Abstract
RIL (product of PDLIM4 gene) is an actin-associated protein that has previously been shown to stimulate actin bundling by interacting with actin-cross-linking protein α-actinin-1 and increasing its affinity to filamentous actin. Here, we report that the alternatively spliced isoform of RIL, denoted here as RILaltCterm, functions as a dominant-negative modulator of RIL-mediated actin reorganization. RILaltCterm is regulated at the level of protein stability, and this protein isoform accumulates particularly in response to oxidative stress. We show that the alternative C-terminal segment of RILaltCterm has a disordered structure that directs the protein to rapid degradation in the core 20 S proteasomes. Such degradation is ubiquitin-independent and can be blocked by binding to NAD(P)H quinone oxidoreductase NQO1, a detoxifying enzyme induced by prolonged exposure to oxidative stress. We show that either overexpression of RILaltCterm or its stabilization by stresses counteracts the effects produced by full-length RIL on organization of actin cytoskeleton and cell motility. Taken together, the data suggest a mechanism for fine-tuning actin cytoskeleton rearrangement in response to stresses.
Keywords: Adapter Proteins, Cytoskeleton, Oxidative Stress, Proteasome, Protein Degradation, Actin Cytoskeleton, Alternative Splicing, RIL/PDLIM4
Introduction
RIL (reversion-induced LIM domain protein) (also known as PDLIM4) is a member of ALP/Enigma family of PDZ and LIM domain-containing adapter proteins found in association with actin cytoskeleton (1, 2). ALP/Enigma genes/proteins are highly conserved throughout evolution with a single member in Caenorhabditis elegans (alp-1/eat-1) and Drosophila melanogaster (tungus) and seven different genes in mammals (3–6). Most of the family members, including the C. elegans prototype, associate with actin cytoskeleton via α-actinins (ALP/PDLIM3 (7–9), CLP-36/PDLIM1 (10–12), Mystique/PDLIM2 (13), RIL/PDLIM4 (12, 14), Enigma Homolog/ENH/PDLIM5 (15), and ZASP/Cypher/PDLIM6) (16–18), or Filamin A (PDLIM2) (13) or β-tropomyosin (Enigma/PDLIM7) (19). The EHN/PDLIM5 has also been reported to affect actin structure by binding the Spine-associated RapGAP (SPAR) (20). Proteins of the PDLIM family mostly function to maintain the structure of Z-discs and are responsible for the integrity of muscle fibers by stabilizing the actin filaments (4, 21, 22). Thus, ablation of Alp or Cypher/ZASP genes leads to cardiomyopathies and/or skeletal myopathies in various animal models from fly to zebrafish to mouse (23–26), whereas mutations in hCypher have been shown to cause similar pathologies in humans (22, 27–29). In nonmuscle cells, PDLIM proteins fulfill similar role in stabilizing actin stress fibers. Specifically, binding to RIL increases the affinity of α-actinin-1 to filamentous actin (F-actin), leading to dramatic rearrangement of the actin cytoskeleton (1). Alternative splicing adds yet another level of complexity and flexibility to fine-tuning the function of ALP/Enigma proteins. With the exception of the CLP-36, the PDLIM genes were reported to express up to six alternatively spliced mRNA species that often include LIM-less isoforms. These splice variants can be tissue type-specific (such as cardiac Zasp/Cypher and skeletal muscle Zasp/Cypher in mice (30) or smooth/cardiac muscle smAlp and skeletal muscle scAlp (31)) or developmental stage-specific (embryonic Enh isoform or adult Enh variants expressed in murine heart (32)) and can be altered in response to stress, e.g. pressure overload in the heart (32). Interestingly, the shorter LIM-less isoforms of Enh have been shown to negatively regulate the longer Enh variant (15, 32). Transcripts from the RIL gene are expressed as two alternatively spliced mRNA species (2). In comparison with the full-length RIL, the shorter isoform lacks the LIM domain at its C terminus, and its function remains unknown (2). Physiological processes affected by RIL function are poorly understood, although its expression has been reported to be altered in various cancers as well as in inflammatory conditions (33–46). Whether alternative splicing plays any role in modulating RIL function in these pathological states has not been addressed.
In this study, we characterize the function of the shorter alternatively spliced isoform of RIL denoted here as RILaltCterm. We report that RILaltCterm is intrinsically unstable due to an unstructured region at its C terminus. Degradation of this isoform is 20 S proteasome-dependent, being modulated by binding to NQO1. We show that RILaltCterm is stabilized in response to oxidative stress and/or UV irradiation. Once stabilized, the RILaltCterm homodimerizes with full-length RIL to change its subcellular localization leading to rearrangement of actin cytoskeleton and attenuation of cell migration.
EXPERIMENTAL PROCEDURES
Plasmids
Constructs expressing N-FLAG and N-HA-tagged full-length RIL, RILaltCterm (Fig. 1A), and control deletion mutant RILΔCterm (lacking both exon 6 and exon 7 sequences that code for the LIM domain and/or alternative C-terminal peptide) were generated by subcloning corresponding PCR-amplified cDNA fragments into XbaI and XhoI sites of pLCMV-N-FLAG or pLCMV-N-HA, respectively. EGFP-RILaltCterm and Luc-RILaltCterm were constructed by subcloning PCR-amplified RILaltCterm cDNA fragment corresponding to amino acids 217–246 (30 C-terminal residues) in-frame with EGFP2 into AvrII and BamHI sites of pLCMV-EGFPfus or in-frame with luciferase gene into XhoI and BamHI sites of pLCMV-Luc. For generation of Luc-RIL-Clike (control construct) a 1-nucleotide frameshift and an in-frame stop codon were introduced into cDNA sequence corresponding to the 30 C-terminal residues of RILaltCterm by PCR, and the resulting fragment was ligated in-frame with the luciferase gene into XhoI and BamHI sites of pLCMV-Luc. pLCMV-Luc-Stop refers to the nonfused luciferase expression construct.
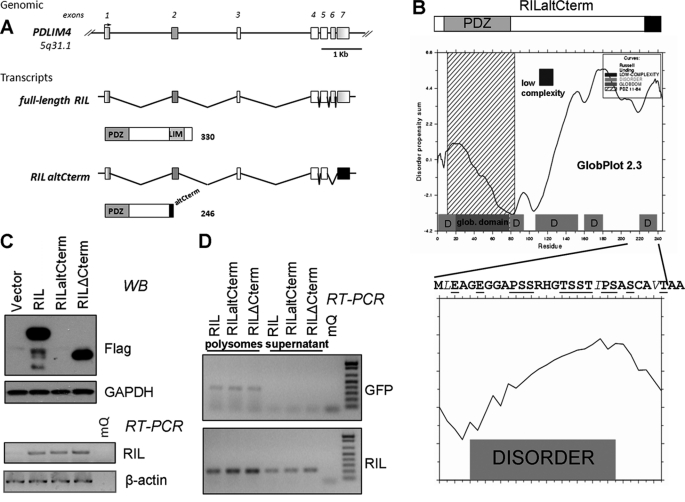
FIGURE 1.
Alternatively spliced isoform of RIL has low stability. A, genomic organization and splice isoforms of the RIL gene. Full-length RIL mRNA includes all seven exons, the corresponding protein features PDZ and LIM domains, and an interdomain linker region. Alternatively spliced isoform of RIL lacks the 6th exon leading a frameshift and a substitution of a short peptide instead of the LIM domain. B, identification of a disordered region within the C-terminal region of RILaltCterm by GlobPlot 2.3 (D denotes disorder) and its sequence. Disorder promoting residues are in boldface; order promoting residues are in italic, and amino acids characteristic for PEST sequences are underlined. C, FLAG-tagged full-length RIL, RILaltCterm, or RIL truncation mutant were lentivirally expressed in A549 cells. RILaltCterm protein was barely detectable (Western blot), although there was no significant difference in mRNA levels as detected by RT-PCR. D, full-length RIL, RILaltCterm, and RIL truncation mutant mRNAs are equally bound by polyribosomes. HeLa cells were transfected by the indicated RIL constructs along with GFP-expressing plasmid as internal control. RNA extracted from polysomal fraction was analyzed by RT-PCR.
Cells and Transient Transfection
293T, HeLa, A549, H1299, MDA-MB-435S, A431, U2OS, and RAW264.7 cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and appropriate antibiotics. Transient transfection was performed using Lipofectamine and Plus reagent (Invitrogen).
