Abstract
Genetic diversity and population structure of Plasmodium vivax parasites are valuable to the prediction of the origin and spread of novel variants within and between populations, and to the program evaluation of malaria control measures. Using two polymorphic genetic markers, the merozoite surface protein genes PvMSP-3α and PvMSP-3β, we investigated the genetic diversity of four Southeast Asian P. vivax populations, representing both subtropical and temperate strains with dramatically divergent relapse patterns. PCR amplification of PvMSP-3α and PvMSP-3β genes detected three and four major size polymorphisms among the 235 infections examined, respectively, while restriction analysis detected 15 and 19 alleles, respectively. Samples from different geographical areas differed dramatically in their PvMSP-3α and PvMSP-3β allele composition and frequency. Samples tended to cluster on the basis of their PCR-RFLP polymorphism. These results indicated that different parasite genotypes were circulating in each endemic area, and that geographic isolation may exist. Multiple infections were detected in all four parasite populations, ranging from 20.5% to 31.8%, strongly indicating that P. vivax populations were highly diverse and multiple clonal infections are common in these malaria-hypoendemic regions of Southeast Asia.
Keywords: Plasmodium vivax, malaria, merozoite surface protein, diversity, mixed strain infection, Asia
1. Introduction
Plasmodium vivax is the most common and globally widespread of four human malaria species. It accounted for at least 70–80 million clinical cases annually (Mendis et al., 2001; Hay et al., 2004; Prajapati et al., 2006). It is the most important species causing human malaria outside of Africa, and is especially prevalent in Southeast Asia and Central and South America (WHO, 2006). In 2007, P. vivax malaria accounted for > 95% and ~25% of all malaria cases reported in China and Myanmar, respectively (Zhou et al., 2008; Moon et al., 2009). Although malaria incidence is low in China, the areas and population at risk are large. In contrast, malaria is highly endemic in some border states of Myanmar, and malaria-related morbidity and mortality are the highest among the countries of the Mekong region (Moon et al., 2009). The emergence of chloroquine resistance in P. vivax, especially in Southeast Asia, raises considerable concern about future control of this parasite (Baird et al., 2004; Guthmann et al., 2008; Lee et al., 2009). As many countries in Southeast Asia including China are entering the phase of malaria elimination (http://apmen.org/), they are facing the challenge of eliminating vivax malaria.
Genetic polymorphism underlies the phenotypic plasticity of the malaria parasites. The P. vivax parasites have exhibited a high degree of phenotypic plasticity, which ranges from relapsing pattern to the ability to invade Duffy-negative erythrocytes (Menard et al., 2010). Understanding the population genetic structure of the malaria parasites may provide us important information about how certain traits such as drug resistance emerge and spread (Cui et al., 2003). Recent studies using mitochondrial genome sequences of world P. vivax populations support the possible origin of this species in Asia, possibly as a result of host switch from macaque monkeys (Jongwutiwes et al., 2005, Mu et al., 2005). The extant P. vivax populations appear to be very diverse (Feng et al., 2003), and significant population differentiation exists among different populations. Studies using neutral nuclear markers such as microsatellites also revealed contrasting genetic structure in P. vivax populations, especially those from different continents (Imwong et al., 2007; Gunawardena et al., 2010). However, little is known about the population genetics of the P. vivax isolates circulating in temperate regions of China and Myanmar.
High levels of genetic diversity in P. vivax and multiple clone infections have been demonstrated in Papua New Guinea, Thailand, India, Colombia, Brazil, Laos, Myanmar, and Sri Lanka despite many of these countries are hypoendemic (Gomez et al., 2003; Imwong et al., 2006, 2007; Kim et al., 2006; Karunaweera et al., 2007; Moon et al., 2009; Gunawardena et al., 2010).
The completion of the P. vivax genome project provides an in-depth overview of the parasite’s genetic blueprint and evolutionary history (Carlton et al., 2008). A number of genes encoding for P. vivax MSPs have been identified and used for genotyping P. vivax field isolates from several different malaria-endemic areas, including PvMSP-1, PvMSP-3α, PvMSP-3β and PvMSP-3γ, PvMSP-4 and PvMSP-5 and PvMSP-9 (Del Portillo et al., 1991; Black et al., 2002; Putaporntip et al., 1997; Galinski et al., 1999, 2001; Mueller et al., 2002; Vargas-Serrato et al., 2002). Pvmsp-3α, Pvmsp-3β and Pvmsp-3γ are members of a multi-gene family of related MSPs (Del Portillo et al., 1991; Galinski et al., 2001). These proteins are comprised of three characteristic domains: an alanine-rich central domain predicted to form a coiled-coil tertiary peptide structure, and highly conserved C- and N-terminal regions (Galinski et al., 1999). This family has similarities with the MSP3 orthologs in multi-allelic homologs (Galinski et al. 2001; Carlton et al., 2008; Singh et al., 2009), and share the same similar structural organization in their C-terminal regions, as described in different malaria parasite species such as P. falciparum and P. knowlesi, and now for P. vivax (Rayner et al., 2002, 2004). The antigenic nature of this protein family has made them candidates for malaria vaccines (Sharma et al., 2008, Singh et al., 2009). This gene family has been under balancing selection, and exhibits high degrees of genetic diversity (Bruce et al., 1999; Cui et al., 2003; Yang et al., 2006; Zakeri et al., 2006). In addition to nucleotide polymorphisms, the central domain has large size variation and can be easily analyzed by using a polymerase chain reaction–restriction fragment length polymorphism (PCR-RFLP) method (Cristiano et al., 2008). Large-scale genetic diversity analysis of P. vivax malaria and the genetic studies of re-emerging P. vivax malaria have been performed with several MSP polymorphic markers, including the Pv-MSP1, PvMSP-3a and PvMSP-3β, using the PCR- RFLP genotyping method (Kirchgatter & del Portillo 1998; Cui et al., 2003; Leclerc et al., 2004; Kim et al., 2006; Yang et al., 2006; Veron et al., 2009; Prajapati et al., 2010; Zeyrek et al., 2010; Rungsihirunra et al., 2011). In this study, we evaluate the extent of diversity in field isolates of four P. vivax populations from China and Myanmar by PCR-RFLP of PvMSP-3a and PvMSP-3β genes. Analysis of allelic variation at multiple independent loci provides the most effective means to determine population structure.
