Abstract
The conserved RCN family of proteins can bind and directly regulate calcineurin, a Ca2+-activated protein phosphatase involved in immunity, heart growth, muscle development, learning, and other processes. Whereas high levels of RCNs can inhibit calcineurin signaling in fungal and animal cells, RCNs can also stimulate calcineurin signaling when expressed at endogenous levels. Here we show that the stimulatory effect of yeast Rcn1 involves phosphorylation of a conservedserine residue by Mck1, a member of the GSK-3 family of protein kinases. Mutations at the GSK-3 consensus site of Rcn1 and human DSCR1/MCIP1 abolish the stimulatory effects on calcineurin signaling. RCNs may therefore oscillate between stimulatory and inhibitory forms in vivo in a manner similar to the Inhibitor-2 regulators of type 1 protein phosphatase. Computational modeling indicates a biphasic response of calcineurin to increasing RCN concentration such that protein phosphatase activity is stimulated by low concentrations of phospho-RCN and inhibited by high concentrations of phospho- or dephospho-RCN. This prediction was verified experimentally in yeast cells expressing Rcn1 or DSCR1/MCIP1 at different concentrations. Through the phosphorylation of RCNs, GSK-3 kinases can potentially contribute to a positive feedback loop involving calcineurin-dependent up-regulation of RCN expression. Such feedback may help explain the large induction of DSCR1/MCIP1 observed in brain of Down syndrome individuals.
Keywords: Calcineurin, calcium signaling, Rcn1p, DSCR1, MCIP, GSK-3
The serine/threonine protein phosphatase calcineurin becomes highly stimulated in response to Ca2+ signals through the direct binding of Ca2+ and calmodulin (Klee et al. 1998; Rusnak and Mertz 2000). The enzyme and its modes of regulation appear to be broadly conserved among all animals and fungi as well as many protozoans (Hilioti and Cunningham 2003a). Animal and fungal calcineurins are sensitive to cyclosporin A and FK506, which bind directly in association with the receptor proteins cyclophilin and FKBP-12. These compounds are used routinely in humans as immunosuppressive drugs to control certain autoimmune disorders and to prevent allograft rejection after transplantation. The chronic side effects of these drugs have generated great interest in understanding the roles of calcineurin in different cell types. The recent realization that, in many pathogenic fungi, calcineurin promotes resistance to a major class of antifungal drugs (Fox and Heitman 2002) has also fueled interest in developing new strategies for more selective calcineurin control. A better understanding of calcineurin regulation in vivo may facilitate these efforts.
Major substrates of calcineurin in vertebrates include the NFAT family of transcription factors (Crabtree and Olson 2002). During the activation of T-lymphocytes, for example, calcineurin dephosphorylates NFATs in the cytoplasm and promotes their import into the nucleus, where they induce the transcription of many genes involved in lymphocyte activation and proliferation. Phosphorylation of NFATs by GSK-3 protein kinases decreases their DNA-binding activity and promotes their export from the nucleus, thereby inhibiting spontaneous activation (Beals et al. 1997). Similar regulatory circuits operate in many other cell types. For example, expression of hyperactive calcineurin in the developing heart can induce hypertrophy in transgenic mice, and this effect can be mimicked by hyperactive NFAT and ameliorated to some degree by hyperactive GSK-3 or the immunosuppressive drugs (Molkentin et al. 1998). Specific inhibitors of calcineurin or NFAT in the heart may therefore become useful therapeutics in the treatment of heart failure.
A family of proteins conserved from yeast to humans, termed RCNs, was recently identified as endogenous regulators of calcineurin (Fuentes et al. 2000; Gorlach et al. 2000; Kingsbury and Cunningham 2000). In budding yeast, overexpression of either Rcn1 or two of its human homologs DSCR1/MCIP1 and ZAKI4/MCIP2 strongly inhibited calcineurin-dependent processes (Kingsbury and Cunningham 2000). Similarly, overexpression of DSCR1/MCIP1 inhibited calcineurin-dependent activation of NFAT and other transcription factors (Fuentes et al. 2000; Rothermel et al. 2000). DSCR1/MCIP1 overexpression in the developing heart also blocked calcineurin-induced cardiac hypertrophy in mouse (Rothermel et al. 2001; Hill et al. 2002). Finally, overexpression of RCN-1 in nematodes produced phenotypes consistent with calcineurin inhibition (Lee et al. 2003). RCNs directly interact with calcineurin in vivo, and purified recombinant DSCR1/MCIP1 can bind to the catalytic subunit of calcineurin in vitro and potently inhibit the protein phosphatase activity of the holoenzyme (Gorlach et al. 2000; Kingsbury and Cunningham 2000; Rothermel et al. 2000; Vega et al. 2002).
The hypothesis that RCNs serve purely as inhibitors of calcineurin is unable to explain several observations in yeast and mouse. Disruption of the RCN1 gene in yeast resulted in significantly lower calcineurin signaling in contrast to the expected increase in calcineurin signaling (Kingsbury and Cunningham 2000). The stimulatory effect of endogenous Rcn1 on calcineurin signaling was not peculiar to yeast because human DSCR1/MCIP1 also stimulated calcineurin signaling when expressed in rcn1 mutants (Kingsbury and Cunningham 2000). Homozygous disruption of the DSCR1/MCIP1 gene in mouse resulted in diminished calcineurin function in the heart under normal circumstances and in response to certain stresses such as pressure overload (Vega et al. 2003). Thus, RCNs seem to stimulate calcineurin signaling when expressed at their physiological levels. Expression of RCN1 and DSCR1/MCIP1 genes is strongly up-regulated in response to calcineurin signaling (Fuentes et al. 2000; Kingsbury and Cunningham 2000; Rothermel et al. 2000), and therefore the accumulation of RCNs may generate either positive or negative feedback depending on the level of expression. Because the DSCR1/MCIP1 gene is overexpressed in the brain of trisomy 21 individuals and is located within the Down syndrome critical region of Chromosome 21 (Fuentes et al. 2000), a clearer understanding of the stimulatory and inhibitory effects of RCNs on calcineurin may shed light on the physiology of this complex disorder.
Here we investigate the stimulatory and inhibitory effects of Rcn1 in greater detail. We show that the stimulatory activity of Rcn1 on calcineurin signaling requires phosphorylation of a conserved serine residue by the protein kinase Mck1, a member of the GSK-3 family of protein kinases. Substituting this serine with alanine in either Rcn1 or DSCR1/MCIP1 abolishes their stimulatory effects on calcineurin and enhances their inhibitory effects. All the biochemical and genetic data fit with a model in which phospho-Rcn1 stimulates and dephospho-Rcn1 inhibits calcineurin signaling. This model is strikingly similar to one proposed for the allosteric regulation of type-1 protein phosphatase (PP1) by inhibitor-2 (Inh2; Cohen 2002). Our findings demonstrate that calcineurin signaling can be significantly modulated by GSK-3 through effects on RCNs and provide new opportunities for the therapeutic control of calcineurin.
Results
A GSK-3 kinase stimulates calcineurin signaling in yeast
The Pmc1 and Vcx1 Ca2+ transporters in yeast are necessary for growth in high-Ca2+ environments but are differentially regulated by calcineurin (Cunningham and Fink 1996). Calcineurin directly or indirectly inhibits Vcx1, which causes diminished growth rates in high Ca2+ conditions, especially in the absence of Pmc1. Consequently, calcineurin-deficient mutants can be easily isolated by selection for Ca2+ resistance in a pmc1 mutant background. Previously we isolated 28 such variants and determined that half of these carried mutations in the CNB1 gene encoding the B subunit of calcineurin (Cunningham and Fink 1994). Of the remaining mutants, 11 were found to carry recessive alleles of the MCK1 gene (see Materials and Methods). Loss-of-function mutations in MCK1 were unexpected because this gene encodes a member of the GSK-3 family of serine/threonine protein kinases (Woodgett 2001), and GSK-3 kinases frequently antagonize calcineurin signaling by phosphorylating calcineurin targets. Disruption of the MCK1 gene increased the Ca2+ tolerance of pmc1 tcn1 double mutants but not of pmc1 tcn1 vcx1 triple mutants (Fig. 1), indicating that Mck1 does not antagonize calcineurin but cooperates with calcineurin to inhibit Vcx1. Complete inhibition of calcineurin with FK506 produced a similar degree of Ca2+ tolerance as the loss of Mck1 and FK506 did not further increase the Ca2+ tolerance of the Mck1-deficient strain (Fig. 1). The Mck1 protein kinase and calcineurin protein phosphatase therefore appear to function cooperatively in a pathway that regulates Vcx1 function.
Figure 1.
