Abstract
Microcin B17 (McB) is a 43-amino acid antibacterial peptide targeting the DNA gyrase. The McB precursor is ribosomally produced and then post-translationally modified by the McbBCD synthase. Active mature McB contains eight oxazole and thiazole heterocycles. Here, we show that a major portion of mature McB contains an additional unusual modification, a backbone ester bond connecting McB residues 51 and 52. The modification results from an N → O shift of the Ser52 residue located immediately downstream of one of McB thiazole heterocycles. We speculate that the N,O-peptidyl shift undergone by Ser52 is an intermediate of post-translational modification reactions catalyzed by the McbBCD synthase that normally lead to formation of McB heterocycles.
Keywords: Bacteria, DNA Gyrase, Enzyme Catalysis, Mass Spectrometry (MS), Post-translational Modification, Microcins
Introduction
Microcins are small (<10 kDa) antibacterial peptides produced by enterobacteria. They are synthesized on the ribosomes, and some are subject to complex post-translational modifications (1, 2). One such post-translationally modified microcin, microcin B17 (McB),3 contains a series of thiazole and oxazole heterocycles that are produced, respectively, from Gly-Cys and Gly-Ser dipeptides of the ribosome-synthesized precursor by a dedicated maturation machinery (3). Recent bioinformatic and biochemical evidence indicates that small peptides containing post-translational-synthesized oxazole and thiazole cycles are widespread in nature. They include, among others, the cyanobactins (4), streptolysin S (5), clostridiolysin (6), goadsporin (7), and trithiazolyl pyridine antibiotics (8–10). The mechanistic details of heterocycle formation from polypeptide backbones are of considerable interest, as compounds containing these structures possess interesting pharmacological properties and include, among others, anticancer drug bleomycin. To date, two types of biosynthetic machines performing heterocyclization have been described. The first type consists of three proteins (zinc-tetrathiolate containing ATP-dependent cyclodehydratase, flavin mononucleotide-dependent dehydrogenase, and a docking scaffold protein) forming a trimeric complex. The best-studied example of such a molecular machine is the McB synthase McbBCD. The docking protein McbD recognizes the leader peptide of microcin precursor McbA and processively moves along the peptide in the N to C direction. Upon encountering a Gly-Ser or Gly-Cys substrate dipeptide, the McbC cyclodehydratase first converts a Ser or a Cys residue into, respectively, oxazoline or thiazoline with a loss of water from the amide backbone. Next, the McbB dehydrogenase removes two electrons and two protons producing aromatic oxazole or thiazole. Because McB variants with unoxidized thiazoline or oxazoline moieties are not detected, aromatization must immediately follow cyclization and probably proceeds without dissociation of the enzyme substrate complex (11–13). Heterocycle-forming enzymes of the second type such as heterocyclases of the cyanobactin pathway PatD and TruD combine substrate recognition and heterocyclization activities in a single polypeptide and lack the dehydrogenation activity. As a result, only peptides with thiazolines and oxazolines are synthesized. They are subsequently partially converted into aromatic heterocycles by uncharacterized cellular enzymes (4).
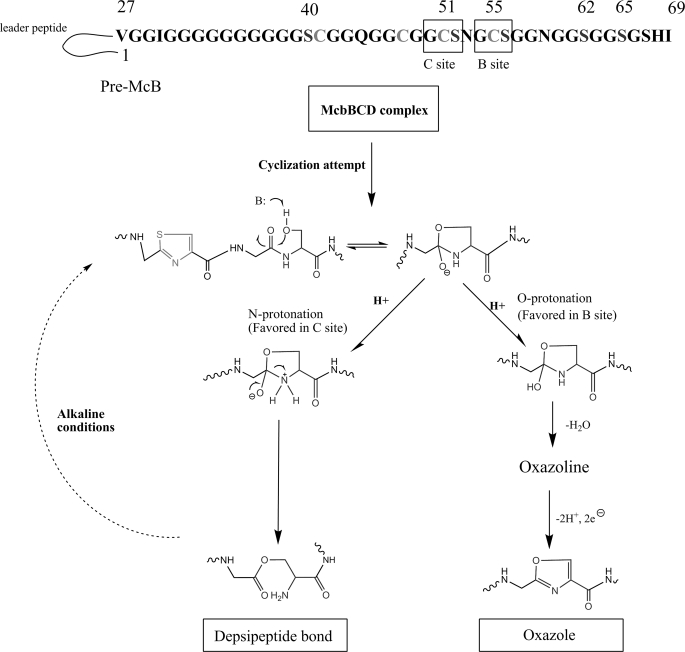
McB is a 3094-Da thiazole/oxazole-containing peptide that inhibits bacterial class II topoisomerase DNA gyrase. McB production and immunity requires seven genes: mcbA, mcbB, mcbC, mcbD, mcbE, mcbF, and mcbG. The mcb genes form an operon that is carried on naturally occurring plasmids. McB is synthesized from a 69-amino acid-long glycine-rich pre-McB, a product of the mcbA gene. Some pre-McB serine residues and all cysteines are converted to oxazole and thiazole rings by the McbBCD complex. After formation of modified pre-McB (pro-McB), the leader peptide, which comprises the first N-terminal 26 amino acids, is cleaved off by the action of TldD/TldE protease, releasing mature McB (14). The McbE and McbF proteins are homologous to ABC-family transporters and are required for the export of McB from the producing cell. The product of mcbG gene, a member of the pentapeptide repeat family of proteins, presumably interacts with DNA gyrase, providing additional immunity to McB.
The major form of McB contains eight rings (four oxazoles and four thiazoles). Incompletely processed intermediates containing seven or six rings are also common (15). These compounds lack, correspondingly, one or two C-terminal oxazole rings but nevertheless possess significant antibacterial activity. Modification of pre-McB Gly-Ser-Cys and Gly-Cys-Ser tripeptides results in the formation of two fused 4,2-oxazole-thiazole and 4,2-thiazole-oxazole bis-heterocycles termed, respectively, the “A” and “B” sites. An additional Gly-Cys-Ser tripeptide is present between the A and B sites and defines the C site. Most McB molecules produced contain a single thiazole cycle at this site. A minor fraction of McB molecules produced contains nine heterocycles. In this compound, an additional serine residue, site C Ser52, is modified to yield a third bis-heterocycle. Such “over-modified” McB was reported to be ∼40% more active than the 8-cycle compound (15). Because the sequences of the B and C sites are identical, the clearly unequal efficiency of bis-heterocycle formation must reflect context dependence of the McB synthase function.
