Abstract
Peroxisome proliferator-activated receptor γ (PPARγ) coactivator 1 α (PGC-1α) activation coordinates induction of the hepatic fasting response through coactivation of numerous transcription factors and gene programs. In the June 15, 2011, issue of Genes & Development, Lustig and colleagues (pp. 1232–1244) demonstrated that phosphorylation of PGC-1α by the p70 ribosomal protein S6 kinase 1 (S6K1) specifically interfered with the interaction between PGC-1α and HNF4α in liver and blocked the coactivation of the gluconeogenic target genes. This demonstrates how independent fine-tuning of gene programs coregulated by the same coactivator can be obtained.
Keywords: PGC-1α, gluconeogenesis, liver, S6K1
One of the main functions of the liver is to maintain whole-body glucose homeostasis by switching from glucose storage to glucose export. In the fed state, the liver stores glucose in the form of glycogen and converts surplus glucose into fatty acids. In contrast, in the fasted state, the liver induces the gluconeogenetic pathway that synthesizes glucose using primarily fatty acids, but also some amino acids. In addition, the liver initiates the synthesis of ketone bodies that spares the use of glucose in extrahepatic tissues. To adequately respond to whole-body nutrient supply, the liver must be able to switch between the fed and the fasted gene programs in response to hormonal and nutrient signals that report the metabolic status to the liver. Thus, in the fed state, the liver activates its lipogenic and glycogenic gene programs in response to insulin and nutrient signals, whereas it activates gene programs involved in gluconeogenesis, fatty acid oxidation, oxidative phosphorylation, and synthesis of ketone bodies in the fasted state in response to glucagon and different nutrient signals. These dramatic switches in gene programs over a relatively short period require a highly coordinated action of many different transcription factors.
The adaptation of the liver to fasting is mediated by several transcription factors, including cAMP-responsive element-binding protein (CREB), hepatocyte nuclear factor 4 α (HNF4α), forkhead box protein O1 (FOXO1), nuclear respiratory factor 1 (NRF1), and peroxisome proliferator-activated receptor α (PPARα), which activate specific but overlapping sets of target genes. CREB, HNF4α, and FOXO1 are particularly important for the induction of gluconeogenesis (Herzig et al. 2001; Yoon et al. 2001; Puigserver et al. 2003; Rhee et al. 2003), whereas PPARα is important for induction of genes involved in mitochondrial and peroxisomal β-oxidation and ketogenesis (Leone et al. 1999; Koo et al. 2004). It is unclear which transcription factors regulate genes involved in oxidative phosphorylation in liver; however, NRF1 and ERRα are likely to be involved, as they induce genes involved in mitochondrial biogenesis and oxidative phosphorylation in other tissues (Wu et al. 1999; Huss et al. 2002; Mootha et al. 2004; Schreiber et al. 2004) and are involved in other components of the fasting response in liver (Handschin et al. 2005; Gaillard et al. 2006). Interestingly, most of these transcription factors are coactivated by PPARγ coactivator 1 α (PGC-1α), which thereby provides a second layer of coordination in the regulation of the fasting response.
PGC-1α was originally discovered as a cold-inducible coactivator of PPARγ required for the activation of genes involved in mitochondrial biogenesis during adaptive thermogenesis in brown adipose tissue (BAT) (Puigserver et al. 1998). However, multiple subsequent studies, including conventional and tissue-specific targeted disruption of the PGC-1α gene, have demonstrated that this cofactor plays a role in several metabolic responses. Across tissues, PGC-1α is critical for the expression of genes involved in fatty acid oxidation and mitochondrial gene expression and biogenesis through its coactivation of PPARα, NRF1, NRF2, and estrogen receptor-related α (ERRα) (Wu et al. 1999; Vega et al. 2000; Huss et al. 2002; Koo et al. 2004; Mootha et al. 2004; Schreiber et al. 2004). PGC-1α also induces tissue-specific processes associated with increased mitochondrial function, such as mitochondrial uncoupling in BAT (Puigserver et al. 1998; Tiraby et al. 2003), fiber type switching in skeletal muscle (Baar et al. 2002; Lin et al. 2002; Norrbom et al. 2004; Koves et al. 2005), and gluconeogenesis in liver (Yoon et al. 2001; Puigserver et al. 2003; Rhee et al. 2003).
