Abstract
Background
Transforming growth factor beta (TGF-β) plays an essential role in a wide array of cellular processes. The most well studied TGF-β response in normal epithelial cells is growth inhibition. In some cell types, TGF-β induces an epithelial to mesenchymal transition (EMT). NMuMG is a nontransformed mouse mammary gland epithelial cell line that exhibits both a growth inhibitory response and an EMT response to TGF-β, rendering NMuMG cells a good model system for studying these TGF-β effects.
Method
A National Institutes of Aging mouse 15,000 cDNA microarray was used to profile the gene expression of NMuMG cells treated with TGF-β1 for 1, 6, or 24 hours. Data analyses were performed using GenePixPro and GeneSpring software. Selected microarray results were verified by northern analyses.
Results
Of the 15,000 genes examined by microarray, 939 were upregulated or downregulated by TGF-β. This represents approximately 10% of the genes examined, minus redundancy. Seven genes previously not known to be regulated by TGF-β at the transcriptional level (Akt and RhoB) or not at all (IQGAP1, mCalpain, actinin α3, Ikki, PP2A-PR53), were identified and their regulation by TGF-β verified by northern blotting. Cell cycle pathway examination demonstrated downregulation of cyclin D2, c-myc, Id2, p107, E2F5, cyclin A, cyclin B, and cyclin H. Examination of cell adhesion-related genes revealed upregulation of c-Jun, α-actinin, actin, myosin light chain, p120cas catenin (Catns), α-integrin, integrin β5, fibronectin, IQGAP1, and mCalpain.
Conclusion
Using a cDNA microarray to examine TGF-β-regulated gene expression in NMuMG cells, we have shown regulation of multiple genes that play important roles in cell cycle control and EMT. In addition, we have identified several novel TGF-β-regulated genes that may mediate previously unknown TGF-β functions.
Keywords: adhesion, cDNA microarray, cell cycle control, epithelial, mammary gland
Introduction
Transforming growth factor beta (TGF-β) is the prototype of a large family of signaling molecules with more than 40 members. TGF-β signals through the type I receptor (TβRI) and the type II receptor (TβRII), and the Smad transcriptional regulators, as well as many other signaling pathways [1]. TGF-β plays an essential role in a wide array of cellular processes, including early embryonic development, cell growth, differentiation, motility, and apoptosis. In nontransformed epithelial cells, the most studied TGF-β response is growth inhibition. In some cell types, TGF-β induces an epithelial to mesenchymal transition (EMT). TGF-β plays an important, albeit complex, role in tumorigenesis. In many tumors of epithelial origin, cells become resistant to TGF-β-mediated growth inhibition. In parallel, TGF-β induces EMT in tumor cells, and increases tumor invasion and metastasis [2]. NMuMG is a nontransformed mouse mammary epithelial cell line that exhibits both a growth inhibitory and EMT response to TGF-β [3]. These properties render NMuMG cells a suitable model system for studying these TGF-β actions.
Previous gene expression profiling studies of TGF-β action on cell lines utilized cDNA microarrays to characterize extracellular matrix-related genes in human dermal fibroblasts. The upregulation of these genes by TGF-β was Smad mediated [4]. Another study examining the genetic programs of epithelial cell plasticity directed by TGF-β utilized the human keratinocyte HaCaT cell line [5]. In this study, HaCaT cells were treated for up to 4 hours, and microarray gene expression profiling was performed using an array of 16,580 human cDNAs. The expression of 728 known genes (10% of those analyzed) was regulated by TGF-β within 4 hours of treatment.
In the present study, we examined TGF-β regulation of gene expression in the NMuMG epithelial cell line. NMuMG cells were chosen as a model system because the results are more relevant to our studies employing transgenic mouse mammary tumor models, and because NMuMG cells are a well-established model system for studying both growth inhibition and EMT induced by TGF-β [3]. EMT represents a complex physiological process that includes dissolution of adherens junctions, a change to spindle-like cell morphology, cytoskeletal reorganization, increased cell motility, loss of epithelial markers, and induction of mesenchymal markers. EMT thus includes several cellular responses that may occur through different mechanisms and at different time points after TGF-β treatment. We consequently examined changes in gene expression at 1, 6, and 24 hours after TGF-β treatment.
In agreement with previous studies with HaCaT cells [5], approximately 10% of the genes examined were regulated by TGF-β in our microarray studies that queried more than 15,000 mouse cDNAs. Our study demonstrates that by 1 hour, TGF-β suppresses the expression of multiple genes that play key roles in regulating cell cycle progression. At 6 and 24 hours, TGF-β enhances the expression of multiple genes involved in regulating cell shape and cell adhesion, and therefore may mediate TGF-β-induced EMT. Additionally, several novel TGF-β-regulated genes were identified that may play important roles in TGF-β responses.
Materials and methods
Cell culture and treatment
NMuMG cells were cultured in Dulbecco's modified Eagle's medium containing 10% serum and 10 μg/ml insulin, until reaching 70–80% confluency. The cells were treated with 4 ng/ml TGF-β1 for 1, 6 or 24 hours. Untreated cells were used as reference samples at each time point, and were referred to as controls in the analyses. Four independent replicates of each experiment were performed. Cell morphology was examined under phase contrast using an Olympus CK40 microscope (Melville, NY, USA).
Growth inhibition assay
3H-thymidine incorporation assays were performed using cells that had been treated for 24 hours with TGF-β. The cells were pulsed with 3H-thymidine for the final 2 hours of the treatment period. 3H-thymidine incorporation was quantitated using a LS 6500 Multi-Purpose Scintillation Counter (Beckman). Four replicates of each experiment were performed, and each 3H-thymidine incorporation experiment was performed in triplicate.
RNA sample preparation
NMuMG cells were lysed and homogenized in Trizol reagent (Invitrogen, Carlsbad, CA, USA), and total RNA was prepared according to the manufacturer's instructions. Two rounds of extraction were performed to isolate pure, high-quality total RNA. Thirty micrograms of total RNA from each sample were used for reverse transcription and cyanine (Cy)-dye incorporation.
cDNA microarray hybridization
The National Institutes of Aging (NIA) 15,000 cDNA microarray was printed on three contiguous slides by the Vanderbilt Microarray Shared Resource. The mouse 15,000 NIA microarray contains 15,247 genes of known and unknown function, developed at the NIA. Detailed descriptions of the gene array list, microarray hardware and procedures are available online http://array.mc.vanderbilt.edu.
