Evidence is presented that the linked histone modifications H2B ubiquitylation and H3 methylation play an important role in preventing the ectopic association of silencing proteins with telomere-distant euchromatic genes important for the cellular response to heat.
Abstract
In Saccharomyces cerevisiae, ubiquitylation of histone H2B signals methylation of histone H3 at lysine residues 4 (K4) and 79. These modifications occur at active genes but are believed to stabilize silent chromatin by limiting movement of silencing proteins away from heterochromatin domains. In the course of studying atypical phenotypes associated with loss of H2B ubiquitylation/H3K4 methylation, we discovered that these modifications are also required for cell wall integrity at high temperatures. We identified the silencing protein Sir4 as a dosage suppressor of loss of H2B ubiquitylation, and we showed that elevated Sir4 expression suppresses cell wall integrity defects by inhibiting the function of the Sir silencing complex. Using comparative transcriptome analysis, we identified a set of euchromatic genes—enriched in those required for the cellular response to heat—whose expression is attenuated by loss of H2B ubiquitylation but restored by disruption of Sir function. Finally, using DNA adenine methyltransferase identification, we found that Sir3 and Sir4 associate with genes that are silenced in the absence of H3K4 methylation. Our data reveal that H2B ubiquitylation/H3K4 methylation play an important role in limiting ectopic association of silencing proteins with euchromatic genes important for cell wall integrity and the response to heat.
INTRODUCTION
Covalent modification of histones plays a vital role in establishing correct patterns of gene expression. Histones are modified by an impressive assortment of moieties that often act in combination to dictate the transcriptional state of a piece of chromatin or to orchestrate other DNA-centric events such as nucleosome assembly and repair of damaged DNA (Murr, 2010). Acetylation, methylation, phosphorylation, and ubiquitylation of histones together establish a complex epigenetic code (Strahl and Allis, 2000) that may be heritable and—by its effect on the transcriptome—can have a profound influence on cellular physiology and development of diseases such as cancer (Varier and Timmers, 2010).
One particularly intriguing covalent histone modification is ubiquitylation. Although all four core histones can be ubiquitylated (Goldknopf and Busch, 1977; West and Bonner, 1980; Chen et al., 1998; Yan et al., 2009), the best-understood example of this type of modification is histone H2B ubiquitylation (H2B–Ub), which has been studied extensively in the genetically tractable yeast Saccharomyces cerevisiae (Chandrasekharan et al., 2010). Monoubiquitylation of H2B at lysine 123 (K123) occurs predominantly in active chromatin (Kao et al., 2004) and is catalyzed by a complex containing the Rad6 ubiquitin (Ub)-conjugating enzyme (Robzyk et al., 2000), the Bre1 Ub-protein ligase (Hwang et al., 2003; Wood et al., 2003a), and the accessory protein Lge1 (Hwang et al., 2003), which are recruited to active genes via the concerted action of the Bur1/Bur2 kinase (Wood et al., 2005) and Paf1 (Wood et al., 2003b) complexes. H2B ubiquitylation facilitates transcriptional elongation (Pavri et al., 2006) and promotes nucleosome stability (Chandrasekharan et al., 2009), whereas its subsequent deubiquitylation is important for multiple steps in transcription, including appropriate RNA polymerase II phosphorylation (Wyce et al., 2007).
In addition to direct effects on chromatin structure and activity, ubiquitylation of H2B is also required for the methylation of histone H3 (Sun and Allis, 2002), a phenomenon referred to as “histone cross-talk” or “trans-histone” modification (Chandrasekharan et al., 2010). Specifically, ubiquitylation of H2B is required for the dimethylation and trimethylation of H3 at lysine residues 4 (K4) and 79 (K79), which are carried out by the Set1/COMPASS (Briggs et al., 2001; Sun and Allis, 2002; Lee et al., 2007) and Dot1 (Ng et al., 2002) methyltransferases, respectively. Consistent with the distribution of H2B–Ub, H3 methylation is a mark of active chromatin and plays an important role in regulating gene activation in response to cryptic transcription (Chu et al., 2007; Pinskaya et al., 2009).
Perhaps the most prominent role for H3 methylation, however, is the maintenance of gene silencing (Ng et al., 2002; Sun and Allis, 2002) at heterochromatic loci. Although H3K4 and H3K79 methylation occur predominantly at active genes, these modifications are absolutely required for heterochromatin silencing, likely because they act as a barrier that prevents the Sir silencing proteins from spreading into euchromatic domains, thereby titrating them away from silent chromatin (Chandrasekharan et al., 2010). Consistent with this notion, the majority of genes dysregulated by loss of Set1 are telomere proximal (Venkatasubrahmanyam et al., 2007), although it has recently been found that simultaneous deletion of SET1 and the histone variant gene HTZ1—but not either alone—allows Sir proteins to interact with telomere-distal loci, silencing their expression. Exactly how both SET1 and HTZ1 collaborate to perform this antisilencing function is unknown.
Despite our understanding of the connection between H2B ubiquitylation and H3 methylation, little is known of the broad physiological impact of this cross-talk or of the precise role of H3 methylation alone in preventing ectopic silencing of euchromatic genes. To address this deficit, we sought to identify novel phenotypes associated with loss of these modifications. We describe several such phenotypes here, one of which is cell lysis at elevated temperatures, and present data arguing that H3K4—but not K79—methylation plays a critical role in keeping Sir proteins from interacting with, and ectopically silencing, euchromatic genes required for the cellular response to heat.
RESULTS
Novel phenotypes associated with loss of H2B–H3 cross-talk
Cross-talk between H2B ubiquitylation and H3 dimethylation and trimethylation is involved in a number of cellular processes, including transcriptional activation, gene silencing, and repair of damaged DNA (Chandrasekharan et al., 2010). To further illuminate the physiological role of this process, we sought to characterize additional phenotypes associated with loss of these linked modifications.
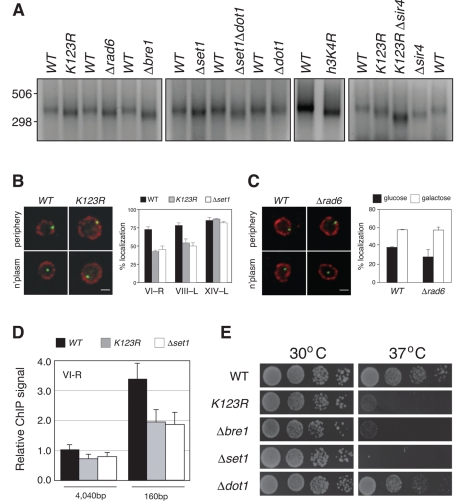
Previous reports demonstrated that deletion of genes required for H2B ubiquitylation (BRE1 and RAD6) and H3K4 methylation (SET1) lead to modest telomere shortening (Nislow et al., 1997; Gatbonton et al., 2006). To ask whether this phenomenon results from loss of H2B–Ub–dependent signaling, we analyzed telomere length in a yeast strain in which the site of H2B ubiquitylation is blocked by substitution with an arginine residue (K123R) and compared the effect of this cis-mutation with effects resulting from loss of Rad6, Bre1, Set1, and Dot1 (Figure 1A). Using a PCR-based method (Kramer and Haber, 1993) to probe telomere length at the right arm of chromosome VI, we found that the VI–R telomere was roughly 25–50 base pairs shorter than wild type (WT) in all mutant cells examined, with the exception of the dot1-null cells, which displayed WT telomere lengths. These effects were also observed with a cis-mutation in the site of Set1-dependent methylation on histone H3 (h3K4R; Figure 1A) and are clearly independent of the loss of silencing that occurs in K123R mutant cells, as simultaneous disruption of H2B ubiquitylation and deletion of SIR4 has additive effects on telomere shortening (K123RΔsir4; Figure 1A). On the basis of these observations, we conclude that H2B-ubiquitylation–dependent dimethylation and trimethylation of H3K4, but not H3K79, contributes to proper telomere length control in S. cerevisiae. Moreover, because of the additive effect of the K123R and Δsir4 mutations on telomere shortening, we conclude that effects of H2B–H3 cross-talk on telomere length are independent of their role in silencing.
FIGURE 1:
Novel phenotypes associated with loss of H2B–H3 cross-talk. (A) PCR was used to determine VI-R telomere length in WT (FGY8.2), htbK123R (FGY8.3), Δrad6 (FGY8.4), Δbre1 (FGY8.5), Δset1 (FGY8.6), Δset1 Δdot1 (FGY8.7), Δdot1 (FGY8.8), hht2K4R (FGY8.17), htbK123R Δsir4 (FGY8.11), and Δsir4 (FGY8.10) cells. Note that the congenic WT strain for hht2K4R (FGY8.18) is different from FGY8.2, which is congenic with all other strains. Representative image of three independent experiments. (B) Fluorescence microscopy analysis, comparing localization of telomere-proximal GFP-LacI (green) vs. Nsp1 (red). Left, representative image of cells, showing those that were scored as GFP-LacI being at the nuclear periphery or the nucleoplasm. Right, quantification of GFP-LacI signals localized to the nuclear periphery for each of the three telomeres in three yeast backgrounds WT (ICY119, ICY123, ICY127), htbK123R (ICY120, ICY124, ICY128), and Δset1 (ICY131, ICY132, ICY133). Error bars indicate standard errors from at least two independent experiments. (C) Fluorescence microscopy analysis, comparing localization of LacO sites integrated proximal to the GAL1 locus in WT (ICY74) or Δrad6 (ICY31) cells. Left, representative image of cells, showing those that were scored as GFP-LacI being at the nuclear periphery or the nucleoplasm. Right, quantification of GFP-LacI signals localized to the nuclear periphery in media with either glucose (off) or galactose (on) as the carbon source. Error bars indicate standard errors from at least two biological replicates. (D) ChIP analysis, examining association of NUPs with sequences proximal to the VI-R telomere. ChIP DNA was sampled with two sets of primers: 4040 and 160 base pairs from the TG repeats of VI-R. Error bars indicate standard errors from four independent experiments. (E) Dilutions (1:9) of yeast strains WT (FGY8.2), htbK123R (FGY8.3), Δbre1 (FGY8.5), Δset1 (FGY8.6), and Δdot1 (FGY8.8) were spotted on YPAD plates and grown for 3 d at either 30 or 37°C.