Mck1 is required for calcineurin-dependent inhibition of Vcx1 function. Vcx1 function was measured by its ability to promote growth of Pmc1- and Tcn1-deficient cells in YPDS media supplemented with high concentrations of CaCl2 as indicated. (A) The Vcx1-proficient pmc1 tcn1 double mutants (circles, strain K1349) and mck1 pmc1 tcn1 triple mutants (triangles, strain K1353) were inoculated at low density into media lacking (solid lines) or containing (dashed lines) FK506 and incubated for 18 h before measuring optical density at 650 nm (OD650). (B) As a reference, the Vcx1-deficient pmc1 tcn1 vcx1 triple mutant (circles, strain K1357) and mck1 pmc1 tcn1 vcx1 quadruple mutant (triangles, strain K1361) were analyzed as in A.
Independent of its effects on Vcx1, calcineurin dephosphorylates and activates the Tcn1/Crz1 transcription factor, which induces expression of target genes such as PMC1, RCN1, and FKS2 (Matheos et al. 1997; Stathopoulos and Cyert 1997; Kingsbury and Cunningham 2000). To test whether Mck1 also cooperates with calcineurin to activate Tcn1/Crz1, the expression of PMC1 and RCN1 genes in wild-type cells and mck1 mutants was monitored by Northern blot analysis. After treatment with 100 mM CaCl2, PMC1 and RCN1 transcripts increased to much higher levels in wild-type cells than in mck1 mutants (Fig. 2A,B). Expression of the CDRE-lacZ reporter gene (derived from the FKS2 promoter) was also diminished in mck1 mutants to only 15% of wild-type levels (Fig. 2D). The residual induction of CDRE-lacZ in mck1 mutants was completely sensitive to FK506 (data not shown), indicating that calcineurin activity persists in mck1 mutants at a very low level that could not be detected in the Vcx1 inhibition assay. The possibility that [Ca2+]c rises to higher levels in wild-type cells than in mck1 mutants was ruled out by several experiments. First, luminescence of the Ca2+-sensitive photoprotein aequorin expressed in the cytoplasm was indistinguishable in wild-type and mck1 mutant cells before and after treatment with high Ca2+ (data not shown). Second, strains that lack both Pmc1 and Vcx1 are very sensitive to high Ca2+ conditions with or without Mck1 (Fig. 1B). Finally, induction of CDRE-lacZ, PMC1-lacZ, and RCN1-lacZ reporter genes in mck1 pmc1 vcx1 triple mutants (strain K1329) was, respectively, only 8.3%, 1.5%, and 3% of that observed in pmc1 vcx1 double mutants (strain K1325). As controls, three Tcn1/Crz1-independent reporter genes (CYC1-lacZ, CTS1-lacZ, and HO-lacZ) were found to be expressed in mck1 mutants at levels only slightly lower than those of wild-type cells (66%, 61%, and 71%, respectively). These experiments indicate that Mck1 plays a strong positive role in calcineurin signaling.
Figure 2.
Mck1 is required for calcineurin-dependent activation of Tcn1/Crz1 function. Expression of (A) RCN1, (B) PMC1, and (C) MCK1 genes in wild type, rcn1 mutants, mck1 mutants, and mck1 rcn1 double mutants (strains K601, K1365, K1305, and K1369) was measured by Northern blot analysis at various times after treatment of cultures with 100 mM CaCl2. (D) Expression of CDRE-lacZ in strains lacking Rcn1 and/or the GSK-3 family kinases Mck1, Mrk1, Rim11, and Ygk3 (strains) was measured after 4 h of growth in YPDS medium with or without 100 mM CaCl2. Bars represent the average of three independent experiments (±SD).
Effects of other GSK-3 kinases in yeast
Yeast expresses four members of the GSK-3 family of protein kinases (Mck1, Mrk1, Rim11, and Ygk3) which may have arisen from relatively recent duplications of an ancestral gene followed by rapid divergence (K.W. Cunningham, unpubl.). To assess the potential contributions of these other GSK-3 family kinases in calcineurin signaling, CDRE-lacZ expression was measured in a panel of mutants lacking one or more of the GSK-3 kinases. In contrast to mck1 mutants, the loss of either Ygk3, Rim11, or Mrk1 had no significant effects on CDRE-lacZ induction relative to wild-type cells (data not shown). Induction of CDRE-lacZ in mck1 mrk1 rim11 ygk3 quadruple mutants was diminished ∼threefold relative to mck1 single mutants, suggesting a weak contribution of Mrk1, Rim11, and/or Ygk3 to calcineurin signaling (Fig. 2D). Triple mutants that express a single GSK-3 family kinase in the absence of the other three revealed no significant contributions of either Mrk1 or Ygk3, a weak stimulatory effect of Rim11, and a nearly wild-type stimulatory effect of Mck1 alone (Fig. 2D). Thus, Mck1 and Rim11 (to a much lesser degree) appeared capable of stimulating calcineurin signaling in yeast.
In vertebrates, GSK-3 is typically very active in resting cells and inactivated by signals that cause either phosphorylation of serine 9, dephosphorylation of tyrosine 216, or sequestration into complexes (Woodgett 2001). Yeast GSK-3 kinases lack the serine 9 pseudosubstrate domain, and their modes of regulation are not fully known. Recently, MCK1 gene expression and function were proposed to increase upon treatment of yeast cells in high Ca2+ conditions (Mizunuma et al. 2001). However, genome-wide expression studies failed to detect any increase in MCK1 expression after treatment with high Ca2+ (Yoshimoto et al. 2002). Northern blot analysis of MCK1 transcripts revealed no significant effects of Ca2+ treatment in either wild-type or rcn1 mutant cells (Fig. 2C). Similarly, Western blot analysis of epitope-tagged Mck1–MYC and Mck1–HA derivatives also failed to show any increase in Mck1 protein levels (data not shown). The original evidence used to infer stimulation of Mck1 function by Ca2+ involved assays of phenomena that themselves were sensitive to calcineurin (Mizunuma et al. 2001). Although we cannot rule out the possibility that Ca2+ stimulates Mck1 function, all the available data are consistent with the model that Mck1 stimulates calcineurin signaling toward multiple targets, including Hsl1 (Mizunuma et al. 2001) and a newly identified Ca2+ channel in the plasma membrane of yeast cells (Bonilla and Cunningham 2003).
Mck1 functions in the same pathway as Rcn1
The stimulatory effect of Mck1 on calcineurin signaling resembles that of Rcn1, a direct regulator of calcineurin (Kingsbury and Cunningham 2000). If Mck1 and Rcn1 act in the same pathway to regulate calcineurin signaling, the defects of mck1 rcn1 double mutants should be similar to those of the mck1 and rcn1 single mutants. The single mutants differ slightly in the induction of CDRE-lacZ, but this small difference may be due to the nonspecific ∼33% reduction in the expression of all such reporter genes in mck1 mutants (see above). Induction of CDRE-lacZ in mck1 rcn1 double mutants was indistinguishable from that of mck1 single mutants (Fig. 2D), indicating that Mck1 is required for the ability of Rcn1 to stimulate calcineurin. Overexpression of Mck1 in rcn1 mutants or wild-type cells had no detectable effect on calcineurin-dependent induction of CDRE-lacZ (data not shown). Northern blot analysis of PMC1 transcripts confirmed equivalent induction in mck1 mutants and mck1 rcn1 double mutants, which was much lower than that of wild type at all times after Ca2+ addition (Fig. 2A). Mck1 and Rcn1 may therefore act within the same regulatory pathway to enhance calcineurin signaling.
To further examine the potential interactions between Mck1, Rcn1, calcineurin, and Tcn1/Crz1, a series of cDNA microarray measurements of genome expression was performed. We compared genome expression in wild-type cells to that of mck1, rcn1, and tcn1 mutants with or without brief treatments with 100 mM CaCl2. Most experiments were repeated independently in two different strain backgrounds. Published data comparing wild type and tcn1 mutants under similar conditions (Yoshimoto et al. 2002) were reprocessed and included in our analysis for comparison (see Materials and Methods). A total of 388 genes were found to exhibit a significant increase or decrease in expression in at least one experiment. Hierarchical clustering and analysis of this subset of data revealed three major groups of coregulated genes.
