Abstract
In the present study, we have exploited the properties of a fibrillar protein for the template synthesis of zinc sulfide (ZnS) nanoparticle chains. The diameter of the ZnS nanoparticle chains was tuned in range of ~30 to ~165 nm by varying the process variables. The nanoparticle chains were characterized by field emission scanning electron microscopy, UV–Visible spectroscopy, transmission electron microscopy, electron energy loss spectroscopy, and high-resolution transmission electron microscopy. The effect of incubation temperature on the morphology of the nanoparticle chains was also studied.
Keywords: Nanoparticle chains, Template, Synthesis, Morphology, One-dimensional nanostructure
Introduction
One-dimensional structures (1D) such as nanowires, nanorods, nanoparticle chains, and nanotubes have attracted much attention in recent years (Cui et al. 2001; Diehl et al. 2002; Huang et al. 2001; Bachtold et al. 2001; Collins et al. 2001; Murray et al. 2000; Kimberly et al. 2002; Johnson et al. 2002; Alivisatos 1996). Their growing importance is due to the unique properties they exhibit. The 1D structures show future promise in a variety of fields such as electronics, optoelectronics, catalysis, and biosensing. Although the advances in the field of nanotechnology are promising, there are few obstacles that need to be overcome. The synthesis of 1D nanostructures by solvothermal process (Chen et al. 2003), thermal evaporation (Meng et al. 2003; Wang et al. 2002), liquid crystal template (Jiang et al. 2001), and electrodeposition (Xu et al. 2006) in porous anodic alumina templates require high temperatures or pressures and the precise control of the process variables. Moreover, after synthesis it is generally difficult to manipulate and position the nanostructures in devices. In turn, biological molecules have the chemical recognition capacity that is promising in allowing for a higher degree of flexibility in their positioning in specific places in nanoelectronic devices. In addition, certain peptides and proteins can self-assemble into chemically reactive readymade shapes that can serve as templates for further growth of inorganic nanostructures. Biological systems possess a high degree of organization from molecular building blocks (peptides, amino acids, proteins, and nucleic acids) and are perfect models for bottom–up strategies for controlled material synthesis. Their molecular recognition capabilities, combined with the specificity toward certain ions and molecules, can be used to precisely control the fundamental processes involved in materials synthesis and processing, such as phase stability, nucleation and growth, pattern formation, and assembly. The electroless deposition, especially on DNA molecules and viruses, has lead to the fabrication of several different 1D structures (Klein et al. 1997; Keren et al. 2003; Claridge et al. 2005; Flynn et al. 2003; Yan et al. 2003; Monson and Woolley 2003; Deng and Mao 2003; Richter et al. 2001; Dong et al. 2007; Mao et al. 2004; Huang et al. 2005; Nam et al. 2006). However, peptides and proteins, with the ability to self-assemble into ordered fibrils have been much less investigated. While several metallic nanowires, as well as CdS nanoparticle chains have been synthesized via protein-directed nucleation and growth in our laboratory (Padalkar et al. 2007, 2008), to our knowledge, there are no reports in literature regarding the use of fibrillar proteins for the template synthesis of zinc sulfide (ZnS) nanowires or nanoparticle chains. ZnS is an II–VI semiconducting material having a band gap of 3.7 eV. It is a particularly interesting material due to its wide range of potential applications. It shows promise in several fields and has applications in electronics and photonics. ZnS has semiconducting, photoluminescent, and field emission properties. These properties have been exploited in many applications such as light converting electrodes, ultraviolet light-emitting diodes, phosphors in cathode ray tubes, flat panel displays, injection lasers, and infrared windows (Kar et al. 2003; Xu et al. 2006; Lu et al. 2007). Several ZnS 1D structures such as nanorods, nanowires, nanobelts, and nanotubes (Zhang et al. 2002; Yin et al. 2005; Ma et al. 2003) have be fabricated. All these structures have been synthesized at high temperatures and require long reaction times. However, the fabrication process described here was carried out at atmospheric conditions and requires a very short synthesis time typically not more than 15 min for the completion of the entire experiment.
Here, we report the synthesis of ZnS nanoparticle chains on a fibrillar protein (α-synuclein) template. Synuclein is a 14.4 kDa amyloidogenic protein, which is found in the human brain (Spillantini et al. 1997). This protein has the ability to self-assemble into fibrillar structures having an approximate diameter of 8 nm and a length between 500 nm and 1 μm (Conway et al. 2000; Hoyer et al. 2002). The presence of protein fibrils in the human brain can lead to different pathologies (Serio et al. 2000; Scheibel and Lindquist 2001; Scheibel et al. 2003; DePace and Weissman 2002). However, when the α-synuclein protein self-assembles into fibrils in vitro, its properties can be potentially useful for the synthesis of inorganic nanostructures. The structure of amyloidogenic fibrillar proteins, such as α-synuclein, is mainly composed of adjacent β-sheets assembled into a twisted fibrillar structure by hydrogen bonding (Vilar et al. 2008; Serpell et al. 2000; Nelson and Eisenberg 2006; Rochet 2007). The charge on the β-sheets can be manipulated during the self-assembly process to obtain fibrillar structures with different charge arrangements, thus making them ideal structural templates for the fabrication of 1D nanostructures.
Experimental details
Self-assembly of α-synuclein fibrils
The expression and purification of α-synuclein were carried out as previously described (Conway et al. 2000; Rochet et al. 2000). The E46K mutant of α-synuclein, was used because it has the ability to rapidly self-assemble into fibrils. The lyophilized protein was dissolved in phosphate-buffered saline (PBS) with pH 7.4, 0.02% (w/v) NaN3 and dialyzed against the same buffer at 4 °C, for 24 h. The protein solution was filtered through a 0.22 μm nylon spin filter followed by a Microcon-100 spin filter, yielding a stock solution depleted of aggregates. The final concentration of protein in PBS was of 100–300 μM [determined by bicinchoninic acid (BCA) assay]. The protein was incubated at 37 °C for 12–96 h in a tissue culture rolling drum to generate fibrils.
