Abstract
Most regions of the central nervous system contain numerous subtypes of inhibitory interneurons that play specialized roles in circuit function. In mammalian retina, the ~30 subtypes of inhibitory interneurons called amacrine cells (ACs) are generally divided into two groups: wide/medium-field GABAergic and narrow-field glycinergic, which mediate lateral and vertical interactions, respectively, within the inner plexiform layer. We used expression profiling and mouse transgenic lines to identify and characterize two closely-related narrow-field AC subtypes. Both arise postnatally and one, surprisingly, is neither glycinergic nor GABAergic (nGnG). Two transcription factors selectively expressed by these subtypes, Neurod6 and Satb2, regulate a postmitotic cell fate choice between them. Satb2 induces Neurod6, which persists in nGnG ACs and promotes their fate, but is down-regulated in the related glycinergic AC subtype. Our results support the view that cell fate decisions made in progenitors and their progeny act together to diversify ACs.
INTRODUCTION
Recent studies have demonstrated a remarkable diversity of inhibitory neurons in many regions of the mammalian central nervous system (CNS), including cortex, hippocampus, spinal cord and retina1-4. Classifying these interneurons is essential for understanding how neural circuits function and learning how they diversify from progenitors is essential for understanding how neural circuits assemble.
Amacrine cells (ACs), the inhibitory interneurons of the retina, are well-suited for addressing these issues. Approximately 30 AC subtypes have been defined by morphological criteria3,5-8, a number similar to that found in other CNS regions. These subtypes are generally divided into two broad classes: wide/medium- and narrow-field ACs, which use γ-aminobutyric acid (GABA) or glycine, respectively, as neurotransmitters, often along with a co-transmitter or neuropeptide6. Wide/medium-field ACs project to individual sublaminae of the inner plexiform layer (IPL) and mediate lateral interactions that shape receptive fields of the retina’s output neurons, retinal ganglion cells (RGCs). Most narrow-field ACs, in contrast, project to multiple IPL sublaminae, mediating vertical interactions across parallel circuits6,9. Subtypes within these broad classes play specific roles in determining the visual features to which the ~20 RGC subtypes selectively respond.
Increasingly, molecular criteria are being paired with morphological criteria to better classify inhibitory interneurons. Here, we used gene expression profiling to identify molecular markers that in turn allowed us to define and characterize two closely-related, diffusely stratified narrow-field AC subtypes. One is glycinergic, but surprisingly, the other is neither glycinergic nor GABAergic. This result is not completely unexpected, in that several studies have shown that GABAergic and glycinergic markers are present in <100% of ACs10-13. Nonetheless, no previous studies have characterized non-GABAergic non-glycinergic (nGnG) ACs.
In the second part of this paper, we consider how these two AC subtypes arise. The competence of retinal progenitors changes over time, such that they sequentially generate the main neuronal types14. Transcription factors acting in progenitors to promote the AC fate include Foxn4, Neurod1, Neurod4 and Ptf1a6,15-18. We and others showed previously that GABAergic ACs are born prior to glycinergic ACs12,13, suggesting that the competence model may also apply to neuronal subtypes. We show here that nGnG ACs are born after glycinergic ACs. We also characterize a transcriptional regulatory network involving Satb2 and Neurod6 that acts postmitotically to determine whether a late-born AC becomes nGnG or the related glycinergic subtype. Together, these results support the view that cell fate decisions made both in progenitors and their progeny act to diversify interneurons14,19,20.
RESULTS
Non-GABAergic non-glycinergic (nGnG) amacrine cells
Amacrine cells (ACs) are conventionally divided into groups that use GABA or glycine as their neurotransmitter. Some studies suggest, however, that these classes do not account for all ACs10-13. To test this idea, we triple-stained sections of adult mouse retina with antibodies to glutamic acid decarboxylase (Gad65/67, abbreviated here as GAD), which label all GABAergic neurons; to glycine cell membrane transporter 1 (GlyT1), which label all retinal glycinergic neurons21,22; and to either Syntaxin-1 (Stx1) or Pax6, both of which label all ACs11,23. The GABAergic and glycinergic AC populations were mutually exclusive and accounted for ~85% of all ACs (Fig. 1a,c and data not shown). Based on this result and on further studies detailed below, we refer to the GAD−GlyT1− AC population as non-GABAergic, non-glycinergic or “nGnG” ACs. To ask whether nGnG ACs were a peculiarity of mice, we performed similar staining on macaque monkey retina; again GAD−GlyT1− ACs were prominent, with a prevalence similar to that in mice (Fig. 1b).
Figure 1. Non-GABAergic, non-glycinergic ACs.
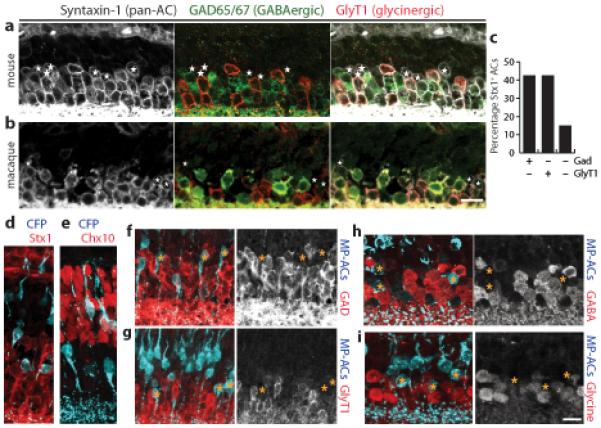
a,b: Mouse (a) and macaque (b) retina sections triple-stained for a pan-AC marker, Syntaxin-1 (white); a glycinergic AC marker, GlyT1 (red); and a GABAergic AC marker, GAD (green). Glycinergic and GABAergic ACs are mutually exclusive. Asterisks mark ACs that do not show GAD or GlyT1 immunoreactivity (nGnG ACs).
c: Percentage of Syntaxin-1 (Stx1)-positive ACs that are GABAergic, glycinergic, or neither in adult mouse retina (n ≥ 200 cells counted for each cell type).
d,e: Retinal sections from the MP transgenic mouse line stained with anti-GFP to reveal CFP+ cells (blue). A subset of Stx1+ ACs (red, d) and Chx10+ bipolar cells (red, e) are CFP+. The CFP+ AC and bipolar populations can be distinguished based on their laminar position within the INL.
f–i: CFP+ MP-ACs (blue) are not immunoreactive for GAD (f, red), GlyT1 (g, red), GABA (h, red) or glycine (i, red). Right panels show marker alone, with asterisks to mark the location of MP-ACs. Mouse tissue from P15 (a,d–g) or adult (>P40) animals (h,i). Scale bar for (a,b) and for (d–i) = 10 μm.
To study nGnG ACs in detail, we sought a marker for them by screening available transgenic mouse lines for fluorescent protein (XFP) expression in AC subsets. Of particular interest were lines in which XFPs were expressed under the control of regulatory elements from the Thy1 gene; neuronal subsets are labeled in some such lines, presumably owing to influences from the genomic site of integration24,25. In one Thy1 line25, denoted MP here, a mitochondrially targeted cyan fluorescent protein (CFP) was expressed by subsets of ACs and bipolar cells26 (Fig. 1d,e). Strikingly, <2% of CFP+ ACs in these mice were GAD+ or GlyT1+ (sections: Fig. 1f,g; dissociated cells: Supplementary Fig. S1). Thus, most if not all MP+ ACs are nGnG ACs.
We used the MP line to further characterize the neurotransmitter phenotype of nGnG ACs. First, we considered the possibility that they might express GlyT2 (Slc6a5), a marker of glycinergic neurons in other CNS regions. No MP+ ACs expressed GlyT2 as judged by in situ hybridization (ISH; not shown). Second, we stained tissue sections with antibodies to the transmitters themselves. Less than 5% of CFP+ cells were immunoreactive for GABA or glycine, even though both neurotransmitters were readily detectable in neighboring ACs (Fig. 1h,i). Thus, a substantial subset of ACs is neither GABAergic nor glycinergic, and we can use the MP transgene to mark this subset.
Neurod6 is selectively expressed by nGnG amacrine cells
To identify endogenous molecular markers of nGnG ACs, we purified them from the MP line at postnatal day (P)6 (Fig. 2a), verified their purity (Fig. 2b), and inventoried the genes they express using Affymetrix microarrays. Gene expression data were compared to those obtained from six other amacrine and bipolar cell types (J.N.K and J.R.S, unpublished), including the MP-CFP+ bipolar cells (Fig. 2a–c), allowing us to identify genes selectively expressed in nGnG ACs (Fig. 2c). Using ISH or immunohistochemistry, we found 8 genes (out of 14 tested) that proved excellent markers of CFP+ ACs in MP retina (Supplementary Table 1). Several were also expressed by other AC subsets (see below), but Neurod6 (previously called NEX or Math-2), encoding a transcription factor of the basic-helix-loop-helix (bHLH) family27,28, was exclusively expressed by CFP+ nGnG ACs (Fig. 2d).