2. Materials and methods
2.1. Sample collection
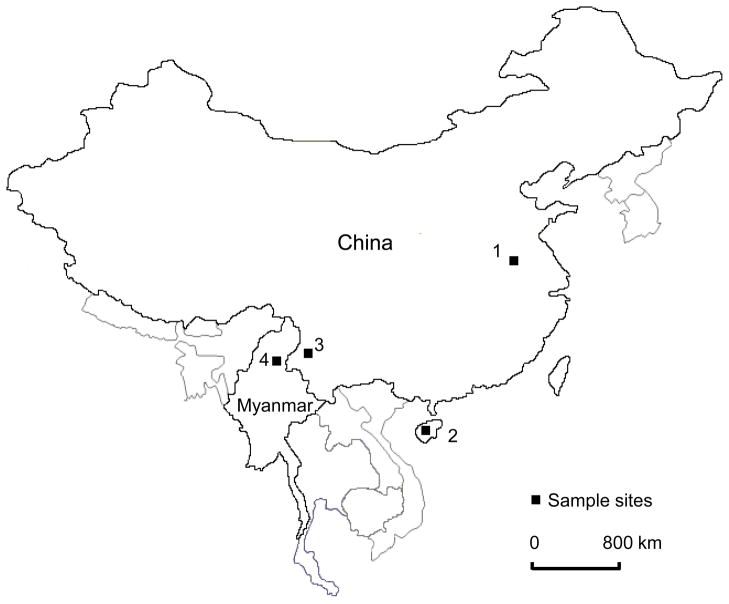
A total of 212 finger-prick blood samples were collected between 2006 and 2008 from symptomatic patients attending hospitals or malaria clinics in four localities of China and Myanmar. Specifically, 72 samples were collected in Kachin State (Myanmar); 46 in Sanya city (Hainan Province, China); 22 in the Autonomous Region of Dehong (Yunnan Province, China) and 72 in Suzhou City (Anhui Province, China) (Fig. 1). Twenty-three archival Giemsa-stained dried blood samples collected in Suzhou city in 2006 were also included in this study. These sampling sites represent regional vivax malaria transmission hot spots: Kachin State was among the most malaria endemic states in Myanmar (WHO, 2007), whereas Anhui, Hainan and Yunnan are three Chinese provinces where the majority of P. vivax malaria cases in 2006 were registered (Zhou et al., 2008).
Fig. 1.
Sample sites: 1, Suzhou city (Anhui Province, China); 2, Sanya city (Hainan Province, China); 3, Dehong (Yunnan Province, China); and 4, Kachin State (Myanmar).
2.2. Parasite DNA extraction
Parasite DNA from dried blood dots stored on filter paper (~100 ul of blood) was extracted using the Saponin/Chelex method (Wooden et al., 1993). Parasite DNA from archival Giemsa-stained blood samples was extracted following the method of Vince et al. (1998). Before extraction, smears were scraped off the glass slides in a Fisher Hamilton Fume Hood with a sterile scalper blade, and the resulting powdered material was transferred to 1.5 ml Eppendorf tubes. The identification of P. vivax was conducted following the published method (Sineo et al., 1998).
2.3. PCR-RFLP genotyping of P. vivax isolates at the PvMSP-3α and PvMSP-3β loci
Allelic diversity of the PvMSP-3a and PvMSP-3β genes was assessed using established PCR-RFLP techniques (Bruce et al., 1999; Yang et al., 2006). Briefly, PvMSP-3a was amplified by nested PCR using primers P1—5′ CAGCAGACACCATTTAAGG 3′ and P2—5′ CCGTTTGTTGATTAGTTGC 3′ for the primary PCR and primers N1—5′ GACCAGTGTGATACCATTAACC 3′ and N2—5′ ATACTGGTTCTTCGTCTTCAGG 3′ for the nest PCR. Primary PCR of PvMSP-3β was performed with primers F1 (5′GTATTCTTCGCAACACTC 3′) and R1 (5′-CTTCTGATGTTATTTCCAG- 3′), while nested reactions were done with primers F2 (5′-CGAGGGGCGAAATTGTAAACC- 3′) and R2 (5′-GCTGCTTCTTTTGCAAAGG- 3′). PCR was performed for 35 cycles using the following conditions: 94 ºC for 20 s, 54 ºC for 30 s, and 68 ºC for 2.5 min. For RFLP analysis, the PCR product of PvMSP-3a and PvMSP-3β were digested with Hha I and Pst I, respectively, and DNA was separated in a 0.8% agarose gel. Multiple strain infections were identified when the summed size of the DNA fragments resulting from the restriction digestion exceeded the size of the uncut PCR product (Yang et al 2006). Alleles were identified based on unique restriction banding patterns. Alleles were considered the same if the restriction banding patterns were measured to be within 20 base-pairs in molecular weight (Cattamanchi et al 2003).
2.4. Sequence analyses of PvMSP-3β
To confirm the RFLP-pattern of 27 samples (19 samples with unique PCR-RFLP patterns, 5 samples of mixed strain infection, and 3 samples with same PCR-RFLP pattern but slight difference in molecular size of the RFLP fragments), we amplified their PvMSP-3β fragment corresponding to nucleotides 1733 – 3176 of PvMSP-3β gene of the Belem strain (AF099662) (Yang et al., 2006). Specifically, first PCR reaction was carried out using primers BF1 (5′ GTATTCTTCGCAACACTC) and BR1 (5′CTTCTGATGTTATTTCCAG), whereas nested PCR was carried out with primers BF2 (5′CGAGGGGCGAAATTGTAAACC) and BR2 (5′GCTGCTTCTTTTGCAAAGG). PCR conditions included an initial denaturing at 95ºC for 3 min, followed by 35 (first PCR) or 29 (nested PCR) cycles of 94ºC for 30 sec, 55ºC for 30 sec, 68ºC for 2.5 min, and a final extension step at 68ºC for 10 min. Products of the nested PCR reaction were cloned in the pCR4-TOPO vector (Invitrogen, Carlsbad, CA, USA) following the supplier’s instructions. Plasmid DNA was extracted using the GeneElute Plasmid Miniprep Kit (Sigma) and sequenced by GENEWIZ San Diego Laboratory (La Jolla, CA, USA). Two to five clones were sequenced in their entirety in both directions for each of the 27 samples. If sequence differences were noted between two clones, the clones were re-sequenced to confirm the sequence identity. To eliminate the possibility that the polymorphisms are resulted from polymerase error, we used a procedure of assessing each single nucleotide polymorphism (SNP) and indel by their recurrence in more than one isolate or by sequencing an additional clone from the original sample (Rayner et al., 2004).
2.5. Statistical analysis
The chi-square (χ2) test was used to compare the frequencies of individual RFLP alleles among the populations using the Minitab Software 15.0 (MINITAB Inc., PA). The test was done for all alleles in the four parasite populations both simultaneously and in pairs. The level of significance was set at P < 0.05. Sequences were deposited in GenBank™ with accession numbers HQ847379 – 408 and JF900763-64. Sequences were assembled and aligned using BioEdit software, version 7.0.9.0 (Isis Pharmaceuticals, Carlsbad, CA). Phylogenetic analyses were conducted using PHYLIP 4.0 (Felsenstein, 1993). Unrooted trees were generated using the neighbor-joining method with the Jukes-Cantor distance option. Support for node was assessed by 1000 bootstrap resampling of the original dataset. The portions of the PvMSP-3β gene sequence corresponding to the N- and C-terminal domains were determined as in Rayner et al. (2004) on the basis of the SalI strain sequence (AY454095). The PvMSP-3β gene sequence of the Belem strain (AF099662) and other available sequences (Rayner et al., 2004; accession numbers AY454080-98) were also included in the analysis.
