Abstract
The Spt–Ada–Gcn5–acetyltransferase (SAGA) complex was discovered from Saccharomyces cerevisiae and has been well characterized as an important transcriptional coactivator that interacts both with sequence-specific transcription factors and the TATA-binding protein TBP. SAGA contains a histone acetyltransferase and a ubiquitin protease. In metazoans, SAGA is essential for development, yet little is known about the function of SAGA in differentiating tissue. We analyzed the composition, interacting proteins, and genomic distribution of SAGA in muscle and neuronal tissue of late stage Drosophila melanogaster embryos. The subunit composition of SAGA was the same in each tissue; however, SAGA was associated with considerably more transcription factors in muscle compared with neurons. Consistent with this finding, SAGA was found to occupy more genes specifically in muscle than in neurons. Strikingly, SAGA occupancy was not limited to enhancers and promoters but primarily colocalized with RNA polymerase II within transcribed sequences. SAGA binding peaks at the site of RNA polymerase pausing at the 5′ end of transcribed sequences. In addition, many tissue-specific SAGA-bound genes required its ubiquitin protease activity for full expression. These data indicate that in metazoans SAGA plays a prominent post-transcription initiation role in tissue-specific gene expression.
Keywords: SAGA, coactivator, histone acetyltransferase, histone deubiquitination, transcription initiation, ubiquitin protease
The Spt–Ada–Gcn5–acetyltransferase (SAGA) complex is a highly conserved transcription coactivator that contains >20 protein subunits (Koutelou et al. 2010). In budding yeast, where SAGA was discovered, its functions in transcription are best characterized. SAGA contains the Gcn5 acetyltransferase, which acetylates nucleosomes at the promoters of genes in vitro and in vivo. SAGA is recruited to target genes through the interaction of its largest subunit, Tra1, with the activation domains of sequence-specific DNA-binding transcription factors (Baker and Grant 2007). Acetylation of promoter nucleosomes by SAGA stabilizes its interactions with promoters and targets promoter nucleosomes for displacement by the SWI/SNF nucleosome remodeling complex (Li et al. 2007). SAGA has many features related to that of the general transcription factor TFIID, including a number of common TAF subunits and the ability to interact with the TATA-binding protein TBP. SAGA functions as a bona fide coactivator by delivering TBP to target genes in vivo and in vitro (Baker and Grant 2007; Rodriguez-Navarro 2009). Thus, the functions of SAGA in histone modification, chromatin remodeling, and preinitiation transcription complex assembly have been well characterized.
The discovery of a second catalytic activity within the SAGA complex has implicated SAGA function in later steps during activation of transcription. SAGA contains ubiquitin protease activity specific for monoubiquitinated histone H2B (ubH2B) (Henry et al. 2003). Ubiquitination of H2B occurs following transcription complex formation and initiation of transcription (Rodriguez-Navarro 2009). The Ubp8 ubiquitin protease within yeast SAGA is involved in the transition of RNA polymerase II (Pol II) from transcription initiation into elongation (Wyce et al. 2007). Specifically, deubiquitination of ubH2B by SAGA facilitates recruitment of the Pol II Ser-2 C-terminal domain (CTD) kinase Ctk1 (Wyce et al. 2007). Thus, SAGA promotes transcription elongation by deubiquitination of ubH2B, which may extend from the 5′ end into the coding region of active genes (Rodriguez-Navarro 2009).
More recently, SAGA has been purified from Drosophila and mammalian cells and was found to contain homologs of most of the yeast SAGA subunits, including the Gcn5 and Ubp8 catalytic subunits. In metazoans, SAGA may have roles in both normal development and cancer (Koutelou et al. 2010). Individual loss of the SAGA subunits Gcn5, Ada2b, Ada3, WDA, Sgf11, and SAF6 results in developmental defects and larval lethality in flies (Weake et al. 2009 and references therein). Similarly, Gcn5 deletion in mice leads to defects in mesoderm development and embryonic lethality (Xu et al. 2000). However, catalytic site mutations in Gcn5 survive longer but suffer neural tube closure defects and exencephaly (Bu et al. 2007). Furthermore, loss of the ubiquitin protease in Drosophila SAGA (Nonstop) leads to defects in photoreceptor axon targeting followed by lethality at late larval stages (Weake et al. 2008).
While loss of SAGA subunits results in embryonic or larval lethality in flies and mice, deletion of SAGA subunits in yeast are not lethal and only show synthetic lethality when combined with other mutations (Howe et al. 2001). Moreover, mouse embryonic stem cells lacking Gcn5 grow and remain partially pluripotent (Lin et al. 2007). Thus, while SAGA does not appear to be essential at the cellular level, it seems to play important roles in the development of metazoans. To better understand the roles of SAGA functions in developmental gene expression, we examined its composition and binding profile and the effects of subunit loss on gene expression in two distinct differentiated cell types in late stage Drosophila embryos: muscle and neurons.
