Abstract
The use and development of post-genomic tools naturally depends on large-scale genome sequencing projects. The usefulness of post-genomic applications is dependent on the accuracy of genome annotations, for which the correct identification of intron–exon borders in complex genomes of eukaryotic organisms is often an error-prone task. Although automated algorithms for predicting intron–exon structures are available, supporting exon evidence is necessary to achieve comprehensive genome annotation. Besides cDNA and EST support, peptides identified via MS/MS can be used as extrinsic evidence in a proteogenomic approach. We describe an improved version of the Genomic Peptide Finder (GPF), which aligns de novo predicted amino acid sequences to the genomic DNA sequence of an organism while correcting for peptide sequencing errors and accounting for the possibility of splicing. We have coupled GPF and the gene finding program AUGUSTUS in a way that provides automatic structural annotations of the Chlamydomonas reinhardtii genome, using highly unbiased GPF evidence. A comparison of the AUGUSTUS gene set incorporating GPF evidence to the standard JGI FM4 (Filtered Models 4) gene set reveals 932 GPF peptides that are not contained in the Filtered Models 4 gene set. Furthermore, the GPF evidence improved the AUGUSTUS gene models by altering 65 gene models and adding three previously unidentified genes.
Keywords: Genome annotation, Mass spectrometry, Plant proteomics, Proteogenomics
1 Introduction
With the increasing number of sequenced genomes available, the need for new computational data mining approaches is evident. Software tools are needed to cover a broad range of applications. One of the biggest obstacles is the correct identification of intron–exon borders in complex genomes of eukaryotic organisms. Prediction of intron–exon boundaries for the identification of open reading frames using genomic data can be performed by numerous software tools [1]. However, those predictions are often erroneous, as highlighted in a recent study. An experimental verification of the C. elegans genome annotation demonstrated that 50% of the predicted genes (about 4000 genes) needed corrections in their intron–exon structures [2]. To improve ab initio annotation of protein-coding genes in eukaryotic genomes, the ENCODE genome annotation assessment project (EGASP) was initiated [3–5]. Ab initio gene prediction is particularly important in genome projects of species where a large fraction of genes cannot be constructed using expressed sequence tag (EST) evidence. In model organisms like human, mouse or Arabidopsis, it is obvious that roughly half of the gene products that are encoded by the genome cannot be annotated from EST data. In Chlamydomonas, only about 60% of the gene models are supported by EST sequences, including partially mapped ESTs [6]. Thus, the need for ab initio gene prediction programs for enhancing genome annotation in these model organisms is critical. Gene prediction programs like AUGUSTUS [7] and Exogean [8] take advantage of EST, protein and genomic alignments for improved gene prediction in the human genome. In the case of AUGUSTUS, additional extrinsic data such as EST and protein sequences have been shown to further improve annotation. Peptide data from proteomic mass spectrometric experiments can also be taken into account. Mass spectrometry (MS) has become a powerful tool for peptide and protein identification since it allows for sensitive, fast and specific measurement and thus allows for recognition of peptides and proteins from complex mixtures [9]. Proteogenomics, a field that has recently originated from the intersection of genomics and proteomics [10], attempts to support genome annotation with peptide hints obtained from proteomic experiments. In 2007, Tanner et al. proposed the construction of an exon splice graph to deduce putative peptides from the genomic DNA level, including spliced peptides [11]. Using this approach, Castellana et al. were able to improve the annotation of the Arabidopsis genome by discovering 778 new genes and correcting the annotation of 695 gene models [12] with the help of the gene prediction program AUGUSTUS [13]. Like the exon splice graph approach, the Genomic Peptide Finder (GPF) [14] can be used to provide exon hints deduced from peptides observed in tandem mass spectra. In principle, this tool allows the deduction of peptides from mass spectrometric data using a genomic sequence, where deduced peptides may be spliced at the genomic level. In a previous study, a high-throughput platform was established in which tandem mass spectrometric (MS/MS) data were analyzed by the Sequest search algorithm [15] and the de novo sequencing algorithm PEAKS [16] in conjunction with GPF to analyze membrane proteins from the green alga Chlamydomonas reinhardtii [17]. The concerted action of Sequest and GPF allowed the identification of 2622 distinct peptides. In total, 448 peptides were identified by GPF analysis alone including 98 spliced peptides, resulting in the identification of novel proteins, improved annotation of gene models and evidence of alternative splicing. Here, we present a new version of GPF that provides higher sensitivity due to a more accurate representation of genes. In addition, the new version provides higher search speed by employing an indexing strategy in a pre-processing step, thus rendering the use of a computing cluster for the purpose of data evaluation unnecessary. In addition, we show how the concerted action of GPF and AUGUSTUS can be used for genome annotation of the C. reinhardtii genome in an automatic fashion. Finally, we compare the JGI C. reinhardtii Filtered Models 4 (FM4) with an annotation created by AUGUSTUS incorporating GPF hints and show that coding peptide sequences as observed by MS/MS are enriched in the AUGUSTUS gene models when compared to the FM4 gene models. The use of GPF alignments as extrinsic hints for AUGUSTUS gene prediction provides a highly unbiased approach toward proteogenomic genome annotation because the amino acid sequences used, including those inferred from spliced alignments, are deduced from MS/MS spectra and the genomic DNA sequence only. The genome of C. reinhardtii contains about 8 exons on average per gene [6] and is therefore an excellent candidate organism for establishing genome annotation workflows, which potentially could later also be applied to higher species.