It was found that 60 genes depend on Tcn1/Crz1 for expression in high-Ca2+ conditions (Fig. 3B, lanes 2,3). As expected, the great majority of these genes also depended on Rcn1 and Mck1 for maximal expression in high-Ca2+ conditions (52 and 47 genes, respectively; lanes 4–7) but not in low-Ca2+ conditions (1 and 21 genes, respectively; lanes 8–11). A second cluster of 168 genes was characterized by a strong dependence on Mck1 for maximal expression in both the presence and absence of Ca2+ (Fig. 3A, lanes 6–9). Most of the Mck1-dependent genes were found in other studies to be targets of the stress-activated transcription factors Msn2 and Msn4 (Gasch et al. 2000; Causton et al. 2001), which depend on Mck1 for maximal transcription activity in a variety of stressful environments (Hirata et al. 2003). Of the 54 members of this group that depended on Mck1 in both strain backgrounds, none was affected reproducibly by the loss of either Tcn1/Crz1 or Rcn1 (lanes 2–5,10,11), suggesting that cross talk from the calcium signaling pathway was not necessary for Mck1 function. To examine potential cross-regulation more carefully, the effects of Rcn1 overexpression were examined. None of the genes within the Mck1-dependent or Tcn1/Crz1-dependent clusters was strongly affected by the forced overexpression of Rcn1 in low-Ca2+ conditions (lane 1). Only 14 genes, many of which are involved in protein folding and stress responses, were significantly induced by Rcn1 overexpression (Fig. 3C). In summary, Mck1 function appeared to have pleiotropic effects that were insensitive to Rcn1, whereas calcineurin function was strongly dependent on both Mck1 and Rcn1.
Figure 3.
Targets of Mck1, Tcn1/Crz1, and Rcn1 determined by genome-wide expression analysis. Total RNA from wild type and tcn1, mck1, and rcn1 mutants in two different strain backgrounds was extracted before and after 20 or 24 min of treatment with 100 mMCaCl2 and analyzed using cDNA microarrays corresponding to the complete yeast genome (see Materials and Methods). In the absence of Ca2+, no genes were expressed at significantly different levels in either tcn1 mutants or rcn1 mutants relative to wild type. Clusters of (A) 168 Mck1-dependent genes, (B) 60 Tcn1/Crz1-dependent genes, and (C) 14 genes induced by overexpression of Rcn1 are shown. Column headings indicate the presence or absence of Ca2+ treatment (+Ca or -Ca), the mutant strain and reference strain in parentheses (WT, wild type), followed by the strain background (B, BY4741; W, W303-1A). For comparison with a previous study of the response to 200 mM CaCl2 (Yoshimoto et al. 2002), the data from the 15- and 30-min time points after treatment of wild type and tcn1 (crz1) mutants in the DBY7286 strain background were averaged and included in column 2 (blue highlighting).
Mck1 phosphorylates Rcn1 and phospho-Rcn1 stimulates calcineurin signaling
The central region of Rcn1 contains a perfect match to the GSK-3 consensus phosphorylation site (Woodgett 2001) within the highly conserved FLISPPxSPP signature motif of the RCN family (Fig. 4A). To examine the possible phosphorylation of Rcn1 by Mck1, we first analyzed the migration of epitope-tagged Rcn1 proteins extracted from tcn1 mutants and tcn1 mck1 double mutants using SDS-PAGE and Western blotting (tcn1 mutants were used to avoid strong up-regulation of Rcn1 in response to high-Ca2+ conditions). Rcn1 proteins migrated as a tight doublet after extraction from tcn1 mutants, but migrated as a single species after extraction from mck1 tcn1 double mutants (Fig. 4B). The lower abundance of Rcn1 evident in the mck1 tcn1 strain was not reproducible in other experiments, but the doublet migration was always absent in mck1 tcn1 double mutants even after longer exposures. A mutant form of Rcn1 in which serine 113 was mutated to alanine (Rcn1S113A) migrated as a single band similar in mobility to that of Rcn1 in mck1 mutants. These data are consistent with the possibility that Mck1 phosphorylates Rcn1 at the GSK-3 consensus site in vivo. To test this hypothesis directly, GST-Rcn1 and GST-Rcn1S113A fusion proteins were expressed in Escherichia coli, purified, incubated with [γ-32P]ATP with or without purified Mck1 from yeast, and analyzed by SDS-PAGE and autoradiography. Neither of the recombinant proteins was phosphorylated by Mck1 Fig. 4C, lanes 5,6), although both were phosphorylated to a similar degree by purified p42 MAP kinase (lanes 3,4). Because Mck1 and GSK-3 kinases strongly prefer substrates that have been phosphorylated at a nearby site, we tested if prephosphorylation by p42 affected phosphorylation by Mck1. GST-Rcn1 and GST-Rcn1S113A proteins bound to glutathioneagarose beads were first incubated with p42 plus ATP, washed, and then incubated with Mck1 and [γ-32P]ATP as before. After phosphorylation by p42, GST-Rcn1 was efficiently phosphorylated by Mck1 (lane 7) in striking contrast to GST-Rcn1S113A (lane 8). Additionally, the Mck1-phosphorylated GST-Rcn1 was efficiently dephosphorylated by bovine calcineurin (Fig. 4D). Thus, Mck1 phosphorylated the GSK-3 consensus site of Rcn1 only after prephosphorylation(s) by a suitable priming kinase.
Figure 4.
Rcn1 is phosphorylated by Mck1 and dephosphorylated by calcineurin. (A) The highly conserved central domain of RCNs contains an absolutely conserved GSK-3 consensus phosphorylation site (underscored). (B) Western blot analysis of epitope-tagged Rcn1 or Rcn1S113A obtained from either tcn1 mutants or mck1 tcn1 double mutants after 4 h of growth in YPDS with or without 100 mM CaCl2. Doublet migration of Rcn1 was abolished after disruption of Mck1 or substitution of serine 113 with alanine in the GSK-3 consensus phosphorylation site. (C) Purified recombinant GST-Rcn1 or GST-Rcn1S113A was incubated with [γ-32P]ATP and no kinase, p42 MAP kinase, or purified Mck1, and analyzed by SDS-PAGE and autoradiography. Preincubation of the proteins with nonradioactive ATP plus p42 MAP kinase followed by extensive washing was necessary for phosphorylation of GST-Rcn1 by Mck1 and [γ-32P]ATP. (D) Mck1-phosphorylated GST-Rcn1 was incubated with 0, 5, or 10 units of purified calcineurin plus calmodulin or 20 units of calf intestinal phosphatases and analyzed as in C.
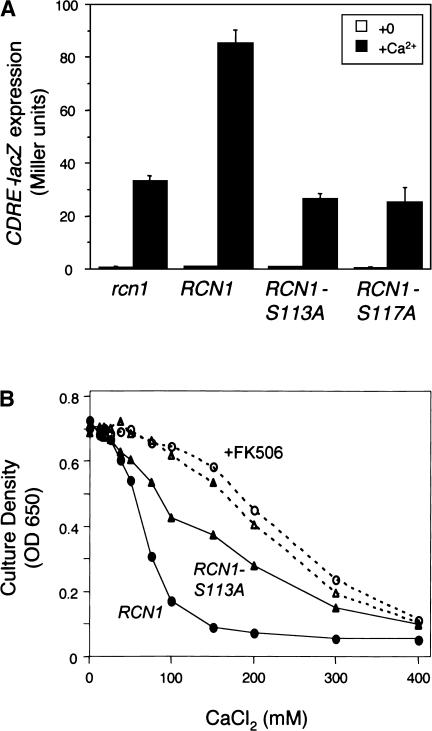
The effects of Rcn1 phosphorylation on calcineurin signaling were examined by comparing the activities of Rcn1 and Rcn1S113A in both the Vcx1-inhibition assay and the Tcn1/Crz1-activation assay. Mild overexpression of Rcn1S113A in pmc1 mutants conferred greater resistance to high-Ca2+ than wild-type Rcn1 (Fig. 5B), suggesting that Rcn1S113A may inhibit calcineurin signaling more effectively than wild-type Rcn1. The stimulatory activities of Rcn1 and Rcn1S113A were assessed by measurements of CDRE-lacZ induction. Wild-type Rcn1 stimulated CDRE-lacZ induction relative to the rcn1-null mutant, whereas the Rcn1S113A mutant had no stimulatory effect (Fig. 5A). Similar results were obtained with a Rcn1 derivative in which the putative priming site serine 117 was mutated to alanine. Therefore, phosphorylation of serine 113 of Rcn1 was necessary for stimulation of calcineurin signaling but not necessary for inhibition of calcineurin signaling.
Figure 5.
Serine 113 of Rcn1 is specifically required for stimulation of calcineurin activity in vivo. Calcineurin-dependent activation of Tcn1 function and inhibition of Vcx1 function were measured using the CDRE-lacZ expression assay and the calcium-resistance assay, respectively. (A) CDRE-lacZ expression in the rcn1 mutant (strain TKY275) transformed with low-dosage vectors bearing RCN1, RCN1-S113A, RCN1-S117A, or no insert (plasmids pZH200, pZH202, pZH203, or pRS415) was measured in each case after 4 h of growth in YPDS medium with or without 100 mM CaCl2. Substitution of either serine 113 or serine 117 with alanine abolished the stimulatory effect of Rcn1 on calcineurin signaling. (B) Growth of the calcium-sensitive pmc1 tcn1 double mutant (strain K1349) transformed with high-dosage plasmids bearing either RCN1 or RCN1-S113A (plasmids pZH100 or pZH101) was measured as described in Fig. 1. Rcn1S113A overexpression diminished calcineurin-dependent inhibition of Vcx1 function more potently than Rcn1 overexpression.
