Abstract
Prediction of transcription-factor target sites in promoters remains difficult due to the short length and degeneracy of the target sequences. Although the use of orthologous sequences and phylogenetic footprinting approaches may help in the recognition of conserved and potentially functional sequences, correct alignment of the short transcription-factor binding sites can be problematic for established algorithms, especially when aligning more divergent species. Here, we report a novel phylogenetic footprinting approach, CONREAL, that uses biologically relevant information, that is, potential transcription-factor binding sites as represented by positional weight matrices, to establish anchors between orthologous sequences and to guide promoter sequence alignment. Comparison of the performance of CONREAL with the global alignment programs LAGAN and AVID using a reference data set, shows that CONREAL performs equally well for closely related species like rodents and human, and has a clear added value for aligning promoter elements of more divergent species like human and fish, as it identifies conserved transcription-factor binding sites that are not found by other methods. CONREAL is accessible via a Web interface at http://conreal.niob.knaw.nl/.
Complexity and dynamics of a living organism are established and maintained largely as a result of differential gene expression, both temporal and spatial. Thus, understanding the mechanisms that regulate gene expression is one of the most challenging objectives in contemporary biology. Although gene expression can be regulated at several levels, the stage of transcription initiation is best understood. Specific transcription of a gene is driven by complexes of regulatory proteins, transcription factors (TFs), which bind to specific regulatory regions of a gene, known as transcription-factor binding sites (TFBSs). A number of experimental procedures to determine TFBSs have been developed, including DNAse I protection (Galas and Schmitz 1978; Gross and Garrard 1988), electrophoretic mobility shift assays (Kadonaga and Tjian 1986), SELEX (Fitzwater and Polisky 1996), and chromatin immunoprecipitation (Kuo and Allis 1999; Nal et al. 2001; Horak and Snyder 2002). With substantial efforts being invested into determination of DNA-binding specificity of specific transcription factors and the availability of completely sequenced genomes, the next question would be whether it is possible to predict all target genes in a genome for a specific transcription factor.
Most transcription factors recognize relatively short (<10 bp)DNA motifs, which often allow degeneration. Computational identification of such motifs is a well-developed method, and depending on the level of degeneracy of the motif, is usually performed by scanning genome sequences with either the consensus pattern or with a position weight matrix (PWM). PWMs are calculated from a set of experimentally defined TF-binding sequences and reflect binding specificity of the transcription factor. The major repository of PWMs is TransFac database (Matys et al. 2003), and several software implementations that use PWMs to predict binding sites are available (Quandt et al. 1995; Lenhard and Wasserman 2002; Matys et al. 2003).
Although pattern-recognition programs do predict most of the known functional TFBSs, they are not very useful for genome-wide analysis of uncharacterized sequences because of a high rate of false-positive predictions, as a result of the nature of binding sites. However, the substantial amount of false positives can be filtered by comparison of orthologous sequences from multiple species. This approach, termed phylogenetic footprinting (Tagle et al. 1988), has been successfully applied for the discovery of a limited number of functional TFBSs (Gumucio et al. 1992; Aparicio et al. 1995; Hardison et al. 1997a,b; Loots et al. 2000; Wasserman et al. 2000). The underlying assumption of the phylogenetic footprinting approach is that functional elements evolve slower than nonfunctional background sequences due to selective pressure. As a result, regulatory regions are expected to be evolutionarily conserved. There are two flavors of phylogenetic footprinting. The first approach relies on multiple alignments of orthologous sequences to identify conserved regions, whereas the second approach does not require a priori alignment, but tries to find motifs that are shared by orthologous sequences and evolve slower than surrounding regions (Blanchette and Tompa 2002). Both approaches have their own advantages and drawbacks (for review, see Ureta-Vidal et al. 2003). For example, discovery of motifs in unaligned orthologous sequences requires relatively large sets of orthologs to make a clear distinction between conserved and nonconserved elements. Only sufficiently conserved motifs can be discovered and only relatively short regions can be analyzed, as performance of the approach decreases dramatically for longer sequences.
Construction of multiple alignments using orthologous sequences, and identification of regulatory elements in the alignment that satisfy certain conservation criteria is a common phylogenetic footprinting approach. The major advantage of this approach is that in principle, only a few orthologous sequences (e.g., human and mouse)are required for the analysis (Hardison et al. 1997a,b; Dubchak et al. 2000; Wasserman et al. 2000; Wu et al. 2001). However, the success of this approach depends largely on the quality of the multiple alignment. Especially for more divergent species, alignment of promoter sequences may become difficult, and the chance that short regulatory elements are aligned incorrectly and remain undetected increases (Cliften et al. 2001; Tompa 2001). This disadvantage is common to all local and global alignment algorithms that can be applied to phylogenetic footprinting, such as DBA (Jareborg et al. 1999), BBA (Zhu et al. 1998), ClustalW (Thompson et al. 1994), Multialign (Corpet 1988), DIALIGN (Morgenstern et al. 1998), BLASTZ (Schwartz et al. 2000), AVID (Bray et al. 2003), and LAGAN (Brudno et al. 2003).
Aligning more closely related species is not a problem using the above-mentioned programs, but as a result of the high-sequence conservation, the alignment is not very informative for phylogenetic footprinting, as no distinction can be made between conserved functional elements and nondivergent nonfunctional background. Unfortunately, although phylogenetic shadowing (Boffelli et al. 2003)addresses this problem efficiently by including at least four to six closely related species in the analysis, this approach cannot be used for genome-wide analysis, due to the lack of complete genome sequences for such species. What is already available are complete or nearly complete genome sequences of human, mouse, rat, fugu, and other organisms like mosquito, Drosophila, and Ciona, with more genomes, such as chimpanzee, chicken, and zebrafish to be released over the next 2 yr. Therefore, further development of phylogenetic footprinting approaches that focus specifically on the discovery of functional regulatory elements is required to fully benefit from all of the emerging genomic data and to obtain an understanding of genome-wide regulatory transcriptional networks.
