Abstract
Cryptococcal meningoencephalitis is an AIDS-defining illness caused by the opportunistic pathogen Cryptococcus neoformans. This organism possesses an elaborate polysaccharide capsule that is unique among pathogenic fungi, and the glycobiology of C. neoformans has been a focus of research in the field. The capsule and other cellular glycans and glycoconjugates have been described, but the machinery responsible for their synthesis remains largely unexplored. We recently discovered Xpt1p, an enzyme with the unexpected activity of generating a xylose-phosphate-mannose linkage. We now demonstrate that this novel activity is conserved throughout the C. neoformans species complex, localized to the Golgi apparatus, and functions in the O-glycosylation of proteins. We also present the first survey of O-glycans from C. neoformans.
Keywords: Carbohydrate Biosynthesis, Carbohydrate Structure, Glycoprotein, Glycosylation, Yeast, Cryptococcus neoformans, O-Glycan Synthesis, Glycosyltransferase, Pathogenic Fungus, Xylosyltransferase
Introduction
Members of the Cryptococcus neoformans species complex are the only organisms in the cryptococcal genus that are commonly associated with human disease. Historically categorized into four serotypes (A-D), these yeasts have more recently been classified into separate species and varieties: C. neoformans var. grubii (serotype A), C. neoformans var. neoformans (serotype D), and Cryptococcus gattii (serotypes B and C) (1). All four serotypes can be isolated from the environment, where they are most commonly found in association with avian excreta and soil (A and D) or certain tree species (B and C). Humans are infected via the inhalation of small yeast cells or basidiospores, which are typically neutralized within the lungs by the immune system without any symptomatic evidence of infection (2). In some instances, however, C. neoformans can disseminate from the lungs to other organs in the body; if it reaches the brain, it causes a fatal meningoencephalitis. Serotypes A and D are responsible for most infections of humans; they are generally associated with disease in immunocompromised individuals and have a worldwide distribution. In contrast, serotypes B and C characteristically infect immunocompetent populations and are typically found in tropical and subtropical regions, although C. gattii is also responsible for an ongoing outbreak in the Pacific Northwest (3).
The capsule is the most distinctive structure of C. neoformans, and because it is required for virulence, has been intensively studied. This intricate structure is primarily composed of two polysaccharides, glucuronoxylomannan (GXM)2 (4) and glucuronoxylomannogalactan (GXMGal) (5, 6). The capsule surrounds a cell wall composed of polymers of glucose, mannose, and N-acetyl-glucosamine, which are cross-linked to each other and to various highly glycosylated proteins. Cryptococcus species synthesize glycoproteins with extensive N- and O-linked modifications that may be comparable in mass to the protein itself (7–10); many of these bear glycosylphosphatidylinositol anchors (11–13) and are subsequently transferred to covalent linkage with the cell wall. The organism also synthesizes lipids modified with mannose, xylose, and galactose (14, 15). C. neoformans thus generates a broad array of cellular glycans and glycoconjugates that are only partially characterized. What we do know about their structures, however, indicates that they differ significantly from the glycan structures of their mammalian hosts as well as from related molecules in model yeasts.
We are interested in glycosyltransferases, enzymes that function in glycan synthesis by catalyzing the transfer of sugar moieties from activated donors (usually nucleotide sugars) to various acceptors (proteins, lipids, or other sugars). Previous studies of nucleotide sugar synthesis pathways in C. neoformans found that cells unable to produce UDP-xylose (UDP-Xyl) had no detectable Xyl residues in capsule polysaccharides, displayed shortened and deformed capsule fibers, and were avirulent in animal models of infection (16, 17). This physiological relevance of Xyl together with its frequent occurrence in cryptococcal glycoconjugates stimulated our interest in Xyl transfer activities. Using radiolabeled UDP-Xyl (UDP-[14C]Xyl) as the donor and α-1,3-linked dimannose (α-1,3-Man2) as the acceptor in an in vitro assay of glycosyltransferase activities, we have characterized two xylosyltransferase reactions in C. neoformans. One of these is the transfer of Xyl from UDP-Xyl to the reducing Man of the disaccharide substrate, forming a β-1,2-linkage (18). This is mediated by either of two proteins: Cxt1p (18) and Cxt2p.3 Cxt1p acts in the synthesis of both of the capsule polysaccharides (GXM and GXMGal) and some cellular glycolipids (19, 20); less is known regarding the function of Cxt2p.
Besides the xylose addition that is mediated by the Cxt proteins, we have discovered a completely distinct and novel activity of C. neoformans that adds xylose-phosphate (Xyl-P) to Man-containing substrates (21). The resulting Xyl-P-Man linkage has not been previously observed in the glycan structures of C. neoformans or any other organism nor has an enzyme with xylosylphosphotransferase activity been described. We have identified and characterized the C. neoformans protein responsible for this activity, xylosylphosphotransferase 1 (Xpt1p (21)). This protein is specific for UDP-Xyl as the reaction donor but readily modifies a variety of Man-containing substrates. A significant gap in our knowledge is the biological function of Xpt1p. Although the moiety it forms has not been detected in the known glycolipid or capsule polysaccharide structures of C. neoformans, many glycan elements of the cell remain uncharacterized. Below, we describe our investigation into the function of Xpt1p and present evidence that this enzyme acts in the O-linked glycosylation of proteins.
EXPERIMENTAL PROCEDURES
Materials
UDP-[U-14C]xylose (UDP-[14C]Xyl; 151 mCi/mmol) was from PerkinElmer Life Sciences, and α-1,3-d-mannobiose (α-1,3-Man2) was from Carbohydrate Synthesis (Oxford, UK). The murine monoclonal anti-GXM antibodies used were 3C2 and 339 (22), F12D2 (23), 1255 (24), and 1326 (25). Unless specified, all other chemicals or reagents were obtained from Sigma.
Strains, Plasmids, and Viability Testing
C. neoformans strains (Table 1) were grown in liquid culture at 30 °C in YPD medium (1% w/v yeast extract, 2% w/v peptone, 2% w/v dextrose) with shaking (230 rpm) or at 30 °C on YPD agar plates (YPD medium with 2% w/v agar). As appropriate, media included 100 μg/ml nourseothricin (Werner BioAgents) and/or Geneticin® (G418; Invitrogen). For immunofluorescence localization, strains were cultured at 30 °C in minimal medium (0.17% yeast nitrogen base without amino acids, 2% w/v dextrose).
TABLE 1.
C. neoformans strains used in these studies
| Namea | Serotype | Origin |
|---|---|---|
| KN99α | A | Nielsen et al. (43) |
| KN99α xpt1Δ | A | Reilly et al. (21) |
| KN99α xpt1Δ pXPT1 | A | Reilly et al. (21) |
| KN99α xpt1Δ pXPT1-HA | A | Reilly et al. (21) |
| KN99α xpt1Δ pXPT1(D) | A | This study |
| KN99α cap59Δ | A | Baker et al. (44) |
| WM276 | B | Warren et al. (45) |
| MMRL2651 | C | Fraser et al. (46) |
| JEC21 | D | Kwon-Chung et al. (47) |
| JEC21 pXPT1 | D | This study |
| JEC21 pXPT1-HA | D | This study |
| JEC21 GMT1-HA | D | This study |
| JEC21 GMT1-HA pXPT1 | D | This study |
| JEC21 GMT1-HA pXPT1-FLAG | D | This study |
a All strains are MATα.
