Abstract
A highly conserved signaling pathway involving insulin-like growth factor 1 (IGF1), and a cascade of intracellular components that mediate its effects, plays a major role in the regulation of skeletal muscle growth. A central component in this cascade is the kinase Akt, also called protein kinase B (PKB), which controls both protein synthesis, via the kinases mammalian target of rapamycin (mTOR) and glycogen synthase kinase 3β (GSK3β), and protein degradation, via the transcription factors of the FoxO family. In this paper, we review the composition and function of this pathway in skeletal muscle fibers, focusing on evidence obtained in vivo by transgenic and knockout models and by muscle transient transfection experiments. Although this pathway is essential for muscle growth during development and regeneration, its role in adult muscle response to mechanical load is less clear. A full understanding of the operation of this pathway could help to design molecularly targeted therapeutics aimed at preventing muscle wasting, which occurs in a variety of pathologic contexts and in the course of aging.
Introduction
Muscle wasting occurs in a variety of conditions, such as cancer cachexia, diabetes, renal failure and heart failure, and aging itself. The survival and quality of life of these patients and of the older person can be improved by counteracting loss of muscle mass and strength, and different approaches to this have been explored, including nutritional supplementation, resistance training and anabolic drugs. Recent advances in understanding the mechanisms responsible for muscle atrophy may pave the way to new and perhaps more effective treatments.
During the past several years, experimental studies based on rigorous genetic approaches have started to dissect the signaling pathways involved in muscle-mass regulation. Although studies on cultured muscle cells have contributed to identify these pathways, definitive evidence of their physiological relevance can only be obtained using in vivo systems, when myofibers have a mature structure, and the integrity of the neuromuscular and musculoskeletal system is preserved.
Two in vivo genetic approaches have been used to understand how muscle mass is regulated. One is based on the generation of transgenic and knockout mice, in which expression of muscle regulatory genes is selectively modified. The potential of the traditional gene overexpression or deletion approaches has been fully exploited with the introduction of the Cre/loxP technique and the use of inducible transgenes, which allows for the modulation of gene expression specifically in muscle tissues and at different developmental stages. It is thus possible to distinguish between the effects on the regulation of muscle growth during development from the effects on the maintenance of muscle mass in adulthood. An alternative approach to address muscle-mass regulation in the adult is based on in vivo transfection of skeletal muscles by electroporation with plasmids coding for specific components of signaling pathways, or for mutants bearing constitutively active or dominant negative properties. Transfection with plasmids able to generate specific small interfering RNAs in muscle fibers is also increasingly being used as a loss-of-function model. The ability of various factors in preventing muscle atrophy can be explored by transfecting denervated muscles.
In this review, we discuss how in vivo studies based on these genetic models have contributed to define the role of a specific signaling pathway, the insulin-like growth factor 1-Akt/protein kinase B (IGF1-Akt/PKB) pathway, in muscle mass regulation. Various aspects of the role of this pathway in skeletal muscle have been previously discussed [1-3]. The IGF1-Akt1 pathway shares most of its components with the insulin-Akt2 pathway, and the two pathways intersect at various levels. For example, insulin can also bind the IGF1 receptor and IGF1 can bind to the insulin receptor; furthermore, hybrids between the IGF1 and insulin receptors are present in skeletal muscle. However, insulin is especially important in glucose homeostasis, whereas IGF1 is mostly active in muscle growth. In this review, we consider exclusively the role of this pathway on growth rather than on metabolism.
Overview of the IGF1-Akt/PKB pathway
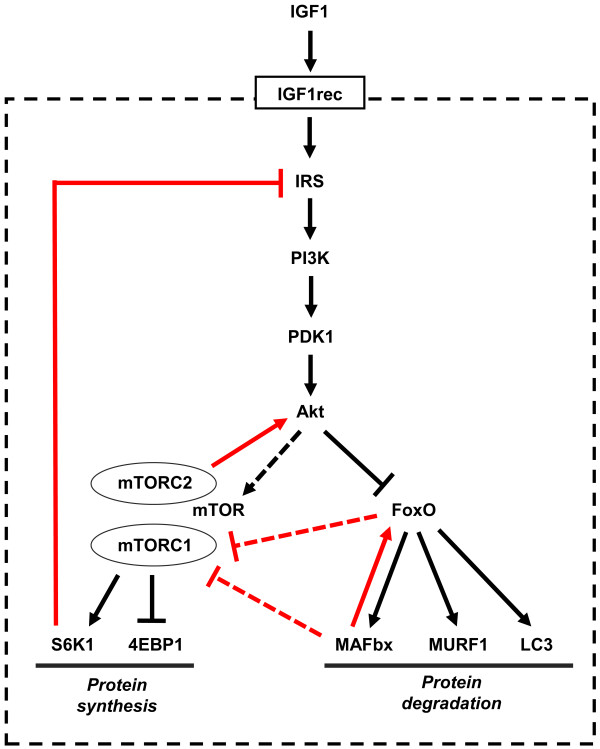
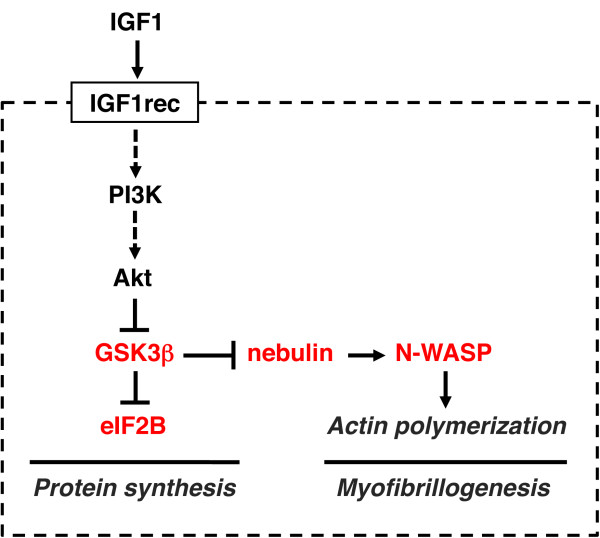
A simplified scheme of the IGF1-Akt pathway is shown in Figure 1. Binding of IGF1 to its receptor leads to activation of its intrinsic tyrosine kinase and autophosphorylation, thus generating docking sites for insulin receptor substrate (IRS), which is also phosphorylated by the IGF1 receptor. Phosphorylated IRS then acts as docking site to recruit and activate phosphatidylinositol-3-kinase (PI3K) which phosphorylates membrane phospholipids, generating phosphoinositide-3,4,5-triphosphate (PIP3) from phosphoinositide-4,5-biphosphate (PIP2). PIP3 acts in turn as a docking site for two kinases, phosphoinositide-dependent kinase 1 (PDK1) and Akt, and the subsequent phosphorylation of Akt at serine 308 by PDK1, leading to Akt activation. All these steps take place at the inner surface of the plasma membrane. Akt inhibits protein degradation by phosphorylating and thus repressing the transcription factors of the FoxO family, and stimulates protein synthesis via the mammalian target of rapamycin (mTOR) and glycogen synthase kinase 3β (GSK3β) [4]. FoxO factors are required for the transcriptional regulation of the ubiquitin ligases atrogin-1, also called muscle atrophy F-box (MAFbx) and muscle ring finger 1 (MuRF1), leading to the ubiquitylation of myosin and other muscle proteins (see below), and their degradation via the proteasome. FoxO factors are also required for the transcriptional regulation of the microtubule-associated protein 1 light chain 3 (LC3), which together with BCL2/adenovirus E1B interacting protein 3 (BNIP3) is essential for the activation of the autophagy-lysosome pathway. The effect of Akt on mTOR is indirect: Akt inhibits the tuberous sclerosis complex (TSC) proteins 1 and 2, which act as a GTPase activating protein (GAP) to inhibit the small G protein Ras homolog enriched in brain (Rheb) which activates mTOR signaling. mTOR forms two different protein complexes, the rapamycin-sensitive mTORC1, when bound to Raptor, and the rapamycin-insensitive mTORC2, when bound to Rictor [5]. TORC2 is required for Akt phosphorylation and activation [6]. mTORC1 phosphorylates S6 kinase (S6K), which in turn phosphorylates the ribosomal protein S6 and other factors involved in translation initiation and elongation, thus stimulating protein synthesis. TORC1 also activates eukaryotic translation initiation factor 4E (eIF4E) by phosphorylating the inhibitory eIF4E-binding proteins (4EBPs). Akt also promotes protein synthesis by phosphorylating and inactivating GSK3β, thus releasing the GSK3β-dependent inhibition of the eukariotic translation initiation factor 2B (eIF2B) (Figure 2).
Figure 1.
The insulin-like growth factor 1 (IGF1)-Akt pathway controls muscle growth via mammalian target of rapamycin (mTOR) and FoxO. The internal feedback loops that control the IGF1-Akt pathway are indicated in red. The dotted line indicates that the effect of Akt on mTOR is indirect, being mediated by the tuberous sclerosis complex (TSC) proteins 1 and 2 and by Rheb (Ras homolog enriched in brain). See text for details.
Figure 2.
The insulin-like growth factor 1 (IGF1)-Akt pathway controls muscle growth also via glycogen synthase kinase β (GSK3β). GSK3β inhibits protein synthesis via eukaryotic initiation factor 2B (eIF2B) and actin filament formation via nebulin and neuronal Wiscott-Aldrich syndrome protein (N-WASP). See text for details.
The activity of the IGF1-Akt pathway is controlled by several feedback loops (Figure 1). Negative feedback involves S6K, which inhibits IRS by phosphorylation at multiple sites, thus inducing its degradation and altered cell localization [7]. Positive feedback involves mTORC2, which phosphorylates Akt at serine 473, a phosphorylation required for maximum activation of Akt in addition to phosphorylation at threonine 308 by PDK1 [8]. Rictor-dependent phosphorylation of Akt at serine 473 is required for Akt-mediated phosphorylation of FoxO but not of TSC2, thus it does not affect activation of S6K [5]. In another feed-forward mechanism, MAFbx, which is activated by FoxO, acts in turn as coactivator of FoxO [9]. MAFbx also appears to control protein synthesis by ubiquitylating and thus promoting the degradation of the eukariotic translation initiation factor 3 subunit F (eIF3F), thus suppressing S6K1 activation by mTOR [10,11]. FoxO factors have been shown to inhibit mTORC1 and activate Akt by inducing the expression of sestrin3 and Rictor in cultured mammalian cells [12], but this pathway has not been characterized in skeletal muscle in vivo.