Rich biochemical evidence proves that McB promotes accumulation of covalent complexes of DNA gyrase with cleaved DNA leading to the formation of double-stranded DNA breaks, inhibition of DNA replication, and SOS response (16–18), thus acting similarly to clinically important quinolones that also target the gyrase. However, the details of the interactions of McB with the gyrase are still obscure (19). No systematic structure-activity analysis of McB was performed. Establishment of the structure-activity relationships in the McB molecule requires exact knowledge of the molecule covalent structure. The accepted primary structure of the major form of McB is represented on Fig. 1. In an effort to prepare defined McB intermediates suitable for structural analysis and fine biochemical and biophysical analyses of interactions between the antibiotic and gyrase or gyrase-DNA complexes, we performed systematic analysis of various McB forms. Here, we report that most McB produced by Escherichia coli cells harboring a plasmid-borne mcb operon is distinct from the structure shown in Fig. 1. The new structure opens ways for specific modification of McB by biochemical and biophysical probes and sheds light into the mechanism of oxazole and thiazole formation.
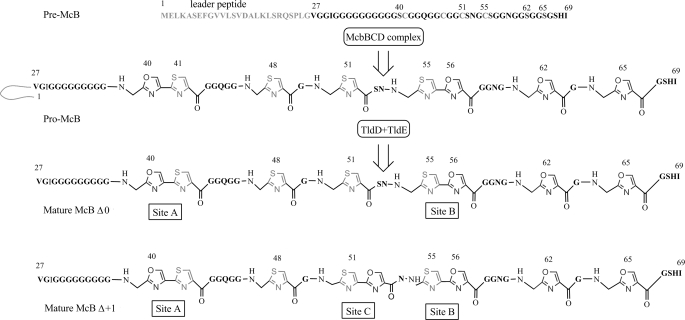
FIGURE 1.
Maturation and processing of McB. Pre-McB peptide, the product of the mcbA gene, is synthesized on the ribosomes. Eight heterocycles (four oxazoles and four thiazoles) are introduced by the McbBCD complex, generating pro-McB. Two McbA tripeptides are converted into fused heterocycles (sites A and B). The leader peptide is cleaved off by the TldD and TldE proteases generating mature McB, McB Δ0. An additional form of McB containing a bis-heterocycle in site C is referred to as McB Δ+1.
EXPERIMENTAL PROCEDURES
Strains
Strain ZK195 (BW25113 ΔsbmA) was obtained from the Keio collection (20). Strain MG1655 (F-λ- ilvG-rfb-50rph-1) harboring the gyrB W751R mutation (ML751) and the wild-type MG1655 strain were provided by Dr. Anthony Maxwell (John Innes Centre). Strain CSH50 λsfiA::lacZ (14) (a CSH50 λ lysogen carrying a transcriptional sfiA::lacZ fusion) was a gift of Dr. Van Melderen.
Plasmids
Plasmid pPY113 (21) harboring the entire mcb operon was provided by Dr. Roberto Kolter (Harvard Medical School).
Chemicals
Propionic anhydride and NHS-biotin were purchased from Sigma and dissolved in DMSO (Panreac). Ciprofloxacin was purchased from Sigma, dissolved in 0.1 m NaOH, and further diluted with water or 50% MeCN. Purified DNA gyrase (A and B subunits) was a gift of Dr. Anthony Maxwell (John Innes Centre). Microcin C was purified in our laboratory and diluted to 100 μm with 50%MeCN.
Preparation of Relaxed DNA Substrate
Supercoiled pBR322 was prepared by alkaline lysis using Qiagen Midi kit. Pure relaxed DNA substrate was prepared by treating supercoiled plasmid with exonuclease T5 (Epicenter Biotechnologies) and E. coli topoisomerase I (New England Biolabs) according to the manufacturer's instructions.
In Vitro DNA Cleavage Assays
DNA cleavage assays were carried using a method based on that described by Heddle et al. (16). 100 nm DNA gyrase was incubated with 7 nm relaxed pBR322 substrate for 90 min at 37 °C in the following reaction buffer: 35 mm Tris-HCl, pH 7.5, 24 mm KCl, 4 mm MgCl2, 2 mm DTT, 1.8 mm spermidine, 1 mm ATP, 6.5% (w/v) glycerol, 0.1 mg/ml albumin. McB “X” and “N” variants were added as DMSO solutions, and final concentration of DMSO was 3%. Equal amounts of DMSO were added to control reactions. Reactions were quenched with 0.2% SDS and 0.3 mg/ml proteinase K and further incubated at 37 °C for 30 min. An equal volume of chloroform and a half-volume of a gel loading buffer (40% sucrose, 100 mm Tris-HCl, pH 7.5, 1 mm EDTA, 2 mg/ml bromphenol blue) was added; next, reaction tubes were vortexed and centrifuged for 5 min at 13,000 rpm. The aqueous phase was loaded onto 0.9% agarose, TAE (40 mm Tris acetate, 1 mm EDTA) gel without ethidium bromide. The gel was run in TAE for 3 h at 5 V/cm and stained for 1 h with 10 μg/ml ethidium bromide then quickly washed with tap water.
McB Purification and HPLC Analysis
Starter 10-ml LB cultures of E. coli DH5α transformed with mcb plasmids were grown overnight at 37 °C and used to inoculate 0.5 liters of M63 supplemented with sodium succinate and appropriate antibiotics. Cultures were grown for 22 h at 37 °C. Cells were collected by centrifugation and boiled in 20 ml of 100 mm acetic acid, 1 mm EDTA for 10 min. The clarified supernatant containing crude McB was applied onto a SepPack C18 cartridge (ThermoScientific), which was extensively washed with 10% acetonitrile, 0.1% trifluoroacetic acid (TFA) to remove contaminants. McB-containing fraction was eluted in ∼2 ml of 25% AcCN, 0.1% TFA and vacuum-dried. The resulting pellet was dissolved in DMSO and applied onto HPLC XTerra C18 column (Waters) in 10% DMSO, 0.1% TFA. The column was equilibrated in 0.1% TFA. The bound material was eluted with a linear gradient of AcCN, 0.1% TFA (from 0 to 20% in 10 min, then from 20 to 27% in 20 min). McB was eluted between 18 and 25 min. Individual peaks were collected, vacuum-dried, and resuspended in 50% AcCN, 0.1% TFA.
