Abstract
Under “permissive” conditions at 25°C, the chaperonin substrate protein DM-MBP refolds 5–10 times more rapidly in the GroEL/GroES folding chamber than in free solution. This has been suggested to indicate that the chaperonin accelerates polypeptide folding by entropic effects of close confinement. Here, using native-purified DM-MBP, we show that the different rates of refolding are due to reversible aggregation of DM-MBP while folding free in solution, slowing its kinetics of renaturation: the protein exhibited concentration-dependent refolding in solution, with aggregation directly observed by dynamic light scattering. When refolded in chloride-free buffer, however, dynamic light scattering was eliminated, refolding became concentration-independent, and the rate of refolding became the same as that in GroEL/GroES. The GroEL/GroES chamber thus appears to function passively toward DM-MBP.
Keywords: DM-MBP, MBP, GroEL, GroES, aggregation
1. Introduction
The GroES-encapsulated ring of the chaperonin GroEL provides a folding chamber that is essential for proper folding of many polypeptide substrates under what are called “nonpermissive conditons”, where there is relatively high substrate protein concentration and elevated temperature, resembling physiologic conditions [1–4]. Under such conditions, polypeptide substrates cannot reach the native state in free solution, being subject to wholesale aggregation. Yet under conditions of reduced substrate concentration and lower temperature, so-called “permissive” conditions which disfavor aggregation in free solution, many of these proteins can reach the native form either inside the chaperonin chamber or free in solution [2,5,6]. In some cases under permissive conditions, the rate of folding to native form by the chaperonin system is many fold greater than in free solution [5,7]. This has been suggested to reflect an ability of the chaperonin chamber to intrinsically accelerate the rate of folding of encapsulated substrate proteins, e.g. by entropic effects of close confinement [7,8]. On the other hand, certain experiments have shown that the difference in rates is due to reversible off-pathway steps of aggregation of substrate proteins attempting to fold in free solution [9].
A substrate for which considerable testing has been carried out under permissive conditions is a secretion signal-deleted (cytosolic) double mutant form of maltose binding protein (DM-MBP; V8G/Y283D; 41 kDa; e.g. refs. 7–9). Most recently, it has been reported that this protein, expressed at low temperature in E. coli and purified in native form using amylose affinity resin, exhibited no evidence of aggregation in free solution following its dilution from guanidine-HCl [8]. This is in contrast to earlier studies observing aggregation of inclusion body-purified DM-MBP after dilution from guanidine-HCl into free solution [9]. Here we report refolding studies using low temperature-expressed, native-purified, DM-MBP, as in ref.8.
2. Materials and Methods
2.1. Proteins
GroEL and GroES were expressed and purified as described in Rye et al [10]. The mature forms of wild-type MBP and DM-MBP (V8G/Y283D) were expressed as soluble proteins in E. coli grown at 25°C and purified on amylose resin (New England Biolabs) as described previously [7,8].
2.2. MBP refolding
For refolding free in solution containing chloride, DM-MBP or wild-type MBP was diluted 100-fold from a solution containing 6 M guanidine-HCl, 100 mM Tris-HCl (pH 7.5), 20 mM KCl, into buffer containing 20 mM Tris-HCl (pH 7.5), 200 mM KCl, 5 mM Mg(OAc)2 at 25°C. For refolding under chloride-free conditions, DM-MBP was diluted from a 9.5 M urea solution into buffer containing 100 mM Hepes (pH 7.5), 20 mM KOAc, 5 mM Mg(OAc)2. For chaperonin-mediated refolding, DM-MBP unfolded in 6 M guanidine-HCl or 9.5 M urea was diluted 100-fold into buffer with or without chloride, respectively, containing a 2.5-fold molar excess of GroEL over the final DM-MBP concentration to form binary complexes. GroES in a two-fold molar excess to GroEL and 5 mM ATP were subsequently added to commence folding.
2.3. Assays
Refolding of MBP and DM-MBP was followed by an increase of tryptophan fluorescence (excitation, 295 nm; emission, 345 nm) using a PTI QuantaMaster fluorescence spectrometer. Rates of refolding of MBP were determined by fitting the change in fluorescence to a single exponential equation using Origin. Results from at least three separate experiments were averaged for each data point. Light scattering experiments were carried out on a DynaPro dynamic light scattering instrument (Wyatt Technology; 830 nm light, 90° scattering angle), using buffer conditions identical to those used for refolding.
