Abstract
Receptor-mediated activation of protein kinase (PK) C is a central pathway regulating cell growth, homeostasis, and programmed death. Recently, we showed that calpain-mediated proteolytic processing of PKCα in ischemic myocardium activates PKC signaling in a receptor-independent manner by releasing a persistent and constitutively active free catalytic fragment, PKCα-CT. This unregulated kinase provokes cardiomyopathy, but the mechanisms remain unclear. Here, we demonstrate that PKCα-CT is a potent regulator of pathological cardiac gene expression. PKCα-CT constitutively localizes to nuclei and directly promotes nucleo-cytoplasmic shuttling of HDAC5, inducing expression of apoptosis and other deleterious genes. Whereas PKD activation is required for HDAC5 nuclear export induced by unprocessed PKCs activated by phorbol ester, PKCα-CT directly drives HDAC cytosolic relocalization. Activation of MEF2-dependent inflammatory pathway genes by PKCα-CT can induce a cell-autonomous transcriptional response that mimics, but anticipates, actual inflammation. Because calpain-mediated processing of PKC isoforms occurs in many tissues wherein calcium is increased by stress or injury, our observation that the catalytically active product of this interaction is a constitutively active transcriptional regulator has broad ramifications for understanding and preventing the pathological transcriptional stress response.
Keywords: Calpain, Histone Deacetylase, Ischemia, Protein Kinase C (PKC), Transcription
Introduction
Protein kinase Cs (PKC)3 are activated by Gq/phospholipase C-coupled receptors and regulate a myriad of cellular responses, including growth, mobility, and programmed death (1). Of cardiac-expressed PKCs, PKCα is expressed at higher levels than other isoforms, is dynamically regulated in cardiac disease, and is implicated in functional decompensation and development of cardiomyopathy (2–6). PKCϵ is also highly expressed, but is cardio-protective (7, 8). Accordingly, there is a need to better understand isoform-specific actions of cardiac-expressed PKCs and to develop isoform-specific pharmacological PKC activators and inhibitors as therapeutics.
PKCs regulate proteins directly through substrate phosphorylation on serines or threonines, or indirectly by phosphorylating positive and negative regulators of protein expression or function. An example of indirect PKC control is transcriptional up-regulation of the cell death factor, Nix, which mediates programmed cell death and the transition to dilated cardiomyopathy in Gq-mediated and pressure overload hypertrophy: PKC is activated via the canonical Gq/phospholipase C signaling pathway and activates the transcription factor SP1, which binds to and induces the Nix promoter (9). Transcriptional changes induced by protein kinase C are implicated in other cardiac syndromes (10).
Because PKCs are typically activated downstream of Gq-coupled receptors, angiotensin, α1-adrenergic, and endothelin receptor blockade can been used to interrupt PKC signaling (11). We recently described a mechanism for PKC activation in cardiac ischemia that bypasses the receptor-G protein signaling pathway (12): Calpain proteases are activated by increased cytosolic calcium in ischemic myocardium and cleave PKC at the hinge region, generating free N-terminal regulatory and C-terminal kinase fragments. The free N-terminal (NT) fragment of PKCα had no biological activity in cultured cells or mouse hearts, whereas the free C-terminal (CT) fragment of PKCα exhibited constitutive kinase activity and promiscuous substrate targeting. Forced PKCα-CT expression induced a dilated cardiomyopathy that progressed more rapidly and was more severe than that produced by overexpression of unprocessed PKCα at similar levels (3, 12).
Normal substrate phosphorylation by PKCs is critically dependent upon proper subcellular localization. As PKCα-CT lacks the N-terminal sites that direct normal PKC subcellular translocation (13), it is mistargeted, which may contribute to its promiscuous kinase activity. Previously, we established that receptor-independent, calpain-mediated PKCα signaling through PKCα-CT increased myocardial phosphoserine/threonine protein content and lead to an aggressive dilated cardiomyopathy (12). However, the changes in the cardiac phosphoproteome of PKCα-CT hearts relative to that of unprocessed PKCα-expressing hearts were modest in comparison to the severity and rapid progression of the PKCα-CT-induced cardiomyopathy. We hypothesized that one or a few atypical effects of PKCα-CT, rather than the general increase in kinase activity, produced the deleterious effects.
Here, using microarray RNA expression profiling and RNA-sequencing, we reveal that PKCα-CT is a powerful transcriptional activator in otherwise normal mouse hearts. Transcriptional activity derives in part from partial constitutive nuclear localization of the free PKCα-CT peptide, leading to HDAC5 nuclear export that de-represses expression of MEF2-dependent pathological genes. These results uncover a receptor-independent pathway for transcriptional regulation by an ischemia-induced, PKCα-derived rogue kinase. The data provide further support for therapeutic use of direct kinase inhibitors in cardiac ischemia and other conditions wherein PKC can be proteolytically activated.
EXPERIMENTAL PROCEDURES
Transgenic Mice
The generation and phenotypic characterization of mice conditionally overexpressing human PKCα and fragments encoding peptides corresponding to the calpain N-terminal cleavage fragment (PKCα-NT) and C-terminal cleavage fragment (PKCα-CT) have been described previously (12). Mice were housed and studied according to procedures approved by Animal Studies Committee at Washington University School of Medicine.
Immunoblot Analysis
SDS-PAGE and immunoblotting of myocardial fractions used standard techniques: Cardiac homogenates were clarified at 200 × g for 20 min, followed by centrifugation at 1000 × g for 10 min to pellet nuclei. The supernatant was centrifuged at 100,000 × g for 1 h to generate a microsomal membrane pellet fraction and a cytoplasmic supernatant fraction. Nuclear and microsomal pellets were resuspended in 50 mm Tris (pH 7.5), 2.5 mm EDTA, 25 mm NaCl, 0.2% Nonidet P-40, 10 mm EGTA, 20 mm sodium fluoride, 2 mm sodium orthovanadate, and Complete Mini Protease Inhibitor Mixture Tablet (Roche). Antibodies to label fractions were: anti-LDH (Invitrogen) for cytosolic protein, anti-transferrin receptor (Invitrogen) for microsomal protein, and anti-lamin A/C (Abcam) for nuclear protein.
