Abstract
Recent studies distinguish the biological and pharmacological effects of nitroxyl (HNO) from its oxidized/deprotonated product nitric oxide (NO), but lack of HNO detection methods limits understanding its in vivo mechanisms and the identification of endogenous sources. We previously demonstrated that reaction of HNO with triarylphosphines provides aza-ylides and HNO-derived amides, which may serve as stable HNO biomarkers. We now report a kinetic analysis for the trapping of HNO by phosphines, ligations of enzyme-generated HNO, and compatibility studies illustrating the selectivity of phosphines for HNO over other physiologically relevant nitrogen oxides. Quantification of HNO using phosphines is demonstrated using an HPLC-based assay and ligations of phosphine carbamates generate HNO-derived ureas. These results further demonstrate the potential of phosphine probes for reliable biological detection and quantification of HNO.
Keywords: Nitroxyl, triarylphosphine, aza-ylide, chemical ligation, Angeli’s salt, heme protein, carbamate ligation
Introduction
Nitroxyl (HNO) has attracted increasing attention in the past two decades as evidence continues to emerge demonstrating its unique physiology and chemistry from its more well-known one-electron reduced and deprotonated product nitric oxide (NO). Most notable are the findings that HNO donors increase cardiac inotropy and lusitropy and elicit arterial and venous dilation without building tolerance, making these compounds intriguing candidates as prodrugs for congestive heart failure, a condition that affects 5.8 million Americans.1–8 The unique ability of HNO donors to increase systolic force and decrease diastolic pressure appears to occur through specific and covalent thiol modification that facilitates myocardial calcium cycling and enhances the calcium sensitivity of myofilament.9–11 These findings motivate increased efforts for the definition of new HNO donors and identification of endogenous sources.12–22
In addition to thiols, HNO reacts with ferric and ferrous heme proteins and is a potent electrophile toward amine and phosphine nucleophiles.23–26 In the absence of these reactive partners, HNO rapidly dimerizes (k = 8 × 106 M−1 s−1) to yield highly labile hyponitrous acid, which dehydrates to nitrous oxide (N2O), illustrating the short lifetime of free HNO and necessitating the use of donor compounds in chemical and biochemical studies.27 Dimerization and the rapid reaction with various biomolecules challenge HNO detection and limit the conclusive identification of endogenous sources, the understanding of HNO mechanisms in vivo, and the development of its full therapeutic potential.
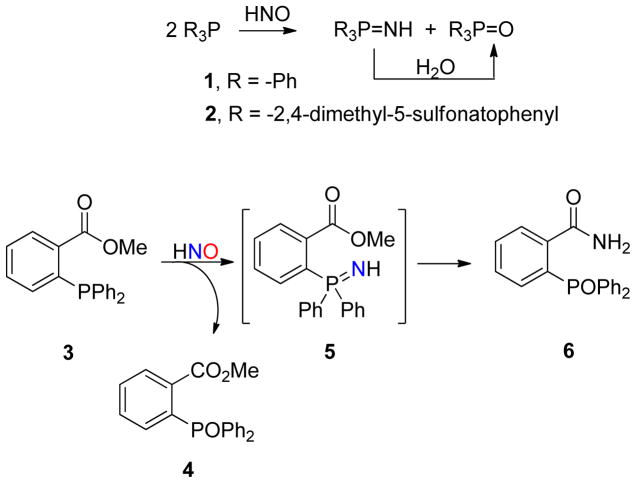
We previously reported that the reaction of donor-provided HNO with triarylphosphine nucleophiles (1–2, Scheme 1) provides equimolar amounts of phosphine oxide and aza-ylide.26 The newly formed HNO-derived ylide hydrolyzes to generate phosphine oxide and presumably ammonia at a rate determined by the steric and electronic environment of the phosphorus atom. In the presence of an intramolecular electrophile (3, Scheme 1), the aza-ylide subsequently undergoes a Staudinger ligation to generate an amide with an HNO-derived nitrogen atom (6). These first phosphine-mediated ligations provide a unique and thermodynamically stable HNO-derived product. Herein, we further characterize the kinetics and compatibility of a phosphine-based method for HNO detection and quantification to overcome impediments to understanding HNO’s biological chemistry. We report a kinetic analysis, aqueous ligations to form HNO-derived amides and ureas, an HPLC-based method to quantify HNO, and the use of triarylphosphines to trap and ligate heme protein-generated HNO. These results highlight the potential of this chemistry for the detection and understanding of HNO.
Scheme 1.
Experimental Methods
General
Chemicals
Reagents were obtained from commercial sources and used without additional purification. Angeli’s salt (AS) used in HPLC quantification experiments was purchased from Cayman Chemical and stored at −20°C. AS used in all other experiments was prepared as described and stored at −20°C.28 Reaction solvents such as dichloromethane, acetonitrile, and ethyl acetate were dried and distilled over calcium hydride. Extraction, silica, and preparative reverse phase chromatography solvents were technical grade. LC-MS, ESI-MS, and HPLC solvents were HPLC grade. Deionized water was obtained in-house and filtered for HPLC experiments.
Analytical Determinations
Analytical TLC was performed on silica gel plates (normal phase) or C18 silica gel plates (reverse phase), and visualization was accomplished with UV light. 1H NMR spectra were recorded on Bruker Avance DPX-300 and DRX-500 instruments at 300.13 and 500.13 MHz, respectively. 13C NMR spectra were recorded on the described instruments operating at 75.48 and 125.76 MHz, respectively. 31P NMR spectra were recorded on the described instruments operating at 121.49 MHz and 202.46 MHz, respectively. 31P chemical shifts are referenced to 85% H3PO4 (δ= 0 ppm) in a concentric internal capillary (Wilmad). NMR spectra were obtained using Bruker 5 mm BBO and QNP probes held at 25°C unless otherwise stated. UV spectra were obtained on a Cary Bio UV-Visible Spectrophotometer equipped with a temperature controller. Measurements of pH were performed on an Oakton pH meter, model pH 11, with a combination electrode. Two point calibrations with commercially available standards were performed before pH values were recorded. Low-resolution mass spectra were obtained using an Agilent Technologies 1100 LC/MSD ion trap mass spectrometer equipped with an atmospheric pressure electrospray ionization source and was operated in positive ion mode unless otherwise noted. High resolution mass spectrometry for 15a and 19 was performed at the University of North Carolina at Chapel Hill.
Liquid chromatography-mass spectrometry
Liquid chromatography-mass spectrometry experiments were performed on an Agilent Technologies 1100 LC/MSD Trap instrument. Separations were achieved using an Agilent Zorbax Rapid Resolution SB-C18 reverse phase column (2.1 × 30 mm/3.5 μm) held at 25°C. Eluents: 0.1% ammonium formate in water (AFW); acetonitrile. Flow rate: 0.4 mL/min. Gradients for conditioning of column and for analyses: see specific experimental protocols. The ion trap mass spectrometer was equipped with an atmospheric pressure electrospray ionization source, and was operated in smart mode. Nebulization was achieved with N2 pressure of 50 psi, and solvent evaporation assisted by a flow of He drying gas (11 L/min, 325°C). Mass spectra were obtained in positive ion mode unless otherwise noted. Retention times and masses of extracted ions were compared to standards (either commercially available or synthesized as described herein) run on the described instrument.
Kinetic analysis
Kinetics studies were performed using 5 mm O.D. thin-walled precision NMR tubes (Wilmad). All 31P NMR experiments were performed on a Bruker DRX-500 instrument operating at 202.46 MHz and equipped with a BBO probe held at 37°C. Data was acquired using the standard Bruker zgpg pulse program and all spectra were processed in an identical fashion.
A calibration plot relating 31P NMR integral to TXPTS ylide concentration was determined for TXPTS ylide standards, which were prepared via reaction of TXPTS (133 mg, 0.20 mmol) and hydroxylamine O-sulfonic acid (26 mg, 0.23 mmol).26 The reaction was incubated at 37°C for one hour to provide an ylide stock solution (215 mM). For NMR analysis, seven calibration samples were prepared ranging from 10 to 88 mM ylide in 0.5 mL of 20% D2O/Tris buffer (500 mM Tris, 0.2 mM EDTA, pH 7.4). TXPTS ylide integrals were determined relative to the integral for the H3PO4 standard (set to 100).
The rate constant for the reaction of TXPTS and AS-derived HNO at 37°C was determined by direct in situ competition with the well-known reaction of glutathione (GSH) with HNO at the same temperature.23 All competition reactions contained TXPTS (52 mg, 0.08 mmol, 160 mM) and AS (5 mg, 0.04 mmol, 80 mM) in 0.5 mL of 20% D2O/Tris buffer (500 mM Tris, 0.2 mM EDTA, pH 7.4). A GSH stock solution (325 mM) was prepared in the previously mentioned Tris buffer, and reaction concentrations of GSH ranged from 0 to 90 mM. To initiate the reactions, TXPTS was dissolved in the appropriate volume of Tris buffer, followed by addition of the corresponding volume of GSH stock. AS was quickly added, and the reaction was incubated at 37°C overnight. The mixture was then diluted with D2O (0.1 mL) and analyzed by 31P NMR. TXPTS ylide concentrations were calculated using the 31P NMR calibration curve and compared to the maximum amount of ylide formed in the absence of competitor to determine the GSH concentration at which ylide formation is reduced to 50%.