2 Materials and methods
2.1 Sample preparation and measurement
For the generation of intron/exon hints, MS/MS fragmentation scans have been collected from a total of 19 experiments on C. reinhardtii, including isolated chloroplasts and mitochondria as well as whole cells, grown on different types of media (Tris acetate phosphate or high salt medium) and under various conditions (iron deficiency (Höhner et al., manuscript in preparation) and anaerobiosis [18]). The cell wall-less strain CC124 (CW15) or the arginine auxotrophic strain CC424 was used for all experiments. Proteins were fractionated via SDS-PAGE, yielding a total of 949 bands. The resulting protein bands were excised, tryptically digested and analyzed by reverse phase LC coupled online to an LTQ Orbitrap mass spectrometer as described [18].
2.2 Reimplementation of GPF
Since its first publication, GPF has been redesigned to provide for increased sensitivity and higher search speed (software available at http://github.com/specht/gpf). The new search strategy consists of two steps: (i) given a query peptide, determine all loci of interest, i.e. all positions within the genome where the query peptide could be encoded, and for each of these loci, (ii) find unspliced and spliced peptide alignments that match the fragmentation scan’s precursor mass within a user-defined mass tolerance.
For the first step, we assume that although the de novo predicted peptides are probably not completely correct over their entire lengths, partial short sequence tags of a few amino acids are correctly predicted for most scans. Highly precise precursor masses for these short, correctly predicted sequence tags can be determined in the Orbitrap. We assume that any location in the genome that encodes for a tryptic peptide with a tag in the correct position is determined by the mass of either the N- or C-terminal tryptic fragment and is thus a valid candidate to explain the fragmentation scan. To quickly determine all such locations of interest, the new GPF version uses an indexing strategy which records the position of every possible amino acid n-mer in the six-frame translation of the genomic DNA sequence in a pre-processing step. As proposed in [17], the default tag size is defined as n=5. In addition to the position, the masses of the N- and C-terminal fragments toward the adjoining tryptic cleavage sites are recorded. During the search, all n-mers are extracted from the de novo predicted sequence and subsequently located within the genomic DNA sequence.
In the second step, peptides are assembled from previously determined locations of interest by association with either N- or C-terminal tags. In the case of an N-terminal tag (defined by a tryptic cleavage site, an N-terminal fragment mass and an amino acid pentamer), the initial tag-genome alignment is extended toward the C-terminus until the mass exceeds the highest possible precursor mass for the given scan. If an alignment can be deduced that is within an acceptable mass range, an unspliced alignment is added to the output. In addition, the deduction of spliced alignments is attempted by searching for user-defined splice donor/acceptor site consensus sequences within a user-defined maximum intron length (see Fig. 1). A resulting spliced alignment is added to the output if the mass of a resulting peptide is within the user-defined mass tolerance of the scan’s precursor mass. This search strategy also allows for the identification of alignments in which a single nucleotide triplet is spliced.
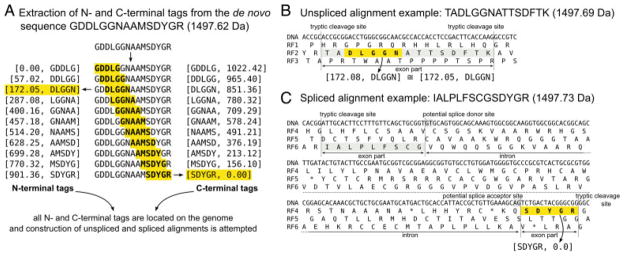
Figure 1.
GPF peptide alignment procedure. (A) From the de novo predicted amino acid sequence GDDLGGNAAMSDYGR, all possible amino acid pentamers are extracted with both their N- and C-terminal tryptic fragment masses. Each of these pentamer/fragment mass pairs is located within the genomic DNA sequence with the help of a previously compiled index. From the determined locus, the deduction of unspliced and spliced alignments is attempted. (B) From the N-terminal tag [172.05, DLGGN], a matching peptide can be constructed by N- and C-terminal extension toward the adjoining tryptic cleavage sites. The mass of the resulting peptide TADLGGNATTSDFTK is sufficiently similar for the peptide to be considered a valid GPF peptide candidate. (C) For the C-terminal tag [SDYGR, 0.0], an unspliced alignment cannot be deduced because N-terminal extension of the tag would result in the peptide QSDYGR, which is too light to match the target mass. Deduction of spliced alignments is attempted by searching for all dinucleotide pairs of GT and AG, a common splice donor/acceptor motif in C. reinhardtii, within a user-defined maximum intron length. In this example, the spliced peptide IALPLFSCGSDYGR is deduced and added to the list of GPF peptide candidates. Note: In the right hand side of the figure, an amino acid inferred from a nucleotide triplet is shown centered to the three corresponding nucleotides.
