Abstract
In the United States, candidemia is one of the most common hospital-acquired infections and is estimated to cause 10,000 deaths per year. The species Candida albicans is responsible for the majority of these cases. As C. albicans is capable of developing resistance against the currently available drugs, understanding the molecular basis of drug resistance, finding new cellular targets, and further understanding the overall mechanism of C. albicans pathogenesis are important goals. To study this pathogen it is advantageous to manipulate its genome. Numerous strategies of C. albicans gene manipulation have been introduced. This review evaluates a majority of these strategies and should be a helpful guide for researchers to identify gene targeting strategies to suit their requirements.
Keywords: Pathogenic fungi, Candida albicans, gene manipulation, selectable marker, homologous recombination
1. Introduction
Candida species are serious human fungal pathogens among immune compromised individuals (Odds, 1994). Susceptible patients include those undergoing cancer chemotherapy or organ transplantation, patients infected with HIV, and pre-mature infants (Schelenz et al., 2011; Epstein et al., 2003; Muller et al., 1999). Candida blood stream infections (candidemia) are life threatening among hospitalized immune-compromised patients, including neonates (Fridkin et al., 2006). The incidence rate of hospital-acquired candidemia is as high as 8 in every 100, 000 people (Kao et al., 1999). Today, candidemia is one of the leading bloodstream infections in the United States with an annual cost of treatment approaching $1 billion (Miller et al., 2001). Although infections with non-albicans Candida species have emerged in recent years (Miceli et al., 2011), the species, C. albicans is still responsible for the majority of cases (Horn et al., 2009; Ruhnke, 2006).
Researchers have studied this pathogen for decades. As a result, there are several clinically available drugs to treat C. albicans infections (De Rosa et al., 2009; Shao et al., 2007; Lai et al., 2008). However, C. albicans has the ability to acquire resistance against many of these drugs (Cannon et al., 2007; Casalinuovo et al., 2004). Therefore, even today there is a need for additional drugs to inhibit this pathogen.
Molecular analysis of C. albicans has been particularly challenging for many reasons. C. albicans has no clearly defined sexual cycle (Noble and Johnson, 2007). Moreover genetic manipulation of C. albicans is not simple. First, the organism is diploid (Barnett, 2008; Jones et al., 2004) and in most cases both alleles of a gene must be manipulated. Second, compared to the highly studied yeast, Saccharomyces cerevisiae, C. albicans lacks natural plasmids, such as the 2-micron plasmid, for use in transformation and shows lower transformation and recombination frequencies making the manipulation of both alleles of a gene even more challenging (De Backer et al., 2000; Magee et al., 2003). Third, C. albicans shows non-conventional codon usage where the CUG codon is decoded as serine, not leucine (Santos and Tuite, 1995). Therefore, when using marker gene sequences from other organisms for gene manipulation in C. albicans, the codons need to be optimized. Fourth, the use of selectable markers has been particularly challenging because some markers have effects on virulence of the pathogen. In addition, C. albicans shows natural resistance to drugs such as G418, hygromycin B and cycloheximide, which are used for selection in S. cerevisiae (De Backer et al., 2000). This review highlights the milestones achieved for effective methods to manipulate the C. albicans genome.
2. Nutritional Markers for Selection
Early researchers introduced mutations in the C. albicans genome by exposure to UV or chemical mutagens. These methods left behind undesirable mutations that could not be traced or repaired (Fonzi and Irwin, 1993).
Fonzi and Irwin (1993) used a clinical isolate of C. albicans (SC5314) to make a strain (CAI4) that was auxotrophic for growth on uracil-deficient media. To do this, they disrupted both copies of the URA3 gene in C. albicans using the imm434 region of the λgt10 bacterial phage. This was the first time in the field of C. albicans research that a reliable parent strain was available to introduce mutations into the genome of C. albicans.
In C. albicans, the URA3 gene encodes the enzyme, orotidine 5'-monophosphate decarboxylase that catalyzes the conversion of orotidine 5'-monophosphate to uridine 5'-monophosphate in the de-novo pyrimidine biosynthesis pathway (Lay et al., 1998). The resulting CAI4 strain is unable to grow in the absence of uracil, hence URA3 gene could be used as a dominant nutritional selectable marker in C. albicans.
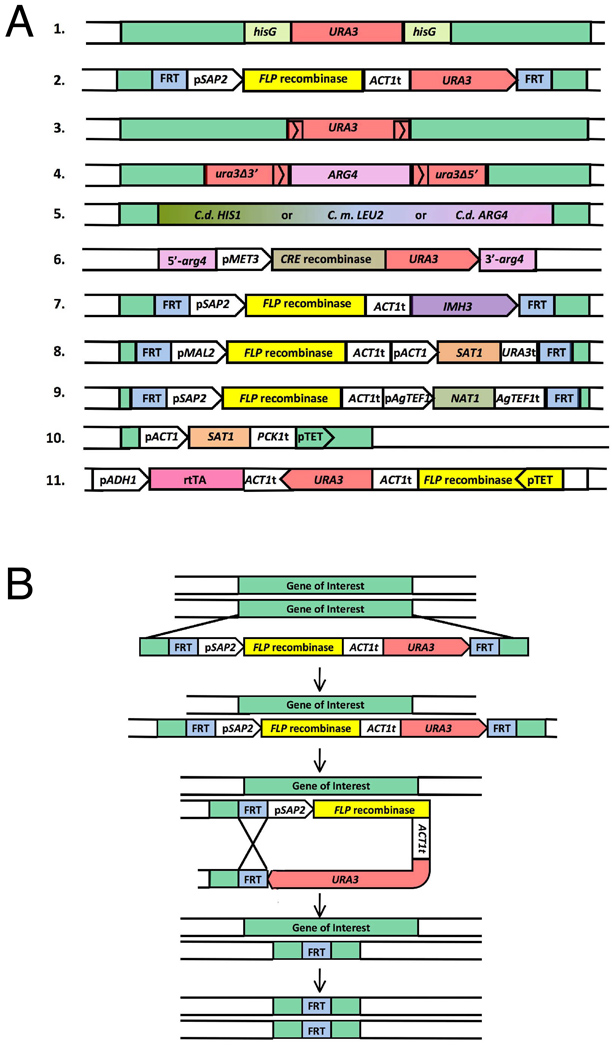
Because of the use of URA3 as a selectable marker to disrupt genes, the method is popularly known as the “URA Blaster method”. Here, the gene of interest is disrupted using a cassette that carries the URA3 gene flanked by two hisG sequences from Salmonella typhimurium (Fig. 1A-1). After the initial transformation, the transformants are selected on uracil-deficient media. Because the placement of two direct repeats of the hisG sequence at close proximity in the genome is unstable, spontaneous recombination events can occur between these hisG sequences. Such recombination events will loop out the URA3 gene and leave behind one copy of the hisG sequence in the genome. The loopout event allows the re-cycling of the URA3 marker for the disruption of the second allele. The use of URA3 is specifically advantageous because it can also be counter selected. Therefore, in the next step the cells that excise the URA3 gene by a recombination event between the hisG direct repeats can be selected on 5-FOA (5-Fluoroorotic acid). An additional second transformation step potentially generates cells that are null for the gene of interest, if recombination occurs at the wild-type allele.
