Abstract
Anticancer drugs are effective against tumors that depend on the molecular target of the drug. Known targets of cytotoxic anticancer drugs are involved in cell proliferation; drugs acting on such targets are ineffective against nonproliferating tumor cells, survival of which leads to eventual therapy failure. Function-based genomic screening identified the coatomer protein complex ζ1 (COPZ1) gene as essential for different tumor cell types but not for normal cells. COPZ1 encodes a subunit of coatomer protein complex 1 (COPI) involved in intracellular traffic and autophagy. The knockdown of COPZ1, but not of COPZ2 encoding isoform coatomer protein complex ζ2, caused Golgi apparatus collapse, blocked autophagy, and induced apoptosis in both proliferating and nondividing tumor cells. In contrast, inhibition of normal cell growth required simultaneous knockdown of both COPZ1 and COPZ2. COPZ2 (but not COPZ1) was down-regulated in the majority of tumor cell lines and in clinical samples of different cancer types. Reexpression of COPZ2 protected tumor cells from killing by COPZ1 knockdown, indicating that tumor cell dependence on COPZ1 is the result of COPZ2 silencing. COPZ2 displays no tumor-suppressive activities, but it harbors microRNA 152, which is silenced in tumor cells concurrently with COPZ2 and acts as a tumor suppressor in vitro and in vivo. Silencing of microRNA 152 in different cancers and the ensuing down-regulation of its host gene COPZ2 offer a therapeutic opportunity for proliferation-independent selective killing of tumor cells by COPZ1-targeting agents.
Keywords: cancer targets, genetic suppressor elements
Developing drugs against tumor-specific molecular targets is a primary goal of current cancer pharmacology. Target-specific agents are effective against tumors that depend on the function of the gene targeted by the drug, e.g., trastuzumab in breast cancers that overexpress human epidermal growth factor receptor 2 (1), gefitinib in tumors with EGFR mutations (2), and all-trans retinoic acid in acute promyeolocytic leukemia, a disease that involves a translocation of retinoic acid receptor α (3). Tumor dependence on a gene that is necessary for tumors but not for normal cells has been termed “oncogene addiction” (4), but there are genes that are required selectively by tumor cells but cannot be classified as oncogenes (5). Most existing anticancer drugs act either on DNA or on protein targets involved in cell proliferation. Such drugs kill proliferating tumor cells but are ineffective against the nondividing tumor cell population, which includes growth-arrested cells that secrete various tumor-promoting factors (6), dormant cells capable of eventual reentry into cell cycle (7), and resting tumor stem cells (8). Proliferating and nondividing tumor cells, however, share common genetic and epigenetic changes, and some of the tumor-specific targets in proliferating cells also may be required for the survival of nondividing tumor cells. With an increase in the number of proteins targeted by drugs in the preclinical pipeline and the improvement in methods for target-based de novo drug design (9), identification of additional categories of tumor-specific targets can guide the development of new classes of anticancer agents.
Genes playing essential roles in tumor cells can be revealed through the selection of phenotypically active transdominant genetic inhibitors, gene-derived DNA sequences that block the function of their cognate gene. These sequences include antisense cDNAs (10), genetic suppressor elements (GSEs, short cDNA fragments that express dominant negative protein fragments or antisense RNA segments) (5, 11, 12), and shRNAs that inhibit gene expression through RNA interference (13–16). In the present article, we report the identification, through GSE selection, of a tumor-specific gene target, inhibition of which kills both proliferating and nondividing tumor cells. This gene, coatomer protein complex ζ1 (COPZ1), encodes one of two isoforms of the ζ subunit of coatomer protein complex 1 (COPI), a secretory vesicle coat protein complex involved in Golgi apparatus and endoplasmic reticulum traffic, endosome maturation, and autophagy (17, 18). We show that tumor cell dependence on COPZ1 is caused by the widespread down-regulation of its isoform coatomer protein complex ζ2 (COPZ2) in different types of cancer, as a corollary of the silencing of a tumor-suppressive microRNA (miR), miR-152, encoded within the COPZ2 gene.
Results
Selection of Growth-Inhibitory GSEs.