The reference RNA samples from nontreated NMuMG cells were labeled with Cy5, and the test RNA samples from NMuMG cells treated with 4 ng/ml TGF-β for 1, 6, and 24 hours were labeled with Cy3. The Cy3-labeled and Cy5-labeled samples were hybridized simultaneously to the same array. Four independent replicates of TGF-β treatment, RNA isolation and labeling, and microarray hybridization were performed. To estimate 'system noise' that may be due to differences in Cy-dye labeling between samples, we performed a 'self-to-self' hybridization, where the same control sample was labeled separately with Cy3 and Cy5, and then hybridized.
Oligo dT for reverse transcription was synthesized by Invitrogen. SuperScript II reverse transcriptase (Invitrogen) was used. A Qiagen PCR purification kit was used for purification of probes (Qiagen, Valencia, CA, USA). Slides were prehybridized to eliminate nonspecific interactions in prehybridization solution: 1% BSA, 5 × sodium chloride/sodium citrate buffer (SSC), 0.1% SDS at 65°C for 45 min. Hybridizations were performed at 65°C for 14–16 hours in a humidified hybridization chamber (Corning, Acton, MA, USA). After hybridization, the slides were washed once each in solutions 1–3 (wash solution 1, 2 × SSC, 0.1% SDS; wash solution 2, 1 × SSC; wash solution 3, 0.1 × SSC) for 5 min at 55°C with gentle stirring/agitation. Washed slides were centrifuged in a conical tube for 5 min at 1600 rpm to dry them. Microarray slides were scanned using a Genepix 4000 B scanner (Axon Instruments, Union City, CA, USA) at a resolution of 10 μm, and the original data files were generated by GenePixPro software (version 4.0; Axon Instruments).
Data analysis
A merged 15,000 data file was constructed by combining the three original 5000 data files generated from the three contiguous slides on which the total 15,000 gene clones were printed. The GenBank accession number and the full annotation gene name for each gene were incorporated. Data analyses were performed using the GeneSpring 4.1.5 software package (Silicon Genetics, Redwood City, CA, USA).
The normalization for the two-color cDNA microarray data was designed as follows. The net signal intensity in each channel (Cy3 or Cy5) was determined by subtracting the local background from signal intensity values. Each gene's measured intensity was divided by its control channel value (the reference RNA sample channel of Cy5). A Cy3/Cy5 ratio represents the relative abundance of a target transcript in TGF-β-treated and nontreated samples respectively. When the control channel value was below 1000.0, the data point was considered too weak and was discarded. Each sample was normalized to the 50th percentile of all measurements. The bottom 10th percentile was considered background, and was subtracted from all the other values.
Each experimental result was interpreted as the average of the four replicates of each time point. Time-course gene expression profiles were required to be present in two of the four datasets, and to be present in at least two of the three time points. A signal to control channel ratio > 2.5 and a probability t test P < 0.05 was defined as upregulation by TGF-β. A signal to control channel ratio < 0.4 and a probability t test P < 0.05 was defined as downregulation. The annotated gene lists were constructed from all genes represented in the normalized dataset. The differentially expressed genes were classified into functional subgroups using Simplified Ontology in GeneSpring. For each pathway, pathway images were saved as GIF files and loaded into GeneSpring. Each pathway element was identified in the normalized data.
Northern hybridization
All cDNAs of the NIA mouse 15,000 microarray were cloned into the NotI/SalI sites of the ampicillin-resistant pSPORT1 vector (Invitrogen). The insert sizes ranged from 0.5 to 3 kb, with an average insert size of 1.5 kb. The Escherichia coli host strain used was DH10B. Plasmid DNA was isolated using the High Pure Plasmid Isolation Kit (Roche, Indianapolis, IN, USA). The inserts were verified for their correct annotation by DNA sequencing. The reverse pSPORT1 primer was used as the sequencing primer. To prepare cDNA probes, cDNA inserts were amplified using pSPORT1 primers: forward, GTTTTCCCAGTCACGACGTTG; reverse, TGA-GCGGATAACAATTTCACACAG.
Plasmid DNA was used as the template for PCR amplification under the following conditions: initial denaturation at 95°C for 1 min; then 40 cycles of denaturation at 94°C for 30 s, annealing at 54°C for 30 s, and extension at 72°C for 3.5 min. A final extension was performed at 72°C for 7 min. To examine the quality and quantity of PCR products, they were separated on 1.2% agarose gels. Specific cDNA bands were recovered from the gels using QIAEX II Gel Extraction Kits (Qiagen). Seventy-five nanograms of cDNA was labeled with 32P using a Random Primed DNA Labeling Kit (Roche).
For northern blot analysis, 10 μg total RNA was separated by electrophoresis on a 1% agarose, 0.66 M formaldehyde gel and transferred to Hybrid Nylon transfer membranes (Amersham Biosciences, Piscataway, NJ, USA). After transfer, the membrane was UV cross-linked (Stratalinker; Stratagene, La Jolla, CA, USA). All cDNA probes were hybridized overnight at 42°C in ULTRAhyb solution (Ambion, Austin, TX, USA). Each filter was washed twice for 5 min in 2 × SSC and 0.1% SDS at 42°C, followed by two 15 min washes each in 0.5 × SSC and 0.1% SDS at 42°C, followed by two 15 min washes in 0.1 × SSC, 0.1% SDS at 42°C. Equal loading of gel lanes was confirmed by hybridization with the house keeping gene 1B15. Results were visualized and quantitated using a FUJIFILM-FLA-5000 Phosphoimager (Fuji Photo Film Co. Ltd, Stamford, CT, USA).
Results
Biological and gene expression responses of NMuMG cells to TGF-β
NMuMG cells react to TGF-β treatment with both an EMT response and a growth inhibitory response (Fig. 1). These characteristics render NMuMG cells an appropriate model system to study the signaling pathways mediating these TGF-β effects. To examine TGF-β regulation of genes involved in EMT and cell cycle control, we treated NMuMG cells with TGF-β for 1, 6, or 24 hours, isolated the total RNA, and performed gene expression profiling by cDNA microarray. The relative transcript abundance was expressed as Cy3/Cy5 ratios of signal intensities after background subtraction in each channel. Data analysis and quality control procedures are described in detail in Materials and methods. TGF-β treatment resulted in the upregulation or downregulation of 939 genes out of the 15,247 genes examined. Subtracting redundancy or signals too weak to score, this represents approximately 10% of the genes analyzed.