3. Results
3.1. Genetic diversity of the PvMSP-3α gene
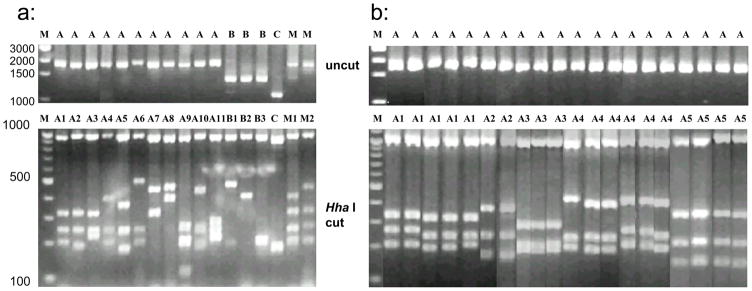
PvMSP-3α gene was successfully amplified in 226 of 235 P. vivax samples tested (96.2%). Based on the size of the PCR products, three different allele-types were detected and classified as type A (~1.9 kb), type B (~1.5 kb) and type C (~1.1 kb) (Cui et al., 2003). Most of the samples (>70%) were type A with frequencies in the four regions ranging from 92% in Anhui to 70.9% in Yunnan (Table 1), whereas the least frequent allele type was type C in Anhui (3%) and type B in the other three populations. The PCR-RFLP analysis revealed a high level of polymorphism in PvMSP-3α gene. Hha I digestion identified a total of 15 patterns (Fig. 2a). All samples exhibited a ~1.0 kb size band in Hha I digestion. Because of fluctuation in allele size determination from agarose gels, a bin of 20 bp in the size of restriction digestion fragments of the PCR products was allowed in allele calling (Fig. 2b), and this is consistent with the previous published allele designation scheme (Cui et al. 2003). Allele variants A1-A9, B1, B2 and C were reported earlier (Cui et al. 2003), whereas allele variants A10, A11 and B3 were new alleles identified in this study. Samples from different geographical areas differed dramatically in their PvMSP-3α allele composition and frequency. The highest number of alleles was found in Myanmar (14), while the lowest was found in Yunnan (7). In Anhui, the most common allele variant was A9 (29.3%). In Hainan there was a more homogeneous distribution of variants among the samples, with variant A8 accounting for 16.7% of the samples, and variants A1, A3 and A7 accounting each for 14.6% of the samples. In Hainan, 70.8% of the samples showed the allele variants A1, A3, A6, A7 and A8. In Yunnan, the alleles A1, A4 and C were found in 75% of the samples. In Myanmar, the most prevalent alleles were A1, A4, A6 and C. The χ2 test detected significant differences in allele frequency among the four populations (type A: χ2 = 98.30, df = 33, P < 0.0001; type B: χ2 = 14.23, df = 6, P < 0.05; type C: χ2 = 14.62, df = 3, P < 0.01). The overall differences in allele frequency among the four regions are highly significant (χ2 = 124.22, df = 45, P < 0.0001).
Table 1.
PvMSP-3α PCR-RFLP allele types and frequencies in four Asian Plasmodium vivax populations.
| Allele (PCR-RFLP) | Anhui (n = 92) | Hainan (n = 44) | Yunnan (n = 21) | Myanmar (n = 69) |
|---|---|---|---|---|
| A1 | 12.1 | 14.6 | 29.2 | 19.7 |
| A2 | 3.0 | 2.1 | 4.2 | 3.9 |
| A3 | 9.1 | 14.6 | 8.3 | 2.6 |
| A4 | 17.2 | 6.3 | 25.0 | 13.2 |
| A5 | 0 | 4.2 | 0 | 2.6 |
| A6 | 9.1 | 10.4 | 0 | 14.5 |
| A7 | 0 | 14.6 | 4.2 | 7.9 |
| A8 | 1.0 | 16.7 | 0 | 6.6 |
| A9 | 29.3 | 0 | 0 | 1.3 |
| A10 | 5.1 | 4.2 | 0 | 0 |
| A11 | 6.1 | 0 | 0 | 2.6 |
| B1 | 0 | 0 | 0 | 1.3 |
| B2 | 5.1 | 4.2 | 8.3 | 2.6 |
| B3 | 0 | 0 | 0 | 5.3 |
| C | 3.0 | 8.3 | 20.8 | 15.8 |
| Multi-infection | 7.6 | 9.1 | 14.3 | 10.2 |
| Total number of alleles | 11 | 11 | 7 | 14 |
Fig. 2.
PCR-RFLP analysis of PvMSP-3α gene in Plasmodium vivax samples obtained from four Asian populations. a: Examples of major alleles and mixed genotypes; and b: reperesentive samples showing various restriction digestion patterns and classification of alleles. Upper panel, undigested PCR products, and Lower panel, PCR products digested with Hha I. Lane marker: M – DNA size marker; A, B, C – allele types; M1 and M2 - mixed strain infections; A1, A2, …A10, B1, B2… - alleles identified by PCR-RFLP.
Size polymorphism associated with the PvMSP-3α products allowed easy detection of mixed genotype infections. Mixed infections were assigned when a single sample resulted in more than one PCR products of different sizes (Fig. 2, lanes M1 uncut) or when the summed size of the restriction fragments of a single PCR band exceeded the size of the uncut PCR band (Fig. 2, lanes M1 and M2). PCR alone detected mixed infections of PvMSP-3α between 2.6%–4.5% of samples among the four populations analyzed. RFLP analysis detected more mixed infections, ranging from 7.6% to 14.3% among the four populations (Table 1). The highest percentage of mixed infection was detected in Yunnan (14.3%), followed by Myanmar (10.2%), Hainan (9.1%) and Anhui (7.6%). There was no significant difference among the four populations in the percentage of mixed infection (χ2 = 0.99, df = 3, P = 0.804).