Synthesis of ZnS nanoparticle chains
The synthesis of ZnS nanoparticle chains was carried out by using zinc chloride (ZnCl2; 2 mM) as the salt solution, and hydrogen sulfide (H2S) gas as the sulfur source. A stock solution of ZnCl2 was prepared and its pH value was adjusted to be in the acidic regime by the addition of concentrated hydrochloric acid. This was done to avoid precipitation of zinc hydroxide in solution. For the synthesis of ZnS nanoparticle chains a p-type silicon (Si) (111) wafer was used as a substrate to prepare a field emission scanning electron microscopy (FESEM) sample. The same synthesis procedure was performed on a 3-mm-diameter carbon-coated gold grid as a substrate to obtain a transmission electron microscopy (TEM) sample. A volume of 10 μL of α-synuclein fibrils suspended in the PBS buffer was pipetted onto the, Si wafer, substrate and dried in a desiccator. The ZnCl2 solution (10 μL) was deposited on to the dried protein solution, followed by an incubation time of 5 min. The substrate with the protein and the ZnCl2 solution was then exposed to H2S gas for 5 min. Later, the substrate was rinsed using deionized water and dried under a jet of air. A similar procedure was carried out for the preparation of a TEM sample. A similar, ZnS nanoparticle chain, TEM sample was prepared on a carbon-coated gold grid.
Characterization of α-synuclein fibril and ZnS nanoparticle chains
The diameter and morphology of the α-synuclein fibril were studied, with TEM, by using the Philips CM-10 operated under 80 kV accelerating voltage. A carbon-coated copper TEM grid was used as the substrate. The protein solution (3 μL) was pipetted out on to the TEM grid and was stained using 2% uranyl acetate solution for 1 min. The excess solution on the grid was then blotted and the sample was used for imaging.
The average diameter and morphology of the ZnS nanoparticle chains were analyzed, with FESEM, by using a Hitachi S4800 field emission scanning electron microscope and, with TEM, by using an FEI Titan 80/300 transmission electron microscope. High-resolution transmission electron microscopy (HRTEM) images were registered to investigate the crystalline nature of the sample. Further, an electron diffraction pattern was obtained to study the crystal structure of the sample. Electron energy loss spectra (EELS) were obtained from the ZnS nanoparticle chains to verify the presence of zinc (Zn) and sulfur (S) in the samples. Further, elemental mapping of Zn and S were also obtained to study the distribution of Zn and S in the samples. Finally, UV–visible (UV–Vis) absorption spectra were obtained from the ZnS sample and from the ZnS colloidal sample having a particle size of 10 μm, purchased from Sigma Aldrich to compare the absorption peaks. The FESEM analyses were performed on a Hitachi S4800. TEM imaging was performed on either Philips CM-10 operating at 80 kV or on an FEI Titan 80/300 transmission electron microscope having a Gatan Imaging Filter (GIF) and a 2 k CCD, which operated at 300 kV. EELS and HRTEM images were registered on the FEI Titan. The UV–Vis absorption spectra of the ZnS nanoparticle chains and colloidal ZnS were recorded with a molecular device microplate reader.
Results and discussion
The α-synuclein fibril formation
The formation of α-synuclein fibrils is believed to occur through a stepwise mechanism (Conway et al. 2000; Rochet et al. 2000). The incubation of the α-synuclein protein in PBS, under the conditions described in ‘Experimental details’, leads to the formation of small oligomers after a period of approximately 12 h. These oligomers transform into protofibrils after an incubation of 36 h in PBS. The fully formed α-synuclein fibrils are obtained after an incubation of 96 h. The relationship between the oligomers, protofibrils, and the fully formed α-synuclein fibrils is still unclear. The TEM image of one such α-synuclein fibril is shown in Fig. 1a. The twisted morphology of the fibril can be observed in the image. Figure 1b is another TEM image of the α-synuclein fibrils. Here, two fibrils appear to have wound around each other.
Fig. 1.
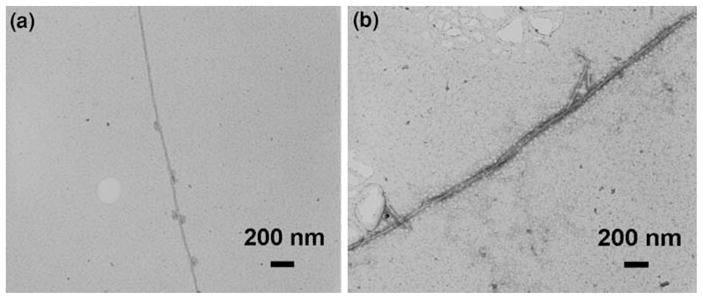
a A single α-synuclein fibril. b Two α-synuclein fibrils twisted around each other
Synthesis and characterization of ZnS nanoparticle chains
The morphology and average diameter of the nanoparticle chains were obtained after analyzing the FESEM and TEM images of the ZnS samples. Figure 2a and b shows one FESEM and one TEM image, respectively, of ZnS nanoparticle chains. The average diameter for these samples was approximately in the range of 60–65 nm. The inset in the TEM image shows the α-synuclein template between he two ZnS nanoparticles, possibly stained by the metal salt. The information garnered from the inset confirms the formation of these nanoparticles on the α-synuclein template.
Fig. 2.
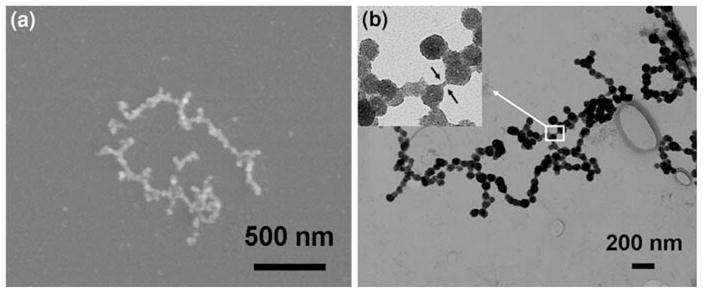
a FESEM image of ZnS nanoparticle chains. b TEM image of ZnS nanoparticle chains. The inset is a zoomed-in image that shows the highlighted area where what may be a single α-synuclein fibril can be clearly seen between two ZnS nanoparticles
High-resolution transmission electron microscopic imaging was carried out on the ZnS nanoparticle chains, to study the nanocrystalline nature of the samples. The HRTEM images indicate that the nanoparticles are composed of several nanocrystals which have an approximate dimension of ~2 nm. Figure 3 is a HRTEM image of a ZnS sample. The lattice fringes are clearly visible, which indicate the nanocrystalline nature of the ZnS sample, shown in the inset at the bottom-right of the image. The inset is a zoomed-in image of a, 2–3 nm sized, ZnS nanoparticle from the highlighted region. It is viewed along a 110 zone axis and the lattice fringes can be clearly resolved. The other inset at the top-left of the image is a Fast Fourier transform (FFT) from the same highlighted region. This FFT has been indexed based on the symmetry and lattice spacing and can be assigned to the FCC pattern along a 110 zone axis, indicating the zinc blende structure of ZnS.
Fig. 3.