Anti-PKC antibodies used were: anti-PKCα C terminus (Santa Cruz Biotechnology, sc-208; recognizes an epitope at the C terminus of human PKCα) and anti-PKCα (Santa Cruz Biotechnology, sc-80; recognizes an epitope within the N-terminal hinge region (residues 292–317) of human PKCα).
GFP-HDAC5 Nuclear Export Studies
COS7 cells were transfected using lipofection with plasmids expressing GFP-HDAC5 (14) and either empty pcDNA3 (vector control) or pcDNA3-human PKCα, -PKCα-CT, or -PKCα-NT. Forty-eight hours after transfection, cells were treated with vehicle or 100 nm PMA, and examined 30 min later under a confocal fluorescence microscope. Kinase inhibitors were added to cell cultures at the indicated concentrations ∼18 h before assay. Nuclei were counterstained with Hoescht dye. Each condition is the result of 4–6 individual experiments performed on separate days; each experiment evaluated ∼100 cells.
HDAC5 Phosphorylation Studies
Lysates of HEK293 cells transfected with the plasmids described above were incubated with purified active full-length HDAC5 protein (Abcam) for 15 min in the absence or presence of PKD inhibitor CID755673, the reactions stopped by boiling in SDS sample buffer, and phosphorylation of PAGE-separated proteins assessed by immunoblotting with rabbit polyclonal anti-HDAC5 (phosphor-S498) (Thermo Scientific).
Histological Studies
Mouse heart sections were fixed in formalin and assayed by TUNEL assay for programmed cell death or Masson's trichrome stain for myocardial fibrosis using standard methods.
Cardiac mRNA Profiling using Affymetrix Microarrays
Total cardiac RNA was isolated from flash-frozen ventricular tissue harvested from 4-week-old hearts using Trizol (Invitrogen) and quantified using a NanoDrop spectrophotomer (Thermo Scientific). Affymetrix Mouse Gene 1.0 ST arrays were used to assess individual mRNA levels. To facilitate comparison with RNA sequencing data, we excluded data pertaining to ESTs and rRNAs (15). Selection of genes expressed above median values, hierarchical clustering, and computation of significant changes were performed as described (16, 17).
Cardiac mRNA Profiling by RNA Sequencing
Preparation of cardiac cDNA libraries and DNA bar-coding was as previously described (15). Four barcoded libraries were combined in equimolar (10 nm) amounts and diluted to 6 pm for cluster formation on a single Illumina Genome Analyzer II flowcell lane. Single-end sequencing, base calling of DNA clusters and sorting of sequence data by bar code was as previously described (15). Cufflinks was used with Ensembl gene annotation files to calculate overall gene expression in terms of Fragments Per Kb of exon per Million mapped reads (FPKM), equivalent to RPKM (Reads Per Kb of exon per Million mapped reads). We analyzed only RNA elements with FPKM expression data in at least 2 of 3 biological replicates, and excluded ESTs and rRNAs from our analyses. An FPKM of 3 was assumed to correspond to 1 copy/cell, as per (18).
Prediction of Mef2 Binding Sites
The Transcription Element Search System (TESS; (19)) located at was used to examine 1 kb regions upstream of known transcriptional start sites in the mouse genome. A 1 kb upstream region gives fewer false positives, with a lower cost of missing true signals, than a search conducted over a larger region (20).
Statistical Methods
For gene expression data on mouse hearts, a p value of <0.001 was defined as significant and false discovery rates (FDR) calculated for each group comparison using q-value methods in Partek Genomics Suite 6.5 software (Partek, St Louis, MO) (16, 17). FDRs for microarray comparison of PKCα transgenic groups to tTA control mice were as follows: -CT, <0.01; -FL, <0.2; NT, >0.3. FDRs for RNA-sequencing comparison of PKCα transgenic groups to control mice were as follows: -CT, <0.01; -FL, <0.08; NT, <0.18. Unsupervised hierarchical clustering using Euclidean distance and average distance was performed using Partek. Biological Networks Gene Ontology (BiNGO; (21)) was used to assign gene-ontology categories to each of the regulated genes. Over-representation of Gene Ontology categories was assessed at p < 0.05 using a hypergeometric test with Benjamini-Hochberg false discovery rate correction (21).
RESULTS
The Free C-terminal Fragment of PKCα Activates a Unique Profile of Gene Expression
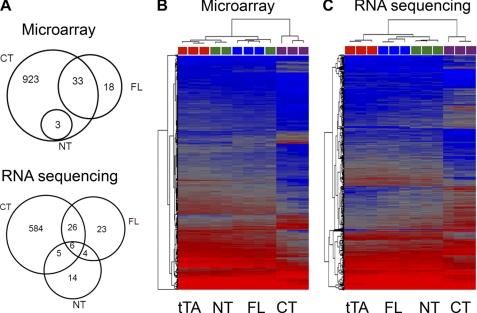
Altered gene expression is characteristic of cardiomyopathies, and PKCs activated through conventional receptor signaling pathways are potent modifiers of cardiac gene expression (10, 22). We used Affymetrix GeneChip Mouse Gene 1.0ST microarrays to examine the mRNA signature of hearts conditionally expressing PKCα-CT and PKCα 4 weeks after transgene induction, which is before heart failure develops. The mRNA expression profiles of hearts expressing these two kinases were compared with those of same-age hearts expressing the tetracycline transcriptional activator alone (tTA) or the other daughter peptide of calpain-mediated PKCα processing, catalytically inactive PKCα-NT. Using the control tTA transgenic hearts as a normal baseline, 959 mRNAs were regulated at least 1.3-fold (p < 0.001) in PKCα-CT hearts, compared with only 51 in unprocessed PKCα-expressing hearts, and 3 in PKCα-NT hearts (Fig. 1A, top and supplemental Table S1). Unsupervised hierarchical clustering of relative transcript expression levels (Fig. 1B) revealed a transcriptional signature in PKCα-CT-expressing hearts that was distinct from the other three groups.
FIGURE 1.
PKCα-CT reprograms cardiac gene expression. A, Venn diagrams illustrating the pattern of regulated mRNAs in unprocessed full-length PKCα (FL), PKCα-CT (CT), and PKCα-NT (NT) expressing hearts, compared with tTa-expressing controls hearts. Top is result of microarray-based profiling; bottom is result of deep RNA sequencing. B and C, heat maps showing results of unsupervised hierarchical clustering of mRNA expression assayed by microarrays (B) or RNA sequencing (C). Each column represents expression of the 500 consensus most regulated mRNAs in a single heart; red is high expression, blue is lower expression. Color code for transgene categories (squares at top of each column) is: tTA control, red; PKCα-NT, green; full-length PKCα, blue; PKCα-CT, purple.