2.3 Data evaluation
All recorded MS/MS scans were passed to PEAKS, yielding de novo predicted amino acid sequences. These sequences were in turn passed to GPF, which aligned the query peptides to the genomic DNA sequence of C. reinhardtii. For the spliced peptide alignment procedure of GPF, two parameters had to be chosen carefully: the maximum intron length and the considered splice donor/acceptor consensus sequences.
For the determination of the highest allowed intron length, we analyzed introns in C. reinhardtii genes using previous annotations of the genome. According to Merchant et al., the average C. reinhardtii intron has a length of 373 base pairs [6]. Our analysis of the FM4 models shows that the intron length follows a log-normal distribution. A histogram of logarithmic intron lengths could be fitted to a normal distribution with μ=2.35 and σ=0.215, indicating that 50% of all introns have a length of less than 224 base pairs. Using this average intron length as a starting point, two concurrent effects have to be taken into consideration when choosing the highest allowed intron length for spliced GPF alignments: On the one hand, more false spliced peptides will be found by GPF as the maximum intron length increases. On the other hand, as becomes evident fromthe intron length distribution, more and more correct spliced alignments will be missed as the maximum intron length is decreased. As a compromise, we chose a maximum log intron length of μ+2 · σ, roughly comprising 97% of all naturally occurring intron lengths in C. reinhardtii and corresponding to a maximum intron length of 602 base pairs.
An analysis of the FM4 gene set revealed a splice donor/acceptor dinucleotide pair of GT/AG in 85%, and GC/AG in another 7.8% of all splice junctions. However, we chose GT/AG as the only allowed splice donor/acceptor site consensus sequence for GPF in C. reinhardtii because the low probability of a true GC/AG intron is in strong contrast to the high G/C content of ~66% in the genome of the alga.
Using a GPF tag size of five amino acids, a maximum intron length of 602 base pairs and a splice junction dinucleotide pair of GT/AG; all PEAKS candidate peptides were aligned to the genomic DNA of C. reinhardtii. The resulting set of GPF candidate peptides is completely DNA-deducible and therefore should contain less incorrectly predicted peptide candidates than the peptide sequences predicted by PEAKS. However, because there are generally several GPF peptide candidates for one MS/MS scan, we use OMSSA [19] for the identification of the correct GPF peptide candidate. OMSSA, like other database search programs, occasionally reports multiple peptide/spectral matches (PSM) for a single scan if several, ambiguous candidate peptides are available to explain the MS/MS scan. Due to the inherent similarity of GPF peptides stemming from the de novo predictions of one MS/MS scan, it can be expected that this happens often for GPF peptides, and especially for spliced GPF peptides, because a peptide which combines two exon parts potentially gives rise to a multitude of similar peptides. To resolve these ambiguous cases, we applied a hit distinctiveness filter, which calculates the distinctiveness of every OMSSA hit in the following way: Considering the best and the second best PSM E-values e1 and e2, we calculate the hit distinctiveness of the best hit as d = log(e2) − log(e1). The resulting hit distinctiveness d represents the order of magnitude of the difference between the two best-scoring hits. As a filtering threshold, we chose a minimum hit distinctiveness of 2, corresponding to a factor of 100 between both E-values.
To provide a way of assessing the confidence of PSM, a target/decoy approach [20] was implemented. In contrast to the approach described by Elias and Gygi, we were faced with the challenge that an unknown subset of incorrect peptides is contained in the list of GPF candidate peptides, and a false discovery rate (FDR) cannot be estimated without knowledge of the relative amount of correct peptides in the database. To solve this problem, we chose to create a target/decoy database from the JGI C. reinhardtii gene models v3.1 via sequence reversal and add the GPF peptides afterwards (see Fig. 2). Although we do not assume the JGI annotation to be complete, we still consider it sufficient for providing target peptides in a target/decoy strategy. This setup enabled the determination of an E-value threshold based on the JGI v3.1 gene models alone. Because the target/decoy sequences and GPF peptides were combined prior to the database search, the resulting E-value threshold can also be applied to the GPF peptide hits, thus enabling the identification of putative GPF peptides at a user-defined FDR. We estimated the FDR as FDR=(2 · decoys)/(targets+decoys), as described in [20]. As an additional filtering step, the OMSSA results were filtered in such a way that all PSM with a precursor mass deviation of more than 5ppm were discarded.