MALDI-MS and MS/MS Analysis
Mass spectra were recorded on Ultraflex II MALDI-ToF-ToF mass spectrometer (Bruker Daltonik) equipped with an Nd laser (355 nm). The MH+ molecular ions were measured in reflector mode; the accuracy of monoisotopic mass peak measurement was 0.01%. Aliquots (1 μl) of the sample were mixed on a steel target with 0.5 μl of 2,5-dihydroxybenzoic acid (Aldrich) solution (20 mg/ml in 30% AcCN + 0.5% TFA), and the droplet was left to dry at room temperature. Fragment ion spectra were generated by laser-induced dissociation in Lift mode; the accuracy of mass peak measurement was 1 Da. Correspondence of the found masses to the microcin B peptides and to MS/MS peptide fragments was manually interpreted with the help of GPMAW 4.04 software (Lighthouse data). For direct MALDI analysis of cell components, McB-producing cells grown in 1 ml of M63 medium for 24 h were pelleted, and ∼1 μl of cell pellet was resuspended in 10 μl of 70% AcCN, 0.5% TFA and mixed with 10 μl of matrix solution (20 mg/ml 2,5-dihydroxybenzoic acid in 30% AcCN + 0.5% TFA). 1 μl of the resulting mix was dropped onto a steel target and dried at room temperature. The MH+ of whole proteins was acquired in linear mode; the accuracy of average mass peak measurement was 2 Da.
Chemical Modification
Compounds diluted to 100 μm with 50% AcCN, 0.1% TFA were mixed with 10 mm propionic anhydride or NHS-biotin in 0.1 m acetate buffer, pH 4.0, or 0.1 m phosphate buffer, pH 7.4, and incubated at 37 °C overnight (pH 4.0 reactions) or at 50 °C for 1 h (pH 7.4 reactions). For alkaline hydrolysis, a 0.5 volume of 20% NH3 water solution was added to the reactions, and incubation was continued for 0.5 h at 50 °C. Reactions were terminated by the addition of glacial acetic acid and loading on Zip-Tip C18 columns (Millipore). Reaction products were eluted from Zip-Tips in 50% AcCN, 0.1% TFA.
Determination of Antibiotic Activity and SOS Response
Concentration of purified McB X and N was measured according to Sinha Roy et al. (15). Samples then were diluted to 1–100 μm with 50% MeCN, and 2 μl drops were deposited on freshly made lawns of various E. coli strains. 100 μm ciprofloxacin diluted with 50% MeCN was used as the control. The lawns were prepared by overlaying M63 agar plates with 5 ml of 0.6% agar solution in distilled water containing 100 μl of overnight E. coli culture grown in M63 medium. Plates were incubated at room temperature overnight.
For the demonstration of SOS response, lawns of lysogenized CSH50 sfiA::lacZ cells (14) were prepared on the surface of MacConkey agar (Difco) as described above. 2-μl drops of 10 μm McB N, X, ciprofloxacin, and 100 μm microcin C were deposited on the lawns. Plates were incubated for 24 h at room temperature.
RESULTS
Discovery of an McB Fraction with Unusual MS-MS Fragmentation Pattern
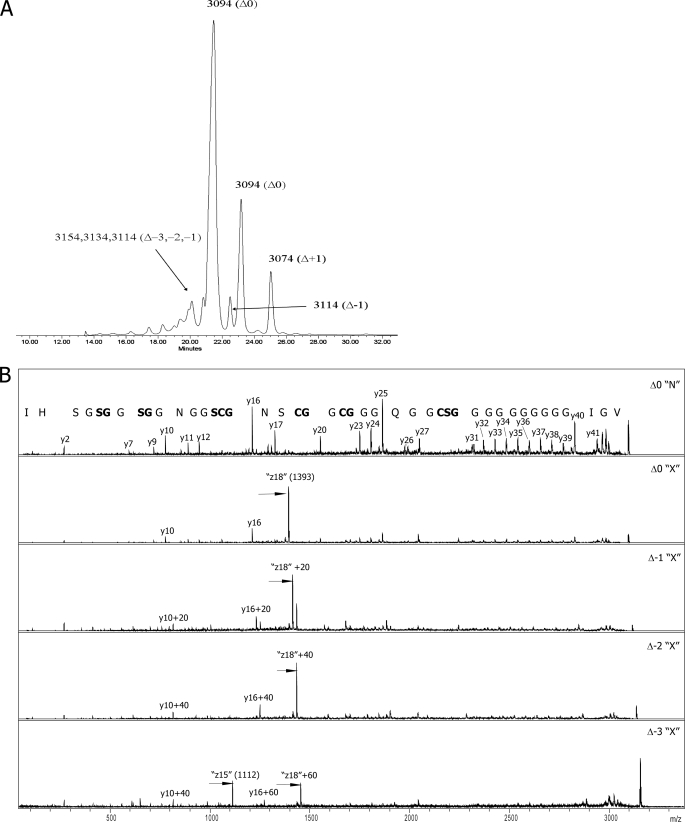
A typical result of the final step of McB purification, reverse-phase HPLC, is presented in Fig. 2A. As can be seen, three major peaks are detected. The profile was highly reproducible and was observed during purification of multiple independent McB samples. MALDI-MS analysis revealed that the first two HPLC peaks contained mass-ions with m/z = 3094 and thus correspond to fully matured 8-cycle compound shown in Fig. 1 and previously termed McB Δ0 (15). The third peak contained a 3074-Da compound corresponding to McB Δ+1 with a bis-heterocycle in site C. In addition, several minor poorly resolved peaks that eluted earlier than the major peaks contained mass-ions corresponding to not fully matured McB species (Δ-1, Δ-2, and Δ-3). Another minor peak that eluted between major peaks 2 and 3 also contained mass-ions corresponding to the Δ-1 mass. Overall, the profile of McB elution is very similar to that observed by Sinha Roy et al. (15). In their case, the material eluted in the first peak contained a mixture of Δ0, Δ-1, Δ-2, and Δ-3, and the second peak contained pure Δ0, whereas the third peak contained Δ+1. Sinha Roy (15) and others (22) concentrated their attention on McB forms from the second and third HPLC peaks.
