Abstract
Maintaining physical connections between the nucleus and the cytoskeleton is important for many cellular processes that require coordinated movement and positioning of the nucleus. Nucleo-cytoskeletal coupling is also necessary to transmit extracellular mechanical stimuli across the cytoskeleton to the nucleus, where they may initiate mechanotransduction events. The LINC (Linker of Nucleoskeleton and Cytoskeleton) complex, formed by the interaction of nesprins and SUN proteins at the nuclear envelope, can bind to nuclear and cytoskeletal elements; however, its functional importance in transmitting intracellular forces has never been directly tested. This question is particularly relevant since recent findings have linked nesprin mutations to muscular dystrophy and dilated cardiomyopathy. Using biophysical assays to assess intracellular force transmission and associated cellular functions, we identified the LINC complex as a critical component for nucleo-cytoskeletal force transmission. Disruption of the LINC complex caused impaired propagation of intracellular forces and disturbed organization of the perinuclear actin and intermediate filament networks. Although mechanically induced activation of mechanosensitive genes was normal (suggesting that nuclear deformation is not required for mechanotransduction signaling) cells exhibited other severe functional defects after LINC complex disruption; nuclear positioning and cell polarization were impaired in migrating cells and in cells plated on micropatterned substrates, and cell migration speed and persistence time were significantly reduced. Taken together, our findings suggest that the LINC complex is critical for nucleo-cytoskeletal force transmission and that LINC complex disruption can result in defects in cellular structure and function that may contribute to the development of muscular dystrophies and cardiomyopathies.
Keywords: Cytoskeleton, Muscular Dystrophy, Nuclear Membrane, Nuclear Structure, Nucleus, Intracellular Force Transmission, LINC Complex, Nesprin, Nucleo-cytoskeletal Coupling, SUN Proteins
Introduction
A stable connection between the nucleus and cytoskeleton is required for a wide range of physiological functions such as cell migration or nuclear positioning. Two recently discovered major molecular components involved in nucleo-cytoskeletal coupling are nesprin and SUN proteins, nuclear envelope transmembrane protein families that form a bridge across the nuclear envelope. SUN1 and SUN2 are retained at the inner nuclear membrane by their interaction with lamins, nuclear pore complex proteins, and the nuclear interior, whereas their conserved C-terminal SUN domain extends into the perinuclear space (1–3). Here, they interact with the highly conserved C-terminal KASH domain of nesprins located at the nuclear envelope. Four nesprin genes have been identified to date, many of them containing diverse isoforms as a result of alternate initiation and splicing sites. The largest isoforms of nesprins-1 and -2 contain an N-terminal actin-binding domain, enabling them to interact with cytoplasmic actin filaments (4, 5). Through spectrin-repeat-mediated interactions with kinesin and/or dynein subunits, nesprins-1 and -2 can also connect to microtubules (6–8). Nesprin-3 can bind to intermediate filaments via plectin (6), and nesprin-4, which is only expressed in secretory epithelial cells, is a kinesin-binding protein that connects the nucleus to microtubules (7). Thus, SUN proteins and nesprins can form a physical connection between the nuclear interior and the cytoskeleton that is referred to as the LINC2 complex (Linker of Nucleoskeleton and Cytoskeleton)(1).
Previous studies suggest that the LINC complex could be well suited to transmit forces between the nucleus and the cytoskeleton; however, it remains to be determined if the interaction between SUN and nesprin proteins is essential for intracellular force transmission in mammalian cells or if other nuclear envelope proteins are sufficient to carry out this function. This question is particularly relevant in light of recent reports that nesprin mutations can give rise to a variety of diseases, including Emery-Dreifuss muscular dystrophy and some forms of dilated cardiomyopathy (7, 8). In addition, it is now well established that defects in intracellular force transmission, for example by mutations in the genes encoding dystrophin (10, 11), desmin (9), or myosin heavy chain (16) can cause muscular dystrophy and dilated cardiomyopathy. Therefore, we decided to explore whether LINC complex disruption by functional loss of nesprins (or SUN proteins) could also result in disturbed intracellular force transmission and modulate critical cellular functions. We applied custom-developed biophysical assays to assess intracellular force transmission and related cellular functions in cells after disruption of the LINC complex by dominant negative nesprin and SUN protein constructs. LINC complex disruption resulted in defects in cell polarization and migration, disturbed cytoskeletal organization, and impaired force transmission between the cytoskeleton and the nuclear interior. Our data provide direct evidence for a critical role of the LINC complex in nucleo-cytoskeletal force transmission and suggest that mutations that disrupt the LINC complex could have severe effects on intracellular force transmission and cytoskeleton-mediated cellular functions, thereby contributing to the development of muscular dystrophy and dilated cardiomyopathy.
EXPERIMENTAL PROCEDURES
Cell Culture
Immortalized mouse embryonic fibroblasts (MEFs) were cultured in Dulbecco's Modified Eagle's Medium (DMEM; Invitrogen), high glucose (Invitrogen) supplemented with 10% fetal bovine serum (Aleken Biologicals), and 5% penicillin/streptomycin (Sigma); primary human skin fibroblasts (HG090; Progeria Research Foundation) were grown in Eagle's Minimal Essential Medium (Lonza) supplemented with 15% fetal bovine serum (Aleken Biologicals), 5% penicillin/streptomycin (Sigma), and 5% l-glutamine (Invitrogen).
Dominant Negative Nesprin KASH (DN KASH) Plasmid Construction and Transduction
To generate DN KASH fluorescently labeled with an N-terminal mCherry domain, mCherry was amplified from pCDH-EF1-MCS1-puro-mCherry (kindly provided by Dr. P. Patwari (10)) by PCR (see supplemental data for primer sequences) and then subsequently subcloned into pcDNA4/His-Max vector (Invitrogen) to create mCherry- pcDNA4/His-Max. The KASH domain from GFP-mouse nesprin-1α (11) was amplified by PCR (see supplemental data for primer sequences) and inserted downstream of the mCherry sequence into the mCherry- pcDNA4/His-Max plasmid. mCherry-Nesprin-1αKASH was subcloned from pcDNA4/His-Max mCherry-Nesprin-1αKASH, into pCDH-EF1-MCS1-puro vector (System Biosciences) to create pCDH-EF1-MCS1-puro-mCherry-Nesprin-1αKASH. The resulting plasmid is referred to as DN KASH in this work. As a mock control (mCherry alone), pCDH-EF1-MCS1-puro-mCherry was used and is referred to as mCherry control in this manuscript. As an additional control, EGFP-nesprin-2αext was used. This construct, based on wild-type nesprin-2α, has a modified KASH domain containing additional C-terminal amino acids from the expression vector. It correctly localizes to the nuclear envelope and endoplasmic reticulum but cannot interact with SUN proteins in the perinuclear space (12). We also performed experiments with a modified form of nesprin-2 giant (which is too large for ectopic expression), referred to as mini-nesprin-2G. This GFP-tagged chimeric construct, kindly provided by Dr. Gundersen's laboratory (13), consists of the N-terminal actin-binding domain with two adjacent spectrin repeats fused to the C-terminal KASH domain with two spectrin repeats preceding it, and was recently shown to rescue nuclear positioning in nesprin-depleted cells (14).