Figure 1. (A) Concepts underlying gene disruption constructs of C. albicans.
The figure shows the various constructs used for gene manipulation in C. albicans. The gene of interest and/or flanking sequences are shown in green. All sequences coming from different species have been codon optimized to be used in C. albicans. Line 1. URA blaster (Fonzi and Irwin,1993) 2. URA flipper cassette. p; promoter region of given gene, t; termination sequence of given gene. (Morschhauser et al.,1999) 3. PCR amplifiable URA3 disruption cassette (Wilson et al., 2000) 4. UAU1 cassette (Enloe et al., 2000). 5. PCR amplifiable marker cassettes from non-albicans species. C. d.; Candida dubliniensis, C. m.; Candida maltosa. (Noble and Johnson, 2005) 6. Cre-loxP system (Dennison et al., 2005), note that when this construct integrates, the arg4 sequences will be flanked by loxP sites. 7. “MPAR flipper” cassette (Wirsching et al., 2000) 8. SAT1 flipper cassette (Reuss et al., 2004) 9. CaNAT1-FLP cassette. Ag; Ashbya gossypii. (Shen et al., 2005) 10. “Tet-Off” system (Roemer et al., 2003) 11. “Tet-On” system (Park and Morschhäuser, 2005). (B) Schematic representation of the URA-flipper method, adapted from Morschhauser et al. (1999). Step 1: Exogenously added insert DNA replaces one copy of the gene of interest by homologous recombination. Step 2: Expression of FLP recombinase promotes loss of URA3, leaving behind one copy of FRT. Steps 3–4: Repeating steps 1 and 2 enables the replacement of the remaining allele and results in a knockout of the gene of interest.
The “URA Blaster” method was the method of choice for not only genetic manipulation of C. alibicans, but also for constructing other parent strains that lacked additional nutritional markers (Negredo et al., 1997; Wilson et al., 1999). However, during the construction of the parent strain CAI4, the deletion construct also removed a portion of the 3’ end of the adjacent gene, IRO1 (García et al., 2001). IRO1 is a gene associated with iron utilization (García et al., 2001). Although there is no direct evidence that the loss of the 3’ portion of the IRO1 gene affects the virulence of the pathogen, iron uptake is known to be important for growth of C. albicans (Ramanan and Wang, 2000). Further, the CAI4 strain is known to show defective expression of 14 different proteins when compared to its parent, SC5314 (Brand et al., 2004).
When using the “URA Blaster” method for gene manipulation, the recombination between hisG repeats leaves a scar (one copy of the hisG sequence) in the genome. In instances where a promoter region of an adjacent gene is within or just following the termination region of the gene of interest, leaving behind a scar could be detrimental as it might affect the expression of the adjacent gene. Foreign DNA sequences can also cause rearrangements in the genome or make further manipulations or evaluations more complex (Fehér et al., 2008). For these reasons, gene manipulation without leaving a scar has become a goal in other organisms as well (Storici et al., 2001; Cox et al., 2007; Liang and Liu, 2010).
Another drawback of using the “URA Blaster” method is that exposure to 5-FOA in the second step is potentially mutagenic and can introduce chromosomal rearrangements (Wellington et al., 2006). In addition, this method leaves a single copy of URA3 at the locus of the gene of interest. Unfortunately almost half a decade later, researchers discovered that the expression of URA3 at ectopic loci in the genome can affect the virulence of C. albicans (Lay et al., 1998; Brand et al., 2004). URA3 gene expression in some strains disrupted by the “URA Blaster” method, hence carrying URA3 in an ectopic location of the genome, showed a 2 to 18 fold reduction in orotidine 5'-monophosphate decarboxylase enzyme activity (Lay et al., 1998). Overall the change in the chromosomal location of the URA3 gene affected the C. albicans Ura3 activity, hyphal morphogenesis, adherence and lethality in mice (Cheng et al., 2003). Phenotypic changes in about 30% of the published papers at that time were, upon reevaluation, due to the expression of URA3 at an ectopic location in the genome and not due to the deletion of the gene of interest per se (Brand et al., 2004).
To overcome this problem, the single remaining copy of URA3 was removed from the locus of the gene of interest and placed at a highly expressed locus such as, RPS1, ENO1, ARG4 or at the native locus of URA3 itself (Murad et al., 2000; Brand et al., 2004; Sundstrom et al., 2002; Davis et al., 2000; Ramon and Fonzi, 2003). It was shown that 40% expression from URA3 was adequate for full virulence affects of C. albicans on mice (Lay et al., 1998; Brand et al., 2004). Therefore, replacing one copy of URA3 was considered adequate for a reliable experiment. It was around this time that “reconstitution” of a mutant strain by introduction of a wild-type copy of the gene to rescue the mutant phenotype was considered an essential part of a C. albicans experiment (Magee et al., 2003; Brand et al., 2004).
As a result of the problems associated with the “URA Blaster” method, many researchers developed alternative versions of the “URA Blaster” cassette or entirely new deletion cassettes. Morschhauser et al. (1999) introduced the use of FLP recombinase system to make a recyclable URA3 cassette to disrupt both copies of the gene of interest in the parent strain CAI4 (URA Flipper). In this method, the URA3 marker is flanked by two direct repeats of the Flp recombinase recognition site, FRT (Fig. 1A-2). The expression of the FLP recombinase gene is controlled by the inducible SAP2 promoter (secreted aspartyl proteinase family 2). After transformation, the SAP2 promoter is activated and FLP is expressed. To activate the SAP2 promoter the cells should be grown in YCB-BSA (Yeast Carbon Base and Bovine Serum Albumin, pH4.0) media. The expressed Flp recombinase then recognizes the FRT sites and loops out the URA3 gene by homologous recombination, making the auxotrophic marker available for the second transformation (Fig. 1B). This method avoids the use of 5-FOA and its associated mutagenic potential to counter select for URA3 gene. However, similar to the “URA Blaster” cassette, the “URA Flipper” cassette also leaves behind a scar (a single FRT sequence) in the genome, at the locus of the gene of interest. Nevertheless the scar left behind is very small - only 34 base pairs. In addition, the method requires that the cells are grown in YCB-BSA media to express Flp recombinase, which could be undesirable for specific gene manipulations, such as genes associated with certain metabolic processes and should be carefully considered.
Both “URA Blaster” and “URA Flipper” cassettes require several cloning steps to be customized for the use of one’s own gene of interest. Wilson et al. (1999) tested the use of relatively rapid PCR-amplified deletion cassettes to disrupt genes of interest in parent strains that were derivatives of CAI4. They were, RM1000, auxotrophic for URA3 and HIS1 (Negredo et al., 1997) and BWP17, auxotrophic for URA3, HIS1 and ARG4 (Wilson et al., 1999). A 50–60 base pair overhang of homologous sequences of the gene of interest on either side of the selectable marker was found to be sufficient for disruption. Because this gene cassette was not recyclable, they used two nutritional markers from the available URA3, HIS1 or ARG4 to disrupt the two alleles.