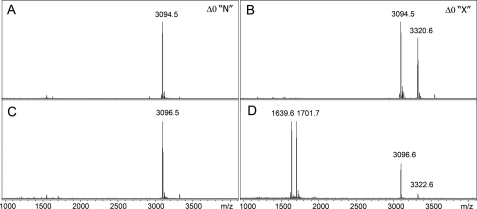
FIGURE 2.
Detection of a new form of McB. A, shown is a reverse-phase HPLC elution profile of McB at the final stage of purification. Numbers indicate m/z values of mass-ions detected in different HPLC peaks by MALDI-MS analysis. For each m/z value, the corresponding form of McB is identified (the nomenclature is that of Sinha Roy et al. (15)). B, shown are MS-MS spectra of McB Δ0 from HPLC peak 2 (top) and 1 (second from the top) and of indicated X-type McB maturation intermediates.
What is unexpected about the HPLC elution profile shown in Fig. 2A is that compound Δ0 (and Δ-1) elutes as two distinct HPLC peaks. To determine the reason(s) for such an unusual chromatographic behavior, material from various HPLC peaks was analyzed by MS-MS. MS-MS analysis unequivocally identified the material from peak 2 as McB Δ0 (Fig. 1). In agreement with earlier data, material from peak 2 fragmented well at peptide bonds and formed the b- and y-ion series (Fig. 2B, top). Ions from the latter series were better represented, likely due to protonation of His68. The y-ion series was complete except for the absence of fragmentation products of peptide bonds incorporated into the heterocycles as well as conjugated peptide bonds lying directly downstream from some of the cyclized residues. These bonds were previously observed to resist fragmentation (23). Also in agreement with published data, material from peak 3 corresponded to the Δ+1 species (data not shown).
The fragmentation spectrum of material from peak 1, which was the most abundant, is shown in Fig. 2B, bottom. The overall pattern of fragmentation matches that of McB Δ0 with two significant differences. First, the m/z = 1324 y-ion present in peak 2 fragmentation spectrum and resulting from cleavage between Ser52 and Asn53 was absent from peak 1 fragmentation spectrum. Second, the peak 1 fragmentation spectrum contained a new m/z = 1393 peak of very high intensity. The abnormally high intensity of this peak was probably responsible for suppression of other mass peaks in fragmentation spectrum of material from HPLC peak 1. The m/z = 1393 peak could not be assigned to any product of conventional fragmentation. Its closest match was a z-ion resulting from breakage of the N-Cα (Ser52) bond. However, this z-ion should have a mass of 1394 [M]+• or 1395 [MH]+. Despite a mass mismatch, we will refer to this peak as a product of N-Cα (Ser52) bond fragmentation, as results presented below confirm this assignment. A possible N-terminal paired ion arising from fragmentation at Ser52 with an m/z value of 1702 and another possibly related ion with m/z = 1696 was also observed in MS-MS spectrum of peak 1 material.
The preferred fragmentation site indicates that McB in HPLC peak 1 contains a significantly more labile bond that is absent from the conventional McB Δ0 structure shown in Fig. 1. For the purposes of clarity, we will refer to the novel form of McB and its maturation intermediates as “X-type” compounds, whereas the conventional form and its maturation intermediates will be referred to as “N-type,” for “normal.”
Mass-ions corresponding to McB Δ-1 variants with distinct chromatographic behavior were also subjected to MS-MS. In both cases, the C-terminal oxazole heterocycle was absent as evidenced by a 20-mass unit shift of the y7 mass-ion (Fig. 2B and data not shown). The later eluting Δ-1 material had a fragmentation pattern expected from normal McB structure (data not shown). The earlier eluting material displayed the X-type fragmentation pattern. The only difference from Δ0 X-type fragmentation was a shift of the prominent 1393 mass-ion by 20 mass units (m/z = 1413), reflecting the lack of the terminal heterocycle (Fig. 2C, compare the top spectrum of X-type McB Δ0 and the lower spectrum of McB Δ-1). In contrast, the likely paired ions (m/z = 1702 and 1696) remained unchanged, confirming their N-terminal nature.
The early eluting Δ-2 and Δ-3 material was also analyzed and found to lack, respectively, two or three terminal oxazole cycles, as evident from 20 (Δ-2) and 40 (Δ-3) mass unit shifts of the y10 ion (Fig. 2C, bottom two spectra). Both Δ-2 and Δ-3 material fragmented generating X-type-specific peaks shifted by, respectively, 40 and 60 mass units from the m/z = 1393 value seen in the case of X-type fragmentation of McB Δ0 (Fig. 2C). Analysis of fragmentation pattern of Δ-3 form revealed an additional prominent m/z = 1112 fragment that can be assigned to an X-type cleavage N-terminal to Ser56 (expected z15 value of 1113 [M]+• and 1114 [MH]+). A corresponding paired ion (m/z = 2043) was also detected.
Ser56 is normally part of site B bis-heterocycle but remains uncyclized in Δ-3. Instead, a single thiazole cycle is present. Therefore, the structure in site B (a thiazole cycle followed by a serine residue) in Δ-3 is identical to site C structure with uncyclized Ser52. It, therefore, appears that during modification of Gly-Cys-Ser tripeptides, the McB synthase produces a structure that is distinct from those known to be present in conventional forms of McB. The new McB form constitutes at least 60% of McB molecules produced at standard conditions. During MS-MS, the new form fragments with very high efficiency generating fragments that do not match expected fragmentation products of peptide bonds.
Chemical Modifications of McB
In principle, isomerization of the peptide bond N-terminal to serine residue in site C or site B Gly-Cys-Ser tripeptides to a depsipeptide bond could generate MS-MS fragments matching those experimentally observed in spectra of X-type fragmenting McB forms. Furthermore, because the depsipeptide bond is an ester bond, it shall fragment more easily than the normal peptide (amide) bond, accounting for the high intensity of X-type MS-MS signal. In what follows we present data that confirm this hypothesis.
