Abstract
Formation and repair of platinum (Pt)-induced DNA adducts is a critical step in Pt drug-mediated cytotoxicity. Measurement of Pt–DNA adduct kinetics in tumors may be useful for better understanding chemoresistance and therapeutic response. However, this concept has yet to be rigorously tested because of technical challenges in measuring the adducts at low concentrations and consistent access to sufficient tumor biopsy material. Ultrasensitive accelerator mass spectrometry was used to detect [14C]carboplatin–DNA monoadducts at the attomole level, which are the precursors to Pt–DNA crosslink formation, in six cancer cell lines as a proof-of-concept. The most resistant cells had the lowest monoadduct levels at all time points over 24 hr. [14C]Carboplatin “microdoses" (1/100th the pharmacologically effective concentration) had nearly identical adduct formation and repair kinetics compared to therapeutically relevant doses, suggesting that the microdosing approach can potentially be used to determine the pharmacological effects of therapeutic treatment. Some of the possible chemoresistance mechanisms were also studied, such as drug uptake/efflux, intracellular inactivation and DNA repair in selected cell lines. Intracellular inactivation and efficient DNA repair each contributed significantly to the suppression of DNA monoadduct formation in the most resistant cell line compared to the most sensitive cell line studied (p < 0.001). Nucleotide excision repair (NER)-deficient and - proficient cells showed substantial differences in carboplatin monoadduct concentrations over 24 hr that likely contributed to chemoresistance. The data support the utility of carboplatin microdosing as a translatable approach for defining carboplatin–DNA monoadduct formation and repair, possibly by NER, which may be useful for characterizing chemoresistance in vivo.
Keywords: chemoresistance, platinum chemotherapy, microdosing, accelerator mass spectrometry, DNA damage, DNA repair
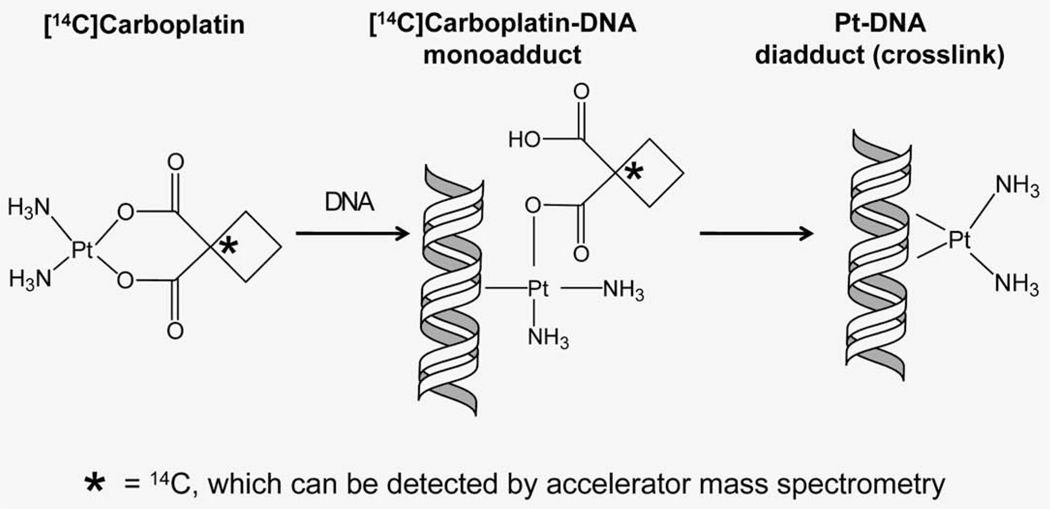
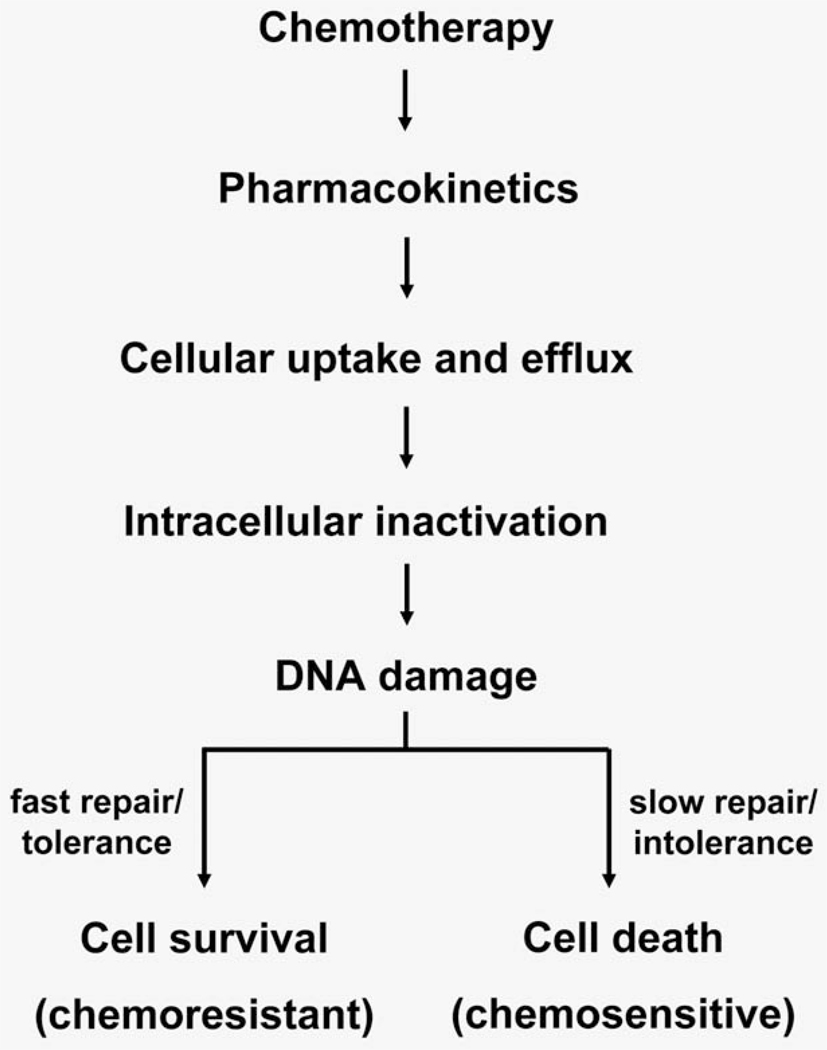
Cisplatin and carboplatin are the most frequently used agents in cancer treatment. The cytotoxic action of Pt therapy is due mostly to DNA damage.1–4 Pt-based drugs react at predominantly guanine nucleotides to form Pt–DNA monoadducts (Fig. 1), which often react with a second purine nucleotide to form more toxic interstrand and intrastrand crosslinks. Carboplatin and cisplatin form the same Pt–DNA crosslinks in vivo because of their identical cis-diamine carrier ligands. Both drugs share similar in vitro chemoresistance spectra and clinical indications, even though cisplatin is possibly more effective in some cancer types.5 Our hypothesis is that quantitation of carboplatin–DNA monoadducts is useful for characterizing several aspects of chemoresistance, including DNA repair (Fig. 2).