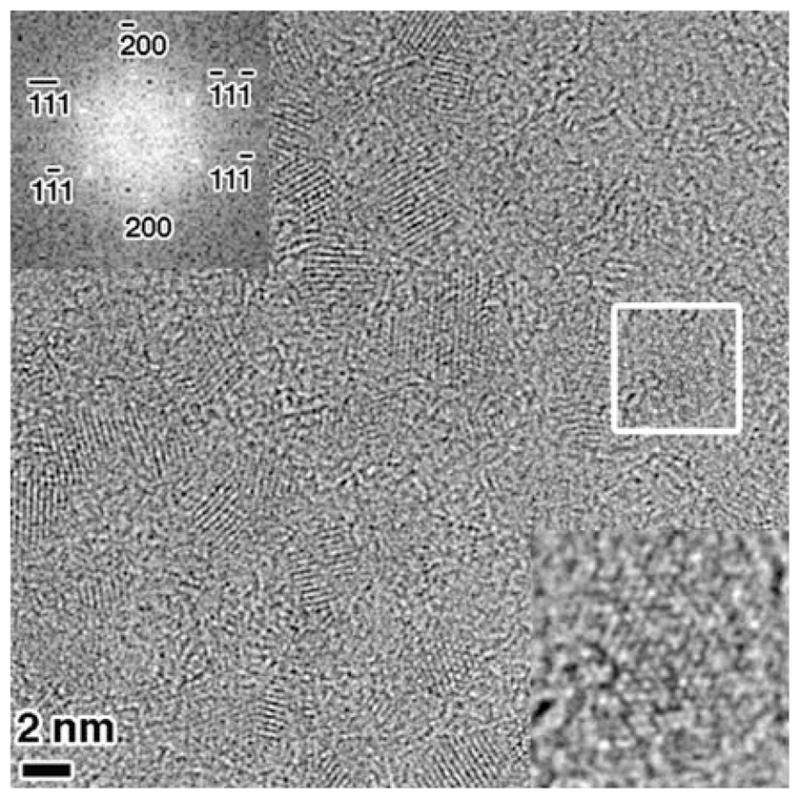
HRTEM image of ZnS nanoparticle chains. The inset at the bottom-right is a zoomed-in image of the highlighted region, showing the lattice fringes and the other inset at the top-left is an indexed FFT from the same highlighted region
In addition to the structural information from a localized area (the single nanoparticle), a selected area diffraction (SAD) pattern were obtained from one of the ZnS nanoparticle chains to confirm the crystal structure. Figure 4 shows a bright field image of a ZnS nanoparticle chain and the inset shows the diffraction pattern obtained. The SAD pattern shows (111), (220), and (311) reflection rings, which match the spacing of corresponding reflections of ZnS zinc blende structure, but the (200) reflection is not quite distinguishable from (111) reflection. However, the first ring in the SAD pattern has very strong intensity and broad intensity distribution, and (111) and (200) lattice spacings of ZnS zinc blende structure from JCPDS are within the strong and broad first ring. This is most probably caused by very small sizes (2–3 nm) of ZnS nanocrystals, as confirmed in Fig. 3. It is very well known that the diffraction ring from nanocrystals should be much broader compared to its bulk form. Therefore, based on HRTEM imaging and SAD pattern, the structure of ZnS nanoparticles in the chain can be assigned as a zinc blende structure with little ambiguity.
Fig. 4.
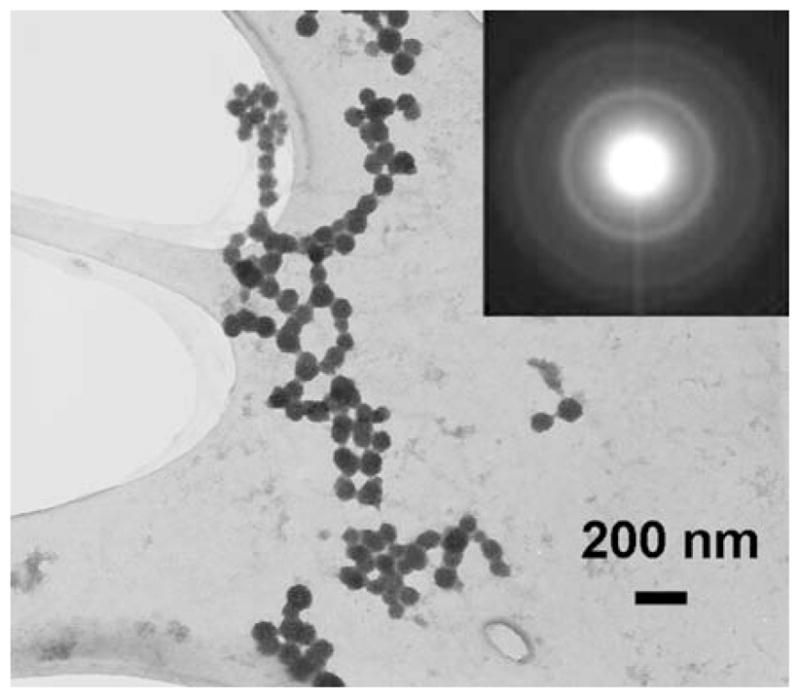
A TEM image of a ZnS nanoparticle chain. The inset shows the SAD pattern obtained from the ZnS sample
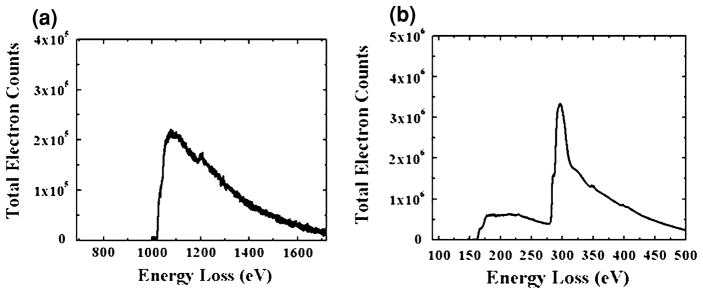
Along with the SAD results, the presence of Zn and S in ZnS nanoparticle chains was ascertained by performing EELS on the ZnS samples. The EELS obtained from the ZnS sample are shown in Fig. 5a and b. Figure 5a shows the Zn L3, L2 edges at 1,020 and 1,043 eV. The Zn L1 edge at 1,194 eV is also clearly visible, even though it is a minor edge. Figure 5b shows the sulfur L2,3 edge at 165 eV. These spectra confirm the presence of Zn and S in the sample, further supporting the SAD results.
Fig. 5.