3. Results and Discussion
Concern about the purity of the inclusion body-isolated DM-MBP used in our previous experiments relative to the native-isolated DM-MBP employed in the recently reported experiments [8] led us to compare the renaturation of the two preparations in free solution. We observed that, at 1 μM protein concentration, the inclusion body preparation of DM-MBP refolded more slowly, at ~50% the rate of the native-isolated DM-MBP. It seemed likely that the slower refolding of the inclusion body-prepared protein was due to contaminants, although no major protein contaminants were apparent in SDS gel analysis (Suppl. Fig. 1) or in gel filtration analysis. In addition, Q-Tof MS analyses of the two protein preparations were identical (not shown).
Nevertheless we worried that lipid or carbohydrate contamination, not detectable with these analytical methods, might be involved. Therefore, we further purified the inclusion body material via anion exchange chromatography in urea, followed by dialysis/renaturation. The rate of refolding of this purified material at 1 μM after dilution from guanidine-HCl was identical to that of the native-isolated DM-MBP (not shown). For ease of comparison, however, in all subsequent experiments we employed native-isolated material prepared exactly as described by Chakraborty et al [8].
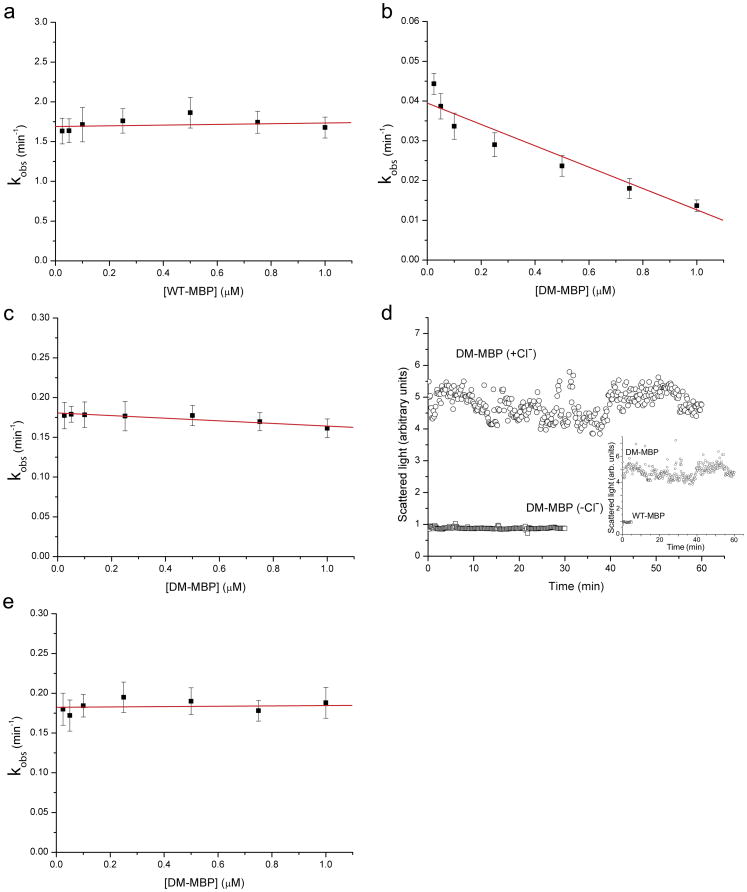
First we compared the rate of refolding of native-isolated wild-type MBP diluted from guanidine-HCl denaturant with that of identically treated native-isolated DM-MBP. The rate of refolding of the wild-type protein was approximately 50-fold greater than that of the mutant protein (Fig. 1, compare panels a and b), in agreement with earlier studies. Whereas the rate of refolding of the wild-type protein did not exhibit a concentration dependence across the range of 25 nM to 1 μM, the rate for the mutant protein exhibited a concentration-dependent decrease of approximately 3-fold across this range, with a steeper decrease at the lower concentrations. This compares with a roughly 5-fold decrease in rate across the same concentration range using inclusion body-purified material [9], suggesting that contaminants in the inclusion body preparation might have increased the aggregation of that protein. (Note, however, that in all cases there is >95% recovery of input material in the native form, consistent with any appreciable aggregation being reversible.)