Results
Isolation of muscle and neuronal cells expressing the SAGA-specific subunit Ada2b
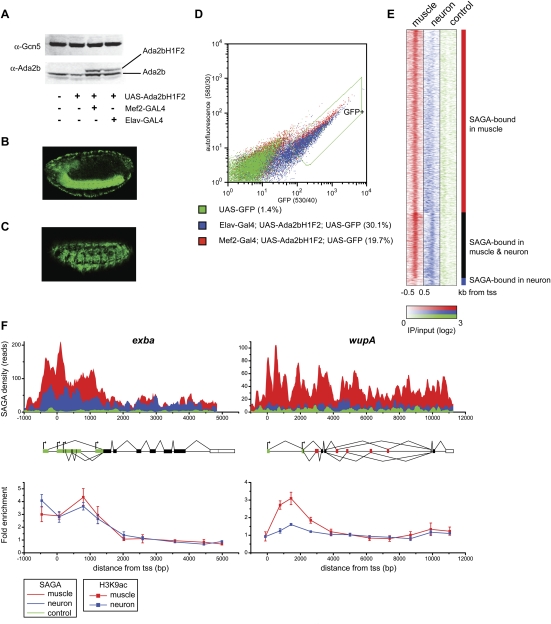
We sought to design a system in which SAGA could be isolated from different cell types in Drosophila embryos so that its composition and localization pattern could be determined in different tissues. To this end, we used the GAL4/UAS system to express a Flag-HA tagged version of the SAGA-specific protein Ada2b (Ada2bH1F2) (Kusch et al. 2003; Weake et al. 2009) in muscle or neuronal cells using the mef2-GAL4 and elav-GAL4 drivers, respectively (Brand and Perrimon 1993; Luo et al. 1994; Ranganayakulu et al. 1996; Elliott and Brand 2008). Expression of Ada2bH1F2 under the control of its genomic enhancer sequences rescues viability of the lethal ada2b1 allele (see the Supplemental Material for details; Qi et al. 2004). Whereas mef2 is expressed in committed mesoderm, the somatic and visceral musculature, and cardiac progenitors (Lilly et al. 1994; Nguyen et al. 1994), elav is expressed prominently in neuronal cells and transiently in glial cells of the embryonic CNS (Campos et al. 1987; Berger et al. 2007). Ada2bH1F2 is expressed at levels similar to those of endogenous Ada2b using this system (Fig. 1A). To enrich for our cell populations of interest that express tagged Ada2b, we labeled muscle and neuronal cells using GFP (Fig. 1B,C) and isolated these cells using fluorescence-activated cell sorting (FACS) (Fig. 1D; Supplemental Fig. 1). To determine whether the purified cells exhibit the characteristic gene expression profiles of each cell type, we first isolated GFP-labeled neuronal and muscle cells from late stage embryos by FACS (Supplemental Fig. 2). We then compared RNA isolated from these tissues with RNA extracted from whole embryos using cDNA microarrays, and identified genes that are differentially expressed in muscle or neurons using significance analysis of microarrays (Supplemental Table 1). The differentially expressed genes identified using our approach were compared with ImaGO terms that describe the expression pattern of individual genes during Drosophila embryogenesis as determined by in situ hybridization (Tomancak et al. 2002). Genes identified as being expressed preferentially in muscle relative to neurons were enriched for ImaGO terms including embryonic/larval muscle system and dorsal prothoracic pharyngeal muscle (Supplemental Fig. 2). In contrast, genes identified as being expressed preferentially in neurons were enriched for ImaGO terms such as ventral nerve cord and dorsal/lateral sensory complexes (Supplemental Fig. 2). We conclude that cells isolated using our FACS approach are enriched for the cell types of interest.
Figure 1.
(A) Western analysis of nuclear extract from Drosophila embryos probed with antibodies against Gcn5 and Ada2b. Extracts were isolated from OregonR (wild type, lane 1), UAS-Ada2bH1F2 in the absence of the GAL4 driver (lane 2), UAS-Ada2bH1F2 expressed under control of mef2-GAL4 (lane 3), and elav-GAL4 drivers (lane 4). (B) Lateral view of GFP expression in the ventral nerve chord and peripheral nervous system of a stage 15 elav-GAL4; UAS-Ada2bH1F2; UAS-GFP embryo. (C) Lateral view of GFP expression in somatic musculature of a stage 15 mef2-GAL4; UAS-Ada2bH1F2; UAS-GFP embryo. (D) GFP was plotted against yellow autofluorescence for single cells. GFP-positive nondebris events are shown within the green line (GFP+). The percentage of GFP-positive events is given in brackets beside each genotype. (E) Region map displaying the density of the SAGA ChIP-seq signal in muscle relative to neurons as IP/input (log2) for muscle-specific SAGA-bound genes (top panel), genes bound by SAGA in both muscle and neurons (middle panel), and genes bound by SAGA only in neurons (bottom panel). SAGA-binding density is shown for individual genes (rows) from −500 base pairs (bp) to +500 bp around the transcription start site (+1) in muscle (red) and neurons (blue) relative to the signal from a control Flag ChIP-seq experiment in untagged neuronal cells (green). (F) Binding profiles for SAGA (top panel) and acetylated H3-Lys9 (bottom panel) in muscle (red) and neurons (blue) are shown for the exba and wupA loci. The gene structure is indicated below the SAGA-binding profile: 5′ and 3′ UTRs are shown in green and white, respectively; constitutively and alternatively spliced exons are shown in black and red, respectively; and alternative transcription start sites are indicated by arrows. The Y-axis in the top panel represents the number of unique reads observed in two biological experiments for the SAGA ChIP-seq experiment for 20-bp windows across the two loci. The control profile in the top panel (green) represents the Flag ChIP-seq signal in untagged neuronal cells. ChIP for acetylated H3-Lys9 was performed on chromatin isolated from muscle and neurons. ChIP data represent mean percent input normalized to the coding region of a gene that is not expressed in embryos. Error bars denote standard error of the mean for three biological experiments.
SAGA localizes differentially in muscle and neuronal cells at sites of histone acetylation
To examine the genome-wide distribution of SAGA in muscle and neuronal cells from late stage Drosophila embryos, we conducted chromatin immunoprecipitation (ChIP) followed by high-throughput sequencing (ChIP-seq). An outline of the experimental protocol used for ChIP analysis is provided in Supplemental Figure 3. Antibodies against Flag were used to immunoprecipitate Ada2bH1F2 from FACS-isolated muscle or neuronal cells. The Flag epitope has been shown to be specific for ChIP-seq analysis (X Zhang et al. 2008), and we confirmed this result by showing that a nonspecific protein tagged with the Flag epitope does not have an affinity for the SAGA-bound loci we examined in S2 cells (Supplemental Fig. 4). As controls, input chromatin samples from each embryonic cell type were sequenced, as was immunoprecipitated DNA from control Flag ChIP in untagged neuronal cells. Our analysis identified 1874 peaks of SAGA localization in late stage embryonic muscle cells, the majority of which were located within the 5′ untranslated region (UTR) of genes (Supplemental Table 2). Due to this gene-biased pattern of SAGA binding, we modified our analysis approach to examine localization patterns on the promoter and coding region of all Drosophila genes. We used a scoring system based on the number of reads per kilobase of gene length to identify genes with a significant level of SAGA bound either at the promoter or coding region relative to both the input chromatin and negative Flag control immunoprecipitation. Using an identical threshold score, we identified 1997 genes in muscles and 586 genes in neurons that were bound by SAGA (Supplemental Table 3). Note that levels of SAGA below the threshold score are observed in neuronal cells at some additional genes, but are not concentrated at promoters (Fig. 1E). Surprisingly, only 59 genes are specifically bound by SAGA in neurons but not in muscle under the threshold conditions used. In contrast, 1470 genes are bound specifically by SAGA in muscle but not in neurons at this late stage of embryogenesis. Thus, SAGA localizes to a total of 14% of genes in muscle and 4% of genes in neuronal cells of late stage Drosophila embryos.