Figure 2. Transcriptional profiling of MP-CFP+ amacrine and bipolar cells.
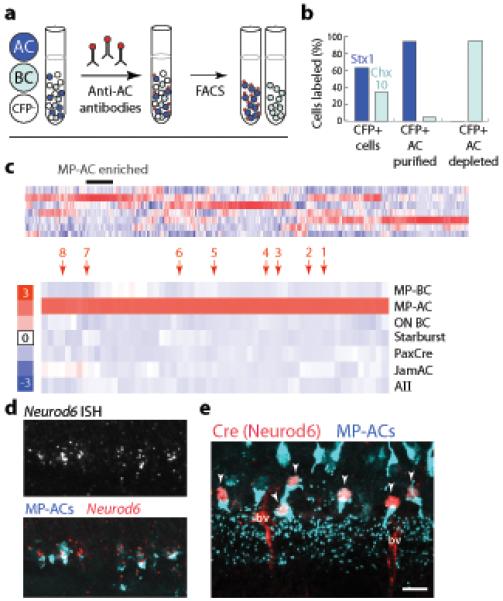
a: Method used to purify CFP+ ACs and bipolar cells (BCs) from MP transgenic mice. Dissociated retina contains CFP− cells (white), CFP+ ACs (dark blue), and CFP+ BCs (light blue). Monoclonal antibody VC1.1, recognizing a cell-surface epitope specific to ACs, was applied to the suspension, allowing a second fluorophore (red) to be introduced onto ACs. Two-color flow cytometry (FACS) was then used to collect CFP+VC1.1+ ACs or CFP+VC1.1− BCs.
b: Purity of the sorted populations. CFP+ cells were plated with or without VC1.1-based sorting and stained for pan-AC (Stx1) or pan-BC (Chx10) markers. Expected cell types were strongly enriched.
c: Top: Heatmap showing genes clustered according to their expression level across the 7 cell types in the microarray dataset. Scale (red = high; blue = low) indicates gene expression level in each cell type relative to the mean for that gene. MP-AC enriched genes are marked. Bottom: Portion of the heatmap containing genes that were strongly expressed by MP-ACs but not other cell types (labeled at right) in the dataset. Genes indicated by arrows: 1: Ebf3; 2: Neurod6; 3: 6430573F11rik; 4: Satb2; 5: Pde5a; 6: Galr2; 7: Frem1; 8: Pkdcc.
d: ISH for Neurod6 in line MP retina at P7. Neurod6+ cells (red) are CFP+ (blue).
e: Neurod6:Cre mice carrying the MP transgene immunostained for CFP and Cre at P15. All Cre+ cells (red) were CFP+ (arrowheads). Second antibodies non-specifically stain blood vessels (bv). Scale bar (d,e) = 10 μm.
Based on this result, we obtained a mouse line in which Cre recombinase had been targeted to the Neurod6 locus by homologous recombination (Neurod6:Cre, initially denoted NEX-Cre)28. We mated these mice to the MP line and stained sections with anti-Cre. Cre immunoreactivity was detectable only in CFP+ nGnG ACs (Fig. 2e). Thus, our cell purification and microarray strategy allowed us to identify an endogenous marker for the nGnG ACs.
nGnG amacrines are a lamina-restricted narrow-field subtype
Morphological classification of AC subtypes centers on the IPL sublaminae in which their processes arborize and the extent of their arbors in the tangential plane. To classify nGnG ACs, we crossed Neurod6:Cre mice to a Cre-dependent YFP reporter line29. An additional subset of glycinergic ACs was also labeled in these Neurod6:Cre x Thy1-stop-YFP (Nd6CY) mice, owing to an earlier phase of Neurod6-driven Cre expression; their identity is discussed below. A small number of cells in other layers was also labeled. We therefore limited initial analysis to Nd6CY+GlyT1− ACs.
nGnG ACs project primarily to the outer (OFF) portion of the IPL (Fig. 3a). Double-staining for calbindin, which marks bands in IPL sublaminae S2, S3, and S4 (using the convention of dividing the IPL into 5 equal sublaminae30), demonstrated that nGnG AC terminals are predominantly confined to sublaminae S1–S3 (Fig. 3b). Analysis of peripheral retina, in which well-isolated cells could be found, revealed that all nGnG ACs were narrow-field (Fig. 3c), with a dendritic field area of ~1100 μm2. As an independent morphological assay, we injected low titer YFP-expressing retrovirus into P0 retina, to label dividing progenitor cells, then identified nGnG ACs among the progeny with subtype-specific markers (see Methods). All nGnG ACs labeled in this manner were narrow-field cells, with arbors that ramified across S1–S3 (Fig. 3d). Thus, nGnG ACs are narrow-field, multistratified, OFF-type ACs.
Figure 3. Morphology and development of nGnG ACs.
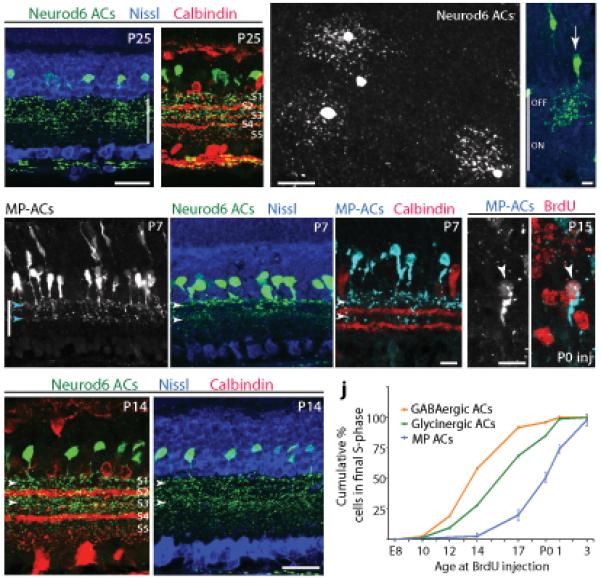
a,b: Morphology of ACs labeled in Nd6CY mice (anti-GFP, green). Anti-calbindin (red, b) marks IPL sublaminae S2, S3, and S4. Arbors of YFP+ ACs stratify predominantly in S1–S3. Nissl counterstain (blue, a) reveals retinal layers.
c: Wholemount adult Nd6CY retina stained with anti-GFP, viewed en face. Narrow-field YFP+ cells are evident in this retinal region with sparse YFP labeling.
d: Single nGnG cell from P20 retina (green), labeled with YFP-retrovirus, identified as nGnG using markers described in Methods. Blue = Nissl counterstain. nGnG ACs are multistratified in S1–S3.
e–g: Morphology of nGnG ACs in MP (e,g) and Nd6CY (f) mice at P7. Note bistratified projection to IPL strata S1 and S3 (arrowheads).
h: Morphology of nGnG ACs at P14 (Nd6CY mice; green). Co-labeling with calbindin (red) or Nissl (blue). Projections to S1 and S3 are still prominent (arrowheads) but S2 is also labeled.
i: Bromodeoxyuridine (BrdU) immunolabeling (red) in MP mice injected with BrdU at P0. Many nGnG ACs (left = gray; right = blue) are BrdU+ (arrowhead), indicating that they undergo their final cell cycle at P0.
j: Birthdate curve for nGnG ACs, constructed by injecting BrdU at 9 time points (P5 not shown) and counting cells double-positive for CFP and BrdU at P15 (as in i). Plot shows cumulative percentage of MP-AC population born by a given age. For comparison, birthdate curves of GABAergic and glycinergic ACs are replotted from ref. 12.
Scale bars (a–c,h) = 25 μm; (d–g,i) = 10 μm. Vertical bar in a,d,e = IPL.
AC somata of single subtypes are clustered at restricted levels within the INL12. Analysis of sections from MP, Neurod6:Cre and Nd6CY mice (CFP+, Cre+ and YFP+ cells, respectively) indicated that nGnG AC somata are aligned at the outermost edge of the AC zone, just proximal to the region of bipolar cell somata (Supplementary Fig. S2). This result supports the idea that nGnG ACs may comprise just one or a few subtypes.
Late birth and development of nGnG amacrine cells
We used the MP and Nd6CY lines to analyze the development of nGnG ACs. First, we asked whether nGnG AC processes target appropriate IPL sublaminae from the outset or whether their arbors remodel as development proceeds. Some AC subtypes are restricted to their definitive sublaminae during the first few postnatal days31. In contrast, projections of MP+ and Neurod6+ ACs were bistratified in S1 and S3, but absent from S2 at P4–P7 (Fig. 3e–g and data not shown). At P14, however, soon after eye opening, arbors filled S1–S3 (Fig. 3h), in a pattern indistinguishable from that seen in adults (Fig. 3a,b). Thus, nGnG AC arbors remodel postnatally.