To test whether Xpt1 from serotype D complements the defect in a serotype A xpt1Δ strain, the coding sequence of the serotype D gene, designated here as XPT1(D), was PCR-amplified from JEC21 genomic DNA (prepared as described in Wickes et al. (26)) using primers MSPE-003 (CACCGCGCCCCGCCCGAAAATGCCGCCGACCGCGCTGC) and MSPE-004 (CTCCTTCTACGTTCTATGTCTAGTCATGATATCTGTCTTTTACAGG). Regions flanking the serotype A XPT1 gene were PCR-amplified from the plasmid pXPT1 (21) using primers MSPE-001 (GCTCAGTGTCTCGCATATGGT) and MSPD-002 (GCTGCAGGAATTCGATATCAAGC) for the upstream sequence and primers MSPD-005 (GCTCTCCAGCTCACATCCTCGCAGCGTTCGAGAGTTGCTTTTCACGG) and MSPE-006 (CTTACAACGGCTGGTACTCTGG) for the downstream sequence. The three amplicons (upstream sequence, coding region, and downstream sequence) were next assembled by three-way fusion PCR using primers MSPE-001 and MSPE-006, and the resulting product was digested with HpaI and NheI and cloned into pXPT1 that had been digested with HpaI and NheI and treated with calf intestinal alkaline phosphatase. The resulting pXPT1(D) plasmid, differing from pXPT1 only in the XPT1 coding sequence, was transformed into DH5α Escherichia coli cells, isolated, linearized with I-SceI, and transformed into the C. neoformans strain KN99α xpt1Δ by electroporation (26). G418-resistant transformants were screened by PCR for the presence of serotype D XPT1 sequence, and positive clones were assayed for xylosyltransferase activity.
For some experiments an Xpt1p overexpression strain was used. This was generated by transforming pXPT1 linearized with I-SceI into KN99α by electroporation and selecting and maintaining transformants in YPD containing 100 μg/ml G418.
To test C. neoformans viability under various growth conditions, cells from an overnight culture were washed in water and adjusted to 2 × 106 cells/ml; 5 μl (104 cells) of this suspension and of four 10-fold serial dilutions were spotted on YPD agar or YPD stressor plates (YPD agar with 0.5 mg/ml caffeine, 0.05% sodium dodecyl sulfate (SDS), 1 mm sodium nitrite, or 0.5 mm hydrogen peroxide) and then incubated at 30 °C or 37 °C.
Total Membrane Preparation and Detergent Extraction
C. neoformans membranes and their detergent extracts were prepared as in Reilly et al. (21). Briefly, cells from an overnight culture were washed in Tris-EDTA buffer (100 mm Tris-HCl, pH 8.0, 0.1 mm EDTA) and broken with glass beads, and the resulting lysate was subjected to a clearing centrifugation step. Total membranes were isolated from the resulting supernatant fraction by ultracentrifugation, resuspended in Tris buffer (100 mm Tris-HCl, pH 8.0), and stored at 4 °C. Protein concentration of the total membrane sample was determined using the Bio-Rad Protein Assay. For some studies, membranes were extracted with 1% Triton X-100 and subjected to a second round of ultracentrifugation, and the resulting supernatant fraction was stored at 4 °C. Protein concentration of the Triton X-100 extract sample was determined using the Bio-Rad Detergent Compatible Protein Assay.
Xylosyltransferase Activity Assays
Enzyme activity was assayed by monitoring the transfer of [14C]Xyl from a UDP-[14C]Xyl donor to an α-1,3-Man2 acceptor as in (21). Briefly, reactions containing 625 μg of protein (from C. neoformans total membranes or Triton X-100 extracts), 1 μm UDP-[14C]Xyl, 8.8 mm α-1,3-Man2, and 7.5 mm MnCl2 in 100 mm Tris-HCl, pH 6.5, were incubated overnight at 20 °C. Unreacted UDP-Xyl was removed with anion exchange resin, and the reaction products were resolved by thin layer chromatography (TLC) and visualized by autoradiography.
Growth in Mice
For each strain tested, eight 4–6-week-old female C57Bl/6 mice (The Jackson Laboratory) were anesthetized with a combination of Ketaset-HCl and xylazine and inoculated intranasally with 50 μl of a 2.5 × 105 cells/ml suspension in phosphate-buffered saline, pH 7.4 (PBS). Three animals from each cohort were sacrificed at 1 h post-inoculation; the remaining five were sacrificed at 7 days post-inoculation. After sacrifice, lungs were harvested and homogenized in PBS, and serial dilutions of the homogenate were plated on YPD agar for determination of colony-forming units. Initial inocula were also plated to confirm colony-forming units.
Capsule Staining
Cells from an overnight culture of C. neoformans were washed twice with PBS, resuspended at 107 cells/ml in PBS containing 1% bovine serum albumin (PBS + 1% BSA), and rotated for 1 h at room temperature with a final concentration of 8 μg/ml anti-GXM antibody. Cells were washed twice with PBS and then incubated for 1 h at room temperature with rotation in PBS + 1% BSA containing 3.2 μg/ml Alexa Fluor® 546 goat anti-mouse IgG (Invitrogen). Cells were again washed twice with PBS and suspended in PBS for visualization. Bright field and fluorescence images were acquired simultaneously on a Zeiss Axioskop 2 MOT Plus wide-field fluorescence microscope; all samples were imaged with identical acquisition settings.
Capsule Induction
Cells from an overnight culture of C. neoformans were resuspended at 105 cells/ml in 50 ml of YPD media plus appropriate antibiotics and again cultured overnight. The subcultured cells were resuspended at 107 cells/ml in 10 ml of Dulbecco's modified Eagle's medium, transferred to a 25-cm2 tissue culture flask, and incubated at 37 °C with 5% CO2 for 20 h. 1 ml of cells was harvested, washed once with water, and resuspended in 100 μl of India ink for visualization. Bright field images were acquired on a Zeiss Axioskop 2 MOT Plus wide-field fluorescence microscope as above.
Lipid Glycosylation
Reactions of 625 μg of total membrane protein, 1 μm UDP-[14C]Xyl, and 7.5 mm MnCl2 in 100 mm Tris-HCl, pH 6.5, were incubated overnight at 20 °C. Chloroform and methanol were added for a final ratio of 10:10:3 (chloroform:methanol:buffer); the resulting mixture was incubated for 30 min at room temperature with occasional vortex mixing and then centrifuged to remove particulate material (16,000 × g; 2 min). The supernatant fraction was transferred to a fresh tube and dried under a stream of nitrogen gas. The sample was then resuspended in n-butanol, vortexed with an equal volume of water, and centrifuged as before, and the resulting organic phase was transferred to a fresh tube. The remaining aqueous phase was again extracted with an equal volume of n-butanol, and the pooled organic phases were washed with an equal volume of water, transferred to a fresh tube, and dried as before. The sample was resuspended in 1:1 methanol:water, applied to a Silica Gel 60 plate (EM Sciences), and resolved by TLC in a solvent system of 10:10:3 chloroform:methanol:water. The dried plate was sprayed with En3Hance® Spray (PerkinElmer), and radiolabeled products were visualized by autoradiography.
Protein Glycosylation
Xylosyltransferase reactions were performed as in the previous section. After overnight incubation, some reactions were treated with 1 unit of Protease Type XIV (from Streptomyces griseus) or a buffer control (0.01 m sodium acetate, 0.005 m calcium acetate, and 0.01 m CaCl2) for 24 h at 37 °C. All samples were mixed with sample buffer, heated for 15 min at 100 °C, and resolved by SDS-PAGE on a 10% gel according to standard methods (27). The gel was fixed for 1 h in 5:4:1 methanol:water:acetic acid, incubated in Enlightning Rapid Autoradiography Enhancer (PerkinElmer) for 30 min, dried onto 3MM chromatography paper (Whatman), and visualized by autoradiography.
Preparation of Protein-linked Glycans for Analysis
A 50-ml overnight culture of C. neoformans was used to inoculate 1 liter of YPD with or without 100 μg/ml G418 at 105 cells/ml. The sub-cultured cells were grown for 24 h (to ∼107 cells/ml), harvested by centrifugation, and washed with Tris-EDTA buffer. Cells were resuspended in 10 ml of the same buffer and disrupted with glass beads, and the resulting lysate was subjected to a clearing centrifugation step as described in Reilly et al. (21) for the preparation of membrane proteins. The supernatant fraction was transferred to a fresh tube, and CHAPS was added to a final concentration of 1%; the sample was incubated at 4 °C with rocking for 2 h and then subjected to ultracentrifugation (75,000 × g; 45 min). The detergent-soluble fraction was transferred to cellulose dialysis tubing (8000 Mr cut-off; from Fisher) and dialyzed against 2 liters of 50 mm ammonium bicarbonate buffer at 4 °C; the buffer was changed every 12 h for a total of 8 liters over 48 h. The dialyzed sample was transferred to a 50-ml conical tube, frozen at −80 °C, and lyophilized. The lyophilized protein material was washed by precipitation using ice-cold 80% acetone to reduce residual detergent and other polymeric contaminants that can interfere with mass spectrometric detection (a polyhexose ladder of unknown origin, also observed in similar analyses of non-cryptococcal samples, remains at low levels and is indicated on spectra below). Protein content of the washed, lyophilized material was determined by BCA assay to be 169 ± 42 or 198 ± 27 μg of protein/mg of lyophilized material for KN99α or KN99α xpt1Δ, respectively (n = 3).