Figure 1.
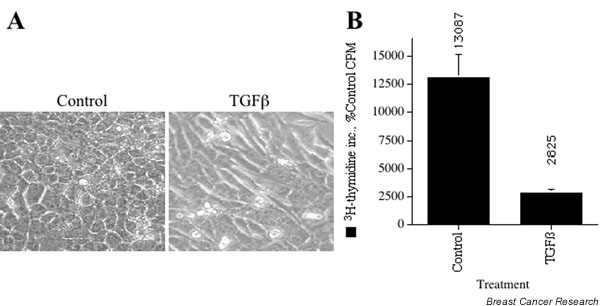
Biological responses of NMuMG cells to transforming growth factor beta (TGF-β) treatment. (A) NMuMG cells were treated for 24 hours with 4 ng/ml TGF-β1 or left untreated (control), and were examined by phase-contrast microscopy. TGF-β1-treated cells exhibited an epithelial to mesenchymal transition. (B) NMuMG cells were treated for 24 hours with 4 ng/ml TGF-β1 or left untreated (control). The cells were pulsed with 3H-thymidine for the final 2 hours of the treatment period. CPM, counts per minute.
The time course of TGF-β1-dependent changes in gene expression demonstrates a progressively greater alteration of gene expression over time (Fig. 2). In general, TGF-β1 regulation of cell cycle-related genes occurred with an onset of less than 1 hour, while TGF-β1 regulation of EMT-related genes occurred with a later onset, with changes first becoming apparent at 6 or 24 hours (see later).
Figure 2.
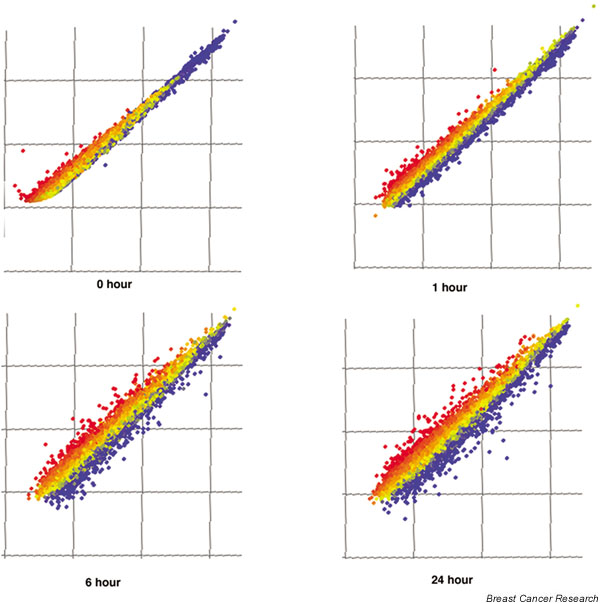
Time course of transforming growth factor beta (TGF-β)1-dependent changes in gene expression. The Cy3 signal is shown on the y axis and the Cy5 signal is shown on the x axis. The 0 hours graph represents the self–self hybridization for Cy-dye labeling noise control, and demonstrates a minimal level of systemic noise. TGF-β induces a progressively larger alteration of gene expression over time.
Figure 3 and Table 1 present the distribution of downregulated genes and of upregulated genes, and the sum of upregulated and downregulated genes at each time point. The total number of differentially expressed genes at each time point does not differ significantly. The 6 and 24 hour time points shared more TGF-β1-regulated genes than were shared between the 1 and 6 hour time points, or the 1 and 24 hour time points. Full lists of TGF-β-regulated genes at each time point can be accessed on our website http://array.mc.vanderbilt.edu/supl/tgfb_mouse.
Figure 3.

Venn diagrams demonstrating the distribution of downregulated genes, of upregulated genes, and of all regulated genes at each time point. The expression of genes present within all three circles (white) is altered at all three time points. Genes present in two circles (purple, yellow and light blue) are regulated at the two time points indicated. The expression of genes only present in one circle (red, green and blue) are only altered at one time point. The number of differentially expressed genes at each time point does not differ significantly. The 6 and 24 hour time points shared more transforming growth factor beta1-regulated genes than were shared between the 1 and 6 hour time points, or the 1 and 24 hour time points.
Table 1.
Genes previously reported to be regulated by transforming growth factor beta (TGF-β)
| Response | Response | ||||||
| Gene | 1 hour | 6 hours | 24 hours | Gene | 1 hour | 6 hours | 24 hours |
| Cellular communication | Cell cycle regulator | ||||||
| Platelet-derived growth factor | No | Up** | Up*** | CDK2 interacting protein (CINP) | Down* | No | Down* |
| Fibroblast growth factor regulated | Up** | Up*** | Up*** | Cyclin D2 | No | Down* | Down* |
| Cadherin 1 (Cdh1) | No | Down* | No | Cyclin D3 | Up* | No | No |
| Signal transduction | CDK9 | Up* | Up** | Up* | |||
| Nfkb inducing kinase | Up** | No | No | Cdk4 | Down* | No | No |
| RhoB | Up*** | Up*** | Up** | Cyclin A2 | Down* | No | Down* |
| Serine/threonine kinase 25 | No | Up* | No | Cyclin B1 | No | No | Down** |
| Integrin β4 | No | Up** | No | Cyclin B2 | Down* | No | Down* |
| Bmp-1 | No | Up* | Up* | Cdc25a | No | Up* | No |
| Janus kinase 2 (Jak2) | No | No | Up* | Cyclin H | No | No | Down* |
| Integrin α3A | No | No | Up* | Cancer | |||
| Integrin β1 | No | Up* | Up** | c-myc | Down** | Down* | No |
| Integrin-linked kinase (Ilk) | No | Up* | Up* | Akt | Up** | No | No |
| Integrin β5 | Up* | Up** | Up*** | GRO1 oncogene | Down** | Down** | Down** |
| Tyrosine phosphatase PTPT9 | No | Up* | Up* | Jun oncogene | Up** | No | Up* |
| Ephrin-A1 (EFNA1) | Up* | No | No | E2 oncogene (Ets1) | Up** | Up** | Up* |
| BMP-6 | No | Up* | Up** | Transport | |||
| TGF-β1-induced transcript | Up* | Up** | Up*** | Glucose transport protein | Up* | No | No |
| Colony stimulating factor 1 | Up** | Up** | Up*** | Ubiquitins and elongation factors | |||
| Intracellular components | Elongation factor 2 (ef-2) | No | Up** | Up** | |||
| TG interacting factor (Tgif) | Up* | Up** | Up** | Ubiquitin conjugating enzyme | Up* | Up* | Up* |
| α-actinin | No | Up** | Up** | Enzyme | |||
| β-tropomyosin 2 | No | Up* | Up*** | Glutathione S-transferase | Down* | Down* | Down* |
| Catenin src (Catns) | No | Up* | Up* | Protein tyrosine phosphatase | Up* | No | No |
| Extracellular components | Protease serine 15 (PRSS15) | Up* | Up* | No | |||
| α1 type IV collagen | No | No | Up* | Aldose reductase | No | Up** | Up** |
| Fibronectin | No | Up* | Up*** | Disulfide isomerase (ERp59) | No | Up* | No |
| Nucleic acid binding | Casein kinase II (Csnk2a2) | No | Up* | No | |||
| Helix-loop-helix DNA binding (Id) | Up*** | Down*** | Down*** | Ornithine decarboxylase | No | Up** | Up** |
| Inhibitor of DNA binding (Idb3) | No | Down* | Down* | RNA polymerase II | No | No | Up* |
| Translation initiation factor 2 | No | No | Up* | Bruton's tyrosine kinase (Btk) | No | Up* | Up* |
| Id2 | No | Down* | Down* | Cytochrome b-5 reductase (DIA1) | No | No | Up* |
The genes presented were upregulated (up) or downregulated (down) at least 2.5-fold (* 2.5-fold to fourfold, ** fourfold to sixfold, *** above sixfold) and were found in a Medline search in which the gene name and TGF-β1 were used as keywords. The genes are grouped according to their involvement in specific cellular functions.