Given that H2B–H3 cross-talk is important for maintenance of telomere length, we next asked whether other aspects of telomere biology are influenced by this process. In yeast, telomeres are clustered and associated with the nuclear periphery (Sexton et al., 2007), a process that is thought to facilitate silencing by creating local concentrations of silencing proteins at chromosome ends. We therefore asked whether telomere association with the nuclear periphery is disrupted by mutations that disrupt H2B ubiquitylation and H3K4 methylation. For this purpose, we integrated Lac operator (LacO) sites adjacent to telomeres on either the right arm of chromosome VI (VI-R) or the left arms of chromosomes VIII (VIII-L) and XIV (XIV-L). We monitored the location of these sites by expression of a green fluorescent protein (GFP)-LacI fusion protein and scored their location relative to the nuclear periphery (Brickner et al., 2010), as measured by immunofluorescence visualization of the nuclear pore protein Nsp1 (Figure 1B).
In wild-type yeast, we scored localization of all three telomeres at the nuclear periphery in ∼70–80% of yeast cells examined (Figure 1B, black bars), consistent with previous reports (Hediger et al., 2002; Sexton et al., 2007). In the htbK123R, and Δset1 mutants, however, the perinuclear localization of VI-R and VIII-L was reduced to roughly 50%. This phenotype was strongest in the htbK123R (p = 0.001 for VI-R and p = 0.015 for VIII-L using two-tailed t test) and set1Δ mutants (p = 0.002 for VI-R and p = 0.005 for VIII-L), indicating that these telomeres disengage from the nuclear periphery in the absence of H2B ubiquitylation and H3K4 methylation. Similar results were obtained in rad6 cells (unpublished data). Of interest, the effect of disrupting the H2B–H3 nexus on localization is telomere specific, as XIV-L remained associated with the nuclear periphery in all strains examined (Figure 1B) and appears to be confined to telomeric chromatin, as association of the active GAL1 gene with the perinuclear space (Cabal et al., 2006) is not affected by loss of H2B ubiquitylation (Figure 1C). Chromatin immunoprecipitation (ChIP) confirmed that telomere VI-R fails to associate robustly with nuclear pore proteins in the htbK123R and Δset1 cells (Figure 1D). On the basis of these data, we conclude that H2B ubiquitylation and H3K4 methylation are required for the association of a specific subset of telomeres with the periphery of the nucleus.
Finally, we sought to identify a phenotype associated with loss of H2B–H3 cross-talk that might be amenable to genetic screening. We tested viability of htbK123R mutant yeast under a variety of conditions and observed that, when placed at elevated temperatures (37°C), htbK123R cells fail to proliferate (Figure 1E). This phenotype was not strain specific, as it was also observed in htbK123R mutant yeast with a different genetic background (unpublished data). Of importance, we also found that a failure to grow at elevated temperature was shared by congenic yeast lacking BRE1 and SET1 but not in yeast deleted for DOT1 (Figure 1E), supporting the notion that H2B-dependent H3K4 methylation is required for survival of yeast at elevated temperatures. The robust and simple nature of this phenotype gave us the opportunity to genetically interrogate the contribution of H2B–H3 cross-talk to yeast cell physiology.
Isolation of high-copy suppressors of the htbK123R mutation
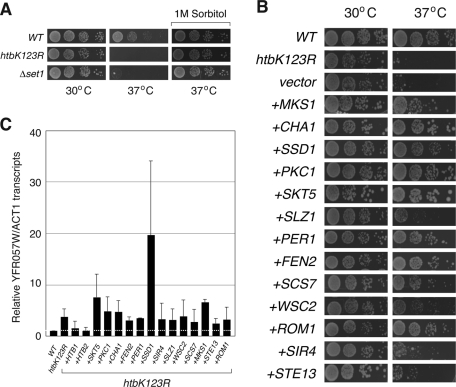
Before beginning a genetic screen, we first asked why htb1K123R mutant yeast fail to proliferate at 37°C. We observed that htb1K123R cells lyse at the restrictive temperature (unpublished data), a phenotype characteristic of mutations in the cell wall integrity (CWI) pathway (Levin, 2005). Consistent with other CWI mutants, the temperature sensitivity of htb1K123R and Δset1 cells was osmoremedial and was rescued by growth of yeast on media containing 1 M sorbitol (Figure 2A). From this preliminary analysis, we conclude that loss of H2B–H3 cross-talk affects cellular integrity, most likely via defects in CWI and the response to osmotic and heat stresses.
FIGURE 2:
Suppression of heat-sensitive phenotype of htbK123R cells. (A) Dilutions (1:9) of yeast strains WT (FGY8.2), htbK123R (FGY8.3), or Δset1 (FGY8.6) were spotted onto either YPAD or YPAD containing 1 M sorbitol and grown for 3 d at either 30 or 37°C. (B) Spotting assay was performed on either WT yeast (FGY8.2) or on htbK123R cells (FGY8.3) that were untransformed, carried an empty pVV215 vector (vector), or carried a pVV215-based vector expressing the indicated open reading frames (ORFs). Wild-type HTB1 and HTB2 are not included here. (C) Reverse-transcriptase quantitative PCR was used to measure transcript levels from the normally silent YFR057W locus in the same yeast strains described in B. YFR057W cDNA levels were normalized to those of ACT1. Error bars show standard errors from three independent experiments.
Because CWI mutants have been subject to extensive analysis by high-copy suppression strategies (Levin, 2005), we performed an overexpression screen for yeast genes capable of rescuing the temperature-sensitive-growth phenotype of htb1K123R cells. For this purpose, we used the recently developed systematic high-copy-array library (Jones et al., 2008), in which tiled ∼10-kb segments of the genome are carried on high-copy (2 μM) plasmids, permitting comprehensive screening of >95% of the yeast genome. We identified and validated 15 suppressor clones from this library, carrying a total of 72 genes. Each of these clones suppressed temperature-sensitive growth of both htbK123R and Δset1 cells (unpublished data), supporting an intimate connection between H2B ubiquitylation and H3K4 methylation in the ability of yeast to grow at elevated temperatures. We subsequently analyzed all 72 genes individually for their ability to suppress the growth phenotype of htbK123R cells (unpublished data) and found that each clone contained just one gene capable of suppressing the phenotype when overexpressed (Figure 2B), resulting in a total of 15 high-copy suppressors of the htbK123R mutation.
Analysis of these suppressors (Table 1) allowed us to place them into three categories. First, as expected, each copy of the wild-type H2B gene (HTB1 and HTB2) was capable of rescuing defects associated with the K123R mutation, confirming that temperature-sensitive growth of htbK123R cells was due to the lysine-to-arginine substitution at K123 in H2B. Second, 11 of the suppressors either had direct links to the cell wall integrity pathway or are flagged as being important for the response to osmotic or heat stress, consistent with the nature of the phenotype under investigation. Finally, two of the suppressors—STE13 and SIR4—were not readily linked to cell wall integrity or the stress response. STE13 encodes a Golgi-associated dipeptidyl-aminopeptidase required for α-factor maturation (Anna-Arriola and Herskowitz, 1994; Johnston et al., 2005), whereas SIR4, as discussed, encodes a silent information regulator (Rine and Herskowitz, 1987) that, together with Sir2 and Sir3, assembles silent chromatin at telomeres and the mating-type loci. Despite the links between Sir4 and silencing, the suppression did not appear to result from alterations in telomeric silencing, as none of the suppressors—with the exception of the wild-type H2B genes—restored transcriptional suppression at the telomere-proximal YFR057W gene (Figure 2C). Finally, we note that none of the suppressors—again with the exception of wild-type H2B genes—restored H3K4 or H3K79 methylation in htbK123R yeast (unpublished data), revealing that their effects on resistance to heat are exerted downstream of H2B–H3 cross-talk.
Table 1: .
High-copy suppressors of htbK123R temperature sensitivity.
| Suppressor | Molecular function | Relevant physiological role |
|---|---|---|
| HTB1 | Histone H2B | Chromatin |
| HTB2 | Histone H2B | Chromatin |
| WSC2 | Sensor transducer of the stress-activated protein kinase C–mitogen-activated protein kinase-1 pathway | Cell wall integrity |
| ROM1 | GDP/GTP exchange protein | Cell wall integrity |
| PKC1 | Protein kinase C | Cell wall integrity |
| PER1 | Activates glycosylphosphatidylinositol-phospholipase A2 in the endoplasmic reticulum | Cell wall integrity |
| SSD1 | RNA-binding protein | Cell wall integrity |
| FEN2 | Plasma membrane H+-pantothenate symporter | Cell wall integrity |
| MKS1 | Negative transcriptional regulator | Cell wall integrity |
| CHA1 | l-Serine (l-threonine) deaminase | Transcriptional target of cell wall integrity pathway |
| SKT5 | Activator of chitin synthase III | Response to osmotic stress |
| SCS7 | Sphingolipid α-hydroxylase | Membrane synthesis; osmotic stress |
| SLZ1 | Leucine-zipper protein | Resistance to heat stress |
| STE13 | Dipeptidyl-aminopeptidase | Regulates α-factor maturation |
| SIR4 | Silent information regulator | Assembly of silent chromatin at telomeres and mating-type loci |
Results of this screen support the notion that H2B–H3 cross-talk is involved in the maintenance of cell wall integrity and the cellular response to heat or osmotic stress. The results also reveal a novel role for STE13 in this response and suggest that interactions between H3K4 methylation and the Sir4 protein—independent of silencing—are also involved in these processes. Because of the established connections between H3K4 methylation and silent information regulators, we chose to investigate the mechanism through which overexpression of Sir4 rescues the growth of htbK123R and Δset1 yeast at elevated temperatures.
