lim CG, Lim KH, Koffas MA. Strain improvement of recombinant Escherichia coli for efficient production of plant flavonoids. Mol Pharm. 2008;5:257–265. doi: 10.1021/mp7001472. [DOI] [PubMed] [Google Scholar]
- 75.Li SJ, Du J, Sun J, Galazka JM, Glass NL, Cate JHD, Yang XM, Zhao H. Overcoming glucose repression in mixed sugar fermentation by co-expressing a cellobiose transporter and a beta-glucosidase in Saccharomyces cerevisiae. Mol BioSyst. 2010;6:2129–2132. doi: 10.1039/c0mb00063a. [DOI] [PubMed] [Google Scholar]
- 76.Linger JG, Adney WS, Darzins A. Heterologous expression and extracellular secretion of cellulolytic enzymes by Zymomonas mobilis. Appl Environ Microbiol. 2010;76:6360–6369. doi: 10.1128/AEM.00230-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lynd LR, Cushman JH, Nichols RJ, Wyman CE. Fuel ethanol from cellulosic biomass. Science. 1991;251:1318–1323. doi: 10.1126/science.251.4999.1318. [DOI] [PubMed] [Google Scholar]
- 78.Lynd LR, Laser MS, Brandsby D, Dale BE, Davison B, Hamilton R, Himmel M, Keller M, McMillan JD, Sheehan J, Wyman CE. How biotech can transform biofuels. Nat Biotechnol. 2008;26:169–172. doi: 10.1038/nbt0208-169. [DOI] [PubMed] [Google Scholar]
- 79.Lynd LR, van Zyl WH, McBride JE, Laser M. Consolidated bioprocessing of cellulosic biomass: an update. Curr Opin Biotechnol. 2005;16:577–583. doi: 10.1016/j.copbio.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 80.Matsushika A, Inoue H, Murakami K, Takimura O, Sawayama S. Bioethanol production performance of Wve recombinant strains of laboratory and industrial xylose-fermenting Saccharomyces cerevisiae. Bioresour Technol. 2009;100:2392–2398. doi: 10.1016/j.biortech.2008.11.047. [DOI] [PubMed] [Google Scholar]
- 81.Mazumdar S, Clomburg JM, Gonzalez R. Escherichia coli strains engineered for homofermentative production of D-lactic acid from glycerol. Appl Environ Microbiol. 2010;76:4327–4336. doi: 10.1128/AEM.00664-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.McDaniel R, Weiss R. Advances in synthetic biology: on the path from prototypes to applications. Curr Opin Biotechnol. 2005;16:476–483. doi: 10.1016/j.copbio.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 83.Menzella HG, Reeves CD. Combinatorial biosynthesis for drug development. Curr Opin Microbiol. 2007;10:238–245. doi: 10.1016/j.mib.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 84.Minami H, Kim JS, Ikezawa N, Takemura T, Katayama T, Kumagai H, Sato F. Microbial production of plant benzylisoquinoline alkaloids. Proc Natl Acad Sci USA. 2008;105:7393–7398. doi: 10.1073/pnas.0802981105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Miyahisa I, Funa N, Ohnishi Y, Martens S, Moriguchi T, Horinouchi S. Combinatorial biosynthesis of flavones and flavonols in Escherichia coli. Appl Microbiol Biotechnol. 2006;71:53–58. doi: 10.1007/s00253-005-0116-5. [DOI] [PubMed] [Google Scholar]
- 86.Moon TS, Dueber JE, Shiue E, Prather KLJ. Use of modular, synthetic scaffolds for improved production of glucaric acid in engineered E. coli. Metab Eng. 2010;12:298–305. doi: 10.1016/j.ymben.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 87.Moon TS, Yoon SH, Lanza AM, Roy-Mayhew JD, Prather KLJ. Production of glucaric acid from a synthetic pathway in recombinant Escherichia coli. Appl Environ Microbiol. 2009;75:589–595. doi: 10.1128/AEM.00973-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Muntendam R, Melillo E, Ryden A, Kayser O. Perspectives and limits of engineering the isoprenoid metabolism in heterologous hosts. Appl Microbiol Biotechnol. 2009;84:1003–1019. doi: 10.1007/s00253-009-2150-1. [DOI] [PubMed] [Google Scholar]