Functional class distribution of the novel and known TGF-β1-regulated genes
Of the 939 genes regulated by TGF-β1, approximately one-third (350 genes) can be classified into functional groups. The classified genes were further separated into two tables of genes previously reported or not previously reported to be regulated by TGF-β. This was determined by performing a Medline search using the gene name and TGF-β as keywords. Tables of the complete dataset with each gene in its functional category are presented on our website http://array.mc.vanderbilt.edu/supl/tgfb_mouse. Tables 1 and 2 present some selected examples of known or unknown TGF-β-regulated genes, respectively. The genes involved in cell cycle regulation and cell adhesion (see later) are included.
Table 2.
Genes not previously reported to be regulated by transforming growth factor beta (TGF-β)
| Response | Response | ||||||
| Gene | 1 hour | 6 hours | 24 hours | Gene | 1 hour | 6 hours | 24 hours |
| Cellular communication | Cell cycle regulator | ||||||
| Gadd45g | Up*** | Up* | Up* | Calcyclin binding protein (CACYBP) | Up* | No | Up* |
| cdc2 | No | Up* | Up* | E2F transcription factor 5 (E2f5) | Down* | Down* | Down* |
| Hsp84-1 | No | Up* | Up* | Cancer | |||
| Junctional adhesion molecule (Jam) | No | Up* | Up* | Suppression of tumorigenicity 13 | No | Up* | Up* |
| Catenin α1 (Catna1) | No | Up* | Up* | Candidate tumor suppressor p33 ING1 | No | Up* | Up* |
| Signal transduction | Transport | ||||||
| NFKBIL2 gene for IκBR | No | Down*** | Down** | Solute carrier family 4 | Up** | Up** | Up* |
| GTPase-activating protein GIT1 | Up* | Up* | No | ATPase, H+ transporting (ATP6J) | No | Up* | Up* |
| Ras-GTPase-activating SH3 binding | Up** | No | Up* | Ca2+/calmodulin-dependent kinase I | Up* | Up*** | Up** |
| Ikki | Down* | Down* | Down* | Microtubular dynamics | |||
| Iqgap1 | Up* | Up** | Up** | Kinesin light chain 1 (Klc1) | Up* | No | Up* |
| Milk fat globule-EGF factor 8 protein | No | Up* | Up** | Microtubule-associated protein 4 | Up** | Up** | Up** |
| Tnfrh1 gene | Up* | No | Up* | Ubiquitin and elongation factors | |||
| Intracellular components | Ubiquitin-specific protease 5 (Usp5) | Up** | Up* | Up* | |||
| Actinin α3 (Actn3) | Up* | Up** | Up* | Elongation factor 1-alpha (EF 1-α) | No | Up** | Up*** |
| Actin-related protein 2/3 complex | Up* | Up* | Up* | Chaperone | |||
| Unconventional myosin-15 gene | Up* | No | Up** | Ccth gene for chaperonin | Up* | Up* | Up** |
| Nonmuscle tropomyosin 5 | No | Up* | Up* | Chaperonin subunit 7 (Cct7) | Up* | Up* | Up** |
| Melanoma X-actin (Actx) | No | Up* | Up** | Enzyme | |||
| Tropomyosin isoform 2 | No | Up* | Up** | Phosphoserine aminotransferase | No | Down** | Down** |
| Actin-related protein 1 | No | Up* | Up** | α-methylacyl coenzyme (Macr1) | No | Down** | Down** |
| Karyopherin β3 (KPNB3) | No | No | Up* | PLC-L2 for phospholipase C-L2 | No | Down** | Down** |
| β-tropomyosin 2 | No | Up* | Up*** | Fatty acid coenzyme A ligase (FACL4) | No | Down** | Down** |
| TM-4 gene for fibroblast tropomyosin 4 | No | Up** | Up** | Phospholipase A2, group 4 (Pla2g4) | No | No | Up* |
| Calpain | No | Up** | Up** | Serine/threonine kinase (Unc51.1) | No | Up* | Up** |
| Myosin light chain 2 (Mlc2) | No | Up* | No | Proteasome (Psmd13) | Up** | Up* | Up* |
| Extracellular components | Na, K-ATPase α-1 | No | Up** | Up* | |||
| Cis-Golgi matrix protein GM130 | Up** | No | Up* | Aldehyde dehydrogenase 5 (ALDH5) | Up* | Up* | No |
| α-1(XVIII) collagen (COL18A1) | No | Up* | Up** | Protein phosphatase 2A (PR 53) | Up* | No | Up* |
| Smarce1 | No | Up* | Up* | Casein kinase 1 (Csnk1e) | Up* | Up* | Up*** |
| Nucleic acid binding | Serine/threonine phosphatase-2C | Up* | No | No | |||
| HNRPM | Up* | Up* | No | Coatomer protein (Copg1) | Up* | Up* | Up* |
| Hnrpu | No | Up* | Up* | MAP kinase kinase 3b | Up** | No | No |
| Hnrnpc | Up* | Up* | Up* | Protein phosphatase 1 (Ppp1cc) | No | Up* | No |
| Damage-specific DNA binding (Ddb1) | Up* | Up** | Up*** | Na, K-ATPase β-subunit | No | Up** | Up* |
The genes presented were upregulated (up) or downregulated (down) at least 2.5-fold (*2.5-fold to fourfold, ** fourfold to sixfold, *** above sixfold) and were not found in a Medline search in which the gene name and TGF-β1 were used as keywords. The genes are grouped according to their involvement in specific cellular functions.