SAGA contains the histone acetyltransferase Gcn5; therefore, we asked whether the presence of SAGA correlates with histone acetylation in the two different cell types. We examined two genes that are bound differentially by SAGA in muscle and neurons for enrichment of acetylated H3-Lys9: exba, which encodes the translation factor eIF-5C, is bound by SAGA in both tissues, whereas wupA, encoding troponin I, is bound by SAGA specifically in muscle cells (Fig. 1F, top panel). Both genes exhibit peaks of acetylated H3-Lys9 toward the 5′ region of the gene body in muscle cells (Fig. 1F, bottom panel). However, in neurons, a peak of acetylated H3-Lys9 is observed at exba but not at wupA. Thus, the tissue-specific localization of SAGA correlates with tissue-specific enrichment of one of its associated histone modifications: acetylated H3-Lys9. Additional examples of the tissue-specific localization of SAGA are provided in Supplemental Figure 5.
SAGA associates with different sets of transcription factors in different cell types
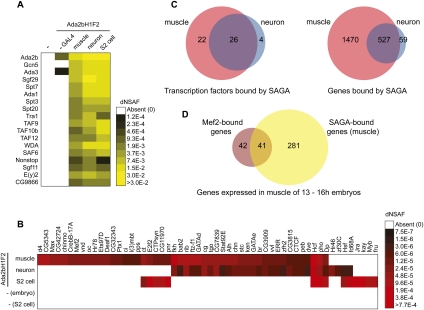
To determine whether the different SAGA localization pattern in muscle and neurons was caused by differences in the composition of the complex in these two cell types, we examined the composition of SAGA in both tissues. Tissue-specific forms of chromatin modifying and remodeling complexes such as TFIID and SWI/SNF have previously been identified (Hiller et al. 2001, 2004; Wu et al. 2007; Yoo et al. 2009). To determine whether tissue-specific forms of SAGA could contribute to its different localization pattern, we affinity-purified SAGA from stage 10–17 embryos expressing the tagged Ada2b subunit in muscle or neurons and analyzed the immunoprecipitated proteins using multidimensional protein identification technology (MudPIT) (Fig. 2A; Supplemental Table 4). An outline of the experimental procedure is provided in Supplemental Figure 3. Flag purifications from wild-type untagged embryos and embryos that lack the GAL4 driver were compared as controls (Fig. 2A). All SAGA subunits were present in purifications from both muscle and neuronal cells, as compared with purifications from cultured Drosophila cells, indicating that Ada2bH1F2 effectively incorporates into the endogenous SAGA complexes in these embryonic tissues and that the composition of the core complex does not differ in these three cell types.
Figure 2.
(A) Heat map showing the relative spectral abundance of SAGA subunits in purifications using antibodies against Flag from Drosophila embryos or tandem Flag-HA purifications from S2 cells, expressed as distributed normalized spectral abundance factor (dNSAF). Purifications were conducted from the following genotypes: OregonR (column 1), UAS-Ada2bH1F2 in the absence of the GAL4 driver (column 2), UAS-Ada2bH1F2 expressed under control of mef2-GAL4 (column 3), and elav-GAL4 drivers (column 4). MudPIT data from a Flag-HA purification of Ada2bH1F2 from S2 cells are shown in column 5 for comparison. The dNSAF scale represents the abundance of each protein on a scale ranging from yellow (highly abundant) to black (less abundant), with white squares representing proteins that were not identified in a given purification. Mean dNSAF values are shown for muscle and neurons representing three biological experiments. (B) Heat map showing the relative spectral abundance of transcription factors associated at substoichiometric levels with SAGA purifications from Drosophila embryos or S2 cells. The dNSAF scale represents the abundance of each protein on a scale ranging from red (more abundant) to black (less abundant), with white squares representing proteins that were not identified in a given purification. Mean dNSAF values are shown for muscle and neurons representing three biological experiments. (C) Venn diagrams showing the overlap between (1) transcription factors associated with SAGA in muscle versus neuron and (2) genes bound by SAGA in muscle versus neuron. (D) Venn diagram showing the overlap between genes that are closest to a defined Mef2-binding site in 10- to 12-h embryos and genes bound by SAGA in muscle. Only those genes with defined expression in muscle of 13- to 16-h embryos were included in this analysis.
The core subunits of SAGA are all present in both muscle and neurons; therefore, we reasoned that the interaction of SAGA with additional proteins could regulate its association with different gene targets in embryonic tissues. Notably, in addition to the 18 core SAGA subunits, we identified a large number of proteins associated with SAGA at substoichiometric levels in muscle, neurons, and cultured S2 cells. These include many proteins that have been identified as having sequence-specific DNA-binding transcription factor activity. Drosophila SAGA has been previously shown to interact with the transcriptional activators VP16 and p53 (Kusch et al. 2003). Thus, we hypothesized that differential interactions of SAGA with transcription factors in embryonic muscle and neurons could direct its association with different target genes. If we compare the putative transcription factors that copurify with SAGA in muscle, relative to neurons and S2 cells (Fig. 2B,C; Supplemental Table 4), we observe that many more transcription factors are associated with SAGA uniquely in muscle relative to neurons. The low number of transcription factors associated with SAGA in neurons is not due to a deficiency in the sensitivity of the SAGA purification in neurons, as a higher number of peptide spectra for both SAGA subunits and total proteins associated with SAGA are observed in the neuronal purifications relative to muscle (Supplemental Fig. 6). The higher number of transcription factors associated with SAGA in muscle correlates with the increased number of genes bound by SAGA specifically in muscle relative to neurons (Fig. 2C). Thus, at this stage of development, SAGA is bound to more genes in muscle than in neurons and was found to interact with more transcription factors in muscle than in neurons.