Loss of Sir protein function rescues growth defects of htbK123R mutant yeast
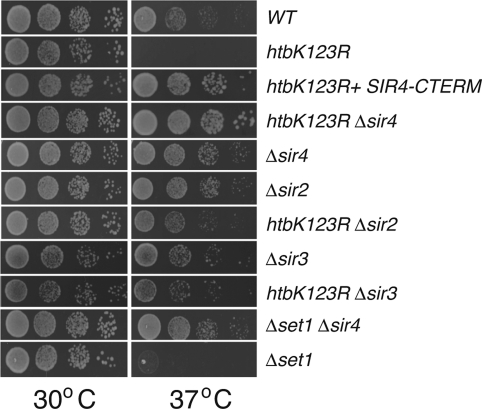
One explanation for how overexpression of Sir4 rescues the temperature sensitivity of htbK123R yeast (Figure 2B) would be that, at high levels, Sir4 tempers the expression of a gene or set of genes that inhibit growth at 37°C. Alternatively, as overproduction of Sir4 has been shown to inhibit silencing (Maillet et al., 1996), likely via titration of members of the Sir complex away from chromatin, it was also possible that Sir4 suppresses the K123R mutation via antagonistic effects on Sir activity. To distinguish between these possibilities, we asked whether the silencing function of Sir4 is required for suppression of the K123R phenotype. We expressed in htbK123R cells a silencing-deficient carboxy-terminal fragment of Sir4 (SIR4–CTERM) that has been shown to dominantly inhibit Sir repression in vivo (Marshall et al., 1987). This analysis (Figure 3) showed that SIR4–CTERM efficiently suppressed temperature-sensitive growth of htbK123R mutant yeast, suggesting that inactivation of SIR function is the basis through which the high-copy Sir4 suppression occurs. To test this notion directly, we examined genetic interactions between the htbK123R allele and the SIR genes (Figure 3). Deletion of either SIR2, SIR3, or SIR4 alone had no effect on the viability of yeast at 37°C, providing further evidence that it is not loss of silencing per se that leads to a failure of htbK123R or Δset1 cells to grow at elevated temperatures. When we combined these deletions with the htbK123R mutation, however, we found that all three double mutants were able to grow robustly at 37°C, demonstrating that deletion of SIR genes is epistatic to disruption of H2B ubiquitylation. We also found that deletion of SIR4 suppressed the temperature sensitivity of Δset1 yeast, suggesting that its actions are directed against H3K4 methylation. Together, these results demonstrate that inactivation of the SIR complex is a bona fide way to suppress growth defects associated with loss of H2B–H3 cross-talk.
FIGURE 3:
Loss of SIR function rescues the heat-sensitive phenotype of htbK123R and Δset1 cells. Dilutions 1:9 of yeast were plated on YPAD and grown for 3 d at either 30 or 37°C. Strains used here are WT (FGY8.2), htbK123R (FGY8.3), htbK123R Δsir4 (FGY8.11), Δsir4 (FGY8.10), Δsir2 (FGY8.12), htbK123R Δsir2 (FGY8.13), Δsir3 (FGY8.14), htbK123R Δsir3 (FGY8.15), Δset1 Δsir4 (FGY8.16), and Δset1 (FGY8.6). To analyze the effects of expressing a dominant-negative version of Sir4 (SIR4–CTERM), htbK123R cells (FGY8.3) carried a pVV214-based plasmid expressing the carboxy-terminal 167 residues of Sir4.
Ectopic silencing of telomere-distal genes occurs in the absence of H2B ubiquitylation
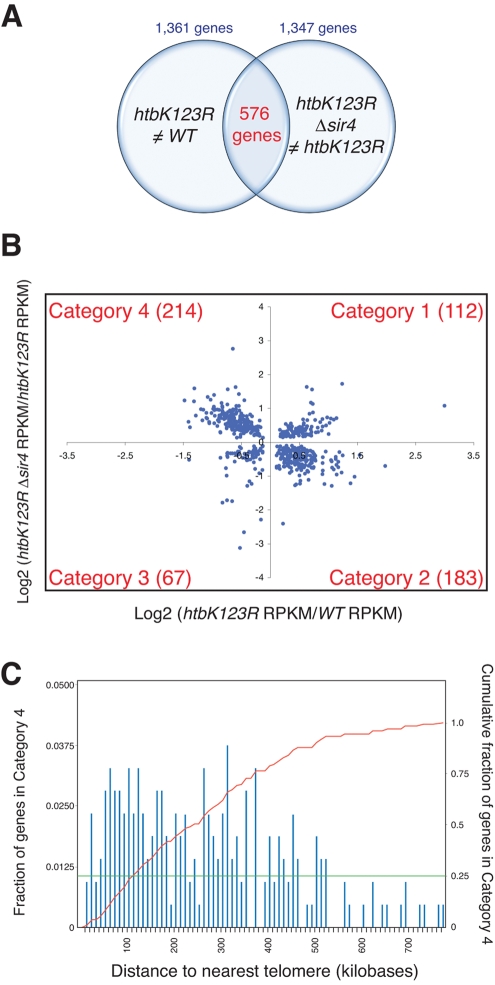
How does disruption of SIR function suppress growth defects associated with loss of H2B ubiquitylation? Given that SIR proteins have recently been found to inhibit transcription of a set of euchromatic genes in the absence of SET1 and HTZ1 (but not either individually [ Venkatasubrahmanyam et al., 2007]), we speculated that there might be a subset of genes inhibited by loss of H3K4 methylation alone, and that SIR-dependent suppression of these genes is responsible for the failure of htbK123R and Δset1 yeast to grow at elevated temperatures. To test this notion, we used RNA sequencing (RNA-seq) (Nagalakshmi et al., 2010) to compare transcriptomes of four yeast strains—WT, htbK123R, Δsir4, and htbK123R Δsir4—specifically looking for genes whose expression is decreased by the K123R mutation but reactivated by loss of Sir4.
We obtained 3.1 M unique (nonribosomal) reads for the WT and htbK123R strains, 2.6 M reads for the Δsir4 mutant, and 3.5 M reads for htbK123R Δsir4 mutant cells. DEGseq software (Wang et al., 2010) was used to identify differentially expressed genes between the different mutants with a significance cutoff of p < 0.001. The reads per kilobase of open reading frame per million reads (RPKM) was used to quantify transcript level changes between the different stains. Using this approach, we identified a total of 1361 genes differentially expressed in the htbK123R mutants, compared with wild type, 109 of which are altered by more than twofold (Supplemental Table S1). Thirty of these genes overlap with results of previous microarray analysis of K123R cells (Mutiu et al., 2007). In sir4-null cells, we found that 1266 genes were deregulated compared with wild type, with expression of a total of 157 genes differing by more than twofold between the strains (Supplemental Table S2). In htbk123R Δsir4 cells, we saw the most pronounced changes in transcript levels, with 1347 genes statistically different from WT cells, 287 of which differed by a factor of two or more (Supplemental Table S3). In all cases, differentially expressed genes were distributed throughout the genome (unpublished data), highlighting the broad impact of H2B ubiquitylation, and SIR protein function, on patterns of gene transcription in yeast cells.
To identify genes that are affected by loss of H2B ubiquitylation and subsequently affected by deletion of SIR4, we compared genes that are differentially expressed in htbK123R mutants, relative to WT, with those that are differentially expressed in htbK123R Δsir4 cells, relative to the htbK123R mutant. As diagrammed in Figure 4, A and B, this comparison yielded a set of 576 genes (Supplemental Table S4), which could be grouped into four categories as follows. Category 1, expression increased in htbk123R cells and further increased upon deletion of SIR4 (112; Figure 4B, top right). Category 2, expression increased in htbk123R cells and decreased upon deletion of SIR4 (183; Figure 4B, bottom right). Category 3, expression decreased in htbk123R cells and decreased further upon deletion of SIR4 (67; Figure 4B, bottom left). Category 4, expression decreased in htbk123R cells and increased by deletion of SIR4 (214; Figure 4B, top left). These data are represented in heat map format in Supplemental Figure S1. Because we were specifically interested in testing the notion that a set of genes involved in growth at 37°C are ectopically suppressed by Sirs in the absence of H2B ubiquitylation, we focused our subsequent analysis on those 214 genes in category 4—genes whose expression is inhibited by the K123R mutation but subsequently restored by loss of Sir4.
FIGURE 4:
Comparative transcriptomics reveals a set of euchromatic genes suppressed by Sir4 in the absence of H2B ubiquitylation. RNA sequencing was performed on yeast strains WT (FGY8.2), htbK123R (FGY8.3), Δsir4 (FGY8.10), and htbK123R Δsir4 (FGY8.11) and transcript levels expressed as RPKM values. (A) Venn diagram, comparing the 1361 genes differentially expressed between WT and htbK123R cells, and the 1347 genes differentially expressed between htbK123R and htbK123R Δsir4 cells. A total 576 of these genes overlap. (B). Scatterplot comparing the relative expression of the 576 genes highlighted in A. Category 4 genes (top left) are those whose expression is decreased in htbK123R cells but increased upon subsequent deletion of SIR4. (C) Histogram of the fraction of category 4 genes as a function of their distance to the nearest telomere. The red line shows the cumulative fraction of category 4 genes as a function from distance to the nearest telomere. The green line indicates that 25% of category 4 genes lie within 100 kb of a telomere.
We analyzed the confirmed and predicted physiological roles of genes in category 4 by querying their Gene Ontology (GO) annotations (Dwight et al., 2002). This analysis (Table 2) revealed highly significant enrichment of category 4 genes in 10 GO terms, three of which were directly relevant to the temperature-sensitive phenotype under consideration: 1) response to temperature stimulus, 2) response to heat, and 3) cellular response to heat. Twenty-nine category 4 genes are linked to the cellular response to heat, and no significant enrichment in these heat-related GO terms were observed in categories 1–3 (unpublished data), indicating a unique involvement of the interaction between H2B ubiquitylation and SIR4 regulation in the physiological response of yeast to elevated temperatures.