The appearance of depsipeptide bonds in X-type McB shall be accompanied by the formation of primary amino groups inside the polypeptide chain. The presence of such groups could be revealed with chemical modification. Two types of chemical reagents were used for modification of McB Δ0 X and N forms. The first reagent was propionic anhydride, which can react with primary amino and sulfhydryl groups and, to a lesser extent, with hydroxyl groups. The second reagent used, N-hydroxysuccinimide biotin ester, is a more selective N-acylation reagent. Because depsipeptide bonds are pH-labile, chemical modification reactions were carried out at pH 4.0 and at pH 7.4. The former condition is less favorable for acylation but should prevent back-isomerization of the putative depsipeptide bond to an amide bond.
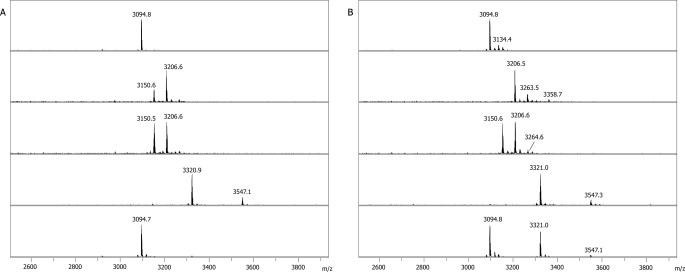
In the case of the N-type compound, we expected to obtain mono-modified derivatives with acylated N terminus. This expectation was only partially fulfilled. With propionic anhydride, a mixture of mono- and diacyl derivatives was revealed at both pH conditions (Fig. 3A), with diacyl derivative being more abundant at high pH. With N-hydroxysuccinimide biotin ester, mostly mono-modified derivative was observed at pH 7.4 with trace amounts of double-modified species. No N-hydroxysuccinimide biotin ester modification was detected at pH 4.0. For Δ+1, which was used as a control, only mono-modified derivatives were detected (data not shown), suggesting that uncyclized Ser52 is important for double modification with both reagents. Δ+1 as well as all mono-modified derivatives of N-type Δ0 appeared to be modified exclusively at the N terminus, as judged by MS-MS analysis (data not shown). Double-modified derivatives of N-type McB Δ0 were modified at the N terminus and at Ser52 (data not shown). The latter finding is somewhat unexpected as it reveals unusually high reactivity of Ser52.
FIGURE 3.
Modification of McB Δ0 with propionic anhydride and NHS-biotin. A, shown are MALDI-MS spectra of (from top to bottom) unmodified N-type McB Δ0, N-type McB Δ0 modified with propionic anhydride at pH 7.4 or pH 4.0, or NHS-biotin at pH 7.4 or pH 4.0 are shown. B, MALDI-MS spectra of (from top to bottom) unmodified X-type McB Δ0, X-type McB Δ0 modified with propionic anhydride at pH 7.4 or pH 4.0, or NHS-biotin at pH 7.4 or pH 4.0 are shown.
Modification of X-type McB Δ0 was performed next. With propionic anhydride, di- and triacylated products (but no monoacylated products) were detected at pH 7.4 (Fig. 3B). At lower pH, mono- and diacylated products were obtained with the overall MS profile of modification products appearing identical to that seen with the N-type compound at these conditions (compare the corresponding panels in Figs. 3, A and B). Diacylated X-type Δ+0 derivatives contained modifications at the N terminus and at Ser52. Triacylated species contained an additional modification at Ser67. In addition, Ser62 and Ser65 were modified in McB Δ-1 and Δ-2 (data not shown). The result confirms that McB serine side chains can be subject to acylation with propionic anhydride at our conditions, albeit with different efficiency.
Modification of X-type Δ+0 with N-hydroxysuccinimide biotin ester at pH 7.4 yielded mono- and diacylated products, whereas modification at pH 4.0 resulted in a monoacylated compound, with a significant fraction of the starting compound remaining unmodified (Fig. 3B, bottom panel).
Overall, comparison of the data presented in Figs. 3, A and B, reveals that on average, the X form of McB is much more reactive than the N form. Moreover, comparison of bottom panels in Figs. 3, A and B, reveals that at pH 4.0 a mono-modified X-type McB biotin derivative containing a modification that is specific for X-type McB can be obtained.
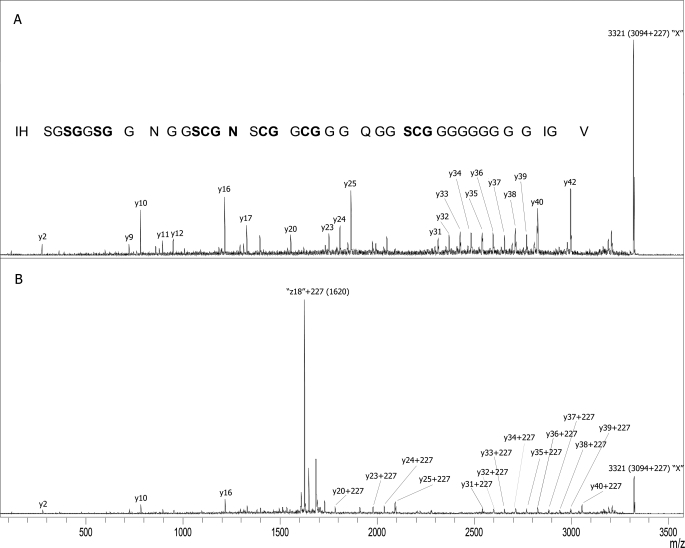
MS-MS spectra of monoacylated products of X-type McB modified with N-hydroxysuccinimide biotin ester at both pH values are shown in Fig. 4. Unexpectedly, a fragmentation pattern corresponding to N-terminal-modified N-type McB was observed with the compound modified at pH 7.4 (Fig. 4A), suggesting that an X to N conversion took place during modification. A derivative modified at pH 4.0 exhibited the X-type fragmentation pattern (Fig. 4B). However, the unusual z-ion characteristic for X-type fragmentation was shifted by 227 mass units, reflecting acylation of Ser52 with biotin. Because no modification of Ser52 hydroxyl was observed at these conditions for N-type compound (Fig. 3), the result implies the existence of a highly reactive group at this position of X-type McB. At the conditions of the experiment, this group was more reactive than the N-terminal amino group.
FIGURE 4.