The DN KASH construct was overexpressed in MEFs and HG090 primary human skin fibroblasts by transduction with a vector based on replication-deficient human immunodeficiency virus. Lentiviral particles were produced by cotransfecting the DN KASH or mCherry control plasmids with packaging gag-pol and VSVG-envelope plasmids 2.5:1:1 ratio into 293TN cells (ATCC) using Lipofectin transfection reagent (Invitrogen). Virus-containing medium was collected 2–7 days after transfection, spun at 3,000 rpm for 10 min, 0.45-μm filtered, and applied to ∼1 × 105 cells plated the previous day. Cells were transduced with the virus every 10 h, for 30 h to increase the percentage of gene-modified cells. Experiments were performed on cells identified by mCherry expression. Nesprin-2αext and mini-nesprin-2G were transiently transfected into MEFs using Lipofectamine PLUS (Invitrogen), following the manufacturer's instructions.
Dominant Negative SUN1 Luminal Domain (DN SUN1L) Plasmid Construction and Transfection
The 5′-end of SS-HA-Sun1L-KDEL was amplified from pcDNA3.1SS-HA-Sun1L-KDEL (1) by PCR (see supplemental data for primer sequences). The SS-HA-Sun1L-KDEL sequence was ligated into the pCDH-CMV-MCS-EF1-copGFP-T2A-puro vector (System Biosciences). The resulting plasmid was pCDH-CMV-MCS-EF1-copGFP-T2A-puroSS-HA-Sun1L-KDEL and is referred to as DN SUN1L throughout this work. As a mock control (GFP alone), pCDH-CMV-MCS-EF1-copGFP-T2A-puro was used. As an additional control, we used GFP modified with the SS-KDEL sequence (SS-GFP-KDEL) to recruit the protein to the perinuclear space and endoplasmic reticulum, just as in the DN SUN1L construct. Lastly, we used the expression of GFP-tagged wild-type SUN1 to assess the effect of increased SUN1 expression on nucleo-cytoskeletal coupling. Because stable expression of DN SUN1L for more than a week appeared cytotoxic, resulting in a selection bias toward cells with very low expression levels, we transiently transfected the plasmid into cells using Mirus TransIT-LT1 Transfection reagent MIR 2300 (Mirus Bio Corp.) according to the manufacturer's instructions and conducted experiments 72 h after transfection. At this time point, all cells were viable and proliferated normally. SS-GFP-KDEL and GFP-tagged wild-type SUN1 were transiently transfected into MEFs using Mirus TransIT-LT1 Transfection reagent MIR 2300 (Mirus Bio Corp.) and Lipofectamine PLUS (Invitrogen), respectively, following the manufacturer's instructions.
Immunofluorescence Analysis
Cells were grown on glass coverslides coated with 0.5 μg/ml fibronectin (Millipore) and fixed in 4% paraformaldehyde/phosphate-buffered saline for 10 min, followed by permeabilization for 10 min with 0.1% Triton X-100 in phosphate-buffered saline and then blocked with 5% horse serum for 1 h. Cells were probed with primary rabbit polyclonal antibody against nesprin-3 (kindly provided by Dr. A. Sonnenberg, dilution 1:500), primary rabbit polyclonal antibody against nesprin-2G (kindly provided by Dr. D. Hodzic, dilution 1:1000), primary rabbit polyclonal antibody vimentin (kindly provided by Dr. T. Magin, dilution 1:500), primary mouse monoclonal antibody β-tubulin (E7, Developmental Studies Hybridoma Bank, dilution 1:400), and primary mouse monoclonal antibody HA-tag (HA.11 Clone 16B12, Covance, dilution 1:1000). Cells were stained with secondary antibodies conjugated to Alexa Fluor 488 or Alexa Fluor 568 (dilution 1:200) or Alexa Fluor 488 phalloidin (dilution 1:50) and Hoechst 33342 nuclear stain (dilution 1:1000) (all Invitrogen). Fluorescence images were acquired with 20× (0.4 N.A., Plan-Achromat, Olympus) or 60× oil immersion (1.42 N.A., Plan-Achromat, Olympus) objectives using an Olympus IX-70 microscope with a digital charge-coupled device camera (CoolSNAP EZ, Roper Scientific) using IPLab version 4.0 (Scanalytics) software. Images were analyzed by an observer, blinded by treatment. Cytoskeletal structures were scored as either normal or abnormal. For actin organization, the presence of short actin fragments near the nucleus was considered abnormal. For vimentin organization, a well-defined intermediate filament cage surrounding the nucleus was considered normal while the absence or disruption of the perinuclear cage-like network was considered abnormal.
Microneedle Manipulation Experiments
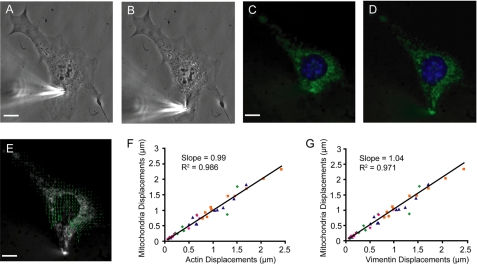
Cells were grown on 0.5 μg/ml fibronectin-coated glass dishes. Thirty minutes prior to the experiments, cells were incubated with MitoTracker Green or Red mitochondrial stain and Hoechst 33342 nuclear stain (both Invitrogen), washed and imaged in Hank's Buffered Salt Saline (Invitrogen). Microneedles with tip diameters of 1 to 3 μm were pulled with a pipette puller (Sutter Instrument Company). The microneedle, controlled by an InjectMan NI 2 micromanipulator (Eppendorf), was carefully inserted into the cell 5 μm away from the nuclear periphery and moved 10 μm toward the cell periphery at 1 μm/second while collecting fluorescence (Alexa Fluor 488 or 568 and 350) and phase contrast images every 10 s with a 60× objective (0.70 N.A., Plan-Achromat, Olympus). Displacement maps were computed by tracking fluorescently labeled features of the nucleus and cytoplasm using custom-written MATLAB (MathWorks) algorithms (see “Generation of Displacement Maps and Deformation Measurements” below). Induced nuclear strain was computed by measuring nuclear elongation before and during strain application. To validate fluorescently labeled mitochondria as valid fiducial markers for cytoskeletal displacements, we also conducted experiments on cells transiently expressing GFP- or mCherry-actin (kindly provided by Dr. F. Gertler) and GFP-vimentin (kindly provided by Dr. R. Goldman) and double labeled with MitoTracker Red or Green (Invitrogen) and then correlated the measured displacements.