The striking difference in mRNA expression profile between PKCα-CT and PKCα hearts was unexpected. Therefore, we performed a technical replication of the comparative transcriptional profiling using deep RNA sequencing, which provides greater sensitivity and specificity than microarray profiling for quantifying mRNAs expressed at fewer than 20 copies per cell (15). Assessed by RNA-sequencing, 621 mRNAs were regulated at least 1.3-fold (p < 0.001) in PKCα-CT hearts, 59 in PKCα hearts, and 29 in PKCα-NT hearts (compared with tTA hearts) (Fig. 1A, bottom and supplemental Table S2). Again, unsupervised hierarchical clustering revealed a distinct transcriptional signature induced by PKCα-CT (Fig. 1C). The microarray and RNA-sequencing transcriptional profiling results agreed for 276 uniquely PKCα-CT-regulated transcripts. These results demonstrate genetic reprogramming of hearts expressing PKCα-CT.
The Free C-terminal Fragment of PKCα Is Constitutively Localized to Cardiac Myocyte Nuclei
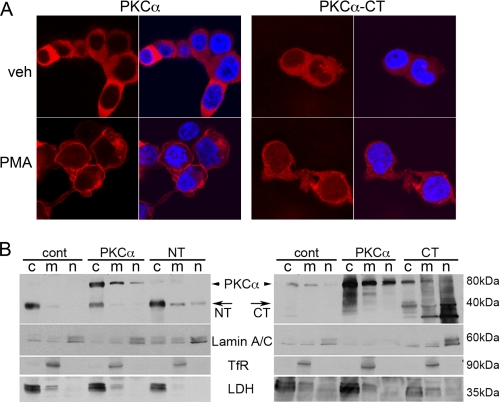
Distinct cellular effects of PKC isoforms are determined by their ability to phosphorylate specific targets, which is dependent upon subcellular co-localization of the kinase and its substrates (13). The ability of PKCα-CT to regulate gene expression might therefore accrue not only from constitutive kinase activity, but also from nuclear mis-localization. We assessed subcellular compartmentalization of PKCα-CT compared with full-length PKCα in transfected HEK293 cells. Confocal analysis showed that full-length PKCα is almost entirely cytosolic and rapidly translocates to cell membranes after activation by 100 nm phorbol myristate acetate (PMA) (Fig. 2A, left). In contrast, PKCα-CT localizes throughout the cytosol and nucleus, and shows minimal redistribution after PMA treatment; a substantial fraction remains in the nucleus (Fig. 2A, right). (We previously found that PKCα-NT appears as cytosolic punctae and does not undergo translocation after PMA stimulation (12)).
FIGURE 2.
PKCα-CT localizes to nuclei in HEK293 cells and cardiac myocytes. A, fluorescent confocal microscopy of PKCα (left) and PKCα-CT (right) expression at baseline (top) and 1 h after (bottom), treatment with 100 nm PMA. PKC is red; nuclei in right merged panels are stained blue with DAPI. B, immunoblot analysis of PKCα and peptides corresponding to its NT or CT calpain fragments in transgenic myocardial subcellular fractions. Lamin A/C labels nuclear fraction (n), transferrin receptor (TfR) labels microsomal membrane fraction (m), and lactate dehydrogenase (LDH) labels cytoplasmic fraction. Arrows indicate 40 kDa PKCα fragments. Left blots use anti-human PKCα antibody that recognizes the N terminus, right blot uses anti-PKCα antibody that recognizes the C terminus.
To examine the possibility that partial constitutive nuclear localization of PKCα-CT in cardiac myocytes induced transcription in hearts, we performed immunoblot analysis of fractionated myocardium from the same groups of transgenic mice used for the transcriptional profiling studies: tTA controls, full-length human PKCα, PKCα-NT, or PKCα-CT. Full-length human PKCα predominantly co-fractionated with cytosolic lactate dehydrogenase (LDH); minor proportions were detected in the transferrin receptor (TfR)-rich microsomal and lamin A/C-enriched nuclear fraction (Fig. 2B). These findings were consistent using antibodies that recognize either the N terminus of PKCα (Fig. 2B, left) or its C terminus (Fig. 2B, right). The free N-terminal PKCα calpain-derived peptide was detected almost exclusively in the LDH-rich cytosolic fraction (Fig. 2B, left), whereas PKCα-CT was observed primarily in the microsomal and nuclear-rich fractions (Fig. 2B, right). These results show that the catalytically active calpain proteolysis fragment PKCα-CT is, in part, constitutively localized within cardiomyocyte nuclei.
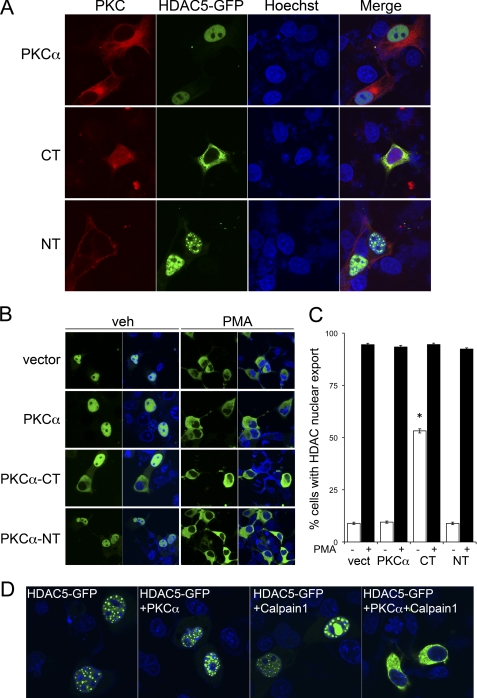
The Free PKCα C-terminal Peptide Directly Induces Nuclear Export of HDAC5
Activated protein kinase C regulates transcription in multiple cell types by promoting the phosphorylation and nucleo-cytoplasmic shuttling of HDAC5 (14, 23). Because PKCα-CT exhibits unregulated kinase activity, is constitutively nuclear, and has pronounced effects on gene expression, we examined its effects on HDAC5 nuclear export in COS7 cells, which exhibit the same partial nuclear localization of PKCα-CT as seen in HEK293 cells (Fig. 3A). Green fluorescent protein (GFP)-tagged HDAC5 was co-expressed with PKCα, PKCα-NT, or PKCα-CT in cultured COS7 cells, and the subcellular localization of HDAC5 was examined by confocal microscopy. HDAC5 was nuclear in the vast majority of cells co-transfected with empty vector, PKCα or PKCα-NT (Fig. 3, A and B), and was exported from nuclei after treatment with PKC-activating phorbol ester (Fig. 3, A–C). This confirms the previously described response of HDAC5 to activation of endogenous cellular PKCs (14, 24, 25). The normal pattern of HDAC5 nucleo-cytoplasmic redistribution by PMA was not altered by transfection with full-length PKCα or PKCα-NT (Fig. 3, B and C). Strikingly however, transfection with PKCα-CT induced PMA-independent HDAC5 nuclear export (Fig. 3, A–C).