Oligosaccharides were released from the C. neoformans samples by reductive β-elimination as detailed in Aoki et al. (28). Briefly, the lyophilized C. neoformans material (2.5 mg) was resuspended in a solution of 100 mm sodium hydroxide and 1 m sodium borohydride and incubated for 18 h at 45 °C. The reaction mixture was neutralized with 10% acetic acid on ice and then loaded onto a column of AG 50W-X8 cation-exchange resin (Bio-Rad) for desalting. The column run-through and a subsequent wash with three bed volumes of 5% acetic acid, both containing released oligosaccharides, were combined and lyophilized to dryness. To remove borate from the sample, a solution of 10% acetic acid in methanol was added, and the sample was then dried under a stream of nitrogen gas at 37 °C; this was repeated for a total of 5 times. The sample was then resuspended in 5% acetic acid and loaded onto a C18 cartridge column (Mallinckrodt Baker) that was previously washed with acetonitrile and pre-equilibrated with 5% acetic acid. Flow-through from the column was collected after loading; the column was then washed a total of five times with 5% acetic acid. The flow-through and washes were combined and evaporated to dryness.
Analysis of Glycans by Nanospray Ionization Mass Spectrometry (NSI-MSn)
Samples were analyzed as in Aoki et al. (28). Briefly, the released oligosaccharide sample was permethylated (29), dissolved in 1 mm sodium hydroxide in 50% methanol, and infused into a linear ion trap mass spectrometer equipped with an Orbitrap FT detector (LTQ-Orbi Discoverer, Thermo-Fisher). MS analysis was performed in positive ion mode. The total ion mapping (TIM) function of the Xcalibur software package (Thermo-Fisher) was utilized to detect and quantify the prevalence of individual glycans in the total glycan profile; peaks were considered quantifiable if they were 2-fold or greater above background. The generation of TIM profiles includes automated acquisition of MS/MS spectra in overlapping collection windows (Δm/z = 2.8 mass units, window-to-window overlap set at 0.8 mass units) across user-defined mass ranges (generally from m/z = 200–2000). These fragmentation spectra highlighted the presence of cryptococcal glycans at particular m/z values, providing guidance for subsequent experiments in which glycans of interest were manually fragmented (MSn) to assign structure.
Protein Localization
pXPT1-FLAG, a construct for the expression of a C-terminal FLAG-tagged Xpt1p, was generated exactly as pXPT1-HA (described in Reilly et al. (21)) except that primers MCR136 (TTACTTGTCATCGTCATCCTTGTAATCATCATTATACCTATCTTTTACAGGATCCC) and MCR137 (GATTACAAGGATGACGATGACAAGTAAACATAGAACGTAGAAGGAGATGGAGG) were used in place of MCR120 and MCR121, respectively. The pXPT1-HA or pXPT1-FLAG plasmids were linearized with I-SceI and transformed into the C. neoformans strain JEC21 by electroporation (26), and transformants were selected by growth on G418 plates.
Cells were cultured overnight in minimal medium (final density ∼108 cells/ml), mixed with formaldehyde (4% final), and incubated for 30 min at 30 °C with shaking. All subsequent steps were performed at room temperature. Cells (4 × 108) were harvested by centrifugation (400 × g; 1 min), washed once with PBS, resuspended in 1 ml of a fresh solution of 4% formaldehyde in PBS, and incubated for 30 min with rotation. The sample was then washed 3 times with PBS, resuspended in 1 ml of lysis buffer (50 mm sodium citrate pH 6.0, 1 m d-sorbitol, 35 mm β-mercaptoethanol) plus 25 mg/ml Lysing Enzymes (from Trichoderma harzianum), and incubated for 1 h at 30 °C with occasional inversion. After digestion, the cells were washed twice with HS Buffer (100 mm HEPES, pH 7.5, 1 m d-sorbitol) and resuspended in 200 μl of HS Buffer.
The washed cell suspension was spotted in 20-μl aliquots on a glass microscope slide coated with 0.1% poly-l-lysine and incubated for 20 min at room temperature, and the buffer was removed by aspiration. All subsequent treatments and washes were performed by the application of 20-μl volumes and incubation at room temperature and were followed by aspiration. The slides were first treated with HS Buffer containing 1% Triton X-100 and incubated for 10 min; they were then washed 3 times with HS Buffer and 3 times with PBS. The slides were next treated with Blocking Buffer (5% goat serum, 0.02% Triton X-100 in PBS) for 1 h followed by either a high affinity rat anti-HA monoclonal antibody (20 ng/ml in Blocking Buffer; from Roche Applied Science) or Blocking Buffer alone overnight in a moist chamber at 4 °C. Samples were then washed six times with Blocking Buffer and stained with Alexa Fluor® 594 goat anti-rat IgG (1 μg/ml in Blocking Buffer; from Invitrogen) for 1 h in the dark; next, slides were washed 6 times with Blocking Buffer, 3 times with PBS, and then stained with DAPI (5 μg/ml in PBS) for 30 min in the dark. Finally, samples were washed three times with PBS and allowed to air-dry. Prolong Gold was applied to each sample followed by a glass coverslip, and the slide was stored for less than 24 h at −20 °C in the dark. Bright field and fluorescence images were acquired as for the capsule staining assays above.
RESULTS
Xylosyltransferases in C. neoformans
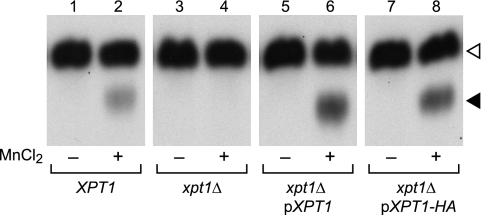
We have identified several xylosyltransferase activities in C. neoformans by assaying the transfer of radiolabeled Xyl from the nucleotide sugar donor UDP-[14C]Xyl to an α-1,3-Man2 oligosaccharide acceptor (see “Experimental Procedures”). When membranes prepared from the wild-type strain KN99α were used as the source of enzyme activity in this assay, two distinct products were observed (Fig. 1, track 2). The upper product (open arrowhead) is Xyl-linked β-1,2 to the reducing Man of the α-1,3-Man2 substrate (18); this is synthesized primarily by Cxt1p, which generates a Man-α(1→3)[Xyl-β(1→2)]-Man motif that occurs in one of the major cryptococcal glycosylinositol phosphorylceramides, GXM, and GXMGal (19, 20). The lower product (closed arrowhead) is Xyl-P-linked to the non-reducing Man of α-1,3-Man2, a completely novel linkage that is generated by Xpt1p (21). Strains in which XPT1 has been replaced with a drug resistance cassette no longer generate this product (Fig. 1, track 4), a defect that can be complemented by the episomal expression of XPT1 (Fig. 1, track 6; Ref. 21). Modifying the episomal copy to incorporate an HA epitope tag at the protein C-terminus does not alter enzyme activity in this assay (Fig. 1, track 8).
FIGURE 1.
Xylosyltransferase activities in C. neoformans. Total membranes prepared from wild-type KN99α (XPT1), mutant (xpt1Δ), or episomally-complemented mutant (xpt1Δ pXPT1 and xpt1Δ pXPT1-HA) strains were assayed with UDP-[14C]Xyl and α-1,3-Man2 in the absence (−) or presence (+) of MnCl2, as described under “Experimental Procedures.” An autoradiograph of the products resolved by TLC is shown; no signal was detected in other regions of the plate beyond minor amounts of free Xyl. The filled triangle indicates the product of the manganese-dependent xylosyltransferase activity, and the open triangle indicates the product of the unrelated xylosyltransferase Cxt1p (see “Results”). No signal was detected in the absence of the α-1,3-Man2 acceptor (not shown).