- 89.Mutka SC, Bondi SM, Carney JR, Da Silva NA, Kealey JT. Metabolic pathway engineering for complex polyketide biosynthesis in Saccharomyces cerevisiae. FEMS Yeast Res. 2006;6:40–47. doi: 10.1111/j.1567-1356.2005.00001.x. [DOI] [PubMed] [Google Scholar]
- 90.Mutka SC, Carney JR, Liu Y, Kennedy J. Heterologous production of epothilone C and D in Escherichia coli. Biochemistry. 2006;45:1321–1330. doi: 10.1021/bi052075r. [DOI] [PubMed] [Google Scholar]
- 91.Nair NU, Zhao H. Evolution in reverse: engineering a D-xylose-specific xylose reductase. ChemBioChem. 2008;9:1213–1215. doi: 10.1002/cbic.200700765. [DOI] [PubMed] [Google Scholar]
- 92.Nair NU, Zhao H. Selective reduction of xylose to xylitol from a mixture of hemicellulosic sugars. Metab Eng. 2010;12:462–468. doi: 10.1016/j.ymben.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 93.Nakamura CE, Whited GM. Metabolic engineering for the microbial production of 1, 3-propanediol. Curr Opin Biotechnol. 2003;14:454–459. doi: 10.1016/j.copbio.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 94.Nguyen KT, He X, Alexander DC, Li C, Gu JQ, Mascio C, Van Praagh A, Mortin L, Chu M, Silverman JA, Brian P, Baltz RH. Genetically engineered lipopeptide antibiotics related to A54145 and daptomycin with improved properties. Antimicrob Agents Chemother. 2010;54:1404–1413. doi: 10.1128/AAC.01307-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nguyen KT, Ritz D, Gu JQ, Alexander D, Chu M, Miao V, Brian P, Baltz RH. Combinatorial biosynthesis of novel antibiotics related to daptomycin. Proc Natl Acad Sci USA. 2006;103:17462–17467. doi: 10.1073/pnas.0608589103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nicolaou KC, Yang Z, Liu JJ, Ueno H, Nantermet PG, Guy RK, Claiborne CF, Renaud J, Couladouros EA, Paulvannan K, et al. Total synthesis of taxol. Nature. 1994;367:630–634. doi: 10.1038/367630a0. [DOI] [PubMed] [Google Scholar]
- 97.Olano C, Mendez C, Salas JA. Antitumor compounds from actinomycetes: from gene clusters to new derivatives by combinatorial biosynthesis. Nat Prod Rep. 2009;26:628–660. doi: 10.1039/b822528a. [DOI] [PubMed] [Google Scholar]
- 98.Olano C, Mendez C, Salas JA. Post-PKS tailoring steps in natural product-producing actinomycetes from the perspective of combinatorial biosynthesis. Nat Prod Rep. 2010;27:571–616. doi: 10.1039/b911956f. [DOI] [PubMed] [Google Scholar]
- 99.Pfeifer BA, Admiraal SJ, Gramajo H, Cane DE, Khosla C. Biosynthesis of complex polyketides in a metabolically engineered strain of E. coli. Science. 2001;291:1790–1792. doi: 10.1126/science.1058092. [DOI] [PubMed] [Google Scholar]
- 100.Holton RA, Biediger RJ, Boatman PD. Taxol: science and applications. CRC; Boca Raton: 1995. [Google Scholar]
- 101.Raab AM, Gebhardt G, Bolotina N, Weuster-Botz D, Lang C. Metabolic engineering of Saccharomyces cerevisiae for the biotechnological production of succinic acid. Metab Eng. 2010;12:518–525. doi: 10.1016/j.ymben.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 102.Rao Z, Ma Z, Shen W, Fang H, Zhuge J, Wang X. Engineered Saccharomyces cerevisiae that produces 1, 3-propanediol from D-glucose. J Appl Microbiol. 2008;105:1768–1776. doi: 10.1111/j.1365-2672.2008.03868.x. [DOI] [PubMed] [Google Scholar]
- 103.Rathnasingh C, Raj SM, Jo JE, Park S. Development and evaluation of efficient recombinant Escherichia coli strains for the production of 3-hydroxypropionic acid from glycerol. Biotechnol Bioeng. 2009;104:729–739. doi: 10.1002/bit.22429. [DOI] [PubMed] [Google Scholar]
- 104.Ro DK, Ouellet M, Paradise EM, Burd H, Eng D, Paddon CJ, Newman JD, Keasling JD. Induction of multiple pleiotropic drug resistance genes in yeast engineered to produce an increased level of anti-malarial drug precursor, artemisinic acid. BMC Biotechnol. 2008;8:83. doi: 10.1186/1472-6750-8-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ro DK, Paradise EM, Ouellet M, Fisher KJ, Newman KL, Ndungu JM, Ho KA, Eachus RA, Ham TS, Kirby J, Chang MC, Withers ST, Shiba Y, Sarpong R, Keasling JD. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature. 2006;440:940–943. doi: 10.1038/nature04640. [DOI] [PubMed] [Google Scholar]