Figure 1.
Formation of carboplatin–DNA damage. The first step of the carboplatin and DNA interaction is the formation of carboplatin–DNA monoadducts in which only one bond is formed between Pt and DNA. Monoadducts can further react to form interstrand or intrastrand crosslinks (diadducts) in which the leaving group is irreversibly lost. The 14C label (asterisk) is located at the leaving group, which is displaced from the Pt–DNA complex upon repair or crosslink formation. Therefore, AMS can only measure carboplatin–DNA monoadducts from purified DNA.
Figure 2.
Pathways leading to chemotherapy-induced cell death and resistance. A broad overview of Pt-based cytotoxicity and cellular resistance is shown in this diagram in sequential order. DNA damage is the critical step in the cytotoxic response. Cells with low chemotherapy-induced DNA damage will survive chemotherapy and are chemoresistant. Besides DNA damage, other steps, such as drug metabolism, cell uptake and efflux of drug, intracellular inactivation and DNA repair can also affect the levels of drug–DNA damage.
In cancer patients treated with Pt drugs, positive correlations between levels of Pt-induced DNA adducts in peripheral blood mononuclear cells, as surrogates for tumor tissue, and good clinical outcomes are reported,6–13 but with some inconsistencies.14–16 The insufficiently sensitive methods used in these studies required patients to receive toxic full doses of chemotherapy before DNA damage and chemoresistance could be assessed—a considerable disadvantage for clinical applications.
Currently, one of the most sensitive techniques for DNA adduct detection is the 32P postlabeling assay, which has measurement sensitivity of one adduct in 107–108 nucleotides for detecting Pt–DNA crosslinks,17,18 about tenfold more sensitive than ELISA-based quantitative assays.19–21 However, neither the postlabeling nor ELISA-based assays are useful for quantitating carboplatin–DNA monoadducts. The 32P postlabeling assay requires sample processing that is incompatible with monoadducts. The antibodies used in ELISA that are specific for monoadducts were developed against cisplatin–DNA adducts, which do not contain the cyclobutane dicarboxylate (CBDCA, Fig. 1) ligand present in carboplatin. Therefore, only a subset of the possible carboplatin–DNA monoadduct structures are detected by ELISA. Atomic absorption mass spectroscopy (AAS) measures total platinum (Pt) but lacks sufficient sensitivity for routine clinical applications.8,13,19,21,22 Inductively coupled plasma mass spectrometry (ICP-MS) also measures total Pt, but with higher sensitivity, allowing clinical applications.14 Both AAS and ICP-MS are not specific for Pt–DNA monoadducts.
To address some of these limitations, we have used ultrasensitive accelerator mass spectrometry (AMS), which quantifies 14C at attomole levels per sample with high accuracy and precision.23–26 AMS is increasingly being used in Phase 0 microdose trials to determine pharmacokinetics (PK) after patients receive small doses of 14C-labeled drugs.27–30 In case of [14C]carboplatin, AMS can measure one carboplatin–DNA monoadduct per 109 nt and requires far less technically demanding sample processing protocols than the other adduct-specific methods described above.23 Our assay is specific for detecting [14C]carboplatin–DNA monoadducts because the 14C label, located on the CBDCA ligand of carboplatin, is irreversibly displaced from the drug–DNA conjugate upon crosslink formation or DNA repair (Fig. 1). Therefore, any increase above background of radiocarbon bound to DNA after exposure to [14C]carboplatin is due to the presence of CBDCA-containing Pt–DNA monoadducts. AMS can also be used to analyze some of the known underlying chemoresistance mechanisms, such as drug metabolism, cell uptake/efflux and DNA repair, after microdosing in patients and for cell line studies.
Here, for the first time, we used AMS to quantify carboplatin–DNA monoadduct formation in cancer cells to characterize chemoresistance. We determined if carboplatin–DNA monoadducts levels are associated with carboplatin resistance in cancer cells, are substrates for DNA repair and whether AMS can be used to assess chemoresistance at very low subtoxic “microdoses.”
Material and Methods
Carboplatin
Carboplatin solutions were prepared immediately before use. Mixtures of 53 mCi/mmol [14C]carboplatin (GE Healthcare, Waukesha, WI) and nonlabeled carboplatin (Novaplus, Irving, TX) were used to minimize the usage of radiocarbon and achieve the different specific activities required for microdoses and therapeutic doses.
Cell lines
Human cell lines were purchased from ATCC and cultured with the recommended medium unless otherwise specified. A549 cells were transfected with pcDNA3 vector alone or pcDNA-siRNA targeting ERCC1. ERCC1 expression was confirmed with Western blots using anti-ERCC1 (Novus Biologicals, Littleton, CO), anti-GAPDH (Abcam, Cambridge, MA) or anti-α-tubulin clone TU-01 (Invitrogen) antibodies. The ovarian cancer cell line 2008 was generously provided by Stephen Howell, MD, at the University of California, San Diego. H23 2A and 2B cell lines were generously provided by Gerold Bepler, MD, PhD, of the H. Lee Moffitt Cancer Center and Research Institute, Tampa, FL. The expression of RRM1 was confirmed with quantitative RT-PCR by Response Genetics (Los Angeles, CA).
Carboplatin treatment and AMS analysis
Cells were seeded in 60-mm dishes at a density of 1 × 106 cells per dish and allowed to attach overnight in a 37°C humidified atmosphere containing 5% CO2. At hour 0, cells were dosed and incubated with 1 µM carboplatin (microdose) or 100 µM carboplatin (therapeutic dose), each supplemented with 0.3 µM [14C]carboplatin. The 4-hr incubation was used to mimic the in vivo carboplatin half-life (1.3–6 hr) in patients.31,32 The cells were then washed twice with PBS and maintained thereafter with [14C]carboplatin-free culture media. DNA was harvested at 0, 2, 4, 8 and 24 hr using a Qiagen® Genomic DNA Purification Kit according to the manufacturer’s instruction. Ten micrograms of DNA per sample was converted to graphite and measured by AMS for 14C quantification as previously described.33 Triplicate sets of AMS experiments were performed, and the data were plotted as time vs. carboplatin–DNA monoadducts per 108 nt. Details of calculation of carboplatin–DNA monoadducts can be found in Supporting Information S1.
MTT assay to determine IC50
Carboplatin IC50 values were determined after incubating cells for 3 days with a variety of carboplatin doses as previously described.34
mRNA quantification by real-time RT-PCR
RNA isolation from cell line samples was performed according to a proprietary procedure defined by Response Genetics (Los Angeles, CA; United States Patent # 6,248,535). Briefly, after RNA isolation, cDNA was prepared from each sample. Quantitation of ERCC1 and an internal reference (β-actin) cDNA used a fluorescence-based real-time detection method (ABI PRISM 7900 Sequence detection System (TaqMan®) Perkin-Elmer (PE) Applied Biosystem, Foster City, CA).35 For each sample, parallel TaqMan PCR reactions were performed for each gene of interest and the β-actin reference gene to normalize for input cDNA. The obtained ratio between the values provides relative gene expression levels for the gene locus investigated.