Clearly, there is a paradox in phylogenetic footprinting. To be able to recognize conserved (regulatory)elements, one needs enough evolutionary distance, but at the same time, this evolutionary distance makes it difficult to recognize short conserved elements such as, for example, TFBSs. To address this paradox, we have developed a novel variation of phylogenetic footprinting termed CONREAL (CONserved Regulatory Elements anchored ALignment). The algorithm is designed to find conserved transcription-factor binding sites in a pair of orthologous promoters, without prior alignment of the promoters. Evaluation of CONREAL performance for prediction of conserved TFBSs, on the basis of a reference set of known functional sites, indicates that the algorithm performs comparably with other global-alignment approaches like AVID and LAGAN when applied for less divergent species (human and mouse), but is particularly useful for phylogenetic footprinting in evolutionarily more distant species (human and fugu), as it can identify potential counterparts of functional human elements in the fugu genome that are not detected by other approaches. Furthermore, CONREAL has been made accessible via a clear Web-based interface that allows users to find conserved TFBSs in any promoter pair of interest.
RESULTS
CONREAL Algorithm
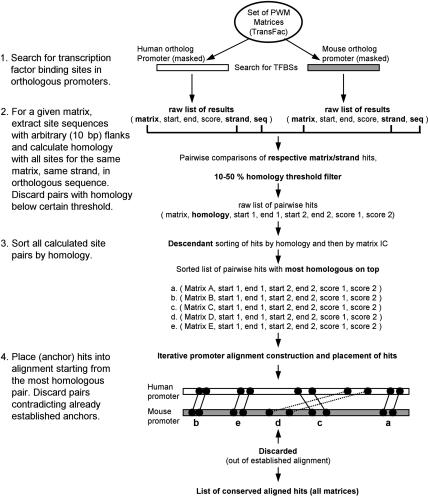
The assumptions behind CONREAL are that the sequence and order of functional regulatory elements are mainly conserved in orthologous promoters. The general principle of the method is outlined in Figure 1. CONREAL uses orthologous sequences that can optionally be masked for repeat regions as input and applies a defined set of PWMs for the analysis. This set should be large enough to produce comprehensive results. In our analysis, we used all human-, mouse-, and rat-derived PWMs (409)available in TransFac 7.2 Pro database. These PWMs represent ∼30% of all transcription factors present in a mammalian genome. As the first step, the orthologous sequences are searched independently with the PWMs set, and raw lists of predicted TFBSs are generated for each promoter. For each matrix, a hit start, end, strand, and score are recorded. Next, for a given matrix, all predicted TFBSs from one orthologous sequence are compared with all hits for the same matrix in the same orientation from the second orthologous sequence. Sequence identity is calculated for the region spanning a hit, including predefined flanking sequences on each side (5–20 bp). Results of these pairwise comparisons are used to generate a list that is sorted first by descending percentage of identity and then by matrix information content. Finally, a process similar to anchoring (Bray et al. 2003; Brudno et al. 2003)is performed; hits from the sorted list, starting from the most homologous one, are placed on the sequences, and pairs that disrupt already established anchors are discarded as inconsistent. As a result, CONREAL produces a list of consistent conserved TFBSs for all PWMs used in the analysis. This list essentially represents an alignment of orthologous sequences on the basis of conservation of potential regulatory elements, and conventional clustalw-like sequence representation of the alignment can easily be produced from this list. Because the goal of CONREAL algorithm is to establish an ordered chain of conserved TFBSs rather than to produce a complete global alignment of two sequences, regions in which no conserved TFBSs were found remain unaligned.
Figure 1.
Outline of the CONREAL algorithm.
Construction of the Reference Set
To evaluate the performance of CONREAL in relation to other phylogenetic footprinting approaches for the prediction of functional TFBSs, it is necessary to have a reference set of known regulatory elements. To maximize such a set, while keeping uniformity of data handling, we restricted ourselves to five vertebrate organisms for which assembled and annotated genomes are available in the Ensembl database (Clamp et al. 2003)as follows: human, rat, mouse, fugu, and zebrafish. We used data from TransFac database (Matys et al. 2003)to build a reference set of regulatory sites that satisfy the following criteria: (1)The site is experimentally verified in either human, mouse, or rat; that is, the site annotation indicates that functional analysis has been performed on these sites; (2)the site has been used to construct a PWM of relevant TF and can be found back in the genomic sequence of the respective organism using this PWM with at least 75% threshold parameter; (3)the site can be unambiguously mapped to an Ensembl gene in the respective organism within 3 kb upstream and 1 kb downstream of the start of the gene as annotated in Ensembl, and (4)the Ensembl gene containing the site should have an annotated Ensembl orthologous gene in at least one other organism (human, mouse, rat, fugu, or zebrafish). The resulting reference set consists essentially of three lists grouped by the organism, in which the site is experimentally confirmed to be functional. Each entry in the list contains the site ID, linked PWM, coordinates of the site in the reference gene and IDs of orthologous genes (Supplementary Table S1, available online at www.genome.org). In total, 88 sites were included in the reference set (Table 1).
Table 1.
Composition of the Reference Site Set
|
Experimentally verified in:
|
|||
|---|---|---|---|
| Human | Mouse | Rat | |
| TransFac matrices | 24 | 11 | 14 |
| Ensembl genes | 35 | 14 | 15 |
| TransFac sites | 52 | 17 | 19 |
TFBSs that are experimentally verified in human, mouse or rat gene with the target gene having an annotated ortholog in at least one other organism (human, mouse, rat, fugu and zebrafish) constitute the reference set. Because some reference sites are represented by the same PWMs and can occur in several genes, and because several different reference sites can occur in the same gene, the numbers of TransFac sites and matrices and Ensembl genes are different. A detailed list of the reference sites is provided in Supplemental Table S1.
Comparison of CONREAL With Other Approaches
To compare results of the standard alignment-based phylogenetic footprinting approach with CONREAL predictions, we selected two recently developed tools, AVID (Bray et al. 2003)and LAGAN (Brudno et al. 2003), to generate alignments for phylogenetic footprinting. Both of these programs were developed for fast, but sensitive global alignment of large sequences and are currently the methods of choice for the whole-genome alignment. There are three parameters that can be varied in the CONREAL algorithm, threshold for PWMs, length of site flanks to calculate homology, and threshold for homology. Homology is calculated as a mere percentage of identical positions over the complete region of the TFBS, including equal flanks to the left and right of the site.