Wilson et al. (2000) introduced a recyclable URA3 cassette that could be efficiently PCR amplified. Since the cassette was recyclable a single nutritional marker could be used for gene manipulation in C. albicans. In addition, this cassette saved an enormous amount of time as it could be rapidly PCR amplified. The cassette is similar to the “Ura Blaster” cassette with the exception that it does not carry the bulky hisG sequences, but instead carries a 200 base pair region of the 3’ end of the URA3 gene, at its 5’ end (Fig. 1A-3). However, the method does leave behind a scar (200bp) after the recombination between the repeating 3’ end sequences of the URA3 gene and also requires the use of 5-FOA.
Enloe et al. (2000) constructed a UAU1 cassette as a further development of this cassette and implemented it on the parent strain, BWP17 (Fig. 1A-4). The importance of the UAU1 cassette is that it allows the disruption of both copies of the gene of interest with a single transformation step. UAU1 cassette has the URA3 gene disrupted by the ARG4 gene, but the 5’ end sequence of the URA3 gene also carries a 530 base pair sequence homology to its 3’ end sequence. Therefore, when transformed into the genome, recombination loops out the ARG4 gene and makes a functional copy of URA3. This unique feature of the UAU1 cassette allows selection of rare instances where both alleles of the gene are replaced by mitotic recombination or gene conversion, in a single transformation step. These rare double-delete cells are detected due to the presence of both, ARG4 and URA3 markers. However, these potential homozygous deletion mutants need to be confirmed to rule out possible allelic triplications, in which a wild-type allele is retained in spite of two alleles been replaced by the cassette. This condition may arise due to known triplicated alleles in the genome, increase in ploidy, tandem duplications or translocations (Enloe et al., 2000).
It is known that C. albicans has remarkable tolerance for aneuploidy (Rustchenko, 2007). Aneuploidy refers to the gain or loss of full or parts of chromosomes that leads to an alteration in the normal complement of chromosomes. Aneuploidy can affect the virulence of C. albicans (Chen et al., 2004). The chromosome alterations were initially tested through contour-clamped homogenous electric field gels (CHEF gels) and quantitative Southern blot hybridization, two fairly laborious methods (Thrash-Bingham and Gorman, 1992; Navarro-García et al., 1995; Chen et al., 2004). Selmecki et al. (2005) tested aneuploidy in C. albicans laboratory strains using comparative genome hybridization (CGH). CGH allows comprehensive analysis of genomic alterations across over 6000 ORFs of C. albicans in a microarray format. Aneuploidy of the laboratory strains (CAI4 and BWP17) were tested compared to the clinical isolate SC5314. The results suggested that the laboratory strain, CAI4 had an unstable trisomy at chromosome 2 as it was not seen in the successive strains derived from it. In addition, the laboratory strain BWP17 and its parent (RM1000#6) showed a heterozygous deletion of the distal portion of right arm of chromosome 5. Thereafter, more attention was paid to constructing parents strain that had no or minimal karyotypic changes.
Noble and Johnson (2005) constructed parent strains that were auxotrophic for LEU2, HIS1 and ARG4 nutritional markers. The main strains constructed were, SN87 (auxotrophic for LEU2 and HIS1), SN95 (auxotrophic for HIS1 and ARG4) and SN152 (auxotrophic for LEU2, HIS1 and ARG4). Unlike many of the previously constructed parent strains, these showed no karyotypic changes. All these strains have one copy of URA3 expressed at the native locus. Using these strains, C. albicans genes are disrupted without the use of the URA3 marker. These markers are not recyclable and are left at the locus of the gene of interest (Fig. 1A-5). However, extensive tests indicate that expression of any of the above nutritional markers in ectopic genomic locations did not affect the virulence of C. albicans. They also initiated the use of heterologous marker genes from C. maltosa or C. dubliniensis strains when disrupting genes in C. albicans to decrease recombination events at the endogenous locus of the marker gene.
Dennison et al. (2005) adapted the popular Cre-loxP system for C. albicans gene disruption in the BWP17 parent strain. In two separate transformation steps, the two copies of the gene of interest are disrupted by nutritional markers such as HIS1 and ARG4, which are flanked by loxP sequences. In a third step, a fragment flanked by arg4 sequences that carry both the Cre recombinase under the control of the MET3 promoter and the nutritional marker URA3 is transformed into C. albicans (Fig. 1A-6). This fragment integrates into the ARG4 gene that is disrupting one copy of the gene of interest. Next, activation of MET3 promoter produces Cre recombinase, which catalyses the recombination between loxP sites, looping out both the HIS1 and the Cre recombinase-URA3 fragment. The method requires three transformation steps and leaves the null strain auxotrophic for all three nutritional markers. The method also leaves behind only a 34 base pair scar (a single loxP sequence) at the locus of the gene of interest.
Many deletion cassettes that were developed since the “URA blaster” method, allow one to remove URA3 from the locus of the gene of interest and replace it at its native locus. It should be noted that in addition to the multiple steps required to create the null mutant (gene deletion and the subsequent step to loop out the URA3 gene), the URA3 gene replacement step requires an additional transformation step. Unlike these methods, the UAU1 cassette leaves the URA3 gene at the locus of the gene of interest (Enloe et al., 2000). This method can still be useful so long as a reconstituted strain is created and shown to restore the wild-type phenotype. Further, this method is faster (creates C. albicans null mutants with a single transformation step) and will not leave behind any chromosomal rearrangements that may arise due to the use of, 5-FOA since it avoids the use of this drug. Therefore, the UAU1 cassette seems to be prominent among the gene disruption methods that use URA3. However, if one prefers to avoid the URA3 marker for selection altogether, the use of parent strains that are auxotrophic for LEU2, HIS1 and ARG4 nutritional markers may be useful (Noble and Johnson, 2005). This method uses two transformation steps, but does not require additional steps to loop out the nutritional markers. To date, no drawbacks of retaining the LEU2, HIS1 or ARG4 markers at ectopic loci have been identified. Nevertheless the resulting null mutant phenotypes should be validated by comparing phenotypes with an isogenic strain that is reconstituted for the gene of interest.
In 2004, C. albcians was selected as the first eukaryotic pathogen for genome sequencing. Jones et al. (2004) used the heterozygous diploid genome of the widely-used clinical isolate, SC5314 to sequence the genome of C. albcians by using a whole-genome shotgun approach. Thereafter, sequences of a majority of the C. albcians genes were available making gene targeting in this organism much easier, faster, more efficient and, precise. Soon after, d’Enfert et al. (2005) established the Candida Genome Database (CGD) using the genome sequence of C. albicans and also enriched it with published literature on C. albicans. The CGD (www.candidagenome.org) is a freely available valuable resource for researches in the field because it is a reliable source of organized data, tools for data analysis and information about the current research community.
3. Drug Markers for Selection
Some researchers have chosen to avoid entirely the use of nutritional markers to manipulate the C. albicans genome. This is necessary if the genes of interest are in pathways affected by nutritional status. Instead, drug resistance markers can be used. The use of drug makers also opens way for testing clinical isolates as is, without creating any auxotrophic mutations. This advantage heavily benefits the researchers that test C. albicans clinical isolates. For example, clinical isolates can be tested to identify mechanisms of drug resistance, which will be another phase of addressing the current problem of drug resistance seen in this pathogen (Heilmann et al., 2010). The milestones in the development of these drug markers are discussed below.