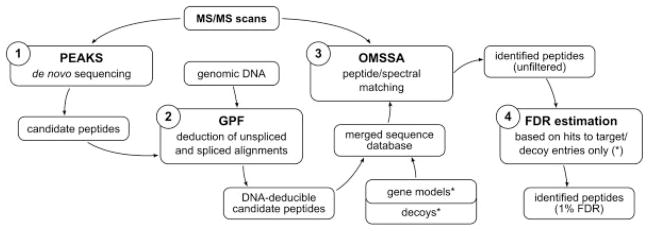
Figure 2.
Peptide/spectral matching of GPF peptides. Because multiple GPF-aligned peptides usually result from a single MS/MS scan, a mechanism is required which allows for the statistical assessment of peptide/spectral matches to GPF peptides. The identification of putative GPF-aligned peptides consists of four steps: (1) PEAKS is used for de novo peptide sequencing on the input MS/MS scans, resulting in a list of candidate peptides, which could be possible explanations for the MS/MS scans. (2) GPF uses the genomic DNA of C. reinhardtii to construct unspliced and spliced alignments of the PEAKS peptides, resulting in a list of candidate peptides, which can actually be deduced from the genomic DNA sequence. (3) A target/decoy protein database is created from the JGI gene models v3.1 of C. reinhardtii and subsequently merged with the GPF peptides, which are neither denoted as targets nor as decoys. (4) In the filtering step, a score threshold is determined to yield an estimated FDR of 1% as described in [20]. Because the FDR estimation is performed on the gene model hits alone, the FDR is not affected by the presence of the putative GPF peptides. However, because the GPF peptides have been assigned OMSSA scores in the same run as the gene model peptides, the resulting score threshold can be applied to the GPF peptides as well.
The genomic alignments of all resulting PSM to GPF peptides were then further processed by the following filters before being passed as extrinsic peptide hints to AUGUSTUS (see Fig. 3):
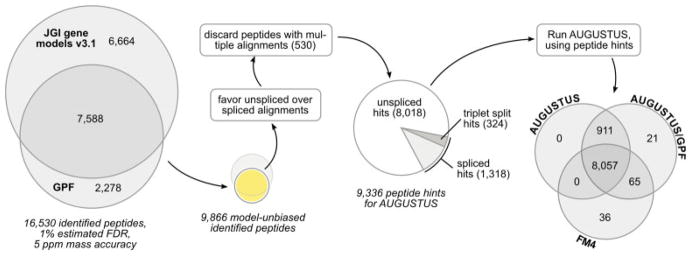
Figure 3.
Processing of OMSSA results. The OMSSA results show that 53% of the identified gene model peptides can be independently confirmed via PEAKS/GPF. Additionally, a set of 2278 peptides identified via PEAKS/GPF alone is available. The set of peptides identified via PEAKS/GPF is used to provide a list of peptide hints for AUGUSTUS. The following filtering steps are applied to this set: (i) whenever an unspliced alignment is available for a peptide, all spliced alignments of the same peptide are discarded because the unspliced variant is the most probable explanation. (ii) Peptides with multiple alignments on the genomic DNA are discarded in order to provide AUGUSTUS with unambiguous peptide hints. A comparison of the resulting AUGUSTUS gene sets with and without the GPF peptide hints to the FM4 gene set reveals an increased amount of coding sequences in the AUGUSTUS gene sets. The AUGUSTUS/GPF gene set confirms most peptides already present in the AUGUSTUS gene set without the peptide hints but introduces 21 new peptides. In addition, 65 peptides that were present in FM4 but not in AUGUSTUS are retained in the AUGUSTUS/GPF gene set.
Favor unspliced over spliced alignments. Whenever an unspliced alignment existed for a peptide, all spliced alignments were discarded because the unspliced alignment is the most likely explanation of the peptide.
Discard peptides with multiple alignments. To provide unambiguous peptide hints to AUGUSTUS, all peptides with more than one alignment to the genome were discarded. This filtering step affected 530 of 9866 peptides (5.4%).
The alignments of the remaining 9336 peptides were passed as extrinsic peptide hints to AUGUSTUS.
2.4 Gene prediction using peptide alignments
A peptide was either aligned contiguously from genomic position a to position b or, when it was spliced, to two regions [c,d] and [e,f] with a putative intron in between. In the first case, this was considered evidence that the complete region [a,b] is protein coding; in the second case, the alignment was considered evidence that [c,d] and [e,f] are protein coding as well as that an intron goes from exactly d+1 to e−1. Further, the reading frame and the strand are given by GPF. This evidence was incorporated into AUGUSTUS using a probabilistic model of extrinsic evidence as described before [7, 12]. In the terminology of AUGUSTUS above, individual pieces of evidence are hints of types CDSpart and intron, respectively, which are specified in the GFF file format. The CDSpart hints are shortened at the ends by 3 base pairs with respect to the alignment range because in some cases a spliced peptide with one very short exon fragment may coincidentally also be aligned perfectly contiguously.