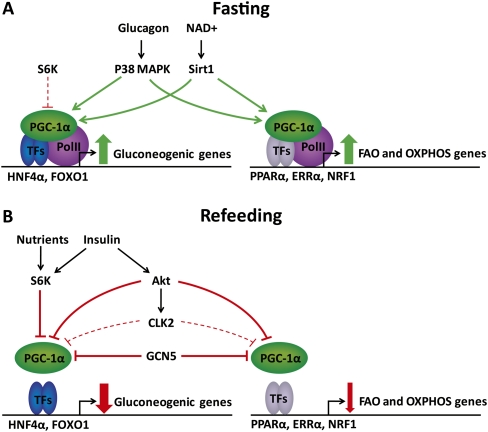
The fasting response in the liver involves the direct activation of the transcription factors in play, but PGC-1α itself is also under hormonal and nutrient regulation. Thus, the expression level of PGC-1α is induced by CREB, which is activated via glucagon-stimulated activation of protein kinase A and MAPK p38 signaling (Herzig et al. 2001; Cao et al. 2005). In addition, loss of insulin-mediated phosphorylation of transducer of regulated CREB activity 2 (TORC2) and CREB-binding protein (CBP) contributes to the activation of CREB during fasting (Zhou et al. 2004; Koo et al. 2005). Importantly, the activity of PGC-1α is also subject to extensive post-translational regulation, such as MAPK p38-mediated phosphorylation, which potentiates activity by stabilizing PGC-1α and displacing the strong transcriptional inhibitor p160 myb (Knutti et al. 2001; Puigserver et al. 2001; Fan et al. 2004; Cao et al. 2005). Furthermore, NAD+-stimulated Sirtuin 1 (SIRT-1)-mediated deacetylation is required for PGC-1α coactivation of gluconeogenesis in liver and fatty acid oxidation and mitochondrial function in muscle during fasting (Rodgers et al. 2005; Gerhart-Hines et al. 2007; Rodgers and Puigserver 2007). In skeletal muscle, the AMP-activated kinase (AMPK) plays an important role in PGC-1α activation (Jager et al. 2007); however, the role of this kinase in the liver is unclear, and AMPK activity is only slightly induced in liver during fasting (Gonzalez et al. 2004). Conversely, upon refeeding, induction of the expression of steroid receptor coactivator 3 (SRC3) and general control of amino acid synthesis 5 (GCN5) causes acetylation and inactivation of PGC-1α (Lerin et al. 2006; Coste et al. 2008). Furthermore, PGC-1α activity is inhibited by insulin-induced phosphorylations by Akt and Cdc2-like kinase 2 (CLK2) (Li et al. 2007; Rodgers et al. 2010). Thus, PGC-1α integrates multiple cellular signals in the regulation of the fasting response in the liver (Fig. 1).
Figure 1.
Post-translational regulation of PGC-1α activity in liver during fasting and refeeding. (A) During fasting, glucagon and NAD+ activate PGC-1α through MAPK p38-mediated phosphorylation and Sirt1-mediated deacetylation. Low levels of S6K may be involved in feedback regulation of gluconeogenesis by specifically interfering with PGC-1α coactivation of HNF4α. (B) After refeeding, increased nutrient levels activate S6K1, which may lead to an acute, specific repression of gluconeogenic gene expression by phosphorylation of PGC-1α. Furthermore, the nutrient-induced rise in insulin activates the Akt kinase, which phosphorylates PGC-1α, leading to repression of gene programs involved in gluconeogenesis, fatty acid oxidation (FAO), and oxidative phosphorylation (OXPHOS). In addition, Akt phosphorylates the CLK2 kinase, which promotes sustained repression of predominantly gluconeogenic genes. Finally, refeeding leads to increased expression of steroid receptor coactivator 3 (SRC3) and general control of amino acid synthesis 5 (GCN5), which acetylates and thereby inactivates PGC1-α.