- 106.Roberts SC. Production and engineering of terpenoids in plant cell culture. Nat Chem Biol. 2007;3:387–395. doi: 10.1038/nchembio.2007.8. [DOI] [PubMed] [Google Scholar]
- 107.Roca C, Haack MB, Olsson L. Engineering of carbon catabolite repression in recombinant xylose fermenting Saccharomyces cerevisiae. Appl Microbiol Biotechnol. 2004;63:578–583. doi: 10.1007/s00253-003-1408-2. [DOI] [PubMed] [Google Scholar]
- 108.Runquist D, Fonseca C, Radstrom P, Spencer-Martins I, Hahn-Hagerdal B. Expression of the Gxf1 transporter from Candida intermedia improves fermentation performance in recombinant xylose-utilizing Saccharomyces cerevisiae. Appl Microbiol Biotechnol. 2009;82:123–130. doi: 10.1007/s00253-008-1773-y. [DOI] [PubMed] [Google Scholar]
- 109.Runquist D, Hahn-Hagerdal B, Bettiga M. Increased ethanol productivity in xylose-utilizing Saccharomyces cerevisiae via a randomly mutagenized xylose reductase. Appl Environ Microbiol. 2010;76:7796–7802. doi: 10.1128/AEM.01505-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rutherford BJ, Dahl RH, Price RE, Szmidt HL, Benke PI, Mukhopadhyay A, Keasling JD. Functional genomic study of exogenous n-butanol stress in Escherichia coli. Appl Environ Microbiol. 2010;76:1935–1945. doi: 10.1128/AEM.02323-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Saha BC. Hemicellulose bioconversion. J Ind Microbiol Biotechnol. 2003;30:279–291. doi: 10.1007/s10295-003-0049-x. [DOI] [PubMed] [Google Scholar]
- 112.Sanchez AM, Bennett GN, San KY. Efficient succinic acid production from glucose through overexpression of pyruvate carboxylase in an Escherichia coli alcohol dehydrogenase and lactate dehydrogenase mutant. Biotechnol Prog. 2005;21:358–365. doi: 10.1021/bp049676e. [DOI] [PubMed] [Google Scholar]
- 113.Sanchez C, Mendez C, Salas JA. Engineering biosynthetic pathways to generate antitumor indolocarbazole derivatives. J Ind Microbiol Biotechnol. 2006;33:560–568. doi: 10.1007/s10295-006-0092-5. [DOI] [PubMed] [Google Scholar]
- 114.Sanchez C, Salas AP, Brana AF, Palomino M, Pineda-Lucena A, Carbajo RJ, Mendez C, Moris F, Salas JA. Generation of potent and selective kinase inhibitors by combinatorial biosyn-thesis of glycosylated indolocarbazoles. Chem Commun (Camb) 2009:4118–4120. doi: 10.1039/b905068j. [DOI] [PubMed] [Google Scholar]
- 115.Sato F, Hashimoto T, Hachiya A, Tamura K, Choi KB, Morishige T, Fujimoto H, Yamada Y. Metabolic engineering of plant alkaloid biosynthesis. Proc Natl Acad Sci USA. 2001;98:367–372. doi: 10.1073/pnas.011526398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sato F, Inui T, Takemura T. Metabolic engineering in isoquinoline alkaloid biosynthesis. Curr Pharm Biotechnol. 2007;8:211–218. doi: 10.2174/138920107781387438. [DOI] [PubMed] [Google Scholar]
- 117.Schafer H, Wink M. Medicinally important secondary metabolites in recombinant microorganisms or plants: progress in alkaloid biosynthesis. Biotechnol J. 2009;4:1684–1703. doi: 10.1002/biot.200900229. [DOI] [PubMed] [Google Scholar]
- 118.Schirmer A, Rude MA, Li XZ, Popova E, del Cardayre SB. Microbial biosynthesis of alkanes. Science. 2010;329:559–562. doi: 10.1126/science.1187936. [DOI] [PubMed] [Google Scholar]