HPLC quantification of phosphine-mediated HNO trapping
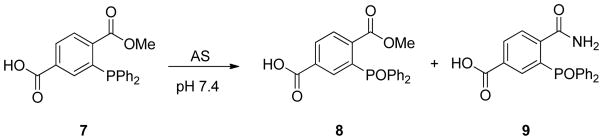
A solution of 7 in NaOH (0.5 M) was added to an aqueous solution of 80% anion phosphate buffer (0.1 M, 50 μM DTPA, pH 7.4). Solid AS was added to this solution to provide final concentrations of 7 (5 mM) and AS (1 mM). The reaction was stirred at 37°C in a sealed vial. Aliquots were removed periodically and chromatographically separated and analyzed using a Waters 2695 HPLC with an autosampler, a temperature-controlled carousel, and a Waters 996 photodiode array detector monitoring at 254 nm. Separations were performed on a Restek Pinnacle II reverse phase C18 250 × 4.6 mm/5 μm column and effected by means of a H2O/CH3CN gradient. Flow rate: 1.0 mL/min. Gradient: 2% CH3CN to 25% CH3CN over 10 min, 25% CH3CN to 50% CH3CN over 2 min, hold for 6 min, 50% CH3CN to 2% CH3CN over 1 min, and hold for 11 min for a total run time of 30 min. Products 8 and 9 were identified using authentic standards with the yields being calculated from standard curves. Control experiments involved the incubation of 7 with nitric oxide (NO) or sodium nitrite in aqueous phosphate buffer at 37°C monitoring for the formation of 9.
Formation of TXPTS ylide via catalase-mediated HNO generation
A solution of β-D(+)glucose (10 mM), cyanamide (50 mM), and TXPTS (50 mM) in Tris buffer (500 mM, 0.2 mM EDTA, pH 7.4, 0.5 mL) was incubated at 37°C for 5 minutes, then glucose oxidase (20 units, type II from Aspergillus niger) was added. After 15 seconds, bovine hepatic catalase (9 μM) was added to initiate the reaction, which was maintained at 37°C. UV-Vis spectra obtained at 37°C reveal formation of the cat(FeIII-CN) complex within 15 minutes. At 4 hours, a portion of the reaction mixture (0.35 mL) was diluted with D2O (0.10 mL) and the resulting solution analyzed by 31P NMR (202 MHz) to reveal formation of TXPTS ylide. Control experiments performed by exclusion of any one of the reaction components do not lead to formation of the ylide. An additional control experiment involving the reaction of TXPTS (16 mg, 50 mM) and cyanamide (50 mM) in 15% D2O/Tris buffer (500 mM, 0.2 mM EDTA, pH 7.4, 0.5 mL) suggests no reaction of the phosphine over 24 hours.
The cat(FeIII-CN) complex was prepared via treatment of bovine hepatic catalase (2 μM) with potassium cyanide (0.02 mM) in Tris buffer (500 mM, 0.2 mM EDTA, pH 7.4). A UV-Vis spectrum obtained at 37°C reveals the immediate formation of cat(FeIII-CN) which appears irreversible over the course of several hours. Treatment of this complex with AS (0.02 mM) does not result in a subsequent reaction. A separate control experiment involving the incubation of catalase (2 μM) with cyanamide (0.02 mM) in the same buffer at 37°C suggests no reaction of the ferric heme with cyanamide over several hours.
Formation of TXPTS ylide via HRP-mediated HNO generation
A deoxygenated solution of TXPTS (50 mM), hydroxylamine hydrochloride (50 mM), and horseradish peroxidase type I (23 μM) in Tris buffer (500 mM, 0.2 mM EDTA, pH 7.4, 0.45 mL) was prepared at ambient temperature in a septum-sealed flask. Hydrogen peroxide (5.2 μL, 11.5 mM) was added to initiate the reaction. After a minimum of three hours at room temperature, the reaction was diluted with D2O (0.10 mL) and the resulting solution analyzed by 31P NMR (202 MHz) to reveal formation of TXPTS ylide. Control experiments performed by exclusion of any one of the reaction components do not lead to formation of the ylide. An additional control experiment involving the reaction of TXPTS (26 mg, 89 mM) and hydroxylamine hydrochloride (4 mg, 102 mM) in 15% D2O/Tris buffer (500 mM, 0.2 mM EDTA, pH 7.4, 0.45 mL) provided no reaction of the phosphine over 4 days.
Ligation of HNO with phosphine biotin (10)
With Angeli’s salt-derived HNO
Phosphine biotin (10, Cayman Chemicals) was stored at −20°C as a solution in DMSO (2.5 mM). For ligation with AS-derived HNO, a solution of AS (7.6 μL, 0.625 mM) in sodium hydroxide (0.25 M) was added to a solution of 10 (0.5 mL, 0.625 mM) in Tris buffer (100 mM, 0.2 mM EDTA, pH 7.4, 1.5 mL) at room temperature. After six hours, an aliquot of the reaction mixture was diluted 10-fold with 0.1% ammonium formate in water (AFW) for analysis by LC/MS. A phosphine biotin standard (20 μM) was prepared by dilution of phosphine biotin with AFW. A phosphine oxide standard was prepared by treatment of 10 (16 μL, 20 μM) with a 10-fold excess of hydrogen peroxide in AFW.
Gradient for conditioning of column: Held at 80% CH3CN for 30 minutes, decreased to 40% CH3CN over 5 minutes and held for 30 minutes, then decreased to 5% CH3CN over 5 minutes and held for 20 minutes. Gradient for analyses: Held at 5% CH3CN for 2 minutes, increased to 20% CH3CN over one minute, then increased to 100% CH3CN over 2 minutes. The column was held at 100% CH3CN for 3 minutes, then decreased to 20% CH3CN over one minute, and returned to 5% CH3CN over one minute for a total run time of 10 minutes. The post-run time was 3 minutes, held at 5% CH3CN. Target masses (m/z) were set at 793 (phosphine biotin), 808 (phosphine oxide), and 794 (ligation reaction mixture).
With HRP-generated HNO
Hydroxylamine hydrochloride (10 mM) was added to a solution of horseradish peroxidase type I (23 μM) in Tris buffer (100 mM, 0.2 mM EDTA, pH 7.4, 0.745 mL) and the resulting solution and headspace deoxygenated. Phosphine 10 (0.25 mL, 0.625 mM) was then added, followed by the addition of hydrogen peroxide (4.6 μL, 2.3 mM). The reaction proceeded at ambient temperature for four hours and was quenched by addition of excess hydrogen peroxide. HRP was removed using a 7 mL Apollo spin column (Orbital Biosciences) equilibrated with Tris buffer (100 mM, 0.2 mM EDTA, pH 7.4). The sample was eluted with AFW (4 mL) then an aliquot was diluted 5-fold with AFW for analysis by LC/MS.
Gradient for conditioning of column: Held at 80% CH3CN for 30 minutes, decreased to 45% CH3CN over 5 minutes and held for 30 minutes, then decreased to 25% CH3CN over 5 minutes and held for 20 minutes. Gradient for analyses: Held at 25% CH3CN for 2 minutes, increased to 60% CH3CN over one minute, then increased to 100% CH3CN over 2 minutes. The column was held at 100% CH3CN for 3 minutes, then decreased to 60% CH3CN over one minute, returned to 25% CH3CN over one minute, and held at 25% CH3CN for 2 minutes for a total run time of 10 minutes. The post-run time was 3 minutes, held at 25% CH3CN. Target masses (m/z) were set at 793 (phosphine biotin), 808 (phosphine oxide), and 794 (ligation reaction mixture).
Phosphine carbamates
4-nitrophenyl(2-(diphenylphosphino)ethyl)carbamate (15a)
2-(Diphenylphosphino)ethylamine 13 (360 mg, 1.6 mmol) was added dropwise to a solution of 4-nitrophenylchloroformate 14a (295 mg, 1.5 mmol) in dichloromethane (5 mL) and the reaction was stirred for 1 hour at room temperature under inert atmosphere. The mixture was diluted with sodium phosphate buffer (5 mL, 500 mM, pH 6.3) and extracted with dichloromethane. The organic extracts were combined, dried over magnesium sulfate, filtered, and concentrated to provide a light yellow solid in 86% yield. The crude material contained a 7% phosphine oxide impurity (by 31P NMR) but was not purified as previous purification efforts had resulted in additional oxidation. Rf = 0.4 (30% EtOAc/hexanes). 1H NMR (300 MHz, CDCl3,δ) 8.19 (d, J = 9 Hz, 2H), 7.47-7.34 (m, 10H), 7.24 (d, J = 9 Hz, 2H), 3.45 (m, 2H), 2.45 (t, J = 6 Hz, 2H); 13C NMR (75 MHz, CDCl3, δ) 163.0, 155.9, 153.1, 144.8, 136.8, 132.8 (d, JP-C = 18.8 Hz), 129.2, 128.8 (d, JP-C = 9.8 Hz), 125.1, 122.0, 115.7, 38.7 (d, 1JP-C = 20.3 Hz), 28.6 (d, 2JP-C = 11.3 Hz); 31P NMR (121 MHz, CDCl3, δ) −21.4; ESI-MS (m/z): 395.3 [M + H]+; HRMS-ESI+ (m/z): [M + H]+ calcd for C21H20N2O4P, 395.1161; found, 395.1199.