Background subtracted EELS obtained from ZnS nanoparticle chains, showing a the Zn L3, L2, and L1 edges at 1,020, 1,043, and 1,194 eV, respectively, and b the S L2,3 edge at 165 eV
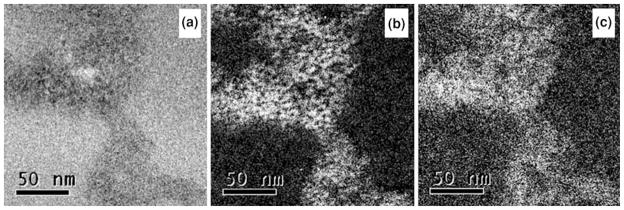
Elemental mapping using the Zn L3 edge at 1,020 eV and the S L2,3 edge at 165 eV was also performed on one of the ZnS nanoparticle chains to study the distribution of Zn and S in the nanoparticle chains. Figure 6a shows a zero energy loss image, followed by the sulfur (Fig. 6b) and zinc (Fig. 6c) maps. These elemental maps show that Zn and S are uniformly distributed through the whole nanoparticle chains.
Fig. 6.
a Zero energy loss image, b sulfur map using L2,3 edge at 165 eV, and c zinc map using L3 edge at 1,020 eV
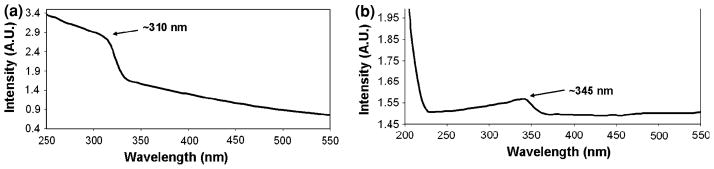
The next step in the characterization of the ZnS nanoparticle chains was to perform UV–Vis spectroscopy on the ZnS nanoparticle chain sample and on ZnS colloidal sample having a particle size of 10 μm, purchased from Sigma Aldrich for comparison of the absorption peaks. To obtain a UV–Vis spectrum, the sample was prepared in the solution form. The α-synuclein fibrils, suspended in phosphate buffer, were pipetted in an Eppendorf tube. To this the ZnCl2 solution was added and incubated for 5 min. The H2S gas was then made to pass through the solution mixture for 2 min. Another absorption spectrum was obtained from the colloidal ZnS sample. Figure 7 shows two UV–Vis spectra obtained from ZnS nanoparticle chains (a) and ZnS powder (b). The spectrum for 2 min H2S gas exposure showed an absorption peak at ~310 nm. The absorption peak for the colloidal ZnS sample was obtained at ~345 nm. The absorption peak at ~310 nm for the 2 min H2S exposure appears to have blue shifted (Jovanovic et al. 2007; Yu et al. 2005). This shift is consistent with the quantum confinement effect. The UV–Vis results were confirmed with the help of HRTEM images obtained from the ZnS nanoparticle chain sample. The HRTEM images indicate that the nanoparticle chains are composed of nanocrystals, having a dimension of ~2 nm (20 Ǻ).
Fig. 7.
UV–Vis spectra obtained from a ZnS nanoparticle chain sample. a An absorption peak at ~310 nm corresponds to the 2 min H2S gas exposure time. b The next absorption spectrum was obtained from the colloidal ZnS sample, showing an absorption spectrum of ~345 nm
Nanoparticles assembled into functional structures hold promise for applications in nanocircuitry and therefore designing nanoparticle chains with controlled electrical properties is of practical and scientific interest. However, to realize nanoscale interconnects based on biological templates as building blocks for nanocircuits, it is critical to achieve control on the diameter of the synthesized nanostructures. A change in the nanoparticle chains’ lateral dimension has a dramatic influence on the resistance per unit length. Because of this relationship between size and resistance, the inability to control the nanoparticle diameter can limit the application of these functional materials in nanoscale electronic devices. In an attempt to achieve control of the nanoparticles’ diameters a series of samples were prepared with varying process conditions. In the first set of experiments, three samples were prepared with varying H2S gas exposure times. The first sample was exposed to H2S gas for 2 min, followed by two more samples exposed for 5 and 10 min, respectively. All the samples were prepared with a concentration of the salt solution (ZnCl2) at 2 mM and a pH value of ~5.0 for the salt solution. Figure 8 shows TEM images of ZnS nanoparticle chains obtained at varying H2S gas exposure times. From the TEM images, it can be clearly seen that the diameter of the ZnS nanoparticles increases with an increase in the H2S gas exposure time. The diameter of the nanoparticles thus can be varied from ~30 to ~165 nm.
Fig. 8.
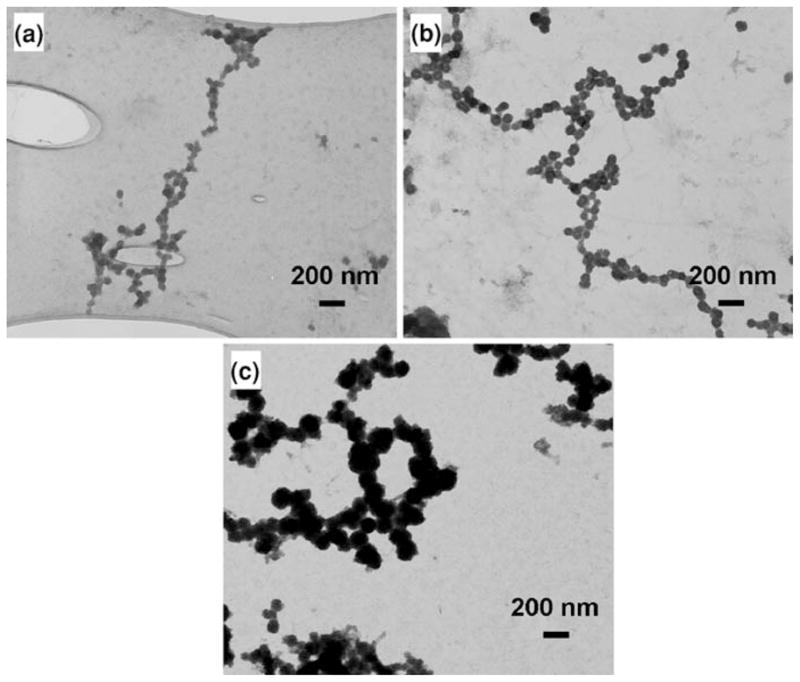
TEM images of ZnS nanoparticle chains obtained after a 2 min, b 5 min and c 10 min of H2S gas exposure
A similar set of experiments was carried out to investigate the effect of the pH value of the salt solution (ZnCl2) on the size and morphology of the ZnS nanoparticle chains. For this experiment three samples were prepared with varying pH values of the salt solution. The first sample was prepared with a pH of 4 followed by the next two samples prepared with a pH value of 5 and 6. All the samples were synthesized with a concentration of the salt solution at 2 mM, and the H2S gas exposure time was 2 min. Figure 9 shows TEM images of the ZnS nanoparticle chains obtained by varying the pH values of the salt solution. From the TEM images, it is evident that the diameter of the nanoparticles increases with an increase in the pH value of the salt solution. Thus, the diameter of the nanoparticle chains can be tuned by varying the process variables such as H2S gas exposure time and the pH value of the salt solution.