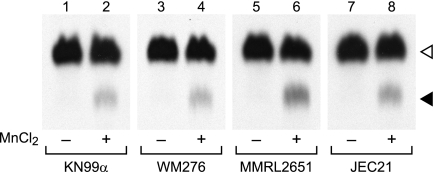
We began our investigation into the biological function of Xpt1p by exploring its conservation among members of the C. neoformans species complex, which differ in terms of ecological niche, host preference, and disease outcome (1). To do this, we performed xylosyltransferase activity assays on membranes prepared from cryptococcal strains representing the four major historically defined serotypes: A (KN99α, H99), B (WM276, R265), C (NIH312, MMRL2751), and D (JEC21, KN355α, KN433α). In each strain tested, we observed a xylosylated species that co-migrated with the Xpt1p product made by KN99α membranes and was similarly manganese-dependent (Fig. 2 and data not shown). These results were consistent with our identification of sequences homologous to the serotype A XPT1 (locus CNAG_04860, from the C. neoformans var. grubii H99 data base maintained by the Broad Institute) in the genomes of serotypes B (locus CNBG_5687; from the C. gattii R265 data base maintained by the Broad Institute) and D (locus CNJ02890; from the C. neoformans var. neoformans JEC21 data base maintained by TIGR).4
FIGURE 2.
Xpt1p activity in members of the C. neoformans species complex is comparable. Total membranes prepared from the indicated strains were assayed with UDP-[14C]Xyl and α-1,3-Man2 in the absence (−) or presence (+) of MnCl2, as described under “Experimental Procedures.” An autoradiograph of the products resolved by TLC is shown; no signal was detected in other regions of the plate beyond minor amounts of free Xyl. The filled triangle indicates the product of the manganese-dependent xylosyltransferase activity, and the open triangle indicates the product of the unrelated xylosyltransferase Cxt1p (see “Results”). No signal was detected in the absence of the α-1,3-Man2 acceptor (not shown).
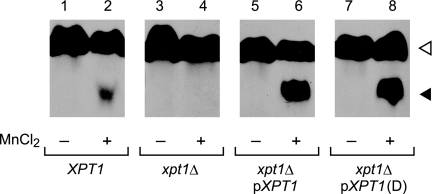
We next investigated whether the XPT1 sequence from one serotype could complement deletion of the native XPT1 from another. Membranes from serotype A xpt1Δ cells, which exhibit no Xpt1p activity (Fig. 3, track 4) acquired activity upon episomal expression of the serotype D gene (Fig. 3, track 8). This activity was equal to that conferred by expression of the native serotype A gene from the same plasmid backbone (Fig. 3, track 6). Xylosylphosphotransferase activity in the mutant strain complemented with either plasmid exceeded that of wild-type cells (Fig. 3, track 2), likely due to the presence of multiple copies of the plasmids.
FIGURE 3.
Expression of XPT1 from serotypes A and D similarly complements the serotype A xpt1Δ mutant. Total membranes prepared from serotype A wild-type (XPT1), serotype A xpt1Δ (xpt1Δ), serotype A xpt1Δ cells expressing XPT1 from the same serotype (xpt1Δ pXPT1), and serotype A xpt1Δ cells expressing XPT1 from serotype D (xpt1Δ pXPT1 (D)) were assayed with UDP-[14C]Xyl and α-1,3-Man2 in the absence (−) or presence (+) of MnCl2, as described under “Experimental Procedures.” An autoradiograph of the products resolved by TLC is shown; no signal was detected in other regions of the plate beyond minor amounts of free Xyl or in the absence of the α-1,3-Man2 acceptor (not shown). The filled triangle indicates the product of the manganese-dependent xylosyltransferase activity, and the open triangle indicates the product of the unrelated xylosyltransferase Cxt1p (see “Results”).
Growth of xpt1Δ Strains
We next assessed the basic growth characteristics of cells deleted for XPT1 (xpt1Δ) compared with wild-type. Cultures of KN99α and KN99α xpt1Δ grew comparably in liquid and on solid rich medium; this held true at both 30 °C (the optimal growth temperature for C. neoformans) and 37 °C (data not shown). Similar results were obtained when the plate medium was supplemented with caffeine, SDS, sodium nitrate, or hydrogen peroxide, which act as cell wall, cell membrane, nitrosative, and oxidative stressors, respectively (data not shown). We also compared the growth of KN99α and KN99α xpt1Δ in a mouse model of cryptococcal infection. The levels of colony-forming units recovered from the lungs of mice after intranasal inoculation were the same for the wild-type and deletion strains (data not shown). Collectively, these studies indicated that the loss of Xpt1p did not influence growth or viability under the conditions tested, but they gave no indication of its cellular function.
Potential Roles of Xpt1p
We examined the potential role of Xpt1p in capsule formation by comparing the capsule morphologies of KN99α, KN99α xpt1Δ, and KN99α xpt1Δ complemented with the native gene on a plasmid (pXPT1). We first assessed the binding of monoclonal anti-GXM antibodies to the cell surface. Each of the antibodies tested (see “Experimental Procedures”) demonstrated uniform staining around the cell periphery (shown for antibody F12D2 in supplemental Fig. 1, panel A), indicating no obvious alteration in the mutant strain at the level of immunofluorescence microscopy. We also tested the ability of mutant strains to increase the radius of their capsule in response to “inducing” growth conditions (30). Both the KN99α and KN99α xpt1Δ strains enlarged the capsule to a comparable extent (supplemental Fig. 1, panel B). For comparison, an acapsular control strain, cap59Δ, showed no capsule staining or induction (supplemental Fig. 1).
Prior studies of capsule polysaccharides did not indicate the presence of Xyl-P in GXM or GXMGal (4–6). To confirm this finding, 31P NMR analysis was performed on GXM prepared from KN99α; no phosphate signal was detected in this analysis (data not shown).5 As this absence is consistent with the lack of capsule alteration in xpt1Δ mutant cells, we did not pursue additional analysis of capsule polysaccharides in the mutant.
We next considered that Xpt1p might participate in the xylosylation of cryptococcal glycolipids. To test this we performed reactions similar to our standard xylosyltransferase assay, but lacking the α-1,3-Man2 substrate such that only endogenous compounds were available as acceptor molecules. Membrane preparations from wild-type, mutant, and complemented strains (KN99α, KN99α xpt1Δ, and KN99α xpt1Δ pXPT1) were assayed in this manner, and the lipids were then extracted from the radiolabeled samples for analysis by TLC. Although we detected the incorporation of [14C]Xyl into several lipid species, this activity was not dependent on the presence of Xpt1p (data not shown). These data indicated that Xpt1p does not act in glycolipid synthesis.
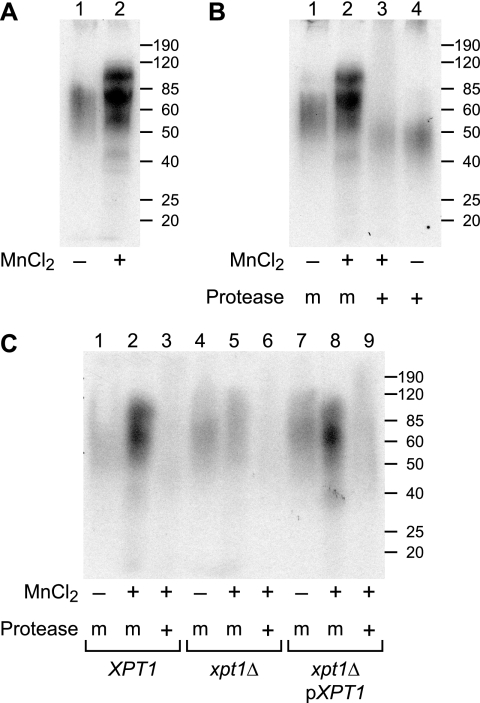
We speculated that another potential role for Xpt1p was in protein glycosylation. Similar to the lipid experiment outlined above, we incubated cryptococcal membranes with UDP-[14C]Xyl (but not α-1,3-Man2) and then analyzed the samples by SDS-PAGE. KN99α membranes assayed in this manner incorporated [14C]Xyl into material that migrated between 50 and 100 kDa (Fig. 4, panel A). Notably, the observed incorporation of [14C]Xyl increased significantly upon the addition of MnCl2 to the reactions, indicating that some xylosylation was due to a manganese-dependent activity (Fig. 4, panel A, compare lanes 1 and 2). Protease treatment of these assay products reduced the [14C]Xyl signal relative to mock-digested controls, demonstrating that the radiolabel was indeed associated with a polypeptide species (Fig. 4, panel B).