Determination of intracellular glutathione levels
Intracellular total glutathione (GSH) level was detected with a colorimetric GSH detection kit per manufacture’s protocol (BioVision, Mountain View, CA). Approximately 107 cells were washed with ice-cold PBS and lysed in GSH lysis buffer. After incubation on ice for 10 min, sulfosalicylic acid solution was added, and supernatant was collected for measurement of absorbance at 410 nm. GSH standard included in the kit was used to generate a standard curve for determining the sample GSH concentrations.
Statistics
We used quantitative and graphical descriptive summaries of the DNA damage, IC50 and AUC values, separately by experiment, cell line and time (mean, standard deviation, range and tests for normality; box plots where appropriate). Differences between sensitive and resistant cell lines across the follow-up times were estimated and tested for each experiment (microdoses, therapeutic doses) using analysis of variance (ANOVA) to test for overall presence of differences between treatments and across time, including an interaction term to test for whether the effect of resistance varied over the follow-up period. We used Tukey’s studentized range test to examine specific patterns of difference for DNA damage. We examined residuals for possible model violations and considered alternative analyses (log transformations) as appropriate. Statistics were calculated with n = 3 for each cell line, except where noted in Supporting Information Tables S1 and S2. ANOVA analysis of IC50 and AUA data was based on a one-sided t-test. All tests were at an experiment-wise error rate of 0.05, and all analyses used SAS/STAT® or MedCalc® software.
Results
Comparision of DNA damage induced by microdoses and therapeutic concentrations of [14C]carboplatin
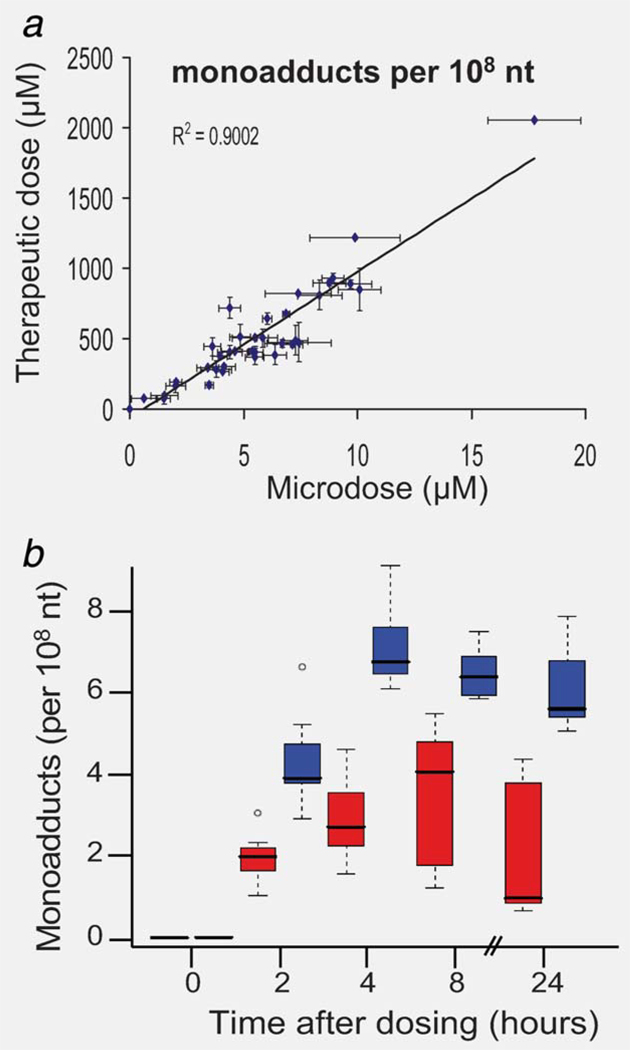
The six cell lines we tested in this project were defined as sensitive and resistant based upon the concentration of carboplatin required to reduce cell viability by 50% (IC50). Sensitive cell lines were defined as having an average IC50 < 100 µM and included H23 2A (NSCLC, IC50 = 6.6 µM), H23 2B (isogenic with H23 2A, but overexpressing RRM1, IC50 = 12 µM) and 2008 (ovarian cancer, IC50 = 37 µM). Resistant cell lines had an average IC50 > 100 µM and included A549 (NSCLC, IC50 = 229 µM), A549B (isogenic with A549, but with siRNA-mediated knockdown of ERCC1, IC50 = 156 µM) and HTB38 (colon cancer, IC50 = 105 µM). The sensitivity definitions were set considering that the maximum achievable carboplatin concentration in human blood plasma is ~100 µM during chemotherapy.36 The half-life of carboplatin in human serum at 37°C is ~33 hr, which is clearly stable enough for microdosing studies in which samples are taken over 24 hr in cell culture medium.37 Figure 3 shows carboplatin–DNA monoadduct levels at each time point for the six cell lines for the microdoses and therapeutic doses of [14C]carboplatin. Monoadducts could be detected in all cell lines at all time points for both drug concentrations (Supporting Information Table S2). The concentration of monoadducts induced by microdoses was linearly proportional to that produced by therapeutic carboplatin (Fig. 3a, p < 0.0001, R2 = 0.90). The damage ranged from ~1 to 10 monoadducts per 108 nucleotides (nt) for the microdose and ~100–1,000 monoadducts per 108 nt for the therapeutic dose—an approximate 100-fold difference. The linear dose-response for DNA adducts suggests that nontoxic microdoses of carboplatin can be used to predict the levels of DNA damage induced by therapeutic concentrations of Pt-based chemotherapy.
Figure 3.
Microdose-induced carboplatin–DNA monoadduct levels predict therapeutic adduct levels and correlate to chemoresistance. (a) Linear regression of carboplatin–DNA monoadduct levels induced by microdosing and therapeutic carboplatin. Cells were dosed for 4 hr followed by washing and incubation in carboplatin-free medium as described in Material and Methods. A linear relationship was observed (R2 = 0.90, p < 0.0001) for DNA monoadduct levels between microdoses and therapeutic doses in all cell lines and at all time points. This suggests that DNA monoadduct levels induced by therapeutic carboplatin can be predicted by the adduct levels induced by nontoxic microdosing carboplatin treatment. (b) Box plot comparing monoadduct levels in carboplatin-sensitive and -resistant cell lines over 24 hr. Resistant cell lines (IC50 > 100 µM, red) had lower levels of Pt–DNA adducts than sensitive cell lines (IC50 < 100 µM, blue). Panel b shows means of N = 3 for each data point (bars), standard deviation (boxes) and range (dashed lines) between sensitive and resistant cell lines.