Fig. 9.
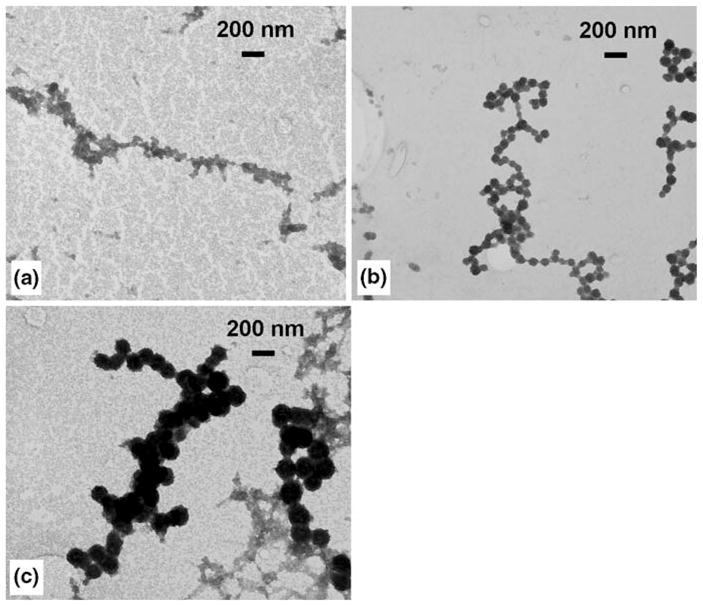
TEM images of ZnS nanoparticle chains obtained for different pH values of the salt solution: a pH 4, b pH 5, and c pH 6
Controlling the size and packing density of nanocrystals on a biological scaffold can be an effective way of tuning the electrical properties of the nanostructures. To achieve controlled growth kinetics of nanocrystals on the self-assembled polypeptide scaffold, it is important to understand the parameters that influence their formation. The reduction of ionic silver to a metallic form in the presence of proteins and DNA was first described by Merril et al. (1981) and Merril (1990). The method is widely applied for the detection of proteins and nucleic acids and is based on the differences between the redox potentials of the biomolecules and those of the matrix. A similar chemical mechanism, in which the metal ions are selectively reduced to a metallic form in the presence of biomolecules was previously used in our laboratory for the fabrication of inorganic nanoparticle chains on biological scaffolds, both metallic and semiconducting (Padalkar et al. 2007, 2008). During the synthesis process, the cations from a salt source [e.g., AgNO3, CdCl2, aned Pb(NO3)2] that reacts with the negatively charged aminoacyl side chains of the protein, at basic pH (Merril et al. 1981; Merril 1990). When these cations are subsequently reduced to the elemental state, metallic nanoparticle chains grow on the protein template. The negatively charged C-terminal domain of α-synuclein contains five aspartate and ten glutamate negatively charged side chains and therefore has the potential of forming complexes with metal cations and subsequently nucleating nanocrystals on the fiber surface. A number of studies have revealed that these negatively charged side chains into the fibril is not known with precision, based on our results that show formation of ZnS nanoparticle chains on the protein fiber scaffold, it can be speculated that some of these negatively charged aminoacyl side chains are exposed at the fiber’s surface rather than buried within the fiber (Qin et al. 2007; Chen et al. 2007; Heise et al. 2005; Murray et al. 2003). For semiconductor compounds, such as ZnS, the metal cations bind to the same negatively charged aminoacyl side chains of α-synuclein. After the introduction of the sulfide anion, semiconductor nanoparticles are expected to nucleate on the protein fiber surface. It could be speculated that there are several regions along the protein fiber where the protein side groups have a significant affinity for the semiconductor nanoparticles, leading to their stabilization.
The same generic mechanism could be expanded to other polypeptide scaffolds and can therefore be of significant potential importance for the field of designing bottom–up strategies for nanomaterials fabrication on biomolecular templates. Our results prove the biomineralization capacities of the α-synuclein protein, and can be extended to other fibrillar proteins or polypeptides.
In an additional experiment, the effect of incubation temperature of the salt solution on the morphology of the nanoparticle chains was studied (the previous studies described in this report were carried out at 22 °C). Here, two separate experiments were carried out where the salt solution was heated to a temperature of 45 and 85 °C for 30 min and then used in the synthesis process. The pH value of the salt solution was kept at 5 and the H2S gas exposure time was fixed to 2 min. Figure 10 shows a TEM image of a ZnS nanoparticle chain obtained by using a ZnCl2 solution at 45 °C. When the ZnS nanoparticle chains obtained at 45 °C were compared with the ZnS nanoparticle chains obtained by varying the H2S exposure time (Fig. 8a) and the pH of the salt solution (Fig. 9b), it was observed that the ZnS nanoparticles, obtained at 45 °C, were very well defined. Figure 10 has several highlighted regions (also shown magnified in the inset). The left two zoomed-in images show regions where, may be, the α-synuclein template can be seen. Although the ZnS nanoparticles look very well connected, there are a few regions along the length of the nanoparticle chains that look disconnected thus exposing the α-synuclein template. However, there are many other regions that clearly show the formation of the neck between the ZnS nanoparticles (shown in the inset to the right).
Fig. 10.