The evidence from MS was used simultaneously with evidence from the alignments of 224 049 ESTs, from genomic conservation with Volvox carteri and from repeat masking. Further, AUGUSTUS has an ab initio model that allows to identify genes (or parts thereof) in absence of extrinsic evidence using statistical evidence. All above types of evidence are weighted in a probabilistic fashion and none of the evidence is trusted unconditionally. In the case of peptide alignments, this accommodates the fact that some fraction of peptides are wrong or aligned incorrectly or ambiguously and it should therefore be possible to override these hints in the presence of sufficient contradicting evidence. The extrinsic weighting parameters that were used in this study are available online (http://augustus.gobics.de/predictions/chlamydomonas/).
3 Results
For the employment of GPF, a GPF index file (2.6 GiB) has been compiled from the genomic DNA sequence (120 MiB) in a pre-processing step. The index file was copied to flash memory, resulting in a search speed of 20 queries per second on average using a single CPU core, rendering the use of a computing cluster unnecessary, as was required with the previous GPF version.
Mass spectrometric data from published [18] and unpublished experiments were subjected to the GPF pipeline. The MS/MS data were generated from either whole cell extracts or isolated organelles (chloroplast and mitochondria). Protein samples were fractionated by SDS-PAGE and individual SDS-PAGE bands (approximately 50 per SDS-PAGE) were excised and digested by trypsin. The resulting peptides were analyzed via LC-MS/MS. A total of 949 bands were investigated.
In the de novo prediction step, a set of ten peptide sequences was created by PEAKS for every scan. From these ten sequences, GPF deduced varying amounts of DNA-aligned peptides, with 3.4 peptides on average, using a maximum intron length of 602 base pairs and the splice site consensus sequence GT/AG for the construction of spliced alignments. Twenty-seven percent of all MS/MS scans yielded no GPF peptides, 61% yielded one to ten GPF peptides and more than ten GPF peptides were deduced for the remaining 12% of MS/MS scans. A compound database consisting of GPF aligned peptides and the JGI C. reinhardtii v3.1 gene models as a target/decoy database was created for every single SDS-PAGE band and used for the generation of PSM via OMSSA. Overall, a set of 16 530 peptides could be identified. From all identified gene model peptides, a total of 53% could be confirmed independently via PEAKS and GPF (see Fig. 3), demonstrating the improved search sensitivity of the new GPF implementation in contrast to the previous version, which confirmed 18% of the gene model peptides [17].
After the removal of peptides for which multiple alignments could be deduced, a total of 9336 peptides identified via PEAKS/GPF remained and were used as extrinsic peptide hints by AUGUSTUS. Unspliced alignments were identified for 8018 of these peptides (85.9%), the remaining 1318 peptides were recognized as spliced alignments (14.1%). From these spliced peptides, 324 peptides (24.5%) could only be deduced via an intron split within a single coding nucleotide triplet.
In the following, three gene sets are compared: (i) FM4 – the JGI FM4 gene set (data available online: http://genome.jgi-psf.org/Chlre4/Chlre4.download.ftp.html); (ii) AUGUSTUS – an AUGUSTUS gene set using extrinsic information from EST evidence, conservation with Volvox and repeat masking (data available online: http://gbrowse.-gobics.de/cgi-bin/gbrowse/chlamydomonas), (iii) AUGUSTUS/GPF – an AUGUSTUS gene set using the same extrinsic hints as the AUGUSTUS gene set plus the GPF peptide hints.
To elucidate the immediate impact of the GPF extrinsic hints, the experimentally deduced set of 9336 peptides was compared to the AUGUSTUS and FM4 gene sets. Most interestingly, it appears that 932 peptides (including 132 spliced peptides) that are present in the AUGUSTUS/GPF gene set are absent in the FM4 gene set. From the 9336 peptides that have been passed to AUGUSTUS, a set of 9054 peptides (97%) was incorporated into the AUGUSTUS/GPF gene model set. Comparing the FM4 and AUGUSTUS gene sets, 101 peptides that are present in FM4 are absent in AUGUSTUS (see Fig. 3). The fact that 65 of these peptides are re-introduced by the AUGUSTUS/GPF gene set suggests high orthogonality of the extrinsic GPF peptide hints relative to the other hint sources used. There is also a small set of 21 peptides that are contained in AUGUSTUS/GPF but neither in FM4 nor in the AUGUSTUS set lacking GPF hints.
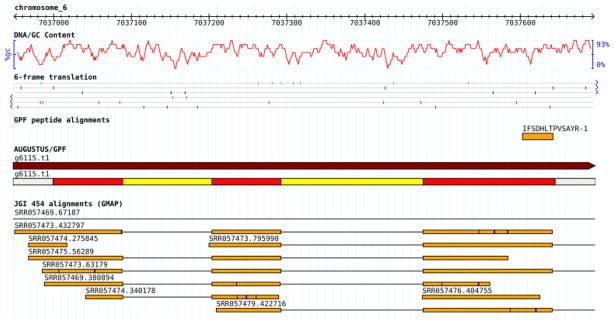
A detailed analysis of the differences between AUGUSTUS and AUGUSTUS/GPF reveals that the incorporation of the GPF hints leads to a modification of 65 gene models and the prediction of three previously unidentified genes (see Supporting Information). Among the modified gene models, gene extension could be observed in 66% of the cases, followed by internal gene adjustments (28%). An example is given in Fig. 4, where the identification of the peptide IFSDHLTPVSAYR via PEAKS/GPF leads to the prediction of a novel gene model, which is also supported by the JGI 454 alignments.