Ligation of HNO with phosphine carbamate 15a
1. LC-MS analysis
Tris buffer (100 mM, 0.2 mM EDTA, pH 7.4, 0.5 mL) was added dropwise (with agitation) to a solution of 15a (16 mg, 0.04 mmol) in acetonitrile (1 mL). AS (5 mg, 0.04 mmol) was swiftly added to initiate the reaction, resulting immediately in a bright yellow solution. The reaction proceeded at ambient temperature for one hour, then an aliquot was diluted 600-fold with HPLC-grade acetone for analysis by LC-MS.
2. Preparative scale
Ammonium formate buffer (700 mM, pH 7.1, 3 mL) was added dropwise to a stirred solution of 15a (408 mg, 1.0 mmol) in acetonitrile (7 mL). AS (127 mg, 1.0 mmol) was swiftly added and the bright yellow solution was stirred at room temperature for 30 minutes. The reaction was then quenched with a solution of NaOH (0.3 M, 2 mL) and concentrated. The resulting residue was taken up in CHCl3, filtered, and purified by silica chromatography (10% MeOH in CHCl3) to provide 19 (80% yield). Rf = 0.3 (10% MeOH/CHCl3). 1H NMR (300 MHz, CDCl3, δ) 7.71-7.65 (m, 4H), 7.51-7.40 (m, 6H), 6.64 (bs, 1H), 5.19 (s, 2H), 3.55-3.44 (m, 2H), 2.60-2.52 (m, 2H); 13C NMR (126 MHz, CDCl3, δ) 159.7, 132.6 (d, 1JP-C = 99.3 Hz), 132.0 (d, 4JP-C = 2.4 Hz), 130.6 (d, JP-C = 9.5 Hz), 128.8 (d, JP-C = 11.8 Hz), 34.0 (d, 2JP-C = 2.5 Hz), 30.1 (d, 1JP-C = 71.0 Hz); 31P NMR (121 MHz, CDCl3, δ) 33.3; ESI-MS (m/z) 289.2 [M + H]+, 311.2 [M + Na]+; HRMS-ESI+ (m/z): [M + Na]+ calcd for C15H17N2O2PNa, 311.0920; found, 311.0901.
phenyl (2-(diphenylphosphino)ethyl)carbamate (15b)
2-(Diphenylphosphino)ethylamine 13 (315 mg, 1.4 mmol) was added dropwise to a solution of phenylchloroformate 14b (0.175 mL, 1.4 mmol) in dichloromethane (5 mL), and the reaction was stirred for 1 hour at room temperature under inert atmosphere. The mixture was then diluted with sodium phosphate buffer (5 mL, 500 mM, pH 6.3) and extracted with dichloromethane. The organic extracts were combined, dried over magnesium sulfate, filtered, and concentrated to provide 15b in 91% yield. The crude material contained a 12% phosphine oxide impurity (by 31P NMR) but was not purified as previous purification efforts had resulted in additional oxidation. Rf = 0.5 (30% EtOAc/hexanes). 1H NMR (300 MHz, CDCl3, δ) 7.41-7.37 (m, 5H), 7.25-7.22 (m, 5H), 7.11-7.03 (m, 4H), 6.77 (d, J = 6.0 Hz, 1H), 5.63 (bs, 1H), 3.34-3.24 (m, 2H), 2.26 (t, J = 6 Hz, 2H); 13C NMR (75 MHz, CDCl3, δ) 156.7, 154.6, 150.9, 137.5 (d, JP-C = 12.0 Hz), 132.6 (d, JP-C = 18.8 Hz), 132.5 (d, JP-C = 9.8 Hz), 129.5, 129.3, 129.2, 128.7, 128.5 (d, JP-C = 7.5 Hz), 125.2, 121.6, 120.2 (d, 1JP-C = 93 Hz), 115.4, 38.4 (d, JP-C = 22.5 Hz), 28.5 (d, JP-C = 13.5 Hz); 31P NMR (121 MHz, CDCl3, δ) −21.8; ESI-MS (m/z): 350.2 [M + H]+ major, 366.2 [oxide]+ minor.
Ligation of HNO with phosphine carbamate 15b
1. LC-MS analysis
Tris buffer (100 mM, 0.2 mM EDTA, pH 7.4, 0.5 mL) was added dropwise (with agitation) to a solution of 15b (14 mg, 0.04 mmol) in acetonitrile (1 mL). AS (5 mg, 0.04 mmol) was swiftly added to initiate the reaction, which proceeded at ambient temperature for one hour, then an aliquot was diluted 600-fold with HPLC-grade acetone for analysis by LC-MS.
2. Preparative scale
Ammonium formate buffer (700 mM, pH 7.1, 3 mL) was added dropwise to a stirred solution of 15b (320 mg, 0.9 mmol) in acetonitrile (6 mL). AS (111 mg, 0.9 mmol) was swiftly added and the resulting solution was stirred at room temperature for 1 hour. The solution was then concentrated and the resulting residue purified by reverse phase chromatography (C18 column) with separation effected by a 0 to 60% MeOH in H2O gradient to provide urea 19 (23% yield). Rf = 0.5 (C18 plate; 50% MeOH/H2O). 1H NMR (300 MHz, CDCl3, δ) 7.73-7.67 (m, 4H), 7.54-7.43 (m, 6H), 6.44 (s, 1H), 4.88 (s, 2H), 3.59-3.51 (m, 2H), 2.60-2.53 (m, 2H); 13C NMR (75 MHz, CDCl3, δ) 159.2, 132.6 (d, 1JP-C = 98.3 Hz), 132.0 (d, 4JP-C = 3.0 Hz), 130.6 (d, JP-C = 9.0 Hz), 128.8 (d, JP-C = 11.3 Hz), 34.1 (d, 2JP-C = 3.8 Hz), 29.9 (d, 1JP-C = 71.3 Hz); 31P NMR (121 MHz, CDCl3, δ) 33.9; ESI-MS (m/z): 289.2 [M + H]+.
LC-MS for ligations with phosphine carbamates
Gradient for conditioning of column: Held at 100% B for 30 minutes, decreased to 50% B over 5 minutes and held for 30 minutes, then decreased to 25% B over 5 minutes and held for 20 minutes. Gradient for analyses: Held at 15% B for 1 minutes, increased to 100% B over 2 minutes, held at 100% B for 3 minutes, then returned to 15% B over 4 minutes for a total run time of 10 minutes. The post-run time was 3 minutes, held at 15% B. Target masses (m/z) were set at 395 (15a), 350 (15b), 411 (16a), 366 (16b), and 289 (ligation reaction mixture).
UV-Visible spectroscopy of 15a and 18a
A stock of a p-nitrophenol (2 mM) was prepared in ammonium acetate buffer (700 mM, pH 7.1) resulting in a bright yellow solution. Standards (10–50 μM) were analyzed over the 200 – 800 nm range, resulting in a λmax at 400 nm (ε= 9.44 mM-1). A solution of 15a in acetonitrile (50 μM) was scanned over the identical range, revealing a stable λmax at 270 nm.
Ligations
Carbamate 15a (2 μL, 30.5 mM in CH3CN) was dissolved in acetate buffer (100 mM, pH 7, 988 μL) followed by the rapid addition of a solution of AS (10 μL, 10 mM) in NaOH (10 mM) to provide final concentrations of 15a (50 μM) and AS (100 μM). A hydrolysis control sample was prepared containing 15a (2 μL, 30.5 mM in CH3CN) in acetate buffer (100 mM, pH 7, 1 mL). A similar set of experiments were performed at pH 6.3 using NaPi buffer (50 mM, 100 mM NaCl, 1 mM EDTA). To investigate the effect of nitrite, 15a (2 μL, 30.5 mM in CH3CN) was dissolved in acetate buffer (100 mM, pH 7, 988 μL) followed by the addition of NaNO2 (100 μM). Each reaction was scanned by UV-Vis spectroscopy every 3 minutes for 3 hours.
Results and Discussion
Nitroxyl (HNO) has gained increased attention over the past two decades as evidence builds indicating that HNO and NO offer discrete chemical biology and do not rapidly interconvert under physiological conditions.29 With a pKa of 11.4, protonated HNO dominates at physiological pH.30 Deprotonation of singlet ground state HNO to generate the triplet nitroxyl anion requires a forbidden electron spin flip, kinetically retarding this transformation and contributing to the chemical distinction of HNO and NO.31 While endogenous NO formation from the NOS-catalyzed oxidation of L-arginine is well-established, endogenous HNO production remains unclear. Mitochondrial NO reduction to HNO may occur with the aid of cytochrome c, manganese superoxide dismutase, and xanthine oxidase.32-34 Other potential biosynthetic sources of HNO include generation by NOS in the absence of tetrahydrobiopterin, heme-mediated oxidation of N-hydroxy-L-arginine and other nitrogen oxides, and thiol-mediated fragmentation of S-nitrosothiols.35–37 The bioavailability of HNO from these sources has yet to be indisputably demonstrated and remains under investigation. The definitive identification of these sources is challenged by a lack of specific HNO detection methods capable of distinguishing HNO from NO in vitro.