The coordinated regulation of the multiple gene programs during fasting through PGC-1α activation provides an efficient mechanism for the regulation of the hepatic fasting response (Spiegelman and Heinrich 2004). However, it is likely that additional, independent control of individual pathways is required to fine-tune the individual pathways. This pathway-specific regulation is, in part, governed by specific combinations of transcriptions factors. However, in the June 15, 2011, issue of Genes & Development, Spiegelman and colleagues (Lustig et al. 2011) demonstrate that the cofactor PGC-1α can change pathway specificity in response to cellular signals. They demonstrate that phosphorylation of PGC-1α by the p70 ribosomal protein S6 kinase 1 (S6K1) specifically inhibited the ability to coactivate gluconeogenic genes, whereas it had no effect on the ability to coactivate genes involved in β-oxidation and oxidative phosphorylation (Lustig et al. 2011). S6K1 is activated by the mammalian target of rapamycin (mTOR) pathway, which is induced by insulin and nutrients like amino acids (Patti et al. 1998; Nobukuni et al. 2005; Sengupta et al. 2010). Consistent with an important role of S6K1 in repressing the hepatic fasting gene program, inhibition of mTOR by rapamycin has been shown to induce gluconeogenic gene expression and cause insulin resistance and glucose intolerance in mice (Houde et al. 2010). Furthermore, inhibition of mTOR by loss of the regulatory-associated protein of mTOR (raptor), which is essential to mTOR function, has been shown to impair suppression of ketone body synthesis upon refeeding (Sengupta et al. 2010).
Using mass spectrometry analysis of PGC-1α ectopically expressed in primary hepatocytes, the Spiegelman group (Lustig et al. 2011) identified two novel phosphorylation sites: S568 and S572. The phosphorylation of these two residues was catalyzed by S6K1 in vitro and was stimulated by amino acids in cultures of primary hepatocytes. Interestingly, these phosphorylations seemed to specifically inhibit the ability of PGC-1α to activate gluconeogenic genes, since ectopic expression of a mutant PGC-1α where both serines have been mutated was more effective than wild-type PGC-1α in the induction of gluconeogenic genes. In contrast, induction of genes involved in fatty acid oxidation and oxidative phosphorylation were induced to a similar extent by wild-type and mutant PGC-1α. Consistent with a role of S6K1 in inducing PGC-1α phosphorylation, ectopic expression of a constitutively active form of S6K1 in primary hepatocytes specifically inhibited the induction of gluconeogenic genes, and rapamycin, or a dominant-negative S6K1, specifically promoted PGC-1α induction of gluconeogenic but not fatty acid oxidation or mitochondrial genes. The importance of these phosphorylations in vivo was confirmed by intravenous injection of adenoviral vectors expressing either mutant or wild-type PGC-1α. Consistent with the cell culture studies, mutant PGC-1α increased gluconeogenic gene expression in liver of fasting mice significantly more than wild-type PGC-1α. Consequently, expression of mutant, but not wild-type, PGC-1α in the liver led to increased plasma glucose levels.
At the mechanistic level, phosphorylation of PGC-1α by S6K1 appears to specifically interfere with the interaction between HNF4α and PGC-1α. Coimmunoprecipitation experiments in HEK293T cells showed that the interaction of HNF4α with PGC-1α, but not with ERRα or PPARα, was increased by rapamycin treatment, and that mutant PGC-1α interacted more strongly with HNF4α than wild-type PGC-1α. Furthermore, a constitutively active form of S6K1 inhibited the ability of wild-type PGC-1α, but not mutant PGC-1α, to coactivate HNF4α in transient transfection assays. Taken together, these data indicate that S6K1 phosphorylation specifically interferes with the ability of PGC-1α to coactivate gluconeogenic gene expression at least in part by disrupting the interaction with HNF4α.