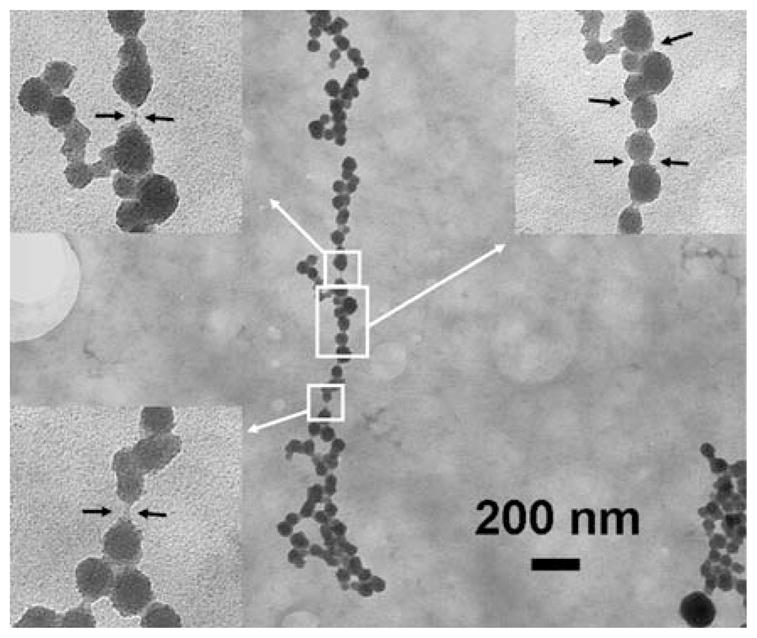
TEM image of a ZnS nanoparticle chain obtained at 45 °C. The insets in the image are zoomed-in images of the highlighted regions. The left two insets show what may be the α-synuclein template between the ZnS nanoparticles. The inset to the right shows the formation of neck regions between the nanoparticles
To improve the connectivity of the ZnS nanoparticles, the ZnCl2 salt solution was heated to a temperature of 85 °C for 30 min prior to its use in the synthesis process. The other variables used in the synthesis process were kept constant, for a better comparison with the sample obtained at 45 °C. Figure 11 is a TEM image of a ZnS nanoparticle chain obtained at 85 °C. When compared with Fig. 10, it can be clearly seen that the nanoparticles are well connected and there are no regions where the α-synuclein template can be seen. The neck regions that can be seen between ZnS nanoparticles are shown in the inset for clarity. The neck regions look very well defined and the ZnS nanoparticle chain appears more like a nanowire. Thus, by varying the incubation temperature the connectivity between the ZnS nanoparticles can be improved.
Fig. 11.
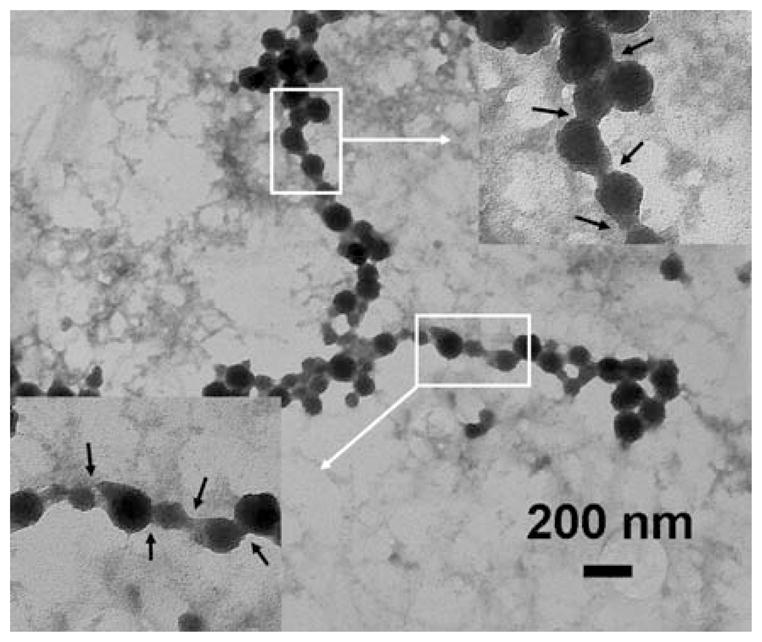
A TEM image of a ZnS nanoparticle chain. The insets are zoomed-in images of the highlighted regions. The inset clearly shows well developed neck regions between ZnS nanoparticles
The changes in the process variables help in varying the size of the nanoparticle chains and also help in varying the connectivity between the nanoparticles, thus making the nanoparticle chains more smoothly connected.
Conclusions
In summary, we report the use of α-synuclein fibrils as biological templates for the synthesis of ZnS nanoparticle chains. The size of the nanoparticle chains can be controlled by varying the process variables. This result was confirmed by TEM imaging carried out on the ZnS samples. The nanoparticles are composed of several nanocrystals having a dimension of ~2 nm. The diffraction pattern reveals the zinc blende structure of ZnS. The EELS confirm the presence of Zn and S in the ZnS nanoparticle chains. Further, elemental mapping of Zn and S shows uniform distribution of both the elements on the nanoparticle chains.
Contributor Information
S. Padalkar, Email: spadalka@purdue.edu, School of Materials Engineering, Purdue University, West Lafayette, IN 47906, USA. Birck Nanotechnology Center, Purdue University, West Lafayette, IN 47906, USA
J. Hulleman, Department of Medicinal Chemistry and Molecular Pharmacology, Purdue University, West Lafayette, IN 47906, USA
S. M. Kim, School of Materials Engineering, Purdue University, West Lafayette, IN 47906, USA. Birck Nanotechnology Center, Purdue University, West Lafayette, IN 47906, USA
T. Tumkur, School of Materials Engineering, Purdue University, West Lafayette, IN 47906, USA
J.-C. Rochet, Department of Medicinal Chemistry and Molecular Pharmacology, Purdue University, West Lafayette, IN 47906, USA
E. Stach, School of Materials Engineering, Purdue University, West Lafayette, IN 47906, USA. Birck Nanotechnology Center, Purdue University, West Lafayette, IN 47906, USA
L. Stanciu, School of Materials Engineering, Purdue University, West Lafayette, IN 47906, USA. Birck Nanotechnology Center, Purdue University, West Lafayette, IN 47906, USA
References
- Alivisatos AP. Semiconductor clusters, nanocrystals, and quantum dots. Science. 1996;271(5251):933–937. [Google Scholar]
- Bachtold A, Hadleym P, Nakanishi T, Dekker C. Logic circuits with carbon nanotube transistors. Science. 2001;294(5545):1317–1320. doi: 10.1126/science.1065824. [DOI] [PubMed] [Google Scholar]
- Chen X, Xu H, Xu N, Zhao F, Lin W, Lin G, Fu Y, Huang Z, Wang H, Wu M. Kinetically controlled synthesis of wurtzite ZnS nanorods through mild thermolysis of a covalent organic-inorganic network. Inorg Chem. 2003;42:3100–3106. doi: 10.1021/ic025848y. [DOI] [PubMed] [Google Scholar]
- Chen M, Margittai M, Chen J, Langen R. Investigation of α-synuclein fibril structure by site-directed spin labeling. J Biol Chem. 2007;282(34):24970–24979. doi: 10.1074/jbc.M700368200. [DOI] [PubMed] [Google Scholar]
- Claridge SA, Goh SL, Frechet JMJ, Williams SC, Micheel CM, Alivisatos AP. Direct assembly of discrete gold nanoparticle groupings using branched DNA scaffolds. Chem Mater. 2005;17(7):1628–1635. [Google Scholar]
- Collins PG, Arnold MS, Avouris P. Engineering carbon nanotubes and nanotube circuits using electrical breakdown. Science. 2001;292(5517):706–709. doi: 10.1126/science.1058782. [DOI] [PubMed] [Google Scholar]
- Conway KA, Harper JD, Lansbury PT., Jr Fibrils formed in vitro from alpha-synuclein and two mutant forms linked to Parkinson’s disease are typical amyloid. Biochemistry. 2000;39(10):2552–2563. doi: 10.1021/bi991447r. [DOI] [PubMed] [Google Scholar]