The rapid dimerization of HNO dictates a very short lifetime and requires the use of donor compounds such as the widely used Angeli’s salt, which decomposes at pH 4–8 (k = 4.6 × 10−4 s−1) to provide equimolar amounts of HNO and nitrite.27,38 Nitroxyl dimerization and reactivity with other biomolecules further complicates simple HNO detection strategies. The quantification of the dimerization/dehydration product N2O by gas chromatography remains a commonly used method of HNO detection, but becomes cumbersome in vivo and cannot exclude other physiological mechanisms of N2O generation.16,39,40 Addition of thiols, such as glutathione (GSH), to suspected HNO sources quenches N2O formation providing evidence for HNO intermediacy and in some cases generating a sulfinamide (RS(=O)NH2) as a unique HNO-derived marker.39,41,42 The thiol-based detection strategy is problematic in that these reactions also produce disulfides, which may arise from multiple oxidative pathways, and furthermore the mechanism and yield of sulfinamide formation remain poorly defined.41,42
Heme-containing proteins, such as metmyoglobin (FeIII), are often used to trap HNO leading to the UV-Vis and EPR-detectable FeII-NO product.43,44 While suitable for the assay of new HNO donors, this method suffers from multiple pathways of FeII-NO formation in vivo and the general instability of these products to oxygen. Synthetic CoII and MnIII porphyrins have recently been examined as compounds capable of discriminating HNO from NO.45,46 Reductive nitrosylation of a MnIII porphyrinate incorporated in a xerogel film allows for the anaerobic quantification of HNO from HNO donors.47 Organic nitronyl nitroxides, such as 2-(4-carboxyphenyl)-4,4,5,5-tetramethylimidazoline-1-oxyl 3-oxide (C-PTIO), well-established to form imino nitroxides upon reaction with NO, also react with HNO forming the respective imino nitroxide and hydroxylamine, but only discriminate between NO and HNO at very low NO concentrations.48 Recently, CuII-based fluoresecent complexes have emerged as new selective HNO detectors and CuII[BOT1] and CuII[COT1] probes illustrated fluorescence-based detection of HNO in HeLa cells and A375 human malignant melanoma cells treated with AS.49,50 While promising, these reagents remain susceptible to other cellular reductants and generate stoichiometric amounts of NO upon CuII reduction, which may lead to kinetic complications in HNO detection. Here, we report further development of our phosphine-based method for HNO detection that addresses both kinetic and selectivity concerns by rapidly and specifically reacting with HNO to yield unique and quantifiable products.
Kinetic Analysis
Because of the inherent instability of free HNO, a useful method for detection and quantification must be sufficiently swift to minimize the loss of HNO to dimerization and other reaction pathways. Preliminary experiments indicated that the presence of the physiologically-relevant HNO scavenger glutathione (GSH, 1 eq.) did not quench aza-ylide formation, suggesting that phosphines trap HNO with a rate constant comparable to the reaction of HNO with GSH (k = 2 × 106 M−1 s−1).23 Direct kinetic measurements for HNO reactions are inherently challenging due to the required use of donor compounds, whose decomposition is rate-limiting, and the unavoidable dimerization pathway. As a result, rate constants for the reactions of HNO are determined computationally,51 using the steady state approximation,46,52 or using competition experiments.53
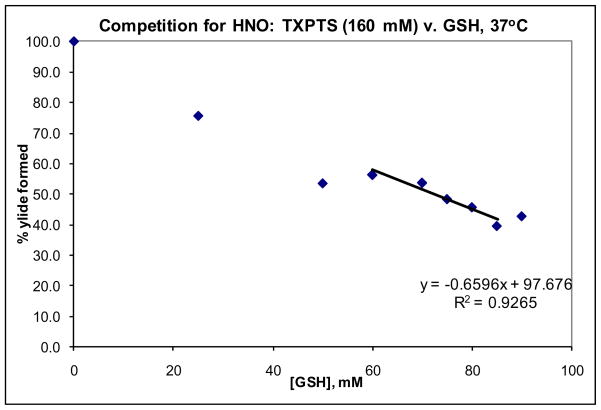
Given these challenges, the rate constant for the aqueous trapping of AS-derived HNO by water-soluble phosphine TXPTS was determined using the competitor nucleophile GSH. The relative affinities of these two nucleophiles for HNO was determined in situ by identifying the concentration of competitor [C]0.5 resulting in 50% maximal aza-ylide formation.23 Aza-ylide generation resulting from the reaction of TXPTS (160 mM) and AS (80 mM) in the presence of varying amounts of GSH (0–90 mM) at 37°C and pH 7.4 was quantified by the TXPTS ylide 31P NMR signal integral relative to an external H3PO4 standard. From a plot of % ylide formation v. [GSH], [C]0.5 was determined to be 72.3 mM (Figure 1).
Figure 1.
31P NMR quantification of TXPTS ylide formed in the presence of GSH at 37°C.
At this concentration of competing reactant, the reaction rates of HNO with TXPTS and GSH are equal:
Use of the known rate constant kGSH for the reaction of GSH and HNO at 37°C results in a rate constant kTXPTS for TXPTS and HNO at 37°C and pH 7.4 of 9 × 105 M−1 s−1, in accordance with previous results suggesting that TXPTS and GSH have similar affinities for HNO. A recent report details the reactions of HNO with numerous reductants and includes the computational analysis of the reaction of tris(2-carboxyethyl)phosphine (TCEP) with HNO where the derived second order rate constant was determined to be 8.4 × 106 M−1 s−1.51 That the rate constant computationally determined for TCEP and HNO is nearly an order of magnitude greater than kTXPTS is not altogether surprising. Aside from significant differences in experimental kinetic methods, the two phosphines have stark differences in reactivity. Steric and electronic effects predict that TCEP, a trialkylphosphine, offers a much more localized and exposed lone electron pair on phosphorus than does TXPTS. Control experiments involving TCEP and AS rapidly lead to phosphine oxide without detection of any other phosphorus-based intermediates or products (data not shown). Additionally, we suspect the involvement of the TCEP carboxylate groups in facilitating aza-ylide hydrolysis in these reactions.
Compatibility
As a robust HNO detection method requires selectivity over other nitrogen oxide species, we set out to examine the reactivity of phosphine nucleophiles with other physiologically-relevant nitrogen oxides at pH 7–7.4. Previous experiments indicated that phosphines do not react with nitrite at ambient temperature and physiological pH.26 Similarly, exposure of TXPTS to equimolar NaNO3 in D2O at ambient temperature did not yield phosphine oxide over 4 days by 31P NMR (see Supporting Information). The addition of a slight excess of NO donor diethylammonium (Z)-1-(N, N-diethylamino)diazen-1-ium-1,2-diolate (DEA/NO) to TXPTS in 20% D2O/Tris buffer (pH 7.4) slowly generated TXPTS oxide with no other phosphorus-containing products (see Supporting Information), in agreement with a previous study.54 Treatment of TXPTS with peroxynitrite only yields TXPTS oxide, and exposure of TXPTS to AS under basic conditions does not produce any new phosphorus-containing products (Supporting Information). In a separate experiment, the TXPTS-derived ylide did not react with a 100-fold excess of H2O2 after 20 hours as judged by 31P NMR further demonstrating the oxidative stability of this compound. Overall, these experiments demonstrate the significant selectivity of phosphines for HNO compared to other nitrogen oxides, most importantly NO.
While TXPTS does not significantly react with other nitrogen oxides, phosphine reactivity with S-nitrosothiols (RSNO) has been reported to generate various products depending on reaction conditions and the structures of the phosphine and RSNO.55 Investigations in aqueous/organic solvent mixtures using phosphine probes with various intramolecular electrophiles provide SNO-specific ligation products.56–58 In contrast, under aqueous conditions, the reaction of TXPTS and S-nitrosoglutathione generated the TXPTS-derived aza-ylide and an S-alkylthiophosphonium ion, which could complicate HNO detection.59 However, the use of careful control experiments and 31P NMR spectroscopy to identify all phosphorus-containing products should allow the clear differentiation of HNO and RSNO-based final products.
Trapping of enzymatically-generated HNO
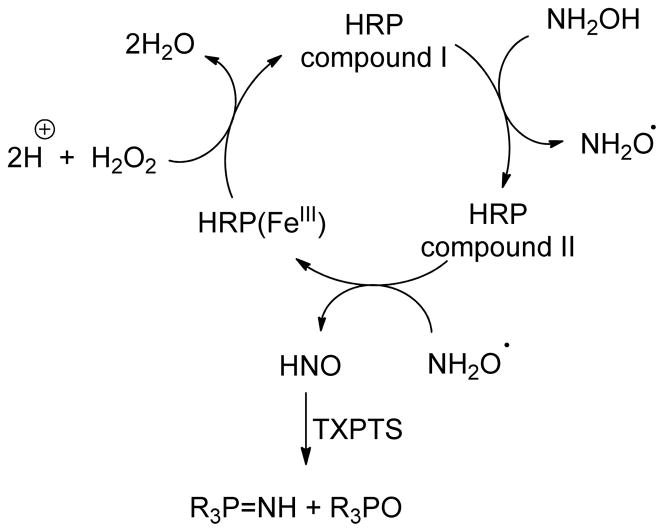
The use of the TXPTS-based method to detect HNO generated enzymatically was investigated using the heme proteins catalase and horseradish peroxidase (HRP) in vitro. Ferric HRP catalyzes the oxidation of numerous organic and inorganic substrates using hydrogen peroxide and the use of NH2OH as substrate yields HNO. This nascent HNO reacts preferentially with GSH over the ferric HRP heme, illustrating the ability of HRP-generated HNO to diffuse from the heme without full dimerization.60 Catalase, a heme enzyme involved in the crucial detoxification of peroxide species in mammalian systems, has a well-established catalytic cycle that also oxidizes lower oxidation state nitrogen oxides to generate HNO.61 Cyanamide, an HNO prodrug most notably used as an aldehyde dehydrogenase inhibitor, is bioactivated by catalase to N-hydroxycyanamide, which fragments to HNO and cyanide ion.62 The ability of cyanamide-derived HNO to subsequently serve as an enzyme inhibitor illustrates its ability to diffuse from the catalase heme pocket without immediate dimerization.