A general model of calcineurin regulation by RCNs
Several other findings suggest that RCNs in all species may regulate calcineurin by mechanisms similar to that of Rcn1 in yeast. A homolog of Rcn1 in the pathogenic fungus Cryptococcus neoformans was dephosphorylated in vivo by a calcineurin-dependent mechanism, and mutants lacking Cbp1 exhibited a partial calcineurin-deficient phenotype (Gorlach et al. 2000). The homologous serine 108 of DSCR1/MCIP1 was phosphorylated by GSK-3 and dephosphorylated by calcineurin in vitro (Vega et al. 2002). Based on the phenotypes of knockout mice, DSCR1/MCIP1 appeared to stimulate and inhibit calcineurin signaling in the heart (Vega et al. 2003). A general model of calcineurin regulation by RCNs can be inferred from all these observations. We suggest that RCNs cycle between inhibitory and stimulatory conformations in vivo in concert with phosphorylation by GSK-3 and dephosphorylation by calcineurin or other protein phosphatases (Fig. 6A, top). This model is strikingly similar to one proposed for the regulation of PP1 by Inh2 (Fig. 6A, bottom). In that case, threonine 72 of Inh2 is phosphorylated by GSK-3 (or other kinases), and subsequent conformational changes allosterically regulate PP1 activity (Cohen 2002). A groove exposed on the surface of PP1 that is necessary for allosteric regulation (Lohse et al. 1995) shows unusually strong conservation among calcineurins from many species (Hilioti and Cunningham 2003b). RCNs may function as phosphorylation-sensitive allosteric regulators of calcineurin much like Inh2s on PP1.
Figure 6.
A working model of calcineurin regulation by RCNs and GSK-3 and its validation by computer simulation. (A) RCNs such as Rcn1 in yeast and DSCR1/MCIP1 are phosphorylated by GSK-3 and, in high-[Ca2+]c conditions, dephosphorylated by calcineurin. Phospho-RCNs bind and stimulate calcineurin activity, whereas dephospho-RCNs bind and inhibit calcineurin activity (top panel). This model closely follows one proposed for PP1 regulation by Inh2 and GSK-3 (bottom panel) except that some RCNs are strongly up-regulated by calcineurin signaling (data not shown). (B) A series of ordinary differential equations describing the calcineurin model shown in A was compiled and solved for total calcineurin activity at equilibrium over a wide range of RCN concentrations using Cellerator software (see Materials and Methods). The calcineurin/phospho-RCN and calcineurin/dephospho-RCN interactions occurred at identical rates in the computation, but the specific activities of the calcineurin complexes were assumed to be, respectively, threefold higher and fivefold lower than that of unbound calcineurin. GSK-3 activity was set 10-fold higher than calcineurin activity, although similar effects were observed at threefold and onefold. Total calcineurin activity initially increased and eventually decreased as RCN levels increased. In the absence of phosphorylation by GSK-3, however, equilibrium calcineurin activity simply declined as RCN levels rose.
To study the feasibility and dynamics of an allosteric model of calcineurin regulation, each reactive element shown in Figure 6A was described by a series of ordinary differential equations that were solved computationally to equilibrium (see Materials and Methods). Because the reaction rate constants of GSK-3 and calcineurin for RCNs have not been experimentally determined, reasonable estimates of these values were derived from other substrates. The catalytic rate constant for calcineurin was assumed to increase threefold and decrease fivefold upon binding of phospho-RCN and dephospho-RCN, respectively, to a common regulatory site on calcineurin (KD = 5 nM). A stimulation of this magnitude has not yet been observed in reconstitution experiments involving GSK-3, Inh2, and PP1, but this high value is consistent with the stimulatory effects of phospho-Inh2 observed in vivo (Tung et al. 1995). Because Rcn1 and DSCR1/MCIP1 can be strongly up-regulated in response to calcineurin signaling (Fuentes et al. 2000; Kingsbury and Cunningham 2000; Yang et al. 2000) and the relative activity of GSK-3 in vivo has not been determined, we varied the concentrations of RCN and GSK-3 over wide ranges while holding calcineurin constant at 0.3 μM. At all such concentrations, calcineurin activity (defined as the scaled sum of all calcineurin species free to engage in catalytic reactions) was calculated after the simulation reached steady state, normalized to the initial condition (no RCN) and plotted as a function of RCN concentration.
In the absence of GSK-3, calcineurin activity rapidly declined to the fully inhibited state as the total RCN concentration increased (Fig. 6B, open circles). When GSK-3 was included at concentrations comparable to or higher than that of calcineurin, calcineurin activity increased, peaked, and eventually declined to low levels as total RCN concentration increased (closed circles). This biphasic response persisted over a broad range of GSK-3 owing to the combination of two factors, the allosteric inhibition of calcineurin by dephospho-RCN (primarily at low GSK-3 concentrations) and the competitive inhibition of calcineurin by phospho-RCN substrates (primarily at high GSK-3 concentrations). Similar biphasic patterns were observed if concentrations of reactants were lowered 10-fold or if RCN modulated the interactions between calcineurin and substrate rather than catalytic efficiency (data not shown).
The biphasic relationship between RCN concentration and calcineurin activity was tested experimentally in yeast using CDRE-lacZ induction as a read-out. Wild-type Rcn1 and nonphosphorylatable derivatives were expressed at a variety of different concentrations by replacing the promoters with a methionine-repressible MET25 promoter and growing the cells in media containing different levels of methionine (Fig. 7A). CDRE-lacZ induction increased, peaked, and declined as Rcn1 levels increased from zero (Fig. 7B, circles). The nonphosphorylatable Rcn1S113A and Rcn1S113D mutants inhibited CDRE-lacZ induction at all levels of expression (triangles). The inability of Rcn1S113D mutants to mimic the phosphorylated state has also been observed for the corresponding mutation in Inh2 (Tung et al. 1995). Similar patterns were observed if human DSCR1/MCIP1 or the nonphosphorylatable mutant DSCR1/MCIP1S108A derivative were expressed instead of Rcn1 (Fig. 7C). These experiments agree very well with the computer simulations and confirm the hypothesis that RCNs can serve as both stimulators and inhibitors of calcineurin in vivo. Based on the calcineurin-deficient phenotypes of rcn1 and mck1 mutants, Rcn1 primarily functions as a stimulator of calcineurin signaling in yeast.
Figure 7.
Rcn1 and DSCR1/MCIP1 stimulate calcineurin at low concentrations and inhibit calcineurin at high concentrations in vivo. Epitope-tagged Rcn1, Rcn1S113A, or Rcn1S113D was expressed in rcn1 mutants at increasing levels (lanes a–f) using the methionine-repressible MET25 promoter (see Materials and Methods) and analyzed for (A) expression levels by Western blotting and (B) effects on CDRE-lacZ expression after addition of CaCl2. (C) Human DSCR1/MCIP1 and DSCR1/MCIP1S108A mutant lacking the GSK-3 consensus phosphorylation site were expressed at increasing levels in rcn1 mutants and analyzed for effects on CDRE-lacZ expression after addition of CaCl2 as described in B. Rcn1 and human DSCR1/MCIP1 stimulated calcineurin signaling at low concentration but inhibited calcineurin signaling when expressed at very high concentrations and at all concentrations after mutation of the GSK-3 consensus site.
Discussion
Previous studies on fungal and animal RCNs have revealed a general ability of these proteins to bind and inhibit calcineurin (Fuentes et al. 2000; Gorlach et al. 2000; Kingsbury and Cunningham 2000; Rothermel et al. 2000, 2001; Hill et al. 2002; Lee et al. 2003). Paradoxically, fungal and animal RCNs also appeared to stimulate calcineurin or to transmit its signals (Gorlach et al. 2000; Kingsbury and Cunningham 2000; Vega et al. 2003). Here we define a molecular mechanism that helps explain the complex behavior of the RCN family of calcineurin regulators.