FIGURE 3.
Calpain 1-generated PKCα-CT induces constitutive nuclear export of HDAC5. A, live-cell confocal fluorescent microscopy of COS7 cells transiently expressing GFP-HDAC5 and PKCα or its CT or NT calpain proteolysis fragments (B) As in A, treated with vehicle (veh) or 100 nm PMA. Nuclei are stained blue with Hoechst dye in merged figures. C, group quantitative data for GFP-HDAC5 nuclear export; means ± S.E. of 21 studies per group. *, p < 0.05 versus same treatment vector-transfected cells. D, effects of calpain cleavage of full-length PKCα on HDAC nucleo-cytoplasmic shuttling.
The above result shows that expression of the predicted calpain 1-generated C-terminal kinase fragment of PKCα induces HDAC5 nucleo-cytoplasmic shuttling, but does not formally show that calpain-mediated PKCα processing affects HDAC5. To address this issue we co-transfected full-length PKCα with or without calpain 1, and examined subcellular localization of HDAC5 (Fig. 3D). Calpain had no effect on HDAC5 localization in cells transfected with empty vector. Strikingly however, cells co-expressing calpain 1 and PKCα exhibited HDAC5 nuclear export, mimicking the activity of PKCα-CT. Thus, calpain processing of PKCα is sufficient to induce PMA-independent HDAC5 nuclear export.
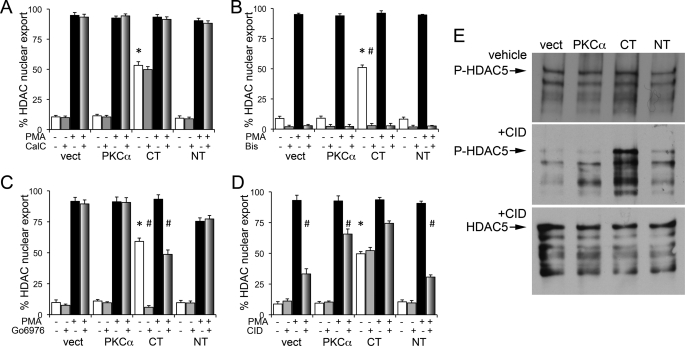
The signaling mechanism for PKCα-CT mediated HDAC5 nuclear export was interrogated using pharmacological inhibitors of PKC and its putative downstream signaling effectors. First, we examined the effects of two different classes of PKC inhibitors, bisindolylmaleimide IX (Bis) that competes with ATP in the PKC C-terminal catalytic domain (26), and calphostin C that interacts at calcium binding sites in the PKC N-terminal regulatory domain (27). Calphostin C did not prevent HDAC5 translocation under any of the experimental conditions (Fig. 4A), whereas Bis prevented HDAC5 nuclear export under all conditions assayed, i.e. in response to PMA and in PKCα-CT cells in the absence of PMA (Fig. 4B). These results show that phorbol ester activation is sufficient (without independent calcium signaling) to activate PKCs and promote HDAC5 nuclear export, and that ATP utilization by the kinases is essential for HDAC5 translocation.
FIGURE 4.
PKCα-CT induced HDAC5 phosphorylation and nuclear export is independent of PKD activation. Quantitative analysis of GFP-HDAC nuclear export before and after PMA treatment in cells pre-treated with PKC-inhibitor calphostin C (CalC 250μmol/liter; A), PKC-inhibitor bisindolylmaleimide IX (Bis 10 μmol/liter; B), PKD inhibitor CID755673 (CID 25μmol/liter; C), and protein kinase inhibitor Go6976 (1μmol/liter; D). *, p < 0.05 versus same treatment in vector-transfected cells; #, p < 0.05 versus PMA-treated cell expressing the same factor. Each study is result of 3–4 independent experiments performed in triplicate. E, in vitro phosphorylation of purified HDAC5 by PKCα or its calpain proteolytic fragments. Anti-phospho S498 HDAC5 immunoblots (top and middle panel) and anti-HDAC5 immunoblot (bottom panel) in the absence (top) or presence (middle, bottom) of PKD inhibitor CID. HDAC5 protein was treated with lysates of HEK293 cells transfected with same constructs as in panel C.
Nuclear export of HDACs can be induced by calcium/calmodulin-dependent kinases (CaM kinase) (29), but CaM kinase inhibition with KN-93 and KN-62 (30, 31) had no effect on HDAC5 translocation in our systems (supplemental Fig. S1). During the course of these studies we identified one pharmacological inhibitor that uniquely affected PKCα-CT-stimulated HDAC5 translocation, the protein kinase inhibitor Go6976 (32). Cells pre-treated with Go6976 exhibited normal PMA-induced HDAC5 nuclear export in vector-, PKCα-, and PKCα-NT-expressing cells. However, the typical basal HDAC5 translocation seen in PKCα-CT expressing cells was abolished, and PMA-induced HDAC5 translocation in these cells was attenuated, by the Go compound (Fig. 4C).