Lentiviral Transduction
Lentiviral particles were packaged as described previously (47) using pGag1 and pRev2 helper plasmid set and pseudotyped with the VSV-G protein. Supernatants containing viral particles were collected every 12 h, pooled, and concentrated by PEG-8000 precipitation. For infection of target cells, viral preparations were diluted in complete growth media supplemented with 4 μg/ml Polybrene. The expression was assayed 48 h post-infection.
Reagents and Antibodies
Translation inhibitor cycloheximide (at 1–5 μg/ml) was from ICN Biomedicals. Proteasome inhibitor MG-132 (N-carbobenzoxyl-l-leucinyl-l-norleucinal, at 20 μm), inhibitor of serine proteases AEBSF (4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride, at 0.4 mm), and calpain inhibitor calpeptin (at 0.1 mm) were from Calbiochem. NQO1 inhibitor dicoumarol (at 400 μm) was from Acros Organics. Cysteine protease inhibitors E-64 (trans-epoxysuccinyl-l-leucylamido-4-guanidinobutane at 5 μm), aspartic protease inhibitor pepstatin A (at 5 μg/ml), serine and cysteine protease inhibitor leupeptin (at 50 μm), hypoxia mimetic deferoxamine mesylate (at 300 μm), H2O2 (0.1–0.5 mm), antioxidants N-acetyl-l-cysteine (at 0.5 mm), Ebselen (organo-selenium compound, at 0.02 mm), and β-Gal substrate o-nitrophenyl β-d-galactopyranoside were from Sigma. LPS from Escherichia coli was purchased from Sigma. FLAG mAb M2 and anti-FLAG rabbit polyclonal IgGs were from Sigma, GAPDH mAb was from Meridian Life Science, and HA mAb clone 12CA5 from rat was from Roche Applied Science. Antibodies against GFP (Ab-2) were from NeoMarkers. mAbs against ubiquitin (P4D1), anti-NQO1 (C-19) rabbit polyclonal IgGs, and anti-PDLIM4/RIL (D-8) mAbs predicted to recognize both isoforms of RIL and anti-actin (C-11) goat polyclonal IgGs were from Santa Cruz Biotechnology. RIL-specific goat polyclonal antibodies were acquired from Abcam, and anti-α-actinin-1 mAbs were from Millipore.
RT-PCR
Total RNA from cells was extracted using TRIzol reagent (Invitrogen) according to the manufacturer's protocol. First cDNA strand was synthesized from oligo(dT)12–18 primer using SuperScript first-strand synthesis system (Invitrogen). For PCR amplification, the following primer pairs were used: RIL (RIL4ex sense 5′-CTCGCTTTCCAGTCCCTCACAAT-3′ and RIL5ex antisense 5′-TCTAGCATGCCCTGCAAGTAGC-3′), β-actin (sense 5′-GCTTGCCATCCAACCACTCAGTCTTG-3′ and antisense 5′-GCGTCTCCTTTGAGCTGTTTGCAGAC-3′), and EGFP (sense 5′-TGACCCTGAAGTTCATCTGCACCA-3′ and antisense 5′-TGTGGCGGATCTTGAAGTTCACCT-3′).
Polyribosome Isolation and Analysis
107 HeLa cells transfected with either full-length RIL, RILaltCterm, or RILΔCterm control truncation mutant were treated with 50 μg/ml cycloheximide for 5 min, harvested by scraping, and washed with ice-cold PBS with 50 μg/ml cycloheximide. Cell pellet was resuspended in 20 volumes of polysome lysis buffer (0.14 m NaCl, 25 mm MgCl2, 10 mm Tris-HCl, pH 8.0, 0.5% Nonidet P-40, 1 mm DTT, 1000 units/ml RNasin (Promega), and 50 μg/ml cycloheximide) and spun down at 2000 × g, and supernatant was applied onto 30% sucrose/lysis buffer cushion and centrifuged at 250,000 × g for 4 h at 4 °C. RNA from supernatant and pellet was isolated with TRIzol reagent (Invitrogen), reverse-transcribed, and analyzed by PCR.
Reporter Assays
H1299 cells were transfected with Luc-stop, Luc-RILaltCterm, or Luc-RIL-Clike and a plasmid constitutively expressing β-galactosidase for transfection efficiency normalization. Cells were briefly washed in PBS and lysed in either Reporter Lysis Buffer (Promega) for subsequent assessment of luciferase luminescence with luciferase assay substrate or in β-Gal staining buffer (1 mm MgCl2, 250 mm Tris-HCl, pH 7.4, 0.02% Nonidet P-40, 2 mg/ml o-nitrophenyl β-d-galactopyranoside in PBS), incubated at 37 °C, and assessed by photometry at 405 nm.
Co-immunoprecipitation and Western Blotting
For straight immunoblotting, cells were lysed in RIPA buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1% sodium deoxycholate, 1% Nonidet P-40, 0.1% SDS with protease inhibitor mixture (Roche Applied Science)), and lysate was pre-cleared by centrifugation (10,000 × g, 4 °C for 10 min). For co-immunoprecipitation studies, cells expressing the necessary constructs were washed with ice-cold PBS, harvested by scraping, and lysed on ice for 10 min in NET buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 5 mm EDTA, and 1% Nonidet P-40 supplemented with protease inhibitors). Cell lysates were pre-cleared by centrifugation as described above, and 500-μg aliquots were subjected to immunoprecipitation with anti-FLAG M2-agarose (Sigma). Detergent-soluble fractions were obtained by harvesting the cells on ice by scraping and subsequent lysis in ice-cold CSK buffer (10 mm PIPES, pH 6.8, 100 mm NaCl, 300 mm sucrose, 3 mm MgCl2, 1 mm EGTA, 0.5% Triton X-100) supplemented with protease inhibitors for 10 min. Detergent-insoluble material was spun down at 10,000 × g, and pellets were resuspended in 2% SDS, 50 mm Tris-HCl, pH 7.5. For Western blotting, protein lysates and prestained molecular mass standards were subjected to SDS-PAGE and transferred to nitrocellulose membrane. The membranes were blocked with 5% nonfat dry milk in PBS with 0.1% Tween 20 (PBS-T), incubated with primary antibodies, extensively washed in PBS-T, and stained with horseradish peroxidase-conjugated secondary species-specific IgG (1:10,000) (Santa Cruz Biotechnology). Immunoreactivity was visualized by enhanced chemiluminescence (ECL) kit (GE Healthcare).
Immunocytochemistry
U2OS cells expressing FLAG- or HA-tagged RIL, HA- or FLAG-tagged RILΔCterm, or both were grown on glass coverslips, fixed in 3.7% formaldehyde for 15 min, permeabilized in 0.5% Triton X-100 in PBS for 15 min, blocked in 3% BSA/PBS for 1 h and then incubated with primary rabbit anti-FLAG (1:500) and rat anti-HA (1:500) antibodies in blocking solution, washed in PBS, incubated with secondary AlexaFluor 350- or AlexaFluor 594-conjugated antibodies (1:1000, Invitrogen), and counterstained with FITC-labeled phalloidin in blocking buffer. Slides were extensively washed in PBS and mounted in Fluoromount G (Southern Biotech). Fluorescent pictures were taken by a Qimaging camera using Leica 1.25NA ×40 objective. Images were processed and assembled using Photoshop (Adobe).
F-actin Content Measurement
U2OS cells transduced with RILaltCterm or empty vector were grown in triplicate wells of 24-well plates, subjected to 0.5 mm H2O2 treatment for 4 h where indicated, and fixed by 4% formaldehyde (methanol-free) in cytoskeleton buffer (10 mm MES, pH 6.1, 138 mm KCl, 3 mm MgCl2, 2 mm EGTA, and 0.32 m sucrose) to prevent actin depolymerization. After permeabilization by 0.1% Triton X-100 in PBS, cells were blocked in 3% BSA and stained with FITC-conjugated phalloidin. F-actin content was quantified using a spectrofluorimeter (Wallac Victor2). Data were normalized by counterstaining with DAPI to quantify DNA content.