The inhibition of PGC-1α activity by S6K1 adds to the previously reported repression of PGC-1α activity in the fed state by CLK2- and Akt-mediated phosphorylations (Fig. 1; Li et al. 2007; Rodgers et al. 2010). The intriguing and novel finding of this new work is that the phosphorylation of S568 and S572 by S6K1 specifically disrupts the interaction with HNF4α and the gluconeogenic gene program (Lustig et al. 2011), whereas Akt phosphorylation of S570 results in a general repression of PGC-1α activity on all gene programs regulated (Li et al. 2007). The differential effects are particularly notable since these kinases phosphorylate adjacent serines in the serine arginine (SR) domain of PGC-1α. The Puigserver group (Rodgers et al. 2010) previously reported that CLK2, which also targets the SR domain, inhibits PGC-1α coactivation of both HNF4α and FOXO1 in transient transfection studies, whereas NRF coactivation was unaffected. Furthermore, CLK2 knockdown or overexpression primarily affected the gluconeogenic gene program in primary hepatocytes overexpressing PGC-1α. These results suggest that also CLK2-mediated phosphorylation(s) result in differential effects on the ability to coactivate specific transcription factors (Rodgers et al. 2010), although in a less specific manner than the S6K1 phosphorylation (Lustig et al. 2011). Unfortunately, however, the exact phosphorylation site was not mapped in the study by the Puigserver group (Rodgers et al. 2010). Taken together, the SR domain of PGC-1α appears to act as a critical switch determining the coactivation abilities of PGC-1α in different gene programs. Further investigation is needed to fully understand the role and interdependencies of the specific phosphorylations of this domain in the regulation of PGC-1α coactivation of gene programs.
In addition to their differential role in activating specific gene programs, the S6K1, Akt, and CLK2 kinases appear to repress the gluconeogenic genes at different time points during the refeeding response. Thus, investigations by the Puigserver group (Rodgers et al. 2010) indicated that whereas Akt-mediated phosphorylation is involved in the acute repression of PGC-1α target genes following refeeding, phosphorylation by CLK2 represses PGC-1α at later time points. Using hepatic knockdown of CLK2, they demonstrated that this kinase is involved in repression of gluconeogenic gene expression 4 h after refeeding but not in acute repression (Rodgers et al. 2010). In contrast, Akt was acutely but transiently activated, indicating that Akt- and CLK2-mediated phosphorylation of PGC-1α facilitates acute and sustained repression of gluconeogenic genes, respectively. Recently, Sabatini and coworkers (Sengupta et al. 2010) showed that S6K1 is phosphorylated prior to Akt during the refeeding response, suggesting that S6K1 activity is initially directly regulated by nutrients via the mTOR pathway, rather than through increased plasma insulin levels. Thus, the S6K1-mediated repression of PGC-1α, as described by the Spiegelman group (Lustig et al. 2011), may mediate repression of gluconeogenic genes even more acutely than Akt.
The experiments by Sabatini and coworkers (Sengupta et al. 2010) also showed that fasted tuberous sclerosis complex (TSC)−/− mice, which have significantly increased mTOR activity and therefore increased S6K1 activity, display a decreased hepatic expression of genes involved in fatty acid oxidation and ketogenesis compared with wild-type mice. Surprisingly, the expression of the key gluconeogenic gene phosphoenolpyruvate carboxykinase (PEPCK) was not compromised (Sengupta et al. 2010), indicating that increased mTOR activity by itself is insufficient to repress gluconeogenesis in vivo, but is sufficient to decrease expression of fatty acid oxidation genes. This may seem contradictory to findings by Spiegelman and coworkers (Lustig et al. 2011), who demonstrated that ectopic expression of a constitutively active form of S6K1 in primary hepatocytes in vitro specifically repressed gluconeogenic genes. However, it is possible that additional stimuli normally provided during refeeding, but missing in the fasting state, are required for full activation of S6K1, for the specific phosphorylation of PGC-1α, or for inactivation of the gluconeogenic program in the TSC−/− mice. Furthermore, it is possible that the repression of fatty acid oxidation genes in these mice is independent of S6K1 activity.