To examine whether these transcription factors might be responsible for directing the tissue-specific recruitment of SAGA to different genes, we compared the binding sites of the muscle-specific transcription factor Mef2 with SAGA-bound genes in muscle (Fig. 2D). This analysis showed that 49% of genes that are closest to a defined Mef2-binding site in 10–12 embryos and are expressed in muscle of 13- to 16-h embryos also have SAGA bound in muscle (Zinzen et al. 2009). However, only 13% of the genes bound by SAGA in muscle that are expressed in muscle of 13- to 16-h embryos have a nearby Mef2 peak in 10- to 12-h embryos, suggesting that additional muscle-specific transcription factors likely play a role in recruiting SAGA to Mef2-independent genes in muscle of late stage embryos.
SAGA colocalizes with Pol II at both promoters and coding regions
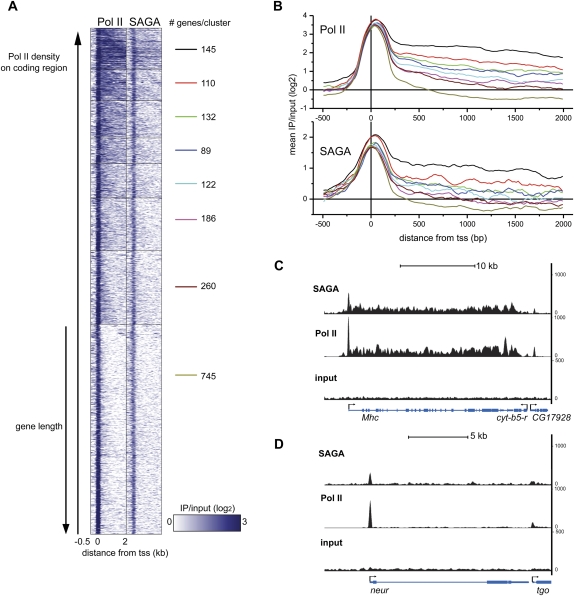
The presence of SAGA is often associated with gene activation in yeast, and acetylation of histone H3 correlates with active transcription in Drosophila (Schubeler et al. 2004). Furthermore, we observe an interaction of SAGA with a large number of different transcription factors in different Drosophila cell types, suggesting that it might be involved in regulating transcription of many different genes. To examine the role of SAGA in Pol II-mediated transcription, we asked where SAGA binds genome-wide relative to Pol II by examining the distribution of Pol II in muscle cells using ChIP-seq. Using the same ranking criteria that we used to identify SAGA-bound genes, we identified 4653 genes that are bound by Pol II in embryonic muscle cells (Supplemental Table 5). If we examine the overlap in the genes that are bound by Pol II and SAGA, we find that 92% of SAGA-bound genes are also bound by Pol II. Many of the remaining SAGA-bound genes that lack detectable Pol II binding are tRNA genes that are transcribed by Pol III.
Although our peak analysis showed that the majority of SAGA is promoter-localized, we found that 14% of SAGA-bound genes in muscle have significant levels of SAGA present on the coding region (score >3) (see the Supplemental Material). To examine whether the presence of SAGA on the coding region correlates with the presence of Pol II, we clustered the genes bound by both Pol II and SAGA based on the density of Pol II present on the coding region of each gene. We then plotted Pol II and SAGA ChIP-seq signals across the promoter and first 2 kb of the coding region for each gene within the cluster (Fig. 3A). If we examine the mean binding density within each cluster (Fig. 3B), we find that SAGA distribution across the gene body correlates with the density of Pol II. Genes that have a high density of Pol II across the coding region, such as Mhc (Fig. 3C), also show high occupancy of SAGA across the coding region (Fig. 3B, black line). Conversely, genes at which Pol II is primarily localized at the promoter, such as neur (Fig. 3D), have SAGA also restricted to this region of the gene (Fig. 3B, dark-yellow line). We conclude from our analysis that SAGA may have a broad role in transcription at both the promoter and coding regions in flies.
Figure 3.
(A) Region map displaying the density of Pol II and SAGA binding in muscle for genes bound by SAGA and Pol II that are longer than 500 bp. SAGA and Pol II binding is plotted from −500 bp to +2000 bp around the transcription start site (columns) as log2 ratios of IP/input. Pol II-bound genes in muscle were clustered into eight groups according to the density of Pol II on the coding region. The number of genes in each cluster is shown to the right of each cluster. Rows (genes) in each cluster were ordered by increasing gene length. (B) The mean Pol II (top panel) and SAGA (bottom panel) ChIP-seq signal intensity was plotted as log2 ratios of IP/input for each of the eight clusters described in A from −500 bp to +2000 bp around the transcription start site (+1). (C,D) Binding profiles for SAGA (top panel) and Pol II (middle panel) relative to input chromatin (bottom panel) are shown for the Mhc (C) and neur (D) loci. The Y-axis represents the number of unique reads observed in two biological experiments for the SAGA and Pol II ChIP-seq experiments.
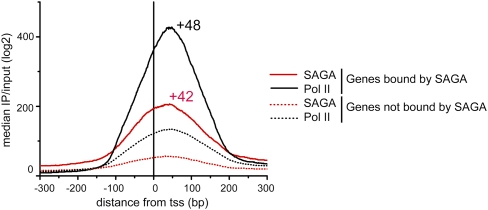
Notably, a peak of SAGA is observed at the promoters of genes that lack detectable levels of Pol II in the gene body, suggesting that SAGA remains bound at genes that are stalled or infrequently transcribed. Consistent with this observation, SAGA localizes near the position of the promoter-proximal pause site, ∼50 nucleotides downstream from the transcription start site, in embryonic muscle cells (Fig. 4). In addition, whereas most SAGA-bound genes were also bound by Pol II, only 40% of Pol II-bound genes show levels of SAGA binding above our threshold criteria. However, much higher levels of Pol II are present at the promoters of SAGA-bound genes when compared with the other 60% of Pol II-bound genes that lack detectable SAGA (Fig. 4). This observation suggests that SAGA might be important for Pol II occupancy at pause sites, even at genes that are not being actively transcribed.