Parameters, which usually apply to alignment-based approaches, are threshold for PWMs and level of sequence conservation over a certain region. There are several methods to calculate the sequence conservation within multiple alignments that are based on different underlying assumptions, but all of them produce similar results when parameters are adjusted properly (Stojanovic et al. 1999). Therefore, we decided to calculate homology of sites in alignments in the same way as for CONREAL, centered on the TFBS with fixed flanks length.
To evaluate the effect of different parameter combinations on the results, we performed profiling of the approaches; for every site and gene pair combination from the reference set, the total number of conserved sites found for the reference matrix was counted, and positive identification of the reference site was scored. Performance of CONREAL, AVID-, and LAGAN-based methods was evaluated separately for human–mouse–rat gene pairs and for mammalian versus fugu or zebrafish gene pairs, to estimate influence of sequence divergence level on the sensitivity of the methods. It appeared that all of the methods have similar parameter profiles (Supplementary Table S2); hence, it is justified to use the same parameters for comparing the approaches. For further investigation, we selected parameter stringencies that result in the prediction of a large number of sites from the reference set to be conserved, while keeping the total number of aligned sites reasonably low (75% PWM threshold, 50% homology threshold, 5-bp flank length). At these parameter settings, CONREAL predicts slightly more reference TFBSs to be conserved in gene pairs than other approaches do, in both intramammalian and mammalian–fish ranges (93 for CONREAL, 90 for LAGAN, and 85 for AVID in mammals, and 12, 7, and 5, respectively, in mammals–fish).
Apart from the number of reference sites found to be conserved, the second parameter to compare is the total number of additional aligned hits found by each of the methods, which might be an indication of false positives. To calculate the total number of hits, first, for every reference site/gene pair combination, all additional aligned hits for respective PWM in the gene pair were calculated, and then the numbers for all gene pairs in a reference set were summed. It appeared that CONREAL finds slightly more additional hits compared with AVID and LAGAN. However, the ratio between confirmed and additional sites remains relatively constant, meaning an equal increase in sensitivity for finding real sites as for additional sites. If sensitivity had been decreased, the ratio of confirmed-to-additional sites should decrease, which is not the case. Furthermore, interpretation of the number of additional hits is complicated in the case of phylogenetic footprinting, because it is difficult to distinguish false positive-aligned hits from potentially functional aligned hits. On the one hand, it is reasonable to assume that a certain fraction of predictions represent false positives, and therefore, the larger the total number of aligned hits, the larger the number of false predictions. On the other hand, it is also reasonable to argue that the more hits that are aligned by a method, the better, as it allows the identification of more, potentially true regulatory elements. At the present time, a real comparison of specificities for the different methods is not possible due to the lack of well-established controls, especially for the mammal–fish data set.
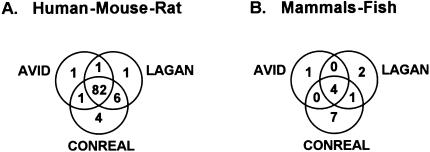
Taken together, sheer calculation of the number of conserved reference TFBSs and the number of additional TFBSs found by different approaches does not clearly indicate that one method is better or worse than another. For further clarification, we performed site-by-site analysis of predictions made by different methods (Table 2; Supplementary Table S3). First, we investigated the extent of overlap between the predictions made by the three different programs. We consider an aligned hit to be common to two methods when both of them align a reference site to exactly the same region in the gene pair. It appeared that for human, mouse, and rat gene pairs, from the total of 96 aligned reference TFBSs, 82 hits (or 85%)are commonly predicted by all three methods (Fig. 2A), and the fraction of method-specific sites is very low (one for AVID and LAGAN and four for CONREAL). The 96 hits represent 51 of 88 TFBSs from the reference set (or 59%), and the remaining 27 reference TFBSs were not found to be conserved in any gene pair by any method. This number is in good agreement with results of Dermitzakis et al. (2002), who estimated that ∼60%–68% of TFBSs are functionally conserved between human and rodents. Therefore, it is reasonable to think that all of the approaches tested, including CONREAL, with the parameters used (75% PWM threshold, 10% homology threshold, and 5-bp flank length), are capable of predicting most, if not all, conserved regulatory elements between human and rodents.
Table 2.
Conservation of TFBSs from the Reference Set Among Different Organisms Identified by Different Approaches
| Site | Human | Mouse | Rat | Fugu | Zebrafish |
|---|---|---|---|---|---|
| R11477 | Ref | ALC | ALC | --C | ALC |
| R08296 | Ref | --C | --- | n/a | --- |
| R08092 | Ref | --C | --- | --- | --- |
| R10227 | Ref | A-C | ALC | --- | n/a |
| R11626 | Ref | -LC | ALC | --- | --C |
| R08293 | Ref | -L- | --C | n/a | --C |
| R03187 | Ref | ALC | -LC | n/a | n/a |
| R13001 | Ref | -LC | --- | n/a | n/a |
| R13048 | Ref | --C | --- | --- | --- |
| R11015 | ALC | Ref | ALC | --- | --C |
| R08112 | ALC | ALC | Ref | --- | --C |
| R08512 | ALC | ALC | Ref | --C | -LC |
Analysis parameters: 75% PWM threshold, 50% homology threshold, 5-bp flank length. (Ref) the site is a reference in the given organism, that is, known to be functional; (A) found by AVID; (L) found by LAGAN; (C) found by CONREAL; (-) not found; (n/a) the reference gene does not have an annotated ortholog in a given organism. The complete and detailed dataset is available as Supplemental Table S3.
Figure 2.
Intersection of conserved reference sites found by AVID, LAGAN, and CONREAL approaches in (A) human–mouse–rat and (B) mammals–fish gene pairs. The analysis parameters are 75% PWM threshold, 50% homology threshold, and 5-bp flank length.