Migration Assays
For the chemotactic migration assay, RAW264.7 macrophage-like cells expressing FLAG-tagged RILaltCterm or empty vector were starved overnight in 1% FBS, treated with 0.5 mm H2O2 for 3 h where indicated, and detached, and 5 × 104 cells were seeded in triplicate into the upper chamber of a transwell system (8-μm membrane pore size). 1 μg/ml LPS added to the lower chamber was used as chemoattractant. Cells were allowed to migrate for 6 h, and then nonmigrated, cells were removed from the upper chamber with a cotton swab; membranes were fixed in methanol, and the cells were stained with DAPI. Five random fields were photographed per well, and nuclei of migrated cells were automatically counted using ImageJ software (National Institutes of Health). For the haptotactic migration analysis, U2OS cells were transfected with RILaltCterm and RILΔCterm constructs or empty vector control and 2 × 105 cells were plated per well of a 6-well plate. Cell were left to attach overnight, and spontaneous motility was followed by time-lapse microscopy for 5 h using a Leica DMI 6000B microscope equipped with 5× NA 0.12 Leica objective and heated motorized stage with humidified CO2 chamber operated by Leica LAS AF software. Images were taken every 5 min, and average migration velocity was calculated using ImageJ Manual Tracking plugin (National Institutes of Health). Approximately 20 cells per experimental group were tracked.
Statistical Analysis
Data were analyzed by Student's t test or one-way analysis of variance (ANOVA) (Holm-Sidak method) where indicated with the help of SigmaStat 3.5 software. p values less than 0.05 were regarded as significant.
RESULTS
Alternatively Spliced Isoform of RIL Contains Unstructured C-terminal Segment
Unlike the major full-length isoform of RIL transcript, minor alternatively spliced isoform originally described by Bashirova et al. (2) and denoted here as RILaltCterm lacks the sequence of exon 6. The exon skipping results in a frameshift in exon 7, leading to deletion of a C-terminal segment (106 amino acids) containing the LIM domain and its substitution with a short (22 amino acids) peptide (Fig. 1A). Analysis of the amino acid composition in the alternative product of RIL revealed a high content of proline, glutamate, serine, and threonine (PEST) residues at its C terminus. Such regions tend to be unstructured, which is characteristic of protein-destabilizing PEST sequences (48). Indeed, analysis by the globular domain/disorder predicting machines DisEMBL (49), GlobPlot (50), and PONDR (51) also suggests that the C terminus of RILaltCterm forms a nonfolded disordered structure (Fig. 1B).
It has been previously suggested that PEST sequences are present in proteins with high turnover rate and can signal to degradation (52, 53). To test if RILaltCterm is unstable, we expressed by lentiviral transduction into A549 cells FLAG-tagged proteins corresponding to full-length RIL, RILaltCterm, and RILΔCterm the truncation mutant lacking the frame-shifted segment. Although either of the constructs was equally expressed at the RNA level, and on Western blots there were strong bands corresponding to the full-length and RILΔCterm proteins, the level of FLAG-RILaltCterm protein was undetectable (Fig. 1C) suggesting that the unfolded protein segment might be responsible for the effect.
To check for possible differences in efficiency of polyribosome entrance, we analyzed the mRNA content of different RIL isoforms bound by polyribosomes in transfected HeLa cells. As shown in Fig. 1D, there was no significant variation in polyribosome entrance of different species of RIL mRNAs and mRNA of GFP used as loading/transfection control.
Fusion with the Alternative C Terminus of RIL Destabilizes Reporter Proteins
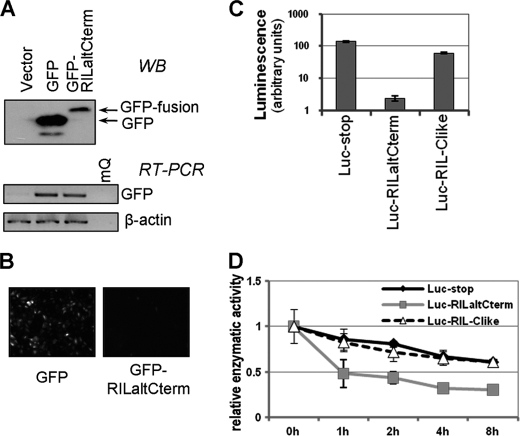
To check the ability of the alternatively spliced C terminus of RIL to destabilize heterologous proteins, we appended the last 30 amino acids of RILaltCterm to C termini of either EGFP or luciferase to generate EGFP-RILaltCterm or Luc-RILaltCterm, respectively. Western analysis indicates that the abundance of the EGFP-RILaltCterm fusion protein was significantly lower compared with the unmodified EGFP, although corresponding mRNA levels were not significantly different (Fig. 2A). Similarly, A549 cells transduced with the EGFP-RILaltCterm construct demonstrated a dramatically decreased fluorescence, compared with the A549 cells transduced with a similar construct carrying control EGFP (Fig. 2B). The luciferase fusion construct was tested by expression in H1299 cells, in parallel with the nonfused luciferase construct and an additional control construct Luc-RIL-Clike. The latter construct has similar to the Luc-RILaltCterm structure, except for one nucleotide insertion leading to an unrelated frame-shifted C-terminal peptide. Fusion of the alternative C-terminal segment of RIL to luciferase resulted in a dramatic (almost 100-fold) decrease in enzymatic activity, whereas the RIL-Clike unrelated peptide had no significant effect on luminescent signal (Fig. 2C).
FIGURE 2.
Alternative C-terminal peptide of RIL destabilizes heterologous proteins. RIL alternative C-terminal peptide was fused in-frame with GFP and lentivirally expressed in A549 cells (A and B). A, abundance of modified GFP protein was significantly decreased (by Western blot (WB)), although mRNA levels remain comparable (RT-PCR). B, decreased fluorescence of cells expressing GFP fused to RIL alternative C-terminus compared with nonmodified GFP, as examined by fluorescence microscopy. C and D, luciferase was fused with RIL alternative C terminus in-frame (Luc-RILaltCterm) or with a frameshift (Luc-RIL-Clike) as a control and introduced into H1299 cells by transfection. C, Luc-RILaltCterm activity was severely impaired compared with controls (Luc-stop and Luc-RIL-Clike). Transfection efficiency was normalized for by β-galactosidase. D, cells were treated with 1 μg/ml cycloheximide for 0, 1, 2, 4, or 8 h, and stability of modified luciferase was assessed based on its enzymatic activity. Luc-RILaltCterm has a lower half-life time relative to controls.
Next, we decided to test whether fusion with the RILaltCterm peptide changes stability of luciferase protein. Cells expressing Luc-RILaltCterm along with two control constructs were treated with protein synthesis inhibitor cycloheximide, and protein decay was monitored by measuring luciferase activity at different time points. Indeed, activity of Luc-RILaltCterm decayed steeply with a half-life of ∼1 h, although the unmodified protein along with the Luc-RIL-Clike control demonstrated a much slower decline (Fig. 2D). The results suggest that RIL alternative C terminus contains a PEST motif that can destabilize heterologous target proteins.
RILaltCterm Is Degraded by the Core 20 S Proteasome
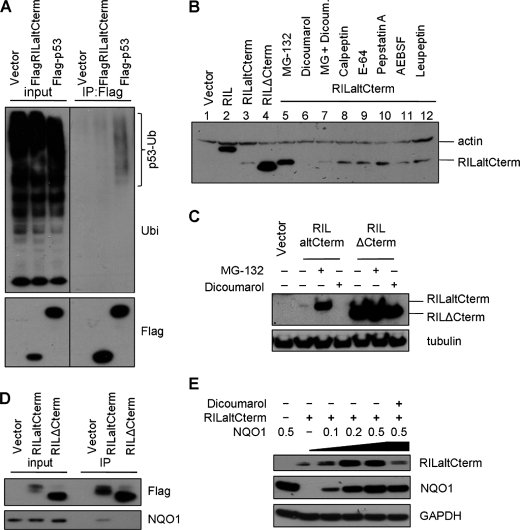
It has been previously reported that PEST sequences may be recognized by ubiquitin E3 ligases and target the protein to 26 S proteasome-dependent degradation. To check possible ubiquitination of RILaltCterm, lysates from 293T cells transiently expressing FLAG-RILaltCterm and treated with proteasome inhibitor MG-132 were immunoprecipitated with anti-FLAG-agarose and separated on gels, and Western blots were probed with anti-ubiquitin antibody. Here, FLAG-p53 was used as positive control as it is well established that p53 is ubiquitinated by HDM2 (54, 55). High molecular weight smears characteristic of polyubiquitination could only be detected in case of p53 but not RILaltCterm (Fig. 3A). These results indicate that RILaltCterm is not ubiquitinated, and hence its degradation could be ubiquitin-independent.
FIGURE 3.