We suggest that the GSK-3 family of protein kinases phosphorylates a conserved site in RCNs and converts them from inhibitors to stimulators of calcineurin by an allosteric mechanism. Recombinant yeast Rcn1 (Fig. 4) and human DSCR1/MCIP1 (Vega et al. 2002) were phosphorylated by purified Mck1 and GSK-3 at the consensus site only after prior phosphorylation of other sites by a priming kinase. The GSK-3 consensus site is absolutely conserved in all fungal and animal homologs of Rcn1 (Hilioti and Cunningham 2003b) and apparently phosphorylated in vivo in cells of yeast (Fig. 4), Cryptococcus neoformans (Gorlach et al. 2000), and mammals (Genesca et al. 2003). Mutants lacking Mck1 or the phosphorylation site within Rcn1 exhibit much lower calcineurin signaling, as detected through assays of at least two independent targets (Tcn1/Crz1 and Vcx1). Mutants lacking Mck1 also exhibit weaker calcineurin-dependent effects on two other targets, Cch1 and Hsl1 (Mizunuma et al. 2001; Bonilla and Cunningham 2003). Nonphosphorylatable derivatives of Rcn1 and human DSCR1/MCIP1 inhibited calcineurin signaling in yeast more potently than the wild-type proteins (Fig. 7). Although Mck1 was involved in other cellular functions such as the regulation of gene expression, no strong roles of Rcn1 were observed outside of its effects on calcineurin-sensitive genes. Calcineurin efficiently dephosphorylated the GSK-3 consensus sites of Rcn1 (Fig. 4) and DSCR1/MCIP1 (Vega et al. 2002). Cycles of RCN dephosphorylation and rephosphorylation may therefore modulate calcineurin signaling by similar mechanisms in a wide range of species.
These effects of RCNs on calcineurin signaling are seemingly parallel to those of the Inh2/Glc8 regulatory protein on PP1. Genetic studies in yeast indicate that Glc8 primarily stimulates PP1 activity toward physiological targets. Overexpression, deletion, and mutation of the conserved noncanonical GSK-3 phosphorylation site of Glc8 caused a significant decrease in PP1 signaling toward multiple targets (Tung et al. 1995). Detailed biochemical experiments have demonstrated Inh2 can undergo conformational changes upon phosphorylation and dephosphorylation that allosterically regulate PP1 activity (Cohen 2002). The groove on the surface of PP1 thought to encompass the allosteric regulatory site is exposed on the surface of the calcineurin catalytic subunit (Lohse et al. 1995) and is unusually well conserved among calcineurins from diverse species (Hilioti and Cunningham 2003a) as expected for sites that bind a conserved regulatory factor. The Inh2 and RCN families of proteins retain no obvious sequence similarity, but several features are common to both. For example, the highly elongated Inh2 molecules contain several conserved domains that independently bind different surfaces of PP1. RCNs are similar in size and composition, and also contain multiple conserved domains capable of independent interactions with calcineurin (Fuentes et al. 2000; Vega et al. 2002). Inh2s may regulate PP1 function in other ways, such as modulating PP1 interactions with substrates or targeting subunits and chaperoning the folding of nonnative PP1 (Cohen 2002). The ability of human DSCR1/MCIP to stimulate and inhibit calcineurin signaling in yeast argues against a general role of RCNs as specific targeting subunits because Tcn1/Crz1 is not conserved in animals. The complex regulation of RCNs and Inh2s by phosphorylation and dephosphorylation is not well explained by a molecular chaperone model for these proteins. For these reasons, we favor an allosteric model of calcineurin regulation by RCNs analogous to ones proposed for PP1 regulation by Inh2s.
The strong induction of Rcn1 and DSCR1/MCIP1 expression by calcineurin signaling adds complexity to calcineurin regulation not seen for PP1 regulation. To comprehend these dynamics, we performed computer simulations of calcineurin regulation over a wide range of RCN concentration. The computational model assumed the phosphatase activity of phospho-RCN–calcineurin complexes is 15-fold higher than that of dephospho-RCN–calcineurin complexes, that the activity of free calcineurin is intermediate between these values, and that the activities of all other regulators (Ca2+, calmodulin, priming kinases, etc.) were not limiting. The simulations revealed a biphasic response of calcineurin activity at equilibrium to increasing RCN concentrations. These simulations closely matched the experimental data obtained from the controlled expression of either Rcn1 or DSCR1/MCIP1 in yeast (cf. Figs. 6 and 7). Interestingly, two modes of calcineurin inhibition were revealed in the simulations at high RCN concentrations. At low GSK-3 activity, total calcineurin activity declined because dephospho-RCN accumulated and competed with phospho-RCN for the regulatory site on calcineurin. At high GSK-3 activity in the simulation, the active phospho-RCN–calcineurin complexes predominated, but the total calcineurin activity was low because most catalytic sites were occupied with phospho-RCN substrates, thereby removing active calcineurin from the pool available to other substrates (substrate competition). The mode and extent of calcineurin inhibition in cells would therefore depend largely on the level of RCN as long as GSK-3 activity exceeded that of calcineurin. In vegetatively growing yeast cells, Mck1 and Rcn1 levels must be within their stimulatory ranges, but this situation probably varies in other cell types. RCNs may therefore link calcineurin signaling to other regulatory pathways that modulate its expression and phosphorylation.
Little is known about the pathways that regulate the GSK-3 activities in yeast. In standard growth medium, calcineurin is inactive, but Mck1 appeared to be active as indicated by the decreased expression of many genes in mck1 mutants relative to wild type (Fig. 6A). A large fraction of these Mck1-dependent genes was found to be targets of the Msn2 and Msn4 transcription activators (Gasch et al. 2000; Causton et al. 2001) that are highly dependent on Mck1 in response to a variety of cellular stresses (Hirata et al. 2003). A recent study suggested high Ca2+ conditions increase MCK1 gene expression and Mck1 function (Mizunuma et al. 2001). We have repeatedly failed to detect any significant effect of Ca2+ on the accumulation of MCK1 transcripts or Mck1 protein levels. The main assays used to infer Ca2+ stimulation of Mck1 function (Ca2+ resistance and Hsl1 stability) are also sensitive to calcineurin (Mizunuma et al. 2001) and therefore may be interpreted as additional support for our conclusion that Mck1 can stimulate calcineurin signaling. Unambiguous assays of Mck1 activity in yeast cells will be necessary to determine its modes of regulation. Other roles of Mck1 in yeast that are potentially independent of calcineurin include regulation of gene expression in meiosis (Neigeborn and Mitchell 1991), regulation of centromere function in mitosis (Shero and Hieter 1991), and inhibition of protein kinase A (Rayner et al. 2002). Of the three other GSK-3 family kinases in yeast, functional redundancy with Mck1 was noted only for Rim11 (Puziss et al. 1994; Hirata et al. 2003), which weakly stimulated calcineurin signaling in the absence of Mck1 (Fig. 3).
GSK-3 stimulation of calcineurin signaling has not yet been demonstrated in vitro or in mammalian cells. Because GSK-3 phosphorylates NFAT and antagonizes calcineurin activation of this substrate (Beals et al. 1997; Graef et al. 1999; Ohteki et al. 2000; Antos et al. 2002; Vyas et al. 2002), other readouts of calcineurin signaling may be necessary to reveal the potential stimulatory effects of GSK-3 and RCNs. Removing or depleting the endogenous RCNs may also be necessary for measuring the activities of transfected RCNs and their nonphosphorylatable derivatives in mammalian cells (Vega et al. 2002; Genesca et al. 2003). Nevertheless, several phenotypes of DSCR1/MCIP–/– knockout mice suggest that stimulatory roles of RCN may exist in mammals (Vega et al. 2003). RCNs may therefore serve as a nexus for coupling calcineurin signaling to other regulatory pathways that modulate the function of GSK-3 and the unidentified priming kinases that are prerequisite for GSK-3 function on RCNs.
If phospho-RCNs do indeed stimulate calcineurin signaling in animals, a potential positive feedback loop may exist in some cell types resulting from calcineurin- and NFAT-dependent up-regulation of DSCR/MCIP expression. Consistent with this idea, trisomy 21 led to a larger than expected increase in DSCR1/MCIP1 expression in brain samples from adults with Down syndrome (Fuentes et al. 2000). DSCR1/MCIP1 expression was also greatly elevated in cerebellum from individuals with Alzheimer's disease (Ermak et al. 2001). In neither case, however, was the dependence on calcineurin or NFAT assessed. Finally, DSCR1 expression was massively elevated in mouse hypertrophic heart as a consequence of calcineurin hyperactivation (Yang et al. 2000). Several types of induced hypertrophy were blocked by forced overexpression of DSCR1/MCIP1 in the developing heart (Rothermel et al. 2001; Hill et al. 2002). If this phenomenon is a consequence of calcineurin inhibition, our findings predict that the nonphosphorylatable DSCR1/MCIP1S108A will be much more potent than DSCR1/MCIP1 at suppressing cardiac hypertrophy and heart failure. Additional biochemical, genetic, and physiological studies of the RCN–calcineurin interaction may therefore yield important insights into the causes and treatment of several human maladies.