Figure 4.
The median log2 ratio of IP/input was plotted for SAGA (red) and Pol II (black) from −300 bp to +300 bp around the transcription start site at 639 SAGA-bound promoters, relative to 2455 promoters bound by Pol II that lack detectable levels of SAGA binding. The position of the peak summit relative to the transcription start site (+1) is shown above the graph for SAGA and Pol II.
The ubiquitin protease activity of SAGA is important for transcription of muscle-specific developmental genes
Our observation that the presence of SAGA correlated with high levels of Pol II at pause sites indicated that SAGA might have additional functions following recruitment of polymerase in transcription activation. In yeast, studies have shown that ubiquitination of histone H2B occurs post-transcription initiation and is important for the subsequent trimethylation of Lys4 of H3 (Weake and Workman 2008). Thus, we would expect that the deubiquitination activity of SAGA on histone H2B would also occur post-transcription initiation. This model is supported by observations in yeast that deubiquitination by SAGA is important for the release of paused Pol II at GAL1 (Wyce et al. 2007).
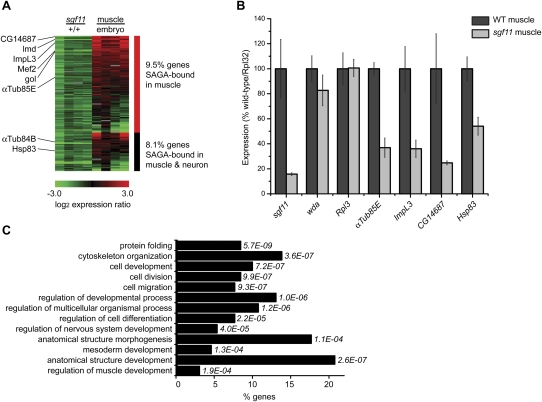
To identify genes that require the ubiquitin protease activity of SAGA for activation of transcription, we examined gene expression in muscle cells from sgf11 embryos that have reduced SAGA ubiquitin protease activity (Weake et al. 2008). Transcript levels in muscle cells from sgf11 embryos isolated using FACS (Supplemental Fig. 2) were compared with wild-type muscle cells using cDNA expression arrays (n = 4). Using this approach, we identified 9.5% of genes bound by SAGA in muscle that are down-regulated twofold or more in sgf11 muscle (Fig. 5A; Supplemental Table 6). In addition, 8.1% of genes bound by SAGA in both muscle and neurons are down-regulated twofold or more in sgf11 muscle (Fig. 5A; Supplemental Table 6). To confirm the changes in expression observed in the sgf11 muscle cells, we examined transcript levels of four of these genes by quantitative RT–PCR (qRT–PCR) in wild-type or sgf11 muscle cells (Fig. 5B). As expected, muscle cells from sgf11 embryos show very low levels of sgf11 transcripts, but do not show decreases in the level of transcripts of another subunit of SAGA, wda, or the ribosomal gene Rpl3. We examined transcript levels of four genes identified as being down-regulated in our microarray analysis and found that all of these genes are indeed down-regulated in sgf11 muscle (Fig. 5B).
Figure 5.
(A) Heat map showing the log2 ratio of expression of SAGA-bound genes that are down-regulated in sgf11 muscle. Expression ratios are plotted for genes (rows) in sgf11 muscle relative to wild-type muscle, and wild-type muscle relative to dissociated cells from whole embryos. Ratios for four separate biological experiments are shown for each comparison. Genes bound by SAGA in muscle only are shown in the top panel (red bar), and genes bound by SAGA in muscle and neurons are shown in the bottom panel (black bar). (B) qPCR was performed on cDNA isolated from wild-type muscle (n = 5) and sgf11 muscle (n = 3) cells. Mean expression levels were normalized to Rpl32 and plotted as percentage wild type. Error bars denote standard error of the mean. (C) GO terms enriched in the set of SAGA-bound genes that are down-regulated in sgf11 muscle. The percentage of down-regulated SAGA-bound genes in each GO category is shown on the X-axis, with P-values shown to the right of each bar. Only a subset of GO terms that show differential enrichment in SAGA-bound sgf11 down-regulated genes are presented. A complete list of enriched GO terms with P-values <0.001 is provided in Supplemental Table 7.
When we compared the expression of SAGA-bound genes down-regulated in sgf11 muscle with the relative expression of those same genes in muscle relative to the whole embryo, we noticed that genes down-regulated in sgf11 muscle tended to be expressed preferentially in muscle relative to the whole embryo (Fig. 5A). Thus, the ubiquitin protease activity of SAGA appears to be important for activating expression of genes that are preferentially expressed in muscle. Although in yeast SAGA-activated genes are often highly regulated and induced in response to environmental stresses (Huisinga and Pugh 2004), we do not find any Gene Ontology (GO) term enrichment in stress response genes when we examine those SAGA-bound genes that are down-regulated in sgf11 muscle (Supplemental Table 7). Instead, when we compare GO term enrichment of SAGA-bound genes that are down-regulated in sgf11 muscle relative to the whole genome, we identify GO terms including cell division and regulation of muscle and mesoderm development as being enriched in this gene population (Fig. 5C). Thus, genes that are especially sensitive to the loss of the ubiquitin protease activity of SAGA tend to be preferentially expressed in muscle cells and have roles in developmental processes. Notably, genes with paused Pol II in early Drosophila embryos also tend to be tissue-specifically expressed and have roles in development (Muse et al. 2007; Zeitlinger et al. 2007).
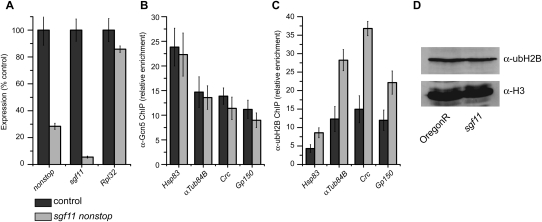
We then asked whether ubH2B levels were increased at individual genes that were down-regulated in sgf11 muscle relative to the whole embryo. To do this, we turned to RNAi in cultured Drosophila cells and examined ubH2B levels at the 5′ end of four genes that are down-regulated in sgf11 muscle (Fig. 6A–C). Knockdown of two components of the ubiquitin protease module of SAGA, sgf11 and not, does not affect recruitment of Gcn5 but does result in increased levels of ubH2B at the 5′ end of these genes (Fig. 6C). However, an increase in global ubH2B levels in sgf11 mutants is not apparent by stage 15 of embryogenesis (Fig. 6D); thus, we cannot exclude the possibility that the ubiquitin protease activity of SAGA may have roles in activation of gene expression that function through targets in addition to ubH2B.