Figure 4.
Example of a new gene deduced by AUGUSTUS via a novel GPF peptide. The peptide IFSDHLTPVSAYR, which has been identified on chromosome 6 of C. reinhardtii and is contained in neither the FM4 nor the AUGUSTUS gene sets, leads to the prediction of a novel gene model, which is also supported by the JGI 454 data.
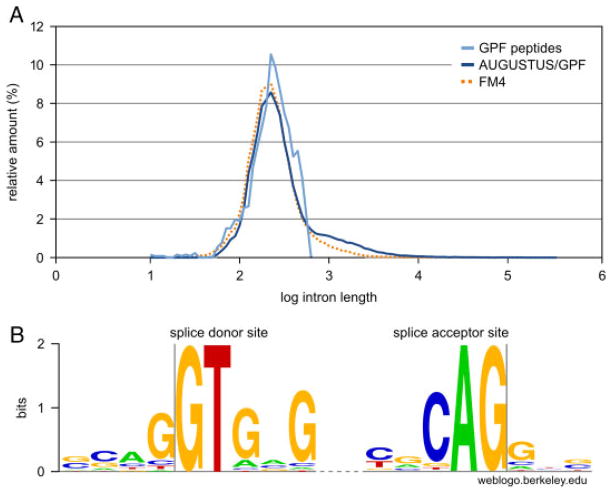
An assessment of intron lengths of the gene sets and the 1318 spliced GPF peptides provided to AUGUSTUS is shown in Fig. 5A. It can be seen that the intron length distribution of the AUGUSTUS/GPF gene set is very similar to the FM4 gene set. The intron lengths of the spliced GPF peptides are also similar to the FM4 gene set, although slightly more coarse, which can be attributed to the small sample size of the spliced peptide set. The WebLogo plot depicted in Fig. 5B has been calculated from the splice donor and acceptor sites of the 1318 spliced GPF peptides and is in line with previously published data [21].
Figure 5.
Assessment of intron lengths and splice site motifs. (A) The distribution of log intron lengths in the FM4 gene set follows a normal distribution with μ=2.35 and σ=0.215. Intron lengths in the AUGUSTUS/GPF gene set are comparable to those in FM4. The histogram of the spliced GPF peptide intron lengths is more coarse due to the low number of peptides (1318) but nevertheless is comparable to both gene sets except for the drop at 2.78, which corresponds to the maximum intron length of 602 base pairs. (B) The WebLogo of the splice donor/acceptor sites of the 1318 spliced GPF peptides shows, among the requirement of the GT/AG splice site, increased probabilities for three guanine residues at the splice donor site and a cytosine residue at the splice acceptor site. This data are in line with previously reported results [21].
4 Discussion
The indexing strategy employed in the new version of GPF allows for high-throughput alignment of de novo predicted amino acid sequences, performing 20 queries per second for C. reinhardtii on average using a typical desktop computer. The ability to deduce peptides that are spliced within a coding nucleotide triplet and the consideration of user-defined splice donor/acceptor site consensus sequences improve the tool in terms of sensitivity and specificity. GPF provides a straightforward way to use information obtained from MS/MS experiments for the annotation of genome sequences by providing alignments of identified peptides as extrinsic evidence to AUGUSTUS. Using de novo predicted amino acid sequences, GPF infers candidate peptides from a genomic DNA sequence. The advantage of this approach is that peptides are not coming from a set of predicted gene models, but are deduced based on the MS/MS spectra and the genome alone. In particular, the GPF approach may be considered more evidence-based than the exon splice graph approach, as it does not require gene prediction as a first step. In addition, no specialized version of a search program is required to search the generated exon splice graph, but any of the available search programs may be used [19, 15, 22, 23]. After the search was completed, the resulting PSM have been filtered according to a minimum hit distinctiveness of 2, corresponding to a 100-fold better E-value for the best match as compared to the second best match, if any, and an estimated FDR of 1%. This filtering strategy allows for the statistically robust identification of model-unbiased PEAKS/GPF peptides.
The question arises whether in addition to the GPF peptides, the six-frame translation of the genome could be used as an additional source of candidate peptides for the OMSSA database search step. While the idea sounds promising because many more unspliced peptides that have escaped detection due to lack of appropriate de novo predictions from the MS/MS scans could be detected, an analysis of the results stemming from such a setup shows that this approach is not feasible (see Fig. 6). Although the database search is embedded into a target/decoy approach, which allows for statistically significant identification, this procedure of significance assessment seems to be less appropriate for peptides stemming from the six-frame translation. Naturally, the six-frame translation contains at least and probably more than 83% false positives because only a single reading frame can be expected to encode for a gene in any genomic locus. While false-positive peptides are also contained in the GPF peptide set, the peptides deduced via GPF tend to produce clusters of highly similar peptides with almost equal precursor masses and common sequence tags. This high level of concurrency between similar GPF peptides and the hit distinctiveness filtering step ensure that a GPF peptide, which has been identified by OMSSA, can be considered highly significant because it has been identified via de novo prediction, and the average set of three to four GPF hits per MS/MS scan provide sufficiently competing peptide candidates.