Anaerobic incubation of horseradish peroxidase (HRP, 11.5–23 μM) with varying amounts of hydrogen peroxide (1.15–50 mM) in the presence of hydroxylamine (50 mM) for one hour at ambient temperature resulted in a 6% yield of N2O by gas chromatography. Introduction of TXPTS (50 mM) into the reaction mixture before H2O2 and HRP were added reduced the yield of dimerization product N2O to 0.3% and resulted in the formation of the aza-ylide by 31P NMR (Scheme 2, Supporting Information). Concentrations of H2O2 below 1.15 mM were not sufficient to initiate the heme-mediated reaction. Raising the H2O2 concentration increased aza-ylide formation but peroxide levels above 11.5 mM resulted in phosphine oxidation as the dominant reaction pathway. Treatment of TXPTS with an equimolar amount of NH2OH at pH 7.4 provides no evidence of reaction by 31P NMR, indicating that aza-ylide formation in this system is enzyme-dependent. Control experiments excluding H2O2 from the incubation mixture resulted in no reaction of TXPTS. Additional control experiments withholding HRP or NH2OH provided TXPTS oxide as the only phosphorus-based product.
Scheme 2.
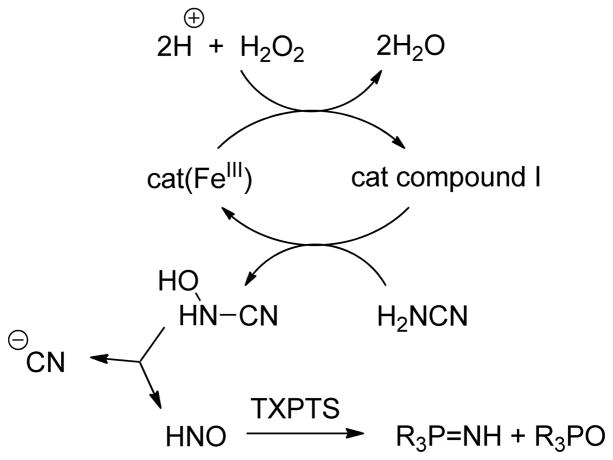
Aerobic incubation of bovine hepatic catalase (9 μM) with cyanamide (50 mM) at 37°C in the presence of a glucose/glucose oxidase (10 mM and 20–30 units, respectively) peroxide-generating system resulted in the formation of low levels of N2O as detected by gas chromatography. The addition of TXPTS (50–100 mM) just before catalase addition to the pre-incubation mixture fully quenched N2O formation and resulted in the formation of the aza-ylide, visible by 31P NMR (Scheme 3, Supporting Information). Exposure of TXPTS to an equimolar amount of cyanamide (50 mM) at 37°C and pH 7.4 did not result in reaction of the phosphine, indicating that aza-ylide formation in the catalase experiments is not derived from direct reaction of TXPTS and cyanamide. Further control experiments illustrate no reaction of TXPTS in this system in the absence of glucose, glucose oxidase, catalase, or cyanamide.
Scheme 3.
Previous comparisons of HNO generation by these heme enzymes have illustrated that HNO produced by catalase is trapped by the heme while HRP-generated HNO is better able to escape the heme pocket to dimerize or otherwise react.61 Experiments with catalase using hydroxyurea as substrate result in ferric heme trapping of HNO, but the use of cyanamide as a prodrug inhibitor of aldehyde dehydrogenase suggested HNO diffusion from the heme pocket.62–64 Since cyanamide is an established HNO prodrug, we hypothesized that cyanide, the by-product of cyanamide bioactivation, was involved in HNO generation and release by catalase.
UV-Vis analysis of the catalase heme upon incubation with glucose, glucose oxidase, and cyanamide as described above reveals a Soret band shift from 405 to 409 nm within 15 minutes at 37°C and pH 7.4 (Supporting Information). Examination of this red-shifted band over the course of several hours does not reveal reversibility back to 405 nm. Incubation of ferric catalase (2 μM) with potassium cyanide (20 μM) at 37°C and pH 7.4 formed the FeIII-CN heme, revealing immediately a Soret band at 409 nm, which was stable over several hours under these conditions (Supporting Information). Control experiments involving the incubation of ferric catalase (2 μM) and cyanamide (20 μM) did not result in the Soret band shift to 409 nm, indicating that FeIII-CN is formed during the concurrent generation of HNO from cyanamide, not from cyanamide itself. An additional control experiment indicated that the FeIII-CN complex formed from the reaction of ferric catalase and KCN is stable toward treatment with AS (20 μM) under identical conditions. These experiments demonstrate that HNO escapes the heme pocket as the by-product cyanide binds irreversibly to the catalase heme faster than the heme traps HNO. Cyanide heme binding would deactivate the enzyme,65 explaining the small amount of HNO generated by catalase and perhaps justifying the lack of cyanide poisoning in patients treated with cyanamide. While competing reactions of TXPTS with excess hydrogen peroxide, enzymatically generated NO, or oxidized enzyme intermediates likely lessen the amount of aza-ylide formed, these experiments clearly illustrate the efficiency of triarylphosphines in trapping HNO generated enzymatically, likely mimicking low levels of free HNO that may be synthesized in vivo. The ability to identify aza-ylide under such complicated reaction conditions further strengthens the use of triarylphosphines for HNO detection.
HNO Quantification by HPLC
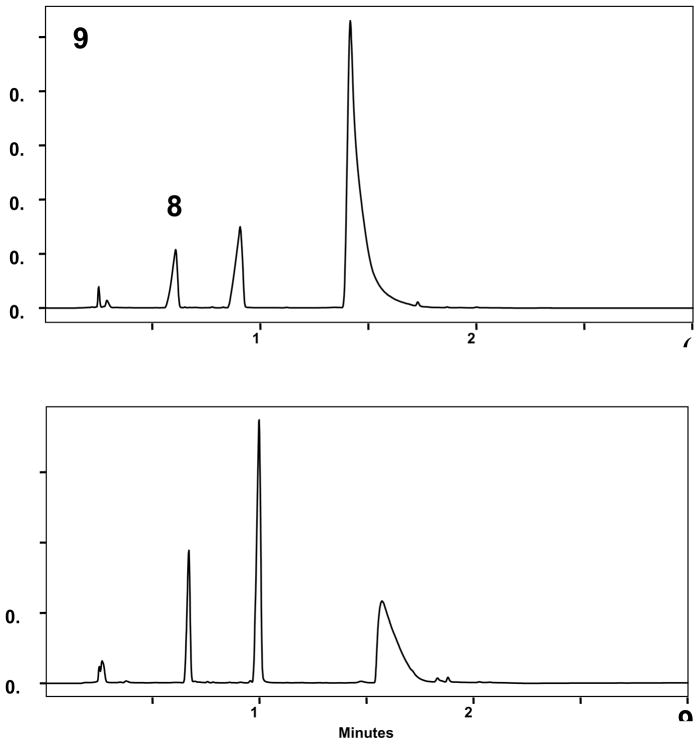
In an effort to quantify HNO via aza-ylide formation and subsequent ligation, an HPLC-based method was developed using an aqueous-soluble phosphine conjugated to an ester electrophile. Initial experiments indicated that phosphine 3 did not possess sufficient aqueous solubility for HPLC, and the carboxylated analog 7 was prepared as previously described.66–68 Trapping of HNO derived from AS by 7 yielded both 8 and 9, whereas reaction of NO and nitrite with 7 produced only oxide 8 (Scheme 4). Phosphine oxide 8 and ligation product 9 were separated, detected, and quantified by HPLC-UV. Identification was confirmed by co-elution under two different chromatography conditions and spiking with authentic standards, and quantitation was effected by interpolation from three point standard curves. Figure 2 shows a typical chromatogram and HPLC quantification indicates greater than 90% HNO release from AS as quantified by yield of amide 9. The high yield of HNO as quantified by the formation of 9 illustrates both the ability of AS to generate >90% yield of HNO and the efficiency of phosphine 7 as an HNO trapping reagent. Table 1 displays a summary of the yield for HNO from AS at various times.
Scheme 4.
Figure 2.
Panel (A) Typical HPLC chromatogram with UV/Vis detection for the reaction of 7 with Angeli’s Salt, 37°C incubation for 1 hr, 0.1 M 80% anion phosphate buffer, pH 7.4. Panel (B) HPLC chromatogram with UV/Vis detection for the separation of standards 7, 8, and 9 prepared in CH3CN with retention times of 16 min, 10 min, and 6 min, respectively. Slight discrepancies in retention times are attributed to differences in sample preparation and pH between the reaction mixture and pure standards.