Table 2: .
GO terms for category 4 genes.
| Gene ontology term | p value | Gene list |
|---|---|---|
| Generation of precursor metabolites and energy (GO:0006091) | 1.95E-11 | MRPL22, NDI1, CYC7, MAM33, HOR2, RSM24, IDH2, ENO1, COX11, PYK2, ACO1, TPS2, FRE1, IDH1, UGP1, TRX2, GCR1, PGM2, CYB5, RIP1, QCR7, TPI1, GLC3, CIT2, MRPL1, APT2, EMI2, YLR345W, ADH4, PIG2, NDE1, SHP1, GLC8, MDH1, GLK1, RMD9, MRPS35, GPM1, COQ5 |
| Energy derivation by oxidation of organic compounds (GO:0015980) | 6.30E-09 | NDI1, MRPL22, CYC7, MAM33, HOR2, RSM24, IDH2, COX11, ACO1, TPS1, IDH1, UGP1, PGM2, RIP1, QCR7, GLC3, CIT2, EMI2, MRPL1, ADH4, PIG2, NDE1, SHP1, GLC8, MDH1, RMD9, MRPS35, COQ5 |
| Oxidation reduction (GO:0055114) | 6.30E-09 | YIR035C, YDL124W, GOR1, PHB2, NDI1, CYC7, CBR1, GPD1, IDH2, FRE1, IDH1, MCR1, YHB1, RSM26, TRX2, CYB5, RIP1, ARA1, QCR7, FET3, AHP1, IMD2, GRE2, YME2, BDH1, ERG4, ATP2, ADH4, NDE1, ZWF11, PIC2, CPR3, MDH1,CTT1, PHB1, DLD2, ERG3, ERG11, LEU2, COQ5 |
| Response to temperature stimulus (GO:0009266) | 5.30E-08 | AHA1, GOR1, CYC7, TPS2, YMR090W, TSL1, TPS1, PNC1, YBR056W, GAD1, UBI4, PGM2, ARA1, YDC1, BDH1, HSP42, NBP2, MRPS16, YLR345W, SSA1, YDL124W, GLK1, STF2, CTT1, LSP1, MSC1, HSP104, YAP1, CPR6 |
| Mitochondrion organization (GO:0007005) | 1.83E-07 | PHB2, MAS2, MRPL22, MEF1, MRPS5, RSM24, YTA12, HSP82, TIM54, ACO1, Mas1, RSM26, MDM35, MRPS18, RPN11, QCR7, MIA40, NAM9, YME2, TIM9, MRP17, RSM25, MRPS16, MRPL1, COA1, SSA1, MRF1, PHB1, MRPS35, MRPL15, CLU1, TIM17 |
| Vacuolar protein catabolic process (GO:0007039) | 5.62E-07 | YDC1, DCS2, GLC3, PEP4, PIG2, PIC2, TSL1, TPS1, CTT1, STF2, NMA2, GLK1, UGP1, HSP104, GGA1, TRX2, GAD1, PGM2, ARA1 |
| Response to heat (GO:0009408) | 6.52E-07 | AHA1, GOR1, CYC7, YMR090W, TPS1, TSL1, PNC1, YBR056W, GAD1, UBI4, ARA1, YDC1, BDH1, MRPS16, NBP2, YLR345W, SSA1, GLK1, YDL124W, STF2, CTT1, MSC1, HSP104, LSP1, YAP1, CPR6 |
| Alcohol metabolic process (GO:0006066) | 1.30E-06 | YLR345W, SEC53, GPD1, HOR2, NSG1, ENO1, PYK2, UGP1, MCR1, CYB5, GCR1, PGM2, TPI1, GRE2, BDH1, EMI2, ERG4, ADH4, OSH7, ERG8, NDE1, ZWF1, MDH1, GLK1, IDI1, ERG3, ERG11, GPM1 |
| Response to abiotic stimulus (GO:0009628) | 2.02E-06 | GOR1, AHA1, CYC7, MSN1, HOR2, GPD1, TPS2, HSP82, YMR090W, TSL1, TPS1, PNC1, YBR056W, GAD1, UBI4, PGM2, ARA1, YDC1, BDH1, HSP42, NBP2, MRPS16, YLR345W, SSA1, YDL124W, GLK1, STF2, CTT1, LSP1, MSC1, HSP104, YAP1, CPR6, AIP1 |
| Cellular response to heat (GO:0034605) | 2.09E-05 | YLR345W, YMR090W, YDL124W, YBR056W, GOR1, AHA1, BDH1, CYC7, MRPS16, SSA1, TSL1, TPS1, CTT1, STF2, GLK1, HSP104, MSC1, PNC1, UBI4, GAD1, ARA1, CPR6 |
Finally, because loss of H2B ubiquitylation/H3K4 methylation would be expected to predominantly influence the expression of telomere-proximal genes (by spreading of SIR proteins from telomeres into adjacent chromatin), we examined the genomic localization of all genes in category 4 (Figure 4C). Although we did find a significant number of telomere-proximal genes in this category, we were surprised to learn that more than 75% of genes in this category are located more that 100 kb away from the nearest telomere, with a handful of genes located more that 500 kb from a chromosome end. The profound bias of category 4 genes away from telomeres indicates that a significant role of H2B ubiquitylation is to prevent silencing proteins from limiting expression of euchromatic genes.
Ectopic association of Sir proteins with euchromatic genes
Given the known function of Sir proteins as transcriptional silencers, the simplest interpretation of our RNA-seq data is that, in the absence of H2B–H3 cross-talk, Sir proteins can ectopically associate with category 4 genes, attenuating their expression. To test this concept, we used DNA adenine methyltransferase identification (DamID) (van Steensel and Henikoff, 2000) to probe the association of Sir3 and Sir4 with select loci in congenic WT or Δset1 yeast cells. In this procedure, Sir3 and Sir4 are fused to the Escherichia coli DNA adenine methyltransferase, with the result being that DNA sequences that encounter Sir proteins become methylated, which can then be detected by cleavage of DNA with a methylation-sensitive restriction enzyme. We selected this method because it has previously been shown to provide a sensitive and robust readout of the interaction of Sir proteins with euchromatic chromatin in the absence of both SET1 and HTZ1 (Venkatasubrahmanyam et al., 2007).
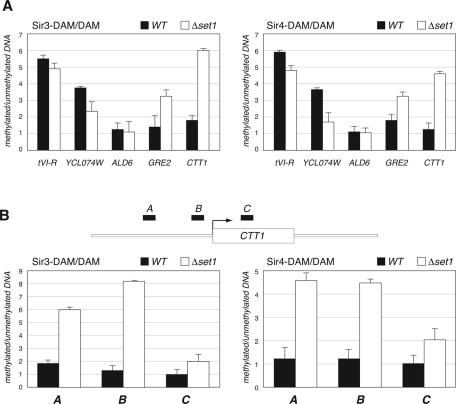
For this analysis, we selected three types of target sequences for analysis: 1) telomere-proximal sites—telomere VI-R (tVI-R) and YCL074W, which is located ∼4 kb from telomere III-L); 2) a category 2 gene—ALD6, whose expression was increased in the htbK123R mutant cells and subsequently decreased in the htbK123R Δsir4 double mutants; and 3) two category 4 genes—GRE2, which is located 43 kb from the left arm of chromosome XV and encodes a stress-induced methylglycoxal reductase, and CTT1, which is located 435 kb from the right arm of chromosome VII and encodes a cytosolic catalase required for resistance to heat and osmotic stress (Wieser et al., 1991). Results of these experiments are presented in Figure 5A. As expected, telomere-proximal sites tVI-R and YCL074W showed reduced association with both Sir3 and Sir4 in Δset1 cells, consistent with the idea that silencing proteins move away from heterochromatic domains when H3K4 methylation is lost (Chandrasekharan et al., 2010). At ALD6, loss of Set1 had no detectable effect on association with either Sir protein, demonstrating that differences we see in Sir3 and Sir4 binding are locus specific. Finally, at both category 4 genes (GRE2 and CTT1) we observed significant (p < 0.05) enrichment of both Sir3 and Sir4 in the absence of Set1, demonstrating that Sir proteins do interact with genes whose expression is attenuated when H3K4 methylation is lost. Moreover, we surveyed available sites within the CTT1 gene and found that enrichment of both Sir2 and Sir3 is greatest within the 5′ nontranscribed region of this locus (Figure 5B), again supporting the concept that, in the absence of H3K4 methylation, Sir proteins limit expression of this gene.
FIGURE 5:
Ectopic association of Sir3 and Sir4 with category 4 genes in the absence of H3K4 methylation. DamID chromatin profiling was performed on cells expressing Sir3-Dam or Sir4-Dam fusion proteins in an otherwise WT (black bars) or Δset1 (open bars) background. The percentage of methylated DNA was determined by using real-time PCR—with primers that flank candidate methylation sites—to compare relative signals for each amplicon in digested vs. undigested DNA. The percentage of methylation by Sir3-Dam or Sir4-Dam in each strain was normalized to their respective strain expressing the unfused Dam protein. Strains YM2302, YM2312, YM2304, YM2313, YM2306, and YM2314 were used in these experiments. (A) DamID comparison of Sir3 and Sir4 association with the promoter regions of the indicated genes. (B) DamID comparison of Sir3-Dam and Sir4-Dam association with candidate methylation sites [amplicons A (−964 to −836), B (−344 to −266), and C (+541 to +642) across the CTT1 gene; CTT1 is a category 4 gene involved in the cellular response to heat. Error bars show standard errors from three independent experiments.
On the basis on these data, we conclude that H3K4 methylation plays a role in preventing the inappropriate association of silencing proteins with select euchromatic loci. In addition, on the basis of the phenotypes we observe, the nature of our high-copy suppressors, and the predicted function of category 4 genes, we posit that a cohort of these ectopically regulated genes is involved in the cellular response to heat or osmotic stress.