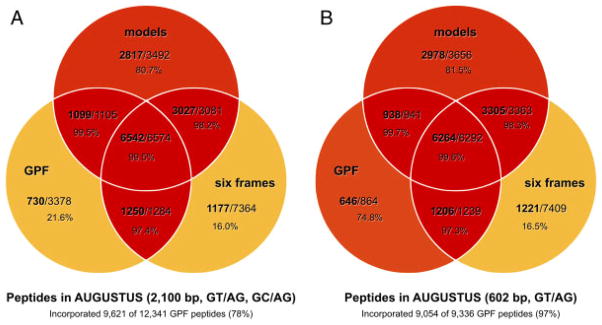
Figure 6.
The effects of applying less stringent GPF search criteria and adding six-frame translated peptides to the list of candidate peptides. Venn diagrams depict the amount of peptides incorporated by AUGUSTUS in relation to peptides provided to AUGUSTUS, for each of three peptide candidate sources: JGI gene models v3.1, GPF peptides, and, in addition, the six-frame translation of the C. reinhardtii genome. (A) Less stringent GPF filtering criteria (max. intron length of 2100 base pairs, GT/AG and GC/AG introns) result in a low peptide incorporation rate for those peptide identified via GPF only (21.6%). In addition, peptides only identified via the six-frame translation show a similar, low incorporation rate of 16%. (B) More stringent GPF filtering criteria, as used in this study, lead to an increase of the incorporation rate of peptides identified via GPF only. Overall, the amount of GPF peptides incorporated by AUGUSTUS increases from 78 to 97%. On the other hand, the incorporation rate of unspliced peptides from the six-frame translation remains low as they are unaffected by the changes.
In addition, Fig. 6 shows how the amount of GPF peptides identified by OMSSA is affected by relaxing the stringency criteria for the construction of spliced peptides via GPF. Using a longer maximum intron length of 2100 base pairs and allowing the additional splice site consensus sequence GC/AG, the amount of peptides identified via GPF increases by 32% from 9336 to 12 341. The number of peptides incorporated by AUGUSTUS is affected less severely. While for the stringent search criteria, 9054 GPF peptides are incorporated, using the relaxed stringency criteria, the number of incorporated GPF peptides is only increased by 6% to 9621. These numbers indicate that AUGUSTUS is robust enough to cope with uncertain extrinsic hints. Thus, it is obvious that the more permissive search criteria for GPF probably lead to more false-positive identifications. On the other hand, the more stringent criteria might result in the loss of correct spliced peptides. In this context, it is interesting to note that, as mentioned above, AUGUSTUS significantly limited the number of included GPF peptide hints, implying that AUGUSTUS itself can be considered as a potent filter to lower the extent of false-positive peptide alignments. For future experiments using GPF, it will therefore be important to find a balance between stringency and sensitivity, which is definitely strongly influenced by the intron length chosen and the number of splice site consensus sequences considered.
The increased GPF peptide confirmation rate of 53% can be considered high, pointing to the fact that the GPF pipeline in conjunction with AUGUSTUS might be a particular useful tool for the alignment of peptides from mass spectrometric data to genomes where the complete genomic sequence is available but only a preliminary gene model set exists. From the data presented, this peptide information will be highly suitable for the validation and annotation of gene models in such a context. This could be especially relevant for proteomic datasets from comparative quantitative analyses since the application of GPF can be expected to increase the number of peptides that can be legitimately considered for quantitation. For the successful employment of GPF, the definition of search parameters such as maximum intron length and splice site motifs is crucial and should be carefully determined from already available, possibly preliminary gene sets. Moreover, while defining these parameters, it should be taken into account that AUGUSTUS intrinsically restricts the number of incorrectly incorporated peptide hints, especially when multiple extrinsic hint sources are available.
We have shown that the concerted action of GPF and AUGUSTUS in an annotation pipeline is a powerful alternative data evaluation approach that results in extrinsic intron/exon hints that may be used for automatic genome annotation. The higher sensitivity, increased speed and more compact memory footprint of the new GPF version make it a promising candidate for use in the proteogenomic annotation of higher organisms such as human and mouse.
Supplementary Material
Acknowledgments
M. Sp. would like to thank Jens Allmer for helpful discussions regarding GPF. The authors are grateful to Susan Hawat for conducting mass spectrometric measurements. Funding: M. H. acknowledges support from BMBF (BMBF 0315265 C, GOFORSYS partner).