A GSE library comprising cDNA fragments of the human transcriptome (average length 135 bp) was prepared from a mixture of normalized (reduced-redundance) cDNA preparations of 18 cell lines derived from different types of cancer and leukemia (SI Methods). The GSE library was cloned into a tetracycline/doxycycline-inducible lentiviral vector. As recipients for the selection of growth-inhibitory GSEs, we used four tumor cell lines: MDA-MB-231 (breast cancer), PC3(prostate cancer), T24 (bladder carcinoma), and HT1080 (fibrosarcoma), as well as human telomerase reverse transcriptase (hTERT)-immortalized BJ normal foreskin fibroblasts (BJ-hTERT). To enable doxycycline-inducible GSE expression, the cell lines were modified by the introduction of tTR-KRAB, a tetracycline/doxycycline-sensitive repressor. Cells transduced with the GSE library were subjected to selection for doxycycline-dependent resistance to BrdU suicide, a procedure that selects for cells carrying growth-inhibitory GSEs (5, 16). The library-derived cDNA fragments were amplified by PCR from genomic DNA of the unselected and BrdU-selected cells. PCR products were analyzed by high-throughput sequencing. The cDNA fragment sequences were matched to their cognate genes and compared in the unselected and BrdU-selected cells to identify genes that give rise to putative GSEs enriched by BrdU suicide selection. Genes targeted by the enriched sequences then were tested for their role in cell proliferation by testing the effects on cell growth of synthetic siRNAs targeting the same genes. The complete results of this analysis will be presented elsewhere.
COPZ1 Knockdown Causes Golgi Apparatus Collapse, Inhibition of Autophagy, and Tumor-Specific Cell Death Independent of Cell Proliferation.
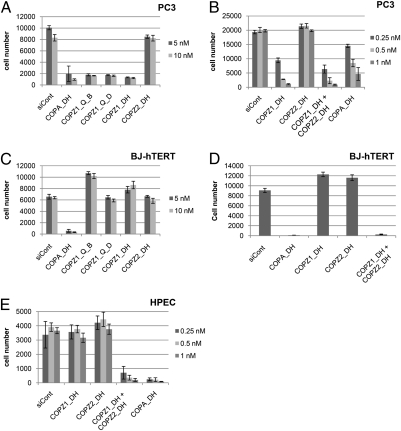
Among the genes enriched by growth-inhibitory GSE selection, we investigated in greater detail COPZ1, which encodes the ζ1 subunit of the coatomer protein complex COPI. COPZ1 was targeted by seven antisense-oriented and one sense-oriented putative GSEs, which were enriched by BrdU selection in all four tumor cell lines but not in immortalized normal BJ-hTERT fibroblasts. We investigated the role of COPZ1 and other COPI subunits in the growth of normal and tumor cells by transfection with different siRNAs that target the genes for these subunits, reducing their RNA levels by >90% (Fig. S1A). Fig. 1 A and B shows that the knockdown of COPZ1 by siRNAs obtained from different sources inhibited the growth of PC3 prostate carcinoma cells, as did the knockdown of another COPI component, COPA. However, no growth inhibition resulted from the knockdown of COPZ2, which encodes ζ2, the isoform of COPZ1 gene product. In contrast to PC3, the growth of immortalized normal BJ-hTERT fibroblasts (Fig. 1 C and D) or normal human prostate epithelial cells (Fig. 1E) was not inhibited by the knockdown of COPZ1 or COPZ2 alone. However, normal cell proliferation was inhibited by simultaneous knockdown of COPZ1 and COPZ2 or by COPA knockdown (Fig. 1 C–E). The siRNA growth-inhibition assays were extended to the other COPI subunit genes (four siRNAs per gene, all from Qiagen), and a panel of tumor cell lines including PC3, HT1080, MDA-MB-231, T24, HeLa, and in vitro-transformed BJ-ELR fibroblasts (19). COPZ1-targeting siRNAs inhibited the growth of all six transformed cell lines but not of BJ-hTERT, whereas none of siRNAs targeting the other COPI proteins exhibited tumor selectivity (Fig. S1B). Hence, COPZ1 is required for cell growth in a wide spectrum of tumor cell lines but not in normal cells.
Fig. 1.
Effects of COPZ1, COPZ2, and COPA knockdown on tumor and normal cells. (A) siRNAs targeting COPA, COPZ1, COPZ2, or no human genes (siCont), obtained from Dharmacon (DH) or Qiagen (Q), were transfected into PC3 cells at the indicated concentrations. Cell numbers were determined 4 d posttransfection by flow cytometry (in three independent transfections) and are expressed as mean ± SD. (B) Experiment similar to A carried out with lower siRNA concentrations, demonstrating dose-dependent inhibition, and including a combination of COPZ1 and COPZ2 siRNAs. Cell numbers were measured 8 d posttransfection (in three independent transfections) and are expressed as mean ± SD. (C) Effects of the siRNAs in A on the proliferation of BJ-HTERT cells. Cell numbers were measured 7 d posttransfection (in triplicate) and are expressed as mean ± SD. (D) Effects of COPA, COPZ1, and COPZ2 siRNAs and a combination of COPZ1 and COPZ2 siRNAs (all at 5-nM concentrations) on the proliferation of BJ-HTERT cells. Cell numbers were measured 7 d posttransfection (six replicates) and are expressed as mean ± SD. (E) Effects of COPA, COPZ1, and COPZ2 siRNAs and a combination of COPZ1 and COPZ2 siRNAs (at the same concentrations as in B on the proliferation of normal human prostate epithelial cells (HPEC). Cell numbers were measured 8 d posttransfection (in triplicate) and are expressed as mean ± SD.