FIGURE 4.
Protein glycosylation studies of xpt1Δ mutant. Total membranes prepared from the indicated strains were assayed with UDP-[14C]Xyl in the presence (+) or absence (−) of MnCl2 as indicated; some samples were subsequently mock-treated (m) or treated with protease (+) as indicated. Panel A shows manganese stimulation of radiolabel incorporation. Panel B shows protease degradation of assay products. Panel C shows Xpt1p dependence of radiolabel incorporation. Autoradiographs of dried SDS-PAGE gels are shown. Molecular weight standards (in kDa) are indicated at the right.
Although [14C]Xyl-labeling of protein in KN99α membranes was clearly stimulated by the addition of MnCl2, this could have been due to synthetic activities of enzymes other than Xpt1p. To test this, we performed similar experiments with membrane preparations from KN99α, KN99α xpt1Δ, and KN99α xpt1Δ pXPT1 cells. The intensity and pattern of radiolabel incorporation was comparable for both the wild-type and complemented strains (Fig. 4, panel C, lanes 1–3 and lanes 6–9); in both strains, the inclusion of MnCl2 increased [14C]Xyl incorporation and protease treatment of the reaction products significantly reduced the resulting [14C]Xyl-labeling. In contrast, membranes from the KN99α xpt1Δ strain showed no increase in protein radiolabeling upon the addition of MnCl2 (Fig. 4, panel C, lanes 4–6). These studies strongly implicated Xpt1p in protein glycosylation.
Analysis of O-Linked Glycans
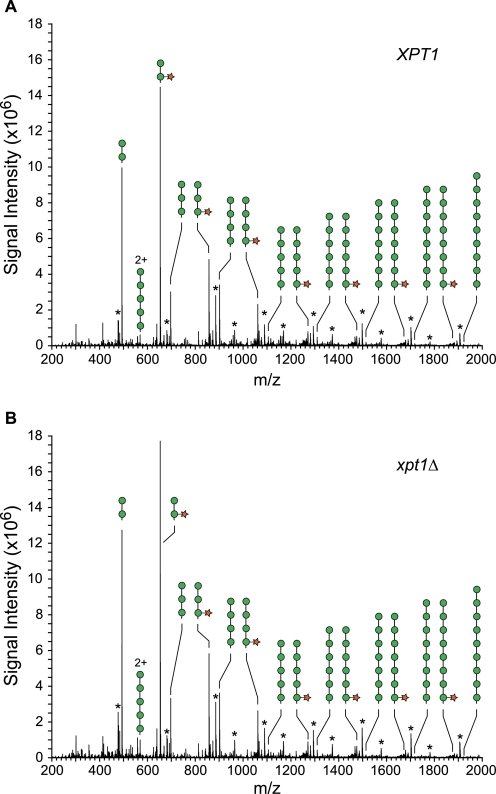
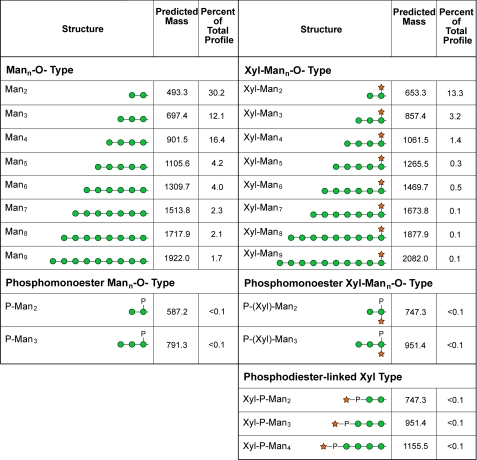
Based on our radiolabeling studies, we next turned to direct analysis of protein-associated glycans. Total cell lysates were prepared from KN99α and KN99α xpt1Δ cells and extracted with 1% CHAPS; O-glycans were then released from the detergent-soluble material by β-elimination, permethylated, and analyzed by NSI-MS (see “Experimental Procedures”). The major species of O-linked glycans in both the wild-type and xpt1pΔ samples were similar in distribution and were consistent with structures of Man2, Man3, Man4, and their corresponding xylosylated forms: Xyl-Man2, Xyl-Man3, and Xyl-Man4 (Fig. 5). MS2 verified the presence of Mann and Xyl-Mann glycans extended to n = 9. The O-glycans of C. neoformans, which have not previously been comprehensively surveyed, also include minor glycans with properties consistent with being Man2 or Man3 cores carrying branching phosphomonoesters with or without Xyl (Fig. 6).
FIGURE 5.
The profiles of major O-linked glycans released from wild-type and xpt1Δ mutant proteins are similar. Full MS scans are shown for glycans released from equivalent amounts of protein harvested from wild-type KN99α (panel A) and the KN99α xpt1Δ mutant (panel B). Mann and Xyl-Mann glycans dominate the O-linked glycan profile. Asterisks denote the presence of a poly-hexose contaminant of unknown origin (non-reduced). 2+ denotes double-charged ion species; circle, Man; star, Xyl.
FIGURE 6.
The mass and prevalence of individual O-linked glycans detected in detergent-extracts of C. neoformans. Protein samples for glycan analysis were prepared from KN99α cells as described under “Experimental Procedures.” After release from protein material by reductive β-elimination, alditols were permethylated and characterized by NSI-MSn as their sodiated structures. Monoisotopic m/z values are given for the permethylated alditol forms of the indicated structures. The prevalence of each glycan is expressed as a percent of the total O-linked glycan profile quantified by measuring signal intensities from full MS spectra. Prevalence values are determined from complete TIM analysis of a single KN99α extract. A prevalence value of <0.1% of total profile indicates that the glycan was detected and characterized by MSn but was present at a level below the threshold for quantification (less than two times background).
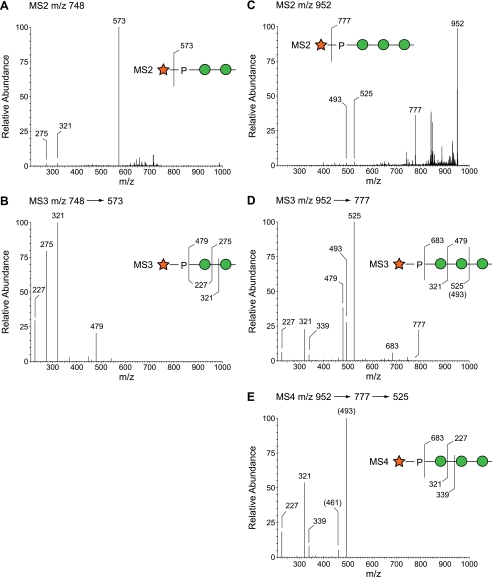
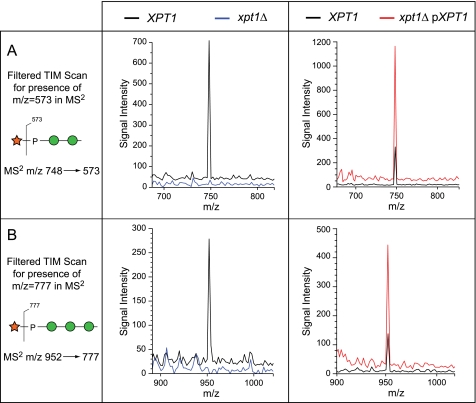
To investigate the potential occurrence of terminal Xyl-P phosphodiester motifs among the minor O-glycans, TIM data were filtered to reveal parent ions that produce MS2 fragments corresponding to the loss of terminal Xyl (Δm/z = 175) or Xyl-1-P (Δm/z = 285). This analysis revealed the presence of low abundance O-glycans with terminal Xyl-1-P at m/z = 747.3 (Xyl-P-Man2), m/z = 951.4 (Xyl-P-Man3), and m/z = 1155.5 (Xyl-P-Man4) (Fig. 6). Subsequent MSn fragmentation in the wild-type KN99α sample supported these structural assignments and unambiguously placed the Xyl-P- substitution on the non-reducing terminal Man residue of the glycan (Fig. 7). We next examined O-glycans from xpt1Δ cells for the presence of these moieties. Extremely low signal intensities were detectable within TIM collection windows of Δm/z ± 1.4 mass units centered around m/z = 748 and = 952 for xpt1Δ glycans (Fig. 8, left profiles). Based on signal intensities in full MS, the reduction in the Xyl-P-Man2 glycan relative to KN99α wild type was between 12- and 20-fold (data from two independent preparations). Fragmentation associated with m/z = 952 (Xyl-P-Man3) and m/z = 1155 (Xyl-P-Man4) failed to yield interpretable spectra in the xpt1Δ glycans, indicating reduction below the threshold for detection.