An interesting aspect of the S6K1-mediated phosphorylation of PGC-1α is that it also appears to modulate PGC-1α activity during fasting: A mutant form of PGC-1α that cannot be phosphorylated by S6K1 is a more potent activator of gluconeogenic genes in the liver of fasting mice (Lustig et al. 2011). This observation indicates that S6K1 is also at least partially active and modulates PGC-1α activity in the liver during fasting. In contrast, phosphorylation by Akt and CLK2 kinases does not appear to be operating during fasting, since S570 phosphorylation of PGC-1α was not detected in the liver of fasted mice, and since expression of a S570A mutant PGC-1α in liver of fasting mice induced gluconeogenic gene expression to similar levels as a wild-type PGC-1α (Li et al. 2007). Consistent with this, CLK2 knockdown did not affect PGC-1α target gene expression during fasting (Rodgers et al. 2010). Hence, it seems that regulation of gluconeogenesis by Akt and CLK2 is restricted to a refeeding situation.
It is unclear what mediates the apparent S6K1 activity during fasting in the studies by the Spiegelman laboratory (Lustig et al. 2011), but it is possible that fasting glucose and insulin levels are sufficient to maintain some mTOR activation of S6K1. In that case, S6K1-mediated phosphorylation of PGC-1α could act as part of the important negative feedback loop regulating gluconeogenesis via glucose itself or via glucose-induced insulin secretion. In any case, the significantly elevated levels of plasma glucose during fasting in mice with ectopic expression of S568A and S572A mutant PGC-1α is consistent with an important role of PGC-1α in feedback regulation of gluconeogenesis. The apparent S6K1 activity during fasting is also consistent with reports showing that branched chain amino acid and S6K1 activity is increased during prolonged fasting (Anand and Gruppuso 2005). Thus, S6K1 activity may be activated by these amino acids, especially following prolonged fasting.
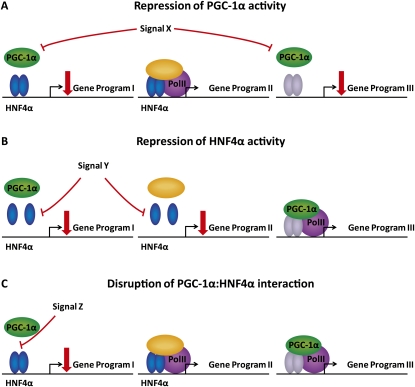
The advantages of regulating PGC-1α and other cofactors by a post-translational modification (PTM) that specifically modifies interactions with selected transcription factors are obvious. Repression of overall PGC-1α activity would affect all PGC-1α-coactivated gene programs (Fig. 2A), whereas repression of HNF4α activity would affect all HNF4α target genes (Fig. 2B). However, by specifically regulating the interaction between the two factors, a much higher degree of specificity is obtained than by targeting overall activity of any of these two factors (Fig. 2C). This ability to specifically feedback-regulate the gluconeogenic gene program without suppressing the expression of genes involved in fatty acid oxidation and ketogenesis is likely to be important during fasting. From a pharmacological point of view, the specificity that can be obtained by PTMs targeting the interaction surface between cofactors and transcription factors is also highly interesting. In the case of PGC-1α, by specifically targeting the interaction with HNF4α, one could decrease hepatic glucose production while leaving the many beneficial effects of PGC-1α in liver and muscle intact.
Figure 2.
Suppression of the HNF4α and PGC-1α gene programs by post-translational modifications at different levels. Repression of overall PGC-1α activity will interfere with all gene programs coregulated by PGC-1α (A), whereas repression of HNF4α activity will affect all gene programs regulated by HNF4α (B). (C) Maximal specificity is achieved by specifically disrupting the interaction between HNF4α and PGC-1α to repress the PGC-1α-coregulated HNF4α gene program.
At present, only a few examples of cofactors that modify their gene program specificities in response to PTMs have been reported. Leff and coworkers (Yang et al. 2001) previously showed that AMPK-mediated phosphorylation of p300 lead to reduced interactions with nuclear receptors but not other transcription factors such as p53 and GATA4; however, they did not identify the physiological importance of this. It is likely that many of the known PTMs of other coactivators have undiscovered differential effects on gene programs. For example, the insulin-induced phosphorylation of the CREB interaction domain of CBP by the phosphatidylinositol 3 kinase (PI3K) pathway is an interesting candidate for gene program-specific PTM (Zhou et al. 2004). The recent advances in proteomic and genomic technologies provide an excellent opportunity for further investigations of coactivator modifications. Proteomics would be applicable for large-scale coimmunoprecipitation interaction studies, whereas transcriptomics and chromatin immunoprecipitation combined with deep sequencing would be useful tools to identify differentially targeted enhancers and gene programs.