3.2. Genetic diversity of the PvMSP-3β gene
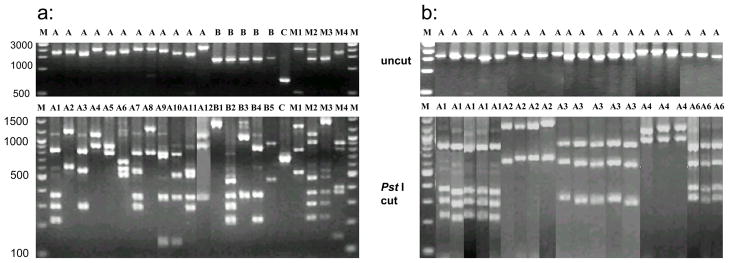
The PvMSP-3β gene was successfully amplified in 216 of 235 malaria samples (91.9%). The PvMSP-3β PCR products showed clear size polymorphism with four allele sizes of 1.7–2.4, 1.4–1.5, and 0.7–0.8, and 0.5–0.6 kb, which were categorized as types A, B, C, and D, respectively, following an earlier scheme (Yang et al., 2006). Type A and B were the most predominant types, accounting for >91% of P. vivax isolates in this study (Table 2). Type D allele was identified in a mixed infection with type A (lane M1 in Fig. 3). Restriction analysis of the PCR products by Pst I revealed 12 variants of for Type A allele and five variants for Type B allele, whereas Type C allele showed one allele variant (Fig. 3). The PCR-RFLP categorization of alleles was confirmed by DNA sequencing of selected fragments (Table 3). Fig. 3b showed representative examples of fragments differing within 20 bp and classified in the same allele class.
Table 2.
PvMSP-3β PCR-RFLP allele types and their distribution frequencies in four Asian Plasmodium vivax populations
| Allele (PCR-RFLP) | Anhui (n = 87) | Hainan (n = 42) | Yunnan (n = 20) | Myanmar (n = 67) |
|---|---|---|---|---|
| A1 | 6.9 | 10.4 | 4.3 | 6.4 |
| A2 | 15.7 | 14.6 | 8.7 | 15.4 |
| A3 | 11.8 | 4.2 | 21.7 | 14.1 |
| A4 | 4.9 | 2.1 | 4.3 | 9.0 |
| A5 | 11.8 | 27.1 | 13.1 | 7.7 |
| A6 | 0 | 0 | 4.3 | 5.1 |
| A7 | 0 | 2.1 | 4.3 | 1.3 |
| A8 | 0 | 0 | 0 | 2.6 |
| A9 | 3.9 | 0 | 4.3 | 0 |
| A10 | 2.9 | 2.1 | 0 | 0 |
| A11 | 0 | 2.1 | 0 | 0 |
| A12 | 0 | 0 | 0 | 1.3 |
| B1 | 33.3 | 20.8 | 21.7 | 12.8 |
| B2 | 6.9 | 10.4 | 8.7 | 9.0 |
| B3 | 0 | 0 | 0 | 1.3 |
| B4 | 0 | 0 | 0 | 3.8 |
| B5 | 0 | 0 | 0 | 1.3 |
| C | 0 | 2.1 | 0 | 7.7 |
| D | 2.0 | 2.1 | 4.3 | 1.3 |
| Multi-infection | 14.7 | 14.3 | 15.0 | 16.4 |
| Total number of alleles | 10 | 12 | 11 | 16 |
Fig. 3.
PCR-RFLP analysis of PvMSP-3β gene in Plasmodium vivax samples obtained from four Asian populations. a: Examples of major alleles and mixed genotypes; and b: reperesentive samples showing various restriction digestion patterns and classification of alleles. Upper panel, undigested PCR products, and Lower panel, PCR products digested with Pst I. Lane marker: M – DNA size marker; A, B, C – allele types; M1…M4 - mixed strain infections; A1, A2, …A12, B1… B5 - alleles identified by PCR-RFLP.
Table 3.
Polymorphism of PvMSP-3β gene detected by PCR and PCR-RFLP assay in Plasmodium vivax population from Southeast Asia.
| Allele | PCR fragment (bp) | PCR-RFLP fragment (bp) | Genbank acc. no | ||||
|---|---|---|---|---|---|---|---|
| A1 | 1731 | 842 | 371 | 294 | 224 | HQ847404 | |
| A2 | 1830 | 1213 | 617 | HQ847398 | |||
| A3 | 1714 | 822 | 596 | 296 | HQ847406 | ||
| A4 | 2034 | 1120 | 914 | HQ847399 | |||
| A5 | 1715 | 912 | 803 | HQ847379 | |||
| A6 | 1812 | 677 | 599 | 536 | HQ847380 | ||
| A7 | 2035 | 799 | 576 | 360 | 300 | HQ847405 | |
| A8 | 2091 | 1291 | 800 | HQ847383 | |||
| A9 | 2068 | 803 | 761 | 360 | 144 | HQ847385 | |
| A10 | 1821 | 782 | 536 | 359 | 144 | HQ847408 | |
| A11 | 1770 | 577 | 536 | 360 | 297 | HQ847400 | |
| A12 | 2365 | 1066 | 883 | 360 | 56 | HQ847403 | |
| B1 | 1461 | 1400 | 61 | HQ847391 | |||
| B2 | 1428 | 480 | 371 | 296 | 225 | 56 | HQ847388 |
| B3 | 1402 | 1031 | 371 | JF900764 | |||
| B4 | 1449 | 854 | 370 | 225 | HQ847389 | ||
| B5 | 1444 | 908 | 480 | 56 | HQ847396 | ||
| C | 710 | 710 | 56 | HQ847402 | |||
| D | 537 | HQ847381 | |||||
Samples from different geographical areas also differed dramatically in their PvMSP-3β allele composition and frequency. The highest number of alleles was found in Myanmar (16), the lowest in Anhui (10). The most frequent allele variant was B1 (33.3%) in Anhui, A5 (27.1%) in Hainan, A3 and B1 (both 21.7%) in Yunnan and A2 (15.4%) in Myanmar. Five allele variants (A8, A12, B3, B4, and B5) were detected only in Myanmar. The allele variant A11 was detected only in Hainan. Chi square test detected significant difference in allele frequency of major alleles among the four populations (type A: χ2 = 46.04, df = 33, P = 0.065; type B: χ2 = 21.25, df = 12, P < 0.05; type C: χ2 = 9.87, df = 3, P < 0.05). The overall differences in allele frequency among the four regions were significant (χ2 = 78.45, df = 57, P < 0.05).
PCR of PvMSP-3β detected mixed infections in 3.9–6.2% of the four populations. RFLP analysis increased the percentage of detected mixed infection (Fig. 3, M1–M4). The highest mixed infection rate was detected in Myanmar (16.4%), followed by Yunnan (15.0%), Anhui (14.7%) and Hainan (14.3%), but this was not statistically significant (χ2 = 0.21, df = 3, P = 0.976) (Table 2).