Second, we used bromodeoxyuridine (BrdU) labeling (Fig. 3i) to ask when nGnG ACs are generated. Most GABAergic ACs are generated prior to most glycinergic ACs12,13. BrdU birthdating revealed that nGnG ACs are born after most GABAergic and glycinergic ACs (Fig. 3j), suggesting that three distinct waves of neurogenesis produce the GABAergic, glycinergic, and nGnG subtypes in succession (Fig. 3j). Thus, nGnG ACs are generated later and refine their arbors later than most other AC subtypes.
Role of Neurod6 in nGnG fate selection
Because Neurod6 is selectively expressed in nGnG ACs from early postnatal life, we wondered if this transcriptional regulator has a role in nGnG development. Homozygotes of the Neurod6:Cre line are Neurod6 null mutants28. The mutants were viable and fertile, the retina was grossly normal in size and lamination, and major retinal cell types were present in normal positions and numbers (Supplementary Fig. S3). Likewise, when we crossed the MP line into the Neurod6 mutant background, CFP+ cells were present in normal numbers in the mutant, they continued to express AC markers, and none were GAD+ (Supplementary Fig. S4 and data not shown). However, unlike wild-type CFP+ cells, which almost never express GlyT1, over half of the mutant CFP+ cells were GlyT1+ (Fig. 4a,b). Consistent with this result, the total number of glycinergic ACs (GlyT1+) was significantly higher (p < 0.01) in mutants than in littermates (Fig. 4c). This small increase (~5%) was the size expected if half of the nGnG ACs became glycinergic. The total numbers of ACs (Pax6+) and GABAergic ACs (GAD+) were unaffected by loss of Neurod6 (Supplementary Fig. S3 and data not shown). These results demonstrate that Neurod6 promotes adoption of the nGnG fate and suggest that when cells fail to become nGnG ACs they become glycinergic ACs instead.
Figure 4. Neurod6 regulates the nGnG vs. glycinergic fate decision.
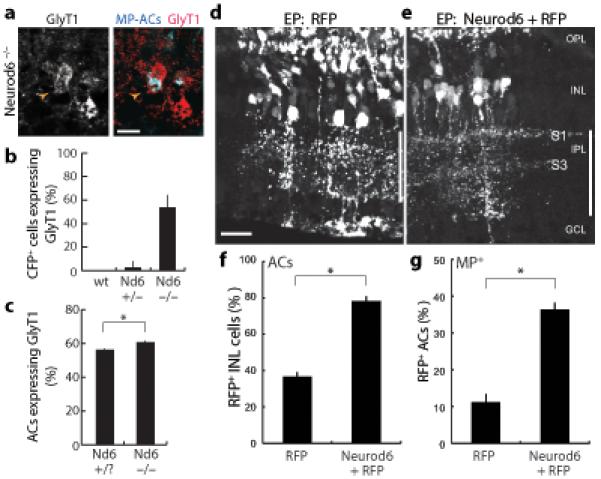
a,b: Many CFP+ MP-ACs adopt glycinergic fate in Neurod6 mutants. Double-labeling of Neurod6−/− sections for CFP (blue) and GlyT1 (red) reveals double-positive ACs (arrowhead). Quantification of CFP+GlyT1+ cells (b) shows large increase in mutants relative to wild-type or Neurod6+/− littermates, in which double-positive cells were almost never observed (p < 0.001; n ≥ 80 MP-ACs per genotype). Scale bar = 10 μm.
c: More ACs (Pax6+) are glycinergic (GlyT1+) in Neurod6 mutants than in littermate controls (p < 0.01).
d,e: Sections from retinas electroporated (EP) with red fluorescent protein (RFP) plasmid (d) or mix of Neurod6 and RFP plasmids (e). Vertical bar marks IPL. In control, cell bodies of transfected neurons are scattered throughout INL, and neuronal processes project throughout IPL. After Neurod6 electroporation, cell bodies of transfected ACs cluster in a single INL stratum and project preferentially to IPL sublaminae S1 and S3. OPL = outer plexiform layer. GCL = ganglion cell layer. Scale bar = 25 μm.
f: Neurod6 promotes AC fate over other INL fates. Graph shows fraction of RFP+ INL cells that became ACs (Stx1+; p < 1×10−6; n ≥ 500 cells/experiment). In both experimental and control retinas, remaining non-AC RFP+ INL cells were bipolars (Chx10+) or Müller glia (Sox9+; not shown).
g: Neurod6 promotes a subtype fate shift among ACs toward the nGnG fate. Graph shows fraction of RFP+ ACs that were MP-CFP+ (p < 0.001; n ≥ 50 CFP+ cells/experiment). Error bars = S.E.M. in all graphs; All animals were P15–17.
To test the idea that Neurod6 controls a switch between nGnG and glycinergic AC fates, we performed gain-of-function experiments. Retinas were electroporated at P0 with a plasmid encoding a red fluorescent protein (RFP), mCherry, alone or with a plasmid encoding Neurod6. Co-electroporated plasmids were regularly coexpressed by the same cells (data not shown and see Ref. 32). Consistent with previous reports32, electroporation at this age led to expression of RFP in late-born cell types: AC, bipolar, photoreceptor, and Müller glial cells (Fig. 4d). Coexpression of Neurod6 led to a striking increase in the proportion of RFP+ ACs, identified by morphology and position, at the expense of bipolar cells and Müller glia (Fig. 4e,f). Thus, overexpression of Neurod6 appears to promote the AC fate.
Does Neurod6 overexpression promote formation of ACs generally or selective formation of nGnG ACs? Three observations suggested that Neurod6-induced supernumerary ACs were predominantly nGnG. First, somata of ACs expressing RFP alone were distributed through the INL, whereas those coexpressing RFP and Neurod6 were arrayed in the position characteristic of nGnG somata (Fig. 4d,e and Supplementary Fig. S2). Second, arbors of ACs expressing RFP alone were distributed through the IPL, whereas arbors of RFP+Neurod6+ ACs were clustered in S1 and S3, the position of nGnG arbors at this age (Figs. 4d,e and 3e–g). Third, in retinas from MP mice, Neurod6 misexpression more than doubled the fraction of RFP+ ACs that was CFP+ (Fig. 4g) and decreased the fraction of RFP+ ACs that was glycinergic (data not shown). CFP+ ACs remained GAD− and GlyT1− after Neurod6 misexpression, confirming that MP-CFP remains a reliable marker for nGnG ACs in these experiments (not shown). Taken together, these loss- and gain-of-function results support the idea that Neurod6 regulates nGnG AC development by controlling a switch between glycinergic and nGnG fates.
Ebf3 and Satb2 mark nGnGs and a related amacrine subtype
To expand our analysis of the transcriptional program that regulates nGnG fate, we investigated Ebf3 and Satb2, two other transcriptional regulators selectively expressed by nGnG ACs (Fig. 2c and Supplementary Table 1). Both regulate cell fate determination20,33 and Ebf class genes have been shown to promote formation of glycinergic ACs34. Immunostaining revealed that Ebf3 and Satb2 are co-expressed in a subset of ACs in the INL (>95% overlap from E18 to P37); they are also expressed, with less overlap, in a subset of RGCs (Brn3a+) in the ganglion cell layer (Fig. 5a–c, Supplementary Table 2, and data not shown). The Ebf3+Satb2+ ACs included all nGnG ACs, as well as ~1/3 of GlyT1+ ACs; none were GAD+. Triple staining of MP tissue confirmed that nGnG (CFP+) and glycinergic ACs together accounted for all Satb2- and Ebf3-positive ACs (Fig. 5c-e and Supplementary Table 2). We refer to the glycinergic group as SEG ACs for Satb2+Ebf3+GlyT1+.
Figure 5. Satb2 and Ebf3 mark nGnG ACs and a related glycinergic subtype.