Acknowledgments
We thank Dr. P. Puigserver for critical reading of this manuscript. This work was supported by grants to S.M. from the NIH NIDDK/ODS (5R01-DK063070).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.2076411.
References
- Anand P, Gruppuso PA 2005. The regulation of hepatic protein synthesis during fasting in the rat. J Biol Chem 280: 16427–16436 [DOI] [PubMed] [Google Scholar]
- Baar K, Wende AR, Jones TE, Marison M, Nolte LA, Chen M, Kelly DP, Holloszy JO 2002. Adaptations of skeletal muscle to exercise: rapid increase in the transcriptional coactivator PGC-1. FASEB J 16: 1879–1886 [DOI] [PubMed] [Google Scholar]
- Cao W, Collins QF, Becker TC, Robidoux J, Lupo EG Jr, Xiong Y, Daniel KW, Floering L, Collins S 2005. p38 mitogen-activated protein kinase plays a stimulatory role in hepatic gluconeogenesis. J Biol Chem 280: 42731–42737 [DOI] [PubMed] [Google Scholar]
- Coste A, Louet JF, Lagouge M, Lerin C, Antal MC, Meziane H, Schoonjans K, Puigserver P, O'Malley BW, Auwerx J 2008. The genetic ablation of SRC-3 protects against obesity and improves insulin sensitivity by reducing the acetylation of PGC-1α. Proc Natl Acad Sci 105: 17187–17192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan M, Rhee J, St-Pierre J, Handschin C, Puigserver P, Lin J, Jaeger S, Erdjument-Bromage H, Tempst P, Spiegelman BM 2004. Suppression of mitochondrial respiration through recruitment of p160 myb binding protein to PGC-1α: modulation by p38 MAPK. Genes Dev 18: 278–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard S, Grasfeder LL, Haeffele CL, Lobenhofer EK, Chu TM, Wolfinger R, Kazmin D, Koves TR, Muoio DM, Chang CY, et al. 2006. Receptor-selective coactivators as tools to define the biology of specific receptor–coactivator pairs. Mol Cell 24: 797–803 [DOI] [PubMed] [Google Scholar]
- Gerhart-Hines Z, Rodgers JT, Bare O, Lerin C, Kim SH, Mostoslavsky R, Alt FW, Wu Z, Puigserver P 2007. Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1α. EMBO J 26: 1913–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez AA, Kumar R, Mulligan JD, Davis AJ, Weindruch R, Saupe KW 2004. Metabolic adaptations to fasting and chronic caloric restriction in heart, muscle, and liver do not include changes in AMPK activity. Am J Physiol Endocrinol Metab 287: E1032–E1037 doi: 10.1152/ajpendo.00172.2004 [DOI] [PubMed] [Google Scholar]
- Handschin C, Lin J, Rhee J, Peyer AK, Chin S, Wu PH, Meyer UA, Spiegelman BM 2005. Nutritional regulation of hepatic heme biosynthesis and porphyria through PGC-1α. Cell 122: 505–515 [DOI] [PubMed] [Google Scholar]
- Herzig S, Long F, Jhala US, Hedrick S, Quinn R, Bauer A, Rudolph D, Schutz G, Yoon C, Puigserver P, et al. 2001. CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature 413: 179–183 [DOI] [PubMed] [Google Scholar]
- Houde VP, Brule S, Festuccia WT, Blanchard PG, Bellmann K, Deshaies Y, Marette A 2010. Chronic rapamycin treatment causes glucose intolerance and hyperlipidemia by upregulating hepatic gluconeogenesis and impairing lipid deposition in adipose tissue. Diabetes 59: 1338–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huss JM, Kopp RP, Kelly DP 2002. Peroxisome proliferator-activated receptor coactivator-1α (PGC-1α) coactivates the cardiac-enriched nuclear receptors estrogen-related receptor-α and -γ. Identification of novel leucine-rich interaction motif within PGC-1α. J Biol Chem 277: 40265–40274 [DOI] [PubMed] [Google Scholar]