3.3. Analysis of PvMSP-3β gene sequences
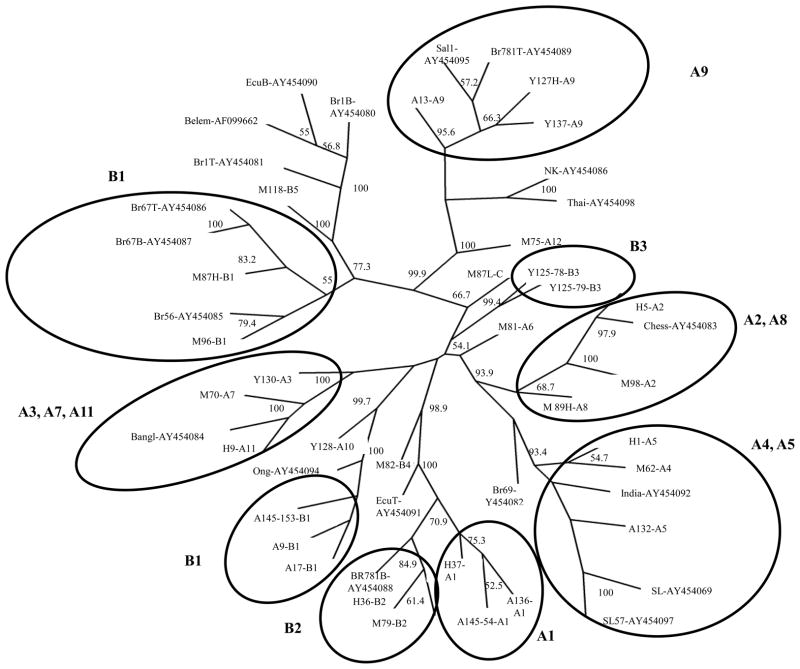
Thirty-two PvMSP-3β sequences were obtained from 27 samples due to mixed strain infections in five samples (i.e. Yunnan 127, clones L and H, Yunnan 125, clones 78 and 79, Myanmar 87, clones L and H, Myanmar 89, clones L and H, Anhui 145, clones 54 and 153) (Fig. 4). Eighteen of these sequences were classified as variants of allele type A, eleven as variants of allele type B, one as allele C, and two of allele type D. Sequencing of the PvMSP-3β gene confirmed the allele type assignments and Pst I restriction patterns (Fig. 3, Table 3). The sequenced PvMSP-3β fragments encompassed the conserved N-terminal part of the protein, a region with high sequence polymorphism previously called “Insert A” (Rayner et al., 2004), the central Ala-rich region expected to harbor deletions and insertions called “Insert B”, and the first 503 bp of the 933 bp the C-terminus (Rayner et al., 2004, Yang et al., 2006). Our sequence data conformed to the expectations of limited polymorphisms in the N- and C-terminal regions and deletions and insertions in the region coding for the Ala-rich domain of the protein (Supplemental file 1, Supplemental file 2). Phylogenetic analyses were performed on the N-terminal and C-terminal domains, separately. From the unrooted neighbor-joining tree based on the N-terminal domain (Fig. 4), 9 major branches were identified, comprising either one RFLP-PCR variant (groups B1, B2, B3, A1, A9) or more than one variant of the same class (i.e. groups: A4/A5, A2/A8, A3/A7/A11). PCR-RFLP variants A6, A10, A12, B4, B5 and C did not group with other variants. Interestingly, samples of variant B1 tended to form two different clusters, one grouping all samples from Anhui, and one comprising samples from Myanmar. Less than half the nodes of the trees based on the sequence of the C-terminal domain had bootstrap values higher than 50%, suggesting that the portion of the C-terminal domain sequenced was not sufficiently polymorphic to provide a meaningful phylogeny (Supplemental file 2). Overall, the four populations were intermingled and not always clustered on the basis of their geographical origins.
Fig. 4.
PvMSP-3β phylogenies based on the N- terminal domain of the PvMSP-3β fragment sequenced from 27 samples from Myanmar (M), Yunnan (Y), Anhui (A) and Hainan (H). The PCR-RFLP allele variant is shown after the sample name. Multiple clone sequences were included only for multiple infections, i.e. sample Y127 (L, H), Y 125 (78, 79), M87 (L, H), M89 (L, H), A145 (153, 54). For the sequences downloaded from NCBI, the GenBankTM accession numbers are showing after the sample name.
4. Discussion
The aim of this study was to assess the level of genetic polymorphism of the PvMSP-3α and PvMSP-3β in P. vivax infections from China and Myanmar. Among the four localities studied, PCR-RFLP analysis identified 15 genotypes of PvMSP-3α among 226 samples, of which 21 were multiclonal. At least eight of the PvMSP-3α alleles detected were common to those from previous studies (Bruce et al., 1999; Cui et al., 2003), suggesting that some parasite genotypes may have a worldwide distribution. Regarding PvMSP-3β, 19 RFLP allele variants were detected from 216 samples analyzed, of which 35 (16.2%) were mixed-strain infections.
It is important to note that the mixed-strain infections were defined by instances when the summed size of the DNA fragments in RFLP analysis was greater than the size of the uncut PCR product, or when more than one size PCR product was seen in the initial amplification. The method for detecting mixed infection likely underestimates the true rate of mixed infections because the summing of RFLP fragments will only identify variants where there is a SNP in the recognition site of the restriction endonuclease being used, and comparison of amplicon size will only identify variants with major indels. Amplicon size will have least power to identify mixed infections, and while RFLP has more power, it is still limited as evidenced by the results that PCR-RFLP analysis of PvMSP-3α and PvMSP-3β identified more mixed infections than PCR amplicon methods. For example, PCR-RFLP detected mixed infections in 7.6–14.3% of infections using the PvMSP-3α gene and 14.7–16.4% infections when measured with the PvMSP-3β gene in the four parasite populations. Combining both PvMSP-3α and PvMSP-3β genes, the PCR-RFLP analysis detected additional mixed genotype infections in these populations, giving a final mixed infection rate of 20.5–31.8%. Sequence analysis of representative allele variants detected 2 out of the 27 (7.4%) samples that showed same PCR product sizes but different nucleotide sequences. Therefore, the PCR-RFLP on single gene yielded conservative estimation of mixed infection rate in comparison to DNA sequencing. The combination of PCR-RFLP on two genes provides higher power to differentiate intra-species genetic variations.
Compared with P. vivax isolates from previous studies conducted in Asia, our result further confirmed the existence of small geographic differentiation among P. vivax populations. An earlier study in western Thailand reported that the B type of PvMSP-3β is more abundant (60.4%) than other types, whereas in the Chinese Bengbu and Guangxi samples, both A and B types were similarly prevalent (Yang et al., 2006). In the present study, the type A allele was the most abundant in all four parasite populations (>57% more abundant than other types). In addition, RFLP analysis also revealed different allele compositions at each sampling site (Yang et al., 2006). In the present study with four Asian populations, a total of 19 alleles (12 A, 5 B, 1 C, and 1 D) was identified. Myanmar had the highest number of alleles (16), whereas the other three populations had 10–12 alleles each. With regard to allele frequency, A5, B1, B2, and B4 were the most abundant alleles in the Thai samples, which accounted for 66% of all isolates. In the Chinese Bengbu P. vivax samples, the frequency of four alleles (A2, A5, B1, and B2) reached 80.8%. In the present study, alleles A2, A3, A5 and B1 were the most abundant alleles, accounting for 50.0–72.5% of all isolates. Interestingly, despite the difference in individual allele types, the four P. vivax populations shared seven alleles (A1–A5, B1, and B2) (Table 2). These results indicated that the PCR-RFLP method for PvMSP-3β could be used to detect genetic structure.