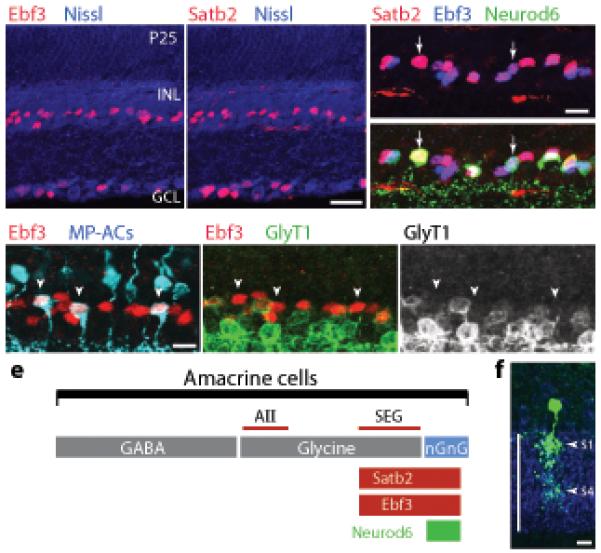
a,b: Retinal sections stained with antisera against Ebf3 (a) or Satb2 (b). Immunoreactivity (red) is observed in nuclei of a subset of ACs in the INL and a subset of RGCs in the ganglion cell layer (GCL). Blue = Nissl counterstain.
c: Staining for Ebf3 (blue), Satb2 (red), and Neurod6 (YFP+ cells, green in bottom panel) in the INL of Nd6CY retina. Top: Ebf3 and Satb2 are expressed by the same AC subset (pink-purple nuclei). Bottom: Virtually all Neurod6+ ACs express Satb2 and Ebf3 (arrows), although Neurod6 is expressed by only a subset of the Satb2+Ebf3+ population.
d: Triple-staining for MP-ACs (blue) Ebf3 (red) and GlyT1 (green) reveals that all Ebf3+ ACs are either nGnG or glycinergic: CFP+Ebf3+ cells (arrowheads) do not express GlyT1 and Ebf3+GlyT1+ cells do not express the MP transgene.
e: Schematic showing AC classes defined by neurotransmitter or by expression of Satb2, Ebf3, and Neurod6 transcription factors. Lines are drawn approximately to scale. The line labeled “SEG” denotes the population of ACs that are Satb2+Ebf3+GlyT1+. The size of the AII population was estimated from refs. 6 and 10.
f. Morphology of SEG ACs (green) in P20 retina, revealed by YFP-retrovirus. Cells were identified by triple staining for YFP, Ebf3 (not shown) and GlyT1 (blue). (see also Supplementary Fig. S5). Vertical line, IPL.
Scale bars (a,b) = 25 μm; (c–f) = 10 μm.
Further analysis revealed that nGnG and SEG ACs are similar in other respects beyond their shared expression of Satb2 and Ebf3. First, they are born at similar times: A birthdate curve obtained previously12 for all Ebf+ ACs, most of which express Ebf3 (data not shown), overlaps substantially with that of nGnG ACs (Fig. 3j). Second, they are morphologically similar, as shown by low-titer retroviral labeling: SEG ACs (Fig. 5f and Supplementary Fig. S5), like nGnG ACs (Fig. 3d), were narrow-field and multistratified. However, SEG AC arbors generally extended into the ON region of the IPL (Fig. 5f and Supplementary Fig. S5), whereas nGnG arbors were confined to S1–S3. Thus, nGnG and SEG ACs are both late-born, narrow-field, multistratified ACs, but they are distinct in their IPL stratification and neurotransmitter phenotype.
Neurod6 controls a postmitotic fate switch
As noted above, Neurod6 appears to control a fate switch between nGnG and glycinergic ACs. The similarities between nGnG and SEG ACs raised the possibility that Neurod6 specifically promotes formation of nGnG ACs at the expense of SEG ACs. Two observations suggest that this is the case.
First, in Neurod6 mutants carrying the MP transgene, many CFP+ cells become glycinergic as described above (Fig. 4b). All of these GlyT1+CFP+ cells expressed Ebf3 and Satb2, the markers that define SEG ACs (Fig. 6a and data not shown). Second, loss of Neurod6 had no effect on the total number of Ebf3+ ACs but increased the fraction of Ebf3+ ACs that was glycinergic (Fig. 6b,c). By contrast, the density of the most abundant Ebf3− glycinergic AC subtype, AII ACs, was unchanged in Neurod6 mutants (Fig. 6d). These results support the idea that Neurod6 plays a role in determining whether Ebf3+Satb2+ ACs adopt the nGnG or the SEG phenotype.
Figure 6. Neurod6 controls a postmitotic choice between nGnG and SEG fates.
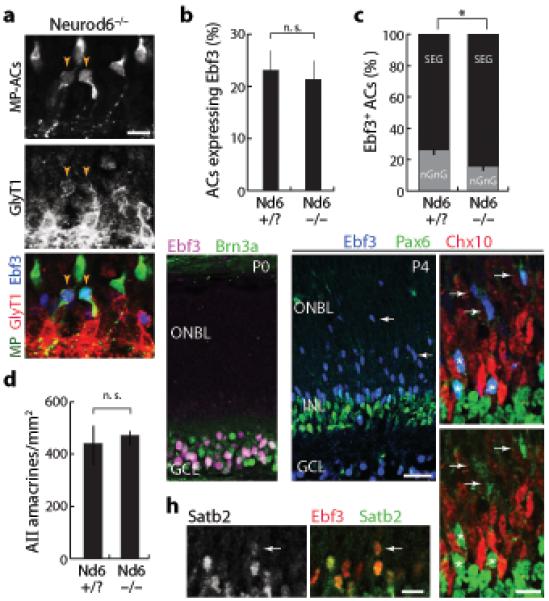
a: Section from MP; Neurod6−/- retina, triple-stained for MP-ACs (anti-GFP; green), GlyT1 (red), and Ebf3 (blue). MP-ACs that adopt glycinergic fate also express Ebf3 (arrowheads), consistent with a fate switch from nGnG to SEG ACs.
b, c: Total size of the Ebf3+ AC population (b) is not changed in Neurod6 mutants relative to littermate controls, but the composition of the Ebf3+ population (c) is altered. Loss of Neurod6 reduces the fraction of Ebf3+ cells that are nGnGs, and increases the fraction that are GlyT1+ SEGs (c; p < 0.002; n > 450 cells).
d: Glycinergic AII ACs (immunoreactive for Disabled1) were unaffected by Neurod6 mutation. Density was calculated from 20μm sections. (n = 4 samples each genotype). Animals were P15 (a–d).
e–g: Ebf3 is expressed exclusively by postmitotic neurons in the developing retina. At P0 (e), Ebf3 (purple) is confined to Brn3a+ RGCs (green) in the ganglion cell layer (GCL). No expression is seen in the outer neuroblast layer (ONBL). At P4 (f,g), Ebf3 (blue) is expressed by migratory newborn Pax6+ ACs (green) in the ONBL (arrows) and by more mature Pax6+ ACs in the INL (asterisks). The Ebf3+ migratory cells do not express bipolar/Müller marker Chx10 (red).
h: Satb2 (green) and Ebf3 (red) are coexpressed in the same AC population at P4, including migrating newborn ACs (arrow).
Error bars = S.E.M. Scale bars (a,g,h) = 10 μm, (e,f) = 25 μm.
When is this determination made? To address this issue, we asked when the three transcriptional regulators are expressed in developing retina. Retinal progenitors reside in the outer neuroblast layer (ONBL); newly postmitotic neurons then migrate through the ONBL before settling in the nascent ganglion cell layer and INL. Neither Ebf3 nor Satb2 was expressed by cells in the ONBL or the forming INL at E18.5 or P0 (Fig. 6e and data not shown); only RGCs were Satb2 or Ebf3 positive at these ages. At P4, however, a subset of cells migrating through the ONBL was Ebf3+ and Satb2+ (Fig. 6f–h). These migrating cells were ACs (Pax6+Chx10−; Fig. 6g). Nearly all of the migrating Pax6+ ACs expressed Ebf3, suggesting that it marks most late-born ACs. By P6, Ebf3 and Satb2 were no longer present in migrating cells; their expression pattern was indistinguishable from that seen in adults (Fig. 5a,b). Thus, Ebf3 and Satb2 expression does not commence in AC progenitors, but rather in newborn nGnG and SEG ACs as they migrate to the INL.
We were unable to determine directly which cells express Neurod6 in early postnatal retina due to non-specific staining by anti-Cre in perinatal Neurod6:Cre mice, the late onset of reporter expression in Nd6CY mice, and lack of suitable Neurod6 antibodies. However, Cre/lox- based lineage-tracing in Nd6CY mice allowed us to infer two important features of the Neurod6 expression pattern. First, Neurod6 is expressed exclusively in postmitotic neurons. If Neurod6 were expressed by progenitors, Cre-mediated excision in retinal progenitors would lead to labeling of radial clones of progeny. Such clones were never observed (Fig. 3a–c,f,h and Supplementary Fig. S6). Second, Neurod6 is transiently expressed by a subset of SEG ACs. Transient expression was detected by comparing Cre immunoreactivity, which reports cells actively expressing Neurod6, to Nd6CY-based YFP expression, which permanently marks all cells with a history of Neurod6 expression. At P14, only nGnG cells were Cre+ (Fig. 2d and Supplementary Table 2) whereas ~1/3 of Nd6CY+ cells were Ebf3+GlyT1+ SEGs (38.1 ± 2.2%; n > 100 cells). Together, these results suggest that Neurod6 is initially expressed by SEG and nGnG ACs, that its expression becomes restricted to nGnGs during an early postnatal period, and that this restriction is critical for promoting the nGnG phenotype at the expense of the SEG phenotype.