Fig. 1. Folding rates and light scattering behavior of DM-MBP and wt-MBP.
a) Rate of refolding of wt-MBP following dilution from guanidine-HCl as a function of final concentration of DM-MBP in the buffer. Tryptophan fluorescence was monitored and the single exponential rise in fluorescence was fit as described in Methods to derive the respective rate constants. There is no appreciable effect of concentration across the range of 25 nM to 1 μM wt-MBP. Error bars represent the standard deviations (n=3).
b) Rate of refolding of DM-MBP following dilution from guanidine-HCl denaturant as a function of final concentration of DM-MBP in the buffer. Rates were determined as described in a) (n=3). The line is illustrative only. There is a 3-fold fall in rate across the same range as tested in a)
c) Rate of refolding of DM-MBP diluted from urea into chloride-free buffer (see Methods) as a function of final concentration of DM-MBP in the buffer. The same concentration range was tested as in a) and b) (n=3). There is a minimal effect of concentration on the rate of refolding (~10% decrease)
d) Dynamic light scattering, in arbitrary units, during refolding of 250 nM DM-MBP in solution, DM-MBP (+Cl−). In contrast, DM-MBP refolding in chloride-free solution did not produce significant light scattering, DM-MBP (−Cl−). Inset, for comparison, shows light scattering when both DM-MBP and wt-MBP are in chloride-containing buffer. Raw light-scattering intensity data, in arbitrary units, is plotted vs time
e) Rate of GroEL-GroES-ATP mediated refolding of DM-MBP as a function of concentration of DM-MBP following dilution from guanidine-HCl into buffer containing GroEL, where a binary complex of DM-MBP/GroEL is formed. GroES and ATP were then added and the refolding of DM-MBP monitored by tryptophan fluorescence (note that GroEL and GroES are devoid of tryptophan). The rate constants were determined as for the protein refolding in free solution (n=3). No appreciable concentration dependence was observed, and the rate corresponded closely to that for DM-MBP refolding in chloride-free buffer (compare with panel c).
In contrast with these results in chloride-containing buffer, when chloride was omitted from the buffer, we now observed an increase in the rate of renaturation (for example, at 25 nM, 0.045 min−1 in chloride buffer to 0.175 min−1 in chloride-free buffer; compare panels 1b and 1c). In addition, there was no appreciable concentration dependence to the refolding of DM-MBP in chloride-free buffer (Fig. 1c). This agrees with the earlier conclusion from the experiments with inclusion-body material that absence of chloride prevents aggregation. This conclusion was directly supported here by light scattering measurements carried out with 250 nM DM-MBP (or with 100 nM, not shown). In chloride-containing buffer, light scattering was observed immediately upon dilution from denaturant, whereas in chloride-free buffer, no significant light scattering was observed (Fig. 1d). The absence of light scattering in chloride-free buffer resembled the behavior of wt-MBP (see Fig. 1d inset). Consistent with the effect of omission of chloride to eliminate reversible off-pathway aggregation steps, the rate of refolding of DM-MBP observed in the chloride-free condition was virtually identical to the rate of GroEL-GroES-ATP-dependent refolding of DM-MBP, 0.18 min−1 (Fig. 1e). The chaperonin reaction was carried out here in the presence of chloride, but the same rate constant is obtained for the chaperonin reaction in chloride-free buffer (not shown).
Thus, these tests further demonstrate that DM-MBP, whether inclusion body-purified or purified as a native protein, is subject to reversible aggregation when attempting to refold in free solution, and this acts to slow the overall rate of recovery of the native state. Notably, such reversible aggregation, with attendant kinetic effects, has been observed for a number of other proteins (e.g. 11–13). By contrast, and for reasons unknown, when chloride is removed from the buffer, the aggregation of DM-MBP is abolished (see the light scattering data in panel d), and this brings the rate constant for refolding free in solution precisely to the rate of renaturation by the chaperonin system (Fig. 1e), where a protein folds in an isolated chamber where it cannot aggregate (see Fig. 13 in ref.14). This supports the conclusion that, at least in the case of DM-MBP, the chaperonin system does not have an intrinsic ability to accelerate the rate of folding of this encapsulated substrate. This suggests that the folding chamber behaves essentially as an infinite dilution chamber. This has been directly supported in recent single molecule FRET studies of Hofmann and coworkers with the substrate protein rhodanese, where its rate of recovery of the native state in solution under infinite dilution conditions was observed to be the same as that inside the chaperonin chamber [15]. These studies of DM-MBP and of rhodanese thus support that the GroEL/GroES chamber serves principally as a passive chamber that prevents aggregation, an “Anfinsen cage” [16].
Supplementary Material
Acknowledgments
A.L.H. and N.K.T. thank Howard Hughes Medical Institute and A.A.D. thanks NIH (GM066833) for support of this work. We are also grateful to members of the Horwich lab for helpful suggestions.