RILaltCterm is degraded by the core 20 S proteasome. A, RILaltCterm is not ubiquitinated. HeLa cells expressing FLAG-tagged RILaltCterm or p53 were treated with 20 μm MG-132 for 6 h. Protein lysates were subjected to immunoprecipitation (IP) with anti-FLAG-agarose beads and analyzed by Western blot with antibodies specific to ubiquitin (Ubi) and FLAG epitope. B, RILaltCterm is degraded by the core 20 S proteasome. Western blot analysis of 293T cells transfected by constructs expressing HA-tagged full-length RIL, RILaltCterm, or RILΔCterm and treated with indicated compounds for 5 h. C, alternative C terminus of RIL targets the protein to the 20 S proteasome. HeLa cells were transfected by constructs expressing HA-tagged RILaltCterm or RILΔCterm and treated as indicated. Protein lysates were analyzed by Western blotting with anti-HA and anti-tubulin antibodies. D, RILaltCterm interacts with NQO1. HeLa cells were transfected with the indicated FLAG-tagged RIL constructs, and protein lysates were subjected to immunoprecipitation with anti-FLAG-agarose. Immune complexes were probed for the presence of NQO1. E, NQO1 protects RILaltCterm from degradation by the 20 S proteasome. HeLa cells were transfected with varying amounts of HA-RILaltCterm and FLAG-NQO1. NQO1 inhibitor dicoumarol was added where indicated. Levels of RILaltCterm, NQO1, and GAPDH proteins were analyzed by immunoblotting with anti-HA, FLAG, and GAPDH antibodies, respectively.
Rapid degradation of RILaltCterm could be mediated either by nonproteasomal degradation or by 20 S proteasomes. There are a number of studies showing that disordered/unfolded protein regions can be exposed and thus targeted to lysosomes (56) or easily accessible by other proteases (57, 58). Specifically, it has been previously reported that PEST sequences can be recognized and cleaved by calpain (59). Also, compelling evidence has recently emerged that proteins harboring unstructured regions can be degraded “by default” by the core 20 S proteasome (60). To probe these protein degradation pathways, we screened a panel of known protease inhibitors with defined specificities in 293T cells to assess their effect on the abundance of transiently expressed RILaltCterm protein. Regarding the 20 S proteasome degradation pathway testing, we used its activator dicoumarol. As shown in Fig. 3B, inhibition of neither calpain, cysteine, serine, nor aspartic proteases had any significant effect on the RILaltCterm protein level (Fig. 3B, lanes 8–11). Proteasomal inhibitor MG-132, however, stabilized RILaltCterm to a high extent (compare Fig. 3B, lanes 3 and 5), yielding as much protein as both full-length RIL and RILΔCterm used as stable positive controls (lanes 2 and 4). This result implies the involvement of proteasomes in RILaltCterm degradation. At the same time, dicoumarol treatment significantly enhanced the degradation making RILaltCterm protein undetectable (Fig. 3B, lane 6). Taken together with the observation that no ubiquitination of RILaltCterm could be detected, this latter result provides strong evidence that the core 20 S proteasome pathway is involved in the degradation process. The protein levels of both full-length RIL (data not shown) and RILΔterm truncation mutant (Fig. 3C) were not significantly affected by inhibition (by MG-132) or activation (by dicoumarol) of 20 S proteasomal degradation, which provides further evidence for the important role of RIL alternative C terminus in regulation of protein stability. NQO1 is known to modulate degradation in the 20 S proteasome by binding to its targets. Indeed, the RILaltCterm protein can interact with endogenous NQO1, as shown by co-immunoprecipitation in HeLa cells (Fig. 3D). Moreover, the alternatively spliced RIL isoform is significantly accumulated upon overexpression of recombinant NQO1 in a dose-dependent manner (Fig. 3E), and the protection is abolished by the NAD(P)H antagonist dicoumarol. The results establish that the stability of RILaltCterm isoform is controlled by the core 20 S proteasome degradation machinery.
RILaltCterm Is Involved in Reorganization of Actin Cytoskeleton
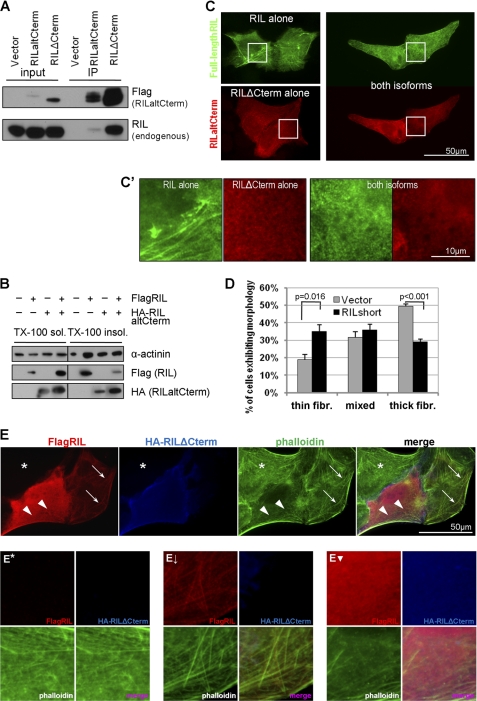
Reorganization of the actin cytoskeleton is a tightly regulated process. As an actin-associated protein, the full-length RIL is known to alter F-actin turnover (1). The alternatively spliced isoform could act as a modulator for fine-tuning the processes. The PDZ domain of RIL is known to associate with the LIM domain of RIL in a yeast two-hybrid system (61). No evidence for such interaction in mammalian cells has been reported so far. By co-immunoprecipitation, we found that both short isoforms of RIL (RILaltCterm and RILΔCterm) are able to bind endogenous full-length RIL in U2OS cells (Fig. 4A). FLAG-RILaltCterm precipitated significantly lower amounts of endogenous RIL because of its very high turnover rate and low abundance. Conversely, the deletion mutant RILΔCterm that lacks the degradation signal was strongly enriched in the precipitate and was able to pull down large amounts of endogenous RIL. We used RILΔCterm as a stable variant of RILaltCterm in subsequent experiments.
FIGURE 4.
RILaltCterm affects actin organization via interaction with full-length RIL. A, RILaltCterm binds to full-length RIL. Lysates from U2OS cells expressing FLAG-RILaltCterm were immunoprecipitated (IP) using anti-FLAG-agarose and probed with anti-RIL antibody. B, RILaltCterm changes distribution of full-length RIL between cytoskeleton-bound and cytosolic fractions and abrogates the effect of RIL on α-actinin-1. 293T cells were transfected as indicated and fractionated based on Triton X-100 solubility, and protein distribution was analyzed by immunoblotting. C, RILΔCterm affects the staining pattern of full-length RIL. U2OS cells were transfected with the indicated RIL constructs, fixed in 4% formaldehyde for 15 min, stained for FLAG and HA tags with suitable primary and fluorescently labeled secondary antibodies, and analyzed by wide field fluorescence microscopy. C′ shows enlarged detail of cell regions outlined in C. D, samples were processed as in C, and cells exhibiting thick fibers (fibr.), meshwork of thin fibers, or mixed staining pattern of full-length RIL in the presence or absence of RILaltCterm were counted. Bars represent an average of four independent experiments ± S.E., and p values were calculated by Student's t test. E, U2OS cells expressing FLAG-tagged full-length RIL and/or HA-tagged short isoform of RIL were grown on glass coverslips, fixed in 4% formaldehyde, and stained with rabbit anti-FLAG and rat anti-HA antibodies and then secondary AlexaFluor594 anti-rabbit and AlexaFluor350 anti-rat antibodies. F-actin was visualized by FITC-conjugated phalloidin. Asterisk indicates nontransfected cell; thick arrows indicate actin/RIL fibers; arrowheads indicate meshwork of thin actin fibers. E*, E↓, and E▾ represent enlarged detail of cells in E marked with asterisk, arrows, and arrowheads, respectively.
Next, we asked if interaction between the full-length and the shorter RIL isoforms would affect its intracellular distribution and the ability to induce formation of actin stress fibers. U2OS cells were co-transfected with constructs expressing FLAG- or HA-tagged full-length RIL and RILaltCterm and subjected to fractionation based on detergent solubility. Overexpression of the RILaltCterm isoform shifted distribution of the full-length RIL from Triton X-100-insoluble (cytoskeleton and nuclei) to the detergent-soluble (cytosol) fraction (Fig. 4B). Similarly, we analyzed distribution of α-actinin-1, an actin cross-linking protein and a marker for fibrillar actin. Upon introduction of RILaltCterm, the full-length RIL failed to increase the content of cross-linked actin fibers as evidenced by α-actinin distribution between cytoskeleton and cytosol. Same results were obtained using RILΔCterm (data not shown) confirming that both short isoforms of RIL are functionally similar.