- 119.Sevrioukova IF, Li H, Zhang H, Peterson JA, Poulos TL. Structure of a cytochrome P450-redox partner electron-transfer complex. Proc Natl Acad Sci USA. 1999;96:1863–1868. doi: 10.1073/pnas.96.5.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Shao Z, Luo Y, Zhao H. Rapid characterization and engineering of natural product biosynthetic pathways via DNA assembler. Mol BioSyst. 2011 doi: 10.1039/c0mb00338g. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Shao Z, Zhao H, Zhao H. DNA assembler, an in vivo genetic method for rapid construction of biochemical pathways. Nucleic Acids Res. 2009;37:e16. doi: 10.1093/nar/gkn991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sieber SA, Marahiel MA. Molecular mechanisms underlying nonribosomal peptide synthesis: approaches to new antibiotics. Chem Rev. 2005;105:715–738. doi: 10.1021/cr0301191. [DOI] [PubMed] [Google Scholar]
- 123.Siewers V, Chen X, Huang L, Zhang J, Nielsen J. Heterologous production of non-ribosomal peptide LLD-ACV in Saccharomyces cerevisiae. Metab Eng. 2009;11:391–397. doi: 10.1016/j.ymben.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 124.Simkhada D, Kim E, Lee HC, Sohng JK. Metabolic engineering of Escherichia coli for the biological synthesis of 7-O-xylosyl naringenin. Mol Cells. 2009;28:397–401. doi: 10.1007/s10059-009-0135-7. [DOI] [PubMed] [Google Scholar]
- 125.Singhvi M, Joshi D, Adsul M, Varma A, Gokhale D. D-(−)-Lactic acid production from cellobiose and cellulose by Lactobacillus lactis mutant RM2–24. Green Chem. 2010;12:1106–1109. [Google Scholar]
- 126.Staunton J, Weissman KJ. Polyketide biosynthesis: a millennium review. Nat Prod Rep. 2001;18:380–416. doi: 10.1039/a909079g. [DOI] [PubMed] [Google Scholar]
- 127.Steen EJ, Chan R, Prasad N, Myers S, Petzold CJ, Redding A, Ouellet M, Keasling JD. Metabolic engineering of Saccharomyces cerevisiae for the production of n-butanol. Microb Cell Fact. 2008;7:36–43. doi: 10.1186/1475-2859-7-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Steen EJ, Kang YS, Bokinsky G, Hu ZH, Schirmer A, McClure A, del Cardayre SB, Keasling JD. Microbial production of fatty-acid-derived fuels and chemicals from plant biomass. Nature. 2010;463:559–562. doi: 10.1038/nature08721. [DOI] [PubMed] [Google Scholar]
- 129.Tang XM, Tan YS, Zhu H, Zhao K, Shen W. Microbial conversion of glycerol to 1, 3-propanediol by an engineered strain of Escherichia coli. Appl Environ Microbiol. 2009;75:1628–1634. doi: 10.1128/AEM.02376-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Tatarko M, Romeo T. Disruption of a global regulatory gene to enhance central carbon flux into phenylalanine biosyn-thesis in Escherichia coli. Curr Microbiol. 2001;43:26–32. doi: 10.1007/s002840010255. [DOI] [PubMed] [Google Scholar]
- 131.Tsai SL, Goyal G, Chen W. Surface display of a functional minicellulosome by intracellular complementation using a synthetic yeast consortium and its application to cellulose hydrolysis and ethanol production. Appl Environ Microbiol. 2010;76:7514–7520. doi: 10.1128/AEM.01777-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Turnbull JJ, Nakajima J, Welford RW, Yamazaki M, Saito K, Schofield CJ. Mechanistic studies on three 2-oxoglutarate-dependent oxygenases of flavonoid biosynthesis: anthocyanidin synthase, flavonol synthase, and flavanone 3β-hydroxylase. J Biol Chem. 2004;279:1206–1216. doi: 10.1074/jbc.M309228200. [DOI] [PubMed] [Google Scholar]
- 133.van Zyl WH, Lynd LR, den Haan R, McBride JE. Biofuels, vol 108. Advances in biochemical engineering/biotechnology. Springer; Berlin Heidelberg New York: 2007. Consolidated bioprocessing for bioethanol production using Saccharomyces cerevisiae; pp. 205–235. [DOI] [PubMed] [Google Scholar]
- 134.Walker GM. Yeast physiology and biotechnology. Wiley; New York: 1998. [Google Scholar]
- 135.Wang Y, Boghigian BA, Pfeifer BA. Improving heterologous polyketide production in Escherichia coli by overexpression of an S-adenosylmethionine synthetase gene. Appl Microbiol Biotechnol. 2007;77:367–373. doi: 10.1007/s00253-007-1172-9. [DOI] [PubMed] [Google Scholar]