Abbreviations
- FDR
false discovery rate
- FM4
Filtered Models 4
- GPF
Genomic Peptide Finder
- PSM
peptide/spectral match
Footnotes
References
- 1.Brent MR, Guigó R. Recent advances in gene structure prediction. Curr Opin Struct Biol. 2004;14:264–272. doi: 10.1016/j.sbi.2004.05.007. [DOI] [PubMed] [Google Scholar]
- 2.Reboul J, Vaglio P, Rual JF, Lamesch P, et al. C. elegans ORFeome version 1.1: experimental verification of the genome annotation and resource for proteome-scale protein expression. Nat Genet. 2003;34:35–41. doi: 10.1038/ng1140. [DOI] [PubMed] [Google Scholar]
- 3.Abbott A. Competition boosts bid to find human genes. Nature. 2005;435:134. doi: 10.1038/435134a. [DOI] [PubMed] [Google Scholar]
- 4.Guigó R, Reese MG. EGASP: collaboration through competition to find human genes. Nat Methods. 2005;2:575–577. doi: 10.1038/nmeth0805-575. [DOI] [PubMed] [Google Scholar]
- 5.Guigó R, Flicek P, Abril JF, Reymond A, et al. EGASP: the human ENCODE genome annotation assessment project. Genome Biol. 2006;7:S2.1–31. doi: 10.1186/gb-2006-7-s1-s2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Merchant SS, Prochnik SE, Vallon O, Harris EH, et al. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science. 2007;318:245–250. doi: 10.1126/science.1143609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stanke M, Diekhans M, Baertsch R, Haussler D. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics. 2008;24:637–644. doi: 10.1093/bioinformatics/btn013. [DOI] [PubMed] [Google Scholar]
- 8.Djebali S, Delaplace F, Crollius HR. Exogean: a frame-work for annotating protein-coding genes in eukaryotic genomic DNA. Genome Biol. 2006;7:S7.1–10. doi: 10.1186/gb-2006-7-s1-s7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Domon B, Aebersold R. Mass spectrometry and protein analysis. Science. 2006;312:212–217. doi: 10.1126/science.1124619. [DOI] [PubMed] [Google Scholar]
- 10.Castellana N, Bafna V. Proteogenomics to discover the full coding content of genomes: A computational perspective. J Proteomics. 2010 doi: 10.1016/j.jprot.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tanner S, Shen Z, Ng J, Florea L, et al. Improving gene annotation using peptide mass spectrometry. Genome Res. 2007;17:231–239. doi: 10.1101/gr.5646507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Castellana NE, Payne SH, Shen Z, Stanke M, et al. Discovery and revision of arabidopsis genes by proteogenomics. Proc Natl Acad Sci USA. 2008;105:21034–21038. doi: 10.1073/pnas.0811066106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stanke M, Steinkamp R, Waack S, Morgenstern B. AUGUSTUS: a web server for gene finding in eukaryotes. Nucleic Acids Res. 2004;32:W309–W312. doi: 10.1093/nar/gkh379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Allmer J, Markert C, Stauber EJ, Hippler M. A new approach that allows identification of intron-split peptides from mass spectrometric data in genomic databases. FEBS Lett. 2004;562:202–206. doi: 10.1016/S0014-5793(04)00212-1. [DOI] [PubMed] [Google Scholar]
- 15.Eng JK, McCormack AL, YatesIII JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 16.Ma B, Zhang K, Hendrie C, Liang C, et al. PEAKS: powerful software for peptide de novo sequencing by tandem mass spectrometry. Rapid Commun Mass Spectrom. 2003;17:2337–2342. doi: 10.1002/rcm.1196. [DOI] [PubMed] [Google Scholar]
- 17.Allmer J, Naumann B, Markert C, Zhang M, et al. Mass spectrometric genomic data mining: Novel insights into bioenergetic pathways in chlamydomonas reinhardtii. Proteomics. 2006;6:6207–6220. doi: 10.1002/pmic.200600208. [DOI] [PubMed] [Google Scholar]
- 18.Terashima M, Specht M, Naumann B, Hippler M. Characterizing the anaerobic response of chlamydomonas reinhardtii by quantitative proteomics. Mol Cell Proteomics. 2010;9:1514–1532. doi: 10.1074/mcp.M900421-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geer LY, Markey SP, Kowalak JA, Wagner L, et al. Open mass spectrometry search algorithm. J Proteome Res. 2004;3:958–964. doi: 10.1021/pr0499491. [DOI] [PubMed] [Google Scholar]
- 20.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 21.Labadorf A, Link A, Rogers MF, Thomas J, et al. Genome-wide analysis of alternative splicing in chlamydomonas reinhardtii. BMC Genomics. 2010;11:114. doi: 10.1186/1471-2164-11-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perkins DN, Pappin DJC, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 23.Craig R, Beavis RC. TANDEM: matching proteins with tandem mass spectra. Bioinformatics. 2004;20:1466–1467. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.