DISCUSSION
To understand better the breadth of the role of H2B–H3 cross-talk in yeast cell physiology, we sought to characterize atypical phenotypes associated with loss of H2B ubiquitylation and H3K4 methylation. We found that, in the absence of these modifications, select telomeres fail to associate with the nuclear periphery and shorten. We also found that yeast deficient in these modifications lyse at elevated temperatures, and we presented evidence that this phenotype results from Sir-dependent silencing of euchromatic heat- and stress-related genes in the absence of H3K4 methylation. Our data underscore the widespread impact of H2B–H3 cross-talk on yeast cell biology and indicate the role of H3K4 methylation in stemming the ectopic silencing of euchromatic genes.
We do not understand why telomeres in these yeast are deregulated as we have described. It is possible that the two phenotypes we describe—telomere deanchoring and shortening—are connected, as other mutations that disengage telomeres from the nuclear periphery, such as deletion of YKU70 (Stellwagen et al., 2003), can result in telomere shortening. If this notion is correct, it implies that the primary effect of loss of H3K4 methylation is to weaken interactions that link telomeres to the nuclear rim. Although there is a role for Sir4 and silencing complexes in telomere tethering (Taddei and Gasser, 2004) and length maintenance (Palladino et al., 1993), we note that the K123R mutation and deletion of SIR4 have additive effects on telomere shortening (Figure 1A), suggesting that H2B ubiquitylation exerts its effects on these processes in a manner independent of Sirs and therefore independent of its effects on telomeric silencing. One possibility is that, in the absence of H2B ubiquitylation or H3K4 methylation, a subset of euchromatic genes required for appropriate telomere maintenance is deregulated, resulting in loss of tethering of select telomeres and shortening of the repeats. We examined gene expression profiles in htbK123R cells and were unable to find any genes that are deregulated more than twofold that could cause such phenotypes. We did observe that expression of the telomere-related gene RIF2 (Hirano et al., 2009) is reduced twofold in htbK123R cells, but if anything this perturbation should have the opposite effect on telomere length, as Rif2 acts to limit telomere elongation (Hirano et al., 2009). Further investigation into the mechanism through which telomeres are perturbed in cells deficient for H2B–Ub and H3K4 methylation is required to resolve this issue.
Regardless of mechanism, however, our data on H2B–H3 cross-talk and telomere maintenance do illuminate another way in which these modifications affect the state of heterochromatic domains. Clustering of telomeres, and their tethering to the nuclear periphery, has been proposed to facilitate silencing by creating high local concentrations of silencing factors at telomere ends (Sexton et al., 2007). The finding that tethering at select telomeres is disrupted in htbK123R and Δset1 cells (Figure 1B) indicates that H2B–H3 cross-talk enforces silencing through at least two distinct processes—the previously described pathway that prevents Sirs from “creeping” away from telomeres (Chandrasekharan et al., 2010), and the process we describe here, which physically constrains telomeres to subnuclear sites where silencing proteins are concentrated.
In this study, we focused primarily on the temperature sensitivity of htbK123R and Δset1 yeast, as this phenotype was robust and gave us the opportunity to genetically interrogate the basis for cell lysis at elevated temperatures. Our high-copy suppression screen identified 13 novel dosage suppressors of both the htbK123R and Δset1 mutations (Table 1), 11 of which have established links to either cell wall integrity or the response to heat or osmotic stress. We note that none of the suppressors restored H3K4 methylation (unpublished data) and that none of them was appreciably down-regulated in the presence of the htbK123R mutation (unpublished data), suggesting that they act downstream of genes directly affected by loss of H2B–H3 cross-talk. We assume that the 11 suppressors in this category rescue growth of htbK123R and Δset1 cells by fortifying yeast for growth at elevated temperatures. STE13, another suppressor, is not flagged as being important for cell wall integrity or related processes, and ste13-null cells do not exhibit temperature-sensitive growth (unpublished data), suggesting that its mechanism of suppression is different from that of the majority of suppressors we identified. Nonetheless, the ability to link 11/13 suppressors to cell wall integrity and the response to stress highlights the importance of H2B–H3 cross-talk to regulation of genes required for these processes.
Given the established links between Sirs and H2B–H3 cross-talk (Chandrasekharan et al., 2010), we focused subsequent efforts on the remaining suppressor Sir4. We showed that dosage suppression by Sir4 is likely due to inactivation of authentic Sir complexes, and we went on to identify a set of 214 genes (category 4) whose expression is attenuated by loss of H2B ubiquitylation and subsequently restored by deletion of SIR4. Many of these genes are involved in the cellular response to heat, and the majority are located more than 100 kb from the nearest telomere. Of importance, we found that both Sir3 and Sir4 associate with representative category 4 genes when H3K4 methylation is lost, implying a direct role for Sir proteins in ectopic repression of this class of genes in the absence of H2B–H3 cross-talk. On the basis of these observations, we present a model (Figure 6) in which the antagonistic relationship between H3K4 methylation and Sir proteins (Santos-Rosa et al., 2004) serves two important functions. First, as previously described (Chandrasekharan et al., 2010), H3 methylation acts at telomere-proximal sites to restrict spreading of Sir complexes away from telomeric heterochromatin, thereby maintaining subtelomeric chromatin in its repressed state (a process that may also be facilitated, as described earlier, by the influence of H3K4 methylation on telomere association with the nuclear periphery). Second, we propose that H3K4 methylation also acts within euchromatic regions to repel Sir complexes and prevent them from association with, and ectopic silencing of, active genes, a subset of which is involved in the cellular response to heat. In this way, the dual action of H3K4 methylation at both heterochromatin and euchromatin acts globally to dictate appropriate patterns of Sir protein association across the entire genome. De novo motif analysis failed to identify common sequence elements in category 4 genes (unpublished data), suggesting that recruitment of Sir proteins to these genes is mediated either by multiple elements or by some epigenetic phenomenon.
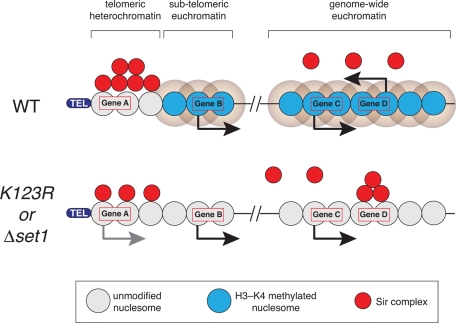
FIGURE 6:
Model for ectopic silencing of euchromatic genes in the absence of H2B–H3 cross-talk. In WT cells, H3K4 methylation at subtelomeric and genome-wide euchromatin keeps Sir proteins localized to telomeric heterochromatin and prevents them from associating from transcribed genes. In the absence of H3K4 methylation (as occurs in htbK123R and Δset1 cells), Sir proteins dissociate from the telomeres, leading to activation of subtelomeric genes (e.g., gene A) and their ectopic association with select euchromatic genes (e.g., gene D), silencing their expression. In this model, gene D would correspond to a category 4 gene.
Finally, we note that the ability of Sir proteins to silence euchromatic genes reveals an important role for H3K4 methylation in maintaining the transcription state of active genes, by preventing ectopic silencing of recently transcribed loci. By extension, it is also possible that selective loss of H3K4 methylation at Sir-susceptible genes could be a mechanism through which expression of select genes could be actively shut down in response to specific signals or during certain states or processes. The negative influence of Sir proteins on active genes, and determination of what renders a gene sensitive to ectopic Sir action, is an area warranting further investigation.
MATERIALS AND METHODS
Yeast strains, plasmids, and growth conditions
Yeast strains used in this study are listed in Table 3. Standard media and growth conditions were used throughout. The majority of experiments were performed in strains derived from FGY8.2 (Δhtb1Δhtb2), in which HTB1 is expressed from a single-copy plasmid under the control of its own promoter (Geng and Tansey, 2008). Deletion mutants were made by PCR-based gene targeting using auxotrophic (LEU2, klTRP, URA3) or dominant resistance markers (KANMX and NATMX4) (Longtine et al., 1998; Goldstein and McCusker, 1999). The 5′ and the 3′ oligonucleotide primers were designed to flank the transcriptional start site and the end of the coding region, respectively. Correct transformants were identified by PCR, screening for 1) loss of targeted gene sequences and 2) integration of targeting cassette at the correct genomic locus. To determine the effects of mutating lysine 4 of histone H3 to arginine (hhtK4R) on telomere length, plasmid pQQ18—which carries the genomic fragments of HTA1-HTB1 and HHF2-HHT2 in the pRS315 (LEU) vector (Ahn et al., 2005)—was first modified by introduction of the hhtK4R mutation via QuikChange XL Site Directed Mutagenesis Kit (Stratagene, Santa Clara, CA) to produce the plasmid pQQ18-hht2–K4R. Then pQQ18 and pQQ18-hht2–K4R were individually transformed into strain JHY205 (Ahn et al., 2005), and plasmid shuffling was used to remove the plasmid expressing wild-type histones. Primer sequences are available on request.
Table 3: .