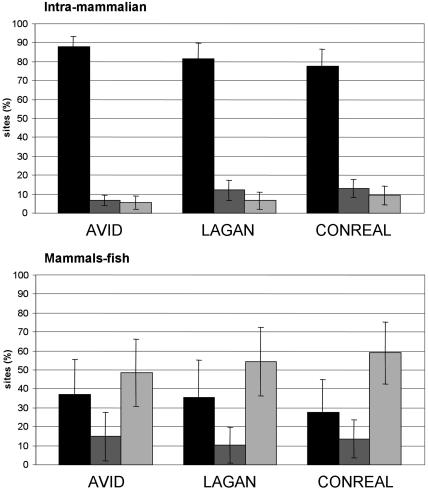
By comparison, for fugu and zebrafish sequences, only four sites are commonly predicted by all three methods, one site is found by CONREAL and LAGAN, and one, two, and seven sites are specific for AVID, LAGAN, and CONREAL, respectively (Fig. 2B). The total number of predicted conserved sites is low in the case of mammals–fish comparisons, and does not allow us to obtain a clear picture of the extent of overlap between sites predicted by different methods. To overcome this problem, we calculated the intersection of all conserved sites (not only sites from the reference set)predicted by different methods on the orthologous gene pairs from the reference set (Fig. 3). It appeared that all three methods have a similar distribution of hits, and in the case of intramammalian comparisons, the fraction of sites commonly predicted by all three methods is ∼80%. Furthermore, the fraction of sites confirmed by one other method and method-specific sites is ∼10%. This distribution changes in mammals–fish comparisons; the fraction of sites common to the three methods drops to 30%, whereas the number of method-specific sites increases to ∼60%. The important observation this analysis provides is that in the case of comparison of evolutionarily distant species, such as human and fugu, more than half of the predictions produced by each method are not confirmed by other approaches, that is, are method specific. This raises the question of which approach is best suited for the comparison of distant species.
Figure 3.
Intersection of the total number of conserved sites found by different approaches in intramammalian (top) and mammal–fish (bottom) gene pairs from the reference set. The percentage of sites found by all three methods is shown in black, sites confirmed by one additional method are dark gray, and method-specific fraction of sites is light gray. Error bars, 95% confidence intervals. The analysis parameters are 75% PWM threshold, 50% homology threshold, and 5-bp flank length.
To get an idea of the properties of sites found exclusively by a specific method, the method-specific reference sites (Fig. 2) were investigated manually. Two properties of the sites were checked, stability of site position in a multiple species sequence alignment and conservation of the site between several species. The first property can be used as a measure of robustness of the alignment in this particular region. For our comparisons, we projected the proper pairwise alignments from human–mouse–rat–fugu–zebrafish multiple alignments produced by MLAGAN (Brudno et al. 2003). The second property addresses the prediction reliability from an evolutionary point of view. If, for example, a human reference site is aligned in the fugu sequence and also found to be conserved in rat and mouse genomes, then it is likely that this human–fugu alignment is correct. If it is not conserved in rodents, then it is more likely that the human–fugu phylogenetic footprint is false.
CONREAL-Specific Sites
From the four CONREAL-specific sites that were found in the human–mouse–rat comparison (Fig. 2A), two are found only in human and mouse by CONREAL and are not conserved in rat and fish orthologs. These reference sites (R08296 and R13048, Supplementary Table S3)are also not identified as conserved in mouse or rat by other methods. Therefore, it is very likely that these two pairs are CONREAL false positives. The third site (R08293, Table 2)is found by CONREAL to be conserved between human, rat, and zebrafish, but not mouse. In contrast, LAGAN identifies this site as conserved only in mouse. Therefore, this TFBS can be functional in both rodents and fish, but a combination of different approaches is required to identify its conservation. The fourth CONREAL-specific reference site (R08092, Table 2)is conserved between mouse and human only. AVID and LAGAN do not align this site, but in the MLAGAN-generated alignment, the site alignment coincides with CONREAL prediction, strongly suggesting that the CONREAL alignment in this case is correct. This example illustrates that the approach is sensitive enough to recognize the correct human–mouse alignment and conserved TFBS from two sequences alone, whereas it requires rat and fish orthologs for MLAGAN to identify the same human–mouse alignment.
Of the seven CONREAL-specific sites from the mammal–fish comparisons (Fig. 2B), six sites are found to be conserved not only between the mammalian organism and fugu or zebrafish, but also in other mammals from the set, strongly suggesting that these CONREAL predictions are very likely to be correct. There is only one CONREAL prediction of the conserved human–zebrafish site (R08294, Supplementary Table S3)not supported by any other evidence.
LAGAN- and AVID-Specific Sites
All three LAGAN-specific site predictions (Fig. 2)are conserved between more than two organisms, providing the evidence that these LAGAN predictions are likely to be correct (sites R02709, R08293, and R01358, Supplementary Table S3). From two AVID-specific sites, one is conserved in multiple species (R11703, Supplementary Table S3), and the other one is conserved only between rat and mouse (R13010, Supplementary Table S3). None of the LAGAN- and AVID-specific sites remains conserved in alignments produced by MLAGAN, indicating that the predictions are not robust.
In summary, the analysis of method-specific sites indicates that all three approaches generate predictions of approximately the same quality, but CONREAL is able to predict more conserved sites in both intramammalian and mammalian–fish ranges of sequence divergence.
Estimation of Noise Predictions Level
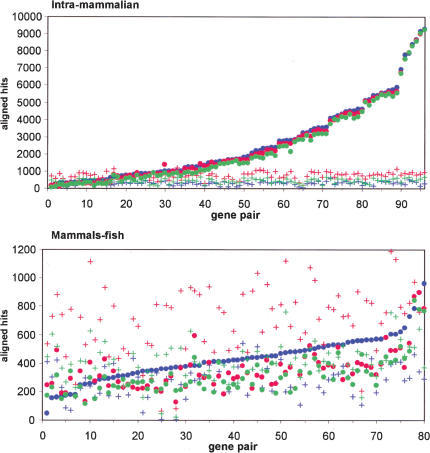
To estimate the level of noise predictions (sites aligned by chance in a pair of sequences)with AVID, LAGAN, and CONREAL approaches, we performed an analysis with the complete reference data set, but with the orthologous sequence in the gene pair randomized by the shuffleseq program from the EMBOSS package (http://www.emboss.org). For every orthologous pair, represented by 4 kb of sequence (3 kb upstream from the gene start and 1 kb downstream), we calculated the total number of aligned hits for all 409 matrices used in the analysis and the total number of aligned hits in a pair with shuffled orthologous sequence. Human–rodent and mammals–fish data sets were evaluated separately to take into account divergence levels between sequences (Fig. 4). For CONREAL, AVID, and LAGAN, 301 ± 69, 436 ± 82, and 741 ± 131 hits, respectively, were found in the randomized sequences using the human–rodent set, and 284 ± 82, 413 ± 93, and 711 ± 159 hits using the mammal–fish data set. From this, it appeared that the average number of hits as a result of noise is similar for both data sets. The same results were obtained in simulations in which orthologous sequences were not randomized, but instead, reversed and not complemented (data not shown). The latter approach has been used successfully for the estimation of spurious matches in BLASTZ alignments (Schwartz et al. 2003).