Correlation of microdose-induced monoadducts with chemoresistance
Figure 3b shows a box plot statistical analysis of the microdose monoadduct data for the resistant (red squares) and sensitive (blue squares) cells. The box plot technique was used as a comprehensive method of rigorously comparing the differences in [14C]carboplatin–DNA adducts for all of the cell lines at all time points, but with a priori classification of the cell lines into sensitive and resistant groupings. The box plot considers carboplatin–DNA monoadduct levels, separated by experiment, cell line and time (mean, standard deviation, range and tests for normality). Differences between sensitive and resistant cell lines across the time points were estimated and tested for each experiment using ANOVA. The average monoadduct concentrations for all cell lines at 2, 4, 8 and 24 hr were 3.14 ± 1.50, 5.06 ± 2.39, 4.96 ± 2.04 and 3.92 ± 2.70 monoadducts per 108 nt. However, highly significant differences existed between resistant and sensitive cells (F1,75 = 186, p < 0.001) and changes over time (F4,75 = 91, p < 0.001). The differences between resistant and sensitive cells also showed some shift over time (F3,75 = 2.91, p = 0.04). The sensitive cells overall were estimated to have a 4.03 adducts per 108 nt higher DNA monoadducts compared to the resistant cells; damage was estimated to increase steadily over time up to 8 hr, but with a significant drop of 1.38 adducts per 108 nt (p = 0.009) at 24 hr in the sensitive cells.
Table 1 shows the correlations of the IC50, the AUC (area under curve, a term used to describe DNA adduct levels over time) of DNA monoadducts and the expression levels of ERCC1 and RRM1 as determined by qRT-PCR. The difference in IC50 values between each individual cell line was statistically significant, as was the overall difference between the sensitive and resistant cell lines (p < 0.001) (Table 1). There were also significant differences (p < 0.01) in AUCs for all of the cell lines (microdose AUCs are shown in Table 1 and in Supporting Information Table S3). There were no significant correlations between the IC50 values and ERCC1 and RRM1 mRNA expression, which is in contrast to clinical studies with larger subject numbers.38,39 In contrast, the AUCs and IC50 data were able to show cell-specific phenotypic correlations between DNA damage and drug resistance.
Table 1.
Correlation of IC50 with area under the adduct curves (AUC) and mRNA levels of ERCC1 and RRM1 relative to β-actin (measured by Response Genetics)
| IC50 (µM) | AUC (hr*adducts per 108 nt) |
mRNA levels (relative to β-actin) |
||
|---|---|---|---|---|
| ERCC1 | RRM1 | |||
| A549 | 229 ± 3 | 56 ± 19 | 1.47 | 9.35 |
| A549B | 156 ± 5 | 74.0 ± 0.1 | 1.84 | 4.72 |
| HTB38 | 105 ± 26 | 93 ± 7 | 5.52 | 26.57 |
| 2008 | 37 ± 9 | 138 ± 2 | 1.52 | 10.02 |
| H232B | 12 ± 4 | 160 ± 4 | 2.53 | 6.64 |
| H232A | 6.6 ± 0.2 | 195 ± 13 | 1.42 | 12.35 |
N = 3 for each experiment.
In summary, these data delineate an association between low levels of DNA monoadducts induced by microdoses of carboplatin and cellular drug resistance. In the following experiments, we set out to determine some of the underlying mechanisms that caused the most resistant (A549) cells to have lower DNA monoadduct formation and resistance to carboplatin.
Comparison of cellular uptake/efflux between chemosensitive and chemoresistant cell lines
Changes in drug uptake and efflux have been attributed to chemoresistance.40 Therefore, cells with decreased cell uptake and/or increased efflux may have low carboplatin–DNA monoadduct levels owing to low intracellular drug concentrations. We selected to compare the drug uptake and efflux of the sensitive H23 2A and the resistant A549 cells.
To determine uptake, cells were incubated with 1 and 100 µM of [14C]carboplatin, washed with ice-cold PBS and harvested as whole cells at various time points for determination of intracellular carboplatin concentration by liquid scintillation counting (LSC). The uptake of carboplatin into the two cell lines plotted as disintegrations per minute (dpm) per million cells at 0, 2, 4, 8 and 24 hr postdosing (Supporting Information Fig. S4). The rate of uptake was linearly proportional between the two carboplatin doses. There was no significant difference in the influx rates for the two cell lines. Microdosing was useful for measuring uptake as a potential factor for resistance, but we conclude that the difference in carboplatin resistance between these two specific cell lines is not mediated by drug uptake.
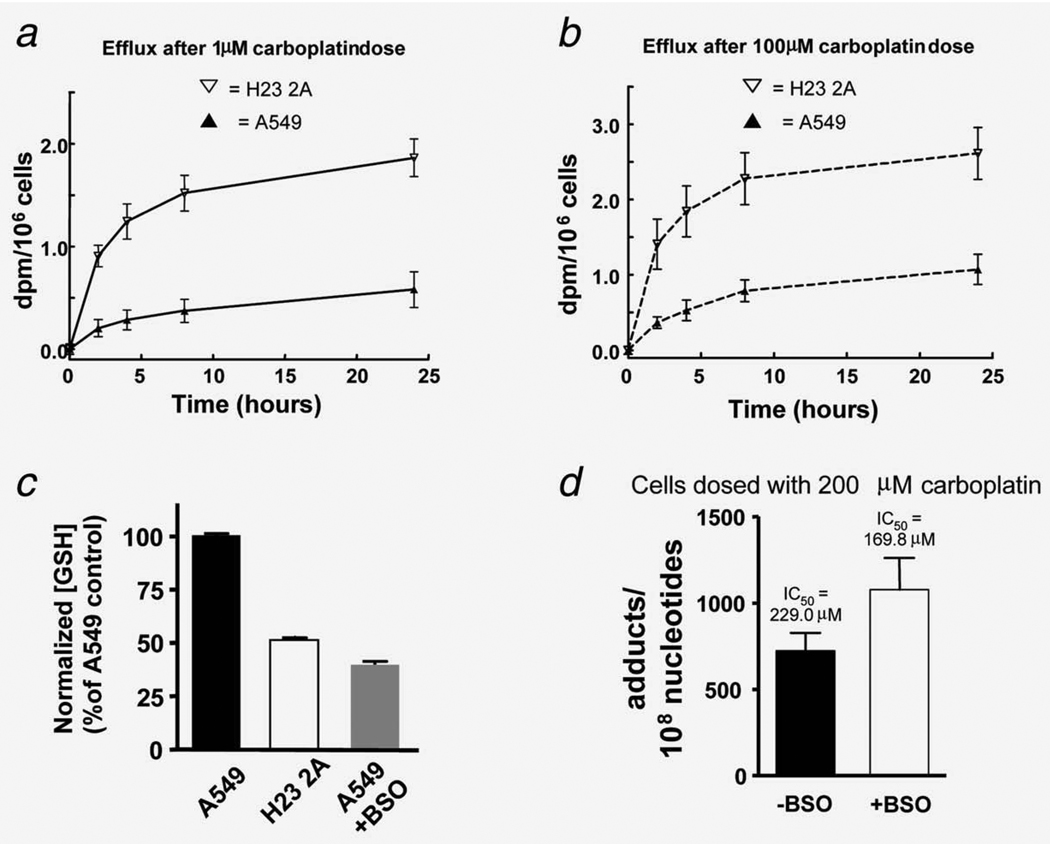
To determine efflux, the two cell lines were incubated with 1 or 100 µM [14C]carboplatin for 4 hr to allow the intracellular accumulation of carboplatin to reach a substantial level. Cells were then washed and cultured in carboplatin-free medium at 37°C. Samples of the extracellular medium were taken at 0, 2, 4, 8 and 24 hr for measurement of cumulative efflux of 14C into the media by LSC (Figs. 4a and 4b). Efflux was rapid at the early time points but did not increase significantly between 8 and 24 hr. Unexpectedly, there was ~3.5-fold (p < 0.001) higher carboplatin efflux for the more sensitive cell line, indicating efflux did not contribute significantly to the resistance differences between these two specific cell lines and may even be a confounding factor.