Abbreviations
- DM-MBP
double mutant of maltose binding protein
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Goloubinoff P, Christeller JT, Gatenby AA, Lorimer GH. Reconstitution of active dimeric ribulose bisphosphate carboxylase from an unfolded state depends on two chaperonin proteins and MgATP. Nature. 1989;342:884–889. doi: 10.1038/342884a0. [DOI] [PubMed] [Google Scholar]
- 2.Schmidt M, Buchner J, Todd MJ, Lorimer GH, Viitanen PV. On the role of GroES in the chaperonin-assisted folding reaction. Three case studies J Biol Chem. 1994;269:10304–10311. [PubMed] [Google Scholar]
- 3.Walter S, Lorimer GH, Schmid FX. A thermodynamic coupling mechanism 0for GroEL-mediated unfolding. Proc Natl Acad Sci USA. 1996;93:9425–9430. doi: 10.1073/pnas.93.18.9425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weissman JS, Rye HS, Fenton WA, Beechem JM, Horwich AL. Characterization of the active intermediate of a GroEL-GroES-mediated protein folding reaction. Cell. 1996;84:481–490. doi: 10.1016/s0092-8674(00)81293-3. [DOI] [PubMed] [Google Scholar]
- 5.Viitanen PV, Lubben TH, Reed J, Goloubinoff P, O’Keefe DP, Lorimer GH. Chaperonin-facilitated refolding of ribulose bisphosphate carboxylase and ATP hydrolysis by chaperonin 60 (groEL) are K+ dependent. Biochemistry. 1990;29:5665–5671. doi: 10.1021/bi00476a003. [DOI] [PubMed] [Google Scholar]
- 6.Buchner J, Schmidt M, Fuchs M, Jaenicke R, Rudolph R, Schmid FX, Kiefhaber T. GroE facilitates refolding of citrate synthase by suppressing aggregation. Biochemistry. 1991;30:1586–1591. doi: 10.1021/bi00220a020. [DOI] [PubMed] [Google Scholar]
- 7.Tang YC, Chang HC, Roeben A, Wischnewski D, Wischnewski N, Kerner MJ, Hartl FU, Hayer-Hartl M. Structural features of the GroEL-GroES nano-cage required for rapid folding of encapsulated protein. Cell. 2006;125:903–914. doi: 10.1016/j.cell.2006.04.027. [DOI] [PubMed] [Google Scholar]
- 8.Chakraborty K, Chatila M, Sinha J, Shi Q, Poschner BC, Sikor M, Jiang G, Lamb DC, Hartl FU, Hayer-Hartl M. Chaperonin-catalyzed rescue of kinetically trapped states in protein folding. Cell. 2010;142:112–122. doi: 10.1016/j.cell.2010.05.027. [DOI] [PubMed] [Google Scholar]
- 9.Apetri AC, Horwich AL. Chaperonin chamber accelerates protein folding through passive action of preventing aggregation. Proc Natl Acad Sci USA. 2008;105:17351–17355. doi: 10.1073/pnas.0809794105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rye HS, Roseman AM, Furtak K, Fenton WA, Saibil HR, Horwich AL. GroEL-GroES cycling: ATP and non-native polypeptide direct alternation of folding-active rings. Cell. 1999;97:325–338. doi: 10.1016/s0092-8674(00)80742-4. [DOI] [PubMed] [Google Scholar]
- 11.Silow M, Oliveberg M. Transient aggregates in protein folding are easily mistaken for folding intermediates. Proc Natl Acad Sci USA. 1997;94:6084–6086. doi: 10.1073/pnas.94.12.6084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Otzen DE, Miron S, Akke M, Oliveberg M. Transient aggregation and stable dimerization induced by introducing an Alzheimer sequence into a water-soluble protein. Biochemistry. 2004;43:12964–12978. doi: 10.1021/bi048509k. [DOI] [PubMed] [Google Scholar]
- 13.Finke JM, Jennings PA. Early aggregated states in the folding of interleukin-1β. J Biol Phys. 2001;27:119–131. doi: 10.1023/A:1013178505077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horwich AL, Fenton WA. Chaperonin-mediated protein folding: using a central cavity to kinetically assist polypeptide chain folding. Q Rev Biophysics. 2009;42:83–116. doi: 10.1017/S0033583509004764. [DOI] [PubMed] [Google Scholar]
- 15.Hofmann H, Hillger F, Pfeil SH, Hoffmann A, Streich D, Haenni D, Nettels D, Lipman EA, Schuler B. Single-molecule spectroscopy of protein folding in a chaperonin cage. Proc Natl Acad Sci USA. 2010;107:11793–11798. doi: 10.1073/pnas.1002356107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saibil HR, Zheng D, Roseman AM, Hunter AS, Watson GM, Chen S, Auf der Mauer A, O’Hara BP, Wood SP, Mann NH, Barnett LK, Ellis RJ. ATP induces large quaternary rearrangements in a cage-like chaperonin structure. Current Biology. 1993;3:265–273. doi: 10.1016/0960-9822(93)90176-o. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.