Goshorn and Scherer (1989) first identified dominant mycophenolic acid (MPA) resistant mutants in C. albicans. MPA inhibits the inosine monophosphate dehydrogenase (Imh3) enzyme that directs de novo synthesis of GMP. These mutants were used to study C. albicans natural variants. Kohler et al. (1997) developed a strategy using IMH3 as a selectable marker for C. albicans genetic manipulations. They overexpressed IMH3 from a plasmid in the strain CAI4 and found that the successful transformants were far more resistant to MPA compared to the wild-type strains. However, the method was not developed to the level of chromosomal integration. Wirsching et al. (2000) used MPA resistance as a dominant selectable marker for chromosomal gene disruptions. Here, they used a form of the IMH3 gene that was mutated so as to avoid re-integration of the fragment into the normal chromosomal copy of IMH3 in C. albicans. They adapted the “URA flipper” strategy to this system whereby FRT recombination sites were positioned to flank the FLP recombinase gene (driven by the SAP2 promoter) and the IMH3 gene (replacing URA3 selectable marker) to generate a “MPAR flipper” cassette (Fig. 1A-7). Using the MPAR flipper method, genes of C. albicans can be disrupted even in a clinical isolate without the use of any other auxotrophic markers. However, the method leaves behind a small 34 base pair FRT sequence at the locus of the gene of interest. In addition, the transformants are slow to appear and recombination is favored at the chromosomal IMH3 locus making the process of screening for successful transformants time consuming and tedious. Deleting the native IMH3 gene would be useful to reduce integration at this locus.
Reuss et al. (2004) used the drug nourseothricin as a dominant selectable marker for gene disruption. They introduced into C. albicans cells the streptothricin acetyltransferase (SAT1) gene (from bacterial transposon Tn1825) that confers resistance to the drug nourseothricin. A FLP-mediated recyclable marker system was used with the FLP recombinase gene regulated by the MAL2 (maltase) promoter and the SAT1 gene regulated by the ACT1 promoter (SAT1 flipper) (Fig. 1A-8). However, the MAL2 promoter that is usually activated when cells are grown in maltose instead of glucose was later found to be a leaky promoter. Therefore FLP recombinase was expressed even when cells were grown in media with glucose.
Shen et al. (2005) used the Streptomyces noursei nat1 (nourseothricin acetyltransferase) gene adapted to suit C. albicans in a CaNAT1-FLP cassette. The CaNAT1 gene (regulated by the Ashbya gossypii TEF1 promoter) and the FLP recombinase gene (regulated by the SAP2 promoter) are flanked by FRT recombination sites, as in the “URA flipper” cassette (Fig. 1A-9). Nourseothricin was found to have minimal affects on filamentation or cell growth. However, the CaNAT1-FLP cassette also leaves a genomic scar in the form of a single FRT sequence.
4. Other Markers for Selection
Limited uses of several other markers were also recorded in C. albicans gene disruption experiments. Morschhauser et al. (1998) used green fluorescence protein (GFP) in a reporter system with promoter and termination sequences from ACT1 gene to disrupt C. albicans genes. The first transformation is selected by the expression of GFP. However the selection of the second transformation depends on the increase in the intensity of GFP signal or the use of a GFP variant with different spectral properties in the second round of transformation. In addition, limited use of codon optimized firefly luciferase and renilla luciferase have been recorded as methods of C. albicans gene disruption (Doyle et al., 2006; Srikantha et al., 1996).
5. Disruption of Essential C. albicans Genes
In most cases, the disruption of non-essential genes is relatively straight forward, compared to disruption of essential genes. Initially, the inability to obtain homozygous deletions was used as evidence that a gene was essential. This was inherently tentative because it is a negative result. Instead, conditional mutants were used to test the essentiality of genes. Here one copy of the gene is deleted and the expression of the second copy is made conditional. The essentiality of the gene is determined by measuring survival of C. albicans after a shift to non-permissive conditions.
For example, Devasahayam et al. (2002) engineered a conditional allele of a gene of interest (ESS1) based on prior work in S. cerevisiae. The mutated C. albicans gene, was first demonstrated to be temperature sensitive (ts) in S. cerevisiae prior to its analysis in C. albicans. The investigators then generated a ts strain of C. albicans by deleting one copy of ESS1 and replacing the other with the engineered allele. The strain grew normally at permissive temperature, but did not grow at restrictive temperature, demonstrating the essentiality of ESS1 in C. albicans.
Roemer et al. (2003) developed the “GRACE method” (Gene Replacement And Conditional Expression) for conditional expression of essential genes. In this method, the first copy of the gene is disrupted by a cassette containing the HIS3 selectable marker (Roemer et al., 2003). This cassette also has two distinct bar codes placed on either side allowing the rapid and simple identification of the strain by PCR amplification. The second copy is placed under the control of a tetracycline (Tet) regulatable promoter system using a codon optimized SAT1 selectable maker (from E. coli) driven by the ACT1 promoter (Fig. 1A-10). Under normal conditions the binding of the transactivator protein to the Tet responsive promoter allows constitutive expression of the essential gene of interest. However, if the cells are provided with tetracycline, the association between the transactivatior protein and the Tet responsive promoter is disrupted leading to repression of the gene. The method is useful to identify a gradient of phenotypes from cidal, to static, to no growth phenotype.
Park and Morschhäuser (2005) also used a tetracycline-regulatable system to make conditional null mutants of essential genes. Instead of the “Tet-Off” system (the gene regulated by tetracycline is turned off in the presence of the drug) explained above, Park and Morschhäuser adapted a “Tet-On” system (the gene regulated by tetracycline is turned on in the presence of the drug). The “Tet-On” system was made possible when Gossen et al. (1995) recognized the reverse Tet repressor. In the “Tet-On” system, the reverse Tet repressor (from E. coli) is fused to an activation domain (GAL4 form S. cerevisiae) to create a reverse tetracycline-controlled transactivator (rtTA), which binds to the Tet responsive promoter in the presence of the drug. This is in contrast to the usual behavior of the transactivator that binds to the Tet responsive promoter in the absence of the drug.
To use this system, the essential gene of interest and the MPA resistance marker flanked by FRT sites is transformed, disrupting one copy of the ACT1 gene. This copy covers the requirement of the essential gene, allowing the native copies to be deleted. Next, a construct that carries the URA3 selectable marker, the rtTA (codon optimized to be used in C. albicans) and the FLP recombinase gene placed under a Tet-responsive promoter is transformed, disrupting one copy of the ADH1 gene (Fig. 1A-11). Park and Morschhäuser used the tetracycline derivative, doxycycline instead of tetracycline in all of their experiments, as the rtTA shows higher sensitivity to this derivative.
Under normal conditions, in the absence of doxycycline the FLP recombinase gene is not expressed. However, if the cells are provided with doxycycline, the rtTA binds the Tet-responsive promoter and transcribes the FLP recombinase gene. Flp recombinase recognizes the FRT sites and loops out the essential gene along with the MPA resistance marker and the loss of the essential gene can be identified by the gain of MPA susceptibility. Deletion of an essential gene gives rise to a nonviable strain, as assessed by the number of colony forming units formed in the presence or absence of doxycycline.