Generation of Displacement Maps and Deformation Measurements
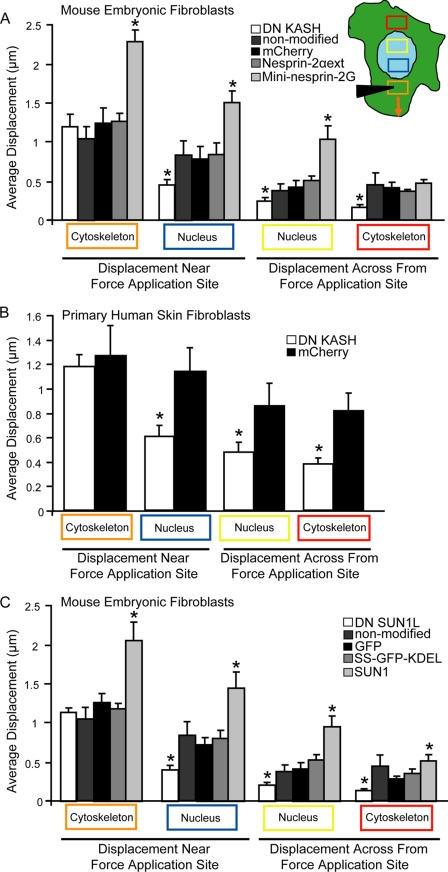
For the microneedle manipulation experiments, displacement maps of nuclear and cytoskeletal deformations were computed using previously developed MATLAB algorithms (15). The approach is based on normalized cross-correlation between small image regions (∼10 μm × 10 μm in size and spaced 5 μm) in subsequent image frames, which identifies corresponding regions within a larger search area in the two frames. For each region center, the displacement is then computed as the shift between the original location and the newly identified position. Regions with low image intensity or insufficient texture (e.g. regions outside the cell) are excluded from the analysis. A median filter was used to eliminate spurious results that can occur from incorrect matches. Subsequently, for each cell, average displacements within predefined regions corresponding to the strain application site, a region of the nucleus toward the strain application site, a nuclear region away from the application site, and a cytoplasmic region across the nucleus (see Fig. 3A, inset) were computed from the displacement maps.
FIGURE 3.
LINC complex disruption impairs intracellular force transmission. A–C, induced cytoskeletal and nuclear displacements during microneedle manipulation, measured in the areas corresponding to the colored rectangles (inset in A). The orange rectangle is the strain application site. Despite similar strain application in the cytoskeleton (orange box), induced nuclear and cytoskeletal displacements (blue, yellow, and red boxes) were significantly smaller in the DN KASH expressing MEFs (A) and human skin fibroblasts (B) and DN SUN1L expressing MEFs (C), compared with non-modified cells and corresponding mCherry or GFP controls. As additional controls, we included experiments with cells expressing nesprin-2α with a modified KASH domain (nesprin-2αext)(A) that cannot bind to SUN proteins (12) and with cells expressing SS-GFP-KDEL that is targeted to the perinuclear space and endoplasmic reticulum, similar to the DN SUNL1 construct (C). We also performed experiments on cells ectopically expressing mini-nesprin-2G (A), which had been previously shown to rescue nuclear movement in cells after nesprin-depletion (14), and in cells expressing wild-type SUN1 (C). In both cases, we observed a marked gain-of-function. For each sample, 15 to 20 cells were analyzed; data are represented as mean ± S.E.; *, p < 0.05. Asterisks shown are relative to corresponding mCherry or GFP controls. Data for non-modified cells from (A) are replotted in (C) for reference. See supplemental Movies S2 and S3.
Nuclear Strain Experiments
Uniaxial strain experiments were carried out as described previously (16). Briefly, cells were plated on fibronectin-coated silicone membranes in phenol red free DMEM, high glucose (Invitrogen) supplemented with 10% fetal bovine serum. Prior to the strain experiments, the cells were incubated with Hoechst 33342 nuclear stain in phenol red free DMEM for 15 min. Membranes were placed on a custom-made strain device mounted on an Olympus IX-70 microscope, with a 60× objective (0.70 N.A., Plan-Achromat, Olympus). Induced nuclear deformations are analyzed by tracking fluorescently labeled nuclei before, during, and after strain application and normalized to membrane strain to compensate for small variations in the applied membrane strain (∼20%) by using custom written image analysis algorithms.
Strain-induced Expression of Mechanosensitive Gene Experiments
Strain-induced expression of mechanosensitive genes was carried out as described previously (17). Briefly, cells were plated on fibronectin-coated silicone membranes. After 48 h of serum starvation, cells were subjected to bi-axial cyclic strain (5% at 1 Hz) for 30 min as previously described (17, 18). Chemical stimulation with PMA (200 ng/ml in DMEM for 30 min, Sigma) served as a positive control. RNA from strained and unstrained control cells was isolated using RNeasy Minikit (Qiagen). Gene expression was then quantified by real-time PCR using probes for mechanosensitive genes Egr-1, Iex-1, Pai-I, Tenascin C, Talin, and Vinculin (see supplemental data for primer sequences). Expression was normalized to an endogenous control, TATA binding protein (see supplemental data for primer sequence) and compared with unstrained controls and strained mCherry controls using the ΔΔCt method.
In Vitro Scratch Assay
A wound was created in serum-starved confluent cell monolayers using a 200 μl-micropipette tip. Subsequently, serum-free medium was replaced with medium containing 3% fetal bovine serum and phase contrast images were acquired at 0 and 3 h post-wound with a 4× objective (0.13 N.A., Plan-Achromat, Olympus). The open wound area was calculated by manually tracing the edge of the wound. Only wounds with an initial width between 53–58 μm were analyzed. For the cell polarization studies, cells were fixed 0 or 3 h post-wounding and probed with primary mouse monoclonal γ-tubulin (Clone GTU-88, Sigma-Aldrich, 2 μg/ml) and secondary antibody conjugated to Alexa Fluor 488 (dilution 1:200) and Hoechst 33342 nuclear stain (dilution 1:1000) to assess centrosome orientation relative to the nucleus and the wound edge as described previously (19). Cells were imaged with a 40× objective (1.15 N.A., Plan-Achromat, Olympus) as described for the wound healing experiment.