MS-MS analysis of monobiotinylated X-type McB Δ0. A, shown is an MS-MS spectrum of X-type McB Δ0 modified at pH 7.4. B, shown is an MS-MS spectrum of X-type McB Δ0 modified at pH 4.0.
Alkaline Hydrolysis of Ser52-modified McB Reveals the Presence of an Ester Bond in X-type McB
The presence of highly reactive chemical group at Ser52 is consistent with an idea of depsipeptide bond formation in X-type McB. N-O-acyl rearrangement that takes place during depsipeptide bond formation should result in accumulation of a compound with an ester bond between Cys51 and Ser52. Such a bond shall be susceptible to mild alkaline hydrolysis. Completed pH 4.0 modification reactions of X- and N-type McB Δ0 with N-hydroxysuccinimide biotin ester (containing unmodified and mono-modified products, see Fig. 3, A and B) were incubated in 10% aqueous NH3, and reaction products were visualized by MALDI-MS (Fig. 5). As can be seen, unreacted N-type McB was not hydrolyzed, although a two-mass unit change indicative of deamidation of asparagine residues was observed (compare Fig. 5, A with C; deamidation was confirmed by MS-MS analysis; data not shown). Such deamidation was described earlier by Parks et al. (19). A more complex pattern was observed after hydrolysis of X-type McB modification reaction products. Residual unmodified X-type McB remained stable but became deamidated (compare Fig. 5, B with D) and, according to MS-MS analysis, converted to N-type (data not shown). The relative intensity of the peak corresponding to mono-modified X-type McB derivative was dramatically reduced, and two new major peaks were observed. One peak (m/z = 1701.7) corresponds to C-terminal-amidated N-terminal McB peptide generated by backbone hydrolysis event after Cys51. The other mass peak (m/z = 1639.6) corresponds to the C-terminal hydrolysis product biotinylated at Ser52. Analogous results were obtained upon alkaline hydrolysis of X-type McB Δ0 modified with propionic anhydride (data not shown).
FIGURE 5.
Mild alkaline hydrolysis of chemically modified McB. N-type (left side) or X-type (right side) McB Δ0 was modified with NCS biotin at pH 4.0. MALDI-MS analysis of completed reactions is shown in panels A and B (see also Fig. 3). Reactions were next subjected to mild alkaline hydrolysis and products analyzed by MALDI-MS (panels C and D).
The results strongly suggest that a mono-modified derivative of X-type McB indeed contains a backbone ester bond connecting residues 51 and 52. Such a bond is not present in N-type McB. It should be noted that efficient hydrolysis of mono-modified X-type McB is not due to modification per se, as none of N-type McB modification products was hydrolyzed (data not shown).
Based on all the data presented above, we conclude that X-type McB, a major form of McB in preparations purified using the standard procedure (15), is distinct from the N-type form whose structure is presented in Fig. 1 and contains a backbone ester bond that is a product of N → O acyl shift of Ser52. As a consequence of the isomerization reaction, X-type McB also contains a primary amino group instead of Ser52 side chain.
Biological Activity of X-type McB and X- to N-type Conversion
A question arises of whether X-type McB is a true intermediate and/or end product of the McB maturation process or is merely an artifact of the purification procedure. Several lines of evidence argue that compound X is produced in situ. First, express MALDI-MS analysis of McB-producing cells treated with 70% acetonitrile and MS matrix solution and immediately deposited on an MS plate (see “Experimental Procedures”) revealed an m/z = 3094 peak corresponding to McB Δ0 (supplemental Fig. S1A). Based on MS-MS analysis, this peak clearly contained X-type fragmenting material (supplemental Fig. S1B). On the other hand, N-type McB failed to convert to X-type McB at conditions used during the standard purification procedure. N-type McB Δ0 was “mock-purified” by 20 min of boiling in 0.1 m acetic acid followed by zip-tip purification and HPLC/MALDI-MS analysis. No evidence of N → X conversion was revealed (supplemental Fig. S2 and data not shown).
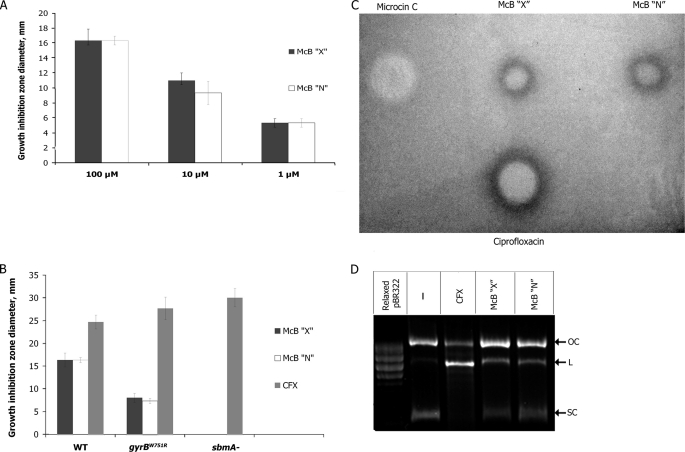
To estimate antibacterial activity of X- and N-type McB and demonstrate that X- and N-type McB have the same target, the following experiments were performed. Using a standard spot-on-lawn plate assay, we demonstrated (Fig. 6A) that growth inhibition zones of equal size were formed around drops of solutions containing matching concentrations of pure X- and N-type McB on lawns of wild-type E. coli, indicating that the two compounds are equally active. As can be seen from Fig. 6B, growth inhibition zones for both compounds decreased to the same extent on lawns of E. coli cells carrying a gyrB mutation that renders the DNA gyrase partially resistant to McB (24). Cells lacking sbmA, a transporter responsible for McB uptake (25), were fully resistant to both compounds (Fig. 6B). As control, we used ciprofloxacin, an unrelated antibiotic that also targets the gyrase but interacts with a different site and does not require SbmA for cell entry (26). As expected, ciprofloxacin was equally active against wild-type and mutant cells (Fig. 6B).
FIGURE 6.