PKCs are thought to promote HDAC5 nuclear export indirectly by activating PKD (also known as PKCμ) that, in turn, phosphorylates HDACs (14, 24, 25). Consistent with this mechanism, the specific PKD inhibitor CID 755673 (28) significantly blunted PMA-induced HDAC5 translocation in vector-, PKCα-, and PKCα-NT-transfected cells (Fig. 4D). Remarkably, CID 755673 did not inhibit HDAC5 nuclear export induced by PKCα-CT in the absence of PMA (Fig. 4D), showing that PKCα-CT directly induces HDAC5 nuclear translocation, independent of PKD. We confirmed the lack of an effect of PKD inhibition on the PKCα-CT HDAC5 interaction by measuring in vitro phosphorylation of HDAC5 protein. Cell lysates expressing PKCα and its fragments all phosphorylated HDAC5, likely due to activation of endogenous kinases and PKCα by cell-derived diacyglycerol. Strikingly, addition of CID 755673 prevented in vitro HDAC5 phosphorylation under all conditions except PKCa-CT (Fig. 4E, middle panel), paralleling the results on in vivo HDAC5 nuclear export. These findings prove that PKCα-CT directly promotes HDAC5 phophorylation and nucleo-cytoplasmic shuttling, mechanistically distinguishing it from the indirect, PKD-mediated effects of its PKCα parent and endogenous PKCs.
Gene Expression Induced by PKCα-CT Is Linked to Cardiac Pathology
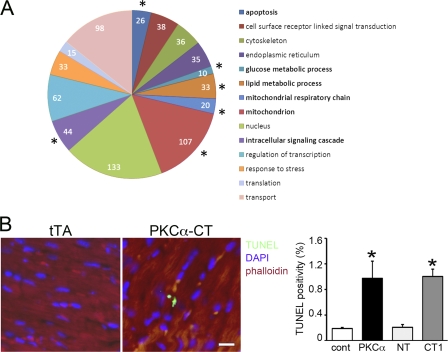
Our prior study showed that calpain-generated PKCα-CT is a constitutively active protein kinase that promiscuously phosphorylates atypical substrates. Ablation of calpain 1 prevented the formation of PKCα-CT and was cardioprotective in myocardial ischemia (12). To establish a pathophysiological connection between transcriptional regulation by PKCα-CT and the pathology it induces in mouse hearts, we performed an analysis of Gene Ontology categories on the PKCα-CT-regulated gene sets. Apoptosis, as well as lipid metabolism and transport and (predictably) intracellular signaling genes were over-represented among PKCα-CT-up-regulated mRNAs (Fig. 5A). Accordingly, we examined PKCα-CT hearts for evidence of programmed cardiomyocyte death. TUNEL positivity was increased in PKCα-CT and PKCα hearts after the onset of cardiomyopathy, although there was no difference in steady-state TUNEL positivity between PKCα- and PKCα-CT-expressing hearts (Fig. 5B). Thus, transcriptional up-regulation of cell death genes likely contributes to the cardiomyopathy of PKCα activation, whether this occurs via receptor-dependent or calapin-mediated receptor-independent pathways.
FIGURE 5.
Cardiac PKCα-CT induces expression of pathological genes that contribute to the cardiomyopathy phenotype. A, schematic depiction of results of gene-ontology analysis of deep RNA sequencing. Size of pie slice is proportional to number of regulated mRNAs in that functional cluster. *, significantly increased proportional expression compared with control tTA-expressing hearts. GO categories by color are listed to the right. Gene lists indexed by gene-ontology category and ID are in supplemental Table S1. B, myocardial TUNEL staining (green), co-stained for sarcomeric actin (phalloidin, red) and DAPI nuclear stain (blue); group quantitative results are on the right (n = 3 12-week-old mice/group). *, p < 0.05 compared with tTA control hearts. #, p < 0.05 compared with PKCα hearts.
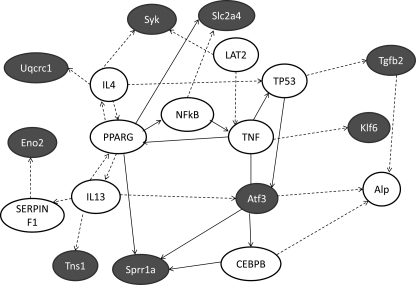
To examine the potential role of the prototypical HDAC-MEF2 interaction (33) on the effects of PKCα-CT, we examined the regulation of bioinformatically identified MEF2 target mRNAs. Our studies reveal 297 predicted MEF2 target genes expressed >1 mRNA copy/cell in mouse hearts. Compared with control tTA hearts, 15 MEF2 target mRNAs were up-regulated in PKCα-CT hearts, only one of which (Rtn4) was also regulated in PKCα hearts; no MEF2 target mRNAs were significantly regulated in PKCα-NT hearts (Table 1). Interestingly, nine of the PKCα-CT-up-regulated MEF2 target mRNAs were classified by Ingenuity Pathways Analysis software within a network of cellular development and inflammatory response signaling activated by tumor necrosis factor (TNF) α (Fig. 6). Thus, the transcriptional signature induced by constitutive PKCα-CT mediated phosphorylation of HDAC5 recapitulates seminal features of genetic reprogramming induced by TNFα-mediated activation of cardiac death receptor pathways.
TABLE 1.
PKCα-CT-regulated MEF2 target genes in transgenic mouse hearts
mRNAs are ordered according to the magnitude of fold-change compared to tTA hearts, and expression values are numbers of copies per cell (mean ± S.E.) determined from RNA sequencing (n = 3 hearts per genotype). All mRNAs are significantly different between PKCα-CT and tTA hearts, p < 0.001.