Satb2 regulates nGnG fate by promoting Neurod6 expression
Because Neurod6 is initially expressed by Satb2+Ebf3+ ACs, we asked whether either Ebf3 or Satb2 regulates Neurod6 expression and nGnG fate. To this end, we electroporated Ebf3 or Satb2 cDNA into the P0 retina, either along with or fused to a fluorescent reporter. Electroporation of Ebf3 produced no detectable effect on Satb2 immunoreactivity, Neurod6 expression or nGnG fate (not shown). By contrast, Satb2 electroporation led to induction of both Ebf3 and Neurod6 in the majority of transfected cells, including many outside the INL (Fig. 7a–b, f–g). Thus, Satb2 positively regulates expression of both Ebf3 and Neurod6. Along with Neurod6, the supernumerary Satb2+XFP+Ebf3+ cells expressed Pax6 and Stx1 (pan-AC markers) but were GlyT1− and GAD−, suggesting that had adopted the nGnG fate (Fig. 7c–g, l–n and data not shown). Moreover, they expressed two other genes enriched in nGnG ACs: Frem1, which encodes an extracellular matrix protein, and 6430573F11rik, which encodes an uncharacterized protein with a methyltransferase domain (Supplementary Table 1 and Fig. 7h–k). Thus, Satb2 appears to have strong nGnG fate-promoting activity.
Figure 7. Satb2 promotes the nGnG fate by inducing Neurod6.
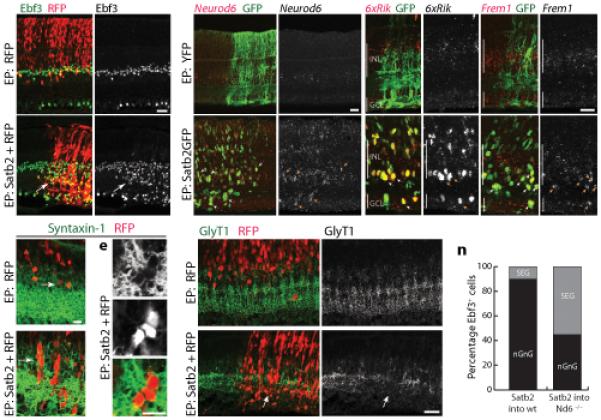
a,b: Satb2 induces Ebf3. Electroporation (EP) with RFP plasmid alone (a) or Satb2 + RFP (b). Overproduction of Ebf3+ cells in Satb2-transfected retinal patches (b, arrow, yelllow cells) is evident relative both to adjacent untransfected territory and to control (a). Satb2+RFP+ cells are also shifted to the AC zone of the INL.
c–e: The AC zone of the INL (Stx1, green) contains more RFP+ cells in Satb2-overexpressing retina (d) than in controls (c). d,e: Clusters of Stx1+RFP+ACs form in Satb2-electroporated retina but not control.
f,g: Electroporation of Satb2, but not YFP, induces Neurod6 expression in transfected cells (arrows). Compared to baseline endogenous expression (f), Neurod6 ISH signal is much higher in Satb2GFP+ retinal patches (g).
h–k: Electroporation of Satb2, but not YFP, induces expression of nGnG markers 6430573F11rik (6xRik; h,i) and Frem1 (j,k). Red in (f–k): = ISH signal; green = transfected cells (anti-GFP). Arrows in (f–k): Satb2GFP+Marker+ cells.
l,m: Excess ACs induced by Satb2 are GlyT1-negative. Clusters of Satb2+RFP+ cells (m) produce striking discontinuities in the normal pattern of GlyT1 immunoreactivity because they are GlyT1− (arrow).
n: The phenotype (nGnG or SEG) of supernumerary Ebf3+ ACs induced by Satb2 (as in b) was scored in wild-type (wt) and Neurod6 (Nd6) mutant mice. Loss of Nd6 function substantially blocked the ability of Satb2 to induce nGnG ACs. Instead, many of the excess Ebf3+ ACs were SEGs. N > 150 cells for each genotype.
Mice were P7–9 (a–m) or P14 (n). Scale bars (a,b,f–m) = 25 μm; (c–e) = 10 μm.
Finally, we asked whether the ability of Satb2 to promote the nGnG fate requires that it upregulate Neurod6. To this end, we electroporated Satb2 into Neurod6 mutants. Satb2 overexpression induced excess Ebf3+Pax6+ACs in Neurod6 mutants as it did in wild-type retinas (not shown). However, whereas >90% of the supernumerary Satb2+XFP+Ebf3+ ACs were nGnG ACs (GlyT1−) in a wild-type background, nearly half were glycinergic (GlyT1+) in the absence of Neurod6 (Fig. 7n). Thus, Neurod6 acts genetically downstream of Satb2 in the production of nGnG ACs, influencing whether Satb2-induced ACs become SEG or nGnG ACs.
DISCUSSION
To understand how neural circuits form and function, it is necessary to inventory the full range of neuronal subtypes and to understand how they arise from a limited set of progenitors. Here, we address these issues with respect to retinal amacrine cells (ACs), which comprise at least 30 subtypes. First, we describe two closely related groups of ACs that we call nGnG (non-GABAergic non-glycinergic) and SEG (Satb2+Ebf3+GlyT1+) ACs. Second, we identify a set of transcription factors (Satb2, Ebf3 and Neurod6) selectively expressed by nGnG and SEG ACs, and present evidence that they control a postmitotic switch that determines which of these two subtypes a late-born AC becomes.
Two new subtypes of narrow-field amacrine cells
Most ACs in mammalian retina use either GABA or glycine as their neurotransmitter, sometimes along with another classical transmitter or a neuropeptide. However, we found that approximately 15% of ACs express neither GABAergic nor glycinergic markers. This result is consistent with several previous reports on mouse, rat and rabbit retina10-13, but no studies have yet characterized this subset and ACs are often claimed to be entirely GABAergic or glycinergic6,30. We found that nGnG ACs are late-born, narrow-field cells with diffuse, multilaminar arbors in the OFF sublaminae of the IPL. It seems likely that nGnG ACs were observed in previous surveys 3,5,6,21,35. For example, they resemble “diffuse bistratified-1” ACs in rabbit3 and “pseudobistratified” ACs in mouse5. Morphological distinctions among narrow-field ACs are subtle6,7, so it is not surprising that they were not recognized as a single type without molecular markers.
The usual function of narrow-field ACs is to provide inhibition across IPL sublaminae, allowing visual circuits that ramify in different strata to silence each other9. Given its morphology, the nGnG AC may serve a similar function for the circuits in S1–S3. However, the nGnG neurotransmitter remains unknown, so these cells could be excitatory. So far, our microarray results and tests of likely candidates have provided no clues to the transmitter that nGnG ACs use (J.N.K. and P.E.V. unpublished). The Neurod6:Cre and MP-CFP lines will allow us to target these cells for stimulation and recording, facilitating physiological and pharmacological analysis of their function and neurotransmitter.
In the course of our molecular analysis of nGnG ACs, we identified a set of three transcription factors – Satb2, Ebf3 and Neurod6 – also expressed by a second group of ACs that we call SEG ACs. SEG ACs resemble nGnG ACs not only in this expression pattern but in being late-born, narrow-field ACs with diffuse arbors that span multiple sublaminae. The two types are clearly distinct, however, in several respects – notably, SEG ACs are glycinergic, extend arbors into ON sublaminae and express Neurod6 only transiently. As with nGnG ACs, it seems likely that SEG ACs were observed before in AC morphological surveys3,5,21,35. The identification of molecular markers for these and other narrow-field ACs will aid in classifying them and studying their role in vision.
Transcriptional control of nGnG and SEG amacrine fate
Several transcriptional regulators have been identified that regulate AC fate choices relative to the main retinal neuronal types15-18. In contrast, much less is known about factors that regulate diversification of ACs into 30 or more subtypes, although recent studies have implicated Bhlhb5 and Nr4a2 in formation of GABAergic ACs, Isl1 in differentiation of an AC subtype that uses both GABA and acetylcholine, and Ebf class genes in formation of glycinergic ACs34,36-38.