Northern analysis verification of microarray data
To validate the microarray results, we subjected nine of the novel TGF-β-regulated genes to northern analysis. Of the nine genes examined, seven exhibited TGF-β1-induced changes in gene expression that matched those observed in the microarray experiments. The seven novel TGF-β1-regulated genes verified by northern analysis include Akt, mCalpain, RhoB, PR53, actinin 3, Ikki (Fig. 4) and the IQ motif-containing GTPase-activating protein 1 (IQGAP1). IQGAP1 showed upregulation in NMuMG cells after 1, 6 and 24 hours of treatment with TGF-β, supporting the microarray data. As reported by other investigators [6], we detected multiple IQGAP1 transcripts. The IQGAP1 northern blot is thus not shown in Fig. 4. Akt and RhoB were reported previously to be regulated by TGF-β at the translational level [7,8], but not at the transcriptional level as we show here. Some of the TGF-β1-induced changes in gene expression were also verified by real-time RT-PCR (data not shown). In further support of the reliability of the microarray experiments, genes that were represented multiple times in the array as different clones exhibited identical expression profiles (examples can be found in the supplementary tables at our website: http://array.mc.vanderbilt.edu/supl/tgfb_mouse. In addition, our expression profiling results are consistent with previously reported expression patterns of TGF-β1-responsive genes [1,5].
Figure 4.
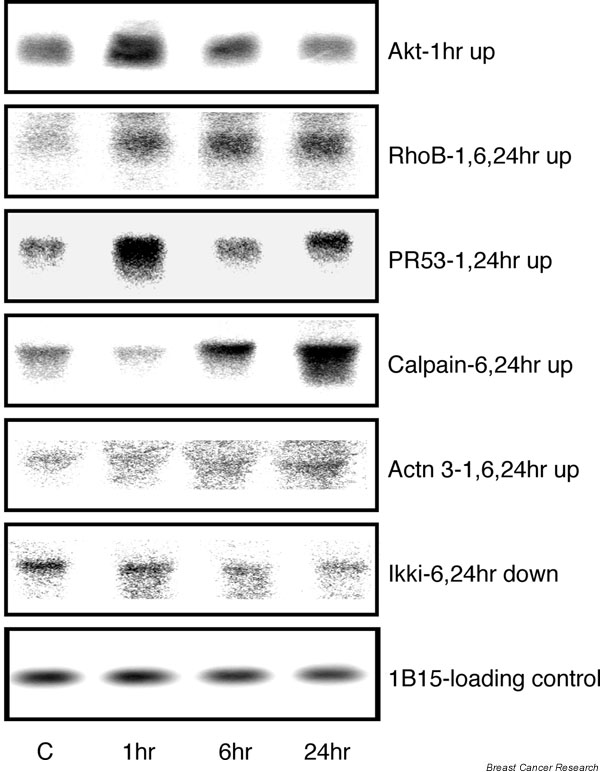
Verification of transforming growth factor beta (TGF-β)1-regulated genes identified in microarray experiments by northern blotting. Ten micrograms of total RNA were loaded in each lane. The resulting blots were hybridized with cDNA probes specific for Akt, RhoB, PR53, calpain, actinin, Ikki, or the house-keeping gene 1B15 as a loading control. The figure shows mRNA expression in NMuMG cells after no treatment (C) or after treatment with 4 ng/ml TGF-β1 for 1, 6, or 24 hours. The gene name and results from microarray data are shown to the right of each blot. up, Upregulated; down, downregulated.
Examination of cell cycle control and cell adhesion-related genes by microarray
There are multiple ways to explore the biological significance of results obtained in microarray experiments. One may organize the genes into functional groups or signaling pathways to examine the regulation of clusters of genes. We were interested in identifying TGF-β-regulated genes involved in the induction of EMT and cell cycle arrest. In our study in NMuMG cells, the expression of many cell cycle regulators was altered by TGF-β. One hour after TGF-β treatment, cyclin D2, cdk4, E2F5, cyclin A, and c-myc were downregulated. Six hours after TGF-β treatment, cyclin D2, p107, E2F5, c-myc, and Id2 were downregulated. Twenty-four hours after TGF-β treatment, cyclin D2, p107, E2F5, cyclin A, cyclin B, cyclin H, and Id2 were downregulated (Fig. 5). These data are consistent with these genes serving as mediators of TGF-β-induced growth arrest.
Figure 5.
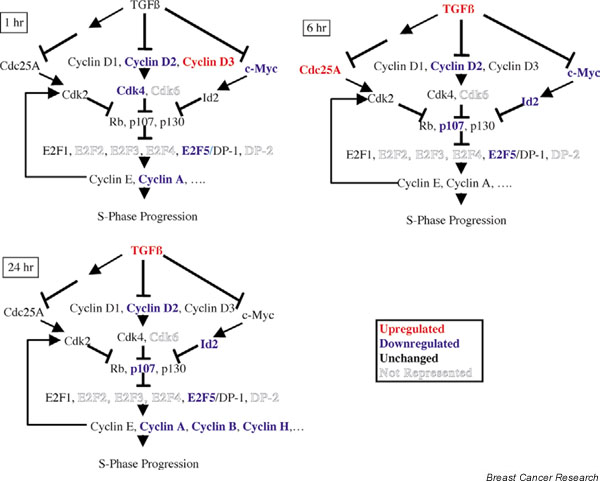
Transforming growth factor beta (TGF-β)1 regulation of gene expression patterns of cell cycle elements. GeneSpring was used to search the normalized 15,000 microarray data. A signal to control ratio above 2.5 was defined as upregulation (red). A signal to control ratio below 0.4 was defined as downregulation (blue). Ratios between 0.4 and 2.5 were considered unregulated (black). Outlined genes were not represented in the 15,000 gene database.