FIGURE 7.
Fragmentation of Xyl-P-Man O-glycans in wild type. Panels A and B, the major MS2 fragment produced from the parent ion at m/z = 748 (Xyl-P-Man2) corresponds to loss of Xyl (m/z = 573). Upon further fragmentation, the m/z = 573 ion produces a P-Hex fragment (m/z = 321), not a P-hexitol, thereby placing the Xyl-P-substitution at the non-reducing terminus of the Man2 disaccharide. Panels C–E, similarly, the major MS2 fragment produced from the parent ion at m/z = 952 (Xyl-P-Man3) corresponds to loss of Xyl (m/z = 777). Further fragmentation produces the P-Hex-Hex (m/z = 525) and P-Hex (m/z = 321) ions, consistent with Xyl-P-substitution at the non-reducing terminus. The placement of the fragmentation line relative to the indicated residue is meant to indicate cleavage on one side or the other of the glycosidic oxygen.
FIGURE 8.
Filtered TIM scans demonstrate Xyl-P-Man2 and Xyl-P-Man3 in O-linked glycans from wild-type, xpt1Δ mutant, and XPT1 overexpressing strains. Total ion mapping scans were filtered for the production of MS2 fragments that indicate the presence of Xyl-P motifs (panel A, m/z = 573 for Xyl-P-Man2; panel B, m/z = 777 for Xyl-P-Man3) in the indicated strains. MS2 is inherently more sensitive than full MS for ion-trap instruments, yet the Xyl-P modified glycans are detected as only minor deflections above base line in the xpt1Δ O-linked glycan preparations. The signal traces for KN99α in the left panels and for xpt1Δ pXPT1 in the right panels are displaced upward on the y axis by 5% of full scale for clarity.
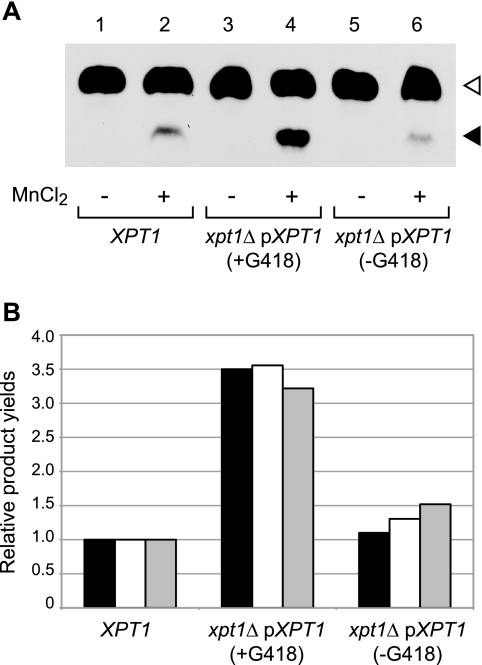
To confirm the relationship between the Xpt1p activity and Xyl-P-modified O-glycans, we analyzed cells expressing increased levels of the transferase. To do this we exploited our observation that episomal expression of Xpt1p increased enzyme activity severalfold over wild-type levels (Fig. 3) and expressed XPT1 in xpt1Δ cells from a plasmid marked with a sequence conferring resistance to G418. Membranes prepared from the resulting strain grown in medium containing G418 showed a 3.5-fold increase in xylosylphosphotransferase activity compared with wild-type cells in which XPT1 is expressed only from its native chromosomal location (Fig. 9A, compare tracks 4 and 2). This activity declined when the plasmid-bearing cells were grown in medium lacking G418 for 24 h, consistent with the expected plasmid loss under non-selective conditions (Fig. 9A, track 6). We also prepared protein O-glycans for analysis from the same cultures. Filtered TIM scans of these samples showed that structures corresponding to Xyl-P-Man2 and Xyl-P-Man3 increased significantly upon Xpt1 overexpression (Fig. 8, right profiles) and that the magnitude of this change matched the assayed enzyme activity in all three cultures assessed (Fig. 9B). The absence of detectable Xyl-P modification in the xpt1Δ mutant together with the parallel increases in both assayed activity and protein modification in the overexpression strain strongly support the conclusion that Xpt1p is responsible for Xyl-P modification of mannose in cryptococcal protein O-glycans.
FIGURE 9.
Xpt1p overexpression yields parallel increases in enzyme activity and O-glycan modifications. Panel A, total membranes were prepared from serotype A wild-type cells or serotype A xpt1Δ cells episomally expressing XPT1 from the same serotype; the latter were cultured either with G418 to maintain the expression plasmid or without it to allow plasmid loss as indicated. Xpt1p activity was assayed with UDP-[14C]Xyl and α-1,3-Man2 in the absence (−) or presence (+) of MnCl2, as described under “Experimental Procedures.” An autoradiograph of the products resolved by TLC is shown; no signal was detected in other regions of the plate beyond minor amounts of free Xyl or in the absence of the α-1,3-Man2 acceptor (not shown). The filled triangle indicates the product of the manganese-dependent xylosyltransferase activity, and the open triangle indicates the product of the unrelated xylosyltransferase Cxt1P (see “Results”). Panel B, shown are Xpt1p products for the strains indicated, normalized to wild-type. Black bars, radiolabel in Xyl-P-Man2 (filled triangle) made in vitro in the reactions shown in panel A, quantified using a BIOSCAN System 200 Imaging scanner; white bars, Xyl-P-Man2 in protein O-glycans, derived from the abundance of MS2 fragments of m/z = 573; gray bars, Xyl-P-Man3 in protein O-glycans, derived from the abundance of MS2 fragments of m/z = 777.
Localization Studies
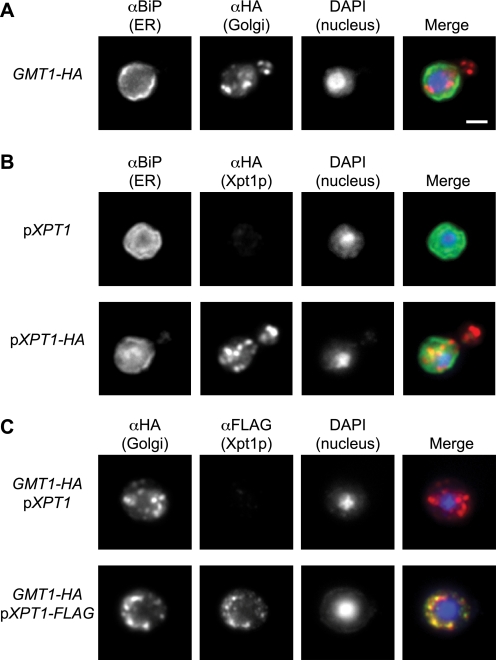
Although most O-glycosylation of proteins occurs in the endoplasmic reticulum (ER) and Golgi complex (31), there are also important examples of cytosolic O-glycosylation in eukaryotes (32). To localize Xpt1p, we first needed to establish markers for the cryptococcal secretory organelles. The C. neoformans genome predicts a protein with 67% homology to Saccharomyces cerevisiae BiP, an ER chaperone (33), so we tested this protein as an ER marker. Polyclonal antibody generated against the S. cerevisiae protein stained cryptococcal cells in a pattern consistent with the expected ER localization, with fluorescence visible both around the nucleus and just within the plasma membrane (Fig. 10, panels A and B). As a Golgi marker we selected Gmt1p, a GDP-mannose transporter that we previously characterized in C. neoformans (34) and showed to functionally complement the corresponding Golgi-localized S. cerevisiae protein (Vrg4p). For these studies we used a C. neoformans strain where we engineered the chromosomal copy of GMT1 to express an HA tag.6 Localization of this protein exhibited a punctate staining pattern consistent with yeast Golgi localization (35) and distinct from that of the ER marker BiP (Fig. 10, panels A and C).