One advantage of the method is that the expression of the FLP recombinase gene is not depended on nutrient changes in the media as in the “URA flipper” (Morschhauser et al. 1999), “Cre-loxP system” (Dennison et al. 2005), “MPAR flipper” (Wirsching et al. 2000) or “CaNAT1-FLP cassette” (Shen et al. 2005) methods, where the recombinase gene is under the regulation of a promoter that shows media-dependent activation. Therefore, in instances where nutritional changes are not desirable, this “Tet-On” system could be an attractive method for gene manipulation in C. albicans. However, the method requires the deletion of both copies of the essential gene before or after the expression of the extra copy at the ACT1 locus, demanding the need for more transformations steps than required for in the “GRACE method”.
6. Gene Manipulation in Other Fungal Species
Similar to C. albicans, gene manipulation in many other clinically important Candida, Cryptococcus and Aspergillus species continues to evolve (Magee et al., 2003; Hull and Heitman, 2002; Brookman and Denning, 2000; Brakhage and Langfelder, 2002). For example, the URA5 gene is a popular marker for gene disruption in Cryptococcus species. In the past, URA5 function was reintroduced at random locations in the genome (Hull and Heitman, 2002), however, today, it can be restored to its native locus. For example, Narasipura et al. (2006) constructed a Cryptococcus gattii parent strain that has a non-functional URA5 gene due to a point mutation, which can subsequently be restored to functionality through rare spontaneous reversion events. Uracil auxotrophs are also used for gene manipulation in Aspergillus species. For this purpose, mutants of pyrG gene that encodes orotidine-5′-monophosphate decarboxylase that is essential for uracil synthesis are used (Brakhage and Langfelder, 2002). Comparable to C. albicans, the use of drug selectable markers has also become popular in both, Cryptococcus and Aspergillus species (Hua et al., 2000; McDade and Cox, 2001; Brakhage and Langfelder, 2002).
RNA interference techniques are also proving to be successful in Cryptococcus neoformans, Aspergillus fumigatus and Aspergillus nidulans giving a new dimension to gene manipulation in these species (Liu et al., 2002; Mouyna et al., 2004; Barton and Prade, 2008). RNA interference techniques are yet to be proven successful in C. albicans (Staab et al., 2011). There is evidence for the presence of RNAi in C. albicans (Drinnenberg et al., 2009) and the presence of a majority of the proteins involved in the RNAi pathway (Drinnenberg et al., 2009; Staab et al., 2011). However, the mere presence of these proteins does not seem sufficient to trigger the RNAi pathway by dsRNA that are exogenously introduced (Staab et al., 2011). It is suggested that the RNAi pathway in C. albicans may have evolved in a species-specific way that it is no longer triggered by exogenous dsRNA (Staab et al., 2011).
7. Concluding Remarks
Over the last decade or so, a variety of methods have been developed to manipulate the C. albicans genome in useful ways. However, special attention should be paid to the method of choice, the selectable markers used, and their collective consequences on virulence or other phenotypes to be studied. Constructing an isogenic strain reconstituted for the gene of interest remains the strongest means of validating the method of choice and mutant phenotypes.
While great progress has been made, it is still discomforting that a scar-less method for disrupting C. albicans genes has not yet been established. A “delitto perfecto” method has been developed to disrupt genes in S. cerevisiae, the highly studied cousin of C. albicans (Storici et al., 2001). Here, a counter selectable reporter (CORE) cassette flanked by homologous sequences flanking the gene of interest is first integrated in to the genome, to replace the gene. In a second step, two 80 bp integrative recombinant oligonucleotides (with 70bp homology to sequence extending from either end of the CORE cassette in the genome) that are designed to have at least a 20 bp overlap between their 3’ end sequences are annealed together, extended and transformed. These molecules will occasionally recombine at the locus of interest replacing the CORE cassette, leaving no scar behind. The cells that are perfectly null at the gene of interest, with no scar left behind are typically counter-selected for using two markers, 5-FOA and a drug selectable marker. The drug selectable markers available in a CORE cassette thus far are, G418 and hygromycin B (Storici and Resnick, 2006).
This method is yet to be adapted for C. albicans gene disruption. The main drawback of implementing the method is that C. albicans is inherently resistant to the drug selection makers (G418 and hygromycin B) that are currently used. However, C. albicans does show susceptibility to the drug, nourseothricin suggesting that it may be possible to implement a different version of this method for C. albicans gene disruption. One can expect that a “delitto perfecto” method for C. albicans gene disruptions will be available in the not too distant future.
The C. albicans research community is benefiting from large-scale and genome-wide approaches developed in S. cerevisiae. Two examples are given. First, Homann et al. (2009) created a homozygous deletion library of 143 non-essential transcriptional regulators using the parent strain SN152 constructed by Noble and Johnson (2005). This deletion library carries two independent isolates for each gene knockout, and is a valuable resource available to study the role of these transcription regulators in drug sensitivity, morphogenetic switching, to identify binding partners to these regulators through chromatin immunoprecipitation and more. By 2010 this homozygous deletion library expanded to include the deletion of 647 genes (Noble et al., 2010), signifying rapid growth in this research field.
Second, Oh et al. (2010) constructed a tagged heterozygous transposon deletion library that included 3633 strains. They modified the Tn5 transposon to include a Gateway conversion cassette, the UAU1 cassette (Enloe et al., 2000) and a kanamycin resistance gene. The modified transposon was introduced to a C. albicans genomic DNA library and the individual mutagenized clones were tested for the disruption of a gene of interest. Each gene insertion was maximized to a have a unique tag to make the process of strain tracking and quantitation easier. Finally, they transformed the transposon flanked by sequences from the gene of interest in to the parent strain BWP17 constructed by Wilson et al. (1999). The correct heterozygous transformants were selected by arginine prototrophy. This publicly available resource is useful to detect haploinsufficient phenotypes, gene function, drug sensitivity and more. Moreover, because the strains are tagged, the experiments can be multiplexed and conducted in a high-throughput manner. Collectively, these resources open the way for comprehensive genome-wide analysis as well as systematic analysis of C. albicans (Noble et al., 2010; Oh et al., 2010).
It is noteworthy that the C. albicans research field is evolving with great caution. Bouchonville et al. (2009) discovered that simple and popular DNA transformation procedures such as, the lithium acetate protocols or electroporation can introduce aneuploidy to the C. albicans strains. It is therefore advisable that all C. albicans mutant strains be tested for aneuploidy before conducting extensive studies (Arbour et al., 2009). Arbour et al. (2009) introduced a simple and straight forward multiplex quantitative PCR assay to test the ploidy of any new C. albicans mutant strain.
In summary, the C. albicans research field is evolving rapidly, and making use of ever more sophisticated and precise genetic tools. One can expect increased use of genome-wide and systematic analysis of C. albicans. Advances in high-throughput data generation and analysis will be a useful step in the development of therapies to manage C. albicans as a human pathogen.