- Jager S, Handschin C, St-Pierre J, Spiegelman BM 2007. AMP-activated protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1α. Proc Natl Acad Sci 104: 12017–12022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutti D, Kressler D, Kralli A 2001. Regulation of the transcriptional coactivator PGC-1 via MAPK-sensitive interaction with a repressor. Proc Natl Acad Sci 98: 9713–9718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo SH, Satoh H, Herzig S, Lee CH, Hedrick S, Kulkarni R, Evans RM, Olefsky J, Montminy M 2004. PGC-1 promotes insulin resistance in liver through PPAR-α-dependent induction of TRB-3. Nat Med 10: 530–534 [DOI] [PubMed] [Google Scholar]
- Koo SH, Flechner L, Qi L, Zhang X, Screaton RA, Jeffries S, Hedrick S, Xu W, Boussouar F, Brindle P, et al. 2005. The CREB coactivator TORC2 is a key regulator of fasting glucose metabolism. Nature 437: 1109–1111 [DOI] [PubMed] [Google Scholar]
- Koves TR, Li P, An J, Akimoto T, Slentz D, Ilkayeva O, Dohm GL, Yan Z, Newgard CB, Muoio DM 2005. Peroxisome proliferator-activated receptor-γ co-activator 1α-mediated metabolic remodeling of skeletal myocytes mimics exercise training and reverses lipid-induced mitochondrial inefficiency. J Biol Chem 280: 33588–33598 [DOI] [PubMed] [Google Scholar]
- Leone TC, Weinheimer CJ, Kelly DP 1999. A critical role for the peroxisome proliferator-activated receptor α (PPARα) in the cellular fasting response: the PPARα-null mouse as a model of fatty acid oxidation disorders. Proc Natl Acad Sci 96: 7473–7478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerin C, Rodgers JT, Kalume DE, Kim SH, Pandey A, Puigserver P 2006. GCN5 acetyltransferase complex controls glucose metabolism through transcriptional repression of PGC-1α. Cell Metab 3: 429–438 [DOI] [PubMed] [Google Scholar]
- Li X, Monks B, Ge Q, Birnbaum MJ 2007. Akt/PKB regulates hepatic metabolism by directly inhibiting PGC-1α transcription coactivator. Nature 447: 1012–1016 [DOI] [PubMed] [Google Scholar]
- Lin J, Wu H, Tarr PT, Zhang CY, Wu Z, Boss O, Michael LF, Puigserver P, Isotani E, Olson EN, et al. 2002. Transcriptional co-activator PGC-1α drives the formation of slow-twitch muscle fibres. Nature 418: 797–801 [DOI] [PubMed] [Google Scholar]
- Lustig Y, Ruas JL, Estall JL, Lo JC, Devarakonda S, Laznik D, Choi JH, Ono H, Olsen JV, Spiegelman BM 2011. Separation of the gluconeogenic and mitochondrial functions of PGC-1a through S6 kinase. Genes Dev 25: 1232–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha VK, Handschin C, Arlow D, Xie X, St Pierre J, Sihag S, Yang W, Altshuler D, Puigserver P, Patterson N, et al. 2004. Errα and Gabpa/b specify PGC-1α-dependent oxidative phosphorylation gene expression that is altered in diabetic muscle. Proc Natl Acad Sci 101: 6570–6575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobukuni T, Joaquin M, Roccio M, Dann SG, Kim SY, Gulati P, Byfield MP, Backer JM, Natt F, Bos JL, et al. 2005. Amino acids mediate mTOR/raptor signaling through activation of class 3 phosphatidylinositol 3OH-kinase. Proc Natl Acad Sci 102: 14238–14243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norrbom J, Sundberg CJ, Ameln H, Kraus WE, Jansson E, Gustafsson T 2004. PGC-1α mRNA expression is influenced by metabolic perturbation in exercising human skeletal muscle. J Appl Physiol 96: 189–194 [DOI] [PubMed] [Google Scholar]
- Patti ME, Brambilla E, Luzi L, Landaker EJ, Kahn CR 1998. Bidirectional modulation of insulin action by amino acids. J Clin Invest 101: 1519–1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM 1998. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell 92: 829–839 [DOI] [PubMed] [Google Scholar]