| Gene symbol | Gene name | tTA copies/cell | PKCα-CT copies/cell |
|---|---|---|---|
| Up-regulated | |||
| Sprr1a | Small proline-rich protein 1A | 3.32 ± 2.99 | 131.31 ± 24.63 |
| Atf3 | Activating transcription factor 3 | 0.78 ± 0.20 | 5.39 ± 0.54 |
| Tgfb2a | Transforming growth factor, beta 2 | 1.39 ± 0.09 | 7.82 ± 0.77 |
| Eno2a | Enolase 2 (gamma, neuronal) | 0.57 ± 0.02 | 2.59 ± 0.44 |
| Srpx2a | Sushi-repeat-containing protein, X-linked 2 | 0.82 ± 0.19 | 3.55 ± 0.64 |
| Palm2 | Paralemmin 2 | 15.44 ± 0.46 | 63.88 ± 3.89 |
| Rtn4a | Reticulon 4 | 6.68 ± 0.15 | 21.82 ± 0.48 |
| Trp53inp1a | Transformation related protein 53 inducible nuclear protein 1 | 1.03 ± 0.20 | 3.23 ± 0.26 |
| Klf6 | Kruppel-like factor 6 | 4.41 ± 0.20 | 11.79 ± 1.88 |
| Ttll7 | Tubulin tyrosine ligase-like family, member 7 | 0.82 ± 0.07 | 2.05 ± 0.12 |
| Fkbp7 | FK506 binding protein 7 | 2.88 ± 0.23 | 6.67 ± 0.89 |
| Acot10 | Acyl-CoA thioesterase 10 | 1.60 ± 0.24 | 3.47 ± 0.28 |
| Syk | Spleen tyrosine kinase | 2.18 ± 0.05 | 4.01 ± 0.44 |
| Zfc3h1 | Zinc finger, C3H1-type containing | 1.16 ± 0.01 | 1.96 ± 0.09 |
| Tns1 | Tensin 1 | 51.00 ± 0.06 | 72.53 ± 1.62 |
| Down-regulated | |||
| R3hdm2a | R3H domain containing 2 | 14.45 ± 0.40 | 10.45 ± 0.57 |
| Phyha | Phytanoyl-CoA 2-hydroxylase | 61.60 ± 3.19 | 34.48 ± 4.64 |
| Cand2 | Cullin-associated and neddylation-dissociated 2 (putative) | 6.89 ± 0.64 | 3.76 ± 0.38 |
| Spega | SPEG complex locus | 8.85 ± 0.12 | 4.79 ± 0.47 |
| Uqcrc1 | Ubiquinol-cytochrome c reductase core protein I | 127.63 ± 7.39 | 68.54 ± 3.16 |
| Slc2a4 | Solute carrier family 2 (facilitated glucose) transporter), member 4 | 18.76 ± 0.77 | 8.97 ± 1.18 |
| Adhfe1a | Alcohol dehydrogenase, iron containing, 1 | 7.11 ± 0.54 | 3.05 ± 0.28 |
| Extl1a | Exostoses (multiple)-like 1 | 2.74 ± 0.22 | 1.13 ± 0.16 |
| Dhrs7ca | Dehydrogenase/reductase (SDR family) member 7C | 22.61 ± 0.84 | 6.41 ± 2.10 |
| Rpl3l | Ribosomal protein L3-like | 15.05 ± 1.13 | 2.04 ± 0.10 |
a Denotes significant difference between PKCα-CT and tTA hearts found by microarray, i.e. concordant results between RNA-Seq and microarray.
FIGURE 6.
Ingenuity pathway analysis of PKCα-CT regulated, MEF2-targetted cardiac mRNAs. Ingenuity Pathways Analysis software was used to depict potential signaling pathways involving the 15 putative MEF2 targets among PKCα-CT up-regulated mRNAs. Nine of 15 mRNAs were included in the network. Lines with arrowheads, molecule acts on a target; lines without arrowheads, binding only. Solid lines, direct interaction; dotted lines, indirect interaction. Blue background, regulated putative MEF2 targets; white background, other signaling network members.
DISCUSSION
We recently described receptor-independent, calpain-mediated proteolytic processing of PKCα in ischemic hearts that generates a stable, constitutively active PKCα-CT kinase fragment, which is sufficient to cause dilated cardiomyopathy (12). However, a mechanistic link between kinase activity and PKCα-CT-induced cardiomyopathy has not been clearly defined. Here, we report constitutive nuclear localization and transcriptional regulation by PKCα-CT that can explain at least some of its detrimental effects on the heart. Our studies further reveal a direct stimulatory effect of PKCα-CT on HDAC5 nuclear export that differentiates it from its parent kinase, PKCα, and that can provoke programmed cell death ensuing from calpain-mediated generation of this rogue kinase.
Signaling pathways mediated via G-protein coupled receptors play important roles in the compensatory responses to cardiac stress, and in functional decompensation that ensues when these mechanisms fail. Gq/phospholipase C signaling initiated by angiotensin, α-adrenergic, and other cardiomyocyte receptors is implicated in pathological hypertrophy and the transition to heart failure (34). Conventional PKC isoforms (α, βI, βII, and γ) are activated by these Gq/phospholipase C-coupled receptors as the consequence of increased intracellular calcium and phospholipid, which bind to specific domains within the regulatory N-terminal half of PKCs (35–38). Calcium and phospholipid binding induce allosteric changes that expose ATP- and substrate-binding domains located within the catalytic carboxy-terminal half of the protein (39). Because PKC unfolding after calcium and phospholipid binding also exposes binding domains for PKC anchoring proteins (RACKs), receptor-mediated PKC activation stimulates PKC translocation from cytosol to particulate cell membranes. Spatial restriction thus produces substrate specificity (13). Because PKC activity has a narrow physiological range, it is limited not only in space, but in time: PKC-activating calcium and phospholipid second messengers are evanescent, lasting only seconds to minutes. As these PKC-activating signals expire, the inactive PKC molecular configuration is restored (temporal restriction). Importantly, post-translational formation of PKCα-CT by calpain-mediated proteolytic processing generates a kinase lacking both of these normal physiological restraints: PKCα-CT lacks the N-terminal targeting domains, and therefore accesses atypical subcellular compartments and abnormal substrates. PKCα-CT also lacks the N-terminal autoinhibitory domains and is therefore constitutively and persistently active (12).
Calpain-generated PKCα-CT corresponds approximately to Nishizuka's description of PKMα and Olson's artificial PKC mutant, PKC-8 (36, 40). Our observation that full-length cardiomyocyte PKCα and PKCα-NT are largely cytosolic, whereas PKCα-CT also resides in the nuclear fraction, echos the findings of James and Olson (41) who reported that: 1. Inactive PKCα in transfected CHO cells and C2 myoblasts was largely cytosolic and underwent nuclear translocation after activation with phorbol ester; 2. A PKCα truncation mutant lacking the catalytic domain (PKC-TM310; analogous to the PKCα-NT calpain proteolysis product) is cytosolic and unstable; and 3. A PKCα truncation mutant consisting only of the PKCα catalytic domain (PKC-8; analogous to the PKCα-CT calpain proteolysis product) is stable and constitutively localized to Triton-insoluble and nuclear fractions. Thus, in vitro mutational analysis of PKCα domains and in vivo analysis of the naturally occurring calpain-generated PKCα fragments agree that the free catalytic kinase domain will localize in part to cell nuclei and regulate gene transcription. It is worth noting that calpain-mediated processing of PKCs has been observed in many tissues, and that an analogous calpain-generated constitutively active PKCζ catalytic domain, designated PKMζ, plays a physiological role in maintaining long-term memory (42). Thus, genetic reprogramming by calpain-generated PKC-derived kinases may be a recurring theme in vertebrate biology.