Acknowledgements
Dhanushki Samaranayake is a Ph.D. student in the laboratory of SDH. We thank the reviewers for constructive advice. This work was supported by a grant from the National Institutes of Health (R01-GM055108) to SDH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arbour M, Epp E, Hogues H, Sellam A, Lacroix C, Rauceo J, Mitchell A, Whiteway M, Nantel A. Widespread occurrence of chromosomal aneuploidy following the routine production of Candida albicans mutants. FEMS Yeast Res. 2009;9:1070–1077. doi: 10.1111/j.1567-1364.2009.00563.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett JA. A history of research on yeasts 12: medical yeasts part 1, Candida albicans. Yeast. 2008;25:385–417. doi: 10.1002/yea.1595. [DOI] [PubMed] [Google Scholar]
- Barton LM, Prade RA. Inducible RNA Interference of brlAβ in Aspergillus nidulans. Euk. Cell. 2008;7:2004–2007. doi: 10.1128/EC.00142-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchonville K, Forche A, Tang KE, Selmecki A, Berman J. Aneuploid chromosomes are highly unstable during DNA transformation of Candida albicans. Euk. Cell. 2009;8:1554–1566. doi: 10.1128/EC.00209-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brakhage AA, Langfelder K. Menacing mold: the molecular biology of Aspergillus fumigatus. Annu. Rev. Microbiol. 2002;56:433–455. doi: 10.1146/annurev.micro.56.012302.160625. [DOI] [PubMed] [Google Scholar]
- Brand A, MacCallum DM, Brown AJP, Gow NAR, Odds FC. Ectopic expression of URA3 can influence the virulence phenotypes and proteome of Candida albicans but can be overcome by targeted reintegration of URA3 at the RSP10 locus. Euk. Cell. 2004;3:900–909. doi: 10.1128/EC.3.4.900-909.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookman JL, Denning DW. Molecular genetics in Aspergillus fumigatus. Curr. Opin. Microbiol. 2000;3:468–474. doi: 10.1016/s1369-5274(00)00124-7. [DOI] [PubMed] [Google Scholar]
- Cannon RD, Lamping E, Holmes AR, Niimi K, Tanabe K, Niimi M, Monk BC. Candida albicans drug resistance – another way to cope with stress. Microbiology. 2007;153:3211–3217. doi: 10.1099/mic.0.2007/010405-0. [DOI] [PubMed] [Google Scholar]
- Casalinuovo IA, Di Francesco P, Garaci E. Fluconazole resistance in Candida albicans: a review of mechanism. Eur. Rev. Med. Pharmacol. Sci. 2004;8:69–77. [PubMed] [Google Scholar]
- Chen X, Magee BB, Dawson D, Magee PT, Kumamoto CA. Chromosome 1 trisomy compromises the virulence of Candida albicans. Mol Microbiol. 2004;51:551–565. doi: 10.1046/j.1365-2958.2003.03852.x. [DOI] [PubMed] [Google Scholar]
- Cheng S, Nguyen MH, Zhang Z, Jia H, Handfield M, Clancy CJ. Evaluation of the roles of four Candida albicans genes in virulence by using gene disruption strains that express URA3 from the native locus. Infect Immun. 2003;71:6101–6103. doi: 10.1128/IAI.71.10.6101-6103.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox MM, Layton SL, Jiang T, Cole K, Hargis BM, Berghman LR, Bottje WG, Kwon YM. Scarless and site-directed mutagenesis in Salmonella enteritidis chromosome. BMC Biotechnol. 2007;7:59. doi: 10.1186/1472-6750-7-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis D, Edwards JE, Jr, Mitchell AP, Ibrahim AS. Candida albicans RIM101 pH response pathway is required for host-pathogen interactions. Infect. Immun. 2000;68:5953–5959. doi: 10.1128/iai.68.10.5953-5959.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Backer MD, Magee PT, Pla J. Recent developments in molecular genetics of Candida albicans. Annu. Rev. Microbiol. 2000;54:463–498. doi: 10.1146/annurev.micro.54.1.463. [DOI] [PubMed] [Google Scholar]
- d'Enfert C, Goyard S, Rodriguez-Arnaveilhe S, Frangeul L, Jones L, Tekaia F, Bader O, Albrecht A, Castillo L, Dominguez A, Ernst JF, Fradin C, Gaillardin C, Garcia-Sanchez S, de Groot P, Hube B, Klis FM, Krishnamurthy S, Kunze D, Lopez MC, Mavor A, Martin N, Moszer I, Onésime D, Perez Martin J, Sentandreu R, Valentin E, Brown AJ. CandidaDB: a genome database for Candida albicans pathogenomics. Nucleic Acids Res. 2005;33(Database issue):D353–D357. doi: 10.1093/nar/gki124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennison PM, Ramsdale M, Manson CL, Brown AJ. Gene disruption in Candida albicans using a synthetic, codon-optimised Cre-loxP system. Fungal Genet. Biol. 2005;42:737–748. doi: 10.1016/j.fgb.2005.05.006. [DOI] [PubMed] [Google Scholar]
- De Rosa FG, Garazzino S, Pasero D, Di Perri G, Ranieri VM. Invasive candidiasis and candidemia: new guidelines. Minerva Anestesiol. 2009;75:453–458. [PubMed] [Google Scholar]
- Devasahayam G, Chaturvedi V, Hanes SD. The Ess1 prolyl isomerase is required for growth and morphogenetic switching in Candida albicans. Genetics. 2002;160:37–48. doi: 10.1093/genetics/160.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle TC, Nawotka KA, Purchio AF, Akin AR, Francis KP, Contag PR. Expression of firefly luciferase in Candida albicans and its use in the selection of stable transformants. Microb. Pathog. 2006;40:69–81. doi: 10.1016/j.micpath.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Drinnenberg IA, Weinberg DE, Xie KT, Mower JP, Wolfe KH, Mower JP, Wolfe KH, Fink GR, Barte lDP. RNAi in budding yeast. Science. 2009;326:544–550. doi: 10.1126/science.1176945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enloe B, Diamond A, Mitchell AP. A single-transformation gene function test in diploid Candida albicans. J. Bacteriol. 2000;182:5730–5736. doi: 10.1128/jb.182.20.5730-5736.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein JB, Hancock PJ, Nante lS. Oral candidiasis in hematopoietic cell transplantation patients: an outcome-based analysis. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2003;96:154–163. doi: 10.1016/s1079-2104(03)00296-8. [DOI] [PubMed] [Google Scholar]
- Fehér T, Karcagi I, Gyorfy Z, Umenhoffer K, Csörgo B, Pósfai G. Scarless engineering of the Escherichia coli genome. Methods Mol Biol. 2008;416:251–259. doi: 10.1007/978-1-59745-321-9_16. [DOI] [PubMed] [Google Scholar]
- Fonzi WA, Irwin MY. Isogenic strain construction and gene mapping in Candida albicans. Genetics. 1993;134:717–728. doi: 10.1093/genetics/134.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridkin SK, Kaufman D, Edwards JR, Shetty S, Horan T. Changing incidence of Candida bloodstream infections among NICU patients in the United States: 1995–2004. Pediatrics. 2006;117:1680–1687. doi: 10.1542/peds.2005-1996. [DOI] [PubMed] [Google Scholar]