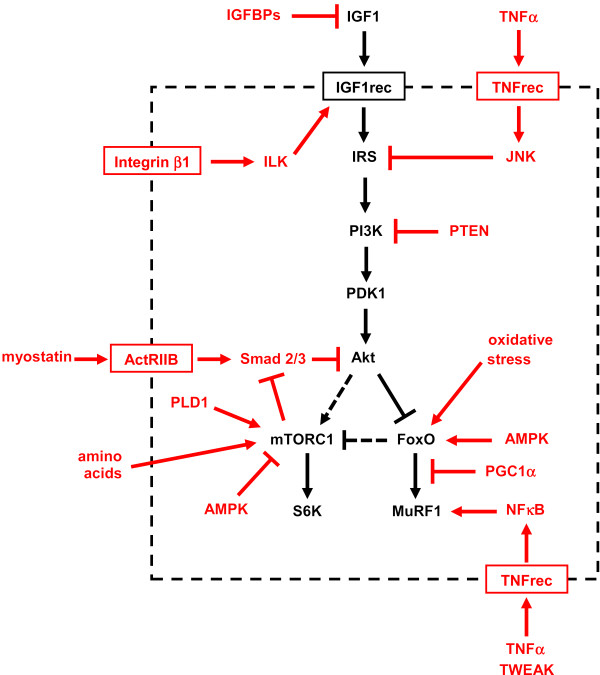
The activity of the IGF1-Akt pathway can be modulated by a variety of other factors and pathways acting on different steps (Figure 3). IGF binding proteins (IGFBPs), the most important probably being IGFBP5, can block IGF1 action by inhibiting its binding to the IGF1 receptor. Mechanical signals via integrin β1 and integrin-linked kinase (ILK) lead to phosphorylation of the IGF1 receptor and activation of the PI3K-Akt pathway in muscle cells [13]. Whether another downstream integrin effector, the integrin-dependent focal adhesion kinase (FAK), affects the IGF1-Akt pathway in skeletal muscle remains to be established. FAK null mice are embryonic lethal, and skeletal muscle-specific conditional mutants have not been reported. Overexpression of FAK by electrotransfer was found to induce slight hypertrophy in adult rat muscles, but the effect on the IGF1-Akt pathway was not investigated [14]. Activation of IRS types is inhibited by phosphorylation of serine residues induced by inflammatory cytokines such as tumor necrosis factor α (TNFα) via Jun N-terminal kinase (JNK) [15]. PTEN (phosphatase and tensin homolog deleted from chromosome 10) is a lipid phosphatase that converts PIP3 to PIP2, thus opposing the action of PI3K, and interfering with Akt docking to the plasma membrane. Myostatin, also called growth and differentiation factor 8 (GDF8), acts as negative regulator of muscle growth, as shown by the hypertrophic phenotype induced by inactivation of the myostatin gene [16]. Myostatin, together with activin A, another member of the TGFβ family, acts via its receptor activin receptor IIB (ActRIIB) on Smad2 and Smad3, inhibitors of Akt; conversely, mTOR inhibits Smads [17,18]. In cultured muscle cells, addition of IGF1 dominantly blocks the effect of myostatin [17]. In adult skeletal muscle, muscle hypertrophy can be induced and muscle wasting prevented by blocking myostatin either via postdevelopmental myostatin gene knockout, or with follistatin (a myostatin antagonist) or anti-myostatin antibodies or a soluble ActRIIB decoy receptor (see [19,20] and references therein).
Figure 3.
Multiple factors and pathways affect insulin-like growth factor 1 (IGF1)-Akt signaling. Various factors and pathways affecting the IGF1-Akt pathway are highlighted in red. See text for details.
mTORC1 integrates growth factor signaling with a variety of signals from nutrients and cellular energy status. Direct activation of mTORC1 by amino acids is mediated by the Rag family of GTPases, which interact with Raptor [21,22]. By contrast, AMP-activated kinase (AMPK), an energy status sensor activated by ATP depletion, inhibits mTORC1 by phosphorylating and inactivating Raptor [23], and by phosphorylating and activating TSC2 [24]. The role of AMPK in the control of muscle fiber size through mTOR inhibition is demonstrated by the phenotype of AMPKα1 knockout and AMPKα1/α2 double knockout mice, which develop muscle hypertrophy [25,26]. mTOR can also be directly activated independently of PI3K/Akt signaling, by phosphatidic acid, generated by phospholipase D (PLD) in response to mechanical signals induced by eccentric muscle contractions [27-29].
AMPK can also impinge on the FoxO-dependent protein degradation pathways. AMPK can activate FoxO factors by phosphorylation at several regulatory sites distinct from Akt phosphorylation sites [30]. FoxO-dependent activation of atrogin-1 and muscle atrophy is inhibited by the peroxisome proliferator activated receptor γ coactivator 1α (PGC1α), a transcriptional coactivator. PGC1α transfection protects adult muscles from FoxO-dependent atrophy, and the higher levels of PGC1α in oxidative muscle fibers can explain their greater resistance to muscle atrophy [31]. Circulating cytokines, such as TNFα and TNF-related weak inducer of apoptosis (TWEAK), can activate MuRF1 via the transcription factor nuclear factor κB (NFκB) [32,33]. Finally, corticosteroids inhibit the IGF1-Akt pathway by acting at multiple levels, such as inducing decreased production of IGF1 and increased production of myostatin [34]. In addition, the glucocorticoid receptor and FoxO1 synergistically activate the MuRF1 gene [35].
Those listed above most probably represent only a minor proportion of the interactions of the IGF1-Akt pathway with other factors and pathways. Indeed, a recent interactome map based on a yeast two-hybrid screen for 33 components of the PI3K-mTOR pathway has identified 802 interactions, including 67 new validated interactions [36]. An additional complicating factor is the fact that various components of the IGF1-Akt pathway exist as multiple isoforms, many of which are coexpressed in skeletal muscle (Table 1). These include different splicing variants of IGF1, two types of IRS (IRS1 and IRS2), different isoforms of both the regulatory and catalytic subunits of PI3K, two types of Akt (Akt1 and Akt2) and S6K (S6K1 and S6K2), and several FoxO factors (FoxO1, FoxO3 and FoxO4). The different roles of some of these isoforms will be discussed below.
Table 1.
Isoforms of major components of the IGF1-Akt pathway
| Component | Isoforms | References |
|---|---|---|
| IGF11 | Two (mouse: 1A, 1B) or three (human: 1A, 1B & 1C) isoforms, differing in the C terminal peptide (E peptide); another two (mouse: 2A, 2B) or three (human: 2A, 2B & 2C) isoforms differing in signal peptide because of utilization of exon 2 instead of exon 1 | [49] |
| IGF1 receptor | Heterotetramer made of two α-subunit (IGF-binding) and two β-subunit (tyrosine kinase). No isoforms, but hybrids with insulin receptor are present in skeletal muscle | [112] |
| IRS2 | IRS1 and IRS2 | [65,113] |
| PDK13 | No isoforms | [114] |
| PI3K4 (class I) | Heterodimer of p85 regulatory and p110 catalytic subunits. p85: three isoforms (p85α, p55α, p50α) encoded by a single gene; two other isoforms (p85β, p55γ) coded by other genes. p55γ is not expressed in muscle. p110: three isoforms (p110α, p110β, p110δ). p110δ is not expressed in muscle | [115] |
| Akt/PKB5 | Akt1/PKBα, Akt2/PKBβ & Akt3/PKBγ. Akt3 is not expressed in muscle | [4,116] |
| mTOR6 | No isoforms, but mTOR can interact with different partners: Raptor in the rapamycin-sensitive complex mTORC1, or Rictor in the rapamycin-insensitive complex mTORC2 | [5] |
| S6K7 | S6K1 and S6K2 | [117-119] |
| 4EBP8 | 4EBP1, 4EBP2, 4EBP3. 4EBP3 is not expressed in muscle | [120] |
| FoxO | 4 isoforms: FoxO1, FoxO3, FoxO4 and FoxO6. FoxO6 is not expressed in muscle | [121] |
1Insulin-like growth factor.
2Insulin receptor substrate.
33-Phosphoinositide-dependent kinase 1, coded by the PDPK1 gene (unfortunately the same abbreviation PDK1 is also used to indicate the pyruvate dehydrogenase kinase isoform 1, coded by the PDK1 gene).
4Phosphoinositide-3 kinase.
5Protein kinase B.
6Mammalian target of rapamycin.
7S6 kinase.
8Eukaryotic initiation factor 4E binding protein.
A large number of transgenic and knockout mouse models involving components of the IGF1-Akt pathway have been generated and are listed in Tables 2 and 3, respectively. In vivo transfection experiments leading to perturbation of IGF1-Akt pathway components in skeletal muscle are listed in Table 4.
Table 2.
Transgenic models of the IGF1-Akt pathway: effect on growth
| Genotype1 | Viability | Phenotype | References |
|---|---|---|---|
| ASA-hIGF1 | Viable | Muscle hypertrophy | [45,47] |
| MLC1-IGF1 | Viable | Muscle hypertrophy | [51] |
| MCK-d.n. IGF1 receptor2 | Viable | Transient delay of postnatal muscle growth; unaffected overload-induced muscle hypertrophy; impaired muscle regeneration | [59-62] |
| HSA-Akt1, inducible | Viable | Muscle hypertrophy | [79] |
| MCK-myrAkt1, inducible | Viable | Muscle hypertrophy | [80] |
| MLC1f-myrAkt1, inducible | Viable | Muscle hypertrophy | [82] |
| HSA-FoxO1 | Viable | Muscle atrophy | [90] |
1Promoters used to drive transgene expression: ASA = avian skeletal actin; HSA = human skeletal actin; MCK = muscle creatine kinase; MLC1 = myosin light chain 1 fast.
2This transgene acts as a dominant negative for both the insulin-like growth factor 1 receptor and the insulin receptor, thus causing diabetes.
Table 3.
Knockout and knock-in models of IGF1-Akt pathway components: effect on growth
| Genotype1 | Viability | Growth phenotype | References |
|---|---|---|---|
| Igf1 null | Severe neonatal lethality | Severe growth retardation | [37-39] |
| Igf1 null in muscle (Mef2c-Cre) | Viable | Normal growth | [63] |
| Igf1 receptor null | Severe neonatal lethality | Severe growth retardation | [37,39] |
| Igf1 receptor null in muscle (Mef2c-Cre) | Viable | Reduced body weight, reduced muscle fiber number and size | [63] |
| IRS1 null | Viable | Reduced growth (weight 30-60% of control) | [64,122] |
| IRS2 null | Viable | Almost normal growth (birth weight 90% of control) | [65] |
| PI3K p85α + p55α + p50α null | Perinatal lethality | [66] | |
| PI3K p85α + p55α + p50α null in heart & muscle (MCK-Cre) | Viable | Normal growth | [68] |
| PI3K p85β null | Viable | Normal growth | [67] |
| PI3K p85α + p55α + p50α null in heart & muscle (MCK-Cre) and p85β null | Viable | Reduced heart size but not muscle size | [68] |
| PTEN null in heart & muscle (MCK-Cre) | Viable | Cardiac hypertrophy but normal skeletal muscle growth; unaffected overload-induced muscle hypertrophy; improved muscle regeneration | [70,71,123] |
| PDK1 null | Embryonic lethality | [124] | |
| PDK1 knock-in mutant unable to bind phosphoinositides | Viable | Reduced growth (weight 35% of control) | [125] |
| PDK1 null in heart & muscle (MCK-Cre) | Lethal at 5-11 weeks | Dilated cardiomyopathy but no change in muscle | [73] |
| Akt1 null | Viable but shorter life span | Mild growth retardation (weight 80% of control) | [74,75] |
| Akt2 null | Viable | Normal growth | [76] |
| Akt1+Akt2 null | Neonatal lethality | Severe growth retardation (birth weight 50% of control), marked muscle atrophy | [77] |
| TSC1 null | Embryonic lethality | [126] | |
| TSC2 null | Embryonic lethality | [127] | |
| mTOR null | Embryonic lethality | [128,129] | |
| mTOR null in muscle (HSA-Cre) | Viable but premature death | Reduced postnatal growth due to reduced fast muscle growth, severe myopathy | [88] |
| Raptor null | Embryonic lethality | [6] | |
| Raptor null in muscle (HSA-Cre) | Viable | Normal growth | [87] |
| Rictor null in muscle (HSA-Cre) | Viable | Reduced postnatal growth with severe myopathy and premature death | [87] |
| S6K1 null | Viable | Reduced growth (birth weight 80% of control), reduced muscle growth (fiber size 80% of control in adult mice) | [89,117] |
| S6K2 null | Viable | Normal growth | [130] |
| S6K1+S6K2 null | Perinatal lethality | Reduced growth | [130] |
| 4EBP1+4EBP2 null | Viable | Normal growth | [131] |
| FoxO1 null | Embryonic lethality | [132,133] | |
| FoxO1 null in muscle (HSA-Cre) | Viable | Normal growth, slow to fast switch in muscle | [134,135] |
| FoxO3 null | Viable but female sterility | Normal growth | [133,36] |
| FoxO4 null | Viable | Normal growth | [137] |
| FoxO3+FoxO4 null | Viable | Normal growth | [137] |
| MAFbx null | Viable | Reduced muscle atrophy after denervation | [92] |
| Murf1 null | Viable | Reduced muscle atrophy after denervation | [92] |
1 Promoters used to drive Cre recombinase expression: HSA = human skeletal actin; MCK = muscle creatine kinase; Mef2c = a promoter that lies 71 kb upstream of the first translated exon of the Mef2c gene and is sufficient to direct expression exclusively to skeletal muscle from embryonic day 8.5.