Yeast strains used in this study.
| Strain | Genotype | Source |
|---|---|---|
| FGY8 | MATa htb1Δ htb2Δ leu2-3,-112 his3-11,-15 trp1-1 ura3-1 ade2-1 can1-100 [Ycp50–HTB1] | Geng and Tansey, 2008 |
| FGY8.2 | FGY8, except [p413-HTB1-HA] replaces [Ycp50–HTB1] | This study |
| FGY8.3 | FGY8, except [p413-htb1K123R-HA] replaces [Ycp50–HTB1] | This study |
| FGY8.4 | FGY8.2, except rad6Δ::klTRP1 | This study |
| FGY8.5 | FGY8.2, except bre1Δ::LEU2 | This study |
| FGY8.6 | FGY8.2, except set1Δ::NatMX | This study |
| FGY8.7 | FGY8.6, except dot1Δ::URA3 | This study |
| FGY8.8 | FGY8.2, except dot1Δ::URA3 | This study |
| FGY8.10 | FGY8.2, except sir4Δ::NatMX | This study |
| FGY8.11 | FGY8.3, except sir4Δ::NatMX | This study |
| FGY8.12 | FGY8.2, except sir2Δ::NatMX4 | This study |
| FGY8.13 | FGY8.3, except sir2Δ::NatMX4 | This study |
| FGY8.14 | FGY8.2, except sir3Δ::NatMX4 | This study |
| FGY8.15 | FGY8.3, except sir3Δ::NatMX4 | This study |
| FGY8.16 | FGY8.6, except sir4Δ::klTRP1 | This study |
| JHY205 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 hht1-hhf1::KAN hhf-2hht2::NAT hta1- htb1::HPH hta2-htb2::NAT pJH33[CEN URA3 HTA1-HTB1 HHT2-HHF2] | Ahn et al., 2005 |
| FGY8.17 | JHY205, except [pQQ18-HTA1-HTB1 hht2K4R-HHF2] | This study |
| FGY8.18 | JHY205, except [pQQ18-HTA1-HTB1 HHT2-HHF2] | This study |
| ICY119 | FGY8.2, except LacI-GFP:LEU2 TELVI-R LacO:TRP1 | This study |
| ICY120 | FGY8.3, except LacI-GFP:LEU2 TELVI-R LacO:TRP1 | This study |
| ICY123 | FGY8.2, except LacI-GFP:LEU2 TELVIII-L LacO:TRP1 | This study |
| ICY124 | FGY8.3, except LacI-GFP:LEU2 TELVIII-L LacO:TRP1 | This study |
| ICY127 | FGY8.2, except LacI-GFP:LEU2 TELXIV-L LacO:TRP1 | This study |
| ICY128 | FGY8.3, except LacI-GFP:LEU2 TELXIV-L LacO:TRP1 | This study |
| ICY131 | FGY8.2, except LacI-GFP:LEU2 TELVI-R LacO:TRP1 set1Δ::NatMX | This study |
| ICY132 | FGY8.2, except LacI-GFP:LEU2 TELVIII-L LacO:TRP1 set1Δ::NatMX | This study |
| ICY133 | FGY8.2, except LacI-GFP:LEU2 TELXIV-L LacO:TRP1 set1Δ::NatMX | This study |
| W303-1A | MATa ade2-1 his3-11,15 leu2-3112,trp1-1 ura3-1 can1-100 | B. Futcher (Stony Brook University, Stony Brook, NY) |
| yWS40 | W303-1A, except rad6Δ::klTRP1 | This study |
| ICY31 | yWS40, except LacI-GFP:LEU2 GAL1:LacO128:URA3 | This study |
| ICY74 | W303-1A, except LacI-GFP:HIS3 GAL1:LacO128:URA3 | This study |
| YM2302 | MATα SIR3-dam::kanMX6 his3Δ1 leu2Δ0 ura3Δ0 lys2Δ0 | Venkatasubrahmanyam et al., 2007 |
| YM2312 | MATα SIR3-dam::kanMX6 set1Δ::HIS3MX6 his3Δ1 leu2Δ0 ura3Δ0 lys2Δ0 | Venkatasubrahmanyam et al., 2007 |
| YM2304 | MATα SIR4-dam::kanMX6 his3Δ1 leu2Δ0 ura3Δ0 lys2Δ0 | Venkatasubrahmanyam et al., 2007 |
| YM2313 | MATα SIR4-dam::kanMX6 set1Δ::HIS3MX6 his3Δ1 leu2Δ0 ura3Δ0 lys2Δ0 | Venkatasubrahmanyam et al., 2007 |
| YM2306 | MATα SIR4pr-dam::kanMX6 SIR4+ his3Δ1 leu2Δ0 ura3Δ0 lys2Δ0 | Venkatasubrahmanyam et al., 2007 |
| YM2314 | MATα SIR4pr-dam::kanMX6 SIR4+ set1Δ::HIS3MX6 his3Δ1 leu2Δ0 ura3Δ0 lys2Δ0 | Venkatasubrahmanyam et al., 2007 |
For imaging telomeres, the indicated yeast strains were transformed with pAFS52-LEXA-LACOR ARS609 (= telVI-R; digested with BbsI), pAFS52-LACOR-VIII-L (digested with SwaI), or pAFS52-LACOR-XIV-L (digested with BstEII) (Hediger et al., 2002; Bystricky et al., 2005) to integrate Lac operator sequences at telomere-proximal positions. For visualization of GAL1 localization, W303-1a or yWS40 was transformed with p6LacO128-GAL1 plasmid (Brickner et al., 2007) that had been digested with NruI. In either case, the gene encoding the GFP-Lac repressor was integrated either at HIS3, using pAFS144 (Straight et al., 1996) digested with NheI, or at LEU2, using pRS305-GFP-LacI digested with AflII. pRS305-GFP-LacI was created by replacing the HIS3 gene from pAFS144 with the LEU2 gene from pRS305 using AatII and SacI sites.
To analyze heat sensitivity of strains, cells were grown overnight in liquid yeast extract/peptone/adenine/dextrose (YPAD) medium, and 1:9 serial dilutions prepared in Tris-EDTA, the lowest dilution corresponding to A600 = 0.5. Dilutions were then spotted on YPAD plates, or YPAD plates supplemented with 1 M d-sorbitol (Sigma-Aldrich, St. Louis, MO), and grown at 30 or 37°C, as indicated.
To construct an expression vector expressing the silencing-inactive carboxy-terminal fragment of Sir4 (CTERM; Marshall et al., 1987), sequences encoding the carboxy-terminal 167 residues of Sir4 were PCR amplified using attB1 and attB2 adapter primers and cloned into the pDONR207 plasmid (Invitrogen, Carlsbad, CA) using the Gateway Cloning System (Invitrogen). The resulting entry clone was then used to transfer SIR4 sequences into the yeast expression vector pVV214 (Van Mullem et al., 2003), which carries the potent PGK1 promoter. Primer sequences are available on request.
Telomere-length determination
Telomere-length was measured by PCR (Kramer and Haber, 1993). Genomic DNA was prepared from the indicated yeast strains and single-stranded telomeric DNA removed by treatment with T4 DNA polymerase. A double-stranded synthetic oligonucleotide (top strand: 5′-CATTTTGCTGCTGCCGGTCATTCGAACCC-3′) was then ligated to the genomic DNA, followed by PCR amplification with an adapter primer (5′-CATTTTGCTGCTGCCGGTCATTCGAACCC-3′) and a telomere VI-R–specific primer (5′-AAATGAGGACTGGGTCATGG-3′). PCR products were resolved by agarose gel electrophoresis and subject to DNA sequencing for validation. Each assay was repeated on at least three different colonies for each strain, with identical results.
Monitoring chromosome location
Imaging of telomeres VI-R, VIII-L, and XIV-L and the activated GAL1 gene with respect to the nuclear envelope was performed exactly as described (Brickner et al., 2010). Cells were fixed in methanol, spheroplasted, detergent extracted, and probed with rabbit polyclonal anti-GFP antibody (A-11122, 1:1000 dilution; Invitrogen) to detect the GFP-Lac repressor and an anti-Nsp1 (MCA-32D6, 1:200 dilution; EnCor Biotechnology, Gainesville, FL) monoclonal antibody to stain nuclear pore complexes. Secondary antibodies were Alexa Fluor 594 goat anti–mouse immunoglobulin (IgG) (A-11032, 1:200 dilution; Invitrogen) and Alexa Fluor 488 goat anti–rabbit IgG (A-11008, 1:200 dilution; Invitrogen). Cells were visualized on a Zeiss LSM510 confocal microscope. A single z-slice through each cell with the brightest and most focused anti-GFP spot was collected. Cells in which this anti-GFP spot colocalized with nuclear pore staining were scored as “peripheral” and all other cells were scored as “nucleoplasmic.” Data were collected from 30–50 cells for each biological replicate. ChIP analysis, examining interaction of telomere VI-R with nucleoporins (NUPs), was performed with anti-NUP antibody mAB414 (Covance, Berkeley, CA) as described (Kuo et al., 1998).
Dosage suppressor screen
The Yeast Tiling Collection Library developed by the G. Prelich laboratory (Jones et al., 2008) was purchased from Open Biosystems (Thermo Biosystems, Huntsville, AL) and DNA isolated on 96-well DNA-binding UNIFILTER plates (Whatman, Piscataway, NJ) as described by the manufacturer. Each clone in the library was individually transformed into FGY8.3 yeast cells using a high-efficiency transformation protocol (Gietz and Schiestl, 2007) in 96-well format. Transformation reactions were pinned onto synthetic complete medium lacking leucine (SCM–LEU) and transformants subsequently grown up and pinned in quadruplicate (384 density) onto YPAD plates for growth at either 30°C (permissive) or 35°C (restrictive). All manipulations were performed using a RoToR HDA Robot (Singer Instrument Company, Watchet, United Kingdom).
Fifty-one library clones appeared to support growth of at least three of four “spots” for each transformation and were scored as positive in the initial screen. These clones were then individually retransformed into FGY8.3 cells and tested in traditional spotting assays for their ability to support growth of yeast at 37°C. Fifteen clones survived this secondary screen. To identify genes within each clone associated with suppression, individual open reading frames (72 in total) from each of the 15 suppressors were PCR cloned using the Gateway System (as described earlier) into vector pVV214 or pVV215 (Van Mullem et al., 2003). These clones were transformed into both FGY8.3 (htbK123R) and FGY8.6 (Δset1) yeast and transformants analyzed for growth at the restrictive temperature by spotting assays. Primer sequences are available on request.
RNA analysis
Total RNA was extracted from the indicated yeast strains using the hot acidic phenol method (Collart and Oliviero, 2001) and processed as described (Geng and Tansey, 2008). Reverse transcription was performed using random hexamer priming, and cDNA levels quantified by real-time PCR. Levels of transcripts from the heterochromatic YFR057W gene were expressed relative to those of the euchromatic ACT1 locus. Primer sequences are available on request.