Here, we have expanded this repertoire by identifying a pair of transcription factors that regulate formation of the SEG and nGnG AC subtypes. Our main results are as follows (Supplementary Fig. 7a): (1) Satb2, Ebf3 and Neurod6 are all selectively expressed by a large group of late-born ACs that includes (and may be limited to) nGnG and SEG ACs. (2) Expression of Satb2 and Ebf3 remains high into adulthood in both populations, whereas Neurod6 is downregulated in SEGs during the first two postnatal weeks. (3) In mice lacking Neurod6, the number of nGnG ACs is greatly reduced, and there is a corresponding increase in the number of SEG ACs. (4) Conversely, ectopic overexpression of Neurod6 promotes formation of supernumerary nGnG ACs. (5) Overexpression of Satb2 induces ectopic expression of Ebf3 and Neurod6 and promotes formation of cells with nGnG properties in a Neurod6-dependent fashion. (Satb2 mutant mice die at birth39, so loss-of-function studies on this gene were infeasible20). (6) Overexpression of Ebf3 had no obvious effect on nGnG development, but Jin et al.34 showed recently that Ebf3 and the closely-related Ebf1 can promote the formation of non-AII glycinergic ACs – a population likely to include SEGs.
Together, these results lead us to propose that Satb2, and perhaps Ebf334, promotes both SEG and nGnG AC fates, whereas Neurod6 controls a fate switch that determines whether late-born ACs become SEG or nGnG ACs. Expression of Neurod6 is, in turn, regulated by Satb2. Because Satb2 can induce SEG as well as nGnG AC phenotypes, we hypothesize the existence of a mechanism that downregulates Neurod6 expression in some Satb2+ ACs, thereby promoting SEG fate. Further evidence in favor of this idea is that Neurod6 is downregulated specifically in cells that become SEGs. There is ample precedent for transcriptional networks in which an upstream factor induces two mutually exclusive cell fates through cross-inhibition40,41.
In that loss of Neurod6 does not completely abolish generation of nGnG ACs, other factors may act in parallel or compensate for its loss. Attractive candidates are the closely related transcription factors Neurod2 and Neurod4. Indeed, Neurod2, 4, and 6 are redundant in the specification of neurotransmitter phenotype in spinal cord40, and both Neurod2 and 4 are expressed in ACs13,16 (J.N.K. unpublished).
Cell fate mechanisms in progenitors and postmitotic cells
Many genes that determine neuronal fate act in progenitors to influence the type of progeny they generate19. In such cases, the competence of progenitors to generate specific progeny may change over time14. With respect to ACs, we find that retinal progenitors pass through three broad phases, first making GABAergic, then glycinergic, then nGnG ACs. Our results imply that, around P1, progenitors begin changing such that they become biased towards producing nGnG ACs and away from GABAergic and early-born glycinergic (e.g., AII) fates (Supplementary Fig. 7b).
Such progenitor-based mechanisms do not, however, fully explain the choice between SEG and nGnG AC fates. We suggest that timing of cell cycle exit specifies the Satb2+Ebf3+ Neurod6+ phenotype initially shared by SEG and nGnG ACs, while postmitotic restriction of Neurod6 to a subset of these cells subdivides the group into Neurod6− SEG ACs and Neurod6+ nGnG ACs. Indeed, expression of Ebf3, Satb2 and Neurod6 commences only after late-born ACs have left the cell cycle, and birthdates of SEG and nGnG ACs overlap substantially. Postmitotic transcriptional networks have recently been shown to influence other neuronal fate choices, and interestingly, Satb2 or Neurod6 are involved in such choices in cortex, spinal cord and sympathetic ganglia20,40,42. The fact that we came upon the same factors through a completely unbiased approach suggests that they may be generally involved in postmitotic mechanisms for generating subtle differences among closely related neuronal subtypes.
Supplementary Material
ACKNOWLEDGEMENTS
We thank R. Reed (Johns Hopkins University) for providing Ebf antibodies, D. Pow (University of Queensland) for anti-glycine antibodies, Louis Reichardt (UCSF) and K. Nave (Goettingen) for NEX:Cre mice, and Xin Duan for advice and assistance with retrovirus. J.N.K. was supported by a Life Sciences Research Foundation postdoctoral fellowship. This work was supported by a grant from NIH to J.R.S and Collaborative Innovation Award #43667 from HHMI.
Appendix
MATERIALS AND METHODS
Animals
CD1 and C57/B6 mice were purchased from Charles River (Wilmington, MA) and Jackson Labs. Transgenic mice in which regulatory elements from the Thy1 gene drive neuron-specific expression of a mitochondrially targeted cyan fluorescent protein (Thy1-MitoCFP Line P, called MP here; were obtained from T. Misgeld and J. Lichtman (Harvard University) and bred in a mixed CD1 and C57/B6 background. NEX-Cre (Neurod6:Cre) knock-in mice28 were obtained from K. Nave (Goettingen) via L. Reichardt (UCSF). Cre-dependent reporter lines in which YFP is driven by the Thy1 promoter or GFP is expressed from the Rosa26 locus, were described previously29,43. Animal procedures were in compliance with the NIH Guide for the Care and Use of Laboratory Animals and approved by the Animal and Care and Use Program at Harvard University.
Macaque retinal tissue was obtained from R. Born and V. Berezovskii (Harvard University).
Immunohistochemistry
Eyes were fixed for 30 minutes in 4% paraformaldehyde (PFA) on ice, then the cornea was cut along the ora serrata and the anterior part of the eye was removed. The retina, preserved in the optic cup, was fixed for 30–45 additional minutes in 4% PFA, incubated for 2h in 30% sucrose/1x phosphate-buffered saline (PBS), frozen in Tissue Freezing Medium (Triangle Biomedical Sciences), and sectioned at 20μm in a cryostat.
Antibody staining was performed as in ref. 12. Briefly, sections were rinsed in PBS and then incubated in blocking solution (3% donkey serum/0.3% Triton X-100/PBS) for 30 min at room temperature, primary antibodies overnight at 4 °C, and Alexa-conjugated secondary antibodies (Molecular Probes or Jackson Immunoresearch) for 1–2 h at room temperature. Sections were then washed thoroughly with PBS, coverslipped with Fluoro-Gel (EMS) or Fluoromount G (Southern Biotech), and imaged on an Olympus FV1000 scanning confocal microscope. Some sections were Nissl-counterstained with NeuroTrace 435 (Invitrogen). Immunohistochemistry on retinal wholemounts was the same except primary antibody incubation was for 6 days at 4°C °with agitation, and secondary antibody was overnight at 4°C.
Cells were dissociated from P0 retinas using papain and plated on poly-D-lysine coated 8-well Permanox chamber slides (Nunc) as described in ref. 44. Cells were fixed with 4% PFA/4% sucrose for 15 minutes and stained as above.
We used the following primary antibodies: mouse anti-Brn-3a (Chemicon), rabbit anti-calbindin (Swant), rat anti-BrdU (Abcam), sheep anti-Chx10 (Exalpha), rabbit anti-DsRed (Promega), mouse anti-syntaxin-1 (HPC-1; Sigma), goat anti-choline acetyl transferase (ChAT; Chemicon), rabbit anti-glutamate decarboxylase (GAD65/67; Chemicon), rabbit anti-GABA (Sigma), goat anti-Glycine Transporter 1 (GlyT1; Chemicon), rabbit anti-Sox9 (Millipore), mouse anti-Pax6 (Developmental Studies Hybridoma Bank), rabbit anti-Disabled1 (Millipore), chicken and rabbit anti-GFP (Aves Labs and Chemicon), rat anti-Glycine (gift from D. Pow), rabbit anti-Ebf and anti-Ebf345 (gift of R. Reed, Johns Hopkins University), mouse anti-Satb2 (Abcam), mouse VC1.1 (Sigma), and mouse anti-Cre (Millipore). Anti-Ebf3 does not cross-react with other Ebf-family transcription factors (R. Reed, personal communication).
In situ hybridization
DNA templates for producing riboprobes were prepared from IMAGE clones or by RT-PCR from retina cDNA. The following IMAGE clones were obtained from Open Biosystems (corresponding gene in parentheses): 6828629 (Pkdcc); 6510171 (Frem1); 40129409 (Pde5a); 40126301 (Galr2); 6336362 (6430573F11Rik); 8861144 (GlyT2; also called Slc6a5). Neurod6 template was PCR-amplified using primers 5′-AATGCTGGTACCCACTTACAGGGA and 5′-CCCAAATACCGTGTTTCCTCCTCT, and cloned into pGEM-T-Easy (Promega). Riboprobes were synthesized from T7, T3, or Sp6 sites in each vector, using the DIG RNA labeling kit (Roche).