X-type McB is biologically active. A, drops of the indicated concentrations of X-type and N-type McB Δ0 were deposited on lawns of McB-sensitive wild-type E. coli cells. The plates were incubated at room temperature overnight, and diameters of growth inhibition zones were recorded. Average values obtained in at least three independent experiments are presented. B, the experiment was performed as in A but with the indicated tester cell lawns. The concentrations of both forms of McB and ciprofloxacin in drops deposited on cell lawns were 10 μm. C, drops of 10 μm solutions of both forms of McB and ciprofloxacin and 100 μm microcin C were deposited on lawns of cells containing LexA-dependent promoter sfiA fused to the lacZ gene grown on McConkey agar. Plates were photographed after overnight growth. D, E. coli DNA gyrase was used to catalyze supercoiling of relaxed pBR322 plasmid in the presence or in the absence of 33 μm of McB or 5 μm ciprofloxacin. The results of separation or reaction products in an agarose gel are shown. SC, L, and OC denote, respectively, supercoiled, linear, and open circular plasmid DNA.
The double-stranded breaks in DNA induced by McB and ciprofloxacin cause a SOS response that is activated when repression by transcriptional repressor LexA is removed (26–28). Using E. coli cells containing a genomic fusion of LexA-dependent promoter sfiA with the lacZ gene (14), we determined the ability of both forms of McB to induce SOS response by following the color around inhibition zones on tester cells lawns grown on McConkey indicator medium. As can be seen from Fig. 6C, both McB forms and ciprofloxacin, but not microcin C, an inhibitor of an aminoacyl-tRNA transferase (1, 2), caused development of intense red color at the outer edges of inhibition zones, indicating that cells receiving sublethal doses of X- and N-type McB and ciprofloxacin underwent SOS response.
The plate assays involve an overnight incubation of McB at the surface of agar plates, pH 7.0, kept at room temperature, and it can be argued that an X to N-type conversion takes place during the bioactivity assay. In fact, the results of chemical modification of X-type McB at pH 7.4 suggest that this is a distinct possibility (see above). HPLC analysis of X-type McB Δ0 incubated overnight at pH 7.4 revealed the appearance of a chromatographic peak with retention time characteristic for N-type McB Δ0 (supplemental Fig. S3, compare the top and bottom panels). We estimate that less than 20% of input X-type McB Δ0 was converted to N-type McB Δ0 during the incubation time. However, accumulation of these relatively small amounts of N-type McB Δ0 cannot account for biological activity of our X-type McB samples, as equal specific activity of N- and X-type McB was observed at subsaturating amounts of both inhibitors (Fig. 6A).
To demonstrate that both types of McB have the same target, in vitro effects of X-type and N-type McB and control ciprofloxacin on DNA gyrase-catalyzed plasmid supercoiling reactions were investigated (Fig. 6D). The addition of ciprofloxacin and both forms of McB caused the expected (16) accumulation of the linear plasmid form (labeled L in Fig. 6D) arising due to stabilization of enzyme complex with cleaved DNA in the presence of drugs (compare the second lane with the third, fourth, and fifth lanes 3, 4, and 5). Both forms of McB demonstrated equal efficiency in stabilizing cleavage complexes. We, therefore, conclude that X-type McB is biologically active and targets the gyrase.
DISCUSSION
In this work we reveal that most of the mature McB Δ0 produced has a structure that is distinct from that established earlier and shown in Fig. 1. The new structure is characterized by the presence of depsipeptide bond between McB residues 51 and 52. The presence of depsipeptide bond follows from 1) results of mild alkaline hydrolysis and 2) chemical modification results that reveal the presence of highly reactive amino group at the place of Ser52. Furthermore, the presence of depsipeptide bond explains the discrepancy of the dominating MS-MS X-type ion with a value calculated based on standard McB structure. Preferential cleaving of the ester bond during MS-MS fragmentation should yield, in the case of X-type McB Δ0, the observed 1393-Da z-ion rather than the 1395 ion expected for fragmentation of the normal peptide bond.
A depsipeptide bond is also present in McB maturation intermediates containing a single thiazole cycle in site B. Evidently, formation of depsipeptide bond occurs when the McbBCD synthase encounters a Gly-Cys-Ser tripeptide and completes the synthesis of N-terminal thiazole (Fig. 7). In the case of site B tripeptide, the McbBCD synthase proceeds to synthesize the fused oxazole cycle with high efficiency followed by the formation of more C-terminal single oxazole cycles. In the case of site C tripeptide, the synthesis of fused oxazole (leading to the formation of McB +Δ1) proceeds with low efficiency. Instead, a competing reaction of N-O-acyl rearrangement takes place, resulting in the accumulation of a compound with an ester bond between Cys51 and Ser52. The choice of pathways during modification of site C and B substrate tripeptides is obviously context dependent, i.e. determined by residues outside the tripeptide substrate. Site-specific mutagenesis of McbA residues around the site B and C tripeptides will be required to understand the choice of the reaction pathway.
FIGURE 7.
A possible mechanism of depsipeptide bond formation in McB. See Discussion for details.
The detection of X-type fragmenting compounds during direct mass spectrometric analysis of McB-producing cells argues that the depsipeptide bond containing McB is physiological. In vitro, the depsipeptide bond-containing form of McB can be spontaneously converted to normal peptide bond containing McB shown in Fig. 1 upon incubation at physiological pH. N-O rearrangements have been reported in serine-rich proteins but only after prolonged boiling with concentrated sulfuric acid (29). However, we failed to observe conversion of N-type McB to X-type under conditions simulating the process of McB purification. Thus, it is possible that most, perhaps all of McB Δ0 produced in situ, contains a depsipeptide bond, with amide peptide bond-containing forms accumulating in the cytoplasm or in the cultured medium (pH ≥ 7.0) after the modification has been completed.
The N-O-acyl rearrangement revealed by our work could be a byproduct of cyclization at Ser52 that normally yields McB Δ+1. The likely mechanism through which depsipeptide bond is formed is shown in Fig. 7. The figure is based on a pioneering work of Walsh and co-workers (11), who noticed a similarity between the McB maturation process and protein splicing and autoproteolysis reactions (30–32). According to the proposed mechanism, initial cyclization involving the serine side chain allows two alternative subsequent protonation events. O protonation should lead to oxazoline formation, whereas alternative N protonation should lead to depsipeptide bond formation. In the case of protein autoproteolysis, the resulting ester bond is cleaved, whereas in the case of McB, the resulting form is stable, possibly due to the presence of upstream thiazole cycle. Detection of depsipeptide bond form of McB provides experimental support to fundamental similarity of heterocycle formation in McB and autoproteolysis.