RNA sequencing and analysis
Total RNA was extracted from parallel cultures of FGY8.2 (WT), FGY8.3 (htbK123R), FGY8.10 (Δsir4), and FGY8.11 (htbK12R Δsir4) cells using the RNeasy Mini Kit (Qiagen, Valencia, CA) as described by the manufacturer. Messenger RNA was isolated using the OligoTex mRNA Isolation Kit (Qiagen) and converted into double-stranded, adapter-ligated cDNA using the Illumina (San Diego, CA) mRNA Sample Prep Kit. Adapter-ligated cDNA libraries were then sequenced on an Illumina GAII Genome Analyzer in a single-end 36–base pair run at the Vanderbilt Genome Technology Core. Bowtie software (Langmead, 2010) was used to align reads to the S. cerevisiae genome (sacCer2 assembly, June 2008), and rRNA and multialigning reads filtered out prior to analysis. Data were compiled from two distinct sets of biological replicates, and DEGseq software (Wang et al., 2010) used to identify statistically significant differentially expressed genes between the mutant and wild-type cells. The level of significance for each gene was calculated using the likelihood ratio test. Genes that had p < 0.001 were defined as differentially expressed in our data set. To analyze the relative change of each gene in the mutants compared with wild-type yeast, we calculated fold changes in RPKM (Mortazavi et al., 2008). RNA-seq data were deposited at the Gene Expression Omnibus database (GSE27934).
GO enrichment analysis (Ashburner et al., 2000) was performed by extracting GO annotations for each gene in the Database for Annotation, Visualization and Integrated Discovery (Huang da et al., 2009) and searching for functional enrichment via the online tool WebGestalt (Duncan et al., 2010). WebGestalt uses a hypergeometric test to examine whether a GO term is significantly enriched within genes of interest by comparing the observed versus the expected number of genes in each category. Multiple-testing correction was performed by using the method of Benjamini and Hochberg (1995).
DNA adenine methyltransferase identification
Wild-type and Δset1 strains expressing E. coli DNA adenine methyltransferase (Dam) alone or fused to Sir3 or Sir4 were generously provided by H. Madhani (University of California, San Francisco, San Francisco, CA) (Venkatasubrahmanyam et al., 2007). Yeast were grown to A600 ∼0.8 and DNA extracted by phenol-chloroform/ethanol precipitation following mechanical disruption. After purification, DNA was either undigested or digested overnight with the restriction enzyme DpnI (New England BioLabs, Ipswich, MA), which cleaves only when the adenine in its consensus site (GATC) is methylated. The percentage of methylated DNA was determined by using real-time PCR—with primers that flank candidate methylation sites—to compare relative signals for each amplicon in digested versus undigested DNA (with methylation of each site scoring as reduced amplification of an amplicon where DpnI-dependent cleavage had occurred). The percentages of methylation by Sir3-Dam or Sir4-Dam in each strain were normalized to their respective strain expressing the unfused Dam protein. Primer sequences are available on request.
Supplementary Material
Acknowledgments
We thank C. D Allis, B. Futcher, S. Gasser, S. Jasperson, and H. Madhani for reagents; S. Levy and C. Schaffer for technical assistance with RNA-seq; and K. Friedman for advice on telomere-length assays. A.L. was a Watson School of Biological Sciences Beckman Scholar. J.H.B. is supported by National Institutes of Health Grant GM080484 and by a W. M. Keck Young Biomedical Scholars Award. W.P.T. is supported by National Institutes of Health Grant GM067728. W.P.T and Z.Z are supported by VICC Cancer Center Grant P30CA68485.
Abbreviations used:
- CWI
cell wall integrity
- DamID
DNA adenine methyltransferase identification
- GO
gene ontology
- H2B–Ub
ubiquitylated H2B
- K4
lysine 4
- K79
lysine 79
- K123
lysine 123
- RPKM
reads per kilobase of open reading frame per million reads
- Ub
ubiquitin
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-05-0426) on June 16, 2011.
REFERENCES
- Ahn SH, Cheung WL, Hsu JY, Diaz RL, Smith MM, Allis CD. Sterile 20 kinase phosphorylates histone H2B at serine 10 during hydrogen peroxide-induced apoptosis in S. cerevisiae. Cell. 2005;120:25–36. doi: 10.1016/j.cell.2004.11.016. [DOI] [PubMed] [Google Scholar]
- Anna-Arriola SS, Herskowitz I. Isolation and DNA sequence of the STE13 gene encoding dipeptidyl aminopeptidase. Yeast. 1994;10:801–810. doi: 10.1002/yea.320100610. [DOI] [PubMed] [Google Scholar]
- Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc. B Methodol. 1995;57:289–300. [Google Scholar]
- Brickner DG, Cajigas I, Fondufe-Mittendorf Y, Ahmed S, Lee PC, Widom J, Brickner JH. H2A.Z-mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PLoS Biol. 2007;5:e81. doi: 10.1371/journal.pbio.0050081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner DG, Light W, Brickner JH. Quantitative localization of chromosomal loci by immunofluorescence. Methods Enzymol. 2010;470:569–580. doi: 10.1016/S0076-6879(10)70022-7. [DOI] [PubMed] [Google Scholar]
- Briggs SD, Bryk M, Strahl BD, Cheung WL, Davie JK, Dent SY, Winston F, Allis CD. Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes Dev. 2001;15:3286–3295. doi: 10.1101/gad.940201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bystricky K, Laroche T, van Houwe G, Blaszczyk M, Gasser SM. Chromosome looping in yeast: telomere pairing and coordinated movement reflect anchoring efficiency and territorial organization. J Cell Biol. 2005;168:375–387. doi: 10.1083/jcb.200409091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabal GG, et al. SAGA interacting factors confine sub-diffusion of transcribed genes to the nuclear envelope. Nature. 2006;441:770–773. doi: 10.1038/nature04752. [DOI] [PubMed] [Google Scholar]
- Chandrasekharan MB, Huang F, Sun ZW. Ubiquitination of histone H2B regulates chromatin dynamics by enhancing nucleosome stability. Proc Natl Acad Sci USA. 2009;106:16686–16691. doi: 10.1073/pnas.0907862106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekharan MB, Huang F, Sun ZW. Histone H2B ubiquitination and beyond: regulation of nucleosome stability, chromatin dynamics and the trans-histone H3 methylation. Epigenetics. 2010;5:460–468. doi: 10.4161/epi.5.6.12314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HY, Sun JM, Zhang Y, Davie JR, Meistrich ML. Ubiquitination of histone H3 in elongating spermatids of rat testes. J Biol Chem. 1998;273:13165–13169. doi: 10.1074/jbc.273.21.13165. [DOI] [PubMed] [Google Scholar]
- Chu Y, Simic R, Warner MH, Arndt KM, Prelich G. Regulation of histone modification and cryptic transcription by the Bur1 and Paf1 complexes. EMBO J. 2007;26:4646–4656. doi: 10.1038/sj.emboj.7601887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collart MA, Oliviero S. Preparation of yeast RNA. Curr Protoc Mol Biol. 2001 doi: 10.1002/0471142727.mb1312s23. Chapter 13, Unit13.12. [DOI] [PubMed] [Google Scholar]
- Duncan D, Prodduturi N, Zhang B. WebGestalt2: an updated and expanded version of the Web-based Gene Set Analysis Toolkit. BMC Bioinformatics. 2010;11:P10. [Google Scholar]
- Dwight SS, et al. Saccharomyces Genome Database (SGD) provides secondary gene annotation using the Gene Ontology (GO) Nucleic Acids Res. 2002;30:69–72. doi: 10.1093/nar/30.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatbonton T, Imbesi M, Nelson M, Akey JM, Ruderfer DM, Kruglyak L, Simon JA, Bedalov A. Telomere length as a quantitative trait: genome-wide survey and genetic mapping of telomere length-control genes in yeast. PLoS Genet. 2006;2:e35. doi: 10.1371/journal.pgen.0020035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng F, Tansey WP. Polyubiquitylation of histone H2B. Mol Biol Cell. 2008;19:3616–3624. doi: 10.1091/mbc.E08-01-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Schiestl RH. Large-scale high-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat Protoc. 2007;2:38–41. doi: 10.1038/nprot.2007.15. [DOI] [PubMed] [Google Scholar]
- Goldknopf IL, Busch H. Isopeptide linkage between nonhistone and histone 2A polypeptides of chromosomal conjugate-protein A24. Proc Natl Acad Sci USA. 1977;74:864–868. doi: 10.1073/pnas.74.3.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein AL, McCusker JH. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 1999;15:1541–1553. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Hediger F, Neumann FR, Van Houwe G, Dubrana K, Gasser SM. Live imaging of telomeres: yKu and Sir proteins define redundant telomere-anchoring pathways in yeast. Curr Biol. 2002;12:2076–2089. doi: 10.1016/s0960-9822(02)01338-6. [DOI] [PubMed] [Google Scholar]
- Hirano Y, Fukunaga K, Sugimoto K. Rif1 and rif2 inhibit localization of tel1 to DNA ends. Mol Cell. 2009;33:312–322. doi: 10.1016/j.molcel.2008.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Hwang WW, Venkatasubrahmanyam S, Ianculescu AG, Tong A, Boone C, Madhani HD. A conserved RING finger protein required for histone H2B monoubiquitination and cell size control. Mol Cell. 2003;11:261–266. doi: 10.1016/s1097-2765(02)00826-2. [DOI] [PubMed] [Google Scholar]