Table 1.
HPLC quantification of HNO derived from AS (1 mM) by triarylphosphine 7 (5 mM)
| Entry | Time (hr) | HNO % Yield (Amide 9) |
|---|---|---|
| 1 | 0.2 | 49 ± 1.4 |
| 2 | 0.7 | 91 ± 1.0 |
| 3 | 1.0 | 93 ± 1.7 |
| 4 | 1.2 | 91 ± 0.7 |
| 5 | 2.0 | 94 ± 3.2 |
Reactions in phosphate buffer with 50 μM DTPA, 37°C, pH 7.4. 1, 2, 4 performed in duplicate. 3 and 5 performed in quadruplicate.
A similar analysis was performed to quantify HNO release from sodium 1-(N-isopropylamino)diazen-1-ium-1,2-diolate (IPA/NO). At physiological pH, IPA/NO provided 65% HNO yield as quantified by formation of amide 9. IPA/NO also generates NO under these conditions,21 and this yield is in accordance with GC experiments performed with IPA/NO at pH 7.4 (data not shown). Increasing the basicity to pH 10.6 provides 86% yield of HNO (Table 2), indicating that IPA/NO is primarily an HNO donor at this pH.
Table 2.
HPLC quantification of HNO derived from IPA/NO (1 mM) by triarylphosphine 7 (5 mM)
| Entry | pH | Time (hr) | HNO % Yield (Amide 9) |
|---|---|---|---|
| 1 | 7.4 | 1.0 | 65 ± 3.5 |
| 2 | 10.6 | 4.0 | 86 ± 6.4 |
Reactions in phosphate buffer with 50 μM DTPA, 37°C. Entries 1 and 2 performed in duplicate.
HNO Ligations
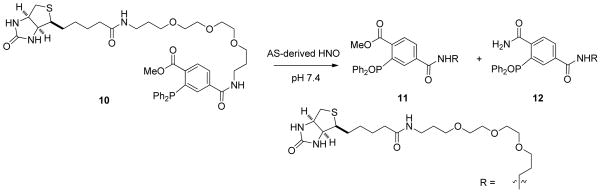
Toward the development of a biologically useful HNO probe, the first fully aqueous ligation was demonstrated using a phosphine biotin derivative (10) with HNO generated from AS at pH 7.4 (Scheme 5). After 6 hours, analysis by LC-MS provided evidence of two phosphine oxides 11 and 12 and un-reacted phosphine biotin (10) as a minor component of the mixture (Scheme 5). This phosphine successfully trapped HNO generated via HRP-mediated oxidation of NH2OH to give the identical ligation products. Specifically, H2O2 (2.3 mM) was added to a deoxygenated buffered solution of NH2OH (10 mM) and HRP (23 μM) in the presence of 10 (0.625 mM). After 4 hours at ambient temperature, HRP was removed via size exclusion chromatography and ligation products 11 and 12 were identified by LCMS. The surprising longevity of the TXPTS ylide over 7–10 days at pH 7.4 has suggested a stabilization effect by the substituted aryl rings and/or by water at this pH. These results with phosphine 10 clearly illustrate the ability of HNO-derived aza-ylides to undergo aqueous Staudinger ligations to generate amides.
Scheme 5.
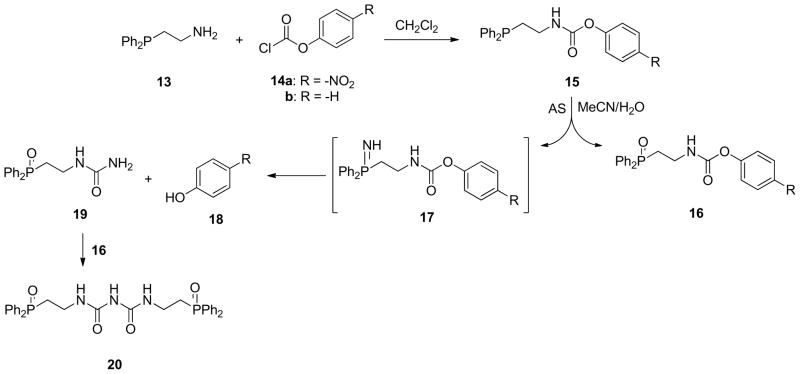
To further apply this phosphine-based HNO detection method to both in vitro and in vivo situations, we designed two phosphines (15a-b) equipped with carbamates as the electrophile for ligation. The carbamate moiety should generally be inert toward cellular esterases, and incorporation of phenolate and p-nitrophenolate leaving groups enhance the electrophilicity of the carbonyl carbon. Furthermore, the bright yellow color of the p-nitrophenolate ion provides the basis of a potential colorimetric HNO detection method. Condensation of 2-(diphenylphosphino)ethylamine (13) and 4-nitrophenylchloroformate (14a) in dichloromethane provided 4-nitrophenyl (2-(diphenylphosphino)ethyl)carbamate (15a) in 86% yield (Scheme 6). Similarly, condensation of 13 and phenylchloroformate (14b) provided the corresponding carbamate (15b) in 91% yield (Scheme 6). The ligations of AS-generated HNO with carbamates 15a-b in 7:3 MeCN:ammonium formate buffer (700 mM, pH 7.1) provided HNO-derived urea 19 in 80% and 23% yields, respectively, with concomitant evidence of phosphine oxide 16a-b. The identification of these products provides strong evidence for the intermediacy of the aza-ylides (17a-b, Scheme 6), which presumably form via the reaction of two equivalents of phosphine (15a-b) with HNO. Ligation of 17a-b generates the urea (19) and the corresponding phenol (18a-b, Scheme 6). Careful NMR analysis of the reaction of 15a with AS revealed the formation of a second urea (20, Scheme 6) from the condensation of 16a and 19.
Scheme 6.
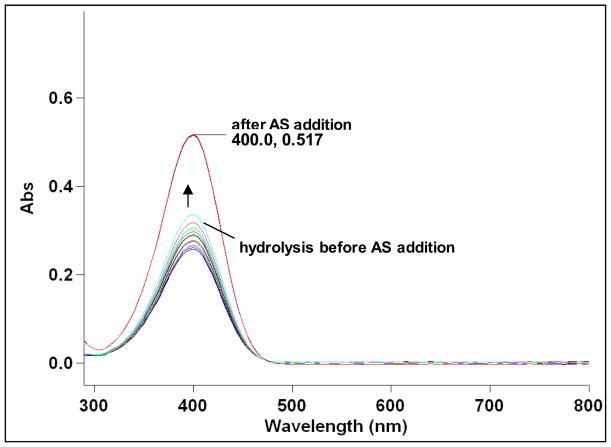
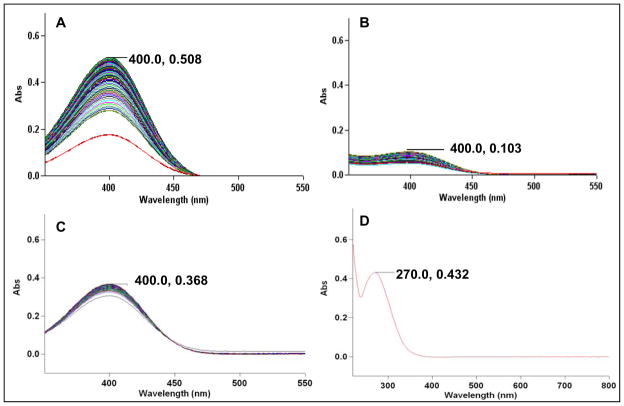
Monitoring the increase in absorption at 400 nm indicates the formation of the HNO ligation product p-nitrophenol (18a) from the reaction of 15a with HNO. UV-Vis spectroscopy indicates that 15a (50 μM) in CH3CN and acetate buffer (100 mM, pH 7) slowly hydrolyzes over several minutes to produce small amounts of 18a (Figure 3) in the absence of HNO. Addition of AS (100 μM) immediately yielded a bright yellow solution with a sharp increase in absorption at 400 nm indicating the complete release of 18a via ligation under these conditions (Figure 3). Figure 4 compares the amount of 18a generated by HNO ligation of 15a (Panel A) versus hydrolysis (Panel B). Based on the mechanism outlined in Scheme 6 that requires two equivalents of phosphine (15a-b) to generate 17a-b, the increase in absorbance at 400 nm during the reaction of 15a with AS reveals the formation of greater than 100% of the expected amount of 18a (Figure 4, Panel A). Figure 4 (Panel B) shows that 15a slowly hydrolyzes to 18a under these conditions in the absence of AS. While these results clearly show HNO-mediated product formation from 15a, the excess formation of 18a indicates that other non-HNO mediated pathways exist that include hydrolysis of 15a and condensation of 16 and 19.
Figure 3.
Formation of p-nitrophenol (18a, λ= 400 nm) from hydrolysis of 15a (50 μM) in acetate buffer (pH 7) with rapid increase in absorbance following addition of AS (100 μM) and subsequent HNO ligation.
Figure 4.