- Conway KA, Rochet JC, Bieganski RM, Lansbury PT., Jr Kinetic stabilization of the α-synuclein protofibril by a dopamine-α-synuclein adduct. Science. 2001;294:1346–1349. doi: 10.1126/science.1063522. [DOI] [PubMed] [Google Scholar]
- Cui Y, Wei QQ, Park HK, Lieber CM. Nanowire nanosensors for highly sensitive and selective detection of biological and chemical species. Science. 2001;293:1289–1292. doi: 10.1126/science.1062711. [DOI] [PubMed] [Google Scholar]
- Deng Z, Mao C. DNA templated fabrication of 1D Parallel and 2D crossed metallic nanowire array. Nano Lett. 2003;3(11):1545–1548. [Google Scholar]
- DePace AH, Weissman JS. Origins and kinetic consequences of diversity in Sup35p yeast prion fibers. Nat Struct Biol. 2002;9(5):389–396. doi: 10.1038/nsb786. [DOI] [PubMed] [Google Scholar]
- Diehl MR, Yaliraki SN, Beckman RA, Barahona M, Heath JR. Self-assembled, deterministic carbon nanotube wiring networks. Angew Chem Int Edit. 2002;41(2):353–356. doi: 10.1002/1521-3773(20020118)41:2<353::aid-anie353>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- Dong L, Hollis T, Connolly BA, Wright NG, Horrocks BR, Houlton A. DNA templated semiconductor nanoparticle chains and wires. J Adv Mater. 2007;19(13):1748–1751. [Google Scholar]
- Flynn CE, Lee SW, Peelle BR, Belcher AM. Viruses as vehicles for growth, organization and assembly of materials. Acta Mater. 2003;51(19):5867–5880. [Google Scholar]
- Heise H, Hoyer W, Becker S, Andronesi OC, Riedel D, Baldus M. Molecular-level secondary structure, polymorphism, and dynamics of full length α-synuclein fibrils studied by solid state NMR. Proc Natl Acad Sci USA. 2005;102(44):15871–15876. doi: 10.1073/pnas.0506109102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyer W, Antony T, Cherny D, Heim G, Jovin TM, Subramaniam V. Dependence of α-synuclein aggregate morphology on solution conditions. J Mol Biol. 2002;322(2):383–393. doi: 10.1016/s0022-2836(02)00775-1. [DOI] [PubMed] [Google Scholar]
- Huang Y, Duan X, Cui Y, Lauhon LJ, Kim KH, Lieber CM. Logic gates and computation from assembled nanowire building blocks. Science. 2001;294:1313–1317. doi: 10.1126/science.1066192. [DOI] [PubMed] [Google Scholar]
- Huang Y, Chiang CY, Lee SK, Gao Y, Hu EL, De Yoreo J, Belcher AM. Programmable assembly of nanoarchitectures using genetically engineered viruses. Nano Lett. 2005;5(7):1429–1434. doi: 10.1021/nl050795d. [DOI] [PubMed] [Google Scholar]
- Jiang X, Xie Y, Lu J, Zhu L, He W, Qian Y. Simultaneous in situ formation of ZnS nanowires in a liquid crystal template by gamma radiation. Chem Mater. 2001;13:1213–1218. [Google Scholar]
- Johnson JC, Choi HJ, Knutsen KP, Schaller RD, Yang P, Saykally RJ. Single gallium nitride nanowire laser. Nat Mater. 2002;1(2):106–110. doi: 10.1038/nmat728. [DOI] [PubMed] [Google Scholar]
- Jovanovic D, Validzic I, Jankovic I, Bibic N, Nedeljkovic J. Synthesis and characterization of shaped ZnS nanocrystals in water, in oil microemulsions. Mater Lett. 2007;61:4396–4399. [Google Scholar]
- Kar S, Biswas S, Chaudhuri S. Catalytic growth and photoluminescence properties of ZnS nanowires. Nanotechnology. 2003;16:737–740. [Google Scholar]
- Keren K, Berman RS, Buchstab E, Sivan U, Braun E. DNA-templated carbon nanotube field-effect transistor. Science. 2003;302(5649):1380–1382. doi: 10.1126/science.1091022. [DOI] [PubMed] [Google Scholar]
- Kimberly HS, Schwartz JJ, Santos AT, Zhang S, Jacobson JM. Remote electronic control of DNA hybridization through inductive coupling to an attached metal nanocrystal antenna. Nature. 2002;415(6868):152–155. doi: 10.1038/415152a. [DOI] [PubMed] [Google Scholar]
- Klein DL, Roth R, Lim AKL, Alivisatos AP, McEuen MP. A single electron transistor made from cadmium selenide nanocrystal. Nature. 1997;389:699–701. [Google Scholar]
- Lu F, Cai W, Zhang Y, Li Y, Sun F. Fabrication and field emission performance of zinc sulfide nanobelt arrays. J Phys Chem C. 2007;111:13385–13392. [Google Scholar]
- Ma C, Moore D, Li J, Wang Z. Nanobelts, nanocombs and nanowindmills of wurtzite ZnS. Adv Mater. 2003;15:228–231. [Google Scholar]
- Mao C, Solis DJ, Reiss BD, Kottmann ST, Sweeney RY, Hayhurst A, Georgiou G, Iverson B, Belcher AM. Virus-based toolkit for the directed synthesis of magnetic and semiconducting nanowires. Science. 2004;303(5655):213–217. doi: 10.1126/science.1092740. [DOI] [PubMed] [Google Scholar]
- Meng XM, Liu J, Jiang Y, Chen WW, Lee CS, Bello I, Lee ST. Structure and size controlled ultrafine ZnS nanowires. Chem Phys Lett. 2003;382:434–438. [Google Scholar]
- Merril CR. Silver staining of protein and DNA. Nature. 1990;343:779–780. doi: 10.1038/343779a0. [DOI] [PubMed] [Google Scholar]
- Merril CR, Miriam L, Dunau ML, Goldman D. A rapid sensitive silver stain for polypeptides in polyacrylamide gels. Anal Biochem. 1981;110:201–207. doi: 10.1016/0003-2697(81)90136-6. [DOI] [PubMed] [Google Scholar]
- Monson CF, Woolley AT. DNA-templated construction of copper nanowires. Nano Lett. 2003;3(3):359–363. [Google Scholar]