Figure 6.
(A) qPCR was performed on cDNA isolated from S2 cells treated with dsRNA against sgf11 and not or the nonspecific control lacZ. Mean expression levels are plotted as a percentage of the control. Error bars denote standard error of the mean for three biological experiments. (B,C) ChIP was performed on chromatin isolated from S2 cells treated with dsRNA against sgf11 and not or the nonspecific control lacZ using antibodies against Gcn5 (B) or ubH2B (C). ChIP data are shown as mean percent input normalized to an intergenic region, and error bars denote standard error of the mean for three biological experiments. (D) Histones were acid-extracted from stage 15–16 wild-type (OregonR) or sgf11 embryos and analyzed by SDS-PAGE and Western blotting against ubH2B and H3.
Discussion
In this study, we examined SAGA composition and localization in muscle and neuronal cells of late stage Drosophila embryos. Surprisingly, we observed extensive colocalization of SAGA with Pol II at both promoters and coding regions in muscle cells. Notably, genes at which SAGA was not detected in our assay have low levels of Pol II bound. We suggest that SAGA might be important for recruitment and/or retention of high levels of Pol II at the promoter-proximal pause site in flies, and perhaps, therefore, more generally in higher eukaryotes. SAGA has been previously observed on the coding sequence of a small number of individual transcribed genes in yeast (Govind et al. 2007; Wyce et al. 2007; Zapater et al. 2007). Recently, low levels of Ada2b were detected on the 3′ region of several different genes during larval development (Zsindely et al. 2009). We note that although some SAGA is present across the coding region of many genes, the peak of acetylated H3-Lys9 is restricted to the 5′ region of the two genes that we examined: wupA and exba. A similar 5′ bias of acetylated H3-Lys9 has been observed previously in genome-wide studies of histone modifications (Li et al. 2007; Roy et al. 2010). We speculate that the acetylation activity of SAGA in the 3′ region of the gene is counteracted by histone deacetylases such as Rpd3S that have been shown to associate with the elongating form of Pol II (Carrozza et al. 2005; Govind et al. 2010).
SAGA localizes to different genes in muscle and neurons of late stage Drosophila embryos, and the number of genes bound by the complex in each tissue correlates with the number of transcription factors associated with the complex. These findings indicate that the differential localization of SAGA may be regulated by its association with different transcription factors in different cell types. A number of studies have found that transcription factor-binding sites tend to be clustered within the fly genome (Moorman et al. 2006; Zinzen et al. 2009). This observed colocalization of transcription factors, together with our data showing the association of SAGA with a large number of different transcription factors, indicates that multiple transcription factors might be involved in recruiting SAGA to its target genes.
We observe that SAGA is present at the promoter-proximal pause site together with Pol II at genes that are stalled or infrequently transcribed. The presence of SAGA together with paused Pol II is consistent with a role for SAGA in post-initiation deubiquitination of H2B, which has been shown in yeast to be important for phosphorylation of Ser-2 of the Pol II CTD and its subsequent transition into transcription elongation (Wyce et al. 2007). In flies, phosphorylation of Ser-2 of the Pol II CTD by P-TEFb is also required for release of the paused polymerase into transcription elongation (Lis et al. 2000; Ni et al. 2008). Hence, the strong colocalization of SAGA with polymerase that has initiated transcription but is paused prior to elongation suggests a prominent function for SAGA in regulating tissue-specific gene expression at a step occurring post-initiation in metazoans. Consistent with the possibility, we observe that the SAGA-bound genes that are most dependent on its ubiquitin protease activity for full expression are preferentially expressed in a specific tissue.
Materials and methods
Additional details of methods are provided in the Supplemental Material.
FACS isolation of cells for RNA and ChIP analysis
Stage 15–16 embryos expressing GFP and Ada2bH1F2 under the control of mef2-GAL4 or elav-GAL4 were dissociated, and cells expressing the tagged protein and GFP were enriched using FACS on a MoFlo high-speed sorter (Beckman Coulter). For RNA experiments, cells were labeled with GFP but did not express Ada2bH1F2. For ChIP, chromatin was cross-linked with 1% formaldehyde prior to FACS isolation.
ChIP
ChIP was conducted on 1 × 107 cell equivalent of chromatin as described previously (Weake et al. 2009) with minor modifications. The following antibodies were used for ChIP: anti-Flag M2 (mouse; F1804, Sigma), anti-Pol II phospho/unphospho-CTD (mouse; 4H8; ab5408, Abcam), anti-acetylated H3-Lys9 (rabbit; 06-942, Upstate Biotechnology), anti-ubH2B (mouse; 05-1312, Millipore), anti-Gcn5 (rabbit) (Kusch et al. 2003). Primer sequences are provided in Supplemental Table 8. Three biological replicates were performed for all ChIP-qPCR analyses.
ChIP analysis
Two separate biological experiments were performed for each ChIP-seq analysis with similar results. Unique reads identified from each biological experiment were merged for further analysis. A single chromatin input sample was sequenced from each cell type as a control experiment. To determine the level of nonspecific antibody binding, a single Flag-ChIP-seq experiment was conducted using chromatin isolated from neuronal cells that do not express a Flag-tagged protein. Preliminary analysis of SAGA binding in muscle cells was performed using model-based analysis of ChIP-seq (MACS) (Y Zhang et al. 2008) with a P-value cutoff of 1 × 10−10. Peaks were mapped to the 5′ UTR and coding region of genes using Galaxy (Goecks et al. 2010). Analysis of SAGA and Pol II distribution on the promoter and coding regions of genes was performed using a modified version of reads per kilobase of gene region (RPKM) analysis (Mortazavi et al. 2008), whereby defined features could be assigned enrichment scores for immunoprecipitation and input samples.
cDNA expression analysis
Two-class unpaired significance analysis of microarrays (SAM) was performed to identify genes expressed preferentially in muscle or neurons using Δ value of 0.811 (false discovery rate percent median <5) (Tusher et al. 2001). ImaGO terms describing the expression pattern of individual genes within the embryo using a controlled vocabulary set (Tomancak et al. 2002) were obtained from FlyMine. Down-regulated genes in sgf11 muscle were identified as being those genes with a mean log2 ratio of −1.0 and valid expression ratios in three of the four cDNA arrays in sgf11 muscle relative to wild-type muscle. GO term enrichment analysis was performed using FuncAssociate 2.0 (Berriz et al. 2009).