Although the PCR-RFLP technique is frequently used for Plasmodium genotyping, it has limitations. First, detection threshold depends on the sensitivity of PCR amplification reaction. Second, resolution for detecting allelic variants is contingent on product size or the size of restriction digestion fragments (Snounou and Beck, 1998). These two major limitations can be especially problematic with low-level parasitemia or when one strain is a minor component of a multistrain infection (Farnert et al., 2001). The heteroduplex tracking assay (HTA) is more sensitive than PCR and is capable of resolving multiple nucleotide sequences in a single sample, but it requires the generation of DNA molecular probes and PCR product clone techniques (Ngrenngarmlert et al., 2005). Finally, direct DNA sequencing can be used to detect sequence and size discrepant variants, but it is costly and unless complemented with cloning, it is ineffective at identifying the number of sequence variants when multiple sequences are present (Snounou and Beck, 1998). The determination of complete sequence of P. vivax and syntenic similarity with other Plasmodium species will accelerate the search for useful genetic markers (Carlton, 2003). Genome-wide analysis technologies such as the next-generation sequencing technique and gene-specific microarray systems may show promising to obtain a more detailed picture of the demographic history and evolution of P. vivax.
Supplementary Material
Sequence alignment of the N-terminal portion of PvMSP-3β used for phylogenetic analyses.
Sequence alignment of the C-terminal portion of PvMSP-3β used for phylogenetic analyses.
Highlights.
We examine genetic diversity of Plasmodium vivax malaria in China and Myanmar
Acknowledgments
This work is supported by grants from the National Institute of Health (R01 AI050243 and U19 AI089672). We thank two anonymous reviewers for their valuable comments.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Daibin Zhong, Email: dzhong@uci.edu.
Mariangela Bonizzoni, Email: mbonizzo@uci.edu.
Guofa Zhou, Email: gzhou@uci.edu.
Guangze Wang, Email: wangguangze63@126.com.
Bin Chen, Email: c_bin@hotmail.com.
Anne Vardo-Zalik, Email: amv12@psu.edu.
Liwang Cui, Email: luc2@psu.edu.
Guiyun Yan, Email: guiyuny@uci.edu.
Bin Zheng, Email: chuner1997@yahoo.com.
References
- Baird JK, Hoffman SL. Primaquine therapy for malaria. Clin Infect Dis. 2004;39:1336–1345. doi: 10.1086/424663. [DOI] [PubMed] [Google Scholar]
- Black CG, Barnwell JW, Huber CS, Galinski MR, Coppel RL. The Plasmodium vivax homologues of merozoite surface proteins 4 and 5 from Plasmodium falciparum are expressed at different locations in the merozoite. Mol Biochem Parasitol. 2002;120:215–224. doi: 10.1016/s0166-6851(01)00458-3. [DOI] [PubMed] [Google Scholar]
- Bruce MC, Galinski MR, Barnwell JW, Snounou G, Day KP. Polymorphism at the merozoite surface protein-3α locus of Plasmodium vivax: global and local diversity. Am J Trop Med Hyg. 1999;61:518–525. doi: 10.4269/ajtmh.1999.61.518. [DOI] [PubMed] [Google Scholar]
- Carlton J. The Plasmodium vivax genome sequencing project. Trends in Parasitol. 2003;19:227–231. doi: 10.1016/s1471-4922(03)00066-7. [DOI] [PubMed] [Google Scholar]
- Carlton JM, Adams JH, Silva JC, Bidwell SL, Lorenzi H, Caler E, Crabtree J, Angiuoli SV, Merino EF, Amedeo P, Cheng Q, Coulson RMR, Crabb BS, del Portillo HA, Essien K, Feldblyum TV, Fernandez-Becerra C, Gilson PR, Gueye AH, Guo X, Kanga S, Kooij TWA, Korsinczky M, Meyer EVS, Nene V, Paulsen I, White O, Ralph SA, Ren Q, Sargeant TJ, Salzberg SL, Stoeckert CJ, Sullivan SA, Yamamoto MM, Hoffman SL, Wortman JR, Gardner MJ, Galinski MR, Barnwell JW, Fraser-Liggett CM. Comparative genomics of the neglected human malaria parasite Plasmodium vivax. Nature. 2008;455:757–763. doi: 10.1038/nature07327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattamanchi A, Kyabayinze D, Hubbard A, Rosenthal PJ, Dorsey G. Distinguishing recrudescence from reinfection in a longitudinal antimalarial drug efficacy study: comparison of results based on genotyping of msp-1, msp-2, and glurp. Am J Trop Med Hyg. 2003;68:133–139. [PubMed] [Google Scholar]
- Cristiano FA, Perez MA, Nicholls RS, Guerra AP. Polymorphism in the Plasmodium vivax msp 3a: gene in field samples from Tierralta, Colombia. Mem Inst Oswaldo Cruz. 2008;103:493–496. doi: 10.1590/s0074-02762008000500015. [DOI] [PubMed] [Google Scholar]
- Cui L, Mascorro CN, Rzomp KA, Fan Q, Khuntirat B, Zhou G, Chen H, Yan G, Sattabongkot J. Genetic diversity and multiple infections of Plasmodium vivax malaria in western Thailand. Am J Trop Med Hyg. 2003;68:613–619. doi: 10.4269/ajtmh.2003.68.613. [DOI] [PubMed] [Google Scholar]
- Del Portillo HA, Longacre S, Khouri E, David PH. Primary structure of the merozoite surface antigen 1 of Plasmodium vivax reveals sequences conserved between different Plasmodium species. Proc Natl Acad Sci USA. 1991;88:4030–4034. doi: 10.1073/pnas.88.9.4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnert A, Arez AP, Babiker HA, Beck HP, Benito A, Bjorkman A, Bruce MC, Conway DJ, Day KP, Henning L, Mercereau-Puijalon O, Ranford-Cartwright LC, Rubio JM, Snounou G, Walliker D, Zwetyenga J, do Rosario VE. Genotyping of Plasmodium falciparum infections by PCR: a comparative multicentre study. Trans Royal Soc Trop Med Hyg. 2001;95:225–232. doi: 10.1016/s0035-9203(01)90175-0. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Phylogeny Inference Package (PHYLIP). Version 3.5. University of Washington; Seattle: 1993. [Google Scholar]