Table 4.
In vivo transfection experiments leading to perturbation of the IGF1-Akt pathway in adult skeletal muscle1
| Transgene | Perturbation | Effect | References |
|---|---|---|---|
| Igf1 (via virus) | Overexpression of Igf1 | Muscle fiber hypertrophy (and muscle regeneration) | [50,106] |
| Igf1 | Overexpression of Igf1 | Prevention of glucocorticoid-induced muscle atrophy | [48,138] |
| RasV12C40 (Ras double mutant) | Activation of the PI3K-Akt pathway | Muscle fiber hypertrophy, which is blocked by rapamycin | [58,69] |
| c.a. PI3K2 | Activation of PI3K | Muscle fiber hypertrophy | [69,78] |
| c.a. Akt | Activation of Akt | Muscle fiber hypertrophy, which is blocked by rapamycin | [69,78] |
| c.a. Akt (via virus) | Activation of Akt | Muscle fiber hypertrophy | [139] |
| c.a.FoxO3 | Activation of FoxO3 | Muscle fiber atrophy and activation of atrogin-1 reporter | [91] |
| Small interfering RNA to FoxO1 and FoxO3 | Knockdown of FoxO1 and FoxO3 by RNA interference | Prevention of atrogin-1 reporter upregulation induced by starvation | [91] |
| Rheb3 | Activation of Rheb and mTORC1 | Muscle fiber hypertrophy | [140] |
| Small interfering RNA to N-WASP | Knockdown of N-WASP4 by RNA interference | Muscle fiber atrophy | [86] |
1Experiments based on intramuscular injection of plasmid DNA or viral vector or small interfering RNA expression vector. Injection of plasmid DNA or siRNA expression vector was followed by electroporation.
2Constitutively active PI3K.
3Ras homolog enriched in brain
4Neuronal Wiscott-Aldrich syndrome protein (N-WASP).
IGF1 and IGF1 receptor
IGF1 was initially considered purely as a circulating growth factor produced by the liver and mediating the effect of growth hormone on body growth. However, subsequent studies showed that IGF1 is also expressed locally in many tissues, including skeletal muscle, suggesting that autocrine/paracrine effects of local IGF1 may be a major mechanism controlling tissue growth. Igf1 null mice exhibit severe growth retardation and most die soon after birth [37-39], whereas targeted ablation experiments showed that liver-derived IGF1, although it is the principal source of IGF1 in the serum, is not required for postnatal body growth [40,41]. However, this interpretation was challenged by the demonstration that conditional expression of IGF1 in the liver of an Igf1 null background contributes to about 30% of the adult body size [42]. This discrepancy was explained by the fact that a residual fraction of circulating IGF1 is detected in the liver-specific knockout [40,41] because of incomplete Igf1 gene excision [43]. In a similar study, elevated serum concentrations of IGF1, induced by overexpressing a rat Igf1 transgene specifically in the liver of Igf1 null mice, were able to rescue the severe growth-retarded phenotype observed in these mice [44]. To define the effects of IGF1 produced by muscle cells, a transgenic construct was generated in which expression of a human IGF1 cDNA was driven by the avian skeletal α-actin gene [45]. IGF1 concentration in the serum was similar in wild-type and transgenic mice, but transgenic mice developed skeletal muscle hypertrophy [45-47]. In these mice, IGF1 overexpression was not sufficient to prevent the decrease in muscle mass induced by hind-limb unloading [46]. However, glucocorticoid-induced muscle atrophy was prevented by IGF1 overexpression via electroporation in adult rats [48].
The Igf1 gene can produce multiple transcripts by alternative RNA processing, thus generating divergent peptides at the carboxyl terminus, called the E peptides. Two isoforms, IGF1A and IGF1B, are found in most mammals, and an additional form, IGF1C, is present in primates and humans [49]. The E peptides appear to be essential for muscle growth regulation because viral delivery of IGF1A or IGF1B promoted functional hypertrophy in mouse muscles, whereas delivery of mature IGF1 devoid of E peptide failed to cause an increase in muscle mass [50]. The effect of an IGF1 isoform (IGF1A) was investigated in a transgenic mouse model, in which expression of this isoform was driven by the myosin light chain 1 fast promoter [51]. The transgenic mice showed postnatal increase in muscle mass and strength, and were protected from age-related muscle atrophy and weakness. Moreover, aging transgenic muscles retained a regenerative capacity comparable with that of young animals. Surprisingly, Akt activation was not detected in these mice [52], suggesting that IGF1 may signal via alternative pathways. A potential alternative pathway could be the serum- and glucocorticoid-responsive kinase 1 (SGK1), a PI3K-dependent kinase with structural homology to Akt, which is strongly expressed in many tissues, including skeletal muscle and heart [53]. SGK1 is activated by IGF1, PI3K and PDK1, and can induce phosphorylation of S6K and GSK3β in cardiomyocytes [54] and phosphorylation of FoxO3 in various cell types [55]. However, this alternative pathway has not been explored in skeletal muscle. IGF1-dependent signaling via the mitogen-activated protein kinase/extracellular signal-regulated receptor kinase (MAPK/ERK) pathway has also been implicated in IGF1-dependent muscle growth regulation [56,57], however a constitutively active Ras double mutant that selectively activates the ERK pathway did not induce hypertrophy of regenerating muscle fibers [58].
Until recently, the only available loss-of-function model for IGF1 and IGF1 receptor was a dominant-negative, kinase-inactive form of the β-subunit of IGF1 receptor driven by a muscle-specific promoter (MKR mice) [59]. Formation of a hybrid of the mutated IGF1 receptor with the endogenous IGF1 and insulin receptors caused impaired insulin and IGF1 receptor signaling pathways, specifically in skeletal muscle. The main phenotype of these mice was the development of peripheral insulin resistance and type 2 diabetes [59]. Muscle growth was transiently delayed from birth to 3 weeks of age, but muscle mass and force was normal in the adult [60]. Compensatory hypertrophy of the plantaris muscle after ablation of the synergistic gastrocnemius muscle, and associated Akt and S6K activation, were unaffected in these mice [61], but muscle regeneration was impaired [62]. More recently, specific loss-of-function mouse models have been generated. Muscle-specific knockout of the Igf1 receptor gene caused impaired skeletal muscle development with reduction in myofiber number and area and reduced numbers of type 1 fibers in gastrocnemius muscle [63]. By contrast, mice lacking Igf1 had no obvious phenotype, and their body weights were indistinguishable from control littermates at all postnatal times (reported as 'data not shown' in [63]).
IRS, PI3K and PDK1
IRS1 null mice have much reduced growth (30-60% of control) [39,64], whereas IRS2 null mice have almost normal growth (about 90% of control), but develop type 2 diabetes [65], therefore IRS1 is thought to be downstream of the IGF1 receptor, and IRS2 downstream of the insulin receptor (Table 3). However, muscle-specific inactivation of the IRS genes has not yet been reported. PI3K p85α null mice have perinatal lethality, apparently due to liver necrosis [66], whereas PI3K p85β null mice are viable and have normal growth [67]. Conditional deletion of the p85α gene in skeletal muscle and heart showed normal growth [68], but when these mice were crossed with p85β null mice, the resulting double-mutant animals showed reduced heart but not reduced skeletal muscle size [68]. By contrast, constitutively active PI3K was found to induce muscle hypertrophy when transfected into regenerating skeletal muscle [69]. Conditional deletion of the PTEN gene in skeletal muscle and heart leads to hypertrophy of cardiac but not skeletal muscle. Overload-induced skeletal muscle hypertrophy is also unaffected by PTEN knockout [70], but maturation of regenerating muscle fibers is accelerated [71]. A skeletal muscle- and cardiac-enriched microRNA, miR-486, has been shown to target and inhibit the PI3K inhibitor PTEN, thus activating the Akt pathway [72]. Expression of miR-486 is regulated by myocardin-related transcription factor A (MRTF-A) and serum response factor (SRF). However, a role of miR-486 on muscle growth regulation has not yet been documented by in vivo genetic approaches. Conditional deletion of PDK1 in heart and muscle leads to dilated cardiomyopathy but no apparent change in muscle [73]. Thus, genetic evidence, based on loss-of-function approaches, supporting a role for PI3K and PDK1 in skeletal muscle growth is still missing.
Akt/PKB
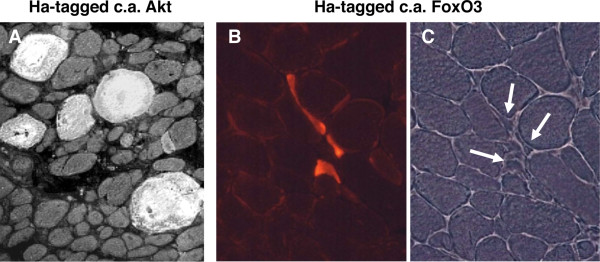
Disruption of the Akt1 gene causes growth retardation and apoptosis [74,75], whereas deletion of Akt2 causes defects in glucose metabolism but not altered growth [76]. When both Akt1 and Akt2 genes were deleted, skeletal muscle atrophy at embryonic day 18.5 was observed, together with dwarfism, impaired skin and bone development, and reduced adipogenesis [77]. The striking effect of Akt1 on muscle size was demonstrated by in vivo transient transfection of a constitutively active Akt1 [69,78] (Figure 4A), and by transgenic mice overexpressing a constitutively active inducible Akt1 transgene in skeletal muscles [79-82]. Muscle hypertrophy was rapidly achieved in all cases when Akt1 expression was induced in the adult animal for a period ranging from 1 to 3 weeks. Downstream mediators of protein synthesis (S6K, S6) were activated, but no incorporation of satellite cells was observed [82]. Akt1 hypertrophic muscles showed increased strength, demonstrating that a functional hypertrophy was induced [80,82]. Moreover, muscle mass was completely preserved in denervated transgenic Akt mice [18]. Akt activation causes multiple changes in muscle gene expression, documented by microarray analyses [80,82,83]. The effects of Akt on muscle mass regulation can be mediated by several different downstream effectors, including GSK3β, mTOR and FoxO.