Figure 4.
Estimation of spurious prediction levels in intramammalian (top) and mammals–fish (bottom) pairwise comparisons. Dots represent total number of aligned hits found in an orthologous pair, whereas crosses represent the number of aligned hits found in the same pair when orthologous sequences are randomized. AVID results are shown in green, LAGAN in red, and CONREAL in blue. The data sets are sorted by the number of CONREAL predictions to improve perception of the graph. The analysis parameters are 75% PWM threshold, 50% homology threshold, and 5-bp flank length.
The number of noise predictions obtained by this simulation indicates that AVID, and in particular, LAGAN, would potentially predict more false-positive hits when applied to substantially diverged sequences. This is probably explained by the fact that LAGAN, in contrast to AVID and CONREAL, attempts to align the whole length of the sequences even if there is little homology.
When comparing real predictions with simulated noise predictions, the majority of aligned hits between mammals are clearly above the noise background for most gene pairs (Fig. 4). More detailed manual analysis of some real cases that end up in the background shows that these predictions are due to incorrect assignment of the orthologous gene, or because most of the sequence was masked as a result of the abundant presence of repetitive sequences (data not shown). In contrast, for most of the mammal–fish gene pairs, the number of aligned hits does not significantly exceed noise background level (Fig. 4). However, this does not mean that fugu and zebrafish sequences cannot be used for phylogenetic footprinting of mammalian regulatory elements, but it indicates that interpretation of the results should be treated with care, and that additional support from other species is needed for firm conclusions.
Example: Analysis of Foxa2 Promoter Region
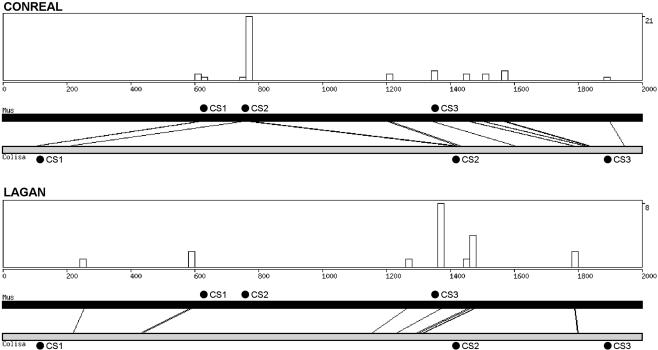
To provide additional evidence that the CONREAL approach can be advantageous for phylogenetic footprinting in divergent species, we searched the literature for experimentally verified enhancer elements conserved between fish and mammalian genomes. Although the available data is very limited, we found a representative example in the Foxa2 gene. The promoter of this gene contains three regulatory elements conserved between mouse, chicken, and dwarf gourami Colisa lalia (Nishizaki et al. 2001). More importantly, these conserved elements were shown to be functional in both mouse and gourami promoters. We analyzed the respective mouse and gourami sequences by CONREAL and LAGAN to find out whether the regulatory regions can be found by these computational approaches. It appeared that CONREAL aligns two of three regions exactly as expected from experimental data (Fig. 5; CS1: positions 617–636 in mouse to positions 108–125 in gourami; CS2: positions 767–783 to positions 1404–1420 in mouse and gourami, respectively. Coordinates are as in GenBank sequences AB050942 and AB050940). The third regulatory element, CS3, was experimentally identified at positions 1347–1360 in mouse and 1870–1883 in gourami. CONREAL does not align these regions together, but the mouse region is still identified as conserved, showing the potential importance of this site.
Figure 5.
Output of the CONREAL webinterface. The example shows the results for the analysis of the mouse and dwarf gourami Foxa2 promoter regions (Accession nos. AB050942 and AB050940, respectively) performed by CONREAL (top) and LAGAN methods (bottom). The graphs show the positions of aligned hits and the distribution/concentration of conserved TFBSs along the sequences. The graphs are followed by sequence-alignment data and tables of conserved TFBSs linked to TransFac entries (data not shown). Black circles above the black bar (mouse sequence) and below the gray bar (gourami sequence) represent positions of the conserved regulatory elements CS1–CS3 that are experimentally confirmed to be functional in mouse and gourami sequences. The analysis parameters are 80% PWM threshold, 50% homology threshold, and 15-bp flank length.
In contrast to CONREAL, LAGAN (Fig. 5) and AVID (data not shown)failed to align any of the respective conserved regions together, clearly demonstrating the usefulness of CONREAL for prediction of functional regulatory elements in evolutionarily distant species.
CONREAL Web Interface
We developed a CONREAL Web interface for pairwise comparison of promoter sequences (http://conreal.niob.knaw.nl). CONREAL accepts as input a pair of masked orthologous sequences in multiple fasta format. The user can define thresholds for PWM and homology, as well as length of flanks. Results are summarized in a graphical output (Fig. 5). In addition, a sequence alignment is produced and a list of conserved TFBSs with positional and statistical information and links to TransFac database for additional TF information is provided. CONREAL Web interface also provides access to LAGAN-based phylogenetic footprinting approach so that the two methods can be easily compared and results intersected if necessary.
DISCUSSION
Genome-wide prediction of transcription-factor binding targets is an important approach for dissecting and understanding gene regulatory networks. The use of orthologous sequences from several species is necessary for reliable prediction of functional conserved binding sites. Whereas alignment of sequences from closely related species is usually obvious, the resulting alignment might be uninformative as a result of the high overall homology between sequences, and comparison of more divergent species may be desirable. However, phylogenetic footprinting on such divergent species can be less sensitive, due to difficulties in establishing the correct alignment harboring functional conserved elements. We tried to address this problem by the development of a new phylogenetic footprinting approach, CONREAL.