Figure 4.
Molecular dissection of chemoresistance: pre-DNA damage events. (a, b) Comparison of efflux of [14C]carboplatin between H23 2A and A549 cells treated with microdoses (a) and therapeutic carboplatin doses (b). A549 cells had 3.5-fold slower efflux (p < 0.001), suggesting that other factors such as intracellular inactivation may be responsible for low carboplatin–DNA adduct levels in A549 cells. (c) Measurement of GSH. The GSH level in A549 cells is 1.9 times that of H23 2A cells. Treatment with 50 µM buthionine sulphoximine (BSO) decreased GSH level by 54% in A549 cells. (d) BSO treatment increased carboplatin–DNA monoadduct levels (p < 0.01) and sensitized A549 cells to carboplatin (IC50 = 229.0 µM without BSO and 169.8 µM with BSO, p < 0.001). Each data point represents the mean from N = 3 ± SD.
Effects of intracellular inactivation on DNA damage and sensitivity to carboplatin
Compared to H23 2A, A549 cells had high intracellular carboplatin (low efflux, Figs. 4a and 4b) but lower DNA monoadduct levels (Table 1), suggesting that some intracellular mechanisms decrease the formation of carboplatin–DNA monoadducts. GSH participates in intracellular inactivation and is involved in chemoresistance.41 Compared to the sensitive H23 2A cell line, A549 had a twofold higher concentration of GSH (p < 0.001, Fig. 4c), suggesting that high GSH might contribute, at least in part, to the resistant phenotype of A549.
To further determine if high GSH concentrations contribute to chemoresistance in A549 cells, we treated these cells with buthionine sulphoximine (BSO). BSO is an inhibitor of gamma-glutamylcysteine synthetase, an enzyme required for biosynthesis of GSH.42 BSO treatment decreased GSH in A549 cells in a dose-dependent manner (Supporting Information Fig. S4). At 50 µM BSO, which is nontoxic to the cells, the GSH concentration was decreased by 54% (Fig. 4c), and the cells became more sensitive to carboplatin (IC50 of 169.8 ± 5.0 µM for cells exposed to BSO, compared to 229.0 ± 3.3µM for A549 control cells, p < 0.001) (Fig. 4d). Carboplatin–DNA monoadducts were increased by 36% in BSO-treated A549 cells when dosed with 200 µM carboplatin (p < 0.01) (Fig. 4d). The data suggest that mitigating some of the pre-DNA damage resistance mechanisms, such as inactivation by GSH or other thiol-containing species, can increase Pt-monoadduct formation and sensitize cancer cells to carboplatin.
The effect of DNA repair on carboplatin–DNA adduct levels and cellular sensitivity to carboplatin
High DNA repair capacity may result in Pt resistance. ERCC1 is one of the key proteins in the multisubunit nucleotide excision repair (NER) complex.43 Ribonucleotide reductase M1 (RRM1, one subunit of the heterodimeric complex) supplies the nucleotide pool with 2′-deoxynucleoside triphosphates that are essential for DNA synthesis and repair.44 Both proteins are involved in resistance to Pt-based chemotherapy.45 We used paired cell lines with differences in the expression of ERCC1 and RRM1 to determine if alterations of these two different DNA repair mechanisms contributed to resistance to carboplatin.
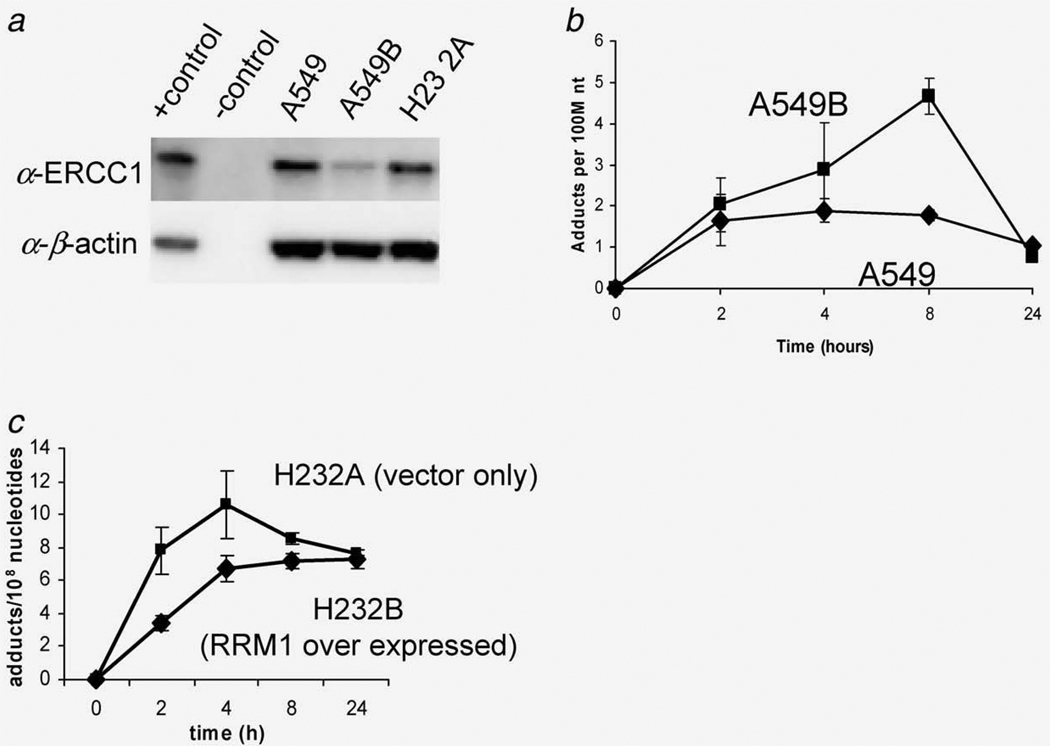
First, downregulated ERCC1 expression was knocked down in A549 cells. Transfection of siRNA targeting ERCC1 in A549B cells resulted in decreased ERCC1 expression as determined with Western blotting (Fig. 5a). The siRNA-expressing cells were sensitized by approximately twofold to carboplatin exposure compared to control cells (Table 1). Consistent with the protein expression results, A549B cells displayed a 32% higher AUC for carboplatin–DNA monoadducts compared to A549 cells transfected with the vector alone (Fig. 5b) (p < 0.001, Table 1). Although A549B cells exhibited a nearly twofold decrease in carboplatin resistance, these cells were still far more resistant than H23 2A cells (IC50 = 156.0 vs. 6.6 µM, respectively, Table 1), indicating that multiple resistant mechanisms contribute to the chemoresistance of A549 cells.
Figure 5.