Figure 4.
Myofiber hypertrophy or atrophy induced by transfection of skeletal muscle with mutants of Akt or FoxO. (A) Regenerating rat soleus muscle transfected with plasmid coding for constitutively active Akt1 linked to a hemagglutinin (HA) epitope. Muscle examined 7 days after transfection; section stained for the HA tag. Note the striking hypertrophy of labeled myofibers compared with untransfected neighbouring fibers. Modified from [69]. (B,C) Adult mouse tibialis anterior muscle transfected by electroporation with plasmid coding for constitutively active Ha-tagged FoxO3. Muscle examined 14 days after transfection; section stained for the HA tag. Note the striking atrophy of labeled myofibers compared with untransfected neighboring fibers. A phase-contrast image of the same field is shown in the right panel. Modified from [91].
GSK3β
A dominant-negative form of GSK3β has been shown to induce hypertrophy in skeletal myotubes [84], and overexpression of wild-type GSK3β in the heart induces a 30% decrease in heart size [85]. However, in vivo studies with GSK3β mutants in skeletal muscle have not been reported. A novel mechanism mediating the effect of GSK3β on muscle growth has been described recently [86]. GSK3β is able to phosphorylate nebulin at two Ser sites in the C-terminal region of nebulin localized to the Z-disk, thus preventing the interaction of nebulin with neuronal Wiscott-Aldrich syndrome protein (N-WASP), a ubiquitously expressed member of the WASP family, which is involved in actin assembly (Figure 2). IGF1-Akt signaling, by inhibiting GSK3β, allows the interaction of N-WASP with the unphosphorylated nebulin; the consequent recruitment of N-WASP to the Z-disk promotes actin nucleation and elongation of actin filaments. This process appears to be relevant to muscle growth in vivo, because long-term knockdown of N-WASP in adult mouse muscles causes atrophy of the transfected muscle fibers [86]. It was therefore suggested that IGF1-Akt signaling controls myofibril growth and maintenance via the GSK3β-nebulin-N-WASP pathway.
mTOR
mTOR is part of two multiprotein complexes, TORC1, which contains contains Raptor and TORC2, which contains Rictor. To define the role of mTOR complexes in skeletal muscle, mice with muscle-specific ablation of Raptor and Rictor were generated [87]. Muscles from Rictor knockout mice were indistinguishable from wild-type controls. By contrast, Raptor-deficient muscles became progressively dystrophic, and were impaired in their oxidative capacity [87]. These changes were accompanied by decreased muscle force and downregulation of genes involved in mitochondrial biogenesis, including PGC-1α. A similar, although not identical, phenotype was observed in muscle-specific mTOR-deficient mice [88]. Like Raptor knockouts, mTOR-deficient muscles develop a myopathy reminiscent of muscular dystrophy, together with impaired oxidative metabolism. The myopathy was more severe in mTOR knockout than in Raptor knockout mice, possibly due to reduction in the content of dystrophin and other components of the dystrophin-glycoprotein complex. It was suggested that mTOR directly controls dystrophin transcription in a rapamycin- and kinase-independent manner [88].
Raptor and Rictor have different susceptibilities to rapamycin treatment, with Raptor activity being blocked by rapamycin, whereas Rictor activity is not. Nevertheless, Raptor inhibition and rapamycin treatment do not cause the same effect on muscles. Rapamycin blocks muscle growth in regenerating or Akt transfected muscles [69,78] but does not cause atrophy or dystrophy in adult mice treated for 15 days [18]. mTOR controls protein synthesis via different targets, including S6K, which exists in two isoforms, S6K1 and S6K2. S6K1 and S6K2 double-knockout mice have reduced muscle fiber size with unchanged number of myonuclei [89]. Deletion of S6K1 is sufficient to reproduce this atrophic phenotype.
FoxO
The transcription factors of the FoxO family have emerged as major regulators of the muscle atrophy program. Mice overexpressing FoxO1 under a muscle-specific promoter have muscle atrophy and increased levels of the lysosomal protease cathepsin L [90]. FoxO3 is induced during muscle atrophy, and its overexpression is able to reduce muscle mass in vivo: the striking effect of a constitutively active mutant of FoxO3 when transfected into skeletal muscle is illustrated in Figure 4(B,C). By contrast, expression of dominant-negative FoxO3 inhibits dexametasone-induced muscle atrophy [91]. FoxO3 acts on the two major pathways of muscle protein degradation, the proteasomal and the autophagic-lysosomal pathways.
Direct evidence for FoxO-dependent activation of the ubiquitin-proteasome pathway was obtained with the demonstration that the muscle-specific ubiquitin ligases atrogin-1/MAFbx and MuRF1, which are induced in various models of muscle atrophy [92,93], are transcriptional targets of FoxO factors, and are upregulated by FoxO3 transfection in adult muscle [91,94]. Their role in muscle-mass regulation is supported by the finding that muscle atrophy induced by denervation is partially prevented in both MuRF1 null and atrogin1/MAFbx null mice [92]. MuRF1 is involved in the degradation of myosin heavy chains and other thick filament proteins, such as myosin light chains and myosin-binding protein C [95,96]. MuRF1 belongs to the muscle-specific RING finger protein family, which also includes MuRF2 and MuRF3. A redundant function of the different MuRFs is suggested by the finding that mice lacking a single MuRF gene do not have a striking skeletal muscle phenotype [92,97,98]. By contrast, MuRF1 and 3 double-knockout mice develop skeletal and cardiac muscle myopathy with myosin accumulation [99], and MuRF1 and 2 double-knockout mice have mild skeletal muscle hypertrophy and have reduced muscle loss during aging [100].
The role of FoxO3 as an inducer of autophagy is supported by the finding that transfection of adult muscle fibers with constitutively active FoxO3 causes accumulation of autophagic vacuoles, whereas fasting-induced autophagy is blocked by dominant-negative FoxO3 and by RNA interference-mediated FoxO3 knockdown [81,101]. FoxO3 is required for the upregulation of autophagy-related genes, such a LC3 and Bnip3 [81]. Inhibition of autophagy by muscle-specific knockout of the autophagy gene Atg7 was found to cause muscle atrophy, accompanied by a decrease in muscle force with accumulation of altered mitochondria and aberrant concentric membranous structures [102]. Moreover, inhibition of autophagy exacerbated muscle loss during denervation and fasting, suggesting that the persistence of dysfunctional organelles affects the progression of muscle atrophy [102]. The normal operation of the autophagic machinery is thus required for the disposal of altered cell organelles and the maintenance of muscle fiber integrity. An unexpected result was the finding that mTOR, which is considered as a major regulator of autophagy in different cell systems, does not appear to play a major role in muscle autophagy, as treatment with the mTOR inhibitor rapamycin has a limited effect on autophagy in skeletal muscle both in vitro and in vivo [81,101].
Open questions
Several open questions on the role of the IGF1-Akt pathway in skeletal muscle remain to be answered, including the role of different isoforms, such as the IGF1 and the FoxO isoforms. Two general points that need to be addressed in future studies will be briefly considered here.
The role of the IGF1-Akt pathway in adult skeletal muscle
There is no doubt that IGF1 is a major regulator of muscle mass during development, thanks to its effect on myogenic cell proliferation and differentiation [103]. As to the role of IGF1 in adult skeletal muscle, several studies indicate that IGF1 can induce hypertrophy and block atrophy. In adult rats, local infusion of recombinant IGF1 results in muscle hypertrophy [104,105] and plasmid-mediated IGF1 gene transfer prevents corticosteroid-induced muscle atrophy [48]. In adult mice, virus-mediated IGF1 gene transfer results in muscle hypertrophy and prevents aging-dependent loss in muscle mass and force [50,106]. However, it is not clear whether IGF1 is involved in mediating the effect of load on adult muscle mass. Overexpression of IGF1 in the skeletal muscle of transgenic mice did not prevent unloading-induced muscle atrophy [46], although IGF1 transgene expression was decreased by unloading in these experiments. Overload hypertrophy was also unchanged in transgenic mice overexpressing a dominant-negative form of the IGF1 receptor [61], although interpretation of this model is complicated by the double effect of the transgene on both insulin and IGF1 signaling. The pathways mediating the effect of unloading and overloading on muscle size have not been identified, and as discussed above, it is possible that the Akt-mTOR pathway is involved either via integrin-ILK or via a direct effect on mTOR.
The role of IGF1 and its downstream effectors in adult skeletal muscle cannot be determined using traditional transgenic and knockout approaches, because developing muscles have greater plasticity than adult muscles, thus compensatory adaptations might occur in response to gene overexpression or ablation that are not seen when the genetic perturbation is induced in adult animals. This might account for the lack of a muscle phenotype in certain knockout models, for example, in the PI3K and PDK1 knockouts. Other knockout models cause maladaptative changes leading to muscle dystrophy rather than atrophy, such as that seen in the mTOR and Raptor knockout models. An understanding of the effect of genetic perturbation in adult rather than developing muscle is especially important for the identification of therapeutic targets and the design of countermeasures to prevent muscle wasting. To obtain useful information in this respect, it is essential to use inducible transgenic approaches, in which transgene overexpression or gene knockout is induced in adult animals. However, to our knowledge, the Akt inducible model is the only inducible model developed to date for the study of the IGF1-Akt pathway. Alternatively, in vivo transfection procedures can be used to explore the effect of transgene overexpression or gene knockdown by RNAi in adult muscles.
The effect of IGF1-Akt pathway activation on satellite cells
Muscle growth and muscle regeneration require the participation of satellite cells, and other cell types, including cells of blood vessels and, in the case of regeneration, inflammatory cells. This aspect must be considered in genetic models involving overproduction of IGF1, as this growth factor is known to act on different cell types, because of the presence of the IGF1 receptor in satellite cells and non-muscle cells. The effect on satellite cells is especially important for muscle hypertrophy. Satellite cells were reported to play a crucial role in the hypertrophic response induced by viral-mediated gene transfer of IGF1 in adult mouse muscles, as shown by the fact that gamma-irradiation, used to block satellite-cell proliferation, was found to reduce the hypertrophic effect of IGF1 overexpression [107]. However, interpretation of this experiment is complicated by the possibility that the reduced hypertrophic response may be due to effects of irradiation on myofiber protein synthesis [108], and perhaps on protein degradation as well. In another study on IGF1 transgenic mice, the finding that IGF1 overexpression causes first an increase in DNA, and only several weeks after birth an increase in protein mass, was interpreted as reflecting a primary effect on satellite-cell proliferation and fusion during the early postnatal stages, when satellite cells undergo active proliferation [47]. This interpretation would be consistent with the view that a growth stimulus is required for IGF1 to induce muscle hypertrophy in vivo [109]. However, satellite-cell proliferation was not directly examined in these studies. Satellite-cell proliferation and fusion leading to increase in myonuclei occurs during postnatal muscle growth [110] and during compensatory hypertrophy induced by ablation of synergistic muscles [111], as demonstrated by electron microscope autoradiographs after 3H-thymidine labeling. However, using an inducible transgenic model of muscle hypertrophy, no significant BrdU incorporation could be detected in satellite cells after short-term Akt activation in adult skeletal muscle, yet muscle hypertrophy was accompanied by increased force generation [82]. Thus, satellite-cell proliferation and fusion is not a prerequisite for a functional hypertrophy induced by Akt activation in adult skeletal muscle.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
SS conceived and designed the study. SS and CM drafted the manuscript.