The two assumptions underlying CONREAL are scientifically motivated. The first assumption is general to all phylogenetic footprinting approaches and says that functional regulatory elements are likely to be conserved between species. In addition, we assume that conservation is likely to expand to some extent beyond the sequences as identified by PWM, although not necessary for the algorithm to work, as the flank length can be set to zero. The second assumption that we introduce in CONREAL is that the order of functional TFBSs in regulatory regions is also conserved between species. It is reasonable to think that in most cases it is necessary to preserve the order of regulatory sites in sequences recognized by TFs, so that transcription factors can interact properly to form a functional regulatory complex. This assumption is supported, for example, by the observation of Jegga et al. (2002), showing that conservation of the order of TFBSs can be a reliable indicator of the presence of a regulatory region.
We compared the performance of CONREAL with two global alignment programs, AVID and LAGAN. Although other programs exist, for example, CLUSTALW, these programs were selected because they outperform other available software both in speed and quality of the alignment (Bray et al. 2003; Brudno et al. 2003). In addition, AVID is the aligner of choice used in the rVISTA system for high-throughput prediction of cis-elements by phylogenetic footprinting (Loots et al. 2002). The principal difference between AVID and LAGAN algorithm is that AVID looks for exact matches to nucleate the alignment, whereas LAGAN uses short inexact words for this purpose. As a result, LAGAN is expected to be more suitable for alignment of distant species. CONREAL algorithm is, in fact, similar to LAGAN and AVID as it first identifies all potential short matches that could be included in the alignment, and then iteratively puts these matches as anchors into the final alignment according to the specified criteria. What makes CONREAL different is that it uses biologically relevant information, that is, potential transcription-factor binding sites, to find the initial inexact short matches that are used as anchors. Therefore, the CONREAL approach should be more suitable for the alignment of regulatory regions compared with the more general alignment programs. Another variant of a motif-based alignment algorithm was reported previously by Cheremushkin and Kel (2003). This algorithm takes into account similarity in distribution of potential binding sites and is a modification of the Needleman-Wunsch dynamic programming algorithm. Although both the CONREAL and motif-based alignment algorithm of Cheremushkin and Kel (2003)use PWMs for inference of alignment, the two approaches are principally different. The motif-based alignment approach was not evaluated in this work, because it was still in the development phase at the time of writing.
Comparison of CONREAL, AVID, and LAGAN using a carefully selected reference set of regulatory sites that are known to be functional in at least one of the reference organisms, revealed that all approaches perform similarly, with CONREAL predicting more sites that are conserved between mammals and fish, whereas the number of total predictions of CONREAL is not substantially elevated. This shows that CONREAL's increased sensitivity in distant species is not due to a general overprediction of conserved sites. Manual investigation of sites specifically predicted by CONREAL indicates that validity of these sites is comparable with validity of sites uniquely predicted by other methods. Moreover, we provide an example of the promoter region of the Foxa2 gene, showing that CONREAL correctly predicts experimentally verified regulatory elements conserved between mouse and gourami promoters of the gene, whereas other methods fail to identify these regions as conserved.
CONREAL utilizes a simplified approach for anchoring conserved TFBSs; the most homologous pairs of TFBSs get priority of placement. The obvious disadvantage of this approach is that in some cases, the incorrect, but high-scoring TFBSs pair could prevent the correct placement of lower-scoring matches in a region affected. We observed this kind of CONREAL behavior for some gene pairs at certain parameter settings. Usually, the problem can be resolved by running CONREAL with different parameters and comparing results. We think that the combination of the CONREAL approach for selection of potential anchors and the LAGAN method for calculation of the optimal chains of anchors would ultimately be a better solution for prediction of conserved transcription-binding sites, and we will concentrate further development of CONREAL in this direction.
Availability of data for conservation of regulatory elements in more than two species should make it possible to estimate rates of turnover of regulatory sites. Previously, it was estimated that 32%–40% of functional transcription-factor binding sites are species specific for human and rodents (Dermitzakis and Clark 2002). Basically, species-specific sites can result from either the creation of a new site in one lineage or the loss of an ancestral site in another lineage after a speciation event. Our preliminary analysis indicates that rates of creation and loss of functional transcription-factor binding sites are nearly identical in the evolutionary interval between human and rodents. At present, however, evolution of regulatory elements is still poorly understood and good working models are lacking (Ureta-Vidal et al. 2003). This prevents us from elaborating on the evolutionary aspects of the data that were acquired in this study.
In summary, we have shown that CONREAL is a powerful tool for phylogenetic footprinting of regulatory elements, especially for evolutionarily more-distant species, thus, it may be an important approach, complementary to existing methods, for dissecting the organization of regulatory regions and for getting closer to building genome-wide transcriptional networks. Although CONREAL does make the most TFBS predictions using a reference set, the highest reliability will be obtained when different methods are combined. Hence, we believe that the different methods compared here are complementary to each other, rather than competitive. Therefore, we already included LAGAN in the CONREAL Web interface to make comparison of results easy. Furthermore, the combined use of more than two orthologous sequences in phylogenetic footprinting may further increase the specificity. Preliminary analysis using MLAGAN, an implementation of LAGAN for multiple alignments that takes into account the phylogenetic relationship of the sequences to be aligned (Brudno et al. 2003), shows that alignment of more divergent species, such as, for example, human and fugu, are improved when mouse and rat orthologous sequences are included in the analysis. With an increased number of whole-genome sequences becoming available, approaches that allow the inclusion of orthologous sequences from as many species as possible will be needed to unravel lineage-specific transcriptional programs, supporting the future development of a multiple alignment CONREAL approach.
METHODS
CONREAL Implementation and Availability
CONREAL is publicly available at http://conreal.niob.knaw.nl/ as a Web-based tool. The stand-alone program is available from the authors upon request. CONREAL is written in Perl with some Inline C-code. PWM-related tasks (storing and retrieving of PWMs from database, searching of sequences)are performed by TFBS Perl modules (Lenhard and Wasserman 2002). Bioperl modules (Stajich et al. 2002)are used for sequence handling, Perl modules GD.pm and CGI.pm are used for generation of graphics and Web interface.