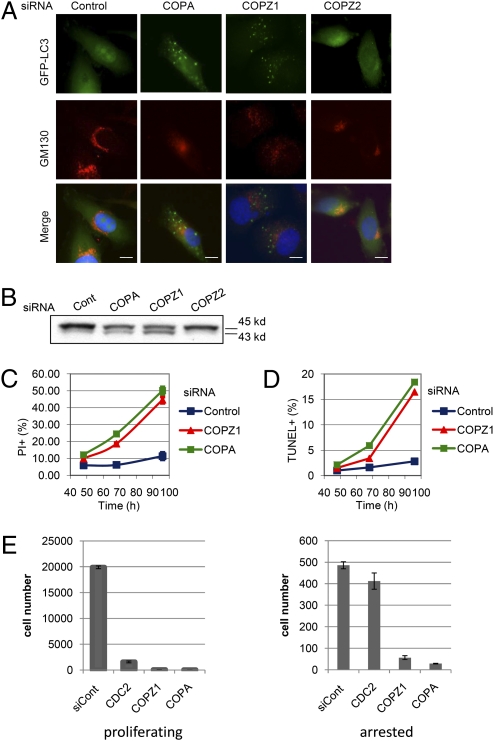
The knockdown of other COPI proteins was reported to arrest cellular traffic, cause a collapse of endoplasmic reticulum and Golgi apparatus (20), and inhibit the maturation of the autophagosome, an essential step in autophagy (18). To determine if COPZ1 knockdown has the same effects, we transfected COPZ1, COPZ2, COPA, and control siRNAs into PC3 cells expressing LC3, an autophagosome marker fused with GFP (GFP-LC3) (21). The knockdown effects were analyzed 72 h later by fluorescence microscopy based on GFP-LC3 localization and on immunofluorescence staining for a Golgi apparatus marker, GM130. COPZ1 and COPA, but not control or COPZ2, siRNAs caused fragmentation and disappearance of GM130+ Golgi structures (Fig. 2A). Knockdown of COPA and COPZ1 (but not of COPZ2) also resulted in the accumulation of GFP-LC3+ puncta (Fig. 2A) and of a 43-kD phosphatidilethanolamine-conjugated form of GFP-LC3 (Fig. 2B), indicative of autophagosome accumulation and inhibition of autophagic flux (22). The same effects were observed in normal BJ-hTERT fibroblasts upon the knockdown of COPZ1 and COPZ2 together (but not alone) and upon COPA knockdown (Fig. S2).
Fig. 2.
Effects of COPZ1 knockdown on Golgi apparatus, autophagosomes, and cell death. (A) PC3 cells expressing the autophagosome marker GFP-LC3 were transfected with control siRNA or siRNAs targeting COPA, COPZ1, or COPZ2 and were analyzed by fluorescence microscopy for GFP fluorescence (green), indirect immunofluorescence staining for Golgi marker GM130 (red), and nuclear DNA staining with DAPI 72 h posttransfection with the indicated siRNAs. (Scale bars: 10 μM.) (B) GFP-LC3 electrophoretic mobility of the cells in A analyzed by immunoblotting with anti-GFP antibody. (C) Changes in the number of membrane-permeable (PI+) PC-3 cells upon transfection with control siRNA or siRNAs targeting COPA or COPZ1, as determined by flow cytometry (mean ± SD, triplicate measurements). (D) Changes in the number of apoptotic (TUNEL+) PC-3 cells upon transfection with control siRNA or siRNAs targeting COPA or COPZ1, as determined by flow cytometry (mean ± SD, triplicate measurements). (E) Effects of COPZ1, COPA, and CDC2 siRNAs on cell number of proliferating and growth-arrested HT1080 cells. HT1080 p21-9 cells with IPTG-inducible expression of the cell-cycle inhibitor p21 were transfected with the indicated siRNAs. Transfected cells were pIated in the absence (proliferating) or in the presence (arrested) of 50 μM IPTG, in triplicate; cell numbers were determined 5 d later. Data are shown as mean ± SD.