UV-Visible spectra illustrating formation of p-nitrophenol (18a) over 3 hours. Panel A: 15a (50 μM), AS (100 μM) at pH 7 (100 mM acetate). Panel B: 15a (50 μM) at pH 7. Panel C: 15a (50 μM), NaNO2 (100 μM) at pH 7. Panel D: 15a (50 μM) in MeCN (non-hydrolytic conditions). SCHEME TITLES.
This reaction provides a rapid colorimetric method for qualitatively indicating the presence of HNO, however these complications, which result from the reactivity of the p-nitrophenol carbamate (15a), limit HNO quantitation by this compound and the use of 15a in the presence of biological nucleophiles. Repeating these HNO-based ligations at pH 6.3 did not affect the extent of 15a hydrolysis or the rate of 18a formation (see Supporting Information). The increased acidity of the ligation mixture was anticipated to diminish the amount of hydrolysis without affecting the rate of AS decomposition, therefore facilitating ligation over hydrolysis and providing a method for determining the amount of 15a hydrolysis. The presence of nitrite (100 μM) did not affect the rate or extent of 15a hydrolysis, and a solution of 15a in CH3CN appeared completely stable (Figure 4, Panels C and D), indicating that carbamate electrophilic moiety is promising for the ligation and quantitation of HNO.
These reactions represent the first reported Staudinger ligations of carbamates and provide water-soluble HNO-derived ureas that may be used for quantifying and detecting HNO at the low micromolar level (in vitro).. These reactions provide strong evidence for the initial reaction of HNO with phosphines (15a-b) to form aza-ylides (17a-b, Scheme 6) that condense onto the carbamate group to yield urea 19 and the corresponding phenol 18a-b. Such compounds allow the colorimetric detection of HNO and form the basis of more sensitive HNO probes for in vivo detection.
Conclusions
In summary, we report expanded investigations into the use of phosphines as selective and efficient agents for the detection and quantitation of HNO. The reaction kinetics of aza-ylide formation from the reaction of HNO and TXPTS reveal that this process occurs at a rate comparable to the reaction of HNO and glutathione, suggesting the utility of phosphines as highly useful HNO traps. In addition, TXPTS reacts with enzymatically-generated HNO to form the aza-ylide, illustrating both the ability of phosphines to trap HNO under various aqueous oxidative conditions and the stability of the aza-ylide product. With appropriately positioned intramolecular electrophiles, HNO was readily ligated to form thermodynamically stable amides and ureas. Using various substrates, an HPLC method was developed to quantify HNO release from donor compounds, fully aqueous ligations using a biotin-derived phosphine were accomplished, and a colorimetric-based HNO detection system was developed. Overall, these results demonstrate progress toward the development of phosphines probes for rapid in vitro and in vivo measurements of HNO.
Supplementary Material
Acknowledgments
This work was supported by the NIH (HL 062198 and R21 087891, S.B.K.), and the Bruker NMR spectrometers used in this work were purchased with partial support from the NSF (CHE-9708077) and North Carolina Biotechnology Center (9703-IDG-1007). J.A.R. and S.B.K. kindly thank Dr. Marcus Wright (Wake Forest University) for helpful discussion regarding NMR kinetics studies. This work was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. C.N.Z. thanks Dr. Sergey Tarasov and Ms. Marzena A. Dyba of the Biophysics Resource in the Structural Biophysics Laboratory, NCI-Frederick, and Dr. Ryan Holland in the Laboratory of Chemical Carcinogenesis, NCI-Frederick, for assistance with the high resolution mass spectrometry studies and Mr. John R. Klose in the Laboratory of Proteomics and Analytical Technologies for assistance with the 31P NMR experiments.
Footnotes
Supporting Information Available: Gas chromatographic and synthetic procedures, compatibility studies, additional figures, analytical data, and full list of authors for references 3, 14, 21. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Irvine JC, Favaloro JL, Widdop RE, Kemp-Harper BK. Hypertension. 2007;49:885–892. doi: 10.1161/01.HYP.0000259328.04159.90. [DOI] [PubMed] [Google Scholar]
- 2.Fukuto JM, Bianco CL, Chavez TA. Free Rad Biol Med. 2009;47:1318–1324. doi: 10.1016/j.freeradbiomed.2009.06.014. [DOI] [PubMed] [Google Scholar]
- 3.Lloyd-Jones D, Adams RJ, Brown TM, Carnethon M, Dai S, De Simone G, Ferguson TB, Ford E, Furie K, Gillespie C, Go A, Greenlund K, Haase N, Hailpern S, Ho PM, Howard V, et al. Circulation. 2010;121:e46–e215. doi: 10.1161/CIRCULATIONAHA.109.192667. [DOI] [PubMed] [Google Scholar]
- 4.Fukuto JM, Wallace GC, Hszieh R, Chaudhuri G. Biochem Pharmacol. 1992;43:607–613. doi: 10.1016/0006-2952(92)90584-6. [DOI] [PubMed] [Google Scholar]
- 5.De Witt BJ, Marrone JR, Kaye AD, Keefer LK, Kadowitz PJ. Eur J Pharmacol. 2001;430:311–315. doi: 10.1016/s0014-2999(01)01289-4. [DOI] [PubMed] [Google Scholar]
- 6.Paolocci N, Katori T, Champion HC, St John ME, Miranda KM, Fukuto JM, Wink DA, Kass DA. Proc Natl Acad Sci USA. 2003;100:5537–5542. doi: 10.1073/pnas.0937302100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zeller A, Wenzl MV, Beretta M, Stessel H, Russwurm M, Koesling D, Schmidt K, Mayer B. Mol Pharmacol. 2009;76:1115–1122. doi: 10.1124/mol.109.059915. [DOI] [PubMed] [Google Scholar]
- 8.Favaloro JL, Kemp-Harper BK. Cardiovasc Res. 2007;73:587–596. doi: 10.1016/j.cardiores.2006.11.018. [DOI] [PubMed] [Google Scholar]
- 9.Tocchetti CG, Wang W, Valdivia HH, Aon MA, Katori T, Zaccolo M, O’Rourke B, Froehlich JP, Cheng HP, Paolocci N. Circulation. 2005;112:U363–U363. [Google Scholar]
- 10.Dai TY, Tian Y, Tocchetti CG, Katori T, Murphy AM, Kass DA, Paolocci N, Gao WD. Journal of Physiology-London. 2007;580:951–960. doi: 10.1113/jphysiol.2007.129254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tocchetti CG, Stanley BA, Murray CI, Sivakumaran V, Donzelli S, Mancardi D, Pagliaro P, Gao WD, van Eyk J, Kass DA, Wink DA, Paolocci N. Antioxid Redox Signal. 2011;14:1687–1698. doi: 10.1089/ars.2010.3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheong E, Tumbev V, Abramson J, Salama G, Stoyanovsky DA. Cell Calcium. 2005;37:87–96. doi: 10.1016/j.ceca.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 13.Stoyanovsky D, Murphy T, Anno PR, Kim YM, Salama G. Cell Calcium. 1997;21:19–29. doi: 10.1016/s0143-4160(97)90093-2. [DOI] [PubMed] [Google Scholar]
- 14.Tocchetti CG, Wang W, Froehlich JP, Huke S, Aon MA, Wilson GM, Di Benedetto G, O’Rourke B, Gao WD, Wink DA, Toscano JP, Zaccolo M, Bers DM, Valdivia HH, Cheng HP, Kass DA, et al. Circ Res. 2007;100:96–104. doi: 10.1161/01.RES.0000253904.53601.c9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sha X, Isbell TS, Patel RP, Day CS, King SB. J Am Chem Soc. 2006;128:9687–9692. doi: 10.1021/ja062365a. [DOI] [PubMed] [Google Scholar]
- 16.Huang ZJ, Velazquez C, Abdellatif K, Chowdhury M, Jain S, Reisz J, DuMond J, King SB, Knaus E. Organic & Biomolecular Chemistry. 2010;8:4124–4130. doi: 10.1039/c005066k. [DOI] [PubMed] [Google Scholar]
- 17.King SB. 7,696,373. USA: US Patent. 2007
- 18.Toscano JP, Brookfield FA, Cohen AD, Courtney SM, Frost LM, Kalish VJ. WO 2007109175. USA: PCT Int Appl. 2007
- 19.Toscano JP, Brookfield FA, Cohen AD, Courtney SM, Frost LM, Kalish VJ. WO 2009042970. USA: PCT Int Appl. 2009