Construction of the Reference Set of TFBSs
We used TransFac 7.2 Pro database to generate a set of reference transcription-factor binding sites. The initial site list was generated by parsing the file “site.dat” and extracting all sites with quality 1 and “functional analysis” entry in the MM field. The site list was further reduced to sites that originate from human, mouse, or rat and have associated matrix (MX field)and EMBL sequences (DR field). The associated EMBL sequence were retrieved from EMBL database and searched with PWM (75% threshold)associated with TFBS annotated in the sequence. Site entries that were not found back in the EMBL sequence at the expected location according to TransFac annotation were discarded along with corresponding EMBL sequence. Retained EMBL sequences were split in three reference groups by the organism (human, mouse, and rat).
To map sites to Ensembl genes, we first generated promoter sets by extracting regions of 3 kb upstream and 1 kb downstream relative to the gene start (as annotated in Ensembl)for all genes in Ensembl human (v.15.33), mouse (v.15. 30), and rat (v.15.2). The EMBL sequence sets were BLASTed against the corresponding promoter sets, BLAST results were parsed semi-automatically, and EMBL sequences were uniquely assigned to Ensembl genes. EMBL sequences without obvious matches to extracted Ensembl regions were discarded. Finally, the exact position of annotated TFBS in the Ensembl sequence was calculated from its position in the EMBL sequence and the position of the EMBL sequence in the Ensembl region. Thus, a list of experimentally verified transcription-factor binding sites that can be found back by associated PWM at certain coordinates in certain Ensembl genes was generated. This list summarizes all of the reference sites in the reference genes.
The list of human, mouse, rat, fugu, or zebrafish genes orthologous to the reference gene was extracted from Ensemblcompara database (v.15.1). In a case in which several genes from the same organism were annotated as orthologs to the reference gene, the orthologous ORFs were blasted against the reference ORF, and the gene with the highest BLASTp score was selected as the true ortholog, and the rest of annotated orthologs were not used in the analysis.
The resulting reference set, which contains matrix ID, reference Ensembl gene ID, coordinates of the site in the reference gene, and Ensembl gene IDs of orthologous genes, is available as Supplementary Table S1.
Acknowledgments
We thank Michael Brudno for helpful comments on the manuscript and Robin May for critically reading the manuscript.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1642804. Article published online before print in December 2003.
Footnotes
[Supplemental material is available online at www.genome.org.]
References
- Aparicio, S., Morrison, A., Gould, A., Gilthorpe, J., Chaudhuri, C., Rigby, P., Krumlauf, R., and Brenner, S. 1995. Detecting conserved regulatory elements with the model genome of the Japanese puffer fish, Fugu rubripes. Proc. Natl. Acad. Sci. 92: 1684-1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette, M. and Tompa, M. 2002. Discovery of regulatory elements by a computational method for phylogenetic footprinting. Genome Res. 12: 739-748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boffelli, D., McAuliffe, J., Ovcharenko, D., Lewis, K.D., Ovcharenko, I., Pachter, L., and Rubin, E.M. 2003. Phylogenetic shadowing of primate sequences to find functional regions of the human genome. Science 299: 1391-1394. [DOI] [PubMed] [Google Scholar]
- Bray, N., Dubchak, I., and Pachter, L. 2003. AVID: A global alignment program. Genome Res. 13: 97-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brudno, M., Do, C.B., Cooper, G.M., Kim, M.F., Davydov, E., NISC Comparative Sequencing Program, Green, E.D., Sidow, A., and Batzoglou, S. 2003. LAGAN and Multi-LAGAN: Efficient tools for large-scale multiple alignment of genomic DNA. Genome Res. 13: 721-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheremushkin, E. and Kel, A. 2003. Whole genome human/mouse phylogenetic footprinting of potential transcription regulatory signals. Pac. Symp. Biocomput. 291-302. [DOI] [PubMed]
- Clamp, M., Andrews, D., Barker, D., Bevan, P., Cameron, G., Chen, Y., Clark, L., Cox, T., Cuff, J., Curwen, V., et al. 2003. Ensembl 2002: Accommodating comparative genomics. Nucleic Acids Res. 31: 38-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cliften, P.F., Hillier, L.W., Fulton, L., Graves, T., Miner, T., Gish, W.R., Waterston, R.H., and Johnston, M. 2001. Surveying Saccharomyces genomes to identify functional elements by comparative DNA sequence analysis. Genome Res. 11: 1175-1186. [DOI] [PubMed] [Google Scholar]
- Corpet, F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16: 10881-10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dermitzakis, E.T. and Clark, A.G. 2002. Evolution of transcription factor binding sites in Mammalian gene regulatory regions: Conservation and turnover. Mol. Biol. Evol. 19: 1114-1121. [DOI] [PubMed] [Google Scholar]
- Dubchak, I., Brudno, M., Loots, G.G., Pachter, L., Mayor, C., Rubin, E.M., and Frazer, K.A. 2000. Active conservation of noncoding sequences revealed by three-way species comparisons. Genome Res. 10: 1304-1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzwater, T. and Polisky, B. 1996. A SELEX primer. Methods Enzymol. 267: 275-301. [DOI] [PubMed] [Google Scholar]
- Galas, D.J. and Schmitz, A. 1978. DNAse footprinting: A simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 5: 3157-3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross, D.S. and Garrard, W.T. 1988. Nuclease hypersensitive sites in chromatin. Annu. Rev. Biochem. 57: 159-197. [DOI] [PubMed] [Google Scholar]