Both Golgi apparatus disruption that leads to endoplasmic reticulum stress (23) and the interference with autophagy can be cytotoxic (24, 25). We have analyzed the ability of COPZ1 and COPA siRNAs to induce cell death and apoptosis. This analysis was conducted by flow cytometric assays for membrane permeability, as defined by the uptake of the fluorescent dye propidium iodide (PI) and by TUNEL staining for apoptotic cells. Transfection with COPZ1 or COPA siRNA into PC3 cells increased the number of membrane-permeable (PI+) (Fig. 2C) and apoptotic (TUNEL+) cells (Fig. 2D) relative to cells transfected with control siRNA. Time-lapse video microscopy showed that cells transfected with COPZ1 siRNA undergo apoptosis (shrinking and blebbing) either directly or after first rounding up and remaining rounded for 10–20 h (Movie S1). Although cell rounding followed by apoptosis superficially resembles mitotic catastrophe (26), DAPI staining did not show chromatin condensation in the rounded cells, indicating that these cells were not in mitosis. Hence COPZ1 knockdown produces the phenotypic effects expected from COPI inhibition, and these effects—inhibition of autophagy and the disruption of Golgi apparatus—lead to apoptotic cell death upon COPZ1 knockdown.
Because the COPI complex affects processes distinct from cell-cycle progression, we asked if COPZ1 knockdown would affect nondividing tumor cells that are resistant to agents targeting cell proliferation. For this analysis, we used HT1080 p21-9, a derivative of HT1080 fibrosarcoma cells expressing the cell-cycle inhibitor protein p21 (CDKN1A) from a promoter inducible by isopropyl β-d-1-thiogalactopyranoside (IPTG) (27). HT1080 p21-9 cells were transfected with control siRNA and siRNAs targeting COPZ1, COPA, or CDC2 (CDK1), an essential mediator of cell-cycle progression (28). The transfected cells were plated in the absence or in the presence of IPTG that induces p21 expression and cell-cycle arrest within 12–16 h. CDC2 siRNA decreased the number of proliferating cells but had almost no effect on the growth-arrested cells. In contrast, the knockdown of COPZ1 or COPA strongly decreased the numbers of both proliferating and growth-arrested cells (Fig. 2E). Hence, inhibition of COPI kills both dividing and nondividing tumor cells.
COPZ1 Dependence of Tumor Cells Results from COPZ2 Down-Regulation in Cancers.
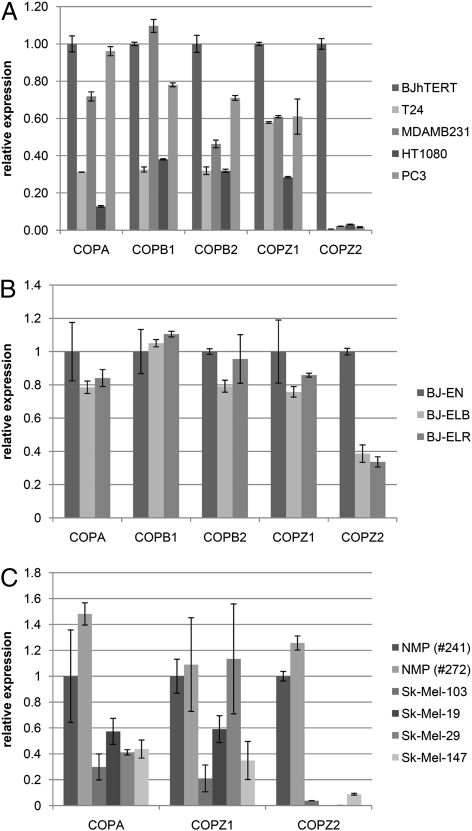
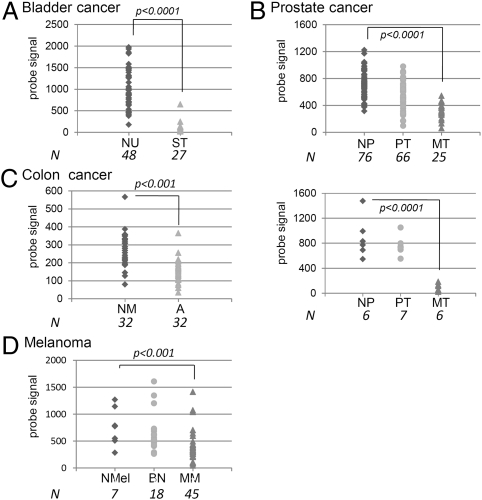
To understand why COPZ1 is the only COPI subunit that is selectively required by tumor cells, we used quantitative RT-PCR (qRT-PCR) to measure mRNA levels of different COPI component proteins in four tumor cell lines and BJ-hTERT (Fig. 3A), in a series of isogenic BJ fibroblast lines that includes normal immortalized BJ-EN, partially transformed BJ-ELB, and fully transformed BJ-ELR (19) (Fig. 3B), and in two samples of normal melanocytes and four melanoma cell lines (Fig. 3C). COPA, COPB1, COPB2, and COPZ1 were expressed at comparable levels in all the cell lines. Strikingly, the expression of COPZ2 was almost undetectable in all the tumor cell lines and was strongly decreased in BJ-ELB and BJ-ELR cells relative to BJ-hTERT cells. We analyzed the expression of COPZ1, COPZ2, and several other COPI subunits in a panel of normal human tissues (Table S1) and expanded the analysis of COPZ2 and COPZ1 mRNA expression to additional tumor and leukemia cell lines (Table S2). COPZ1 expression was relatively uniform among the normal and tumor tissues. COPZ2 showed comparable expression levels among normal tissues (with the lowest levels observed in the thymus, spleen, and ovary) (Table S1), but COPZ2 levels were drastically decreased in 10 tumor cell lines; the only tumor line that maintained COPZ2 expression (Table S2) was a relatively benign WM 793 melanoma (29). We also analyzed the data on COPZ1 and COPZ2 gene expression in microarray studies included in the Gene Expression Omnibus (GEO) database, comparing normal, benign, and malignant tissues of different cancers. COPZ1 expression showed no significant tumor-specific changes, but in several studies COPZ2 expression was decreased significantly in association with carcinogenesis or tumor progression. In particular, COPZ2 was strongly down-regulated in superficial bladder carcinoma relative to normal urothelium (Fig. 4A), in metastatic prostate cancer relative to primary cancers and benign prostatic tissues (Fig. 4B, two studies), in colorectal adenoma relative to normal mucosa (Fig. 4C), and in malignant melanoma relative to benign nevi or normal melanocytes (Fig. 4D). Hence, COPZ2 down-regulation is a broad and general event in different forms of cancer.
Fig. 3.
Down-regulation of the COPZ2 gene in transformed cell lines. (A) qRT-PCR analysis of expression of the indicated COPI subunit genes in tumor cell lines and BJ-hTERT fibroblasts. Expression is presented relative to BJ-hTERT; data are shown as mean ± SD. (B) qRT-PCR analysis of expression of the indicated COPI subunit genes in immortalized normal BJ-EN fibroblasts and their partially transformed (BJ-ELB) and fully transformed (BJ-ELR) derivatives. Expression is presented relative to BJ-EN. (C) qRT-PCR analysis of expression of the indicated COPI subunit genes in two normal melanocyte preparations (NMP) and four melanoma cell lines. Expression is presented relative to NMP 241.
Fig. 4.
COPZ2 expression in normal, benign, and malignant tissues of different tumor types (microarray data from GEO database). P values (student's t test) are indicated for significant differences between the groups. (A) Bladder cancer study (43): normal urothelium (NU) and superficial tumors (ST). (B) Two prostate cancer studies (44, 45): normal prostate (NP), primary tumors (PT), and metastatic tumors (MT). (C) Colon cancer study (46): normal mucosa (NM) and adenocarcinomas (A). (D) Melanoma study (47): normal melanocytes (NMel), benign nevi (BN), and metastatic melanoma (MM).
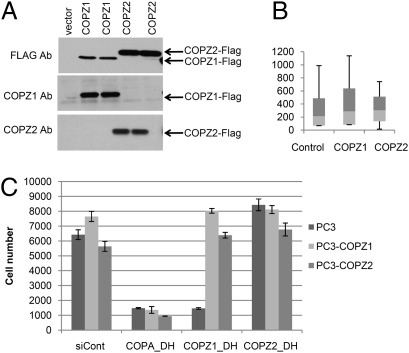
If COPZ1 and COPZ2 isoforms can substitute for each other, COPI complexes should remain functional if either COPZ1 or COPZ2 is expressed. Indeed, the knockdown of COPZ1 or COPZ2 alone had no effect on normal BJ-hTERT or human prostate epithelial cells that express both genes, but simultaneous knockdown of COPZ1 and COPZ2 drastically inhibited the growth of these cells, causing Golgi apparatus collapse and inhibition of autophagy (Fig. 1 C–E and Fig. S2). To determine whether tumor cell sensitivity to COPZ1 knockdown results from the loss of COPZ2 in tumor cells, we asked if reexpression of COPZ2 in PC3 cells would protect them from killing by COPZ1 siRNA. FLAG-tagged COPZ1 and COPZ2 proteins were introduced into PC3 cells by lentiviral transduction, leading to very high expression levels (Fig. 5A). COPZ1 or COPZ2 overexpression had no effect on PC3 cell growth in culture or on the size range of tumors formed by PC3 xenografts in nude mice (Fig. 5B). Transduction with COPZ1 or COPZ2 also had no effect on in vitro growth of HeLa and MDA-MB-231 cell lines. Fig. 5C shows the effects of COPA siRNA, COPZ1 siRNA, and COPZ2 siRNA on PC3 cells transduced with the insert-free vector or with the vectors expressing COPZ1 or COPZ2. COPZ2 siRNA did not inhibit cell growth, whereas COPA siRNA inhibited the proliferation of all three cell populations. COPZ1 siRNA inhibited the proliferation of cells transduced with the insert-free vector, but overexpression of either COPZ1 or COPZ2 rendered cells resistant to COPZ1 knockdown (Fig. 5C). These results demonstrate that tumor cell dependence on COPZ1 is a consequence of tumor-specific COPZ2 silencing.