FIGURE 10.
Xpt1p-HA localization to the Golgi apparatus. Cryptococcal cells (JEC21) were fixed, permeabilized, immunolabeled, stained with DAPI, and visualized by immunofluorescence microscopy as detailed “Experimental Procedures.” Single channel and merged images are shown. Panel A, cells engineered to express an HA-tagged form of Gmt1p (a Golgi marker) were probed with antibodies to BiP (an ER marker) and HA, yielding distinct labeling patterns. Panel B, cells episomally expressing either wild-type or HA-tagged Xpt1p were probed with antibodies to BiP and HA, showing distinct labeling patterns. Panel C, Gmt1p-HA expressing cells (as in panel A) expressing either wild-type or FLAG-tagged Xpt1p were probed with antibodies to HA and FLAG, showing colocalization. All panels are at the same magnification; scale bar, 2 μm.
With these two secretory pathway markers in hand, we proceeded to the localization of Xpt1p. Because immunofluoresence in serotype A strains is technically more challenging,7 we used a serotype D strain (JEC21) episomally expressing either wild-type Xpt1p or the protein modified with a C-terminal HA or FLAG tag. Both untagged and tagged proteins were under control of the same native XPT1 promoter, and we know that C-terminal epitope tagging does not alter Xpt1p activity (Fig. 1, compare tracks 6 and 8). We observed Xpt1p in a punctate distribution (Fig. 10, panels B and C), highly colocalized with the Gmt1p Golgi protein (Fig. 10, panel C) and clearly distinct from the localization of BiP, the nucleus, and the plasma membrane (Fig. 10, panel B). This suggests that Xpt1p participates in the glycosylation of proteins en route through the secretory pathway.
DISCUSSION
We previously identified Xpt1p as a novel glycosyltransferase of C. neoformans that modifies Man with Xyl-P (21). In the current study we focused on determining the role of this unusual enzyme. Based on its biochemical activity, we expected Xpt1p to participate in some aspect of cryptococcal glycan synthesis. Although the capsule polysaccharides and glycosylinositol phosphorylceramides of C. neoformans are known to include Xyl residues (4, 5, 14), as are the O-glycans of a related cryptococcal species, none of these had been reported to contain a Xyl-P modification. However, the possibility that this unusual structure may have been overlooked in previous analyses together with the lack of definitive structures for many cellular glycans in this organism left open multiple potential roles for Xpt1p. In the studies reported here we have determined that this enzyme modifies protein O-glycans.
The data presented in Fig. 6 represent our first insights into the structures of O-linked glycans in C. neoformans. Beyond the Xyl-P-Man-containing species formed by Xpt1p, we demonstrate the presence of O-linked glycans whose mass is consistent with Man- and Xyl-Man-containing structures. This is significantly different from the O-glycans of the model yeast S. cerevisiae, which are composed only of Man residues (36). The Xyl-P-Man motif that we have identified in cryptococcal O-glycans also has not been reported to our knowledge in any system. Phosphate moieties do occur in the O-linked structures of S. cerevisiae and Pichia pastoris as a terminal modification of mannose (37, 38), but these lack the xylose cap that completes the novel motif found in C. neoformans.
The C. neoformans species complex diverged into C. neoformans (serotypes A and D) and C. gattii (serotypes B and C) more than 37 million years ago (39). Genome sequence for strains representing three serotypes (A, B, and D) is available and in each case includes a putative XPT1 gene; these genes encode proteins that are 88% identical and 92% similar at the amino acid level. The conservation of the region represented by residues 407–456 of Xpt1p in serotype A, which has been implicated in sugar-phosphate transfer from a nucleotide sugar donor (21), is especially notable. The sequences from this region are identical in serotypes A and D, with only a conservative (Ser/Thr) change in serotype B. We tested membranes from nine cryptococcal strains, representing the four serotypes, and found robust Xpt1p activity in all of them (Fig. 2 and data not shown). Furthermore, we showed that expression of XPT1 from serotype D complements the loss of enzyme activity in a serotype A xpt1Δ strain (Fig. 3). Together, these results indicate that both the XPT1 gene and enzyme activity are highly conserved among members of the C. neoformans species complex.
We found no difference between the ability of wild-type and xpt1Δ cells to colonize mice, express virulence factors, or grow under stress conditions. It is conceivable that another protein compensates for loss of Xpt1p via some function that is not measured in our activity assay. This model would account for the lack of detectable phenotypic alteration despite the absence of xylose phosphate transfer activity. One potential candidate is the single C. neoformans homolog of Xpt1p, a hypothetical protein that is 35% identical to Xpt1p in serotype A. To test this hypothesis we used RNAi to silence the corresponding gene (CNAG_04705), but we detected no obvious phenotypic change or reduction in Xpt1p activity compared with wild type (21). We also deleted this gene in both wild-type and xpt1Δ strains, but neither the single nor double deletion demonstrated any phenotypic change compared with wild-type.8 Our working model, therefore, supported by evolutionary conservation, is that Xpt1p performs a necessary function for cryptococcal cells under conditions that are as yet unidentified.
Our understanding of the role of Xpt1p will be informed by identifying specific proteins that are modified by Xpt1p. As seen in Fig. 4, the diffuse Xpt1p-dependent [14C]Xyl protein labeling that appears between 50 and 100 kDa sometimes resolves into more focused bands (Fig. 4, panel A), but isolating these few highly radiolabeled polypeptides from total membrane sample preparations would be technically challenging. Furthermore, the radiolabeling of membrane proteins in our in vitro assay may not accurately represent the native targets of the Xpt1p activity. In the future it may be possible to develop an antibody specific for the Xyl-P-Man motif; this could then be used in the purification and identification of the Xpt1p protein substrates.
We previously showed that a single glycosyltransferase acts in multiple distinct synthetic pathways in C. neoformans (19, 20). It is, therefore, possible that Xpt1p modifies N-linked glycans in addition to O-linked structures. In S. cerevisiae, phosphate links mannose residues in some highly mannosylated N-glycan structures (40), perhaps serving as precedent for similar structures involving xylose. In another example of phosphate linking two sugar residues, GlcNAc-P is added to terminal mannose residues of mammalian N-glycans, although this is followed by removal of the GlcNAc to reveal Man-6-phosphate as a lysosomal targeting signal (41, 42) (we have no evidence for such trimming in the cryptococcal system). Future characterization of cryptococcal N-glycans will establish whether they are modified by Xpt1p.
Outstanding questions remain regarding Xpt1p, including the proteins modified by this activity, whether it acts in direct association with other enzymes involved in O-glycosylation, and whether it participates in synthesis of N-glycans. These investigations will help to define the unusual glycans of C. neoformans as well as suggest the biosynthetic pathways required to generate them. Importantly, significant differences in either glycan structure or assembly between this organism and its human host represent potential drug targets for prevention or treatment of cryptococcal infection.
Supplementary Material
Acknowledgments
We are grateful to Christian Heiss for performing 31P NMR studies of GXM and to Mindy Porterfield for protein determinations for glycan analysis. We also thank David Haslam and Jacques Baenziger for helpful insights on this project, Robert Cherniak and Christian Heiss for discussion regarding the structures of GXM and GXMGal, Thomas Kozel for anti-GXM antibodies, and Jeffrey Brodsky for anti-BiP antiserum.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 GM071007 and R21 AI076737 (to T. L. D.) and P01 GM085354 and P41 RR018502 (National Center for Research Resources) (to M. T.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
J. S. Klutts and T. L. Doering, manuscript in preparation.
Genome sequence from a serotype C strain is not currently available.