- Goshorn AK, Scherer S. Genetic analysis of prototrophic natural variants of Candida albicans. Genetics. 1989;123:667–673. doi: 10.1093/genetics/123.4.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossen M, Freundlieb S, Bender G, Müller G, Hillen W, Bujard H. Transcriptional activation by tetracyclines in mammalian cells. Science. 1995;268:1766–1769. doi: 10.1126/science.7792603. [DOI] [PubMed] [Google Scholar]
- García MG, O'Connor JE, García LL, Martínez SI, Herrero E, del Castillo Agudo L. Isolation of a Candida albicans gene, tightly linked to URA3, coding for a putative transcription factor that suppresses a Saccharomyces cerevisiae aft1 mutation. Yeast. 2001;18:301–311. doi: 10.1002/1097-0061(20010315)18:4<301::AID-YEA672>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Heilmann CJ, Schneider S, Barker KS, Rogers PD, Morschhäuser J. An A643T mutation in the transcription factor Upc2p causes constitutive ERG11 upregulation and increased fluconazole resistance in Candida albicans. Antimicrob Agents Chemother. 2010;54:353–359. doi: 10.1128/AAC.01102-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homann OR, Dea J, Noble SM, Johnson AD. A phenotypic profile of the Candida albicans regulatory network. PLoS Genet. 2009;5:e1000783. doi: 10.1371/journal.pgen.1000783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn DL, Neofytos D, Anaissie EJ, Fishman JA, Steinbach WJ, Olyaei AJ, Marr KA, Pfaller MA, Chang CH, Webster KM. Epidemiology and outcomes of candidemia in 2019 patients: data from the prospective antifungal therapy alliance registry. Clin Infect Dis. 2009;48:1695–1703. doi: 10.1086/599039. [DOI] [PubMed] [Google Scholar]
- Hua J, Meyer JD, Lodge JK. Development of positive selectable markers for the fungal pathogen Cryptococcus neoformans. Clin. Diagn. Lab. Immunol. 2000;7:125–128. doi: 10.1128/cdli.7.1.125-128.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull CM, Heitman J. Genetics of Cryptococcus neoformans. Annu. Rev. Genet. 2002;36:557–615. doi: 10.1146/annurev.genet.36.052402.152652. [DOI] [PubMed] [Google Scholar]
- Jones T, Federspiel NA, Chibana H, Dungan J, Kalman S, Magee BB, Newport G, Thorstenson YR, Agabian N, Magee PT, Davis RW, Scherer S. The diploid genome sequence of Candida albicans. Proc. Natl. Acad. Sci. USA. 2004;101:7329–7334. doi: 10.1073/pnas.0401648101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao AS, Brandt ME, Pruitt WR, Conn LA, Perkins BA, Stephens DS, Baughman WS, Reingold AL, Rothrock GA, Pfaller MA, Pinner RW, Hajjeh RA. The epidemiology of candidemia in two United States cities: results of a population-based active surveillance. Clin Infect Dis. 1999;29:1164–1170. doi: 10.1086/313450. [DOI] [PubMed] [Google Scholar]
- Köhler GA, White TC, Agabian N. Overexpression of a cloned IMP dehydrogenase gene of Candida albicans confers resistance to the specific inhibitor mycophenolic acid. J. Bacteriol. 1997;179:2331–2338. doi: 10.1128/jb.179.7.2331-2338.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C, Tan C, Huang Y, Shao P, Hsueh P. Current challenges in the management of invasive fungal infections. J. Infect. Chemother. 2008;14:77–85. doi: 10.1007/s10156-007-0595-7. [DOI] [PubMed] [Google Scholar]
- Lay J, Henry LK, Clifford J, Koltin Y, Bulawa CE, Becker JM. Altered expression of selectable marker URA3 in gene-disrupted Candida albicans strains complicates interpretation of virulence studies. Infect. Immun. 1998;66:5301–5306. doi: 10.1128/iai.66.11.5301-5306.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang R, Liu J. Scarless and sequential gene modification in Pseudomonas using PCR product flanked by short homology regions. BMC Microbiol. 2010;10:209. doi: 10.1186/1471-2180-10-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Cottrell TR, Pierini LM, Goldman WE, Doering TL. RNA interference in the pathogenic fungus Cryptococcus neoformans. Genetics. 2002;160:463–470. doi: 10.1093/genetics/160.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magee PT, Gale C, Berman J, Davis D. Molecular genetic and genomic approaches to the study of medically important fungi. Infect. Immun. 2003;71:2299–2309. doi: 10.1128/IAI.71.5.2299-2309.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDade HC, Cox GM. A new dominant selectable marker for use in Cryptococcus neoformans. Med. Mycol. 2001;39:151–154. doi: 10.1080/mmy.39.1.151.154. [DOI] [PubMed] [Google Scholar]
- Miceli MH, Díaz JA, Lee SA. Emerging opportunistic yeast infections. Lancet Infect Dis. 2011;11:142–151. doi: 10.1016/S1473-3099(10)70218-8. [DOI] [PubMed] [Google Scholar]
- Miller LG, Hajjeh RA, Edwards JE., Jr Estimating the cost of nosocomial candidemia in the united states. Clin Infect Dis. 2001;32:1110. doi: 10.1086/319613. [DOI] [PubMed] [Google Scholar]
- Morschhäuser J, Michel S, Hacker J. Expression of a chromosomally integrated, single-copy GFP gene in Candida albicans, and its use as a reporter of gene regulation. Mol. Gen. Genet. 1998;257:412–420. doi: 10.1007/s004380050665. [DOI] [PubMed] [Google Scholar]
- Morschhäuser J, Michel S, Staib P. Sequential gene disruption in Candida albicans by FLP-mediated site-specific recombination. Mol. Microbiol. 1999;32:547–556. doi: 10.1046/j.1365-2958.1999.01393.x. [DOI] [PubMed] [Google Scholar]
- Mouyna I, Henry C, Doering TL, Latgé JP. Gene silencing with RNA interference in the human pathogenic fungus Aspergillus fumigatus. FEMS Microbiol. Lett. 2004;237:317–324. doi: 10.1016/j.femsle.2004.06.048. [DOI] [PubMed] [Google Scholar]
- Müller FM, Groll AH, Walsh TJ. Current approaches to diagnosis and treatment of fungal infections in children infected with human immuno deficiency virus. Eur J Pediatr. 1999;158:187–199. doi: 10.1007/s004310051047. [DOI] [PubMed] [Google Scholar]
- Murad AM, Lee PR, Broadbent ID, Barelle CJ, Brown AJ. CIp10, an efficient and convenient integrating vector for Candida albicans. Yeast. 2000;16:325–327. doi: 10.1002/1097-0061(20000315)16:4<325::AID-YEA538>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Narasipura SD, Ren P, Dyavaiah M, Auger I, Chaturvedi V, Chaturvedi. S. An efficient method for homologous gene reconstitution in Cryptococcus gattii using URA5 auxotrophic marker. Mycopathologia. 2006;162:401–409. doi: 10.1007/s11046-006-0076-z. [DOI] [PubMed] [Google Scholar]
- Navarro-García F, Pérez-Diaz RM, Magee BB, Pla J, Nombela C, Magee P. Chromosome reorganization in Candida albicans 1001 strain. J Med Vet Mycol. 1995;33:361–366. [PubMed] [Google Scholar]