In situ hybridization (ISH) was performed as described in ref. 46. Briefly, tissue was fixed and sectioned as described above, then sections were post-fixed in methanol, permeabilized with 1% Triton and Proteinase K, and acetylated. Riboprobes, hydrolyzed to ~500bp, were hybridized to sections at 65°C overnight. After extensive washing at 65°C, tissue was incubated with anti-DIG:peroxidase antibody (Roche) and staining was revealed using Cy3-tyramide (Perkin Elmer). For double labeling with anti-GFP, the tissue was washed for at least 2 hours in 4 changes of tris-buffered saline (TBS) following in situ hybridization, then immunostained with chicken anti-GFP as described above.
Electroporation
Retinas were electroporated by minor modifications of methods described in ref. 32. Plasmid DNA (at least 1.5 mg/mL) was injected into the subretinal space of neonatal mice (4–36 hours postpartum). Paddle electrodes (BTX) were used to apply 5 current pulses across the head (50 ms pulse; 950 ms interval; 80 V). In pilot experiments we observed strong expression as early as 24 hours post-electroporation. Neurod6, Satb2, and Ebf3 coding sequences were amplified from retina cDNA using the following primers: Neurod6 (5′-GGTTAAAGAACCATGTTAACACTACCG and 5′-TCATTTTCCTCATTAATTATGAAAAAC); Satb2 (5′-GGTGGGAACTTTGTCTCCAA and 5′-GCCTGCGGAGTTCACATTAT); Ebf3 (5′-ATCATGTTTGGGATTCAGGAG and 5′-CATGAAGGAGTGGGCTTGTT). The resulting PCR products were sequenced and corresponded to RefSeq sequences NM_009717.2, NM_139146.2, and NM_001113414.1 respectively. Each open reading frame was transferred using the Gateway system (Invitrogen) to two separate destination vectors. The first, bearing the CAG promoter, drove expression of the protein alone, while the second, bearing the Ubiquitin-C promoter, drove expression of a GFP-fusion protein. These plasmids gave identical results in electroporation experiments. Expression vectors driving monomeric Cherry (mCherry) fluorescent protein were generated in a similar manner.
Single-cell morphology
To visualize the morphology of single ACs in sections, we used a retrovirus that drives expression of YFP47. The virus was diluted in PBS/polybrene and injected into subretinal space of P0–P2 mice. Pilot experiments determined appropriate levels of dilution at which the morphology of single labeled cells could be clearly distinguished from neighboring labeled cells. Serial sections of virus-labeled tissue were triple-stained for GFP, Ebf3, and GlyT1 to identify nGnG and SEG ACs. Only GFP+ cells that were well-isolated from their neighbors, within the same section and across adjacent sections, were used to score IPL projections. For analysis of single cells in wholemount retina, we used retinas from Neurod6:Cre;reporter mice. Retinas were double-stained with anti-GFP and GlyT1 to distinguish between glycinergic and nGnG ACs.
Quantification of cell number and position
To determine the fraction of cells coexpressing various markers in sections or dissociated cultures, we performed cell counts using methods described previously12. Briefly, confocal image stacks were prepared from representative sections comprising both central and peripheral retina. Double- or triple-labeled cells were scored offline. In gain- and loss-of-function experiments, the Mann-Whitney U-test or t-test were used, as appropriate, to assess the significance of measured differences between experimental and control conditions. For analysis of Neurod6 mutants, counts were performed blind to genotype. Bromodeoxyuridine birthdating curves and soma position plots were constructed as described12.
Cell purification and expression profiling
Transgenic mice expressing XFPs in amacrine or bipolar cells were sacrificed at P6. Retinas were dissociated using Papain (Worthington) and the live cell suspension was passed through a flow cytometer (Mo Flo; Dako) to collect fluorescent cells. Damaged cells were gated out using propidium iodide. The positive cells were sorted either into Neurobasal medium supplemented with BDNF and B27 for plating and purity analysis, or directly into RNA-stabilizing lysis buffer from the PicoPure RNA Isolation Kit (MDS). RNA samples were stored at −80°C until processing for microarrays.
To separately purify CFP+ ACs and bipolar cells from the MP line, we used VC1.1 monoclonal antibody to label ACs48. The live cell suspension was stained with VC1.1 (200 μg/mL) followed by an anti-IgM secondary antibody conjugated to PeCy7 (Southern). Putative MP+ ACs (VC1.1+CFP+) and bipolar cells (VC1.1−CFP+) were sorted separately, using the MoFlo, into RNA lysis buffer. A parallel set of samples was collected from the same preparation to confirm the purity and cellular identity of each fraction (Fig. 2b). Cells were sorted into media, plated, grown for 2 hours at 37°C in a tissue culture incubator, and fixed with 4% PFA. They were then triple-stained with anti-GFP and markers for ACs (Stx1) and bipolar cells (Chx10). In some experiments we replaced the PeCy7 antibody with anti-IgM conjugated to magnetic beads, and used magnetic cell sorting (Miltenyi Biotech) to generate VC1.1-positive and -negative fractions prior to sorting for CFP using the MoFlo. Both methods were effective for separating MP+ ACs from MP+ bipolar cells.
For microarray hybridization, RNA was isolated using the PicoPure kit. Two rounds of RNA amplification were performed with the MessageAmpII system (Ambion/Applied Biosystems), the second resulting in biotin-labeled samples. Affymetrix Mouse 430 2.0 arrays were hybridized according to the manufacturer’s instructions. Two replicates were profiled per transgenic line, each representing a sample of ~1000 cells collected from a single litter. Data was analyzed and cell-type-specific genes identified using Resolver (Rosetta) and dChip software49. Heatmaps were constructed by first filtering all probesets to remove those that did not show at least one 3-fold change between any pair of cell types, then standardizing the expression level of each remaining probeset across samples so that its mean = 0 and standard deviation = 1. Probesets were then subjected to hierarchical clustering using the dChip software, to group them according to their expression levels across the 7 cell types in the dataset.
REFERENCES
- 1.Cossart R. The maturation of cortical interneuron diversity: how multiple developmental journeys shape the emergence of proper network function. Curr Opin Neurobiol. doi: 10.1016/j.conb.2010.10.003. epub 2010. [DOI] [PubMed] [Google Scholar]
- 2.Goulding M, Pfaff SL. Development of circuits that generate simple rhythmic behaviors in vertebrates. Curr Opin Neurobiol. 2005;15:14–20. doi: 10.1016/j.conb.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 3.MacNeil MA, Masland RH. Extreme diversity among amacrine cells: implications for function. Neuron. 1998;20:971–982. doi: 10.1016/s0896-6273(00)80478-x. [DOI] [PubMed] [Google Scholar]
- 4.Klausberger T, Somogyi P. Neuronal diversity and temporal dynamics: the unity of hippocampal circuit operations. Science. 2008;321:53–57. doi: 10.1126/science.1149381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Badea TC, Nathans J. Quantitative analysis of neuronal morphologies in the mouse retina visualized by using a genetically directed reporter. J Comp Neurol. 2004;480:331–351. doi: 10.1002/cne.20304. [DOI] [PubMed] [Google Scholar]
- 6.Vaney DI. The mosaic of amacrine cells in the mammalian retina. Progress in retinal research. 1990;9:49–100. [Google Scholar]
- 7.MacNeil MA, Heussy JK, Dacheux RF, Raviola E, Masland RH. The shapes and numbers of amacrine cells: matching of photofilled with Golgi-stained cells in the rabbit retina and comparison with other mammalian species. J Comp Neurol. 1999;413:305–326. [PubMed] [Google Scholar]
- 8.Jusuf PR, Harris WA. Ptf1a is expressed transiently in all types of amacrine cells in the embryonic zebrafish retina. Neural Dev. 2009;4:34. doi: 10.1186/1749-8104-4-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hsueh HA, Molnar A, Werblin FS. Amacrine-to-amacrine cell inhibition in the rabbit retina. J Neurophysiol. 2008;100:2077–2088. doi: 10.1152/jn.90417.2008. [DOI] [PubMed] [Google Scholar]
- 10.Strettoi E, Masland RH. The number of unidentified amacrine cells in the mammalian retina. Proc Natl Acad Sci U S A. 1996;93:14906–14911. doi: 10.1073/pnas.93.25.14906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marquardt T, et al. Pax6 is required for the multipotent state of retinal progenitor cells. Cell. 2001;105:43–55. doi: 10.1016/s0092-8674(01)00295-1. [DOI] [PubMed] [Google Scholar]