Cell Polarization Assay on Micropatterned Substrates
Cells were plated on fibronectin-coated micropatterned surfaces with crossbow-shaped cell-adhesive areas (CYTOO) according to the manufacturer's instructions. Cells were fixed with 4% paraformaldehyde/phosphate-buffered saline for 15 min, 12 h after plating. Cells were probed with primary mouse monoclonal γ-tubulin (Clone GTU-88, Sigma-Aldrich, 2 μg/ml) and secondary antibody conjugated to Alexa Fluor 488 (dilution 1:200) and Hoechst 33342 nuclear stain (dilution 1:1000). Cell polarization was evaluated as described previously (19). Cells were imaged with 20× (0.40 N.A., Plan-Achromat, Olympus) or 40× (1.15 N.A., Plan-Achromat, Olympus) objectives.
Single Cell Migration Assay
Cells were plated on 0.5 μg/ml fibronectin-coated glass dishes and allowed to adhere for 4 h in phenol red-free DMEM supplemented with 10% fetal bovine serum. Subsequently, cells maintained on a temperature (37 °C) controlled stage were imaged every 5 min for 6 h with a 20× objective (0.40 N.A., Plan-Achromat, Olympus). Cell speed and persistence were analyzed from traces of the cell nucleus as described previously (20).
Statistical Analysis
All experiments were performed at least three independent times. All data are expressed as mean ± S.E. Statistical analysis was performed with PRISM 4.0 (GraphPad Software Inc.). The data were analyzed by unpaired t test (for comparison between two groups) or one-way ANOVA. A two-tailed p value below 0.05 was considered significant.
RESULTS
LINC Complex Disruption Using Dominant Negative Nesprin and SUN Constructs
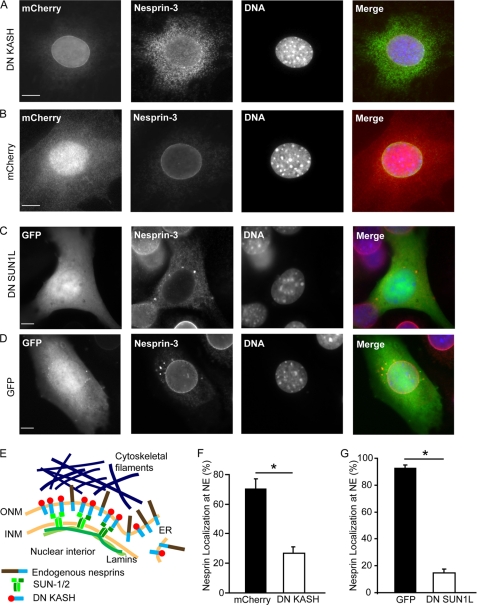
To disrupt the interaction between nesprins and SUN proteins at the nuclear envelope, we stably introduced dominant negative nesprin constructs, containing the C-terminal KASH domain fused to an amino-terminal mCherry, into mouse embryonic fibroblasts (MEFs) and human skin fibroblasts. Overexpression of the dominant negative KASH domain (DN KASH) saturates available binding sites at the nuclear envelope by promiscuously binding to endogenous SUN proteins (12), resulting in the displacement of endogenous nesprins from the nuclear envelope into the endoplasmic reticulum (ER) (Fig. 1, A, B, E). In a parallel approach, we disrupted the LINC complex by introducing a dominant negative SUN1 luminal domain construct (DN SUN1L) targeted to the perinuclear space, co-expressed with GFP and containing a HA tag, into MEFs (Fig. 1, C and D, S1). Overexpression of the DN SUN1L saturates the nesprin KASH domains in the luminal space and prevents binding to endogenous SUN proteins at the inner nuclear membrane (1, 7), causing nesprins to mislocalize from the nuclear envelope to the ER (Fig. 1, C and D). Analysis of intracellular localization of nesprins-2 and -3 confirmed that expression of DN KASH and DN SUN1L resulted in the displacement of endogenous nesprins from the nuclear envelope to the ER (Fig. 1, F and G).
FIGURE 1.
Dominant negative nesprin and SUN constructs displace endogenous nesprins from the nuclear envelope. A and B, immunofluorescence images of MEFs stably expressing DN KASH (A) or mCherry (B). Cells were stained for nesprin-3 (second panel) and DNA (Hoechst 33342, third panel). C and D, immunofluorescence images of MEFs transiently expressing DN SUN1L (C) or GFP alone (D). Cells were stained for nesprin-3 (second panel) and DNA (third panel). Localization of DN SUN1L to the nuclear envelope and endoplasmic reticulum is shown in supplemental Fig. S1. Scale bars, 10 μm. E, scheme of LINC complex disruption by DN KASH and the displacement of endogenous nesprins from the nuclear envelope to the ER. INM, inner nuclear membrane; ONM, outer nuclear membrane. F and G, percentage of cells with normal nuclear envelope (NE) localization of endogenous nesprin-3 in DN KASH (F) and DN SUN1L (G)-expressing cells. More than 100 cells were analyzed for each sample; data are represented as mean ± S.E.; *, p < 0.05.
The LINC Complex Is Essential for Intracellular Force Transmission
To determine whether LINC complex disruption affects intracellular force transmission between the nucleus and cytoskeleton, we developed a microneedle manipulation assay that applies precisely controlled cytoskeletal strain at a defined distance from the nucleus with a computer-controlled microneedle while simultaneously imaging induced nuclear and cytoskeletal deformations across the cell (Fig. 2, A and B and supplemental Movie S1), thereby advancing an earlier approach pioneered by Maniotis et al. (21). Cytoskeletal and nuclear displacements were measured by tracking fluorescently labeled mitochondria and plotted as displacement vector maps (Fig. 2, C–E). Experiments on cells double-labeled with Mitotracker Red or Green and GFP- or mCherry-actin and GFP-vimentin confirmed that mitochondria can serve as reliable fiducial markers for deformations of the actin and vimentin cytoskeleton (Fig. 2, F and G, respectively). Cytoskeletal and nuclear displacements were quantified in selected areas at increasing distances from the strain application site (Fig. 3A, areas corresponding to the colored boxes in inset). Despite comparable cytoskeletal strain application at the microneedle insertion site (orange box), induced nuclear and cytoskeletal displacements (blue, yellow, and red boxes) at other regions were significantly smaller in the DN KASH expressing MEFs than in control cells expressing mCherry alone and in non-modified cells (Fig. 3A and supplemental Movies S2 and S3), indicating that the force transmission between the cytoskeleton and nucleus was disturbed. Similar results to the mCherry expressing MEFs were observed in control cells expressing a modified version of nesprin-2α (i.e. nesprin-2αext) that cannot bind to SUN proteins (Fig. 3A)(12). Nesprin-2αext localizes to the nuclear envelope and does not displace endogenous nesprin-2 (supplemental Fig. S2). As expression of mCherry alone and the nesprin-2αext construct produced nearly identical results in the microneedle manipulation assay (Fig. 3A), subsequent experiments were performed only on MEFs expressing mCherry and DN KASH. In contrast to these control constructs, ectopic expression of mini-nesprin-2G, a chimeric protein consisting of the N-terminal actin-binding domain of nesprin-2 giant and its C-terminal end with the KASH domain (13), which localizes to the nuclear envelope and does not displace endogenous nesprin-2 (supplemental Fig. S2), resulted in significantly increased nuclear and cytoskeletal displacements (Fig. 3A). This observed gain-of-function suggests additional anchoring between the mini-nesprin-2G and the cytoskeleton at the nuclear envelope and increased force transmission between the cytoskeleton and nucleus, further supporting a critical role of the LINC complex in nucleo-cytoskeletal coupling. Experiments in a separate cell system, primary human skin fibroblasts expressing mCherry and DN KASH, produced almost identical results, again revealing markedly reduced induced nuclear and cytoskeletal deformations in regions away from the strain application site (Fig. 3B). To determine if the observed defects in nucleo-cytoskeletal coupling are specific to dominant negative nesprins, we analyzed intracellular force transmission after disrupting the LINC complex with an alternative approach, i.e. expression of the dominant negative SUN1 luminal domain (DN SUN1L). Again, we found that disrupting endogenous SUN-nesprin interactions at the nuclear envelope caused significantly reduced nuclear and cytoskeletal deformations away from the strain application site (Fig. 3C), suggesting that any LINC complex disruption results in disturbed intracellular force transmission. Of note, ectopic expression of wild-type SUN1, which localizes to the nuclear envelope and does not displace endogenous nesprin-2 (supplemental Fig. S2), resulted in increased nuclear and cytoskeletal displacements, i.e. a gain-of-function (Fig. 3C), similar to the results observed for cells expressing mini-nesprin-2G, again supporting a critical role of LINC complex proteins in intracellular force transmission. Because expression of DN SUN1L produced nearly identical results in the microneedle manipulation assay as expression of DN KASH cells, most subsequent experiments were performed only on MEFs expressing DN KASH and appropriate controls.