- Murray CB, Kagan CR, Bawendi MG. Synthesis and characterization of monodisperse nanocrystals and closed packed nanocrystal assembly. Annu Rev Mat Sci. 2000;30:545–610. [Google Scholar]
- Murray IV, Giasson BI, Quinn SM, Koppaka V, Axelsen PH, Ischiropoulos H, Trojanowski JQ, Lee VM. Role of α-synuclein carboxy-terminus on fibril formation invitro. Biochemistry. 2003;42(28):8530–8540. doi: 10.1021/bi027363r. [DOI] [PubMed] [Google Scholar]
- Nam KT, Kim DW, Yoo PJ, Chiang CY, Meethong N, Hammond PT, Chiang YM, Belcher AM. Virus-enabled synthesis and assembly of nanowires for lithium ion battery electrodes. Science. 2006;312(5775):885–888. doi: 10.1126/science.1122716. [DOI] [PubMed] [Google Scholar]
- Nelson R, Eisenberg D. Structural models of amyloid-like fibrils. Adv Protein Chem. 2006;73:235–282. doi: 10.1016/S0065-3233(06)73008-X. [DOI] [PubMed] [Google Scholar]
- Padalkar S, Hulleman J, Deb P, Cunzeman K, Rochet JC, Stach EA, Stanciu L. Alpha-synuclein as a template for the synthesis of metallic nanowires. Nanotechnology. 2007;18:055609. [Google Scholar]
- Padalkar S, Hulleman J, Kim SM, Rochet JC, Stach EA, Stanciu L. Protein-templated semiconductor nanoparticle chains. Nanotechnology. 2008;19:275602. doi: 10.1088/0957-4484/19/27/275602. [DOI] [PubMed] [Google Scholar]
- Qin Z, Hu D, Han S, Hong DP, Fink AL. Role of different regions of α-synuclein in the assembly of the fibrils. Biochemistry. 2007;46(46):13322–13330. doi: 10.1021/bi7014053. [DOI] [PubMed] [Google Scholar]
- Richter J, Mertig M, Pompe W, Monch I, Schackert HK. Construction of highly conductive nanowires on a DNA template. Appl Phys Lett. 2001;78(4):536–538. [Google Scholar]
- Rochet JC. Novel therapeutic strategies for the treatment of protein-misfolding disease. Expert Rev Mol Med. 2007;9(17):1–34. doi: 10.1017/S1462399407000385. [DOI] [PubMed] [Google Scholar]
- Rochet JC, Conway KA, Lansbury PT., Jr Inhibition of fibrillization and accumulation of prefibrillar oligomers in mixtures in human and mouse alpha synuclein. Biochemistry. 2000;39:10619–10626. doi: 10.1021/bi001315u. [DOI] [PubMed] [Google Scholar]
- Scheibel T, Lindquist SL. The role of conformational flexibility in prion propagation and maintenance for Sup35p. Nat Struct Biol. 2001;8(11):958–962. doi: 10.1038/nsb1101-958. [DOI] [PubMed] [Google Scholar]
- Scheibel T, Parthasarathy R, Sawicki G, Lin XM, Jaeger H, Lindquist SL. Conducting nanowires built by controlled self assembly of amyloid fibers and selective metal deposition. Proc Natl Acad Sci USA. 2003;100:4527–4532. doi: 10.1073/pnas.0431081100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serio TR, Cashikar AG, Kowal AS, Sawicki GJ, Moslehi JJ, Serpell L, Arnsdorf MF, Lindquist SL. Nucleated conformational conversion and the replication of conformational information by a prion determinant. Science. 2000;289(5483):1317–1321. doi: 10.1126/science.289.5483.1317. [DOI] [PubMed] [Google Scholar]
- Serpell LC, Berriman J, Jakes R, Goedert M, Crowther RA. Fiber diffraction of synthetic α-synuclein filaments shows amyloid-like cross-beta conformation. Proc Natl Acad Sci USA. 2000;97(9):4897–4902. doi: 10.1073/pnas.97.9.4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spillantini MG, Schmidt ML, Lee VM, Trojanowski JQ, Jakes R, Goedert M. α-Synuclein in Lewy bodies. Nature. 1997;388:839–840. doi: 10.1038/42166. [DOI] [PubMed] [Google Scholar]
- Vilar M, Chou HT, Maji SK, Riek-Loher D, Verel R, Manning G, Stahlberg H, Riek R. The fold of α-synuclein fibrils. Proc Natl Acad Sci USA. 2008;105(25):8637–8642. doi: 10.1073/pnas.0712179105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhang L, Liang C, Wang G, Peng X. Catalytic growth and photoluminescence properties of semiducting single-crystal ZnS nanowires. Chem Phys Lett. 2002;357:314–318. [Google Scholar]
- Xu X, Fei G, Yu W, Wang X, Chen L, Zhang L. Preparation and formation mechanism of ZnS semiconductor nanowires made by electrochemical deposition method. Nanotechnology. 2006;17:426–429. [Google Scholar]
- Yan H, Park SH, Finkelstein G, Reif JH, LaBean TH. DNA-templated self-assembly of protein arrays and highly conductive nanowires. Science. 2003;301(5641):1882–1884. doi: 10.1126/science.1089389. [DOI] [PubMed] [Google Scholar]
- Yin L, Bando Y, Zhan J, Li M, Goldberg D. Self-assembled highly faceted wurtzite-type ZnS single crystalline nanotubes with hexagonal cross-sections. Adv Mater. 2005;17:1972–1977. [Google Scholar]
- Yu J, Joo J, Park H, Baik S, Kim Y, Kim S, Hyeon T. Synthesis of quantum sized cubic ZnS nanorods by the oriented attachment mechanism. J Am Chem Soc. 2005;127:5662–5670. doi: 10.1021/ja044593f. [DOI] [PubMed] [Google Scholar]
- Zhang D, Qi L, Cheng H, Ma J. Preparation of ZnS nanorods by a liquid crystal template. J Colloidal Interface Sci. 2002;246:413. doi: 10.1006/jcis.2001.8081. [DOI] [PubMed] [Google Scholar]