The microarray results also revealed upregulation of a number of genes that play a role in cell–cell and cell–matrix adhesion and EMT. At 1 hour, c-Jun, β-integrin, and IQGAP were upregulated. At 6 hours, actin, calpain, β-integrin, fibronectin, collagen, myosin light chain, α-actinin, β-catenin and IQGAP were upregulated. At 24 hours, c-Jun, actin, calpain, α-integrin, β-integrin, fibronectin, collagen, α-actinin, β-catenin and IQGAP were upregulated. The only gene in this category observed to be downregulated was E-cadherin at 6 hours (Fig. 6). The results are consistent with these genes playing a role in TGF-β1-mediated changes in cell–cell and cell–substratum adhesion.
Figure 6.
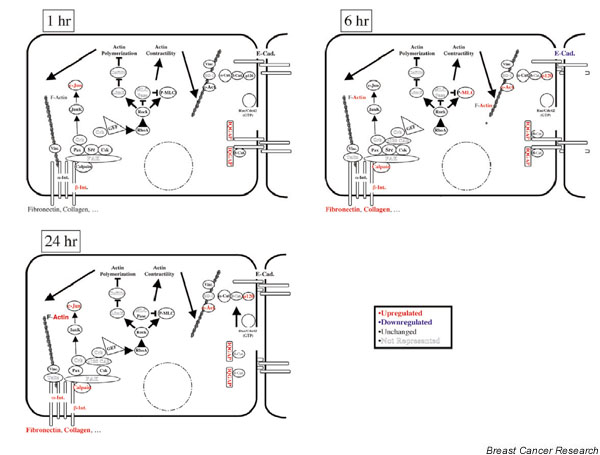
Transforming growth factor beta (TGF-β)1 regulation of the expression of genes involved in cell adhesion. GeneSpring was used to search the normalized 15,000 microarray data using the parameters described in Fig. 5. Cad, cadherin; Int, integrin.
Our results are consistent with previous studies, but we identified several novel TGF-β-regulated genes that could play important roles in both EMT and cell cycle regulation, including RhoB, mCalpain, actinin 3, IQGAP1, Ikki and protein phosphatase 2A (PP2A)-PR53.
Discussion
The mechanisms by which TGF-β signals are not fully understood. In this regard, gene expression profiling has been used successfully by a variety of investigators to explore genetic events involved in TGF-β signaling. Zavadil and colleagues detected genetic programs of epithelial cell plasticity directed by TGF-β [5]. Verrecchia and colleagues identified novel TGF-β/Smad gene targets in dermal fibroblasts [4]. Chen and colleagues found defective repression of c-myc in breast cancer cells, suggesting that this event is at the core of the TGF-β growth arrest program [9]. Shen and colleagues discovered that the activity of the guanine exchange factor NET1 is essential for TGF-β-mediated stress fiber formation [10].
In the present study, we have selected the NMuMG nontransformed mouse mammary gland epithelial cell line as a model system to identify TGF-β-regulated target genes involved in the control of growth inhibition and EMT. To identify TGF-β-regulated genes, we employed a cDNA microarray that represents 15,247 genes of known and unknown function. We found that the expression of 939 of the genes was altered by TGF-β regulation. This represents about 10% of the genes examined. Many of the genes identified were previously determined to mediate key biological responses to TGF-β. In addition, several novel TGF-β-regulated genes were identified. The present report provides a comprehensive view of the coordinated regulation of genetic programs induced by TGF-β during the processes of cell cycle arrest and EMT. A remarkably large number of genes undergo rapid changes in expression after TGF-β treatment. This report adds to the database of hundreds of genes that are regulated by TGF-β [5]. In addition, we have identified several novel TGF-β-regulated genes, and have verified these results in northern blotting experiments.
Table 1 presents previously identified TGF-β-regulated genes, many of which are known to play important roles in TGF-β induction of EMT and cell cycle arrest. Briefly, in the cellular communication category, platelet-derived growth factor was upregulated by TGF-β. Multiple genes involved in cell adhesion were upregulated by TGF-β, including integrins (integrin α3A, integrin β, integrin β5), integrin-linked kinase, and the integrin ligand fibronectin. These results are consistent with TGF-β activating integrin-dependent cell adhesion. TGF-β upregulation of α-actinin, β-tropomyosin 2, and p120 catenin may play a role in the induction of EMT. RhoB was upregulated at 6 and 24 hours. RhoB is an endosomal small GTPase, and it functions as a negative regulator of integrin and growth factor signals [11]. Our previous results [8] and those of other workers [12] suggest a novel mechanism of tumor suppressive regulation by TGF-β, and implicate RhoB as a negative regulator of TGF-β signal transduction.
TGF-β downregulation of the helix–loop–helix DNA binding protein Id2 and of the inhibitor of DNA binding Idb3 were previously identified as important events in TGF-β-regulated signaling. These proteins belong to a family of helix–loop–helix proteins that, in general, function as positive regulators of cell growth and as negative regulators of cell differentiation in many cell types [13].
TGF-β also regulates many genes involved in the progression of cancer. The proto-oncogene Akt is upregulated at 1 hour at the mRNA level. The Gro1 proto-oncogene is downregulated by TGF-β at all the time points examined. The roles that these gene products play downstream of TGF-β during tumorigenesis have not been extensively studied [14,15]. Well-established TGF-β responses such as downregulation of c-myc and upregulation of the Jun oncogene were also observed.
TGF-β is a potent growth inhibitor of epithelial cells. TGF-β-mediated growth inhibition has been associated with effects on G1 phase cyclins, cyclin-dependent kinases (Cdks), Cdk-activating kinase, Cdc25A, and Cdk inhibitors. TGF-β potently inhibits the activities of both the cyclin D–Cdk4/Cdk6 and cyclin E–Cdk2 complexes, resulting in the hypophosphorylation of pRbs and in the decreased transcriptional activity of E2Fs. TGF-β does not affect the expression levels of Cdk4, Cdk6, or Cdk2 in exponentially proliferating cells. TGF-β decreases the expression levels of cyclin A, which is probably the result of cell cycle arrest in the late G1 phase. TGF-β has been associated with upregulation of Cdk inhibitors p15Ink4B, p21Cip1, and p27Kip1. The c-myc proto-oncogene has been implicated in TGF-β-mediated growth inhibition. TGF-β rapidly inhibits c-myc expression in a wide variety of cell types [1]. c-Myc downregulation is a key event in the TGF-β program of growth inhibition [9]. Additional mechanisms mediating TGF-β-induced cell cycle arrest continue to be uncovered. In our study, examination of cell cycle genes demonstrated TGF-β1-induced downregulation of cyclin D2, c-myc, Id2, p107, E2F5, cyclin A, cyclin B, and cyclin H. These data are consistent with these genes serving as mediators of TGF-β-induced growth arrest.