- Johnston HD, Foote C, Santeford A, Nothwehr SF. Golgi-to-late endosome trafficking of the yeast pheromone processing enzyme Ste13p is regulated by a phosphorylation site in its cytosolic domain. Mol Biol Cell. 2005;16:1456–1468. doi: 10.1091/mbc.E04-07-0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones GM, Stalker J, Humphray S, West A, Cox T, Rogers J, Dunham I, Prelich G. A systematic library for comprehensive overexpression screens in Saccharomyces cerevisiae. Nat Methods. 2008;5:239–241. doi: 10.1038/nmeth.1181. [DOI] [PubMed] [Google Scholar]
- Kao CF, Hillyer C, Tsukuda T, Henry K, Berger S, Osley MA. Rad6 plays a role in transcriptional activation through ubiquitylation of histone H2B. Genes Dev. 2004;18:184–195. doi: 10.1101/gad.1149604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer KM, Haber JE. New telomeres in yeast are initiated with a highly selected subset of TG1-3 repeats. Genes Dev. 1993;7:2345–2356. doi: 10.1101/gad.7.12a.2345. [DOI] [PubMed] [Google Scholar]
- Kuo MH, Zhou J, Jambeck P, Churchill ME, Allis CD. Histone acetyltransferase activity of yeast Gcn5p is required for the activation of target genes in vivo. Genes Dev. 1998;12:627–639. doi: 10.1101/gad.12.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B. Aligning short sequencing reads with Bowtie. Curr Protoc Bioinformatics. 2010:17. doi: 10.1002/0471250953.bi1107s32. Chapter 11, Unit 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Shukla A, Schneider J, Swanson SK, Washburn MP, Florens L, Bhaumik SR, Shilatifard A. Histone crosstalk between H2B monoubiquitination and H3 methylation mediated by COMPASS. Cell. 2007;131:1084–1096. doi: 10.1016/j.cell.2007.09.046. [DOI] [PubMed] [Google Scholar]
- Levin DE. Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2005;69:262–291. doi: 10.1128/MMBR.69.2.262-291.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Maillet L, Boscheron C, Gotta M, Marcand S, Gilson E, Gasser SM. Evidence for silencing compartments within the yeast nucleus: a role for telomere proximity and Sir protein concentration in silencer-mediated repression. Genes Dev. 1996;10:1796–1811. doi: 10.1101/gad.10.14.1796. [DOI] [PubMed] [Google Scholar]
- Marshall M, Mahoney D, Rose A, Hicks JB, Broach JR. Functional domains of SIR4, a gene required for position effect regulation in Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:4441–4452. doi: 10.1128/mcb.7.12.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Murr R. Interplay between different epigenetic modifications and mechanisms. Adv Genet. 2010;70:101–141. doi: 10.1016/B978-0-12-380866-0.60005-8. [DOI] [PubMed] [Google Scholar]
- Mutiu AI, Hoke SM, Genereaux J, Liang G, Brandl CJ. The role of histone ubiquitylation and deubiquitylation in gene expression as determined by the analysis of an HTB1(K123R) Saccharomyces cerevisiae strain. Mol Genet Genomics. 2007;277:491–506. doi: 10.1007/s00438-007-0212-6. [DOI] [PubMed] [Google Scholar]
- Nagalakshmi U, Waern K, Snyder M. RNA-Seq: a method for comprehensive transcriptome analysis. Curr Protoc Mol Biol. 2010;Chapter 4:11–13. doi: 10.1002/0471142727.mb0411s89. Unit 4.11.1–4. [DOI] [PubMed] [Google Scholar]
- Ng HH, Feng Q, Wang H, Erdjument-Bromage H, Tempst P, Zhang Y, Struhl K. Lysine methylation within the globular domain of histone H3 by Dot1 is important for telomeric silencing and Sir protein association. Genes Dev. 2002;16:1518–1527. doi: 10.1101/gad.1001502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nislow C, Ray E, Pillus L. SET1, a yeast member of the trithorax family, functions in transcriptional silencing and diverse cellular processes. Mol Biol Cell. 1997;8:2421–2436. doi: 10.1091/mbc.8.12.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palladino F, Laroche T, Gilson E, Axelrod A, Pillus L, Gasser SM. SIR3 and SIR4 proteins are required for the positioning and integrity of yeast telomeres. Cell. 1993;75:543–555. doi: 10.1016/0092-8674(93)90388-7. [DOI] [PubMed] [Google Scholar]
- Pavri R, Zhu B, Li G, Trojer P, Mandal S, Shilatifard A, Reinberg D. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell. 2006;125:703–717. doi: 10.1016/j.cell.2006.04.029. [DOI] [PubMed] [Google Scholar]
- Pinskaya M, Gourvennec S, Morillon A. H3 lysine 4 di- and tri-methylation deposited by cryptic transcription attenuates promoter activation. EMBO J. 2009;28:1697–1707. doi: 10.1038/emboj.2009.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rine J, Herskowitz I. Four genes responsible for a position effect on expression from HML and HMR in Saccharomyces cerevisiae. Genetics. 1987;116:9–22. doi: 10.1093/genetics/116.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robzyk K, Recht J, Osley MA. Rad6-dependent ubiquitination of histone H2B in yeast. Science. 2000;287:501–504. doi: 10.1126/science.287.5452.501. [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H, Bannister AJ, Dehe PM, Geli V, Kouzarides T. Methylation of H3 lysine 4 at euchromatin promotes Sir3p association with heterochromatin. J Biol Chem. 2004;279:47506–47512. doi: 10.1074/jbc.M407949200. [DOI] [PubMed] [Google Scholar]
- Sexton T, Schober H, Fraser P, Gasser SM. Gene regulation through nuclear organization. Nat Struct Mol Biol. 2007;14:1049–1055. doi: 10.1038/nsmb1324. [DOI] [PubMed] [Google Scholar]
- Stellwagen AE, Haimberger ZW, Veatch JR, Gottschling DE. Ku interacts with telomerase RNA to promote telomere addition at native and broken chromosome ends. Genes Dev. 2003;17:2384–2395. doi: 10.1101/gad.1125903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Straight AF, Belmont AS, Robinett CC, Murray AW. GFP tagging of budding yeast chromosomes reveals that protein-protein interactions can mediate sister chromatid cohesion. Curr Biol. 1996;6:1599–1608. doi: 10.1016/s0960-9822(02)70783-5. [DOI] [PubMed] [Google Scholar]
- Sun ZW, Allis CD. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature. 2002;418:104–108. doi: 10.1038/nature00883. [DOI] [PubMed] [Google Scholar]
- Taddei A, Gasser SM. Multiple pathways for telomere tethering: functional implications of subnuclear position for heterochromatin formation. Biochim Biophys Acta. 2004;1677:120–128. doi: 10.1016/j.bbaexp.2003.11.014. [DOI] [PubMed] [Google Scholar]
- Van Mullem V, Wery M, De Bolle X, Vandenhaute J. Construction of a set of Saccharomyces cerevisiae vectors designed for recombinational cloning. Yeast. 2003;20:739–746. doi: 10.1002/yea.999. [DOI] [PubMed] [Google Scholar]
- van Steensel B, Henikoff S. Identification of in vivo DNA targets of chromatin proteins using tethered Dam methyltransferase. Nat Biotechnol. 2000;18:424–428. doi: 10.1038/74487. [DOI] [PubMed] [Google Scholar]
- Varier RA, Timmers HT. Histone lysine methylation and demethylation pathways in cancer. Biochim Biophys Acta. 2010;8515:75–89. doi: 10.1016/j.bbcan.2010.10.002. [DOI] [PubMed] [Google Scholar]
- Venkatasubrahmanyam S, Hwang WW, Meneghini MD, Tong AH, Madhani HD. Genome-wide, as opposed to local, antisilencing is mediated redundantly by the euchromatic factors Set1 and H2A.Z. Proc Natl Acad Sci USA. 2007;104:16609–16614. doi: 10.1073/pnas.0700914104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Feng Z, Wang X, Zhang X. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010;26:136–138. doi: 10.1093/bioinformatics/btp612. [DOI] [PubMed] [Google Scholar]
- West MH, Bonner WM. Histone 2B can be modified by the attachment of ubiquitin. Nucleic Acids Res. 1980;8:4671–4680. doi: 10.1093/nar/8.20.4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieser R, Adam G, Wagner A, Schuller C, Marchler G, Ruis H, Krawiec Z, Bilinski T. Heat shock factor-independent heat control of transcription of the CTT1 gene encoding the cytosolic catalase T of Saccharomyces cerevisiae. J Biol Chem. 1991;266:12406–12411. [PubMed] [Google Scholar]
- Wood A, et al. Bre1, an e3 ubiquitin ligase required for recruitment and substrate selection of rad6 at a promoter. Mol Cell. 2003a;11:267–274. doi: 10.1016/s1097-2765(02)00802-x. [DOI] [PubMed] [Google Scholar]
- Wood A, Schneider J, Dover J, Johnston M, Shilatifard A. The Paf1 complex is essential for histone monoubiquitination by the Rad6/Bre1 complex, which signals for histone methylation by COMPASS and Dot1p. J Biol Chem. 2003b doi: 10.1074/jbc.C300269200. [DOI] [PubMed] [Google Scholar]
- Wood A, Schneider J, Dover J, Johnston M, Shilatifard A. The Bur1/Bur2 complex is required for histone H2B monoubiquitination by Rad6/Bre1 and histone methylation by COMPASS. Mol Cell. 2005;20:589–599. doi: 10.1016/j.molcel.2005.09.010. [DOI] [PubMed] [Google Scholar]
- Wyce A, Xiao T, Whelan KA, Kosman C, Walter W, Eick D, Hughes TR, Krogan NJ, Strahl BD, Berger SL. H2B ubiquitylation acts as a barrier to Ctk1 nucleosomal recruitment prior to removal by Ubp8 within a SAGA-related complex. Mol Cell. 2007;27:275–288. doi: 10.1016/j.molcel.2007.01.035. [DOI] [PubMed] [Google Scholar]
- Yan Q, Dutt S, Xu R, Graves K, Juszczynski P, Manis JP, Shipp MA. BBAP monoubiquitylates histone H4 at lysine 91 and selectively modulates the DNA damage response. Mol Cell. 2009;36:110–120. doi: 10.1016/j.molcel.2009.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.