Fig. 5.
Relationship between COPZ1 and COPZ2 expression and siRNA sensitivity. (A) Immunoblotting of COPZ1 and COPZ2 proteins in PC3 cells transduced with control lentivirus or with lentiviral vectors expressing FLAG-tagged COPZ1 or COPZ2 and probed with FLAG, COPZ1, and COPZ2 antibodies. (B) In vivo growth of PC3 xenograft tumors transduced with a control lentiviral vector or with vectors expressing COPZ1 or COPZ2. Data shown are tumor weights (mean ± SD) at the end of the experiment (41 d postinoculation). (C) Effects of the indicated siRNAs on the proliferation of PC3 cells transduced with control siRNA (siCont) or vectors expressing COPA, COPZ1, or COPZ2 (six replicates). Cell numbers were measured 4 d posttransfection and are expressed as mean ± SD.
COPZ2 Down-Regulation in Cancers Is Associated with the Silencing of a Tumor-Suppressive miRNA Encoded Within COPZ2.
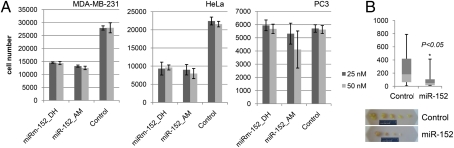
Although the widespread COPZ2 silencing in tumor cells suggests tumor-suppressive activity, high-level COPZ2 expression had no detrimental effect on tumor cell growth in vitro or in vivo, as described above, indicating that COPZ2 is not a tumor suppressor. However, the first intron of the COPZ2 gene encodes the precursor of an miRNA, miR-152 (30). miR-152 was reported to be down-regulated in several types of cancer (31–34) and to display certain tumor-suppressive activities (33, 35). We measured miR-152 expression in different tumor cell lines and found that miR-152, like COPZ2, was down-regulated in all the lines except WM 793 (the only tumor line that expressed COPZ2) (Table S2). We analyzed the effects of miR-152 on tumor cell lines by transfection with two different miR-152 precursors. miR-152 inhibited the growth of HeLa and MDA-MB-231 cells but not PC3 cells (Fig. 6A). Stable miR-152 expression in PC3 cells from a lentiviral vector also failed to inhibit cell growth in culture, but PC3 cells overexpressing miR-152 showed significantly decreased xenograft tumor growth in nude mice relative to PC3 cells transduced with a control vector (Fig. 6B). Hence, miR-152 displays both expression changes and biological activities indicative of a broad-spectrum tumor suppressor, and COPZ2 down-regulation in cancers leads to concurrent silencing of its host gene COPZ2, with the ensuing tumor cell dependence on COPZ1.
Fig. 6.
Effects of miR-152 in tumor cells. (A) Effects of transfection with miR-152 precursors from Dharmacon (DH) or Ambion (AM) and miRNA mimic negative control (Dharmacon) on the proliferation of MDA-MB-231, HeLa, and PC3 cells (in triplicate). Cell numbers were measured 7 d posttransfection and are expressed relative to cells transfected with negative control. Data are shown as mean ± SD. (B) In vivo growth of PC3 xenograft tumors transduced with a control lentiviral vector or with a vector expressing miR-152 precursor. (Upper) Tumor weights (mean ± SD) at the end of the experiment (42 d postinoculation). (Lower) Photographs of tumors.