- Gumucio, D.L., Heilstedt-Williamson, H., Gray, T.A., Tarle, S.A., Shelton, D.A., Tagle, D.A., Slightom, J.L., Goodman, M., and Collins, F.S. 1992. Phylogenetic footprinting reveals a nuclear protein which binds to silencer sequences in the human γ and ε globin genes. Mol. Cell. Biol. 12: 4919-4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardison, R., Slightom, J.L., Gumucio, D.L., Goodman, M., Stojanovic, N., and Miller, W. 1997a. Locus control regions of mammalian β-globin gene clusters: Combining phylogenetic analyses and experimental results to gain functional insights. Gene 205: 73-94. [DOI] [PubMed] [Google Scholar]
- Hardison, R.C., Oeltjen, J., and Miller, W. 1997b. Long human–mouse sequence alignments reveal novel regulatory elements: A reason to sequence the mouse genome. Genome Res. 7: 959-966. [DOI] [PubMed] [Google Scholar]
- Horak, C.E. and Snyder, M. 2002. ChIP-chip: A genomic approach for identifying transcription factor binding sites. Methods Enzymol. 350: 469-483. [DOI] [PubMed] [Google Scholar]
- Jareborg, N., Birney, E., and Durbin, R. 1999. Comparative analysis of noncoding regions of 77 orthologous mouse and human gene pairs. Genome Res. 9: 815-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jegga, A.G., Sherwood, S.P., Carman, J.W., Pinski, A.T., Phillips, J.L., Pestian, J.P., and Aronow, B.J. 2002. Detection and visualization of compositionally similar cis-regulatory element clusters in orthologous and coordinately controlled genes. Genome Res. 12: 1408-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadonaga, J.T. and Tjian, R. 1986. Affinity purification of sequence-specific DNA binding proteins. Proc. Natl. Acad. Sci. 83: 5889-5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo, M.H. and Allis, C.D. 1999. In vivo cross-linking and immunoprecipitation for studying dynamic Protein:DNA associations in a chromatin environment. Methods 19: 425-433. [DOI] [PubMed] [Google Scholar]
- Lenhard, B. and Wasserman, W.W. 2002. TFBS: Computational framework for transcription factor binding site analysis. Bioinformatics 18: 1135-1136. [DOI] [PubMed] [Google Scholar]
- Loots, G.G., Locksley, R.M., Blankespoor, C.M., Wang, Z.E., Miller, W., Rubin, E.M., and Frazer, K.A. 2000. Identification of a coordinate regulator of interleukins 4, 13, and 5 by cross-species sequence comparisons. Science 288: 136-140. [DOI] [PubMed] [Google Scholar]
- Loots, G.G., Ovcharenko, I., Pachter, L., Dubchak, I., and Rubin, E.M. 2002. rVista for comparative sequence-based discovery of functional transcription factor binding sites. Genome Res. 12: 832-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matys, V., Fricke, E., Geffers, R., Gossling, E., Haubrock, M., Hehl, R., Hornischer, K., Karas, D., Kel, A.E., Kel-Margoulis, O.V., et al. 2003. TRANSFAC: Transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 31: 374-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgenstern, B., Frech, K., Dress, A., and Werner, T. 1998. DIALIGN: Finding local similarities by multiple sequence alignment. Bioinformatics 14: 290-294. [DOI] [PubMed] [Google Scholar]
- Nal, B., Mohr, E., and Ferrier, P. 2001. Location analysis of DNA-bound proteins at the whole-genome level: Untangling transcriptional regulatory networks. BioEssays 23: 473-476. [DOI] [PubMed] [Google Scholar]
- Nishizaki, Y., Shimazu, K., Kondoh, H., and Sasaki, H. 2001. Identification of essential sequence motifs in the node/notochord enchancer of Foxa2 (Hnf3β)gene that are conserved across vertebrate species. Mech. Dev. 102: 57-66. [DOI] [PubMed] [Google Scholar]
- Quandt, K., Frech, K., Karas, H., Wingender, E., and Werner, T. 1995. MatInd and MatInspector: New fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 23: 4878-4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, S., Zhang, Z., Frazer, K.A., Smit, A., Riemer, C., Bouck, J., Gibbs, R., Hardison, R., and Miller, W. 2000. PipMaker—A eb server for aligning two genomic DNA sequences. Genome Res. 10: 577-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, S., Kent, W.J., Smit, A., Zhang, Z., Baertsch, R., Hardison, R.C., Haussler, D., and Miller, W. 2003. Human–mouse alignments with BLASTZ. Genome Res. 13: 103-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stajich, J.E., Block, D., Boulez, K., Brenner, S.E., Chervitz, S.A., Dagdigian, C., Fuellen, G., Gilbert, J.G., Korf, I., Lapp, H., et al. 2002. The Bioperl toolkit: Perl modules for the life sciences. Genome Res. 12: 1611-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojanovic, N., Florea, L., Riemer, C., Gumucio, D., Slightom, J., Goodman, M., Miller, W., and Hardison, R. 1999. Comparison of five methods for finding conserved sequences in multiple alignments of gene regulatory regions. Nucleic Acids Res. 27: 3899-3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagle, D.A., Koop, B.F., Goodman, M., Slightom, J.L., Hess, D.L., and Jones, R.T. 1988. Embryonic ε and γ globin genes of a prosimian primate (Galago crassicaudatus). Nucleotide and amino acid sequences, developmental regulation and phylogenetic footprints. J. Mol. Biol. 203: 439-455. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. 1994. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompa, M. 2001. Identifying functional elements by comparative DNA sequence analysis. Genome Res. 11: 1143-1144. [DOI] [PubMed] [Google Scholar]
- Ureta-Vidal, A., Ettwiller, L., and Birney, E. 2003. Comparative genomics: Genome-wide analysis in metazoan eukaryotes. Nat. Rev. Genet. 4: 251-262. [DOI] [PubMed] [Google Scholar]
- Wasserman, W.W., Palumbo, M., Thompson, W., Fickett, J.W., and Lawrence, C.E. 2000. Human–mouse genome comparisons to locate regulatory sites. Nat. Genet. 26: 225-228. [DOI] [PubMed] [Google Scholar]
- Wu, Q., Zhang, T., Cheng, J.F., Kim, Y., Grimwood, J., Schmutz, J., Dickson, M., Noonan, J.P., Zhang, M.Q., Myers, R.M., et al. 2001. Comparative DNA sequence analysis of mouse and human protocadherin gene clusters. Genome Res. 11: 389-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, J., Liu, J.S., and Lawrence, C.E. 1998. Bayesian adaptive sequence alignment algorithms. Bioinformatics 14: 25-39. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://conreal.niob.knaw.nl/; CONREAL Web server.
- http://www.emboss.org; EMBOSS package.