Contributor Information
Stefano Schiaffino, Email: stefano.schiaffino@unipd.it.
Cristina Mammucari, Email: cristina.mammucari@unipd.it.
Acknowledgements
This work was supported by grants from the European Commission (Integrated Project MYOAGE to S.S.), the Italian Space Agency (ASI, project OSMA to S.S.) and the University of Padova ('Progetto di Ateneo' 2009 to C.M.). We thank Marco Sandri for critical reading of the manuscript.
References
- Glass DJ. PI3 Kinase Regulation of skeletal muscle hypertrophy and atrophy. Curr Top Microbiol Immunol. 2010;346:267–278. doi: 10.1007/82_2010_78. full_text. [DOI] [PubMed] [Google Scholar]
- Miyazaki M, Esser KA. Cellular mechanisms regulating protein synthesis and skeletal muscle hypertrophy in animals. J Appl Physiol. 2009;106:1367–1373. doi: 10.1152/japplphysiol.91355.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandri M. Signaling in muscle atrophy and hypertrophy. Physiology (Bethesda) 2008;23:160–170. doi: 10.1152/physiol.00041.2007. [DOI] [PubMed] [Google Scholar]
- Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129:1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guertin DA, Stevens DM, Thoreen CC, Burds AA, Kalaany NY, Moffat J, Brown M, Fitzgerald KJ, Sabatini DM. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCalpha, but not S6K1. Dev Cell. 2006;11:859–871. doi: 10.1016/j.devcel.2006.10.007. [DOI] [PubMed] [Google Scholar]
- Harrington LS, Findlay GM, Gray A, Tolkacheva T, Wigfield S, Rebholz H, Barnett J, Leslie NR, Cheng S, Shepherd PR. et al. The TSC1-2 tumor suppressor controls insulin-PI3K signaling via regulation of IRS proteins. J Cell Biol. 2004;166:213–223. doi: 10.1083/jcb.200403069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- Li HH, Willis MS, Lockyer P, Miller N, McDonough H, Glass DJ, Patterson C. Atrogin-1 inhibits Akt-dependent cardiac hypertrophy in mice via ubiquitin-dependent coactivation of Forkhead proteins. J Clin Invest. 2007;117:3211–3223. doi: 10.1172/JCI31757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagirand-Cantaloube J, Offner N, Csibi A, Leibovitch MP, Batonnet-Pichon S, Tintignac LA, Segura CT, Leibovitch SA. The initiation factor eIF3-f is a major target for atrogin1/MAFbx function in skeletal muscle atrophy. EMBO J. 2008;27:1266–1276. doi: 10.1038/emboj.2008.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csibi A, Cornille K, Leibovitch MP, Poupon A, Tintignac LA, Sanchez AM, Leibovitch SA. The translation regulatory subunit eIF3f controls the kinase-dependent mTOR signaling required for muscle differentiation and hypertrophy in mouse. PLoS One. 2010;5:e8994. doi: 10.1371/journal.pone.0008994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CC, Jeon SM, Bhaskar PT, Nogueira V, Sundararajan D, Tonic I, Park Y, Hay N. FoxOs inhibit mTORC1 and activate Akt by inducing the expression of Sestrin3 and Rictor. Dev Cell. 2010;18:592–604. doi: 10.1016/j.devcel.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HV, Chang LW, Brixius K, Wickstrom SA, Montanez E, Thievessen I, Schwander M, Muller U, Bloch W, Mayer U, Fassler R. Integrin-linked kinase stabilizes myotendinous junctions and protects muscle from stress-induced damage. J Cell Biol. 2008;180:1037–1049. doi: 10.1083/jcb.200707175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durieux AC, D'Antona G, Desplanches D, Freyssenet D, Klossner S, Bottinelli R, Fluck M. Focal adhesion kinase is a load-dependent governor of the slow contractile and oxidative muscle phenotype. J Physiol. 2009;587:3703–3717. doi: 10.1113/jphysiol.2009.171355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draznin B. Molecular mechanisms of insulin resistance: serine phosphorylation of insulin receptor substrate-1 and increased expression of p85alpha: the two sides of a coin. Diabetes. 2006;55:2392–2397. doi: 10.2337/db06-0391. [DOI] [PubMed] [Google Scholar]
- McPherron AC, Lawler AM, Lee SJ. Regulation of skeletal muscle mass in mice by a new TGF-beta superfamily member. Nature. 1997;387:83–90. doi: 10.1038/387083a0. [DOI] [PubMed] [Google Scholar]
- Trendelenburg AU, Meyer A, Rohner D, Boyle J, Hatakeyama S, Glass DJ. Myostatin reduces Akt/TORC1/p70S6K signaling, inhibiting myoblast differentiation and myotube size. Am J Physiol Cell Physiol. 2009;296:C1258–1270. doi: 10.1152/ajpcell.00105.2009. [DOI] [PubMed] [Google Scholar]
- Sartori R, Milan G, Patron M, Mammucari C, Blaauw B, Abraham R, Sandri M. Smad2 and 3 transcription factors control muscle mass in adulthood. Am J Physiol Cell Physiol. 2009;296:C1248–1257. doi: 10.1152/ajpcell.00104.2009. [DOI] [PubMed] [Google Scholar]
- Lee SJ, Lee YS, Zimmers TA, Soleimani A, Matzuk MM, Tsuchida K, Cohn RD, Barton ER. Regulation of muscle mass by follistatin and activins. Mol Endocrinol. 2010;24:1998–2008. doi: 10.1210/me.2010-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Wang JL, Lu J, Song Y, Kwak KS, Jiao Q, Rosenfeld R, Chen Q, Boone T, Simonet WS. et al. Reversal of cancer cachexia and muscle wasting by ActRIIB antagonism leads to prolonged survival. Cell. 2010;142:531–543. doi: 10.1016/j.cell.2010.07.011. [DOI] [PubMed] [Google Scholar]
- Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL. Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol. 2008;10:935–945. doi: 10.1038/ncb1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008;30:214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoki K, Li Y, Zhu T, Wu J, Guan KL. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Biol. 2002;4:648–657. doi: 10.1038/ncb839. [DOI] [PubMed] [Google Scholar]
- Mounier R, Lantier L, Leclerc J, Sotiropoulos A, Pende M, Daegelen D, Sakamoto K, Foretz M, Viollet B. Important role for AMPKalpha1 in limiting skeletal muscle cell hypertrophy. Faseb J. 2009;23:2264–2273. doi: 10.1096/fj.08-119057. [DOI] [PubMed] [Google Scholar]
- Lantier L, Mounier R, Leclerc J, Pende M, Foretz M, Viollet B. Coordinated maintenance of muscle cell size control by AMP-activated protein kinase. FASEB J. 2010;24:3555–3561. doi: 10.1096/fj.10-155994. [DOI] [PubMed] [Google Scholar]
- Hornberger TA, Stuppard R, Conley KE, Fedele MJ, Fiorotto ML, Chin ER, Esser KA. Mechanical stimuli regulate rapamycin-sensitive signalling by a phosphoinositide 3-kinase-, protein kinase B- and growth factor-independent mechanism. Biochem J. 2004;380:795–804. doi: 10.1042/BJ20040274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornberger TA, Chu WK, Mak YW, Hsiung JW, Huang SA, Chien S. The role of phospholipase D and phosphatidic acid in the mechanical activation of mTOR signaling in skeletal muscle. Proc Natl Acad Sci USA. 2006;103:4741–4746. doi: 10.1073/pnas.0600678103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neil TK, Duffy LR, Frey JW, Hornberger TA. The role of phosphoinositide 3-kinase and phosphatidic acid in the regulation of mammalian target of rapamycin following eccentric contractions. J Physiol. 2009;587:3691–3701. doi: 10.1113/jphysiol.2009.173609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Oskoui PR, Banko MR, Maniar JM, Gygi MP, Gygi SP, Brunet A. The energy sensor AMP-activated protein kinase directly regulates the mammalian FOXO3 transcription factor. J Biol Chem. 2007;282:30107–30119. doi: 10.1074/jbc.M705325200. [DOI] [PubMed] [Google Scholar]
- Sandri M, Lin J, Handschin C, Yang W, Arany ZP, Lecker SH, Goldberg AL, Spiegelman BM. PGC-1alpha protects skeletal muscle from atrophy by suppressing FoxO3 action and atrophy-specific gene transcription. Proc Natl Acad Sci USA. 2006;103:16260–16265. doi: 10.1073/pnas.0607795103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai D, Frantz JD, Tawa NE Jr, Melendez PA, Oh BC, Lidov HG, Hasselgren PO, Frontera WR, Lee J, Glass DJ, Shoelson SE. IKKbeta/NF-kappaB activation causes severe muscle wasting in mice. Cell. 2004;119:285–298. doi: 10.1016/j.cell.2004.09.027. [DOI] [PubMed] [Google Scholar]
- Dogra C, Changotra H, Wedhas N, Qin X, Wergedal JE, Kumar A. TNF-related weak inducer of apoptosis (TWEAK) is a potent skeletal muscle-wasting cytokine. Faseb J. 2007;21:1857–1869. doi: 10.1096/fj.06-7537com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schakman O, Kalista S, Bertrand L, Lause P, Verniers J, Ketelslegers JM, Thissen JP. Role of Akt/GSK-3beta/beta-catenin transduction pathway in the muscle anti-atrophy action of insulin-like growth factor-I in glucocorticoid-treated rats. Endocrinology. 2008;149:3900–3908. doi: 10.1210/en.2008-0439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddell DS, Baehr LM, van den Brandt J, Johnsen SA, Reichardt HM, Furlow JD, Bodine SC. The glucocorticoid receptor and FOXO1 synergistically activate the skeletal muscle atrophy-associated MuRF1 gene. Am J Physiol Endocrinol Metab. 2008;295:E785–797. doi: 10.1152/ajpendo.00646.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilot-Storck F, Chopin E, Rual JF, Baudot A, Dobrokhotov P, Robinson-Rechavi M, Brun C, Cusick ME, Hill DE, Schaeffer L. et al. Interactome mapping of the phosphatidylinositol 3-kinase-mammalian target of rapamycin pathway identifies deformed epidermal autoregulatory factor-1 as a new glycogen synthase kinase-3 interactor. Mol Cell Proteomics. 2010;9:1578–1593. doi: 10.1074/mcp.M900568-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JP, Baker J, Perkins AS, Robertson EJ, Efstratiadis A. Mice carrying null mutations of the genes encoding insulin-like growth factor I (Igf-1) and type 1 IGF receptor (Igf1r) Cell. 1993;75:59–72. [PubMed] [Google Scholar]