Despite many years of study, no direct evidence of McB interaction with its target, the DNA gyrase, has been obtained, and the site of McB binding on the gyrase is not known. The discovery of a new form of McB provides a useful handle to study the interactions of McB with its target as well as conformational changes within McB, as availability of sites for site-specific chemical modification at the N terminus and within the molecule at amino acid position 52 should allow to selectively incorporate cross-linkable and fluorescent labels at these positions of McB.
Supplementary Material
This work was supported in part by the Russian Academy of Sciences Molecular and Cell Biology Program grant and Federal Program “Scientific and scientific-pedagogical personnel of innovative Russia 2009–2013,” state contract 02.740.11.0771 (to K. S.), Russian Foundation for Basic Research grant 09-04-00293-a, and Federal Program “Scientific and sicentific-pedagogical personnel of innovative Russia 2009–2013,” state contract P1068 (to I. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- McB
- microcin B17.
REFERENCES
- 1. Duquesne S., Petit V., Peduzzi J., Rebuffat S. (2007) J. Mol. Microbiol. Biotechnol. 13, 200–209 [DOI] [PubMed] [Google Scholar]
- 2. Severinov K., Semenova E., Kazakov A., Kazakov T., Gelfand M. S. (2007) Mol. Microbiol. 65, 1380–1394 [DOI] [PubMed] [Google Scholar]
- 3. Yorgey P., Lee J., Kördel J., Vivas E., Warner P., Jebaratnam D., Kolter R. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 4519–4523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. McIntosh J. A., Schmidt E. W. (2010) Chembiochem 11, 1413–1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mitchell D. A., Lee S. W., Pence M. A., Markley A. L., Limm J. D., Nizet V., Dixon J. E. (2009) J. Biol. Chem. 284, 13004–13012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gonzalez D. J., Lee S. W., Hensler M. E., Markley A. L., Dahesh S., Mitchell D. A., Bandeira N., Nizet V., Dixon J. E., Dorrestein P. C. (2010) J. Biol. Chem. 285, 28220–28228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Onaka H., Nakaho M., Hayashi K., Igarashi Y., Furumai T. (2005) Microbiology 151, 3923–3933 [DOI] [PubMed] [Google Scholar]
- 8. Walsh C. T., Acker M. G., Bowers A. A. (2010) J. Biol. Chem. 285, 27525–27531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Morris R. P., Leeds J. A., Naegeli H. U., Oberer L., Memmert K., Weber E., LaMarche M. J., Parker C. N., Burrer N., Esterow S., Hein A. E., Schmitt E. K., Krastel P. (2009) J. Am. Chem. Soc. 131, 5946–5955 [DOI] [PubMed] [Google Scholar]
- 10. Wieland Brown L. C., Acker M. G., Clardy J., Walsh C. T., Fischbach M. A. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 2549–2553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li Y. M., Milne J. C., Madison L. L., Kolter R., Walsh C. T. (1996) Science 274, 1188–1193 [DOI] [PubMed] [Google Scholar]
- 12. Milne J. C., Roy R. S., Eliot A. C., Kelleher N. L., Wokhlu A., Nickels B., Walsh C. T. (1999) Biochemistry 38, 4768–4781 [DOI] [PubMed] [Google Scholar]
- 13. Kelleher N. L., Hendrickson C. L., Walsh C. T. (1999) Biochemistry 38, 15623–15630 [DOI] [PubMed] [Google Scholar]
- 14. Allali N., Afif H., Couturier M., Van Melderen L. (2002) J. Bacteriol. 184, 3224–3231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sinha Roy R., Kelleher N. L., Milne J. C., Walsh C. T. (1999) Chem. Biol. 6, 305–318 [DOI] [PubMed] [Google Scholar]
- 16. Heddle J. G., Blance S. J., Zamble D. B., Hollfelder F., Miller D. A., Wentzell L. M., Walsh C. T., Maxwell A. (2001) J. Mol. Biol. 307, 1223–1234 [DOI] [PubMed] [Google Scholar]
- 17. Pierrat O. A., Maxwell A. (2003) J. Biol. Chem. 278, 35016–35023 [DOI] [PubMed] [Google Scholar]
- 18. Pierrat O. A., Maxwell A. (2005) Biochemistry 44, 4204–4215 [DOI] [PubMed] [Google Scholar]
- 19. Parks W. M., Bottrill A. R., Pierrat O. A., Durrant M. C., Maxwell A. (2007) Biochimie 89, 500–507 [DOI] [PubMed] [Google Scholar]
- 20. Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K. A., Tomita M., Wanner B. L., Mori H. (2006) Mol. Syst. Biol. 2, 2006–2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Genilloud O., Moreno F., Kolter R. (1989) J. Bacteriol. 171, 1126–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zamble D. B., Miller D. A., Heddle J. G., Maxwell A., Walsh C. T., Hollfelder F. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 7712–7717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kelleher N. L., Belshaw P. J., Walsh C. T. (1998) J. Am. Chem. Soc. 120, 9716–9717 [Google Scholar]
- 24. Vizán J. L., Hernández-Chico C., del Castillo I., Moreno F. (1991) EMBO J. 10, 467–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Laviña M., Pugsley A. P., Moreno F. (1986) J. Gen. Microbiol. 132, 1685–1693 [DOI] [PubMed] [Google Scholar]
- 26. Drlica K., Hiasa H., Kerns R., Malik M., Mustaev A., Zhao X. (2009) Curr. Top. Med. Chem. 9, 981–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Herrero M., Moreno F. (1986) J. Gen. Microbiol. 132, 393–402 [DOI] [PubMed] [Google Scholar]
- 28. Couturier M., Bahassi el-M., Van Melderen L. (1998) Trends Microbiol. 6, 269–275 [DOI] [PubMed] [Google Scholar]
- 29. Elliott D. F. (1952) Biochem. J. 50, 542–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Perler F. B., Xu M. Q., Paulus H. (1997) Curr. Opin. Chem. Biol. 1, 292–299 [DOI] [PubMed] [Google Scholar]
- 31. Perler F. B. (1998) Cell 92, 1–4 [DOI] [PubMed] [Google Scholar]
- 32. Saleh L., Perler F. B. (2006) Chem. Rec. 6, 183–193 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.