- Negredo A, Monteoliva L, Gil C, Pla J, Nombela C. Cloning, analysis and one-step disruption of the ARG5,6 gene of Candida albicans. Microbiology. 1997;143:297–302. doi: 10.1099/00221287-143-2-297. [DOI] [PubMed] [Google Scholar]
- Noble SM, Johnson AD. Strains and strategies for large-scale gene deletion Studies of the diploid human fungal pathogen Candida albicans. Euk. Cell. 2005;4:298–309. doi: 10.1128/EC.4.2.298-309.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble SM, Johnson AD. Genetics of Candida albicans, a diploid human fungal pathogen. Annu. Rev. Genet. 2007;41:193–211. doi: 10.1146/annurev.genet.41.042007.170146. [DOI] [PubMed] [Google Scholar]
- Noble SM, French S, Kohn LA, Chen V, Johnson AD. Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity. Nat Genet. 2010;42:590–598. doi: 10.1038/ng.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odds FC. Candida species and virulence. ASM News. 1994;60:313–318. [Google Scholar]
- Oh J, Fung E, Schlecht U, Davis RW, Giaever G, St Onge RP, Deutschbauer A, Nislow C. Gene annotation and drug target discovery in Candida albicans with a tagged transposon mutant collection. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1001140. pii: e1001140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YN, Morschhäuser J. Tetracycline-inducible gene expression and gene deletion in Candida albicans. Euk. Cell. 2005;4:1328–1342. doi: 10.1128/EC.4.8.1328-1342.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanan N, Wang Y. A high-affinity iron permease essential for Candida albicans virulence. Science. 2000;288:1062–1064. doi: 10.1126/science.288.5468.1062. [DOI] [PubMed] [Google Scholar]
- Ramón AM, Fonzi WA. Diverged binding specificity of Rim101p, the Candida albicans ortholog of PacC. Euk. Cell. 2003;2:718–728. doi: 10.1128/EC.2.4.718-728.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuss O, Vik A, Kolter R, Morschhäuser J. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene. 2004;341:119–127. doi: 10.1016/j.gene.2004.06.021. [DOI] [PubMed] [Google Scholar]
- Roemer T, Jiang B, Davison J, Ketela T, Veillette K, Breton A, Tandia F, Linteau A, Sillaots S, Marta C, Martel N, Veronneau S, Lemieux S, Kauffman S, Becker J, Storms R, Boone C, Bussey H. Large-scale essential gene identification in Candida albicans and applications to antifungal drug discovery. Mol. Microbiol. 2003;50:167–181. doi: 10.1046/j.1365-2958.2003.03697.x. [DOI] [PubMed] [Google Scholar]
- Ruhnke M. Epidemiology of Candida albicans infections and role of non-Candida-albicans yeasts. Curr Drug Targets. 2006;7:495–504. doi: 10.2174/138945006776359421. [DOI] [PubMed] [Google Scholar]
- Rustchenko E. Chromosome instability in Candida albicans. FEMS Yeast Res. 2007;7:2–11. doi: 10.1111/j.1567-1364.2006.00150.x. [DOI] [PubMed] [Google Scholar]
- Santos MA, Tuite MF. The CUG codon is decoded in vivo as serine and not leucine in Candida albicans. Nucleic Acids Res. 1995;23:1481–1486. doi: 10.1093/nar/23.9.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schelenz S, Abdallah S, Gray G, Stubbings H, Gow I, Baker P, Hunter PR. Epidemiology of oral yeast colonization and infection in patients with hematological malignancies, head neck and solid tumors. J Oral Pathol Med. 2011;40:83–89. doi: 10.1111/j.1600-0714.2010.00937.x. [DOI] [PubMed] [Google Scholar]
- Selmecki A, Bergmann S, Berman J. Comparative genome hybridization reveals widespread aneuploidy in Candida albicans laboratory strains. Mol Microbiol. 2005;55:1553–1565. doi: 10.1111/j.1365-2958.2005.04492.x. [DOI] [PubMed] [Google Scholar]
- Shao PL, Huang LM, Hsueh PR. Recent advances and challenges in the treatment of invasive fungal infections. Int. J. Antimicrob. Agents. 2007;30:487–495. doi: 10.1016/j.ijantimicag.2007.07.019. [DOI] [PubMed] [Google Scholar]
- Shen J, Guo W, Köhler JR. CaNAT1, a heterologous dominant selectable marker for transformation of Candida albicans and other pathogenic Candida species. Infect. Immun. 2005;73:1239–1242. doi: 10.1128/IAI.73.2.1239-1242.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikantha T, Klapach A, Lorenz WW, Tsai LK, Laughlin LA, Gorman JA, Soll DR. The sea pansy Renilla reniformis luciferase serves as a sensitive bioluminescent reporter for differential gene expression in Candida albicans. J. Bacteriol. 1996;178:121–129. doi: 10.1128/jb.178.1.121-129.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staab JF, White TC, Marr KA. Hairpin dsRNA does not trigger RNA interference in Candida albicans cells. Yeast. 2011;28:1–8. doi: 10.1002/yea.1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storici F, Lewis LK, Resnick MA. In-vivo site-directed mutagenesis using oligonucleotides. Nat. Biotechnol. 2001;19:773–776. doi: 10.1038/90837. [DOI] [PubMed] [Google Scholar]
- Storici F, Resnick MA. The delitto perfetto approach to in vivo site-directed mutagenesis and chromosome rearrangements with synthetic oligonucleotides in yeast. Methods Enzymol. 2006;409:329–345. doi: 10.1016/S0076-6879(05)09019-1. [DOI] [PubMed] [Google Scholar]
- Sundstrom P, Cutler JE, Staab JF. Reevaluation of the role of HWP1 in systemic candidiasis by use of Candida albicans strains with selectable marker URA3 targeted to the ENO1 locus. Infect. Immun. 2002;70:3281–3283. doi: 10.1128/IAI.70.6.3281-3283.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrash-Bingham C, Gorman JA. DNA translocations contribute to chromosome length polymorphisms in Candida albicans. Curr Genet. 1992;22:93–100. doi: 10.1007/BF00351467. [DOI] [PubMed] [Google Scholar]
- Wellington M, Kabir MA, Rustchenko E. 5-fluoro-orotic acid induces chromosome alterations in genetically manipulated strains of Candida albicans. Mycologia. 2006;98:393–398. doi: 10.3852/mycologia.98.3.393. [DOI] [PubMed] [Google Scholar]
- Wilson RB, Davis D, Mitchell AP. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J. Bacteriol. 1999;181:1868–1874. doi: 10.1128/jb.181.6.1868-1874.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RB, Davis D, Enloe BM, Mitchell AP. A recyclable Candida albicans URA3 cassette for PCR product-directed gene disruptions. Yeast. 2000;16:65–70. doi: 10.1002/(SICI)1097-0061(20000115)16:1<65::AID-YEA508>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Wirsching S, Michel S, Morschhäuser. J. Targeted gene disruption in Candida albicans wild-type strains: the role of the MDR1 gene in fluconazole resistance of clinical Candida albicans isolates. Mol. Microbiol. 2000;36:856–865. doi: 10.1046/j.1365-2958.2000.01899.x. [DOI] [PubMed] [Google Scholar]