- Powell-Braxton L, Hollingshead P, Warburton C, Dowd M, Pitts-Meek S, Dalton D, Gillett N, Stewart TA. IGF-I is required for normal embryonic growth in mice. Genes Dev. 1993;7:2609–2617. doi: 10.1101/gad.7.12b.2609. [DOI] [PubMed] [Google Scholar]
- Baker J, Liu JP, Robertson EJ, Efstratiadis A. Role of insulin-like growth factors in embryonic and postnatal growth. Cell. 1993;75:73–82. [PubMed] [Google Scholar]
- Yakar S, Liu JL, Stannard B, Butler A, Accili D, Sauer B, LeRoith D. Normal growth and development in the absence of hepatic insulin-like growth factor I. Proc Natl Acad Sci USA. 1999;96:7324–7329. doi: 10.1073/pnas.96.13.7324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjogren K, Liu JL, Blad K, Skrtic S, Vidal O, Wallenius V, LeRoith D, Tornell J, Isaksson OG, Jansson JO, Ohlsson C. Liver-derived insulin-like growth factor I (IGF-I) is the principal source of IGF-I in blood but is not required for postnatal body growth in mice. Proc Natl Acad Sci USA. 1999;96:7088–7092. doi: 10.1073/pnas.96.12.7088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratikopoulos E, Szabolcs M, Dragatsis I, Klinakis A, Efstratiadis A. The hormonal action of IGF1 in postnatal mouse growth. Proc Natl Acad Sci USA. 2008;105:19378–19383. doi: 10.1073/pnas.0809223105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z, Yu R, Lu Y, Parlow AF, Liu JL. Age-dependent onset of liver-specific IGF-I gene deficiency and its persistence in old age: implications for postnatal growth and insulin resistance in LID mice. Am J Physiol Endocrinol Metab. 2005;289:E288–295. doi: 10.1152/ajpendo.00494.2004. [DOI] [PubMed] [Google Scholar]
- Wu Y, Sun H, Yakar S, LeRoith D. Elevated levels of insulin-like growth factor (IGF)-I in serum rescue the severe growth retardation of IGF-I null mice. Endocrinology. 2009;150:4395–4403. doi: 10.1210/en.2009-0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman ME, DeMayo F, Yin KC, Lee HM, Geske R, Montgomery C, Schwartz RJ. Myogenic vector expression of insulin-like growth factor I stimulates muscle cell differentiation and myofiber hypertrophy in transgenic mice. J Biol Chem. 1995;270:12109–12116. doi: 10.1074/jbc.270.20.12109. [DOI] [PubMed] [Google Scholar]
- Criswell DS, Booth FW, DeMayo F, Schwartz RJ, Gordon SE, Fiorotto ML. Overexpression of IGF-I in skeletal muscle of transgenic mice does not prevent unloading-induced atrophy. Am J Physiol. 1998;275:E373–379. doi: 10.1152/ajpendo.1998.275.3.e373. [DOI] [PubMed] [Google Scholar]
- Fiorotto ML, Schwartz RJ, Delaughter MC. Persistent IGF-I overexpression in skeletal muscle transiently enhances DNA accretion and growth. FASEB J. 2003;17:59–60. doi: 10.1096/fj.02-0289fje. [DOI] [PubMed] [Google Scholar]
- Schakman O, Gilson H, de Coninck V, Lause P, Verniers J, Havaux X, Ketelslegers JM, Thissen JP. Insulin-like growth factor-I gene transfer by electroporation prevents skeletal muscle atrophy in glucocorticoid-treated rats. Endocrinology. 2005;146:1789–1797. doi: 10.1210/en.2004-1594. [DOI] [PubMed] [Google Scholar]
- Barton ER. The ABCs of IGF-I isoforms: impact on muscle hypertrophy and implications for repair. Appl Physiol Nutr Metab. 2006;31:791–797. doi: 10.1139/H06-054. [DOI] [PubMed] [Google Scholar]
- Barton ER, DeMeo J, Lei H. The insulin-like growth factor (IGF)-I E-peptides are required for isoform-specific gene expression and muscle hypertrophy after local IGF-I production. J Appl Physiol. 2010;108:1069–1076. doi: 10.1152/japplphysiol.01308.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musaro A, McCullagh K, Paul A, Houghton L, Dobrowolny G, Molinaro M, Barton ER, Sweeney HL, Rosenthal N. Localized Igf-1 transgene expression sustains hypertrophy and regeneration in senescent skeletal muscle. Nat Genet. 2001;27:195–200. doi: 10.1038/84839. [DOI] [PubMed] [Google Scholar]
- Barton ER, Morris L, Musaro A, Rosenthal N, Sweeney HL. Muscle-specific expression of insulin-like growth factor I counters muscle decline in mdx mice. J Cell Biol. 2002;157:137–148. doi: 10.1083/jcb.200108071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Deak M, Morrice N, Cohen P. Characterization of the structure and regulation of two novel isoforms of serum- and glucocorticoid-induced protein kinase. Biochem J. 1999;344(Pt 1):189–197. doi: 10.1042/0264-6021:3440189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyama T, Matsui T, Novikov M, Park J, Hemmings B, Rosenzweig A. Serum and glucocorticoid-responsive kinase-1 regulates cardiomyocyte survival and hypertrophic response. Circulation. 2005;111:1652–1659. doi: 10.1161/01.CIR.0000160352.58142.06. [DOI] [PubMed] [Google Scholar]
- Brunet A, Park J, Tran H, Hu LS, Hemmings BA, Greenberg ME. Protein kinase SGK mediates survival signals by phosphorylating the forkhead transcription factor FKHRL1 (FOXO3a) Mol Cell Biol. 2001;21:952–965. doi: 10.1128/MCB.21.3.952-965.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddad F, Adams GR. Inhibition of MAP/ERK kinase prevents IGF-I-induced hypertrophy in rat muscles. J Appl Physiol. 2004;96:203–210. doi: 10.1152/japplphysiol.00856.2003. [DOI] [PubMed] [Google Scholar]
- Shi H, Scheffler JM, Zeng C, Pleitner JM, Hannon KM, Grant AL, Gerrard DE. Mitogen-activated protein kinase signaling is necessary for the maintenance of skeletal muscle mass. Am J Physiol Cell Physiol. 2009;296:C1040–1048. doi: 10.1152/ajpcell.00475.2008. [DOI] [PubMed] [Google Scholar]
- Murgia M, Serrano AL, Calabria E, Pallafacchina G, Lomo T, Schiaffino S. Ras is involved in nerve-activity-dependent regulation of muscle genes. Nat Cell Biol. 2000;2:142–147. doi: 10.1038/35004013. [DOI] [PubMed] [Google Scholar]
- Fernandez AM, Kim JK, Yakar S, Dupont J, Hernandez-Sanchez C, Castle AL, Filmore J, Shulman GI, Le Roith D. Functional inactivation of the IGF-I and insulin receptors in skeletal muscle causes type 2 diabetes. Genes Dev. 2001;15:1926–1934. doi: 10.1101/gad.908001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez AM, Dupont J, Farrar RP, Lee S, Stannard B, Le Roith D. Muscle-specific inactivation of the IGF-I receptor induces compensatory hyperplasia in skeletal muscle. J Clin Invest. 2002;109:347–355. doi: 10.1172/JCI13503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spangenburg EE, Le Roith D, Ward CW, Bodine SC. A functional insulin-like growth factor receptor is not necessary for load-induced skeletal muscle hypertrophy. J Physiol. 2008;586:283–291. doi: 10.1113/jphysiol.2007.141507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heron-Milhavet L, Mamaeva D, LeRoith D, Lamb NJ, Fernandez A. Impaired muscle regeneration and myoblast differentiation in mice with a muscle-specific KO of IGF-IR. J Cell Physiol. 2010;225:1–6. doi: 10.1002/jcp.22218. [DOI] [PubMed] [Google Scholar]
- Mavalli MD, DiGirolamo DJ, Fan Y, Riddle RC, Campbell KS, van Groen T, Frank SJ, Sperling MA, Esser KA, Bamman MM, Clemens TL. Distinct growth hormone receptor signaling modes regulate skeletal muscle development and insulin sensitivity in mice. J Clin Invest. 2010;120:4007–4020. doi: 10.1172/JCI42447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki E, Lipes MA, Patti ME, Bruning JC, Haag B, Johnson RS, Kahn CR. Alternative pathway of insulin signalling in mice with targeted disruption of the IRS-1 gene. Nature. 1994;372:186–190. doi: 10.1038/372186a0. [DOI] [PubMed] [Google Scholar]
- Withers DJ, Gutierrez JS, Towery H, Burks DJ, Ren JM, Previs S, Zhang Y, Bernal D, Pons S, Shulman GI. et al. Disruption of IRS-2 causes type 2 diabetes in mice. Nature. 1998;391:900–904. doi: 10.1038/36116. [DOI] [PubMed] [Google Scholar]
- Fruman DA, Mauvais-Jarvis F, Pollard DA, Yballe CM, Brazil D, Bronson RT, Kahn CR, Cantley LC. Hypoglycaemia, liver necrosis and perinatal death in mice lacking all isoforms of phosphoinositide 3-kinase p85 alpha. Nat Genet. 2000;26:379–382. doi: 10.1038/81715. [DOI] [PubMed] [Google Scholar]
- Ueki K, Yballe CM, Brachmann SM, Vicent D, Watt JM, Kahn CR, Cantley LC. Increased insulin sensitivity in mice lacking p85beta subunit of phosphoinositide 3-kinase. Proc Natl Acad Sci USA. 2002;99:419–424. doi: 10.1073/pnas.012581799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, McMullen JR, Sobkiw CL, Zhang L, Dorfman AL, Sherwood MC, Logsdon MN, Horner JW, DePinho RA, Izumo S, Cantley LC. Class IA phosphoinositide 3-kinase regulates heart size and physiological cardiac hypertrophy. Mol Cell Biol. 2005;25:9491–9502. doi: 10.1128/MCB.25.21.9491-9502.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallafacchina G, Calabria E, Serrano AL, Kalhovde JM, Schiaffino S. A protein kinase B-dependent and rapamycin-sensitive pathway controls skeletal muscle growth but not fiber type specification. Proc Natl Acad Sci USA. 2002;99:9213–9218. doi: 10.1073/pnas.142166599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton DL, Philp A, MacKenzie MG, Baar K. A limited role for PI(3,4,5)P3 regulation in controlling skeletal muscle mass in response to resistance exercise. PLoS One. 2010;5:e11624. doi: 10.1371/journal.pone.0011624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Wang H, Lee IH, Modi S, Wang X, Du J, Mitch WE. PTEN inhibition improves muscle regeneration in mice fed a high-fat diet. Diabetes. 2010;59:1312–1320. doi: 10.2337/db09-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small EM, O'Rourke JR, Moresi V, Sutherland LB, McAnally J, Gerard RD, Richardson JA, Olson EN. Regulation of PI3-kinase/Akt signaling by muscle-enriched microRNA-486. Proc Natl Acad Sci USA. 2010;107:4218–4223. doi: 10.1073/pnas.1000300107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora A, Davies AM, Bertrand L, Sharif I, Budas GR, Jovanovic S, Mouton V, Kahn CR, Lucocq JM, Gray GA. et al. Deficiency of PDK1 in cardiac muscle results in heart failure and increased sensitivity to hypoxia. EMBO J. 2003;22:4666–4676. doi: 10.1093/emboj/cdg469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Thorvaldsen JL, Chu Q, Feng F, Birnbaum MJ. Akt1/PKBalpha is required for normal growth but dispensable for maintenance of glucose homeostasis in mice. J Biol Chem. 2001;276:38349–38352. doi: 10.1074/jbc.C100462200. [DOI] [PubMed] [Google Scholar]