C. Heiss, personal communication.
C. L. Griffith and T. L. Doering, unpublished data.
M. L. Skowyra and T. L. Doering, unpublished observations.
Z. A. Wang and T. L. Doering, unpublished data.
- GXM
- glucuronoxylomannan
- GXMGal
- glucuronoxylomannogalactan
- G418
- Geneticin
- NSI-MS
- nanospray mass spectrometry
- TIM
- total ion mapping
- ER
- endoplasmic reticulum
- Xyl
- xylose
- Xyl-P
- xylose phosphate
- α-1,3-Man2
- α-1,3-linked dimannose
- YPD
- yeast extract/peptone/dextrose.
REFERENCES
- 1. Lin X., Heitman J. (2006) Annu. Rev. Microbiol. 60, 69–105 [DOI] [PubMed] [Google Scholar]
- 2. Heitman J., Kozel T. R., Kwon-Chung K. J., Perfect J. R., Casadevall A. (2011) Cryptococcus, from Human Pathogen to Model Yeast, pp. 373–385, ASM Press, Washington, D. C [Google Scholar]
- 3. Datta K., Bartlett K. H., Baer R., Byrnes E., Galanis E., Heitman J., Hoang L., Leslie M. J., MacDougall L., Magill S. S., Morshed M. G., Marr K. A). (2009) Emerg. Infect. Dis. 15, 1185–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cherniak R., Valafar H., Morris L. C., Valafar F. (1998) Clin. Diagn. Lab. Immunol. 5, 146–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Vaishnav V. V., Bacon B. E., O'Neill M., Cherniak R. (1998) Carbohydr. Res. 306, 315–330 [DOI] [PubMed] [Google Scholar]
- 6. Heiss C., Klutts J. S., Wang Z., Doering T. L., Azadi P. (2009) Carbohydr. Res. 344, 915–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Samuelson J., Banerjee S., Magnelli P., Cui J., Kelleher D. J., Gilmore R., Robbins P. W. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 1548–1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Olson G. M., Fox D. S., Wang P., Alspaugh J. A., Buchanan K. L. (2007) Eukaryot. Cell 6, 222–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schutzbach J., Ankel H., Brockhausen I. (2007) Carbohydr. Res. 342, 881–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Willger S. D., Ernst J. F., Alspaugh J. A., Lengeler K. B. (2009) PLoS One 4, e6321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Franzot S. P., Doering T. L. (1999) Biochem. J. 340, 25–32 [PMC free article] [PubMed] [Google Scholar]
- 12. Djordjevic J. T., Del Poeta M., Sorrell T. C., Turner K. M., Wright L. C. (2005) Biochem. J. 389, 803–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Eigenheer R. A., Jin Lee Y., Blumwald E., Phinney B. S., Gelli A. (2007) FEMS Yeast Res. 7, 499–510 [DOI] [PubMed] [Google Scholar]
- 14. Heise N., Gutierrez A. L., Mattos K. A., Jones C., Wait R., Previato J. O., Mendonça-Previato L. (2002) Glycobiology 12, 409–420 [DOI] [PubMed] [Google Scholar]
- 15. Rhome R., McQuiston T., Kechichian T., Bielawska A., Hennig M., Drago M., Morace G., Luberto C., Del Poeta M. (2007) Eukaryot. Cell 6, 1715–1726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Moyrand F., Klaproth B., Himmelreich U., Dromer F., Janbon G. (2002) Mol. Microbiol. 45, 837–849 [DOI] [PubMed] [Google Scholar]
- 17. Griffith C. L., Klutts J. S., Zhang L., Levery S. B., Doering T. L. (2004) J. Biol. Chem. 279, 51669–51676 [DOI] [PubMed] [Google Scholar]
- 18. Klutts J. S., Levery S. B., Doering T. L. (2007) J. Biol. Chem. 282, 17890–17899 [DOI] [PubMed] [Google Scholar]
- 19. Klutts J. S., Doering T. L. (2008) J. Biol. Chem. 283, 14327–14334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Castle S. A., Owuor E. A., Thompson S. H., Garnsey M. R., Klutts J. S., Doering T. L., Levery S. B. (2008) Eukaryot. Cell 7, 1611–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Reilly M. C., Levery S. B., Castle S. A., Klutts J. S., Doering T. L. (2009) J. Biol. Chem. 284, 36118–36127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Spiropulu C., Eppard R. A., Otteson E., Kozel T. R. (1989) Infect. Immun. 57, 3240–3242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kozel T. R., Levitz S. M., Dromer F., Gates M. A., Thorkildson P., Janbon G. (2003) Infect. Immun. 71, 2868–2875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Eckert T. F., Kozel T. R. (1987) Infect. Immun. 55, 1895–1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Netski D., Kozel T. R. (2002) Infect. Immun. 70, 2812–2819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wickes B. L., Moore T. D., Kwon-Chung K. J. (1994) Microbiology 140, 543–550 [DOI] [PubMed] [Google Scholar]
- 27. Gallagher S. R. (2006) Curr. Protoc. Mol. Biol. Chapter 10, Unit 10.2A [DOI] [PubMed] [Google Scholar]
- 28. Aoki K., Porterfield M., Lee S. S., Dong B., Nguyen K., McGlamry K. H., Tiemeyer M. (2008) J. Biol. Chem. 283, 30385–30400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Anumula K. R., Taylor P. B. (1992) Anal. Biochem. 203, 101–108 [DOI] [PubMed] [Google Scholar]
- 30. Zaragoza O., Casadevall A. (2004) Biol. Proced. Online 6, 10–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wopereis S., Lefeber D. J., Morava E., Wevers R. A. (2006) Clin. Chem. 52, 574–600 [DOI] [PubMed] [Google Scholar]
- 32. Wells L., Hart G. W. (2003) FEBS Lett. 546, 154–158 [DOI] [PubMed] [Google Scholar]
- 33. Nishikawa S., Hirata A., Nakano A. (1994) Mol. Biol. Cell 5, 1129–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cottrell T. R., Griffith C. L., Liu H., Nenninger A. A., Doering T. L. (2007) Eukaryot. Cell 6, 776–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dean N., Zhang Y. B., Poster J. B. (1997) J. Biol. Chem. 272, 31908–31914 [DOI] [PubMed] [Google Scholar]
- 36. Gemmill T. R., Trimble R. B. (1999) Biochim. Biophys. Acta. 1426, 227–237 [DOI] [PubMed] [Google Scholar]
- 37. Jars M. U., Osborn S., Forstrom J., MacKay V. L. (1995) J. Biol. Chem. 270, 24810–24817 [DOI] [PubMed] [Google Scholar]
- 38. Trimble R. B., Lubowski C., Hauer C. R., 3rd, Stack R., McNaughton L., Gemmill T. R., Kumar S. A. (2004) Glycobiology 14, 265-274 [DOI] [PubMed] [Google Scholar]
- 39. Xu J., Vilgalys R., Mitchell T. G. (2000) Mol. Ecol. 9, 1471–1481 [DOI] [PubMed] [Google Scholar]
- 40. Jigami Y., Odani T. (1999) Biochim. Biophys. Acta 1426, 335–345 [DOI] [PubMed] [Google Scholar]
- 41. Kornfeld S. (1987) FASEB J. 1, 462–468 [DOI] [PubMed] [Google Scholar]
- 42. von Figura K., Hasilik A. (1986) Annu. Rev. Biochem. 55, 167–193 [DOI] [PubMed] [Google Scholar]
- 43. Nielsen K., Cox G. M., Wang P., Toffaletti D. L., Perfect J. R., Heitman J. (2003) Infect. Immun. 71, 4831–4841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Baker L. G., Specht C. A., Donlin M. J., Lodge J. K. (2007) Eukaryot. Cell 6, 855–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Warren R. L., Butterfield Y. S., Morin R. D., Siddiqui A. S., Marra M. A., Jones S. J. (2005) Biotechniques 38, 715–720 [DOI] [PubMed] [Google Scholar]
- 46. Fraser J. A., Giles S. S., Wenink E. C., Geunes-Boyer S. G., Wright J. R., Diezmann S., Allen A., Stajich J. E., Dietrich F. S., Perfect J. R., Heitman J. (2005) Nature 437, 1360–1364 [DOI] [PubMed] [Google Scholar]
- 47. Kwon-Chung K. J., Wickes B. L., Stockman L., Roberts G. D., Ellis D., Howard D. H. (1992) Infect. Immun. 60, 1869–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.