- 20.Frost LM, Courtney SM, Brookfield FA, Kalish VJ. WO 2009137717. USA: PCT Int Appl. 2009
- 21.Miranda KM, Katori T, de Holding CLT, Thomas L, Ridnour LA, MeLendon WJ, Cologna SM, Dutton AS, Champion HC, Mancardi D, Tocchetti CG, Saavedra JE, Keefer LK, Houk KN, Fukuto JM, Kass DA, et al. J Med Chem. 2005;48:8220–8228. doi: 10.1021/jm050151i. [DOI] [PubMed] [Google Scholar]
- 22.Andrei D, Salmon DJ, Donzelli S, Wahab A, Klose JR, Citro ML, Saavedra JE, Wink DA, Miranda KM, Keefer LK. J Am Chem Soc. 2010;132:16526–16532. doi: 10.1021/ja106552p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miranda KM, Paolocci N, Katori T, Thomas DD, Ford E, Bartberger MD, Espey MG, Kass DA, Feelisch M, Fukuto JM, Wink DA. Proc Natl Acad Sci USA. 2003;100:9196–9201. doi: 10.1073/pnas.1430507100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bonner FT, Dzelzkalns LS, Bonucci JA. Inorg Chem. 1978;17:2487–2494. [Google Scholar]
- 25.Lemal DM, Rave TW. J Am Chem Soc. 1965;87:393–394. [Google Scholar]
- 26.Reisz JA, Klorig EB, Wright MW, King SB. Org Lett. 2009;11:2719–2721. doi: 10.1021/ol900914s. [DOI] [PubMed] [Google Scholar]
- 27.Shafirovich V, Lymar SV. Proc Natl Acad Sci USA. 2002;99:7340–7345. doi: 10.1073/pnas.112202099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.King SB, Nagasawa HT. Methods in Enzymology: Nitric Oxide, Pt C. 1999;301:211–220. doi: 10.1016/s0076-6879(99)01084-8. [DOI] [PubMed] [Google Scholar]
- 29.Fukuto JM, Carrington SJ. Antioxid Redox Signal. 2011;14:1649–1657. doi: 10.1089/ars.2010.3855. [DOI] [PubMed] [Google Scholar]
- 30.Bartberger MD, Liu W, Ford E, Miranda KM, Switzer C, Fukuto JM, Farmer PJ, Wink DA, Houk KN. Proc Natl Acad Sci USA. 2002;99:10958–10963. doi: 10.1073/pnas.162095599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bartberger MD, Fukuto JM, Houk KN. Proc Natl Acad Sci USA. 2001;98:2194–2198. doi: 10.1073/pnas.041481598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fukuto JM, Dutton AS, Houk KN. Chem Bio Chem. 2005;6:612–619. doi: 10.1002/cbic.200400271. [DOI] [PubMed] [Google Scholar]
- 33.Adak S, Wang Q, Stuehr DJ. J Biol Chem. 2000;275:33554–33561. doi: 10.1074/jbc.M004337200. [DOI] [PubMed] [Google Scholar]
- 34.Sharpe MA, Cooper CE. Biochem J. 1998;332:9–19. doi: 10.1042/bj3320009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cho JY, Dutton A, Miller T, Houk KN, Fukuto JM. Arch Biochem Biophys. 2003;417:65–76. doi: 10.1016/s0003-9861(03)00335-7. [DOI] [PubMed] [Google Scholar]
- 36.Arnelle DR, Stamler JS. Arch Biochem Biophys. 1995;318:279–285. doi: 10.1006/abbi.1995.1231. [DOI] [PubMed] [Google Scholar]
- 37.Reisz JA, Bechtold E, King SB. Dalton Transactions. 2010;39:5203–5212. doi: 10.1039/c000980f. [DOI] [PubMed] [Google Scholar]
- 38.Bonner FT, Ravid B. Inorg Chem. 1975;14:558–563. [Google Scholar]
- 39.Shoman MED, JF, Isbell TS, Crawford JH, Brandon A, Honovar J, Vitturi DA, White CR, Patel RP, King SB. J Med Chem. 2011;54:1059–1070. doi: 10.1021/jm101432z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang ZJ, Velazquez CA, Abdellatif KRA, Chowdhury MA, Reisz JA, DuMond JF, King SB, Knaus EE. J Med Chem. 2011;54:1356–1364. doi: 10.1021/jm101403g. [DOI] [PubMed] [Google Scholar]
- 41.Donzelli S, Espey MG, Thomas DD, Mancardi D, Tocchetti CG, Ridnour LA, Paolocci N, King SB, Miranda KM, Lazzarino G, Fukuto JM, Wink DA. Free Radical Biol Med. 2006;40:1056–1066. doi: 10.1016/j.freeradbiomed.2005.10.058. [DOI] [PubMed] [Google Scholar]
- 42.Shen B, English AM. Biochemistry (Mosc) 2005;44:14030–14044. doi: 10.1021/bi0507478. [DOI] [PubMed] [Google Scholar]
- 43.Doyle MP, Mahapatro SN, Broene RD, Guy JK. J Am Chem Soc. 1988;110:593–599. [Google Scholar]
- 44.Miranda KM, Nims RW, Thomasa DD, Espey MG, Citrin D, Bartberger MD, Paolocci N, Fukuto JM, Feelisch M, Wink DA. J Inorg Biochem. 2003;93:52–60. doi: 10.1016/s0162-0134(02)00498-1. [DOI] [PubMed] [Google Scholar]
- 45.Suarez SA, Fonticelli MH, Rubert AA, de la Llave E, Scherlis D, Salvarezza RC, Marti MA, Doctorovich F. Inorg Chem. 2010;49:6955–6966. doi: 10.1021/ic1007022. [DOI] [PubMed] [Google Scholar]
- 46.Marti MA, Bari SE, Estrin DA, Doctorovich F. J Am Chem Soc. 2005;127:4680–4684. doi: 10.1021/ja044632n. [DOI] [PubMed] [Google Scholar]
- 47.Dobmeier KP, Riccio DA, Schoenfisch MH. Anal Chem. 2008;80:1247–1254. doi: 10.1021/ac702024t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Samuni U, Samuni Y, Goldstein S. J Am Chem Soc. 2010;132:8428–8432. doi: 10.1021/ja101945j. [DOI] [PubMed] [Google Scholar]
- 49.Rosenthal J, Lippard SJ. J Am Chem Soc. 2010;132:5536–5537. doi: 10.1021/ja909148v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou Y, Liu K, Li JY, Fang YA, Zhao TC, Yao C. Org Lett. 2011;13:1290–1293. doi: 10.1021/ol103077q. [DOI] [PubMed] [Google Scholar]
- 51.Jackson MI, Han TH, Serbulea L, Dutton A, Ford E, Miranda KM, Houk KN, Wink DA, Fukuto JM. Free Radical Biol Med. 2009;47:1130–1139. doi: 10.1016/j.freeradbiomed.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bari SE, Marti MA, Amorebieta VT, Estrin DA, Doctorovich F. J Am Chem Soc. 2003;125:15272–15273. doi: 10.1021/ja036370f. [DOI] [PubMed] [Google Scholar]
- 53.Miranda KM, Paolocci N, Katori T, Thomas DD, Ford E, Bartberger MD, Espey MG, Kass DA, Feelisch M, Fukuto JM, Wink DA. Proc Natl Acad Sci USA. 2003;100:9196–9201. doi: 10.1073/pnas.1430507100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lim MD, Lorkovic IM, Ford PC. Inorg Chem. 2002;41:1026–1028. doi: 10.1021/ic0108585. [DOI] [PubMed] [Google Scholar]
- 55.Wang H, Xian M. Curr Opin Chem Biol. 2010;15:32–7. doi: 10.1016/j.cbpa.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang J, Wang H, Xian M. J Am Chem Soc. 2009;131:3854–3855. doi: 10.1021/ja900370y. [DOI] [PubMed] [Google Scholar]
- 57.Zhang JM, Wang H, Xian M. Org Lett. 2009;11:477–480. doi: 10.1021/ol802663q. [DOI] [PubMed] [Google Scholar]
- 58.Zhang D, Devarie-Baez NO, Pan J, Wang H, Xian M. Org Lett. 2010;12:5674–6. doi: 10.1021/ol102491n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bechtold E, Reisz JA, Klomsiri C, Tsang AW, Wright MW, Poole LB, Furdui CM, King SB. ACS Chem Biol. 2010;5:405–414. doi: 10.1021/cb900302u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huang JM, Sommers EM, Kim-Shapiro DB, King SB. J Am Chem Soc. 2002;124:3473–3480. doi: 10.1021/ja012271v. [DOI] [PubMed] [Google Scholar]
- 61.Huang JM, Kim-Shapiro DB, King SB. J Med Chem. 2004;47:3495–3501. doi: 10.1021/jm030547z. [DOI] [PubMed] [Google Scholar]
- 62.Nagasawa HT, Demaster EG, Redfern B, Shirota FN, Goon JW. J Med Chem. 1990;33:3120–3122. doi: 10.1021/jm00174a001. [DOI] [PubMed] [Google Scholar]
- 63.DeMaster EG, Redfern B, Nagasawa HT. Biochem Pharmacol. 1998;55:2007–2015. doi: 10.1016/s0006-2952(98)00080-x. [DOI] [PubMed] [Google Scholar]
- 64.Shoeman DW, Shirota FN, DeMaster EG, Nagasawa HT. Alcohol. 2000;20:55–59. doi: 10.1016/s0741-8329(99)00056-7. [DOI] [PubMed] [Google Scholar]
- 65.Shirota FN, Nagasawa HT, Demaster EG. Federation Proceedings. 1985;44:1123–1123. [Google Scholar]
- 66.Zhou KJ, Li JF, Lu YJ, Zhang GZ, Xie ZW, Wu C. Macromolecules. 2009;42:7146–7154. [Google Scholar]
- 67.Kiick KL, Saxon E, Tirrell DA, Bertozzi CR. Proc Natl Acad Sci. 2002;99:19–24. doi: 10.1073/pnas.012583299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gattas-Asfura KM, Stabler CL. Biomacromolecules. 2009;10:3122–3129. doi: 10.1021/bm900789a. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.