Change in full-length RIL localization in the presence of RILΔCterm was confirmed by immunofluorescent staining (Fig. 4C). When expressed alone, the full-length RIL displayed a fibrillar pattern with characteristic thick fibers and occasional clusters (Fig. 4C, upper left panel), consistent with previous reports (1), although the RILΔCterm isoform stained more diffusely in the cytoplasm with thin fibers forming a dense mesh-like pattern (Fig. 4C, lower left panel). Although occasional fibrillar structures could still be observed, the filaments were short, thin, and disorganized. However, when both isoforms were co-expressed in the same cell, the full-length RIL no longer formed thick fibrillar structures, and its staining had changed to a mesh-work of thin fibers similar to that of the RILΔCterm isoform (Fig. 4C, right panel). The RILaltCterm isoform had a similar effect on the staining pattern of full-length RIL, although fewer RILaltCterm-positive cells could be detected due to the high turnover rate (data not shown). We quantified the proportion of RIL-expressing cells with thin or thick fibers or mixed RIL staining pattern in the presence or absence of the short isoform of RIL. Indeed, expression of RILΔCterm decreased substantially the number of cells with thick RIL fibers from 49.5 ± 2.6 to 29.2 ± 2.8% while increasing the number of cells with thin fibers from 18.8 ± 6.1 to 34.9 ± 7.5% (Fig. 4D).
As a final step, we analyzed the effect of RIL isoforms on F-actin morphology (Fig. 4E). When compared with nontransfected control (Fig. 4E, asterisk), cells expressing the full-length RIL exhibited increased formation of actin cables (Fig. 4E, arrows), and the protein co-localized markedly with the stress fibers. Co-expression with the short isoform dramatically changed the distribution of the full-length RIL to the mesh-work of thin fibers, and the content of F-actin was significantly reduced (Fig. 4E, arrowheads). Collectively, the results provide compelling evidence that RILaltCterm can participate in actin cytoskeleton reorganization by modifying distribution on the full-length isoform of RIL.
RILaltCterm Is Stabilized in Response to Oxidative Stress
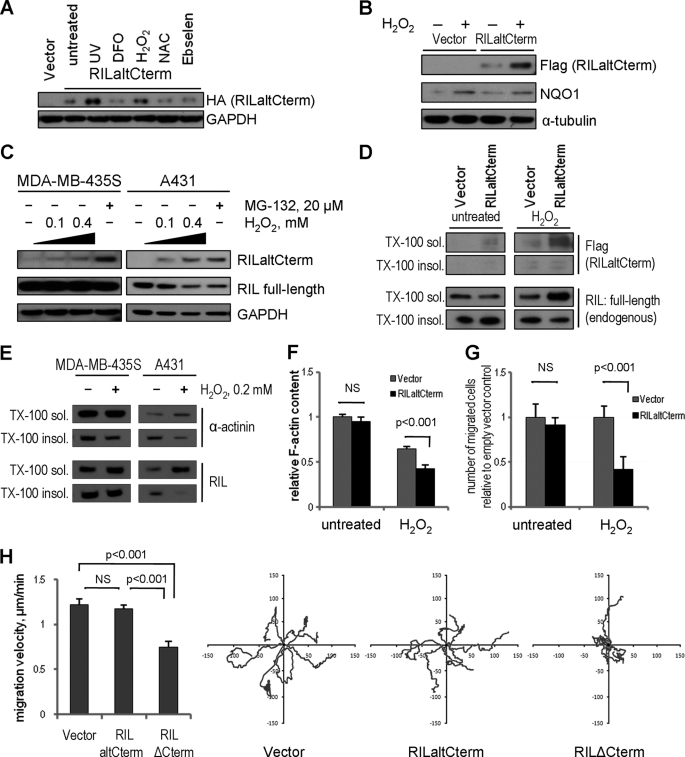
Binding of NQO1 to some of its targets is increased following γ-irradiation leading to protein stabilization (62–64). Ionizing radiation promotes formation of reactive oxygen species (ROS) and reactive nitrogen species that induce oxidative stress response. To find physiological conditions favoring accumulation of the RILaltCterm isoform, we checked whether oxidative stress induced by hydrogen peroxide treatment affects the 20 S proteasomal pathway. Consistent with previous reports, UV irradiation and to a lesser extent H2O2 treatment induced accumulation of the RILaltCterm isoform (Fig. 5A), although ROS and reactive nitrogen species scavengers N-acetyl-l-cysteine and ebselen as well as the hypoxia-mimetic deferoxamine mesylate decreased slightly the abundance of RILaltCterm. In agreement with our previous results (Fig. 3E), stabilization of RILaltCterm after oxidative stress coincided with the increase in expression of NQO1 (Fig. 5B). Immunoblot analysis on nontransfected cells treated with proteasome inhibitor MG-132 or increasing concentrations of H2O2 (using antibodies predicted to recognize both isoforms of RIL) detected an additional band that was identified as RILaltCterm by mass spectrometry analysis of immunoprecipitated material (Fig. 5C). The signal intensity increased dose-dependently with ROS challenge and was comparable with RIL full-length in abundance.
FIGURE 5.
Stabilization of RILaltCterm in response to oxidative stress attenuates cell migration. A, RILaltCterm is stabilized in response to oxidative stress. HeLa cells were transfected with construct expressing HA-RILaltCterm and treated as indicated. Stabilization of RILaltCterm was assessed by immunoblotting with anti-HA antibodies. B, RILaltCterm is stabilized in response to oxidative stress concomitantly with induction of NQO1. U2OS cells expressing FLAG-RILaltCterm were treated with 0.5 mm H2O2 for 4 h and then lysed and probed for FLAG, NQO1, or tubulin by Western blotting. C, oxidative stress induces accumulation of endogenous RILaltCterm. Nontransfected MDA-MB-435S and A431 cells were treated as indicated and protein lysates immunoblotted with anti-RIL antibodies capable of recognizing all isoforms. D, RILaltCterm redistributes full-length RIL from cytoskeleton to cytosol in response to oxidative stress. U2OS cells were treated as in B and fractionated based on Triton X-100 (TX-100 sol.) solubility. Distribution of FLAG-RILaltCterm and endogenous full-length RIL were analyzed by Western blotting. E, oxidative stress induces redistribution of endogenous RIL from cytoskeleton to cytosol. Nontransfected MDA-MB-435S and A431 were treated with 0.2 mm H2O2 where indicated and fractionated based on Triton X-100 solubility. Distribution of endogenous RIL and α-actinin-1 was analyzed by immunoblotting. F, stabilization of RILaltCterm in response to oxidative stress decreases F-actin content. Cells from B were fixed with formaldehyde in cytoskeleton buffer and permeabilized by 0.1% Triton X-100, and F-actin was stained by FITC-labeled phalloidin. Relative F-actin content was assessed by fluorescence intensity and normalized by DNA content (DAPI staining). Bars represent mean ± S.E., relative to empty vector controls. p values were calculated by Student's t test; NS, not significant. G, RAW264.7 monocytic cells were lentivirally transduced to express RILaltCterm and subjected to transwell chemotaxis assay with or without H2O2 treatment in triplicate. Migrated cells were fixed, stained, and automatically counted after 6 h of migration using ImageJ software. Bars represent mean ± S.E., relative to empty vector controls. p values were determined by Student's t test, NS, not significant. H, U2OS cells were transfected with RILaltCterm or RILΔCterm constructs or empty vector, and spontaneous nondirectional haptotactic motility was analyzed by time-lapse microscopy for 5 h. 20 cells in each sample were tracked using ImageJ Manual Tracking plugin. Bars represent mean velocity (μm/min) ± S.E. p values were calculated by one-way ANOVA; NS, not significant. Panels on the right show representative cell trajectories for each experimental group.
Stabilization of RILaltCterm in Response to Oxidative Stress Induces Reorganization of Actin Cytoskeleton
As we found that the short isoform of RIL can modify intracellular localization of full-length RIL, we checked if stabilization of RILaltCterm in response to oxidative stress would produce a similar effect. Indeed, we found that treatment with hydrogen peroxide of U2OS cells expressing RILaltCterm shifted full-length RIL from detergent-insoluble (or cytoskeleton-associated) to detergent-soluble (cytosol) fraction (Fig. 5D). Moreover, accumulation of RILaltCterm after prolonged (4 h) exposure to H2O2 significantly decreased the content of polymerized actin compared with vector-treated cells (Fig. 5F). In keeping with these results, marked redistribution of endogenous RIL from cytoskeleton-bound state to cytosol could be readily detected in nontransfected MDA-MB-435S and A431 cells (Fig. 5E).