- Feng X, Carlton JM, Joy DA, Mu J, Furuya T, Suh BB, Wang Y, Barnwell JW, Su XZ. Single-nucleotide polymorphisms and genome diversity in Plasmodium vivax. Proc Natl Acad Sci U S A. 2003;100:8502–8507. doi: 10.1073/pnas.1232502100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galinski MR, Corredor-Medina C, Povoa M, Crosby J, Ingravallo P, Barnwell JW. Plasmodium vivax merozoite surface protein-3 contains coiled-coil motifs in an alanine-rich central domain. Mol Biochem Parasitol. 1999;101:131–147. doi: 10.1016/s0166-6851(99)00063-8. [DOI] [PubMed] [Google Scholar]
- Galinski MR, Ingravallo P, Corredor-Medina C, Al-Khedery B, Povoa M, Barnwell JW. Plasmodium vivax merozoite surface proteins-3β and-3γ share structural similarities with P. vivax merozoite surface protein-3α and define a new gene family. Mol Biochem Parasitol. 2001;115:41–53. doi: 10.1016/s0166-6851(01)00267-5. [DOI] [PubMed] [Google Scholar]
- Gomez JC, McNamara DT, Bockarie MJ, Baird JK, Carlton JM, Zimmerman PA. Identification of a polymorphic Plasmodium vivax microsatellite marker. Am J Trop Med Hyg. 2003;69:377–379. [PMC free article] [PubMed] [Google Scholar]
- Gunawardena S, Karunaweera ND, Ferreira MU, Phone-Kyaw M, Pollack RJ, Alifrangis M, Rajakaruna RS, Konradsen F, Amerasinghe PH, Schousboe ML, Galappaththy GNL, Abeyasinghe RR, Hartl DL, Wirth DF. Geographic structure of Plasmodium vivax: microsatellite analysis of parasite populations from Sri Lanka, Myanmar, and Ethiopia. Am J Trop Med Hyg. 2010;82:235–242. doi: 10.4269/ajtmh.2010.09-0588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthmann JP, Pittet A, Lesage A, Imwong M, Lindegardh N, Min Lwin M, Zaw T, Annerberg A, De Radiguès X, Nosten F. Plasmodium vivax resistance to chloroquine in Dawei, southern Myanmar. Trop Med Int Health. 2008;13:91–98. doi: 10.1111/j.1365-3156.2007.01978.x. [DOI] [PubMed] [Google Scholar]
- Hay SI, Guerra CA, Tatem AJ, Noor AM, Snow RW. The global distribution and population at risk of malaria: past, present, and future. Lancet Infect Dis. 2004;4:327–336. doi: 10.1016/S1473-3099(04)01043-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imwong M, Nair S, Pukrittayakamee S, Sudimack D, Williams JT, Mayxay M, Newton PN, Kim JR, Nandy A, Osorio L, Carlton JM, White NJ, Day NP, Anderson TJ. Contrasting genetic structure in Plasmodium vivax populations from Asia and South America. Int J Parasitol. 2007;37:1013–1022. doi: 10.1016/j.ijpara.2007.02.010. [DOI] [PubMed] [Google Scholar]
- Imwong M, Sudimack D, Pukrittayakamee S, Osorio L, Carlton JM, Day NPJ, White NJ, Anderson TJC. Microsatellite variation, repeat array length, and population history of Plasmodium vivax. Mol Biol Evol. 2006;23:1016–1018. doi: 10.1093/molbev/msj116. [DOI] [PubMed] [Google Scholar]
- Jongwutiwes S, Putaporntip C, Iwasaki T, Ferreira MU, Kanbara H, Hughes AL. Mitochondrial genome sequences support ancient population expansion in Plasmodium vivax. Mol Biol Evol. 2005;22:1733–1739. doi: 10.1093/molbev/msi168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karunaweera ND, Ferreira MU, Hartl DL, Wirth DF. Fourteen polymorphic microsatellite DNA markers for the human malaria parasite Plasmodium vivax. Mol Ecol Notes. 2007;7:172–175. [Google Scholar]
- Kim JR, Imwong M, Nandy A, Chotivanich K, Nontprasert A, Tonomsing N, Maji A, Addy M, Day N, White N, Pukrittayakamee S. Genetic diversity of Plasmodium vivax in Kolkata, India. Malaria J. 2006;5:71. doi: 10.1186/1475-2875-5-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchgatter K, del Portillo HA. Molecular analysis of Plasmodium vivax relapses using the MSP1 molecule as a genetic marker. J Infect Dis. 1998;177:511–515. doi: 10.1086/517389. [DOI] [PubMed] [Google Scholar]
- Leclerc M, Menegon M, Cligny A, Noyer J, Mammadov S, Aliyev N, Gasimov E, Majori G, Severini C. Genetic diversity of Plasmodium vivax isolates from Azerbaijan. Malaria J. 2004;3:40. doi: 10.1186/1475-2875-3-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KS, Kim TH, Kim ES, Lim HS, Yeom JS, Jun G, Park JW. Chloroquine-resistant Plasmodium vivax in the Republic of Korea. Am J Trop Med Hyg. 2009;80:215–217. [PubMed] [Google Scholar]
- Menard D, Barnadas C, Bouchier C, Henry-Halldin C, Gray LR, Ratsimbasoa A, Thonier V, Carod JF, Domarle O, Colin Y, Bertrand O, Picot J, King CL, Grimberg BT, Mercereau-Puijalon O, Zimmerman PA. Plasmodium vivax clinical malaria is commonly observed in Duffy-negative Malagasy people. Proc Natl Acad Sci U S A. 2010;107:5967–5971. doi: 10.1073/pnas.0912496107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendis K, Sina BJ, Marchesini P, Carter R. The neglected burden of Plasmodium vivax malaria. Am J Trop Med Hyg. 2001;64:97–106. doi: 10.4269/ajtmh.2001.64.97. [DOI] [PubMed] [Google Scholar]
- Moon SU, Lee HW, Kim JY, Na BK, Cho SH, Lin K, Sohn WM, Kim TS. High frequency of genetic diversity of Plasmodium vivax field isolates in Myanmar. Acta Tropica. 2009;109:30–36. doi: 10.1016/j.actatropica.2008.09.006. [DOI] [PubMed] [Google Scholar]
- Mu J, Joy DA, Duan J, Huang Y, Carlton J, Walker J, Barnwell J, Beerli P, Charleston MA, Pybus OG, Su X-z. Host switch leads to emergence of Plasmodium vivax malaria in humans. Mol Biol Evol. 2005;22:1686–1693. doi: 10.1093/molbev/msi160. [DOI] [PubMed] [Google Scholar]
- Mueller I, Kaiok J, Reeder JC, Cortes A. The population structure of Plasmodium falciparum and Plasmodium vivax during an epidemic of malaria in the Eastern Highlands of Papua New Guinea. Am J Trop Med Hyg. 2002;67:459–464. doi: 10.4269/ajtmh.2002.67.459. [DOI] [PubMed] [Google Scholar]