Of the genes that could be placed into functional categories, the expression of the majority of the genes was not previously reported to be regulated by TGF-β. Table 2 presents some examples of genes that we have identified in this study to be regulated by TGF-β that are involved in cell signaling. Gadd45g was upregulated. Gadd45g specifically interacts with the Cdk1-cyclin complex, B1 resulting in inhibition of kinase activity [16]. The IQGAP1 gene is a target molecule of Cdc42 and Rac1. IQGAP negatively regulates the E-cadherin–catenin complex-based cell–cell adhesion by dissociating α-catenin and/or β-catenin, a key molecule that links the E-cadherin-catenin complex to the actin cytoskeleton. These data suggest that TGF-β upregulation of IQGAP may be one of the mechanisms by which TGF-β regulates cell–cell adhesion [17]. Calpains are a family of Ca2+-dependent intracellular cysteine proteases, including the ubiquitously expressed micro-calpains and m-calpains. Calpain activity may be regulated by targeting to specific adhesion-related substrates [18].
Our results also demonstrate that TGF-β regulates multiple genes involved in regulation of the actin cytoskeleton. Some of these components include actinin α3, the Arp2–Arp3 complex, and myosin light chain 2. Four different isoforms of tropomyosin were upregulated. Actinin α3 is a calcium-dependent cytoskeletal protein with an actin-binding domain. Actinin α3 is associated with adherens junctions and desmosomes, together with E-cadherin, α-catenin, β-catenin and γ-catenin, vinculin, α-actinin, and polymerized actin. The genes α-actinin, myosin light chain 1/myosin light chain 2, and tropomyosin are components of myofilaments involved in cell contractility and motility.
During our studies we identified several genes not previously reported to be regulated by TGF-β that are regulated by TGF-β in NMuMG cells. These novel TGF-β-regulated genes include IQGAP1, mCalpain and actinin α3, which were discussed earlier, and also Ikki and PP2A-PR53. For the first time, we show in the present study that Ikki is downregulated by TGF-β within 1 hour of treatment, and that downregulation persisted for at least 24 hours. NF-κB activation depends on the phosphorylation and degradation of its inhibitor protein, IkB. The phosphorylation of IkBα is initiated by an IkB kinase complex that includes a catalytic heterodimer composed of IkB kinase 1 (IKK-1) and IkB kinase 2 (IKK-2) [19]. A novel inducible IkB kinase (IKKi) has recently been described. IKKi is functionally and structurally distinct from the constitutively expressed IKK-1 and IKK-2. In contrast to TGF-β, tumor necrosis factor alpha stimulated IKKi expression, an effect that persisted for at least 24 hours [20]. The biological significance of TGF-β downregulation of IKKi is currently under investigation. PP2A is a multifunctional serine/threonine phosphatase that is critical to many cellular processes, including cell cycle regulation and signal transduction. PR53 belongs to one of the families of PP2A regulatory subunits. The significance of TGF-β regulation of PR53 expression is unknown [21].
Altogether, our results are consistent with the important role of TGF-β in regulating focal adhesions, integrin-based adhesion, actin cytoskeletal architecture, and cell motility. TGF-β plays a profound role in the dedifferentiation of epithelial cells, causing depolarization, disruption of epithelial interactions, altered expression of extracellular matrix proteins, rearrangement of the cytoskeleton, and formation of actin stress fibers. These TGF-β responses provide cells with increased metastatic potential and with increased motility.
Over the past few years, tremendous progress has been made in identifying signal transduction pathways activated by TGF-β family members. In the present study, we have combined microarray analysis with the examination of specific signaling pathways. Because of the important role that TGF-β plays in controlling cell proliferation and cell adhesion, we used the signaling elements involved in these processes as a template for the analysis of our microarray results. Many of the genes that we identified as being regulated by TGF-β were previously reported to be downstream targets of TGF-β. We also discovered several novel TGF-β target genes known to play roles in regulating cell adhesion, and thus EMT, and also cell cycle regulation. Examining the biological functions of these novel TGF-β target genes will increase our knowledge of the mechanisms by which TGF-β mediates its cellular effects. Furthermore, our results indicate several possible points of convergence between TGF-β signaling and other major intracellular and intercellular signaling systems.
Conclusion
A 15,247 cDNA microarray was used to examine TGF-β-regulated gene expression in mouse mammary NMuMG cells. Expression of 10% of the genes examined (939) was altered by TGF-β treatment. We have reported a comprehensive analysis of the coordinated regulation of genetic programs induced by TGF-β in mammary epithelial cells during the processes of cell cycle arrest and EMT. In addition, several genes previously not known to be regulated by TGF-β at the transcriptional level were identified, including Akt, RhoB, IQGAP1, mCalpain, actinin α3, Ikki and PP2A-PR53.
Competing interests
None declared.
Abbreviations
BSA = bovine serum albumin; Cdk = cyclin-dependent kinase; Cy = cyanine; EMT = epithelial to mesenchymal transition; IKKi = inducible IkB kinase; IQGAP1 = IQ motif-containing the GTPase-activating protein 1; NF = nuclear factor; NIA = National Institutes of Aging; PCR = polymerase chain reaction; PP2A = protein phosphatase 2A; RT = reverse transcriptase; SSC = sodium chloride/sodium citrate (buffer); TGF-β = transforming growth factor beta.
Acknowledgments
Acknowledgements
Acknowledgement to Shawn Levy, the director of Vanderbilt Microarray Shared Resources, for technique training and discussions, and to Paul Larsen for data analyzing efforts. The project benefited from a Microarray Chip Scholarship from the Vanderbilt-Ingram Cancer Center.