Affinity purification and MudPIT analysis
Soluble nuclear extracts were prepared and Flag affinity purification and MudPIT analysis were conducted using modified versions of the protocols described for preparation of nuclear extracts from Drosophila S2 cells (Weake et al. 2009). Sequence-specific transcription factors (GO: 0003700) were identified using the GO term definitions provided in FlyBase.
Accession codes
The RAW mass spectrometry and SEQUEST results files for the MudPIT analyses reported in Supplemental Table 4 may be downloaded from ProteomeCommons.org Tranche using the following hash: u3ZMmQTbFnzAYL1frZrXhwMbVFnRDH2AxY/X9dsD09/8wrFWch5tuIleUqy6b4jwjIEAl5pVT6J/FAsF76uhKe2YfgMAAAAAAAAQHg==. These data may be accessed using the passphrase Weake&SAGA. The expression and ChIP-seq data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (Edgar et al. 2002) and are accessible through GEO series accession number GSE29528 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE29528).
Acknowledgments
We thank Bjoern Gaertner and Julia Zeitlinger for assistance in developing the FACS-ChIP protocol. We also thank Madelaine Gogol, Ariel Paulson, and Alexander Garrett for bioinformatics; Kendra Walton, Keith Smith, Anoja Perera, and Karen Staehling for the running of the ChIP-seq samples; and members of the Workman laboratory for helpful comments and suggestions. We thank Mattias Mannervik for providing the ada2b1 allele. Bloomington Drosophila Stock Center resources and information from FlyBase and FlyMine were used in this study. This work was supported by grant R37GM047867-18S1 from the NIGMS to J.L.W. and S.M.A., and funding from the Stowers Institute.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.2046211.
References
- Baker SP, Grant PA 2007. The SAGA continues: expanding the cellular role of a transcriptional co-activator complex. Oncogene 26: 5329–5340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger C, Renner S, Luer K, Technau GM 2007. The commonly used marker ELAV is transiently expressed in neuroblasts and glial cells in the Drosophila embryonic CNS. Dev Dyn 236: 3562–3568 [DOI] [PubMed] [Google Scholar]
- Berriz GF, Beaver JE, Cenik C, Tasan M, Roth FP 2009. Next generation software for functional trend analysis. Bioinformatics 25: 3043–3044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand AH, Perrimon N 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415 [DOI] [PubMed] [Google Scholar]
- Bu P, Evrard YA, Lozano G, Dent SY 2007. Loss of Gcn5 acetyltransferase activity leads to neural tube closure defects and exencephaly in mouse embryos. Mol Cell Biol 27: 3405–3416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos AR, Rosen DR, Robinow SN, White K 1987. Molecular analysis of the locus elav in Drosophila melanogaster: a gene whose embryonic expression is neural specific. EMBO J 6: 425–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, et al. 2005. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123: 581–592 [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30: 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott DA, Brand AH 2008. The GAL4 system: a versatile system for the expression of genes. Methods Mol Biol 420: 79–95 [DOI] [PubMed] [Google Scholar]
- Goecks J, Nekrutenko A, Taylor J 2010. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol 11: R86 doi: 10.1186/gb-2010-11-8-r86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govind CK, Zhang F, Qiu H, Hofmeyer K, Hinnebusch AG 2007. Gcn5 promotes acetylation, eviction, and methylation of nucleosomes in transcribed coding regions. Mol Cell 25: 31–42 [DOI] [PubMed] [Google Scholar]
- Govind CK, Qiu H, Ginsburg DS, Ruan C, Hofmeyer K, Hu C, Swaminathan V, Workman JL, Li B, Hinnebusch AG 2010. Phosphorylated Pol II CTD recruits multiple HDACs, including Rpd3C(S), for methylation-dependent deacetylation of ORF nucleosomes. Mol Cell 39: 234–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry KW, Wyce A, Lo WS, Duggan LJ, Emre NC, Kao CF, Pillus L, Shilatifard A, Osley MA, Berger SL 2003. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev 17: 2648–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller MA, Lin TY, Wood C, Fuller MT 2001. Developmental regulation of transcription by a tissue-specific TAF homolog. Genes Dev 15: 1021–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller M, Chen X, Pringle MJ, Suchorolski M, Sancak Y, Viswanathan S, Bolival B, Lin TY, Marino S, Fuller MT 2004. Testis-specific TAF homologs collaborate to control a tissue-specific transcription program. Development 131: 5297–5308 [DOI] [PubMed] [Google Scholar]
- Howe L, Auston D, Grant P, John S, Cook RG, Workman JL, Pillus L 2001. Histone H3 specific acetyltransferases are essential for cell cycle progression. Genes Dev 15: 3144–3154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisinga KL, Pugh BF 2004. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol Cell 13: 573–585 [DOI] [PubMed] [Google Scholar]
- Koutelou E, Hirsch CL, Dent SY 2010. Multiple faces of the SAGA complex. Curr Opin Cell Biol 22: 374–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusch T, Guelman S, Abmayr SM, Workman JL 2003. Two Drosophila Ada2 homologues function in different multiprotein complexes. Mol Cell Biol 23: 3305–3319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL 2007. The role of chromatin during transcription. Cell 128: 707–719 [DOI] [PubMed] [Google Scholar]
- Lilly B, Galewsky S, Firulli AB, Schulz RA, Olson EN 1994. D-MEF2: a MADS box transcription factor expressed in differentiating mesoderm and muscle cell lineages during Drosophila embryogenesis. Proc Natl Acad Sci 91: 5662–5666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W, Srajer G, Evrard YA, Phan HM, Furuta Y, Dent SY 2007. Developmental potential of Gcn5−/− embryonic stem cells in vivo and in vitro. Dev Dyn 236: 1547–1557 [DOI] [PubMed] [Google Scholar]