Materials and methods
Yeast strains, plasmids, and growth media
All strains (Table 1) were derived from parental strains W303-1A (Wallis et al. 1989) or BY4741 (Giaever et al. 2002) using standard methods of molecular and classical genetics. Spontaneous calcium-resistant mutants were selected in the pmc1 mutant background and placed into complementation groups as described previously (Cunningham and Fink 1994). One major complementation group (termed crm2) consisted of 11 alleles, all of which exhibited a cold-sensitive growth phenotype. The pmc1 crm2-28 strain was transformed with a library of genomic DNA fragments cloned into the low-dosage plasmid pCT3 (gift of R. Young, Whitehead Institute) and plated on selective medium at 14°C. After 10 d of incubation, large colonies were picked, purified, and tested for complementation of the Ca2+-resistant phenotype. Only two plasmids restored Ca2+ sensitivity to this strain, and these plasmids had distinct but overlapping inserts spanning the MCK1 gene. The pmc1 crm2 phenotypes were also reversed by plasmid pJS99 containing a minimal MCK1 locus (Shero and Hieter 1991) and were mimicked by replacing the MCK1 gene in strain K482 with the mck1::HIS3 fragment of plasmid pJS102 (Shero and Hieter 1991) to yield strain K490. Additional strains bearing an mck1::HIS3 or mck1::G418r gene knockout allele were constructed through crosses with isogenic strains or were acquired from commercial suppliers (Research Genetics, Inc.).
Table 1.
Yeast strains used in this study
| Namea | Genotype | Source |
|---|---|---|
| K482-28 | pmc1::TRP1 crm2-28/mck1-28 (MATα) | Cunningham and Fink 1994 |
| K490 | pmc1::TRP1 mck1::HIS3 (MATα) | This study |
| K601 | Wild-type W303-1A | Cunningham and Fink 1996 |
| K1305 | mck1::HIS3 | This study |
| K1325 | pmc1::LEU2 vcxlΔ | This study |
| K1329 | pmc1::LEU2 vcxlΔ mck1::HIS3 | This study |
| K1333 | tcn1::G418r | This study |
| K1337 | tcn1::G418r mck1::HIS3 | This study |
| K1349 | pmc1::LEU2 tcnl::G418r | This study |
| K1353 | pmc1::LEU2 tcn1::G418r mck1::HIS3 | This study |
| K1357 | pmc1::LEU2 tcn1::G418r vcx1Δ | This study |
| K1361 | pmc1::LEU2 tcn1::G418r vcx1Δ mck1::HIS3 | This study |
| K1365 | rcn1::ADE2 | This study |
| K1369 | rcn1::ADE2 mck1::HIS3 | This study |
| TKY275 | rcn1::HIS3 | Kingsbury and Cunningham 2000 |
| ZH35 | mck1::TRP1 mrk1Δ rim11:HIS3 (MATα) | This study |
| ZH38 | mck1::TRP1 mrk1Δ rim11::HIS3 ygk3::LEU2 | This study |
| ZH39 | mck1::TRP1 rim11::HIS3 ygk3::LEU2 | This study |
| ZH40 | mck1::TRP1 mrk1Δ ygk3::LEU2 | This study |
| ZH45 | mrk1Δ rim11::HIS3 ygk3::LEU2 | This study |
| K1251 | Wild-type BY4741 | Research Genetics |
| RG05009 | rcn1::G418r | Research Genetics |
| RG01137 | mck1::G418r | Research Genetics |
| RG05353 | tcn1::G418r | Research Genetics |
Strains K482 through ZH45 are isogenic derivatives of strain K601/W303-1A (MATa ade2-1 can1-100 his3-11, 14 leu2-3, 112 trp1-1 ura3-1). Strains ZH35 through ZH45 were derived from an isogenic cross between K601 and YTA003W (Hirata et al 2003). Strains designated RG are all isogenic derivatives of K1251/BY4741 (MATa his3Δ1 lue2Δ0 met15Δ0 ura3Δ0).
Plasmids carrying the reporter genes CDRE-lacZ (pAMS342), RCN1-lacZ (pKC305), PMC1-lacZ (pKC190), CYC1-lacZ (pLGΔ312), CTS-lacZ (M1820), and HO-lacZ (M1853) have been described previously (Cunningham and Fink 1996; Matheos et al. 1997; Stathopoulos-Gerontides et al. 1999; Kingsbury and Cunningham 2000). Plasmids pKC306 and pZH401 were created by site-directed mutagenesis of pTJK29 and pTJK93 (Kingsbury and Cunningham 2000) using the Quickchange method (Stratagene) to yield plasmids suitable for expression of epitope-tagged Rcn1S113A in yeast and GST-Rcn1S113A in E. coli. The inserts of pTJK29 and pKC306 were subcloned into the high-dosage plasmid pRS425 (Sikorski and Hieter 1989) to yield pZH100 and pZH101 and into the low-dosage plasmid pRS415 (Sikorski and Hieter 1989) to yield pZH200 and pZH202, respectively. Site-directed mutagenesis of pZH200 resulted in plasmid pZH203, which carries epitope-tagged Rcn1S117A. Plasmids pZH300 and pZH301 carrying RCN1-HA and RCN1S113A-HA under control of a methionine-repressible promoter were constructed by subcloning the PstI to XhoI fragments of pTJK29 and pKC306 into p415MET25 (Mumberg et al. 1994). Plasmid pPR001 carrying RCN1S113D-HA under control of the MET25 promoter was constructed by site-directed mutagenesis of pZH300. Plasmid pTJK37 carrying DSCR1/MCIP1 under control of the MET25 promoter was constructed previously (Kingsbury and Cunningham 2000), and plasmid pDG001 carrying DSCR1/MCIP1S108A under control of the MET25 promoter was constructed by subcloning the BamHI to XhoI fragment of DSCR1-S108A (gift of X. Estivill, Center for Genomic Regulation, Barcelona) into p425-MET25 (Mumberg et al. 1994). All site-directed mutagenesis was confirmed by DNA sequencing.
Rich culture medium (YPD) and synthetic complete culture media (SC) lacking one or more nutrients were prepared by standard protocols (Sherman et al. 1986). In experiments involving CaCl2 supplements, YPD was supplemented with 5 mM succinic acid (to pH ∼5.5), yielding YPDS medium. Ca2+ tolerance assays were performed in YPDS medium supplemented with varying amounts of CaCl2 as described previously (Cunningham and Fink 1996).
Northern blot and DNA microarray analysis of gene expression
Total RNA was extracted from log-phase cells growing in YPDS medium using the hot acid phenol method and normalized by absorbance at 260 nm. Total RNA (10 μg/lane) was analyzed by a standard Northern blotting procedure (Jung and Levin 1999), using randomly primed DNA probes complementary to PMC1, RCN1, or MCK1 transcripts. For DNA microarray analysis, total RNA (20 μg) was reverse-transcribed using an anchored oligo(dT) primer and aminoallyl-dUTP, and the resulting cDNA was cross-linked to Cy3 or Cy5 fluorescent dyes using the CyScribe cDNA Post-labeling kit (Amersham Biosciences). Labeled cDNAs were purified using the QiaQuick PCR purification kit (QIAGEN Biosciences), mixed with reference samples, and hybridized overnight at 42°C in 80% DIG Easy Hybe (Roche) to PCR-amplified cDNA microarrays corresponding to 6240 ORFs in the yeast genome (Ontario Cancer Institute). Slides were washed four times in high-salt buffer (1× SSC, 0.2% SDS) and twice in low-salt buffer (0.1× SSC, 0.1% SDS) at 55°C for 4 min each, dried under a stream of 1,1,1,2-tetrafluoroethane, and scanned (5 μm resolution with 2-line averaging) using a GenePix 4000B dual laser scanner (Axon Inc.). A dye-swapped duplicate of each sample was generated in parallel, and data from both slides were normalized and combined using a lowess function in GeneSpring v4.0. Most experiments were performed independently in two strain backgrounds (W303-1A and BY4741). Two published experiments (each comparing tcn1/crz1 mutants to wild type after treatment with 200 mM CaCl2 for 15 or 30 min) were averaged and imported as one data column in our analysis. The complete data set can be viewed in Supplemental Table 1. A subset of 388 genes was expressed at least twofold higher or lower in the mutant relative to wild type in one or more experiments (excluding genes from the imported data set that were induced less than 1.5-fold in our similar experiment). This subset was subjected to hierarchical clustering using Cluster and TreeView (Eisen et al. 1998), and clusters whose members depended on Mck1, Tcn1, or overexpressed Rcn1 are illustrated in Figure 3.