Nuclear-localized PKCα is thought to direct gene transcription by activating PKD, which phosphorylates class II HDACs, thus de-repressing MEF2-dependent genes (14). Our results confirm that PKD is an essential downstream mediator of HDAC5 nuclear export mediated by endogenous COS7 cell PKCs and by unprocessed human PKCα. However, we found that PKCα-CT provoked HDAC5 nucleo-cytoplasmic shuttling independently of PKD by directly inducing HDAC phosphorylation. This describes a new receptor-independent pathway for transcriptional control. Previously described mechanisms for HDAC nuclear export include PKC, PKD, and CaM kinase II, all of which are calcium-dependent factors within G-protein-coupled receptor signaling pathways. HDAC phopshorylation by calpain-generated PKCα-CT links increased cell calcium and MEF2-dependent gene transcription in a manner that bypasses normal receptor signaling pathways, and therefore cannot be prevented by pharmacological receptor antagonism or receptor agonist synthesis inhibition.
Cardiac gene reprogramming by PKCα-CT reproduced aspects of TNFα-stimulated gene expression. Pro-inflammatory cytokines such as TNFα are linked to heart failure by activating their cardiomyocyte receptors and stimulating gene transcription via the well-characterized NFκB pathway (43). Cytokine signaling thus coordinates the response of external inflammatory factors recruited to stressed or damaged myocardium and the cardiomyocytes in the area of stress or injury. Work in other cell types has implicated PKC signaling and HDAC nuclear export in atypical, NFκB-independent transcriptional activation by TNFα (44, 45). Activation of components of the inflammatory gene program by PKCα-CT in viable ischemic and/or reperfused is therefore a mechanism by which the cardiomyocyte transcriptional stress response is rapidly and persistently activated in a manner that anticipates actual mononuclear infiltration and local cytokine production.
Our studies provide additional support for the approach of direct protein kinase inhibition in heart failure. Not only do we observe that an inhibitor of catalytical PKC activity prevented HDAC nuclear export induced by PMA, PKCα, and PKCα-CT, but we also found that Go6976 specifically inhibited constitutive PKCα-CT-induced HDAC nucleoplasmic shuttling. Go6976, which is an inhibitor of calcium-dependent protein kinase C isoforms and has additional activities as well (46), failed to inhibit PMA-stimulated HDAC nuclear export, but abolished that constitutively induced by PKCα-CT. This finding demonstrates the feasibility of developing inhibitors with relative specificity for the strictly pathological free catalytic kinase fragments, but that spare normal receptor-mediated PKC signaling pathways that may have critical essential functions in non-involved tissues.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants R01 HL080008 and R01 HL059888 and by UL1 RR024992 from the National Center for Research Resources (to G. W. D. II).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Fig. S1.
- PKC
- protein kinase C
- NT
- N-terminal
- CT
- C-terminal
- FDR
- false discovery rates
- TUNEL
- terminal deoxynucleotidyltransferase-mediated dUTP nick end-labeling
- tTA
- tetracycline transcriptional activator.
REFERENCES
- 1. Nakashima S. (2002) J. Biochem. 132, 669–675 [DOI] [PubMed] [Google Scholar]
- 2. Hahn H. S., Marreez Y., Odley A., Sterbling A., Yussman M. G., Hilty K. C., Bodi I., Liggett S. B., Schwartz A., Dorn G. W., 2nd (2003) Circ. Res. 93, 1111–1119 [DOI] [PubMed] [Google Scholar]
- 3. Braz J. C., Gregory K., Pathak A., Zhao W., Sahin B., Klevitsky R., Kimball T. F., Lorenz J. N., Nairn A. C., Liggett S. B., Bodi I., Wang S., Schwartz A., Lakatta E. G., DePaoli-Roach A. A., Robbins J., Hewett T. E., Bibb J. A., Westfall M. V., Kranias E. G., Molkentin J. D. (2004) Nat. Med. 10, 248–254 [DOI] [PubMed] [Google Scholar]
- 4. Hambleton M., Hahn H., Pleger S. T., Kuhn M. C., Klevitsky R., Carr A. N., Kimball T. F., Hewett T. E., Dorn G. W., 2nd, Koch W. J., Molkentin J. D. (2006) Circulation 114, 574–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hambleton M., York A., Sargent M. A., Kaiser R. A., Lorenz J. N., Robbins J., Molkentin J. D. (2007) Am. J. Physiol. Heart. Circ. Physiol. 293, H3768–H3771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liu Q., Chen X., MacDonnell S. M., Kranias E. G., Lorenz J. N., Leitges M., Houser S. R., Molkentin J. D. (2009) Circ. Res. 105, 194–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mochly-Rosen D., Wu G., Hahn H., Osinska H., Liron T., Lorenz J. N., Yatani A., Robbins J., Dorn G. W., 2nd (2000) Circ. Res. 86, 1173–1179 [DOI] [PubMed] [Google Scholar]
- 8. Chen L., Hahn H., Wu G., Chen C. H., Liron T., Schechtman D., Cavallaro G., Banci L., Guo Y., Bolli R., Dorn G. W., 2nd, Mochly-Rosen D. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 11114–11119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gálvez A. S., Brunskill E. W., Marreez Y., Benner B. J., Regula K. M., Kirschenbaum L. A., Dorn G. W., 2nd (2006) J. Biol. Chem. 281, 1442–1448 [DOI] [PubMed] [Google Scholar]
- 10. Chien K. R., Knowlton K. U., Zhu H., Chien S. (1991) FASEB. J. 5, 3037–3046 [DOI] [PubMed] [Google Scholar]