Actin polymerization/disassembly balance mirrored by F-actin content is intimately involved in regulation of cell migration. Actin polymerization at the leading edge is the driving force behind lamellipodia protrusion, and actin cables within the cell body function as tracks for myosin during tail retraction (39, 65). Hence, we hypothesized that decreased F-actin content would lead to attenuated cell motility. To address this question experimentally, we subjected RAW264.7 monocytic cells to transwell chemotaxis assay. Monocytes extravasate and migrate along the gradient of chemoattractants while being exposed to oxidative stress associated with inflammation (65). Cells transduced with RILaltCterm or empty vector were preconditioned by 0.5 mm H2O2 for 4 h and then loaded into the upper chamber of a transwell system. Cells were induced to migrate toward the LPS added to lower chamber as a chemoattractant. Consistent with previous data, under oxidative stress conditions RILaltCterm markedly inhibited cell migration compared with empty vector. However, the alternatively spliced isoform of RIL failed to elicit any changes in motility in untreated monocytes (Fig. 5G). Accumulation of the short RIL isoform (mimicked by the stable deletion mutant RILΔCterm) decreased mean migration velocity in a model of spontaneous haptotactic cell motility by almost 40% in U2OS cells (Fig. 5H) corroborating previous results. These data allowed us to conclude that actin cytoskeleton rearrangement mediated by stabilization of RILaltCterm in response to oxidative stress might attenuate cell migration thus contributing to cellular stress response mechanisms.
DISCUSSION
Results of this study suggest a function for an alternatively spliced isoform of the RIL (PDLIM4) gene product. The isoform originates from skipping exon 6 sequences during splicing, which results in an 84-amino acid shorter protein, compared with major full-length RIL (2). The alternative splicing joining exon 5 and exon 7 removes the LIM domain and produces frameshift leading to a totally unrelated stretch of 22 amino acids at the C terminus. Functional characterization of the RIL gene has been carried out in a number of laboratories with cDNA constructs corresponding to the major full-length protein. RIL was implicated in regulation of actin stress fiber turnover. RIL protein binds to α-actinin, enhances its ability to interact with actin filaments, and promotes formation of actin stress fibers, although details of this mechanism are not clearly understood (1).
No information regarding functional properties of the alternative isoform of RIL has been reported to date, although numerous examples for other genes suggest that alternative splicing can substantially modify functions of the genes (66–68). RILaltCterm is extremely unstable, and the protein can hardly be identified by Western analysis even when overexpressed from recombinant constructs. To reveal the basis for its instability, we analyzed protein structure of the C-terminal segment that is translated from an alternative frame of exon 7 and found a PEST-like disordered protein structure (69) commonly associated with low protein stability (52, 53, 59, 60, 70, 71) because of its preferred targeting by E3 ubiquitin ligases, calpain, or by degradation in the core 20 S proteasomes. Placing the C-terminal 30-amino acid segment of RILaltCterm to GFP or luciferase was capable of reducing dramatically the protein half-life (from 48 and 12 h, respectively, to less than 1 h), suggesting that this 30-amino acid segment could be used as a model protein-destabilizing instrument.
In eukaryotes, the vast majority of short lived proteins are degraded by the 26 S proteasome complex following the process of polyubiquitination that marks the proteins for destruction. Regulatory 19 S subunit of the proteasome recognizes the polyubiquitin chain conjugated to the target protein, unfolds the protein, and passes it on to the catalytic core 20 S subunit where it is hydrolyzed to short peptides or single amino acids (72). We found no evidence of polyubiquitinylation of RILaltCterm in transfected cells, although transfected control p53 produced characteristic high molecular weight smears on Western blots developed with anti-ubiquitin antibody.
In bacteria and archaea, the proteasome lacks both the regulatory subunit and ubiquitin-conjugating machinery (73), and a similar ubiquitin-independent 20 S proteasome pathway has been found in eukaryotes (74, 75). It is generally accepted that the 20 S proteasome can degrade unfolded proteins (76) that may be partially denatured as a result of stresses or protein “aging” (57, 77). However, a large subclass of proteins feature intrinsically disordered regions that can either be folded upon binding to their targets or function as flexible linkers and thus remain unstructured (71). The intrinsically disordered proteins tend to be inherently unstable, being rescued from degradation through masking unstructured parts by certain binding partners (78). Degradation in the 20 S proteasomes can be regulated by NAD(P)H quinone oxidoreductase-1 (NQO1). The ubiquitously expressed NQO1 associates with the 20 S proteasome (63) and acts as its gatekeeper (60) by preventing degradation of proteins as diverse as ornithine decarboxylase (79) and p53 (64). We found that dicoumarol, an inhibitor of NQO1, enhances substantially the degradation of RILaltCterm. In addition, by co-immunoprecipitation we found that RILaltCterm interacts with endogenous NQO1. Although overexpression of recombinant NQO1 results in accumulation of the alternative isoform, treatment with dicoumarol abolishes the protection. Collectively, the results indicate that fast degradation of RILaltCterm is achieved by the 20 S proteasome machinery.
Previous observations in a yeast two-hybrid system have revealed binding of the C-terminal LIM domain of RIL with the PDZ domain located close to the N terminus (61). Here, we have shown by direct immunoprecipitation that the two isoforms of RIL (the full-length and RILaltCterm) can hetero-oligomerize (Fig. 4A). The interaction suggested possible mutual functional modulation of the two protein isoforms, such as a dominant-negative effect produced by the alternative isoform. In agreement with the previous report (1), we observed localization of full-length RIL along actin stress fibers where it interacts with the actin-bundling protein α-actinin-1 and assists in the formation of thick actin fibers and actin aggregates. We found that the alternatively spliced isoform of RIL displaces the full-length RIL from the actin fibers leading to reorganization of the cytoskeleton. Ectopic expression of the RILaltCterm isoform induced the release of full-length RIL from the cytoskeleton to the cytosol. The changes in subcellular localization proceeded in parallel with compromised association of RIL with the polymerized actin and α-actinin, leading to a decreased formation of actin fibers. Thus, we have shown that the RILaltCterm isoform can act as a dominant-negative regulator of the full-length RIL and thereby participate in modulation of actin cytoskeleton architecture and integrity.
We also show that RILaltCterm is stabilized in response to oxidative stress induced by UV irradiation or by hydrogen peroxide treatment in various cell types. These conditions result in a notable increase in the expression levels of NQO1. The increase in RILaltCterm protein stability following the treatments produced effects similar to those observed following overexpression of the shorter isoform. The release of full-length RIL from actin cytoskeleton is accompanied by a reduction in F-actin content in the cell and reorganization of actin cytoskeleton. As a consequence of its stabilization, the RILaltCterm isoform reduced cell migration rate in the chemotactic transwell assay.
NQO1 belongs to the phase II detoxifying enzymes that are induced to protect against electrophilic insults, oxidative stress, and ionizing radiation (70). Deficiency in NQO1 is observed in various cancers (80). Recently, it has been found that NQO1 plays a role in suppressing inflammatory response and in attenuation of macrophage migration induced by LPS (65), although the mechanism of the effect is poorly understood. We hypothesize that the stabilization of RILaltCterm by NQO1 in response to prolonged exposure to oxidative stress and subsequent cytoskeleton reorganization might contribute to gradual curbing of inflammatory reactions. Following exposure to ROS generated by phagocyte respiratory burst, direct oxidation of actin results in rapid assembly of filaments (81). Additional cytoskeleton rearrangement is induced through redox activation of signaling pathways affecting Src and Rho (82), and induction of myosin light chain phosphorylation leads to increased contractility and monocytic migration (83). Prolonged exposure to oxidants induces NQO1, which might stabilize RILaltCterm thereby assisting in gradual recovery of the changes in actin cytoskeleton and in cell motility.
Taken together, our results indicate that the alternatively spliced isoform of RIL (PDLIM4) can be activated by protein stabilization in response to certain stresses. The induction can modify the ability of full-length RIL to modulate organization of actin cytoskeleton and to affect cell motility.