- Chen WS, Xu PZ, Gottlob K, Chen ML, Sokol K, Shiyanova T, Roninson I, Weng W, Suzuki R, Tobe K. et al. Growth retardation and increased apoptosis in mice with homozygous disruption of the Akt1 gene. Genes Dev. 2001;15:2203–2208. doi: 10.1101/gad.913901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Mu J, Kim JK, Thorvaldsen JL, Chu Q, Crenshaw EB, Kaestner KH, Bartolomei MS, Shulman GI, Birnbaum MJ. Insulin resistance and a diabetes mellitus-like syndrome in mice lacking the protein kinase Akt2 (PKB beta) Science. 2001;292:1728–1731. doi: 10.1126/science.292.5522.1728. [DOI] [PubMed] [Google Scholar]
- Peng XD, Xu PZ, Chen ML, Hahn-Windgassen A, Skeen J, Jacobs J, Sundararajan D, Chen WS, Crawford SE, Coleman KG, Hay N. Dwarfism, impaired skin development, skeletal muscle atrophy, delayed bone development, and impeded adipogenesis in mice lacking Akt1 and Akt2. Genes Dev. 2003;17:1352–1365. doi: 10.1101/gad.1089403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodine SC, Stitt TN, Gonzalez M, Kline WO, Stover GL, Bauerlein R, Zlotchenko E, Scrimgeour A, Lawrence JC, Glass DJ, Yancopoulos GD. Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nat Cell Biol. 2001;3:1014–1019. doi: 10.1038/ncb1101-1014. [DOI] [PubMed] [Google Scholar]
- Lai KM, Gonzalez M, Poueymirou WT, Kline WO, Na E, Zlotchenko E, Stitt TN, Economides AN, Yancopoulos GD, Glass DJ. Conditional activation of akt in adult skeletal muscle induces rapid hypertrophy. Mol Cell Biol. 2004;24:9295–9304. doi: 10.1128/MCB.24.21.9295-9304.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumiya Y, Hopkins T, Morris C, Sato K, Zeng L, Viereck J, Hamilton JA, Ouchi N, LeBrasseur NK, Walsh K. Fast/Glycolytic muscle fiber growth reduces fat mass and improves metabolic parameters in obese mice. Cell Metab. 2008;7:159–172. doi: 10.1016/j.cmet.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammucari C, Milan G, Romanello V, Masiero E, Rudolf R, Del Piccolo P, Burden SJ, Di Lisi R, Sandri C, Zhao J. et al. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 2007;6:458–471. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- Blaauw B, Canato M, Agatea L, Toniolo L, Mammucari C, Masiero E, Abraham R, Sandri M, Schiaffino S, Reggiani C. Inducible activation of Akt increases skeletal muscle mass and force without satellite cell activation. FASEB J. 2009;23:3896–3905. doi: 10.1096/fj.09-131870. [DOI] [PubMed] [Google Scholar]
- Blaauw B, Mammucari C, Toniolo L, Agatea L, Abraham R, Sandri M, Reggiani C, Schiaffino S. Akt activation prevents the force drop induced by eccentric contractions in dystrophin-deficient skeletal muscle. Hum Mol Genet. 2008;17:3686–3696. doi: 10.1093/hmg/ddn264. [DOI] [PubMed] [Google Scholar]
- Rommel C, Bodine SC, Clarke BA, Rossman R, Nunez L, Stitt TN, Yancopoulos GD, Glass DJ. Mediation of IGF-1-induced skeletal myotube hypertrophy by PI(3)K/Akt/mTOR and PI(3)K/Akt/GSK3 pathways. Nat Cell Biol. 2001;3:1009–1013. doi: 10.1038/ncb1101-1009. [DOI] [PubMed] [Google Scholar]
- Michael A, Haq S, Chen X, Hsich E, Cui L, Walters B, Shao Z, Bhattacharya K, Kilter H, Huggins G. et al. Glycogen synthase kinase-3beta regulates growth, calcium homeostasis, and diastolic function in the heart. J Biol Chem. 2004;279:21383–21393. doi: 10.1074/jbc.M401413200. [DOI] [PubMed] [Google Scholar]
- Takano K, Watanabe-Takano H, Suetsugu S, Kurita S, Tsujita K, Kimura S, Karatsu T, Takenawa T, Endo T. Nebulin and N-WASP cooperate to cause IGF-1-induced sarcomeric actin filament formation. Science. 2010;330:1536–1540. doi: 10.1126/science.1197767. [DOI] [PubMed] [Google Scholar]
- Bentzinger CF, Romanino K, Cloetta D, Lin S, Mascarenhas JB, Oliveri F, Xia J, Casanova E, Costa CF, Brink M. et al. Skeletal muscle-specific ablation of raptor, but not of rictor, causes metabolic changes and results in muscle dystrophy. Cell Metab. 2008;8:411–424. doi: 10.1016/j.cmet.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Risson V, Mazelin L, Roceri M, Sanchez H, Moncollin V, Corneloup C, Richard-Bulteau H, Vignaud A, Baas D, Defour A. et al. Muscle inactivation of mTOR causes metabolic and dystrophin defects leading to severe myopathy. J Cell Biol. 2009;187:859–874. doi: 10.1083/jcb.200903131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohanna M, Sobering AK, Lapointe T, Lorenzo L, Praud C, Petroulakis E, Sonenberg N, Kelly PA, Sotiropoulos A, Pende M. Atrophy of S6K1(-/-) skeletal muscle cells reveals distinct mTOR effectors for cell cycle and size control. Nat Cell Biol. 2005;7:286–294. doi: 10.1038/ncb1231. [DOI] [PubMed] [Google Scholar]
- Kamei Y, Miura S, Suzuki M, Kai Y, Mizukami J, Taniguchi T, Mochida K, Hata T, Matsuda J, Aburatani H. et al. Skeletal muscle FOXO1 (FKHR) transgenic mice have less skeletal muscle mass, down-regulated Type I (slow twitch/red muscle) fiber genes, and impaired glycemic control. J Biol Chem. 2004;279:41114–41123. doi: 10.1074/jbc.M400674200. [DOI] [PubMed] [Google Scholar]
- Sandri M, Sandri C, Gilbert A, Skurk C, Calabria E, Picard A, Walsh K, Schiaffino S, Lecker SH, Goldberg AL. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell. 2004;117:399–412. doi: 10.1016/S0092-8674(04)00400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodine SC, Latres E, Baumhueter S, Lai VK, Nunez L, Clarke BA, Poueymirou WT, Panaro FJ, Na E, Dharmarajan K. et al. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science. 2001;294:1704–1708. doi: 10.1126/science.1065874. [DOI] [PubMed] [Google Scholar]
- Gomes MD, Lecker SH, Jagoe RT, Navon A, Goldberg AL. Atrogin-1, a muscle-specific F-box protein highly expressed during muscle atrophy. Proc Natl Acad Sci USA. 2001;98:14440–14445. doi: 10.1073/pnas.251541198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitt TN, Drujan D, Clarke BA, Panaro F, Timofeyva Y, Kline WO, Gonzalez M, Yancopoulos GD, Glass DJ. The IGF-1/PI3K/Akt pathway prevents expression of muscle atrophy-induced ubiquitin ligases by inhibiting FOXO transcription factors. Mol Cell. 2004;14:395–403. doi: 10.1016/S1097-2765(04)00211-4. [DOI] [PubMed] [Google Scholar]
- Clarke BA, Drujan D, Willis MS, Murphy LO, Corpina RA, Burova E, Rakhilin SV, Stitt TN, Patterson C, Latres E, Glass DJ. The E3 ligase MuRF1 degrades myosin heavy chain protein in dexamethasone-treated skeletal muscle. Cell Metab. 2007;6:376–385. doi: 10.1016/j.cmet.2007.09.009. [DOI] [PubMed] [Google Scholar]
- Cohen S, Brault JJ, Gygi SP, Glass DJ, Valenzuela DM, Gartner C, Latres E, Goldberg AL. During muscle atrophy, thick, but not thin, filament components are degraded by MuRF1-dependent ubiquitylation. J Cell Biol. 2009;185:1083–1095. doi: 10.1083/jcb.200901052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis MS, Ike C, Li L, Wang DZ, Glass DJ, Patterson C. Muscle ring finger 1, but not muscle ring finger 2, regulates cardiac hypertrophy in vivo. Circ Res. 2007;100:456–459. doi: 10.1161/01.RES.0000259559.48597.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fielitz J, van Rooij E, Spencer JA, Shelton JM, Latif S, van der Nagel R, Bezprozvannaya S, de Windt L, Richardson JA, Bassel-Duby R, Olson EN. Loss of muscle-specific RING-finger 3 predisposes the heart to cardiac rupture after myocardial infarction. Proc Natl Acad Sci USA. 2007;104:4377–4382. doi: 10.1073/pnas.0611726104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fielitz J, Kim MS, Shelton JM, Latif S, Spencer JA, Glass DJ, Richardson JA, Bassel-Duby R, Olson EN. Myosin accumulation and striated muscle myopathy result from the loss of muscle RING finger 1 and 3. J Clin Invest. 2007;117:2486–2495. doi: 10.1172/JCI32827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witt CC, Witt SH, Lerche S, Labeit D, Back W, Labeit S. Cooperative control of striated muscle mass and metabolism by MuRF1 and MuRF2. Embo J. 2008;27:350–360. doi: 10.1038/sj.emboj.7601952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Brault JJ, Schild A, Cao P, Sandri M, Schiaffino S, Lecker SH, Goldberg AL. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 2007;6:472–483. doi: 10.1016/j.cmet.2007.11.004. [DOI] [PubMed] [Google Scholar]
- Masiero E, Agatea L, Mammucari C, Blaauw B, Loro E, Komatsu M, Metzger D, Reggiani C, Schiaffino S, Sandri M. Autophagy is required to maintain muscle mass. Cell Metab. 2009;10:507–515. doi: 10.1016/j.cmet.2009.10.008. [DOI] [PubMed] [Google Scholar]
- Florini JR, Ewton DZ, Coolican SA. Growth hormone and the insulin-like growth factor system in myogenesis. Endocr Rev. 1996;17:481–517. doi: 10.1210/edrv-17-5-481. [DOI] [PubMed] [Google Scholar]
- Adams GR, McCue SA. Localized infusion of IGF-I results in skeletal muscle hypertrophy in rats. J Appl Physiol. 1998;84:1716–1722. doi: 10.1152/jappl.1998.84.5.1716. [DOI] [PubMed] [Google Scholar]
- Chakravarthy MV, Davis BS, Booth FW. IGF-I restores satellite cell proliferative potential in immobilized old skeletal muscle. J Appl Physiol. 2000;89:1365–1379. doi: 10.1152/jappl.2000.89.4.1365. [DOI] [PubMed] [Google Scholar]