- 136.Wani MC, Taylor HL, Wall ME, Coggon P, McPhail AT. Plant antitumor agents. VI. The isolation and structure of taxol, a novel antileukemic and antitumor agent from Taxus brevifolia. J Am Chem Soc. 1971;93:2325–2327. doi: 10.1021/ja00738a045. [DOI] [PubMed] [Google Scholar]
- 137.Weisshaar B, Jenkins GI. Phenylpropanoid biosynthesis and its regulation. Curr Opin Plant Biol. 1998;1:251–257. doi: 10.1016/s1369-5266(98)80113-1. [DOI] [PubMed] [Google Scholar]
- 138.Weissman KJ, Leadlay PF. Combinatorial biosynthesis of reduced polyketides. Nat Rev Microbiol. 2005;3:925–936. doi: 10.1038/nrmicro1287. [DOI] [PubMed] [Google Scholar]
- 139.Wen F, Nair NU, Zhao H. Protein engineering in designing tailored enzymes and microorganisms for biofuels production. Curr Opin Biotechnol. 2009;20:412–419. doi: 10.1016/j.copbio.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Wen F, Sun J, Zhao H. Yeast surface display of trifunctional minicellulosomes for simultaneous saccharification and fermentation of cellulose to ethanol. Appl Environ Microbiol. 2010;76:1251–1260. doi: 10.1128/AEM.01687-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Williams PA, Cosme J, Sridhar V, Johnson EF, McRee DE. Microsomal cytochrome P450 2C5: comparison to microbial P450 s and unique features. J Inorg Biochem. 2000;81:183–190. doi: 10.1016/s0162-0134(00)00102-1. [DOI] [PubMed] [Google Scholar]
- 142.Woodyer RD, Wymer NJ, Racine FM, Khan SN, Saha BC. Efficient production of L-ribose with a recombinant Escherichia coli biocatalyst. Appl Environ Microbiol. 2008;74:2967–2975. doi: 10.1128/AEM.02768-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Yakandawala N, Romeo T, Friesen AD, Madhyastha S. Metabolic engineering of Escherichia coli to enhance phenylalanine production. Appl Microbiol Biotechnol. 2008;78:283–291. doi: 10.1007/s00253-007-1307-z. [DOI] [PubMed] [Google Scholar]
- 144.Yan Y, Huang L, Koffas MA. Biosynthesis of 5-deoxyflavanones in microorganisms. Biotechnol J. 2007;2:1250–1262. doi: 10.1002/biot.200700119. [DOI] [PubMed] [Google Scholar]
- 145.Yan Y, Liao JC. Engineering metabolic systems for production of advanced fuels. J Ind Microbiol Biotechnol. 2009;36:471–479. doi: 10.1007/s10295-009-0532-0. [DOI] [PubMed] [Google Scholar]
- 146.Yang SH, Pelletier DA, Lu TYS, Brown SD. The Zymomonas mobilis regulator hfq contributes to tolerance against multiple lignocellulosic pretreatment inhibitors. BMC Biotechnol. 2010;10:11. doi: 10.1186/1471-2180-10-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Zhang H, Boghigian BA, Pfeifer BA. Investigating the role of native propionyl-CoA and methylmalonyl-CoA metabolism on heterologous polyketide production in Escherichia coli. Biotechnol Bioeng. 2010;105:567–573. doi: 10.1002/bit.22560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zhang W, Li Y, Tang Y. Engineered biosynthesis of bacterial aromatic polyketides in Escherichia coli. Proc Natl Acad Sci USA. 2008;105:20683–20688. doi: 10.1073/pnas.0809084105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zhang XM, Li Y, Zhuge B, Tang XM, Shen W, Rao ZM, Fang HY, Zhuge J. Construction of a novel recombinant Escherichia coli strain capable of producing 1, 3-propanediol and optimization of fermentation parameters by statistical design. World J Microbiol Biotechnol. 2006;22:945–952. [Google Scholar]
- 150.Zulak KG, Cornish A, Daskalchuk TE, Deyholos MK, Goode-nowe DB, Gordon PM, Klassen D, Pelcher LE, Sensen CW, Facchini PJ. Gene transcript and metabolite profiling of elicitor-induced opium poppy cell cultures reveals the coordinate regulation of primary and secondary metabolism. Planta. 2007;225:1085–1106. doi: 10.1007/s00425-006-0419-5. [DOI] [PubMed] [Google Scholar]