References
- Yue J, Mulder KM. Transforming growth factor-beta signal transduction in epithelial cells. Pharmacol Ther. 2001;91:1–34. doi: 10.1016/s0163-7258(01)00143-7. [DOI] [PubMed] [Google Scholar]
- Akhurst RJ, Derynck R. TGF-beta signaling in cancer – a double-edged sword. Trends Cell Biol. 2001;11:S44–S51. doi: 10.1016/s0962-8924(01)02130-4. [DOI] [PubMed] [Google Scholar]
- Bhowmick NA, Ghiassi M, Bakin A, Aakre M, Lundquist CA, Engel ME, Arteaga CL, Moses HL. Transforming growth factor-beta1 mediates epithelial to mesenchymal transdifferentiation though a RhoA-dependent mechanism. Mol Biol Cell. 2001;12:27–36. doi: 10.1091/mbc.12.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verrecchia F, Chu ML, Mauviel A. Identification of novel TGF-beta/Smad gene targets in dermal fibroblasts using a combined cDNA microarray/promoter transactivation approach. J Biol Chem. 2001;276:17058–17062. doi: 10.1074/jbc.M100754200. [DOI] [PubMed] [Google Scholar]
- Zavadil J, Bitzer M, Liang D, Yang YC, Massimi A, Kneitz S, Piek E, Bottinger EP. Genetic programs of epithelial cell plasticity directed by transforming growth factor-beta. Proc Natl Acad Sci USA. 2001;98:6686–6691. doi: 10.1073/pnas.111614398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Sacks DB. Elucidation of the interaction of calmodulin with the IQ motifs of IQGAP1. J Biol Chem. 2003;278:4347–4352. doi: 10.1074/jbc.M208579200. [DOI] [PubMed] [Google Scholar]
- Kim G, Jun JB, Elkon KB. Necessary role of phosphatidylinositol 3-kinase in transforming growth factor beta-mediated activation of Akt in normal and rheumatoid arthitis synovial fibroblasts. Arthitis Rheum. 2002;46:1504–1511. doi: 10.1002/art.10314. [DOI] [PubMed] [Google Scholar]
- Engel ME, Datta PK, Moses HL. RhoB is stabilized by transforming growth factor beta and antagonizes transcriptional activation. J Biol Chem. 1998;273:9921–9926. doi: 10.1074/jbc.273.16.9921. [DOI] [PubMed] [Google Scholar]
- Chen CR, Kang Y, Massague J. Defective repression of c-myc in breast cancer cells: a loss at the core of the transforming growth factor beta growth arrest program. Proc Natl Acad Sci USA. 2001;98:992–999. doi: 10.1073/pnas.98.3.992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Li J, Hu PP, Waddell D, Zhang J, Wang XF. The activity of guanine exchange factor NET1 is essential for transforming growth factor-beta-mediated stress fiber formation. J Biol Chem. 2001;276:15362–15368. doi: 10.1074/jbc.M009534200. [DOI] [PubMed] [Google Scholar]
- Liu AX, Rane N, Liu JP, Prendergast GC. RhoB is dispensable for mouse development, but it modifies susceptibility to tumor formation as well as cell adhesion and growth factor signaling in transformed cells. Mol Cell Biol. 2001;21:6906–6912. doi: 10.1128/MCB.21.20.6906-6912.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adnane J, Seijo E, Chen Z, Bizouarn F, Leal M, Sebti SM, Munoz-Antonia T. RhoB, not RhoA, represses the transcription of the transforming growth factor beta type II receptor by a mechanism involving activator protein 1. J Biol Chem. 2002;277:8500–8507. doi: 10.1074/jbc.M104367200. [DOI] [PubMed] [Google Scholar]
- Lasorella A, Uo T, Iavarone A. Id proteins at the cross-road of development and cancer. Oncogene. 2001;20:8326–8333. doi: 10.1038/sj.onc.1205093. [DOI] [PubMed] [Google Scholar]
- Shin I, Bakin AV, Rodeck U, Brunet A, Arteaga CL. Transforming growth factor beta enhances epithelial cell survival via Akt-dependent regulation of FKHL1. Mol Biol Cell. 2001;12:3328–3339. doi: 10.1091/mbc.12.11.3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loukinova E, Chen Z, Van Waes C, Dong G. Expression of proangiogenic chemokine Gro 1 in low and high metastatic variants of Pam murine squamous cell carcinoma is differentially regulated by IL-1alpha, EGF and TGF-beta1 though NF-kappaB dependent and independent mechanisms. Int J Cancer. 2001;94:637–644. doi: 10.1002/ijc.1514. [DOI] [PubMed] [Google Scholar]
- Vairapandi M, Balliet AG, Hoffman B, Liebermann DA. GADD45b and GADD45g are cdc2/cyclinB1 kinase inhibitors with a role in S and G2/M cell cycle checkpoints induced by genotoxic stress. J Cell Physiol. 2002;192:327–338. doi: 10.1002/jcp.10140. [DOI] [PubMed] [Google Scholar]
- Shimao Y, Nabeshima K, Inoue T, Koono M. Complex formation of IQGAP1 with E-cadherin/catenin during cohort migration of carcinoma cells. Its possible association with localized release from cell–cell adhesion. Virchows Arch. 2002;441:124–132. doi: 10.1007/s00428-002-0603-3. [DOI] [PubMed] [Google Scholar]
- Arthur JS, Elce JS, Hegadorn C, Williams K, Greer PA. Disruption of the murine calpain small subunit gene, Capn4: calpain is essential for embryonic development but not for cell growth and division. Mol Cell Biol. 2000;20:4474–4481. doi: 10.1128/mcb.20.12.4474-4481.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothwarf DM, Karin M. The NF-kappa B activation pathway: a paradigm in information transfer from membrane to nucleus. Sci STKE. 1999;5:RE1. doi: 10.1126/stke.1999.5.re1. [DOI] [PubMed] [Google Scholar]
- Aupperle KR, Yamanishi Y, Bennett BL, Mercurio F, Boyle DL, Firestein GS. Expression and regulation of inducible IkappaB kinase (IKK-i) in human fibroblast-like synoviocytes. Cell Immunol. 2001;214:54–59. doi: 10.1006/cimm.2002.1885. [DOI] [PubMed] [Google Scholar]
- Janssens V, Jordens J, Stevens I, Van Hoof C, Martens E, De Smedt H, Engelborghs Y, Waelkens E, Goris J. Identification and functional analysis of two Ca2+-binding EF-hand motifs in the B''/PR72 subunit of Protein Phosphatase 2A. J Biol Chem. 2003;278:10697–10706. doi: 10.1074/jbc.M211717200. [DOI] [PubMed] [Google Scholar]