- Barton-Davis ER, Shoturma DI, Musaro A, Rosenthal N, Sweeney HL. Viral mediated expression of insulin-like growth factor I blocks the aging-related loss of skeletal muscle function. Proc Natl Acad Sci USA. 1998;95:15603–15607. doi: 10.1073/pnas.95.26.15603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton-Davis ER, Shoturma DI, Sweeney HL. Contribution of satellite cells to IGF-I induced hypertrophy of skeletal muscle. Acta Physiol Scand. 1999;167:301–305. doi: 10.1046/j.1365-201x.1999.00618.x. [DOI] [PubMed] [Google Scholar]
- Adams GR, Caiozzo VJ, Haddad F, Baldwin KM. Cellular and molecular responses to increased skeletal muscle loading after irradiation. Am J Physiol Cell Physiol. 2002;283:C1182–1195. doi: 10.1152/ajpcell.00173.2002. [DOI] [PubMed] [Google Scholar]
- Shavlakadze T, Chai J, Maley K, Cozens G, Grounds G, Winn N, Rosenthal N, Grounds MD. A growth stimulus is needed for IGF-1 to induce skeletal muscle hypertrophy in vivo. J Cell Sci. 2010;123:960–971. doi: 10.1242/jcs.061119. [DOI] [PubMed] [Google Scholar]
- Moss FP, Leblond CP. Satellite cells as the source of nuclei in muscles of growing rats. Anat Rec. 1971;170:421–435. doi: 10.1002/ar.1091700405. [DOI] [PubMed] [Google Scholar]
- Schiaffino S, Bormioli SP, Aloisi M. The fate of newly formed satellite cells during compensatory muscle hypertrophy. Virchows Arch B Cell Pathol. 1976;21:113–118. doi: 10.1007/BF02899148. [DOI] [PubMed] [Google Scholar]
- Bailyes EM, Nave BT, Soos MA, Orr SR, Hayward AC, Siddle K. Insulin receptor/IGF-I receptor hybrids are widely distributed in mammalian tissues: quantification of individual receptor species by selective immunoprecipitation and immunoblotting. Biochem J. 1997;327(Pt 1):209–215. doi: 10.1042/bj3270209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey JL, Zheng B, Hu Z, Price SR, Mitch WE. Chronic kidney disease causes defects in signaling through the insulin receptor substrate/phosphatidylinositol 3-kinase/Akt pathway: implications for muscle atrophy. J Am Soc Nephrol. 2006;17:1388–1394. doi: 10.1681/ASN.2004100842. [DOI] [PubMed] [Google Scholar]
- Mora A, Komander D, van Aalten DM, Alessi DR. PDK1, the master regulator of AGC kinase signal transduction. Semin Cell Dev Biol. 2004;15:161–170. doi: 10.1016/j.semcdb.2003.12.022. [DOI] [PubMed] [Google Scholar]
- Fruman DA, Meyers RE, Cantley LC. Phosphoinositide kinases. Annu Rev Biochem. 1998;67:481–507. doi: 10.1146/annurev.biochem.67.1.481. [DOI] [PubMed] [Google Scholar]
- Yang ZZ, Tschopp O, Hemmings-Mieszczak M, Feng J, Brodbeck D, Perentes E, Hemmings BA. Protein kinase B alpha/Akt1 regulates placental development and fetal growth. J Biol Chem. 2003;278:32124–32131. doi: 10.1074/jbc.M302847200. [DOI] [PubMed] [Google Scholar]
- Shima H, Pende M, Chen Y, Fumagalli S, Thomas G, Kozma SC. Disruption of the p70(s6k)/p85(s6k) gene reveals a small mouse phenotype and a new functional S6 kinase. EMBO J. 1998;17:6649–6659. doi: 10.1093/emboj/17.22.6649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gout I, Minami T, Hara K, Tsujishita Y, Filonenko V, Waterfield MD, Yonezawa K. Molecular cloning and characterization of a novel p70 S6 kinase, p70 S6 kinase beta containing a proline-rich region. J Biol Chem. 1998;273:30061–30064. doi: 10.1074/jbc.273.46.30061. [DOI] [PubMed] [Google Scholar]
- Saitoh M, ten Dijke P, Miyazono K, Ichijo H. Cloning and characterization of p70(S6K beta) defines a novel family of p70 S6 kinases. Biochem Biophys Res Commun. 1998;253:470–476. doi: 10.1006/bbrc.1998.9784. [DOI] [PubMed] [Google Scholar]
- Tsukiyama-Kohara K, Poulin F, Kohara M, DeMaria CT, Cheng A, Wu Z, Gingras AC, Katsume A, Elchebly M, Spiegelman BM. et al. Adipose tissue reduction in mice lacking the translational inhibitor 4E-BP1. Nat Med. 2001;7:1128–1132. doi: 10.1038/nm1001-1128. [DOI] [PubMed] [Google Scholar]
- Obsil T, Obsilova V. Structure/function relationships underlying regulation of FOXO transcription factors. Oncogene. 2008;27:2263–2275. doi: 10.1038/onc.2008.20. [DOI] [PubMed] [Google Scholar]
- Tamemoto H, Kadowaki T, Tobe K, Yagi T, Sakura H, Hayakawa T, Terauchi Y, Ueki K, Kaburagi Y, Satoh S. et al. Insulin resistance and growth retardation in mice lacking insulin receptor substrate-1. Nature. 1994;372:182–186. doi: 10.1038/372182a0. [DOI] [PubMed] [Google Scholar]
- Crackower MA, Oudit GY, Kozieradzki I, Sarao R, Sun H, Sasaki T, Hirsch E, Suzuki A, Shioi T, Irie-Sasaki J. et al. Regulation of myocardial contractility and cell size by distinct PI3K-PTEN signaling pathways. Cell. 2002;110:737–749. doi: 10.1016/S0092-8674(02)00969-8. [DOI] [PubMed] [Google Scholar]
- Lawlor MA, Mora A, Ashby PR, Williams MR, Murray-Tait V, Malone L, Prescott AR, Lucocq JM, Alessi DR. Essential role of PDK1 in regulating cell size and development in mice. EMBO J. 2002;21:3728–3738. doi: 10.1093/emboj/cdf387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayascas JR, Wullschleger S, Sakamoto K, Garcia-Martinez JM, Clacher C, Komander D, van Aalten DM, Boini KM, Lang F, Lipina C. et al. Mutation of the PDK1 PH domain inhibits protein kinase B/Akt, leading to small size and insulin resistance. Mol Cell Biol. 2008;28:3258–3272. doi: 10.1128/MCB.02032-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski DJ, Zhang H, Bandura JL, Heiberger KM, Glogauer M, el-Hashemite N, Onda H. A mouse model of TSC1 reveals sex-dependent lethality from liver hemangiomas, and up-regulation of p70S6 kinase activity in Tsc1 null cells. Hum Mol Genet. 2002;11:525–534. doi: 10.1093/hmg/11.5.525. [DOI] [PubMed] [Google Scholar]
- Onda H, Lueck A, Marks PW, Warren HB, Kwiatkowski DJ. Tsc2(+/-) mice develop tumors in multiple sites that express gelsolin and are influenced by genetic background. J Clin Invest. 1999;104:687–695. doi: 10.1172/JCI7319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangloff YG, Mueller M, Dann SG, Svoboda P, Sticker M, Spetz JF, Um SH, Brown EJ, Cereghini S, Thomas G, Kozma SC. Disruption of the mouse mTOR gene leads to early postimplantation lethality and prohibits embryonic stem cell development. Mol Cell Biol. 2004;24:9508–9516. doi: 10.1128/MCB.24.21.9508-9516.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami M, Ichisaka T, Maeda M, Oshiro N, Hara K, Edenhofer F, Kiyama H, Yonezawa K, Yamanaka S. mTOR is essential for growth and proliferation in early mouse embryos and embryonic stem cells. Mol Cell Biol. 2004;24:6710–6718. doi: 10.1128/MCB.24.15.6710-6718.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pende M, Um SH, Mieulet V, Sticker M, Goss VL, Mestan J, Mueller M, Fumagalli S, Kozma SC, Thomas G. S6K1(-/-)/S6K2(-/-) mice exhibit perinatal lethality and rapamycin-sensitive 5'-terminal oligopyrimidine mRNA translation and reveal a mitogen-activated protein kinase-dependent S6 kinase pathway. Mol Cell Biol. 2004;24:3112–3124. doi: 10.1128/MCB.24.8.3112-3124.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bacquer O, Petroulakis E, Paglialunga S, Poulin F, Richard D, Cianflone K, Sonenberg N. Elevated sensitivity to diet-induced obesity and insulin resistance in mice lacking 4E-BP1 and 4E-BP2. J Clin Invest. 2007;117:387–396. doi: 10.1172/JCI29528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuyama T, Kitayama K, Shimoda Y, Ogawa M, Sone K, Yoshida-Araki K, Hisatsune H, Nishikawa S, Nakayama K, Ikeda K. et al. Abnormal angiogenesis in Foxo1 (Fkhr)-deficient mice. J Biol Chem. 2004;279:34741–34749. doi: 10.1074/jbc.M314214200. [DOI] [PubMed] [Google Scholar]
- Hosaka T, Biggs WH, Tieu D, Boyer AD, Varki NM, Cavenee WK, Arden KC. Disruption of forkhead transcription factor (FOXO) family members in mice reveals their functional diversification. Proc Natl Acad Sci USA. 2004;101:2975–2980. doi: 10.1073/pnas.0400093101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura T, Kitamura YI, Funahashi Y, Shawber CJ, Castrillon DH, Kollipara R, DePinho RA, Kitajewski J, Accili D. A Foxo/Notch pathway controls myogenic differentiation and fiber type specification. J Clin Invest. 2007;117:2477–2485. doi: 10.1172/JCI32054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki Y, Kamei Y, Sugita S, Akaike F, Kanai S, Miura S, Hirata Y, Troen BR, Kitamura T, Nishino I. et al. The cathepsin L gene is a direct target of FOXO1 in skeletal muscle. Biochem J. 2010;427:171–178. doi: 10.1042/BJ20091346. [DOI] [PubMed] [Google Scholar]
- Castrillon DH, Miao L, Kollipara R, Horner JW, DePinho RA. Suppression of ovarian follicle activation in mice by the transcription factor Foxo3a. Science. 2003;301:215–218. doi: 10.1126/science.1086336. [DOI] [PubMed] [Google Scholar]
- Paik JH, Kollipara R, Chu G, Ji H, Xiao Y, Ding Z, Miao L, Tothova Z, Horner JW, Carrasco DR. et al. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell. 2007;128:309–323. doi: 10.1016/j.cell.2006.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alzghoul MB, Gerrard D, Watkins BA, Hannon K. Ectopic expression of IGF-I and Shh by skeletal muscle inhibits disuse-mediated skeletal muscle atrophy and bone osteopenia in vivo. FASEB J. 2004;18:221–223. doi: 10.1096/fj.03-0293fje. [DOI] [PubMed] [Google Scholar]
- Takahashi A, Kureishi Y, Yang J, Luo Z, Guo K, Mukhopadhyay D, Ivashchenko Y, Branellec D, Walsh K. Myogenic Akt signaling regulates blood vessel recruitment during myofiber growth. Mol Cell Biol. 2002;22:4803–4814. doi: 10.1128/MCB.22.13.4803-4814.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman CA, Miu MH, Frey JW, Mabrey DM, Lincoln HC, Ge Y, Chen J, Hornberger TA. A phosphatidylinositol 3-kinase/protein kinase B-independent activation of mammalian target of rapamycin signaling is sufficient to induce skeletal muscle hypertrophy. Mol Biol Cell. 2010;21:3258–3268. doi: 10.1091/mbc.E10-05-0454. [DOI] [PMC free article] [PubMed] [Google Scholar]