The effect of DNA repair in cellular sensitivity to carboplatin. (a) siRNA targeting of ERCC1 decreased ERCC1 expression as determined by Western blotting. (b) Downregulation of ERCC1 increased carboplatin–DNA monoadduct levels (p < 0.001). (c) Overexpression of RRM1 decreased carboplatin–DNA adduct levels (p < 0.01).
The influence of RRM1 on resistance was tested by comparing IC50 values and monoadduct formation in H23 2A (vector alone) and H23 2B (overexpression of RRM1) cells exposed to carboplatin (Fig. 5c). Compared to H23 2A as control cells, overexpression of RRM1 in H23 2B cells increased the IC50 from 6.6 to 11.6 µM (p < 0.001) and the microdose-induced monoadducts by 25% based upon AUC (Table 1). These data support a clear role for DNA repair in mediating monoadduct levels and chemoresistance. This is the first report that supports the involvement of NER mechanisms in response to carboplatin–DNA monoadduct formation.
Discussion
We characterized carboplatin–DNA monoadduct formation and repair, the cumulative result of the metabolism and uptake of carboplatin, followed by DNA alkylation and the ensuing cellular damage response. This characterization was performed by exposing cells to microdoses and therapeutic doses of carboplatin in an effort to develop and validate a novel methodology for quantifying monoadduct formation and repair at subtoxic doses, which may have utility for in vivo and clinical studies. We showed that in a set of cancer cell lines: (i) microdoses of [14C]carboplatin induced measurable carboplatin–DNA monoadducts; (ii) the concentrations of carboplatin–DNA monoadducts induced by the microdoses were linearly proportional to those formed in response to therapeutic doses; (iii) low monoadduct levels correlated to resistance, whereas the sensitive cell lines had relatively high concentrations of radiolabeled DNA damage and (iv) carboplatin–DNA adducts are repaired, in part, by NER, but chemoresistance involves several additional and likely only partially defined mechanisms.
The impact of DNA repair on cellular carboplatin–DNA monoadduct levels and chemoresistance has not been previously reported. Our observations suggest that carboplatin–DNA monoadducts are at least in part repaired by NER. Related genetic markers such as ERCC1 and RRM1 have been found to be clinically associated with resistance to Pt-based chemotherapy. Approximately 150 proteins have been identified that participate in DNA repair.4,46 Many of these proteins participate in cellular responses to Pt–DNA damage, particularly for NER. Our ERCC1-based cell line data support that carboplatin–DNA monoadducts are recognized by mammalian NER and may be useful as biomarkers of Pt–DNA adduct formation and repair with specificity for NER.
The microdosing strategy is amenable to the analysis of the mechanisms of cellular resistance to carboplatin other than DNA repair. For example, some cells had relatively low initial carboplatin–DNA adduct levels, which indicate that there are resistance mechanisms participating in the pre-DNA damage steps (Fig. 2) to decrease DNA monoadduct formation, such as elevated GSH levels and low intracellular drug concentration (slow influx and/or rapid efflux). Our study showed that the nontoxic microdosing approach could guide further analysis of the underlying resistance mechanisms based on the Pt–DNA adduct levels.
One major concern is whether the levels of monoadducts can potentially predict cellular resistance to carboplatin, as they are less toxic when compared to diadducts (intrastrand and interstrand crosslinks). In our study with just six cell lines, it seems that, compared to RT-PCR analysis of ERCC1 and RRM1 gene expression levels, DNA monoadduct levels correlate better with cellular carboplatin resistance (Table 1). However, this hypothesis requires further testing, and we have opened a Phase 0 microdosing trial for this purpose. In this clinical trial, patients will receive one nontoxic microdose of [14C]carboplatin about 4 hr before scheduled tumor biopsy. Blood samples and tumor biopsies will be taken up to 24 hr postdose, from which peripheral blood mononuclear cells (PBMCs) will be isolated. Drug uptake, efflux and DNA repair of monoadducts in the cells will be measured by AMS. The data will be correlated with endpoints such as toxicity and tumor response to cisplatin- or carboplatin-based chemotherapy. Pt–DNA damage and repair in PBMC are of interest, as such data have previously been correlated tumor response to chemotherapy, but from much more toxic doses than those planned for the current clinical study.6–8,11 Another clinical trial under consideration is to perform a microdosing trial while NSCLC patients undergo surgical resection and select potential responders for adjuvant chemotherapy. Considering the low efficacy of Pt-based chemotherapy in NSCLC, this approach may have great clinical application, but will need to be verified in a separate clinical trial. Although our study design is specific for measuring carboplatin–DNA monoadducts, AMS can measure both mono- and diadducts formed by the reaction of [14C]oxaliplatin with DNA when the radiocarbon label is located on the diaminocyclohexane ring.24
In conclusion, we developed a microdosing approach that enabled defining carboplatin–DNA monoadduct formation and repair, likely by NER, which may be useful for characterizing chemoresistance in vivo.
Supplementary Material
Acknowledgements
AMS samples were prepared at Lawrence Livermore National Laboratory under the auspices of the DOE contract DE-AC52-07NA27344 and supported by NIH/NCRR (KT) Resource for Biomedical Accelerator Mass Spectrometry P41 RR013461 and DOE LDRD grant 08-LW-100 (MM). The authors thank Kurt Hack and Ted Ognibene for preparation of AMS samples. They are grateful to Judy Li and Primo Lara for helpful discussions and advice.