FIGURE 2.
Microneedle manipulation assay to measure intracellular force transmission. Phase contrast (A and B) and fluorescence (C and D) images of a fibroblast labeled with Hoechst 33342 nuclear stain (blue) and MitroTracker Green mitochondrial stain (green). A microneedle was inserted into the cytoskeleton at a defined distance from the nucleus (A and C) and subsequently moved toward the cell periphery (B and D). Cytoskeletal and nuclear displacements were quantified by tracking the fluorescently labeled nucleus and mitochondria using a custom-written cross-correlation algorithm. E, displacement map of the final cytoskeletal (green) deformations computed from fluorescence image series; arrow length is magnified by 2× for better visibility. Scale bars, 10 μm. F and G, to validate that mitochondria was a suitable cytoskeletal marker, microneedle manipulation was conducted on MEFs transfected with GFP- or mCherry actin (F) and GFP-vimentin (G) and fluorescently labeled with Mitotracker Green or Red. Cytoskeletal displacement maps were calculated independently from the fluorescent signal of the mitochondria and the actin (F) or vimentin (G) cytoskeleton. The average absolute displacement was computed for four distinct cytoskeletal regions at increasing distances away from the strain application site. The slope and R-squared values were computed from the linear regression between the measurements obtained from mitochondria and from actin (F) or vimentin (G), respectively. For actin, the slope was 0.99 and the R2 value was 0.986; for vimentin, the slope was 1.04 and the R2 value was 0.971, confirming that mitochondrial displacements serve as reliable indicators for cytoskeletal deformations. See supplemental Movie S1.
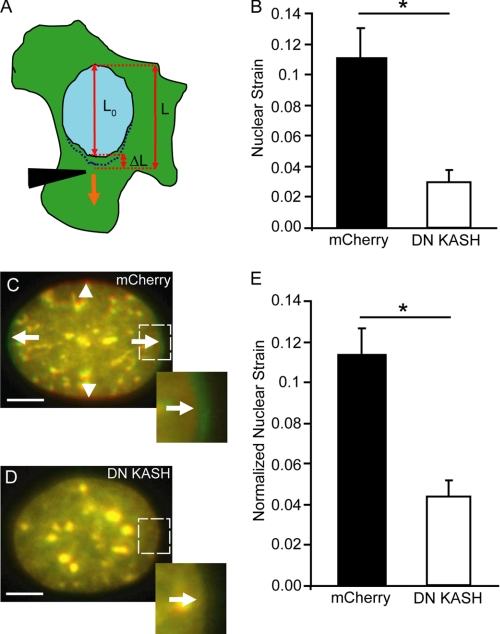
LINC Complex Disruption Reduces Nuclear Deformations in Response to Cytoskeletal or Extracellular Strain Application
To further validate the above results of impaired intracellular force transmission, we applied two independent assays to explore whether LINC complex disruption could alter induced nuclear deformations, which result from cytoskeletal forces acting on the nucleus. In the first assay, we measured induced nuclear elongation during microneedle manipulation (Fig. 4A). As predicted, LINC complex disruption by expression of DN KASH dramatically decreased the induced nuclear strain, defined as the ratio of the change in nuclear length to the initial length (Fig. 4B). In the second approach, designed to mimic the physiological deformations found in mechanically active tissues such as muscle, we subjected cells cultured on fibronectin-coated silicone membranes to uniaxial substrate strain and quantified induced nuclear deformations (supplemental Movies S4 and S5). In this scenario, the cytoskeleton, anchored to the membrane by focal adhesions, closely follows the deformations imposed by the substrate (22). Despite comparable applied substrate strain, MEFs expressing DN KASH once again had significantly decreased nuclear deformations compared with control cells expressing mCherry alone (Fig. 4, C–E), indicating impaired force transmission to the nucleus. Taken together, these experiments suggest that the LINC complex is essential for intracellular force transmission between the cytoskeleton and the nucleus. Consequently, disrupting the LINC complex causes a (partial) uncoupling between the nucleus and cytoskeleton that results in decreased nuclear displacement and deformation in response to cytoskeletal or extracellular strain application.
FIGURE 4.
Strain-induced nuclear deformation is reduced in LINC-disrupted cells. A, computation of nuclear strain induced by microneedle manipulation as shown in B. Nuclear strain was calculated by dividing the nuclear elongation (ΔL = L − L0) by the initial length (L0). L is the final length of the nucleus at the end of strain application. B, DN KASH expressing MEFs show a significant decrease in nuclear strain compared with mCherry alone expressing cells. C–E, nuclear deformation in response to substrate strain application. Overlay of representative pseudo-colored images of fluorescently labeled nuclei of MEFs expressing mCherry (C) or DN KASH (D) before strain (red) and during 20% uniaxial substrate strain application (green). Arrows indicate the nuclear elongation in strain direction; arrowheads indicate the narrowing of the nucleus in the perpendicular direction (C). Inset, detail of nuclear deformation. E, normalized nuclear strain revealed that MEFs expressing DN KASH had a significant decrease in nuclear deformation. See supplemental Movies S4 and S5. Scale bars, 5 μm. For each sample, 15–20 cells were analyzed; data are represented as mean ± S.E.; *, p < 0.05.