- Lis JT, Mason P, Peng J, Price DH, Werner J 2000. P-TEFb kinase recruitment and function at heat shock loci. Genes Dev 14: 792–803 [PMC free article] [PubMed] [Google Scholar]
- Luo L, Liao YJ, Jan LY, Jan YN 1994. Distinct morphogenetic functions of similar small GTPases: Drosophila Drac1 is involved in axonal outgrowth and myoblast fusion. Genes Dev 8: 1787–1802 [DOI] [PubMed] [Google Scholar]
- Moorman C, Sun LV, Wang J, de Wit E, Talhout W, Ward LD, Greil F, Lu XJ, White KP, Bussemaker HJ, et al. 2006. Hotspots of transcription factor colocalization in the genome of Drosophila melanogaster. Proc Natl Acad Sci 103: 12027–12032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628 [DOI] [PubMed] [Google Scholar]
- Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K 2007. RNA polymerase is poised for activation across the genome. Nat Genet 39: 1507–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen HT, Bodmer R, Abmayr SM, McDermott JC, Spoerel NA 1994. D-mef2: a Drosophila mesoderm-specific MADS box-containing gene with a biphasic expression profile during embryogenesis. Proc Natl Acad Sci 91: 7520–7524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Z, Saunders A, Fuda NJ, Yao J, Suarez JR, Webb WW, Lis JT 2008. P-TEFb is critical for the maturation of RNA polymerase II into productive elongation in vivo. Mol Cell Biol 28: 1161–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi D, Larsson J, Mannervik M 2004. Drosophila Ada2b is required for viability and normal histone H3 acetylation. Mol Cell Biol 24: 8080–8089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranganayakulu G, Schulz RA, Olson EN 1996. Wingless signaling induces nautilus expression in the ventral mesoderm of the Drosophila embryo. Dev Biol 176: 143–148 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Navarro S 2009. Insights into SAGA function during gene expression. EMBO Rep 10: 843–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, Ernst J, Kharchenko PV, Kheradpour P, Negre N, Eaton ML, Landolin JM, Bristow CA, Ma L, Lin MF, et al. 2010. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science 330: 1787–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubeler D, MacAlpine DM, Scalzo D, Wirbelauer C, Kooperberg C, van Leeuwen F, Gottschling DE, O'Neill LP, Turner BM, Delrow J, et al. 2004. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev 18: 1263–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomancak P, Beaton A, Weiszmann R, Kwan E, Shu S, Lewis SE, Richards S, Ashburner M, Hartenstein V, Celniker SE et al. 2002. Systematic determination of patterns of gene expression during Drosophila embryogenesis. Genome Biol 3: research0088.1–research0088.14 doi: 10.18186/gb-2002-3-12-research0088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci 98: 5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weake VM, Workman JL 2008. Histone ubiquitination: triggering gene activity. Mol Cell 29: 653–663 [DOI] [PubMed] [Google Scholar]
- Weake VM, Lee KK, Guelman S, Lin CH, Seidel C, Abmayr SM, Workman JL 2008. SAGA-mediated H2B deubiquitination controls the development of neuronal connectivity in the Drosophila visual system. EMBO J 27: 394–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weake VM, Swanson SK, Mushegian A, Florens L, Washburn MP, Abmayr SM, Workman JL 2009. A novel histone fold domain-containing protein that replaces TAF6 in Drosophila SAGA is required for SAGA-dependent gene expression. Genes & Dev 23: 2818–2823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JI, Lessard J, Olave IA, Qiu Z, Ghosh A, Graef IA, Crabtree GR 2007. Regulation of dendritic development by neuron-specific chromatin remodeling complexes. Neuron 56: 94–108 [DOI] [PubMed] [Google Scholar]
- Wyce A, Xiao T, Whelan KA, Kosman C, Walter W, Eick D, Hughes TR, Krogan NJ, Strahl BD, Berger SL 2007. H2B ubiquitylation acts as a barrier to Ctk1 nucleosomal recruitment prior to removal by Ubp8 within a SAGA-related complex. Mol Cell 27: 275–288 [DOI] [PubMed] [Google Scholar]
- Xu W, Edmondson DG, Evrard YA, Wakamiya M, Behringer RR, Roth SY 2000. Loss of Gcn5l2 leads to increased apoptosis and mesodermal defects during mouse development. Nat Genet 26: 229–232 [DOI] [PubMed] [Google Scholar]
- Yoo AS, Staahl BT, Chen L, Crabtree GR 2009. MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature 460: 642–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapater M, Sohrmann M, Peter M, Posas F, de Nadal E 2007. Selective requirement for SAGA in Hog1-mediated gene expression depending on the severity of the external osmostress conditions. Mol Cell Biol 27: 3900–3910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlinger J, Stark A, Kellis M, Hong JW, Nechaev S, Adelman K, Levine M, Young RA 2007. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet 39: 1512–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Guo C, Chen Y, Shulha HP, Schnetz MP, LaFramboise T, Bartels CF, Markowitz S, Weng Z, Scacheri PC, et al. 2008. Epitope tagging of endogenous proteins for genome-wide ChIP–chip studies. Nat Methods 5: 163–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based analysis of ChIP-seq (MACS). Genome Biol 9: R137 doi: 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinzen RP, Girardot C, Gagneur J, Braun M, Furlong EE 2009. Combinatorial binding predicts spatio-temporal cis-regulatory activity. Nature 462: 65–70 [DOI] [PubMed] [Google Scholar]
- Zsindely N, Pankotai T, Ujfaludi Z, Lakatos D, Komonyi O, Bodai L, Tora L, Boros IM 2009. The loss of histone H3 lysine 9 acetylation due to dSAGA-specific dAda2b mutation influences the expression of only a small subset of genes. Nucleic Acids Res 37: 6665–6680 [DOI] [PMC free article] [PubMed] [Google Scholar]