Discussion
The search for tumor-selective molecular targets often involves the concept of “oncogene addiction” (4), in which a tumor-specific target is expected to have oncogenic activity. In the present study, we identified COPZ1 as a promising target, which is required by different tumor cell types but is not essential for normal cells. COPZ1, however, has no known oncogenic activity, and its overexpression does not affect tumor cell growth in vitro or in vivo. Instead, the tumor cell requirement for COPZ1 is the result of the silencing of its isoform COPZ2, which we found to be broadly down-regulated in different cancers, in vitro and in vivo. Our results demonstrate that COPZ1 can substitute functionally for the lack of COPZ2 in tumor cells. This substitution, however, makes COPI function and survival of tumor cells dependent on COPZ1, whereas COPZ2 expression in normal cells allows them to maintain the COPI function and to survive upon COPZ1 inhibition. We suggest a term “isoform addiction” to designate this type of tumor-cell dependence on a specific protein isoform.
Although COPZ2 is down-regulated selectively in tumor cells, its overexpression did not inhibit tumor cell growth either in ;vitro or in vivo. COPZ2 down-regulation in cancers, despite its lack of tumor-suppressive activities, can be explained fully as a consequence of the selection of cells that have silenced the tumor-suppressive miR-152 harbored within the COPZ2 gene. Many miRNAs located within protein-coding genes are linked transcriptionally to the expression of their host genes, because miRNAs and their host genes typically use the same promoter. In the case of miR-152, which is encoded within the first intron of COPZ2, its expression in normal tissues correlates with that of COPZ2 (36), and we show that miR-152 and COPZ2 are silenced coordinately in different tumor cell types. As regulators of gene expression, miRNAs play a key role in cancer, with some miRNAs displaying oncogenic and others tumor-suppressive activities (37). miR-152 on chromosome 17 was shown previously to be down-regulated in clinical samples of breast cancer (31), endometrial serous adenocarcinoma (where decreased expression of miR-152 was a statistically independent risk factor for overall survival) (32), cholangiocarcinoma (33), and gastric and colorectal cancers, where low expression of miR-152 was correlated with increased tumor size and advanced primary tumor stage (34). Furthermore, ectopic expression of miR-152 in cholangiocarcinoma cells decreased cell proliferation (33), and miR-152 overexpression in a placental human choriocarcinoma cell line sensitized the cells to lysis by natural killer cells (35). We found that miR-152 was silenced in >90% of tumor cell lines derived from different types of cancer and that miR-152 overexpression inhibited the growth of HeLa cervical carcinoma and MDA-MB-231 breast carcinoma in vitro and of PC3 prostate carcinoma in vivo. The molecular mechanism for this very broad tumor-suppressive activity of miR-152 remains to be investigated. Regardless of this mechanism, however, our finding that miR-152 down-regulation in tumor cells is associated with concurrent silencing of its host gene COPZ2, with the ensuing tumor cell dependence on COPZ1, potentially can be exploited for cancer therapy.
COPZ1 seems an appealing cancer target, which can be inhibited either by RNA-targeting agents (such as siRNA) or by small-molecule inhibitors that would affect ζ1 encoded by COPZ1 preferentially to ζ2, the COPZ2 gene product. The recently reported solution of the structure of the ζ subunit of COPI (38) may be useful for structure-based design of small-molecule COPZ1 inhibitors. siRNA or small-molecule inhibitors of COPZ1 are expected to be effective against a wide spectrum of cancers in which COPZ2 has been down-regulated. On the other hand, COPZ2 expression in normal tissues suggests that such tissues should not be sensitive to COPZ1 inhibition, as evidenced in our study by the resistance of BJ-hTERT fibroblasts and normal prostate epithelial cells to COPZ1 knockdown. The possibility of achieving an acceptable therapeutic index with COPI-targeting drugs, even those not selective for COPZ1, is suggested by the reports that Brefeldin A, a natural compound that interferes with COPI recruitment to Golgi apparatus (23), showed antitumor efficacy not only in vitro but also in vivo (39). COPZ1-selective inhibitors could be as effective as Brefeldin A, with lower systemic toxicity, especially with the use of selective delivery vehicles, such as targeted nanoparticles or liposomes.
A special appeal of targeting COPZ1 in cancer therapy (and potentially other COPI components, using tumor-specific delivery vehicles) stems from our finding that Golgi apparatus collapse and inhibition of autophagy, which result from COPZ1 or COPA knockdown, cause cell death not only in proliferating but also in growth-arrested tumor cells. Nondividing tumor cells are resistant to agents targeting the process of cell proliferation, as do most of the existing anticancer drugs. The independence of cell death induced by COPI inhibition from cell-cycle progression is indicated further by the observation that cells transfected with COPZ1 siRNA die through apoptosis, without undergoing abnormal mitosis (mitotic catastrophe), the common cause of death in cells treated with conventional anticancer drugs (26, 40). Chemotherapy-resistant nondividing tumor cells include the damaged/senescent cell population that secretes mitogenic, antiapoptotic, angiogenic, and proteolytic factors (6), as well as resting tumor stem cells (8) and dormant cells that can survive for years before reentering the cell cycle (7). The failure to destroy resting and dormant tumor cells is a general cause of tumor relapse after the initial remission. COPI-targeting therapy potentially could bypass this problem, with a greater likelihood of achieving the cure.