Acknowledgments
We thank John Peterson and Eric Diskin (LRI Imaging Core) for help with microscopy.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 CA104903 and AG025276 (to P. M. C.). This work was also supported by Howard Hughes Medical Institute Grant 55005603 (to P. M. C.), Russian Foundation for Basic Research Grants 08-04-00686a (to P. M. C.) and 11-04-01210 (to E. I. F.), a grant from the Russian Academy of Sciences Program on Molecular and Cellular Biology (to P. M. C.), and Government of Russian Federation Grant 11.G34.31.0034 (to P. M. C.).
- EGFP
- enhanced GFP
- ROS
- reactive oxygen species.
REFERENCES
- 1. Vallenius T., Scharm B., Vesikansa A., Luukko K., Schäfer R., Mäkelä T. P. (2004) Exp. Cell Res. 293, 117–128 [DOI] [PubMed] [Google Scholar]
- 2. Bashirova A. A., Markelov M. L., Shlykova T. V., Levshenkova E. V., Alibaeva R. A., Frolova E. I. (1998) Gene 210, 239–245 [DOI] [PubMed] [Google Scholar]
- 3. Jani K., Schöck F. (2007) J. Cell Biol. 179, 1583–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. McKeown C. R., Han H. F., Beckerle M. C. (2006) Dev. Dyn. 235, 530–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Te Velthuis A. J., Isogai T., Gerrits L., Bagowski C. P. (2007) PLoS One 2, e189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Krcmery J., Camarata T., Kulisz A., Simon H. G. (2010) BioEssays 32, 100–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Han H. F., Beckerle M. C. (2009) Mol. Biol. Cell 20, 2361–2370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Xia H., Winokur S. T., Kuo W. L., Altherr M. R., Bredt D. S. (1997) J. Cell Biol. 139, 507–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Klaavuniemi T., Alho N., Hotulainen P., Kelloniemi A., Havukainen H., Permi P., Mattila S., Ylänne J. (2009) BMC Cell Biol. 10, 22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bauer K., Kratzer M., Otte M., de Quintana K. L., Hagmann J., Arnold G. J., Eckerskorn C., Lottspeich F., Siess W. (2000) Blood 96, 4236–4245 [PubMed] [Google Scholar]
- 11. Kotaka M., Kostin S., Ngai S., Chan K., Lau Y., Lee S. M., Li H., Ng E. K., Schaper J., Tsui S. K., Fung K., Lee C., Waye M. M. (2000) J. Cell. Biochem. 78, 558–565 [DOI] [PubMed] [Google Scholar]
- 12. Vallenius T., Luukko K., Mäkelä T. P. (2000) J. Biol. Chem. 275, 11100–11105 [DOI] [PubMed] [Google Scholar]
- 13. Torrado M., Senatorov V. V., Trivedi R., Fariss R. N., Tomarev S. I. (2004) Invest. Ophthalmol. Vis. Sci. 45, 3955–3963 [DOI] [PubMed] [Google Scholar]
- 14. Schulz T. W., Nakagawa T., Licznerski P., Pawlak V., Kolleker A., Rozov A., Kim J., Dittgen T., Köhr G., Sheng M., Seeburg P. H., Osten P. (2004) J. Neurosci. 24, 8584–8594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nakagawa N., Hoshijima M., Oyasu M., Saito N., Tanizawa K., Kuroda S. (2000) Biochem. Biophys. Res. Commun. 272, 505–512 [DOI] [PubMed] [Google Scholar]
- 16. Faulkner G., Pallavicini A., Formentin E., Comelli A., Ievolella C., Trevisan S., Bortoletto G., Scannapieco P., Salamon M., Mouly V., Valle G., Lanfranchi G. (1999) J. Cell Biol. 146, 465–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhou Q., Ruiz-Lozano P., Martone M. E., Chen J. (1999) J. Biol. Chem. 274, 19807–19813 [DOI] [PubMed] [Google Scholar]
- 18. Au Y., Atkinson R. A., Guerrini R., Kelly G., Joseph C., Martin S. R., Muskett F. W., Pallavicini A., Faulkner G., Pastore A. (2004) Structure 12, 611–622 [DOI] [PubMed] [Google Scholar]
- 19. Guy P. M., Kenny D. A., Gill G. N. (1999) Mol. Biol. Cell 10, 1973–1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Herrick S., Evers D. M., Lee J. Y., Udagawa N., Pak D. T. (2010) Mol. Cell. Neurosci. 43, 188–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Klaavuniemi T., Kelloniemi A., Ylänne J. (2004) J. Biol. Chem. 279, 26402–26410 [DOI] [PubMed] [Google Scholar]
- 22. Zheng M., Cheng H., Banerjee I., Chen J. (2010) J. Mol. Cell Biol. 2, 96–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Benna C., Peron S., Rizzo G., Faulkner G., Megighian A., Perini G., Tognon G., Valle G., Reggiani C., Costa R., Zordan M. A. (2009) Cell Tissue Res. 337, 463–476 [DOI] [PubMed] [Google Scholar]
- 24. van der Meer D. L., Marques I. J., Leito J. T., Besser J., Bakkers J., Schoonheere E., Bagowski C. P. (2006) Dev. Biol. 299, 356–372 [DOI] [PubMed] [Google Scholar]
- 25. Pashmforoush M., Pomiès P., Peterson K. L., Kubalak S., Ross J., Jr., Hefti A., Aebi U., Beckerle M. C., Chien K. R. (2001) Nat. Med. 7, 591–597 [DOI] [PubMed] [Google Scholar]
- 26. Zhou Q., Chu P. H., Huang C., Cheng C. F., Martone M. E., Knoll G., Shelton G. D., Evans S., Chen J. (2001) J. Cell Biol. 155, 605–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vatta M., Mohapatra B., Jimenez S., Sanchez X., Faulkner G., Perles Z., Sinagra G., Lin J. H., Vu T. M., Zhou Q., Bowles K. R., Di Lenarda A., Schimmenti L., Fox M., Chrisco M. A., Murphy R. T., McKenna W., Elliott P., Bowles N. E., Chen J., Valle G., Towbin J. A. (2003) J. Am. Coll. Cardiol. 42, 2014–2027 [DOI] [PubMed] [Google Scholar]
- 28. Selcen D., Engel A. G. (2005) Ann. Neurol. 57, 269–276 [DOI] [PubMed] [Google Scholar]
- 29. Xing Y., Ichida F., Matsuoka T., Isobe T., Ikemoto Y., Higaki T., Tsuji T., Haneda N., Kuwabara A., Chen R., Futatani T., Tsubata S., Watanabe S., Watanabe K., Hirono K., Uese K., Miyawaki T., Bowles K. R., Bowles N. E., Towbin J. A. (2006) Mol. Genet. Metab. 88, 71–77 [DOI] [PubMed] [Google Scholar]
- 30. Huang C., Zhou Q., Liang P., Hollander M. S., Sheikh F., Li X., Greaser M., Shelton G. D., Evans S., Chen J. (2003) J. Biol. Chem. 278, 7360–7365 [DOI] [PubMed] [Google Scholar]
- 31. Pomiès P., Macalma T., Beckerle M. C. (1999) J. Biol. Chem. 274, 29242–29250 [DOI] [PubMed] [Google Scholar]
- 32. Yamazaki T., Wälchli S., Fujita T., Ryser S., Hoshijima M., Schlegel W., Kuroda S., Maturana A. D. (2010) Cardiovasc. Res. 86, 374–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Szpirer C., Szpirer J., Rivière M., Hajnal A., Kiess M., Scharm B., Schäfer R. (1996) Mamm. Genome 7, 701–703 [DOI] [PubMed] [Google Scholar]
- 34. Fu S. L., Waha A., Vogt P. K. (2000) Oncogene 19, 3537–3545 [DOI] [PubMed] [Google Scholar]
- 35. Mazurek N., Sun Y. J., Price J. E., Ramdas L., Schober W., Nangia-Makker P., Byrd J. C., Raz A., Bresalier R. S. (2005) Cancer Res. 65, 10767–10775 [DOI] [PubMed] [Google Scholar]
- 36. Boumber Y. A., Kondo Y., Chen X., Shen L., Gharibyan V., Konishi K., Estey E., Kantarjian H., Garcia-Manero G., Issa J. P. (2007) Cancer Res. 67, 1997–2005 [DOI] [PubMed] [Google Scholar]
- 37. Qin T., Youssef E. M., Jelinek J., Chen R., Yang A. S., Garcia-Manero G., Issa J. P. (2007) Clin. Cancer Res. 13, 4225–4232 [DOI] [PubMed] [Google Scholar]
- 38. He M., Vanaja D. K., Karnes R. J., Young C. Y. (2009) Prostate 69, 1643–1650 [DOI] [PubMed] [Google Scholar]