Grant sponsors: Gerry and Susan Knapp Family Fund (PTH), the National Institutes of Health (KWT), American Cancer Society Institutional Research Grant (CXP)
Footnotes
Additional Supporting Information may be found in the online version of this article
Some of the data in this article were presented at the 2010 American Society of Clinical Oncology Genitourinary Cancer Symposium (San Francisco, CA, USA)
Potential conflicts of interest: Nothing to report
References
- 1.Chaney SG, Campbell SL, Temple B, Bassett E, Wu Y, Faldu M. Protein interactions with platinum-DNA adducts: from structure to function. J Inorg Biochem. 2004;98:1551–1559. doi: 10.1016/j.jinorgbio.2004.04.024. [DOI] [PubMed] [Google Scholar]
- 2.Goodisman J, Hagrman D, Tacka KA, Souid AK. Analysis of cytotoxicities of platinum compounds. Cancer Chemother Pharmacol. 2006;57:257–267. doi: 10.1007/s00280-005-0041-4. [DOI] [PubMed] [Google Scholar]
- 3.Unger FT, Klasen HA, Tchartchian G, de Wilde RL, Witte I. DNA damage induced by cis- and carboplatin as indicator for in vitro sensitivity of ovarian carcinoma cells. BMC Cancer. 2009;9:359. doi: 10.1186/1471-2407-9-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang D, Lippard SJ. Cellular processing of platinum anticancer drugs. Nat Rev Drug Discov. 2005;4:307–320. doi: 10.1038/nrd1691. [DOI] [PubMed] [Google Scholar]
- 5.Kartalou M, Essigmann JM. Mechanisms of resistance to cisplatin. Mutat Res. 2001;478:23–43. doi: 10.1016/s0027-5107(01)00141-5. [DOI] [PubMed] [Google Scholar]
- 6.Parker RJ, Gill I, Tarone R, Vionnet JA, Grunberg S, Muggia FM, Reed E. Platinum-DNA damage in leukocyte DNA of patients receiving carboplatin and cisplatin chemotherapy, measured by atomic absorption spectrometry. Carcinogenesis. 1991;12:1253–1258. doi: 10.1093/carcin/12.7.1253. [DOI] [PubMed] [Google Scholar]
- 7.Fichtinger-Schepman AM, van der Velde-Visser SD, van Dijk-Knijnenburg HC, van Oosterom AT, Baan RA, Berends F. Kinetics of the formation and removal of cisplatin-DNA adducts in blood cells and tumor tissue of cancer patients receiving chemotherapy: comparison with in vitro adduct formation. Cancer Res. 1990;50:7887–7894. [PubMed] [Google Scholar]
- 8.Reed E, Ozols RF, Tarone R, Yuspa SH, Poirier MC. Platinum-DNA adducts in leukocyte DNA correlate with disease response in ovarian cancer patients receiving platinum-based chemotherapy. Proc Natl Acad Sci USA. 1987;84:5024–5028. doi: 10.1073/pnas.84.14.5024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reed E, Ozols RF, Tarone R, Yuspa SH, Poirier MC. The measurement of cisplatin-DNA adduct levels in testicular cancer patients. Carcinogenesis. 1988;9:1909–1911. doi: 10.1093/carcin/9.10.1909. [DOI] [PubMed] [Google Scholar]
- 10.Reed E, Parker RJ, Gill I, Bicher A, Dabholkar M, Vionnet JA, Bostick-Bruton F, Tarone R, Muggia FM. Platinum-DNA adduct in leukocyte DNA of a cohort of 49 patients with 24 different types of malignancies. Cancer Res. 1993;53:3694–3699. [PubMed] [Google Scholar]
- 11.Schellens JH, Ma J, Planting AS, van der Burg ME, van Meerten E, de Boer-Dennert M, Schmitz PI, Stoter G, Verweij J. Relationship between the exposure to cisplatin, DNA-adduct formation in leucocytes and tumour response in patients with solid tumours. Br J Cancer. 1996;73:1569–1575. doi: 10.1038/bjc.1996.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van de Vaart PJ, Belderbos J, de Jong D, Sneeuw KC, Majoor D, Bartelink H, Begg AC. DNA-adduct levels as a predictor of outcome for NSCLC patients receiving daily cisplatin and radiotherapy. Int J Cancer. 2000;89:160–166. [PubMed] [Google Scholar]
- 13.Poirier MC, Reed E, Shamkhani H, Tarone RE, Gupta-Burt S. Platinum drug-DNA interactions in human tissues measured by cisplatin-DNA enzyme-linked immunosorbent assay and atomic absorbance spectroscopy. Environ Health Perspect. 1993;99:149–154. doi: 10.1289/ehp.9399149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bonetti A, Apostoli P, Zaninelli M, Pavanel F, Colombatti M, Cetto GL, Franceschi T, Sperotto L, Leone R. Inductively coupled plasma mass spectroscopy quantitation of platinum-DNA adducts in peripheral blood leukocytes of patients receiving cisplatin- or carboplatin-based chemotherapy. Clin Cancer Res. 1996;2:1829–1835. [PubMed] [Google Scholar]
- 15.Fisch MJ, Howard KL, Einhorn LH, Sledge GW. Relationship between platinum-DNA adducts in leukocytes of patients with advanced germ cell cancer and survival. Clin Cancer Res. 1996;2:1063–1066. [PubMed] [Google Scholar]
- 16.Motzer RJ, Reed E, Perera F, Tang D, Shamkhani H, Poirier MC, Tsai WY, Parker RJ, Bosl GJ. Platinum-DNA adducts assayed in leukocytes of patients with germ cell tumors measured by atomic absorbance spectrometry and enzyme-linked immunosorbent assay. Cancer. 1994;73:2843–2852. doi: 10.1002/1097-0142(19940601)73:11<2843::aid-cncr2820731130>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 17.Blommaert FA, Saris CP. Detection of platinum-DNA adducts by 32P-postlabelling. Nucleic Acids Res. 1995;23:1300–1306. doi: 10.1093/nar/23.8.1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Welters MJ, Maliepaard M, Jacobs-Bergmans AJ, Baan RA, Schellens JH, Ma J, van der Vijgh WJ, Braakhuis BJ, Fichtinger-Schepman AM. Improved 32P-postlabelling assay for the quantification of the major platinum-DNA adducts. Carcinogenesis. 1997;18:1767–1774. doi: 10.1093/carcin/18.9.1767. [DOI] [PubMed] [Google Scholar]
- 19.Shamkhani H, Anderson LM, Henderson CE, Moskal TJ, Runowicz CD, Dove LF, Jones AB, Chaney SG, Rice JM, Poirier MC. DNA adducts in human and patas monkey maternal and fetal tissues induced by platinum drug chemotherapy. Reprod Toxicol. 1994;8:207–216. doi: 10.1016/0890-6238(94)90004-3. [DOI] [PubMed] [Google Scholar]
- 20.Giurgiovich AJ, Diwan BA, Olivero OA, Anderson LM, Rice JM, Poirier MC. Elevated mitochondrial cisplatin-DNA adduct levels in rat tissues after transplacental cisplatin exposure. Carcinogenesis. 1997;18:93–96. doi: 10.1093/carcin/18.1.93. [DOI] [PubMed] [Google Scholar]
- 21.Gupta-Burt S, Shamkhani H, Reed E, Tarone RE, Allegra CJ, Pai LH, Poirier MC. Relationship between patient response in ovarian and breast cancer and platinum drug-DNA adduct formation. Cancer Epidemiol Biomarkers Prev. 1993;2:229–234. [PubMed] [Google Scholar]
- 22.Bassett E, King NM, Bryant MF, Hector S, Pendyala L, Chaney SG, Cordeiro-Stone M. The role of DNA polymerase eta in translesion synthesis past platinum-DNA adducts in human fibroblasts. Cancer Res. 2004;64:6469–6475. doi: 10.1158/0008-5472.CAN-04-1328. [DOI] [PubMed] [Google Scholar]