Cells Have Normal Activation of Mechanosensitive Genes Despite Reduced Nuclear Deformation
Induced nuclear deformations have often been proposed as a mechanism for cells to sense their mechanical environment by transducing mechanical deformation into biochemical signals that can activate adaptive cellular responses (23). Consequently, one could speculate that disruption of nucleo-cytoskeletal connections that cause a loss of mechanically induced nuclear deformations (Fig. 4, C–E) should lead to reduced activation of mechanosensitive genes. Nonetheless, cells expressing DN KASH subjected to repetitive mechanical strain responded with the same expression of the mechanosensitive genes Egr-1 and Iex-1 as control cells (supplemental Fig. S3). Similar results were obtained for the strain-induced expression of a broad panel of mechanosensitive genes, including Vinculin, Talin, Tenascin C, and Pai-1 (data not shown), suggesting that at least for these genes, nuclear deformation is not required for mechanically induced expression and that the mechanotransduction pathways are initiated in the cytoskeleton, focal adhesions, or the plasma membrane, which are subjected to the same deformations as in control cells.
LINC Complex Disruption Alters Perinuclear Cytoskeletal Organization
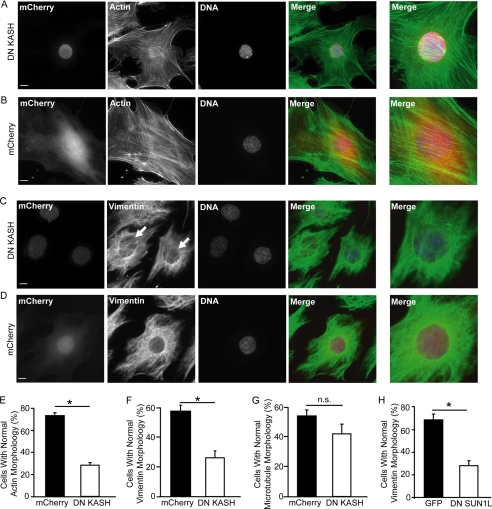
Because nesprins can directly interact with cytoskeletal elements, we hypothesized that LINC complex disruption may affect cytoskeletal structure and organization, which in turn could contribute to the defective nucleo-cytoskeletal force transmission observed in our microneedle manipulation assay and the cellular strain experiments. Immunofluorescence analysis revealed that in DN KASH expressing cells, actin stress fibers in the perinuclear area were discontinuous and fragmented (Fig. 5, A, B, and E), while actin stress fiber organization at the cell periphery was normal (Fig. 5, A and B). Differences in microtubule organization between DN KASH and mCherry alone expressing cells were not quite statistically significant (p = 0.11; Fig. 5G and supplemental Fig. S4), but we observed dramatic effects of DN KASH expression on intermediate filament organization. Whereas control cells displayed a cage-like vimentin intermediate filament network around the nucleus, MEFs expressing DN KASH had disrupted perinuclear vimentin organization with a looser and more irregular network surrounding the nucleus (Fig. 5, C, D, F). Similar defects were observed in cells expressing DN SUN1L (Fig. 5H), suggesting that LINC complex disruption results in disturbed organization of the actin and intermediate filament networks. Importantly, these findings are reminiscent of lamin A/C-deficient cells, which also display defects in perinuclear cytoskeletal organization (23, 29–31). This parallel is particularly relevant since mutations in lamins A and C, just as in nesprins, have been linked to muscular dystrophy and dilated cardiomyopathy (24).
FIGURE 5.
LINC complex disruption alters cytoskeletal organization. A–D, immunofluorescence analysis of MEFs expressing DN KASH (A and C) and mCherry control (B and D). Cells were stained for F-actin (A and B, second panel), vimentin (C and D; second panel), and DNA (A–D, third panel). Last panel, close-up of perinuclear area. Arrows indicate area with disturbed perinuclear vimentin network organization (C). Scale bar, 10 μm. E–H, percentage of cells with normal organization of the perinuclear actin (E), vimentin (F, H), or microtubule (G) network in MEFs expressing DN KASH (E–G) or DN SUN1L (H). More than 100 cells were analyzed for each sample; data are represented as mean ± S.E.; *, p < 0.05. See also supplemental Fig. S4.
Disruption of the LINC Complex Causes Impaired Cell Polarization and Migration
To test whether the disturbed cytoskeletal organization and impaired intracellular force transmission could affect cytoskeleton-mediated functions, we assessed cell migration and polarization in cells after LINC complex disruption by DN KASH expression. DN KASH expressing cells scored significantly lower in an in vitro scratch wound assay (Fig. 6, A and B). In wild-type fibroblasts, migration into the wound is preceded by rearward nuclear movement and positioning of the centrosome toward the wound edge, resulting in polarized cells (25). Consistent with this idea, cells at the wound edge expressing mCherry alone had become polarized at 3 h after wounding; in contrast cells expressing DN KASH remained randomly oriented (Fig. 6C). To investigate whether a similar mechanism could also contribute to polarization in stationary, non-migrating cells, we investigated nuclear positioning and centrosome orientation of cells plated on substrates micropatterned with crossbow-shaped fibronectin-coated areas that induce cell polarization in wild-type cells (26). While control fibroblasts expressing mCherry polarized toward the bowed front (Fig. 6D), cells expressing DN KASH failed to polarize, indicated by a lack of rearward nuclear positioning and random centrosome orientation (Fig. 6, D–F) suggesting that the impaired nucleo-cytoskeletal coupling prevents rearward nuclear movement required for cell polarization. To investigate the consequences of impaired nuclear positioning and cytoskeletal organization in more detail, we analyzed the movement of single cells over a 6-h time course. DN KASH-expressing cells had a dramatically reduced range of movement (Fig. 6, G and H), caused by a combination of decreased migration speed (Fig. 6I) and persistence time (Fig. 6J), indicating that LINC complex disruption causes broad defects in cell migration.
FIGURE 6.