- 12.Voinescu PE, Kay JN, Sanes JR. Birthdays of retinal amacrine cell subtypes are systematically related to their molecular identity and soma position. J Comp Neurol. 2009;517:737–750. doi: 10.1002/cne.22200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cherry TJ, Trimarchi JM, Stadler MB, Cepko CL. Development and diversification of retinal amacrine interneurons at single cell resolution. Proc Natl Acad Sci U S A. 2009;106:9495–9500. doi: 10.1073/pnas.0903264106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cepko CL, Austin CP, Yang X, Alexiades M, Ezzeddine D. Cell fate determination in the vertebrate retina. Proc Natl Acad Sci U S A. 1996;93:589–595. doi: 10.1073/pnas.93.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fujitani Y, et al. Ptf1a determines horizontal and amacrine cell fates during mouse retinal development. Development. 2006;133:4439–4450. doi: 10.1242/dev.02598. [DOI] [PubMed] [Google Scholar]
- 16.Inoue T, et al. Math3 and NeuroD regulate amacrine cell fate specification in the retina. Development. 2002;129:831–842. doi: 10.1242/dev.129.4.831. [DOI] [PubMed] [Google Scholar]
- 17.Li S, et al. Foxn4 controls the genesis of amacrine and horizontal cells by retinal progenitors. Neuron. 2004;43:795–807. doi: 10.1016/j.neuron.2004.08.041. [DOI] [PubMed] [Google Scholar]
- 18.Nakhai H, et al. Ptf1a is essential for the differentiation of GABAergic and glycinergic amacrine cells and horizontal cells in the mouse retina. Development. 2007;134:1151–1160. doi: 10.1242/dev.02781. [DOI] [PubMed] [Google Scholar]
- 19.Batista-Brito R, Fishell G. The developmental integration of cortical interneurons into a functional network. Curr Top Dev Biol. 2009;87:81–118. doi: 10.1016/S0070-2153(09)01203-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Britanova O, et al. Satb2 is a postmitotic determinant for upper-layer neuron specification in the neocortex. Neuron. 2008;57:378–392. doi: 10.1016/j.neuron.2007.12.028. [DOI] [PubMed] [Google Scholar]
- 21.Menger N, Pow DV, Wassle H. Glycinergic amacrine cells of the rat retina. J Comp Neurol. 1998;401:34–46. doi: 10.1002/(sici)1096-9861(19981109)401:1<34::aid-cne3>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 22.Pow DV, Hendrickson AE. Distribution of the glycine transporter glyt-1 in mammalian and nonmammalian retinae. Vis Neurosci. 1999;16:231–239. doi: 10.1017/s0952523899162047. [DOI] [PubMed] [Google Scholar]
- 23.Barnstable CJ, Hofstein R, Akagawa K. A marker of early amacrine cell development in rat retina. Brain Res. 1985;352:286–290. doi: 10.1016/0165-3806(85)90116-6. [DOI] [PubMed] [Google Scholar]
- 24.Feng G, et al. Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron. 2000;28:41–51. doi: 10.1016/s0896-6273(00)00084-2. [DOI] [PubMed] [Google Scholar]
- 25.Misgeld T, Kerschensteiner M, Bareyre FM, Burgess RW, Lichtman JW. Imaging axonal transport of mitochondria in vivo. Nat Methods. 2007;4:559–561. doi: 10.1038/nmeth1055. [DOI] [PubMed] [Google Scholar]
- 26.Schubert T, et al. Development of presynaptic inhibition onto retinal bipolar cell axon terminals is subclass-specific. J Neurophysiol. 2008;100:304–316. doi: 10.1152/jn.90202.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shimizu C, Akazawa C, Nakanishi S, Kageyama R. MATH-2, a mammalian helix-loop-helix factor structurally related to the product of Drosophila proneural gene atonal, is specifically expressed in the nervous system. Eur J Biochem. 1995;229:239–248. doi: 10.1111/j.1432-1033.1995.tb20461.x. [DOI] [PubMed] [Google Scholar]
- 28.Goebbels S, et al. Genetic targeting of principal neurons in neocortex and hippocampus of NEX-Cre mice. Genesis. 2006;44:611–621. doi: 10.1002/dvg.20256. [DOI] [PubMed] [Google Scholar]
- 29.Buffelli M, et al. Genetic evidence that relative synaptic efficacy biases the outcome of synaptic competition. Nature. 2003;424:430–434. doi: 10.1038/nature01844. [DOI] [PubMed] [Google Scholar]
- 30.Haverkamp S, Wassle H. Immunocytochemical analysis of the mouse retina. J Comp Neurol. 2000;424:1–23. [PubMed] [Google Scholar]
- 31.Stacy RC, Wong RO. Developmental relationship between cholinergic amacrine cell processes and ganglion cell dendrites of the mouse retina. J Comp Neurol. 2003;456:154–166. doi: 10.1002/cne.10509. [DOI] [PubMed] [Google Scholar]
- 32.Matsuda T, Cepko CL. Electroporation and RNA interference in the rodent retina in vivo and in vitro. Proc Natl Acad Sci U S A. 2004;101:16–22. doi: 10.1073/pnas.2235688100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang SS, Lewcock JW, Feinstein P, Mombaerts P, Reed RR. Genetic disruptions of O/E2 and O/E3 genes reveal involvement in olfactory receptor neuron projection. Development. 2004;131:1377–1388. doi: 10.1242/dev.01009. [DOI] [PubMed] [Google Scholar]
- 34.Jin K, Jiang H, Mo Z, Xiang M. Early B-cell factors are required for specifying multiple retinal cell types and subtypes from postmitotic precursors. J Neurosci. 2010;30:11902–11916. doi: 10.1523/JNEUROSCI.2187-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heinze L, Harvey RJ, Haverkamp S, Wassle H. Diversity of glycine receptors in the mouse retina: localization of the alpha4 subunit. J Comp Neurol. 2007;500:693–707. doi: 10.1002/cne.21201. [DOI] [PubMed] [Google Scholar]
- 36.Elshatory Y, Deng M, Xie X, Gan L. Expression of the LIM-homeodomain protein Isl1 in the developing and mature mouse retina. J Comp Neurol. 2007;503:182–197. doi: 10.1002/cne.21390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang H, Xiang M. Subtype specification of GABAergic amacrine cells by the orphan nuclear receptor Nr4a2/Nurr1. J Neurosci. 2009;29:10449–10459. doi: 10.1523/JNEUROSCI.3048-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feng L, et al. Requirement for Bhlhb5 in the specification of amacrine and cone bipolar subtypes in mouse retina. Development. 2006;133:4815–4825. doi: 10.1242/dev.02664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alcamo EA, et al. Satb2 regulates callosal projection neuron identity in the developing cerebral cortex. Neuron. 2008;57:364–377. doi: 10.1016/j.neuron.2007.12.012. [DOI] [PubMed] [Google Scholar]
- 40.Brohl D, et al. A transcriptional network coordinately determines transmitter and peptidergic fate in the dorsal spinal cord. Dev Biol. 2008;322:381–393. doi: 10.1016/j.ydbio.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 41.Singh H, Medina KL, Pongubala JM. Contingent gene regulatory networks and B cell fate specification. Proc Natl Acad Sci U S A. 2005;102:4949–4953. doi: 10.1073/pnas.0500480102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Apostolova G, Loy B, Dorn R, Dechant G. The sympathetic neurotransmitter switch depends on the nuclear matrix protein Satb2. J Neurosci. 2010;30:16356–16364. doi: 10.1523/JNEUROSCI.3502-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sousa VH, Miyoshi G, Hjerling-Leffler J, Karayannis T, Fishell G. Characterization of Nkx6-2-derived neocortical interneuron lineages. Cereb Cortex. 2009;19(Suppl 1):i1–10. doi: 10.1093/cercor/bhp038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lefebvre JL, Zhang Y, Meister M, Wang X, Sanes JR. gamma-Protocadherins regulate neuronal survival but are dispensable for circuit formation in retina. Development. 2008;135:4141–4151. doi: 10.1242/dev.027912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Davis JA, Reed RR. Role of Olf-1 and Pax-6 transcription factors in neurodevelopment. J Neurosci. 1996;16:5082–5094. doi: 10.1523/JNEUROSCI.16-16-05082.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamagata M, Weiner JA, Sanes JR. Sidekicks: synaptic adhesion molecules that promote lamina-specific connectivity in the retina. Cell. 2002;110:649–660. doi: 10.1016/s0092-8674(02)00910-8. [DOI] [PubMed] [Google Scholar]
- 47.Duan X, et al. Disrupted-In-Schizophrenia 1 regulates integration of newly generated neurons in the adult brain. Cell. 2007;130:1146–1158. doi: 10.1016/j.cell.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alexiades MR, Cepko CL. Subsets of retinal progenitors display temporally regulated and distinct biases in the fates of their progeny. Development. 1997;124:1119–1131. doi: 10.1242/dev.124.6.1119. [DOI] [PubMed] [Google Scholar]
- 49.Li C. Automating dChip: toward reproducible sharing of microarray data analysis. BMC Bioinformatics. 2008;9:231. doi: 10.1186/1471-2105-9-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