Protein purification and enzymatic reactions
Expression of lacZ reporter genes in yeast was measured using β-galactosidase assays as described previously (Cunningham and Fink 1996). For Western blot analysis of epitope-tagged proteins, log-phase cells were harvested, treated with 10% trichloro-acetic acid for 15 min on ice, neutralized with 50 μL of 1.5 MTris-Cl (pH 8.8), and extracted with 60 μL of 2× sample buffer containing 8 Murea. Solubilized proteins were separated by SDS-PAGE, transferred to Hybond N+, blocked, probed with 12CA5 or 9E10 monoclonal antibodies and HRP-conjugated secondary antibodies, and visualized on X-ray film using the chemiluminescent ECL kit (Amersham).
The hexa-histidine-tagged derivative of Mck1 was expressed in yeast and purified to homogeneity as described (Rayner et al. 2002). Recombinant GST, GST-Rcn1, and GST-Rcn1S113A proteins were expressed in the TOPP2 strain of E. coli from the pGEX-3X (Pharmacia), pTJK93, and pZH401 plasmids and purified using GSTBind kit as described by the manufacturer (Novagen). Briefly, log-phase cultures (OD600 = 0.8) were shifted to 30°C and induced with 1 mM IPTG for 2 h before harvesting and lysis. Purified proteins were dialyzed extensively with buffer A (50 mM Tris-HCl at pH 7.5, 1 mM EDTA, 10% glycerol) using Slide-A Lyzer (Pierce), and protein concentrations were determined by Bradford assays and normalized prior to use as substrates for enzyme reactions. For in vitro kinase assays, protein substrates were mixed with glutathione sepharose-4B beads (Pharmacia) for 1 h at 4°C, and the resin was washed three times in phosphate-buffered saline (PBS) and once in kinase reaction buffer (50 mM Tris-HCl at pH 7.5, 0.1 mM EGTA, 15 mM DTT). The kinase reactions were initiated by adding purified p42 (Upstate) or Mck1, [γ-32P]ATP (10 μCi) to the immobilized substrates in a final volume of 60 μL and incubated for 30 min at 30°C. For dephosphorylation studies, immobilized phosphoproteins were washed and resuspended in phosphatase reaction buffer (100 mM HEPES at pH 7.4, 100 mM NaCl, 20 mM potassium acetate, 2 mM magnesium acetate, 2 mM CaCl2, 2 mM MnCl2), and incubated for 1 h at 30°C in the presence of 0, 5, or 10 units of purified bovine brain calcineurin plus calmodulin (Sigma) or 20 units of purified calf intestinal phosphatase (New England Biolabs). After the kinase and phosphatase reactions, samples were washed in PBS buffer, resuspended in equal volumes of 2× SDS-PAGE sample buffer, boiled for 5 min, and analyzed by SDS-PAGE and autoradiography.
Computational modeling
Cellerator, a Mathematica-based software package developed for simulation of signal transduction and genetic networks (Shapiro et al. 2001), was used to generate and solve a system of ordinary differential equations that describe phosphorylation and dephosphorylation of RCNs by GSK-3 and calcineurin. Calcineurin concentration was fixed at 1 μM and the reaction constants ka, kd, and kcat toward phospho-RCN were set at 0.5 μM–1 sec–1, 0.5 sec–1, and 0.1 sec–1, respectively (KM = 1.2 μM). GSK-3 concentration was fixed at 0, 1, 3, 10, or 30 μMwith reaction constants toward dephospho-RCN set at 1 μM–1 sec–1, 0.4 sec–1, and 0.1 sec–1, respectively (KM = 0.5 μM). Phospho-RCN and dephospho-RCN were assumed to bind with equal affinity to a regulatory site on calcineurin (ka = 10μM–1 sec–1, kd = 0.05 sec–1; KD = 5 nM) and to alter kcat for substrates by factors of 3.0 and 0.2, respectively. These values were estimates based on published kinetic constants for calcineurin (Klee et al. 1998), MAP kinase (Levchenko et al. 2000), and Inh2–PP1 interactions. Simulations were carried out until steady state was achieved, after which the activity of each calcineurin species available for catalytic reactions was summed, normalized to the initial conditions lacking RCN, and plotted as a function of RCN concentration.
Acknowledgments
The authors are grateful to Xavier Estivill, Gerry Fink, Phil Hieter, Tim Rayner, Jeremy Thorner, and Richard Young, for generously providing plasmids. We thank Liz O'Sullivan and Christian Martin for excellent technical assistance. This work was supported by grants from the Whitaker Foundation (to A.L.) and National Institutes of Health (GM053082 to K.W.C.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1159204.
References
- Antos C.L., McKinsey, T.A., Frey, N., Kutschke, W., McAnally, J., Shelton, J.M., Richardson, J.A., Hill, J.A., and Olson, E.N. 2002. Activated glycogen synthase-3 β suppresses cardiac hypertrophy in vivo. Proc. Natl. Acad. Sci. 99: 907–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beals C.R., Sheridan, C.M., Turck, C.W., Gardner, P., and Crabtree, G.R. 1997. Nuclear export of NF-ATc enhanced by glycogen synthase kinase-3. Science 275: 1930–1934. [DOI] [PubMed] [Google Scholar]
- Bonilla M. and Cunningham, K.W. 2003. Mitogen-activated protein kinase stimulation of Ca2+ signaling is required for survival of endoplasmic reticulum stress in yeast. Mol. Biol. Cell 14: 4296–4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causton H.C., Ren, B., Koh, S.S., Harbison, C.T., Kanin, E., Jennings, E.G., Lee, T.I., True, H.L., Lander, E.S., and Young, R.A. 2001. Remodeling of yeast genome expression in response to environmental changes. Mol. Biol. Cell 12: 323–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen P.T. 2002. Protein phosphatase 1—Targeted in many directions. J. Cell Sci. 115: 241–256. [DOI] [PubMed] [Google Scholar]
- Crabtree G.R. and Olson, E.N. 2002. NFAT signaling: Choreographing the social lives of cells. Cell 109 Suppl: S67–S79. [DOI] [PubMed] [Google Scholar]
- Cunningham K.W. and Fink, G.R. 1994. Calcineurin-dependent growth control in Saccharomyces cerevisiae mutants lacking PMC1, a homolog of plasma membrane Ca2+ ATPases. J. Cell Biol. 124: 351–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 1996. Calcineurin inhibits VCX1-dependent H+/Ca2+ exchange and induces Ca2+ ATPases in yeast. Mol. Cell. Biol. 16: 2226–2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen M.B., Spellman, P.T., Brown, P.O., and Botstein, D. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. 95: 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermak G., Morgan, T.E., and Davies, K.J. 2001. Chronic overexpression of the calcineurin inhibitory gene DSCR1 (Adapt78) is associated with Alzheimer's disease. J. Biol. Chem. 276: 38787–38794. [DOI] [PubMed] [Google Scholar]
- Fox D.S. and Heitman, J. 2002. Good fungi gone bad: The corruption of calcineurin. Bioessays 24: 894–903. [DOI] [PubMed] [Google Scholar]
- Fuentes J.J., Genescà, L., Kingsbury, T.J., Cunningham, K.W., Pérez-Riba, M., Estivill, X., and De La Luna, S. 2000. DSCR1, overexpressed in Down syndrome, is an inhibitor of calcineurin-mediated signaling pathways. Hum. Mol. Gen. 9: 1681–1690. [DOI] [PubMed] [Google Scholar]
- Gasch A.P., Spellman, P.T., Kao, C.M., Carmel-Harel, O., Eisen, M.B., Storz, G., Botstein, D., and Brown, P.O. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11: 4241–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genesca L., Aubareda, A., Fuentes, J.J., Estivill, X., De La Luna, S., and Perez-Riba, M. 2003. Phosphorylation of calcipressin 1 increases its ability to inhibit calcineurin and decreases-calcipressin half-life. Biochem. J. 374: 567–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G., Chu, A.M., Ni, L., Connelly, C., Riles, L., Veronneau, S., Dow, S., Lucau-Danila, A., Anderson, K., Andre, B., et al. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418: 387–391. [DOI] [PubMed] [Google Scholar]
- Gorlach J., Fox, D.S., Cutler, N.S., Cox, G.M., Perfect, J.R., and Heitman, J. 2000. Identification and characterization of a highly conserved calcineurin binding protein, CBP1/calcipressin, in Cryptococcus neoformans. EMBO J. 19: 3618–3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graef I.A., Mermelstein, P.G., Stankunas, K., Neilson, J.R., Deisseroth, K., Tsien, R.W., and Crabtree, G.R. 1999. L-type calcium channels and GSK-3 regulate the activity of NF-ATc4 in hippocampal neurons. Nature 401: 703–708. [DOI] [PubMed] [Google Scholar]