LINC complex disruption causes impaired cell migration and polarization. A–C, in vitro scratch wound assay. A, phase contrast images of mCherry and DN KASH expressing MEFs taken at 0 or 3 h post-wound. Scale bar, 80 μm. B, open wound area remaining after 3 h; n = 27 wounds. C, percentage of cells at wound edge with centrosome orientation toward the wound (see scheme) at 0 and 3 h post-wound; n = 127 for mCherry and n = 97 for DN KASH. D–F, cell polarization after culture on micropatterned substrates. D, first panel, red signal, revealing fluorescently labeled fibronectin-coated crossbow pattern and mCherry constructs. Second panel, centrosome labeled by γ-tubulin staining. Third panel, DNA stain. Last panel, merged image; arrows indicate centrosome position. Scale bar, 5 μm. E, percentage of cells with correct polarization. Cells were scored as polarized when the centrosome was located in the forward facing sector (inset, green segment); n = 33 for mCherry and n = 32 for DN KASH. F, average distance of nucleus (Nuc) and centrosome (Cen) from the crossbow pattern center. Control cells display rearward nuclear position and central centrosome position (14); n = 33 for mCherry and n = 32 for DN KASH. G–J, single cell migration analysis of MEFs expressing DN KASH or mCherry. G and H, Rose plots, showing the total distance traveled and the directionality of movement for five representative cells for each cell type during a 6-h period. Average cell speed (I) and persistence time (J) were computed from individual cell traces; n = 138 for mCherry and n = 136 for DN KASH. For each experiment, data are represented as mean ± S.E.; *, p < 0.05.
DISCUSSION
Using custom-developed biophysical assays, we demonstrate that disruption of the LINC complex results in defects in nuclear positioning and centrosome orientation, disturbed perinuclear organization of the actin and vimentin cytoskeleton, impaired cell motility, and reduced propagation of intracellular deformations in cells subjected to internal or external mechanical stress. Our studies offer a mechanistic explanation for these cellular defects by identifying the LINC complex as a critical element for intracellular force transmission between the cytoskeleton and the nucleus in cells, which is highly relevant to nesprin-linked diseases that primarily affect mechanically active tissues such as muscle.
The observed changes in cytoskeletal organization and cell motility and polarization in cells with a disrupted LINC complex are consistent with other studies that report defects in nuclear positioning and centrosome orientation in a scratch wound assay after knockdown of nesprin or SUN proteins or expression of dominant negative LINC complex components (14, 27–29). In migrating cells, the rearward nuclear movement required for cell polarization is caused by retrograde flow of actin cables from the leading edge that engage the nucleus through nesprin-2/SUN2 complexes and drag it backwards (14). Our studies now demonstrate that the role of the LINC complex is not restricted to this specialized function during cell migration; instead, the LINC complex is critically important for intracellular force transmission between the cytoskeleton and the nucleus in general, and that LINC complex disruption causes defects in nuclear positioning, cytoskeletal organization, and propagation of cellular and nuclear deformations in response to cytoskeletal or extracellular strain application.
Induced nuclear deformations have long been thought to contribute to the activation of mechanosensitive genes, for example by causing conformational changes in chromatin structure and organization that could modulate transcription factor binding or transcriptional processes (23). Somewhat surprisingly, our results show that expression of a select panel of mechanosensitive genes is normal in cells with a disrupted LINC complex, despite their markedly reduced nuclear deformation (Fig. 4, D and E). Whereas we cannot exclude the possibility that other mechanosensitive genes are sensitive to LINC complex disruption, our findings suggest that the expression of many classic mechanosensitive genes, such as Egr-1 (30) or Iex-1 (31), is activated by signaling pathways initiated in the cytoskeleton, at the plasma membrane, or at focal adhesions (24, 29). Nonetheless, disturbed nuclear and/or cytoskeletal organization resulting from LINC complex disruption could have indirect effects on transcriptional regulation. For example, C2C12 cells expressing dominant negative nesprin and SUN protein constructs have enhanced activation of NF-κB signaling, regardless of whether the stimulation is mechanical (cyclic strain) or chemical (TNF-α) (32).
Our findings that LINC complex disruption causes disturbed intracellular force transmission could have important implications in understanding the mechanism by which mutations in nesprins can cause muscular dystrophy and dilated cardiomyopathy. Impaired intracellular force transmission is a common finding in other muscular dystrophies; for instance, Duchenne muscular dystrophy is caused by mutations in the dystrophin gene that disrupt the link between the extracellular matrix and cytoskeleton, resulting in mechanically more fragile cells and progressive muscle degeneration (33). Interestingly, Emery-Dreifuss muscular dystrophy and dilated cardiomyopathy can, in addition to nesprins, also arise from mutations in the nuclear envelope proteins lamin A and C, which form a dense protein network underlying the inner nuclear membrane and can directly bind to SUN proteins and nesprins (24). It is noteworthy that cells lacking lamins A and C show similar defects in cytoskeletal organization and impaired cell polarization and migration (19, 28, 34) as cells after LINC complex disruption, suggesting that lamins form an integral part of the LINC complex. Most recently, Folker et al. (35) found that lamin A mutants associated with muscular dystrophy, but not those linked to lipodystrophy, cause similar defects in nuclear positioning and migration. Thus, any disruption of this complex may cause defects in cellular structure and intracellular force transmission that could contribute to the development of muscular dystrophies and cardiomyopathies. While the experiments presented here are based on the disruption of all endogenous nesprins (using dominant negative nesprin and SUN constructs) to demonstrate the general importance of the LINC complex in intracellular force transmission, future studies should be aimed at analyzing the effect of specific nesprin mutations recently linked to muscular dystrophy and cardiomyopathy (7, 8).
In conclusion, we propose that the LINC complex constitutes an essential connection between the cytoskeleton and the nucleus critical for intracellular force transmission. Further studies will be pivotal to improve our understanding of nucleo-cytoskeletal coupling in normal cells and its importance in cellular functions, including cell migration and nuclear anchorage. Insights gained from this work can then potentially provide new targets for the treatment of diseases caused by LINC complex associated mutations, thereby establishing the LINC complex as a potential therapeutic target.
Supplementary Material
Acknowledgments
We thank Dr. A. Sonnenberg, Dr. D. Hodzic, and Dr. T. Magin for providing antibodies; Dr. F. Gertler, Dr. R. Goldman, and Dr. G. G. Gundersen for plasmids; Dr. R. Kakkar and Dr. P. Patwari for discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 NS059348 and R01 HL082792 (to J. L.). and the Brigham and Women's Hospital Cardiovascular Leadership Group Award.

The on-line version of this article (available at http://www.jbc.org) contains supplemental data, Figs. S1–S4 and Movies S1–S5.
- LINC
- linker of nucleoskeleton and cytoskeleton complex
- MEF
- mouse embryonic fibroblasts
- DN
- dominant negative
- ER
- endoplasmic reticulum.