- 23.Hah SS, Stivers KM, de Vere White RW, Henderson PT. Kinetics of carboplatin-DNA binding in genomic DNA and bladder cancer cells as determined by accelerator mass spectrometry. Chem Res Toxicol. 2006;19:622–626. doi: 10.1021/tx060058c. [DOI] [PubMed] [Google Scholar]
- 24.Hah SS, Sumbad RA, de Vere White RW, Turteltaub KW, Henderson PT. Characterization of oxaliplatin-DNA adduct formation in DNA and differentiation of cancer cell drug sensitivity at microdose concentrations. Chem Res Toxicol. 2007;20:1745–1751. doi: 10.1021/tx700376a. [DOI] [PubMed] [Google Scholar]
- 25.Hah SS, Mundt JM, Kim HM, Sumbad RA, Turteltaub KW, Henderson PT. Measurement of 7,8-dihydro-8-oxo-2’-deoxyguanosine metabolism in MCF-7 cells at low concentrations using accelerator mass spectrometry. Proc Natl Acad Sci USA. 2007;104:11203–11208. doi: 10.1073/pnas.0701733104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Turteltaub KW, Felton JS, Gledhill BL, Vogel JS, Southon JR, Caffee MW, Finkel RC, Nelson DE, Proctor ID, Davis JC. Accelerator mass spectrometry in biomedical dosimetry: relationship between low-level exposure and covalent binding of heterocyclic amine carcinogens to DNA. Proc Natl Acad Sci USA. 1990;87:5288–5292. doi: 10.1073/pnas.87.14.5288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sandhu P, Vogel JS, Rose MJ, Ubick EA, Brunner JE, Wallace MA, Adelsberger JK, Baker MP, Henderson PT, Pearson PG, Baillie TA. Evaluation of microdosing strategies for studies in preclinical drug development: demonstration of linear pharmacokinetics in dogs of a nucleoside analog over a 50-fold dose range. Drug Metab Dispos. 2004;32:1254–1259. doi: 10.1124/dmd.104.000422. [DOI] [PubMed] [Google Scholar]
- 28.Li Y, Shawgo RS, Tyler B, Henderson PT, Vogel JS, Rosenberg A, Storm PB, Langer R, Brem H, Cima MJ. In vivo release from a drug delivery MEMS device. J Control Release. 2004;100:211–219. doi: 10.1016/j.jconrel.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 29.Lappin G, Kuhnz W, Jochemsen R, Kneer J, Chaudhary A, Oosterhuis B, Drijfhout WJ, Rowland M, Garner RC. Use of microdosing to predict pharmacokinetics at the therapeutic dose: experience with 5 drugs. Clin Pharmacol Ther. 2006;80:203–215. doi: 10.1016/j.clpt.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 30.Carkeet C, Dueker SR, Lango J, Buchholz BA, Miller JW, Green R, Hammock BD, Roth JR, Anderson PJ. Human vitamin B12 absorption measurement by accelerator mass spectrometry using specifically labeled 14C-cobalamin. Proc Natl Acad Sci USA. 2006;103:5694–5699. doi: 10.1073/pnas.0601251103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fujiwara K, Yamauchi H, Suzuki S, Ishikawa H, Tanaka Y, Fujiwara M, Kohno I. The platelet-sparing effect of paclitaxel is not related to changes in the pharmacokinetics of carboplatin. Cancer Chemother Pharmacol. 2001;47:22–26. doi: 10.1007/s002800000212. [DOI] [PubMed] [Google Scholar]
- 32.Sharma H, Thatcher N, Baer J, Zaki A, Smith A, McAucliffe CA, Crowther D, Owens S, Fox BW. Blood clearance of radioactively labelled cis-diammine 1,1-cyclobutane dicarboxylate platinum (II) (CBDCA) in cancer patients. Cancer Chemother Pharmacol. 1983;11:5–7. doi: 10.1007/BF00257407. [DOI] [PubMed] [Google Scholar]
- 33.Ognibene TJ, Bench G, Vogel JS, Peaslee GF, Murov S. A high-throughput method for the conversion of CO2 obtained from biochemical samples to graphite in septasealed vials for quantification of 14C via accelerator mass spectrometry. Anal Chem. 2003;75:2192–2196. doi: 10.1021/ac026334j. [DOI] [PubMed] [Google Scholar]
- 34.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 35.Gibson UE, Heid CA, Williams PM. A novel method for real time quantitative RT-PCR. Genome Res. 1996;6:995–1001. doi: 10.1101/gr.6.10.995. [DOI] [PubMed] [Google Scholar]
- 36.Jacobs SS, Fox E, Dennie C, Morgan LB, McCully CL, Balis FM. Plasma and cerebrospinal fluid pharmacokinetics of intravenous oxaliplatin, cisplatin, and carboplatin in nonhuman primates. Clin Cancer Res. 2005;11:1669–1674. doi: 10.1158/1078-0432.CCR-04-1807. [DOI] [PubMed] [Google Scholar]
- 37.Gaver RC, George AM, Deeb G. In vitro stability, plasma protein binding and blood cell partitioning of 14C-carboplatin. Cancer Chemother Pharmacol. 1987;20:271–276. doi: 10.1007/BF00262576. [DOI] [PubMed] [Google Scholar]
- 38.Bepler G, Kusmartseva I, Sharma S, Gautam A, Cantor A, Sharma A, Simon G. RRM1 modulated in vitro and in vivo efficacy of gemcitabine and platinum in non-small-cell lung cancer. J Clin Oncol. 2006;24:4731–4737. doi: 10.1200/JCO.2006.06.1101. [DOI] [PubMed] [Google Scholar]
- 39.Lord RV, Brabender J, Gandara D, Alberola V, Camps C, Domine M, Cardenal F, Sanchez JM, Gumerlock PH, Taron M, Sanchez JJ, Danenberg KD, et al. Low ERCC1 expression correlates with prolonged survival after cisplatin plus gemcitabine chemotherapy in non-small cell lung cancer. Clin Cancer Res. 2002;8:2286–2291. [PubMed] [Google Scholar]
- 40.Zhou SF, Wang LL, Di YM, Xue CC, Duan W, Li CG, Li Y. Substrates and inhibitors of human multidrug resistance associated proteins and the implications in drug development. Curr Med Chem. 2008;15:1981–2039. doi: 10.2174/092986708785132870. [DOI] [PubMed] [Google Scholar]
- 41.Godwin AK, Meister A, O’Dwyer PJ, Huang CS, Hamilton TC, Anderson ME. High resistance to cisplatin in human ovarian cancer cell lines is associated with marked increase of glutathione synthesis. Proc Natl Acad Sci USA. 1992;89:3070–3074. doi: 10.1073/pnas.89.7.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Griffith OW, Meister A. Potent and specific inhibition of glutathione synthesis by buthionine sulfoximine (S-n-butyl homocysteine sulfoximine) J Biol Chem. 1979;254:7558–7560. [PubMed] [Google Scholar]
- 43.Westerveld A, Hoeijmakers JH, van Duin M, de Wit J, Odijk H, Pastink A, Wood RD, Bootsma D. Molecular cloning of a human DNA repair gene. Nature. 1984;310:425–429. doi: 10.1038/310425a0. [DOI] [PubMed] [Google Scholar]
- 44.Caras IW, Levinson BB, Fabry M, Williams SR, Martin DW., Jr Cloned mouse ribonucleotide reductase subunit M1 cDNA reveals amino acid sequence homology with Escherichia coli and herpesvirus ribonucleotide reductases. J Biol Chem. 1985;260:7015–7022. [PubMed] [Google Scholar]
- 45.Zheng Z, Chen T, Li X, Haura E, Sharma A, Bepler G. DNA synthesis and repair genes RRM1 and ERCC1 in lung cancer. N Engl J Med. 2007;356:800–808. doi: 10.1056/NEJMoa065411. [DOI] [PubMed] [Google Scholar]
- 46.Helleday T, Petermann E, Lundin C, Hodgson B, Sharma RA. DNA repair pathways as targets for cancer therapy. Nat Rev Cancer. 2008;8:193–204. doi: 10.1038/nrc2342. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.