- Hilioti Z. and Cunningham, K.W. 2003a. Calcineurin: Roles of the Ca2+/calmodulin-dependent protein phosphatase in diverse eukaryotes. Topics Curr. Genet. 5: 73–90. [Google Scholar]
- ____. 2003b. The RCN family of calcineurin regulators. Biochem. Biophys. Res. Commun. 311: 1089–1093. [DOI] [PubMed] [Google Scholar]
- Hill J.A., Rothermel, B., Yoo, K.D., Cabuay, B., Demetroulis, E., Weiss, R.M., Kutschke, W., Bassel-Duby, R., and Williams, R.S. 2002. Targeted inhibition of calcineurin in pressure-overload cardiac hypertrophy. Preservation of systolic function. J. Biol. Chem. 277: 10251–10255. [DOI] [PubMed] [Google Scholar]
- Hirata Y., Andoh, T., Asahara, T., and Kikuchi, A. 2003. Yeast glycogen synthase kinase-3 activates Msn2p-dependent transcription of stress responsive genes. Mol. Biol. Cell 14: 302–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung U.S. and Levin, D.E. 1999. Genome-wide analysis of gene expression regulated by the yeast cell wall integrity signalling pathway. Mol. Microbiol. 34: 1049–1057. [DOI] [PubMed] [Google Scholar]
- Kingsbury T.J. and Cunningham, K.W. 2000. A conserved family of calcineurin regulators. Genes & Dev. 13: 1595–1604. [PMC free article] [PubMed] [Google Scholar]
- Klee C.B., Ren, H., and Wang, X. 1998. Regulation of the calmodulin-stimulated protein phosphatase, calcineurin. J. Biol. Chem. 273: 13367–13370. [DOI] [PubMed] [Google Scholar]
- Lee J.I., Dhakal, B.K., Lee, J., Bandyopadhyay, J., Jeong, S.Y., Eom, S.H., Kim, D.H., and Ahnn, J. 2003. The Caenorhabditis elegans homologue of Down Syndrome Critical Region 1, RCN-1, inhibits multiple functions of the phosphatase calcineurin. J. Mol. Biol. 328: 147–156. [DOI] [PubMed] [Google Scholar]
- Levchenko A., Bruck, J., and Sternberg, P.W. 2000. Scaffold proteins may biphasically affect the levels of mitogen-activated protein kinase signaling and reduce its threshold properties. Proc. Natl. Acad. Sci. 97: 5818–5823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse D.L., Denu, J.M., and Dixon, J.E. 1995. Insights derived from the structures of the Ser/Thr phosphatases calcineurin and protein phosphatase 1. Structure 3: 987–990. [DOI] [PubMed] [Google Scholar]
- Matheos D.P., Kingsbury, T.J., Ahsan, U.S., and Cunningham, K.W. 1997. Tcn1p/Crz1p, a calcineurin-dependent transcription factor that differentially regulates gene expression in Saccharomyces cerevisiae. Genes & Dev. 11: 3445–3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizunuma M., Hirata, D., Miyaoka, R., and Miyakawa, T. 2001. GSK-3 kinase Mck1 and calcineurin coordinately mediate Hsl1 down-regulation by Ca2+ in budding yeast. EMBO J. 20: 1074–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molkentin J.D., Lu, J.R., Antos, C.L., Markham, B., Richardson, J., Robbins, J., Grant, S.R., and Olson, E.N. 1998. A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell 93: 215–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg D., Muller, R., and Funk, M. 1994. Regulatable promoters of Saccharomyces cerevisiae: Comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 22: 5767–5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neigeborn L. and Mitchell, A.P. 1991. The yeast MCK1 gene encodes a protein kinase homolog that activates early meiotic gene expression. Genes & Dev. 5: 533–548. [DOI] [PubMed] [Google Scholar]
- Ohteki T., Parsons, M., Zakarian, A., Jones, R.G., Nguyen, L.T., Woodgett, J.R., and Ohashi, P.S. 2000. Negative regulation of T cell proliferation and interleukin 2 production by the serine threonine kinase GSK-3. J. Exp. Med. 192: 99–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puziss J.W., Hardy, T.A., Johnson, R.B., Roach, P.J., and Hieter, P. 1994. MDS1, a dosage suppressor of an mck1 mutant, encodes a putative yeast homolog of glycogen synthase kinase 3. Mol. Cell. Biol. 14: 831–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayner T.F., Gray, J.V., and Thorner, J.W. 2002. Direct and novel regulation of cAMP-dependent protein kinase by Mck1p, a yeast glycogen synthase kinase-3. J. Biol. Chem. 277: 16814–16822. [DOI] [PubMed] [Google Scholar]
- Rothermel B., Vega, R.B., Yang, J., Wu, H., Bassel-Duby, R., and Williams, R.S. 2000. A protein encoded within the Down syndrome critical region is enriched in striated muscles and inhibits calcineurin signaling. J. Biol. Chem. 275: 8719–8725. [DOI] [PubMed] [Google Scholar]
- Rothermel B.A., McKinsey, T.A., Vega, R.B., Nicol, R.L., Mammen, P., Yang, J., Antos, C.L., Shelton, J.M., Bassel-Duby, R., Olson, E.N., et al. 2001. Myocyte-enriched calcineurin-interacting protein, MCIP1, inhibits cardiac hypertrophy in vivo. Proc. Natl. Acad. Sci. 98: 3328–3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusnak F. and Mertz, P. 2000. Calcineurin: form and function. Physiol. Rev. 80: 1483–1521. [DOI] [PubMed] [Google Scholar]
- Shapiro B.E., Levchenko, A., and Mjolsness, E. 2001. Automatic model generation for signal transduction with applications to MAPK pathway. In Foundations of systems biology (ed. H. Kitano), pp. 145–161. MIT Press, Cambridge, MA.
- Sherman F., Hicks, J.B., and Fink, G.R. 1986. Methods in yeast genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- Shero J.H. and Hieter, P. 1991. A suppressor of a centromere DNA mutation encodes a putative protein kinase (MCK1). Genes & Dev. 5: 549–560. [DOI] [PubMed] [Google Scholar]
- Sikorski R.S. and Hieter, P. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stathopoulos A.M. and Cyert, M.S. 1997. Calcineurin acts through the CRZ1/TCN1 encoded transcription factor to regulate gene expression in yeast. Genes & Dev. 11: 3432–3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stathopoulos-Gerontides A., Guo, J.J., and Cyert, M.S. 1999. Yeast calcineurin regulates nuclear localization of the Crz1p transcription factor through dephosphorylation. Genes & Dev. 13: 798–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung H.Y., Wang, W., and Chan, C.S. 1995. Regulation of chromosome segregation by Glc8p, a structural homolog of mammalian inhibitor 2 that functions as both an activator and an inhibitor of yeast protein phosphatase 1. Mol. Cell. Biol. 15: 6064–6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega R.B., Yang, J., Rothermel, B.A., Bassel-Duby, R., and Williams, R.S. 2002. Multiple domains of MCIP1 contribute to inhibition of calcineurin activity. J. Biol. Chem. 277: 30401–30407. [DOI] [PubMed] [Google Scholar]
- Vega R.B., Rothermel, B.A., Weinheimer, C.J., Kovacs, A., Naseem, R.H., Bassel-Duby, R., Williams, R.S., and Olson, E.N. 2003. Dual roles of modulatory calcineurin-interacting protein 1 in cardiac hypertrophy. Proc. Natl. Acad. Sci. 100: 669–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyas D.R., Spangenburg, E.E., Abraha, T.W., Childs, T.E., and Booth, F.W. 2002. GSK-3β negatively regulates skeletal myotube hypertrophy. Am. J. Physiol. Cell Physiol. 283: C545–C551. [DOI] [PubMed] [Google Scholar]
- Wallis J.W., Chrebet, G., Brodsky, G., Rolfe, M., and Rothstein, R. 1989. A hyper-recombination mutation in S. cerevisiae identifies a novel eukaryotic topoisomerase. Cell 58: 409–419. [DOI] [PubMed] [Google Scholar]
- Woodgett J.R. 2001. Judging a protein by more than its name: GSK-3. Sci. STKE 2001: RE12. [DOI] [PubMed] [Google Scholar]
- Yang J., Rothermel, B., Vega, R.B., Frey, N., McKinsey, T.A., Olson, E.N., Bassel-Duby, R., and Williams, R.S. 2000. Independent signals control expression of the calcineurin inhibitory proteins MCIP1 and MCIP2 in striated muscles. Circ. Res. 87: E61–E68. [DOI] [PubMed] [Google Scholar]
- Yoshimoto H., Saltsman, K., Gasch, A.P., Li, H.X., Ogawa, N., Botstein, D., Brown, P.O., and Cyert, M.S. 2002. Genome-wide analysis of gene expression regulated by the calcineurin/Crz1p signaling pathway in Saccharomyces cerevisiae. J. Biol. Chem. 277: 31079–31088. [DOI] [PubMed] [Google Scholar]