- 11. Clerk A., Sugden P. H. (1999) Am. J. Cardiol. 83, 64H–69H [DOI] [PubMed] [Google Scholar]
- 12. Kang M. Y., Zhang Y., Matkovich S. J., Diwan A., Chishti A. H., Dorn G. W., 2nd (2010) Circ. Res. 107, 903–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dorn G. W., 2nd, Mochly-Rosen D. (2002) Annu. Rev. Physiol. 64, 407–429 [DOI] [PubMed] [Google Scholar]
- 14. Vega R. B., Harrison B. C., Meadows E., Roberts C. R., Papst P. J., Olson E. N., McKinsey T. A. (2004) Mol. Cell Biol. 24, 8374–8385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Matkovich S. J., Zhang Y., Van Booven D. J., Dorn G. W., 2nd (2010) Circ. Res. 106, 1459–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Matkovich S. J., Van Booven D. J., Youker K. A., Torre-Amione G., Diwan A., Eschenbacher W. H., Dorn L. E., Watson M. A., Margulies K. B., Dorn G. W., 2nd (2009) Circulation 119, 1263–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Matkovich S. J., Wang W., Tu Y., Eschenbacher W. H., Dorn L. E., Condorelli G., Diwan A., Nerbonne J. M., Dorn G. W., 2nd (2010) Circ. Res. 106, 166–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mortazavi A., Williams B. A., McCue K., Schaeffer L., Wold B. (2008) Nat. Methods. 5, 621–628 [DOI] [PubMed] [Google Scholar]
- 19. Schug J. (2008) Curr. Protoc. Bioinformatics, Chapter 2, Unit 2.6 [DOI] [PubMed] [Google Scholar]
- 20. Putt M. E., Hannenhalli S., Lu Y., Haines P., Chandrupatla H. R., Morrisey E. E., Margulies K. B., Cappola T. P. (2009) Circ. Cardiovasc. Genet. 2, 212–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Maere S., Heymans K., Kuiper M. (2005) Bioinformatics 21, 3448–3449 [DOI] [PubMed] [Google Scholar]
- 22. Simpson P. C., Kariya K., Karns L. R., Long C. S., Karliner J. S. (1991) Mol. Cell Biochem. 104, 35–43 [DOI] [PubMed] [Google Scholar]
- 23. Ha C. H., Wang W., Jhun B. S., Wong C., Hausser A., Pfizenmaier K., McKinsey T. A., Olson E. N., Jin Z. G. (2008) J. Biol. Chem. 283, 14590–14599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dequiedt F., Van, Lint J., Lecomte E., Van Duppen V., Seufferlein T., Vandenheede J. R., Wattiez R., Kettmann R. (2005) J. Exp. Med. 201, 793–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang S., Li X., Parra M., Verdin E., Bassel-Duby R., Olson E. N. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 7738–7743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Davis P. D., Elliott L. H., Harris W., Hill C. H., Hurst S. A., Keech E., Kumar M. K., Lawton G., Nixon J. S., Wilkinson S. E. (1992) J. Med. Chem. 35, 994–1001 [DOI] [PubMed] [Google Scholar]
- 27. Kobayashi E., Nakano H., Morimoto M., Tamaoki T. (1989) Biochem. Biophys. Res. Commun. 159, 548–553 [DOI] [PubMed] [Google Scholar]
- 28. Sharlow E. R., Giridhar K. V., LaValle C. R., Chen J., Leimgruber S., Barrett R., Bravo-Altamirano K., Wipf P., Lazo J. S., Wang Q. J. (2008) J. Biol. Chem. 283, 33516–33526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Backs J., Song K., Bezprozvannaya S., Chang S., Olson E. N. (2006) J. Clin. Invest. 116, 1853–1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sumi M., Kiuchi K., Ishikawa T., Ishii A., Hagiwara M., Nagatsu T., Hidaka H. (1991) Biochem. Biophys. Res. Commun. 181, 968–975 [DOI] [PubMed] [Google Scholar]
- 31. Hidaka H., Kobayashi R. (1992) Annu. Rev. Pharmacol. Toxicol. 32, 377–397 [DOI] [PubMed] [Google Scholar]
- 32. Gschwendt M., Dieterich S., Rennecke J., Kittstein W., Mueller H. J., Johannes F. J. (1996) FEBS. Lett. 392, 77–80 [DOI] [PubMed] [Google Scholar]
- 33. Olson E. N., Backs J., McKinsey T. A. (2006) Novartis Foundation Symp. 274, 3–12 [PubMed] [Google Scholar]
- 34. Dorn G. W., 2nd, Force T. (2005) J. Clin. Invest. 115, 527–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nishizuka Y. (1988) Nature 334, 661–665 [DOI] [PubMed] [Google Scholar]
- 36. Bright R., Mochly-Rosen D. (2005) Stroke 36, 2781–2790 [DOI] [PubMed] [Google Scholar]
- 37. Mackay H. J., Twelves C. J. (2007) Nat. Rev. Cancer. 7, 554–562 [DOI] [PubMed] [Google Scholar]
- 38. Churchill E., Budas G., Vallentin A., Koyanagi T., Mochly-Rosen D. (2008) Annu. Rev. Pharmacol. Toxicol. 48, 569–599 [DOI] [PubMed] [Google Scholar]
- 39. Newton A. C. (1995) J. Biol. Chem. 270, 28495–28498 [DOI] [PubMed] [Google Scholar]
- 40. Young S., Parker P. J., Ullrich A., Stabel S. (1987) Biochem. J. 244, 775–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. James G., Olson E. (1992) J. Cell Biol. 116, 863–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sacktor T. C. (2011) Nat. Rev. Neurosci. 12, 9–15 [DOI] [PubMed] [Google Scholar]
- 43. Mann D. L. (2001) Heart. Fail. Rev. 6, 71–80 [DOI] [PubMed] [Google Scholar]
- 44. Catley M. C., Cambridge L. M., Nasuhara Y., Ito K., Chivers J. E., Beaton A., Holden N. S., Bergmann M. W., Barnes P. J., Newton R. (2004) J. Biol. Chem. 279, 18457–18466 [DOI] [PubMed] [Google Scholar]
- 45. Kumar A., Lin Z., SenBanerjee S., Jain M. K. (2005) Mol. Cell. Biol. 25, 5893–5903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Koivunen J., Aaltonen V., Koskela S., Lehenkari P., Laato M., Peltonen J. (2004) Cancer Res. 64, 5693–5701 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.