- Ngrenngarmlert W, Kwiek JJ, Kamwendo DD, Ritola K, Swanstrom R, Wongsrichanalai C, Miller RS, Ittarat W, Meshnick SR. Measuring allelic heterogeneity in Plasmodium falciparum by a heteroduplex tracking assay. Am J Trop Med Hyg. 2005;72:694–701. [PubMed] [Google Scholar]
- Prajapati S, Verma A, Adak T, Yadav R, Kumar A, Eapen A, Das M, Singh N, Sharma S, Rizvi M, Dash A, Joshi H. Allelic dimorphism of Plasmodium vivax gam-1 in the Indian subcontinent. Malaria J. 2006;5:1–6. doi: 10.1186/1475-2875-5-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prajapati SK, Joshi H, Valecha N. Plasmodium vivax merozoite surface protein-3 alpha: a high-resolution marker for genetic diversity studies. J Vector Borne Dis. 2010;47:85–90. [PubMed] [Google Scholar]
- Putaporntip C, Jongwuitiwes S, Tanabe K, Thaithong S. Interallelic recombination in the merozoite surface protein1 (MSP1) gene of Plasmodium vivax from Thai isolates. Mol Biochem Parasitol. 1997;84:49–56. doi: 10.1016/s0166-6851(96)02786-7. [DOI] [PubMed] [Google Scholar]
- Rayner JC, Corredor V, Feldman D, Ingravallo P, Iderabdullah F, Galinski MR, Barnwell JW. Extensive polymorphism in the Plasmodium vivax merozoite surface coat protein MSP-3 alpha is limited to specific domains. Parasitology. 2002;125:393 – 405. doi: 10.1017/s0031182002002317. [DOI] [PubMed] [Google Scholar]
- Rayner JC, Huber CS, Feldman D, Ingravallo P, Galinski MR, Barnwell JW. Plasmodium vivax merozoite surface protein PvMSP-3 β is radically polymorphic through mutation and large insertions and deletions. Infect Genet Evol. 2004;4:309–319. doi: 10.1016/j.meegid.2004.03.003. [DOI] [PubMed] [Google Scholar]
- Rungsihirunrat K, Chaijaroenkul W, Siripoon N, Seugorn A, Na-Bangchang K. Genotyping of polymorphic marker (MSP3α and MSP3β) genes of Plasmodium vivax field isolates from malaria endemic of Thailand. Trop Med Int Health, no-no. 2011 doi: 10.1111/j.1365-3156.2011.02771.x. [DOI] [PubMed] [Google Scholar]
- Sharma A, Yogavel M, Akhouri RR, Gill J, Sharma A. Crystal structure of soluble domain of malaria sporozoite protein UIS3 in complex with lipid. J Biol Chem. 2008;283:24077–24088. doi: 10.1074/jbc.M801946200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sineo L, Failli M, Petrocelli P, Martini R. Applications of PCR in the molecular diagnosis of Plasmodium. Int J Anthropol. 1998;13:289–293. [Google Scholar]
- Singh S, Soe S, Weisman S, Barnwell JW, Perignon JL, Druilhe P. A Conserved Multi-gene family induces cross-reactive antibodies effective in defense against Plasmodium falciparum. PLoS ONE. 2009;4:e5410. doi: 10.1371/journal.pone.0005410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snounou G, Beck HP. The use of PCR genotyping in the assessment of recrudescence or reinfection after antimalarial drug treatment. Parasitol Today. 1998;14:462–467. doi: 10.1016/s0169-4758(98)01340-4. [DOI] [PubMed] [Google Scholar]
- Vargas-Serrato E, Barnwell JW, Ingravallo P, Perler FB, Galinski MR. Merozoite surface protein-9 of Plasmodium vivax and related simian malaria parasites is orthologous to p101/ABRA of P. falciparum. Mol Biochem Parasitol. 2002;120:41–52. doi: 10.1016/s0166-6851(01)00433-9. [DOI] [PubMed] [Google Scholar]
- Veron V, Legrand E, Yrinesi J, Volney B, Simon S, Carme B. Genetic diversity of msp3α and msp1_b5 markers of Plasmodium vivax in French Guiana. Malaria J. 2009;8:40. doi: 10.1186/1475-2875-8-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vince A, Poljak M, Seme K. DNA extraction from archival Giemsa-stained bone-marrow slides: comparison of six rapid methods. British J Haematol. 1998;101:349–351. doi: 10.1046/j.1365-2141.1998.00702.x. [DOI] [PubMed] [Google Scholar]
- WHO. Guidelines for the treatment of malaria. World Health Organization; Geneva: 2006. [Google Scholar]
- WHO. Malaria in the Greater Mekong Subregion: Regional and Country Profiles. World Health Organization; Geneva: 2007. [Google Scholar]
- Wooden J, Kyes S, Sibley CH. PCR and strain identification in Plasmodium falciparum. Parasitol Today. 1993;9:303–305. doi: 10.1016/0169-4758(93)90131-x. [DOI] [PubMed] [Google Scholar]
- Yang Z, Miao J, Huang Y, Li X, Putaporntip C, Jongwutiwes S, Gao Q, Udomsangpetch R, Sattabongkot J, Cui L. Genetic structures of geographically distinct Plasmodium vivax populations assessed by PCR/RFLP analysis of the merozoite surface protein 3β gene. Acta Tropica. 2006;100:205–212. doi: 10.1016/j.actatropica.2006.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakeri S, Abouie Mehrizi A, Djadid ND, Snounou G. Circumsporozoite protein gene diversity among temperate and tropical Plasmodium vivax isolates from Iran. Trop Med Int Health. 2006;11:729–737. doi: 10.1111/j.1365-3156.2006.01613.x. [DOI] [PubMed] [Google Scholar]
- Zeyrek FY, Tachibana SI, Yuksel F, Doni N, Palacpac N, Arisue N, Horii T, Coban C, Tanabe K. Limited polymorphism of the Plasmodium vivax merozoite surface protein 1 gene in isolates from Turkey. Am J Trop Med Hyg. 2010;83:1230–1237. doi: 10.4269/ajtmh.2010.10-0353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou SS, Wang Y, Fang W, Tang LH. Malaria situation in the People’s Republic of China in 2007. Zhongguo Ji Sheng Chong Xue Yu Ji Sheng Chong Bing Za Zhi. 2008;26:401–4133. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequence alignment of the N-terminal portion of PvMSP-3β used for phylogenetic analyses.
Sequence alignment of the C-terminal portion of PvMSP-3β used for phylogenetic analyses.