- Puigserver P, Rhee J, Lin J, Wu Z, Yoon JC, Zhang CY, Krauss S, Mootha VK, Lowell BB, Spiegelman BM 2001. Cytokine stimulation of energy expenditure through p38 MAP kinase activation of PPARγ coactivator-1. Mol Cell 8: 971–982 [DOI] [PubMed] [Google Scholar]
- Puigserver P, Rhee J, Donovan J, Walkey CJ, Yoon JC, Oriente F, Kitamura Y, Altomonte J, Dong H, Accili D, et al. 2003. Insulin-regulated hepatic gluconeogenesis through FOXO1–PGC-1α interaction. Nature 423: 550–555 [DOI] [PubMed] [Google Scholar]
- Rhee J, Inoue Y, Yoon JC, Puigserver P, Fan M, Gonzalez FJ, Spiegelman BM 2003. Regulation of hepatic fasting response by PPARγ coactivator-1α (PGC-1): requirement for hepatocyte nuclear factor 4α in gluconeogenesis. Proc Natl Acad Sci 100: 4012–4017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers JT, Puigserver P 2007. Fasting-dependent glucose and lipid metabolic response through hepatic sirtuin 1. Proc Natl Acad Sci 104: 12861–12866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P 2005. Nutrient control of glucose homeostasis through a complex of PGC-1α and SIRT1. Nature 434: 113–118 [DOI] [PubMed] [Google Scholar]
- Rodgers JT, Haas W, Gygi SP, Puigserver P 2010. Cdc2-like kinase 2 is an insulin-regulated suppressor of hepatic gluconeogenesis. Cell Metab 11: 23–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber SN, Emter R, Hock MB, Knutti D, Cardenas J, Podvinec M, Oakeley EJ, Kralli A 2004. The estrogen-related receptor α (ERRα) functions in PPARγ coactivator 1α (PGC-1α)-induced mitochondrial biogenesis. Proc Natl Acad Sci 101: 6472–6477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta S, Peterson TR, Laplante M, Oh S, Sabatini DM 2010. mTORC1 controls fasting-induced ketogenesis and its modulation by ageing. Nature 468: 1100–1104 [DOI] [PubMed] [Google Scholar]
- Spiegelman BM, Heinrich R 2004. Biological control through regulated transcriptional coactivators. Cell 119: 157–167 [DOI] [PubMed] [Google Scholar]
- Tiraby C, Tavernier G, Lefort C, Larrouy D, Bouillaud F, Ricquier D, Langin D 2003. Acquirement of brown fat cell features by human white adipocytes. J Biol Chem 278: 33370–33376 [DOI] [PubMed] [Google Scholar]
- Vega RB, Huss JM, Kelly DP 2000. The coactivator PGC-1 cooperates with peroxisome proliferator-activated receptor α in transcriptional control of nuclear genes encoding mitochondrial fatty acid oxidation enzymes. Mol Cell Biol 20: 1868–1876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, Troy A, Cinti S, Lowell B, Scarpulla RC, et al. 1999. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell 98: 115–124 [DOI] [PubMed] [Google Scholar]
- Yang W, Hong YH, Shen XQ, Frankowski C, Camp HS, Leff T 2001. Regulation of transcription by AMP-activated protein kinase: phosphorylation of p300 blocks its interaction with nuclear receptors. J Biol Chem 276: 38341–38344 [DOI] [PubMed] [Google Scholar]
- Yoon JC, Puigserver P, Chen G, Donovan J, Wu Z, Rhee J, Adelmant G, Stafford J, Kahn CR, Granner DK, et al. 2001. Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature 413: 131–138 [DOI] [PubMed] [Google Scholar]
- Zhou XY, Shibusawa N, Naik K, Porras D, Temple K, Ou H, Kaihara K, Roe MW, Brady MJ, Wondisford FE 2004. Insulin regulation of hepatic gluconeogenesis through phosphorylation of CREB-binding protein. Nat Med 10: 633–637 [DOI] [PubMed] [Google Scholar]