REFERENCES
- 1. Crisp M., Liu Q., Roux K., Rattner J. B., Shanahan C., Burke B., Stahl P. D., Hodzic D. (2006) J. Cell Biol. 172, 41–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Haque F., Lloyd D. J., Smallwood D. T., Dent C. L., Shanahan C. M., Fry A. M., Trembath R. C., Shackleton S. (2006) Mol. Cell. Biol. 26, 3738–3751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Padmakumar V. C., Libotte T., Lu W., Zaim H., Abraham S., Noegel A. A., Gotzmann J., Foisner R., Karakesisoglou I. (2005) J. Cell Sci. 118, 3419–3430 [DOI] [PubMed] [Google Scholar]
- 4. Padmakumar V. C., Abraham S., Braune S., Noegel A. A., Tunggal B., Karakesisoglou I., Korenbaum E. (2004) Exp. Cell Res. 295, 330–339 [DOI] [PubMed] [Google Scholar]
- 5. Zhen Y. Y., Libotte T., Munck M., Noegel A. A., Korenbaum E. (2002) J. Cell Sci. 115, 3207–3222 [DOI] [PubMed] [Google Scholar]
- 6. Wilhelmsen K., Litjens S. H., Kuikman I., Tshimbalanga N., Janssen H., van den Bout I., Raymond K., Sonnenberg A. (2005) J. Cell Biol. 171, 799–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhang Q., Bethmann C., Worth N. F., Davies J. D., Wasner C., Feuer A., Ragnauth C. D., Yi Q., Mellad J. A., Warren D. T., Wheeler M. A., Ellis J. A., Skepper J. N., Vorgerd M., Schlotter-Weigel B., Weissberg P. L., Roberts R. G., Wehnert M., Shanahan C. M. (2007) Hum. Mol. Genet 16, 2816–2833 [DOI] [PubMed] [Google Scholar]
- 8. Puckelwartz M. J., Kessler E. J., Kim G., Dewitt M. M., Zhang Y., Earley J. U., Depreux F. F., Holaska J., Mewborn S. K., Pytel P., McNally E. M. (2010) J. Mol. Cell Cardiol. 48, 600–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Thornell L., Carlsson L., Li Z., Mericskay M., Paulin D. (1997) J Mol Cell Cardiol 29, 2107–2124 [DOI] [PubMed] [Google Scholar]
- 10. Patwari P., Chutkow W. A., Cummings K., Verstraeten V. L., Lammerding J., Schreiter E. R., Lee R. T. (2009) J. Biol. Chem. 284, 24996–25003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang Q., Skepper J. N., Yang F., Davies J. D., Hegyi L., Roberts R. G., Weissberg P. L., Ellis J. A., Shanahan C. M. (2001) J. Cell Sci. 114, 4485–4498 [DOI] [PubMed] [Google Scholar]
- 12. Stewart-Hutchinson P. J., Hale C. M., Wirtz D., Hodzic D. (2008) Exp. Cell Res. 314, 1892–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ostlund C., Folker E. S., Choi J. C., Gomes E. R., Gundersen G. G., Worman H. J. (2009) J. Cell Sci. 122, 4099–4108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Luxton G. W., Gomes E. R., Folker E. S., Vintinner E., Gundersen G. G. (2010) Science 329, 956–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lammerding J., Huang H., So P. T., Kamm R. D., Lee R. T. (2003) IEEE Eng. Med. Biol. Mag. 22, 124–127 [DOI] [PubMed] [Google Scholar]
- 16. Coffinier C., Jung H. J., Li Z., Nobumori C., Yun U. J., Farber E. A., Davies B. S., Weinstein M. M., Yang S. H., Lammerding J., Farahani J. N., Bentolila L. A., Fong L. G., Young S. G. (2010) J. Biol. Chem. 285, 20818–20826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lammerding J., Schulze P. C., Takahashi T., Kozlov S., Sullivan T., Kamm R. D., Stewart C. L., Lee R. T. (2004) J. Clin. Invest. 113, 370–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cheng G. C., Briggs W. H., Gerson D. S., Libby P., Grodzinsky A. J., Gray M. L., Lee R. T. (1997) Circ. Res. 80, 28–36 [DOI] [PubMed] [Google Scholar]
- 19. Lee J. S., Hale C. M., Panorchan P., Khatau S. B., George J. P., Tseng Y., Stewart C. L., Hodzic D., Wirtz D. (2007) Biophys J 93, 2542–2552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Verstraeten V. L., Ji J. Y., Cummings K. S., Lee R. T., Lammerding J. (2008) Aging Cell 7, 383–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Maniotis A. J., Chen C. S., Ingber D. E. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 849–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Caille N., Tardy Y., Meister J. J. (1998) Ann Biomed. Eng. 26, 409–416 [DOI] [PubMed] [Google Scholar]
- 23. Wang N., Tytell J. D., Ingber D. E. (2009) Nat. Rev. Mol. Cell Biol. 10, 75–82 [DOI] [PubMed] [Google Scholar]
- 24. Worman H. J., Fong L. G., Muchir A., Young S. G. (2009) J. Clin. Invest. 119, 1825–1836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gomes E. R., Jani S., Gundersen G. G. (2005) Cell 121, 451–463 [DOI] [PubMed] [Google Scholar]
- 26. Théry M., Racine V., Piel M., Pépin A., Dimitrov A., Chen Y., Sibarita J. B., Bornens M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 19771–19776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chancellor T. J., Lee J., Thodeti C. K., Lele T. (2010) Biophys. J. 99, 115–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hale C. M., Shrestha A. L., Khatau S. B., Stewart-Hutchinson P. J., Hernandez L., Stewart C. L., Hodzic D., Wirtz D. (2008) Biophys. J. 95, 5462–5475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schneider M., Lu W., Neumann S., Brachner A., Gotzmann J., Noegel A. A., Karakesisoglou I. (2011) Cell Mol. Life Sci. 68, 1593–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Morawietz H., Ma Y. H., Vives F., Wilson E., Sukhatme V. P., Holtz J., Ives H. E. (1999) Circ. Res. 84, 678–687 [DOI] [PubMed] [Google Scholar]
- 31. De Keulenaer G. W., Wang Y., Feng Y., Muangman S., Yamamoto K., Thompson J. F., Turi T. G., Landschutz K., Lee R. T. (2002) Circ. Res. 90, 690–696 [DOI] [PubMed] [Google Scholar]
- 32. Brosig M., Ferralli J., Gelman L., Chiquet M., Chiquet-Ehrismann R. (2010) Int. J. Biochem. Cell Biol. 42, 1717–1728 [DOI] [PubMed] [Google Scholar]
- 33. Heydemann A., McNally E. M. (2007) Trends Cardiovasc. Med. 17, 55–59 [DOI] [PubMed] [Google Scholar]
- 34. Broers J. L., Peeters E. A., Kuijpers H. J., Endert J., Bouten C. V., Oomens C. W., Baaijens F. P., Ramaekers F. C. (2004) Hum. Mol. Genet 13, 2567–2580 [DOI] [PubMed] [Google Scholar]
- 35. Folker E. S., Ostlund C., Luxton G. W., Worman H. J., Gundersen G. G. (2011) Proc. Natl. Acad. Sci. U.S.A. 108, 131–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.