- 39. Zhang Y., Tu Y., Zhao J., Chen K., Wu C. (2009) J. Cell Biol. 184, 785–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Carro M. S., Lim W. K., Alvarez M. J., Bollo R. J., Zhao X., Snyder E. Y., Sulman E. P., Anne S. L., Doetsch F., Colman H., Lasorella A., Aldape K., Califano A., Iavarone A. (2010) Nature 463, 318–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Morris M. R., Ricketts C., Gentle D., Abdulrahman M., Clarke N., Brown M., Kishida T., Yao M., Latif F., Maher E. R. (2010) Oncogene 29, 2104–2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mills J. C., Syder A. J., Hong C. V., Guruge J. L., Raaii F., Gordon J. I. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 13687–13692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gonzalez-Juarrero M., Kingry L. C., Ordway D. J., Henao-Tamayo M., Harton M., Basaraba R. J., Hanneman W. H., Orme I. M., Slayden R. A. (2009) Am. J. Respir. Cell Mol. Biol. 40, 398–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kesari A., Fukuda M., Knoblach S., Bashir R., Nader G. A., Rao D., Nagaraju K., Hoffman E. P. (2008) Am. J. Pathol. 173, 1476–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Noble C. L., Abbas A. R., Cornelius J., Lees C. W., Ho G. T., Toy K., Modrusan Z., Pal N., Zhong F., Chalasani S., Clark H., Arnott I. D., Penman I. D., Satsangi J., Diehl L. (2008) Gut 57, 1398–1405 [DOI] [PubMed] [Google Scholar]
- 46. Thurau M., Marquardt G., Gonin-Laurent N., Weinländer K., Naschberger E., Jochmann R., Alkharsah K. R., Schulz T. F., Thome M., Neipel F., Stürzl M. (2009) J. Virol. 83, 598–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Guryanova O. A., Makhanov M., Chenchik A. A., Chumakov P. M., Frolova E. I. (2006) Mol. Biol. (Mosk) 40, 448–459 [PubMed] [Google Scholar]
- 48. Belizario J. E., Alves J., Garay-Malpartida M., Occhiucci J. M. (2008) Curr. Protein Pept. Sci. 9, 210–220 [DOI] [PubMed] [Google Scholar]
- 49. Linding R., Jensen L. J., Diella F., Bork P., Gibson T. J., Russell R. B. (2003) Structure 11, 1453–1459 [DOI] [PubMed] [Google Scholar]
- 50. Linding R., Russell R. B., Neduva V., Gibson T. J. (2003) Nucleic Acids Res. 31, 3701–3708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Romero P., Obradovic Z., Li X., Garner E. C., Brown C. J., Dunker A. K. (2001) Proteins 42, 38–48 [DOI] [PubMed] [Google Scholar]
- 52. Rogers S., Wells R., Rechsteiner M. (1986) Science 234, 364–368 [DOI] [PubMed] [Google Scholar]
- 53. Rechsteiner M., Rogers S. W. (1996) Trends Biochem. Sci. 21, 267–271 [PubMed] [Google Scholar]
- 54. Haupt Y., Maya R., Kazaz A., Oren M. (1997) Nature 387, 296–299 [DOI] [PubMed] [Google Scholar]
- 55. Kubbutat M. H., Jones S. N., Vousden K. H. (1997) Nature 387, 299–303 [DOI] [PubMed] [Google Scholar]
- 56. Hayes S. A., Dice J. F. (1996) J. Cell Biol. 132, 255–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Goldberg A. L. (2003) Nature 426, 895–899 [DOI] [PubMed] [Google Scholar]
- 58. Wickner S., Maurizi M. R., Gottesman S. (1999) Science 286, 1888–1893 [DOI] [PubMed] [Google Scholar]
- 59. Bordone L., Campbell C. (2002) J. Biol. Chem. 277, 26673–26680 [DOI] [PubMed] [Google Scholar]
- 60. Asher G., Reuven N., Shaul Y. (2006) BioEssays 28, 844–849 [DOI] [PubMed] [Google Scholar]
- 61. Cuppen E., Gerrits H., Pepers B., Wieringa B., Hendriks W. (1998) Mol. Biol. Cell 9, 671–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Asher G., Lotem J., Cohen B., Sachs L., Shaul Y. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 1188–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Asher G., Tsvetkov P., Kahana C., Shaul Y. (2005) Genes. Dev. 19, 316–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Gong X., Kole L., Iskander K., Jaiswal A. K. (2007) Cancer Res. 67, 5380–5388 [DOI] [PubMed] [Google Scholar]
- 65. Liu H., Dinkova-Kostova A. T., Talalay P. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 15926–15931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Dubey D., Ganesh S. (2008) Hum. Mol. Genet. 17, 3010–3020 [DOI] [PubMed] [Google Scholar]
- 67. Narla G., Difeo A., Reeves H. L., Schaid D. J., Hirshfeld J., Hod E., Katz A., Isaacs W. B., Hebbring S., Komiya A., McDonnell S. K., Wiley K. E., Jacobsen S. J., Isaacs S. D., Walsh P. C., Zheng S. L., Chang B. L., Friedrichsen D. M., Stanford J. L., Ostrander E. A., Chinnaiyan A. M., Rubin M. A., Xu J., Thibodeau S. N., Friedman S. L., Martignetti J. A. (2005) Cancer Res. 65, 1213–1222 [DOI] [PubMed] [Google Scholar]
- 68. Yea S., Narla G., Zhao X., Garg R., Tal-Kremer S., Hod E., Villanueva A., Loke J., Tarocchi M., Akita K., Shirasawa S., Sasazuki T., Martignetti J. A., Llovet J. M., Friedman S. L. (2008) Gastroenterology 134, 1521–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Radivojac P., Iakoucheva L. M., Oldfield C. J., Obradovic Z., Uversky V. N., Dunker A. K. (2007) Biophys. J. 92, 1439–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Begleiter A., Fourie J. (2004) Methods Enzymol. 382, 320–351 [DOI] [PubMed] [Google Scholar]
- 71. Dyson H. J., Wright P. E. (2005) Nat. Rev. Mol. Cell Biol. 6, 197–208 [DOI] [PubMed] [Google Scholar]
- 72. Glickman M. H., Ciechanover A. (2002) Physiol. Rev. 82, 373–428 [DOI] [PubMed] [Google Scholar]
- 73. Maupin-Furlow J. A., Wilson H. L., Kaczowka S. J., Ou M. S. (2000) Front. Biosci. 5, D837–865 [DOI] [PubMed] [Google Scholar]
- 74. Bercovich Z., Rosenberg-Hasson Y., Ciechanover A., Kahana C. (1989) J. Biol. Chem. 264, 15949–15952 [PubMed] [Google Scholar]
- 75. Sheaff R. J., Singer J. D., Swanger J., Smitherman M., Roberts J. M., Clurman B. E. (2000) Mol. Cell 5, 403–410 [DOI] [PubMed] [Google Scholar]
- 76. Liu C. W., Corboy M. J., DeMartino G. N., Thomas P. J. (2003) Science 299, 408–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Davies K. J., Shringarpure R. (2006) Neurology 66, S93–S96 [DOI] [PubMed] [Google Scholar]
- 78. Buchler N. E., Gerland U., Hwa T. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 9559–9564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Asher G., Bercovich Z., Tsvetkov P., Shaul Y., Kahana C. (2005) Mol. Cell 17, 645–655 [DOI] [PubMed] [Google Scholar]
- 80. Talalay P., Dinkova-Kostova A. T. (2004) Methods Enzymol. 382, 355–364 [DOI] [PubMed] [Google Scholar]
- 81. Fiaschi T., Cozzi G., Raugei G., Formigli L., Ramponi G., Chiarugi P. (2006) J. Biol. Chem. 281, 22983–22991 [DOI] [PubMed] [Google Scholar]
- 82. Chiarugi P., Pani G., Giannoni E., Taddei L., Colavitti R., Raugei G., Symons M., Borrello S., Galeotti T., Ramponi G. (2003) J. Cell Biol. 161, 933–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Haorah J., Knipe B., Leibhart J., Ghorpade A., Persidsky Y. (2005) J. Leukocyte Biol. 78, 1223–1232 [DOI] [PubMed] [Google Scholar]