Methods
Poly(A)+ RNA, extracted from a mixture of 18 cancer and leukemia cell lines, was used to prepare normalized cDNA through duplex-specific nuclease normalization (41), as described in SI Methods. Normalized cDNA fragments were cloned into a lentiviral vector inducible by tetracycline/doxycycline, yielding a GSE library of ∼2.6 × 108 clones, with an average insert length of 135 bp. The recipient cell lines were transduced first with a lentiviral vector expressing tTR-KRAB (42) and then with the GSE library. Twenty-five percent of the transduced cells were used for DNA extraction immediately, and the rest were subjected to selection for doxycycline-dependent resistance to BrdU suicide (16), followed by DNA extraction. Library-derived cDNA inserts were amplified by PCR from genomic DNA using vector specific primers and subjected to 454 massive parallel sequencing. BLAST analysis was used to identify genes giving rise to cDNA fragments enriched by GSE selection.
siRNA assays for all the genes were first conducted using siRNAs from Qiagen (four siRNAs per gene) (Table S3); subsequent assays incorporated additional siRNAs from Dharmacon (SI Methods). miR-152 precursors were from Dharmacon and Ambion. siRNA targeting no known genes (Qiagen) was used as a control. Cell numbers were determined 4–8 d after transfection by flow cytometry or by staining cellular DNA with Hoechst 33342. Full-length COPZ1 and COPZ2 cDNAs (Open Biosystems) were cloned into a lentiviral vector, pLenti6-bsd-FLAG, constructed in our laboratory, which adds a FLAG tag at the C terminus. Retroviral vector expressing GFP-LC3 (21) was obtained from Addgene. Lentiviral vector expressing miR-152 precursor was from SBI Bioscience. The recipient cell populations transduced with these and the corresponding insert-free control vectors were selected with blasticidin or puromycin before analysis.
Gene and miRNA expression were analyzed by qRT-PCR (primers listed in Table S4) and, in some cases, by immunoblotting (SI Methods). Flow cytometric assays were used to measure membrane permeability by PI uptake and for TUNEL assays for apoptosis. Fluorescence microscopy was used for chromatin staining with DAPI, GFP-LC3 expression, and immunofluorescence analysis of GM130. Time-lapse phase-contrast microscopy was used for video analysis of cell death induced by COPI knockdown.
In vivo PC3 xenograft assays were conducted in male NCR nude mice. Mice were inoculated s.c. with 106 cells of each tested PC3 derivative (five mice per group). Tumor size was measured every 4 d using calipers; tumors were excised and weighed at the end of the study.
Supplementary Material
Acknowledgments
We thank Dr. William Hahn for BJ fibroblast-derived cell lines. This study was supported by National Institutes of Health Grants R33 CA95996 and R01 AG028687 (to I.B.R.) and Grant W81XWH-08-1-0070 from the Department of Defense Prostate Cancer Research Program (to M.S.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1103842108/-/DCSupplemental.
References
- 1.Garnock-Jones KP, Keating GM, Scott LJ. Trastuzumab: A review of its use as adjuvant treatment in human epidermal growth factor receptor 2 (HER2)-positive early breast cancer. Drugs. 2010;70:215–239. doi: 10.2165/11203700-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 2.Yoshida T, Zhang G, Haura EB. Targeting epidermal growth factor receptor: Central signaling kinase in lung cancer. Biochem Pharmacol. 2010;80:613–623. doi: 10.1016/j.bcp.2010.05.014. [DOI] [PubMed] [Google Scholar]
- 3.Collins SJ. Retinoic acid receptors, hematopoiesis and leukemogenesis. Curr Opin Hematol. 2008;15:346–351. doi: 10.1097/MOH.0b013e3283007edf. [DOI] [PubMed] [Google Scholar]
- 4.Weinstein IB, Joe A. Oncogene addiction. Cancer Res. 2008;68:3077–3080, discussion 3080. doi: 10.1158/0008-5472.CAN-07-3293. [DOI] [PubMed] [Google Scholar]
- 5.Primiano T, et al. Identification of potential anticancer drug targets through the selection of growth-inhibitory genetic suppressor elements. Cancer Cell. 2003;4:41–53. doi: 10.1016/s1535-6108(03)00169-7. [DOI] [PubMed] [Google Scholar]
- 6.Roninson IB. Tumor cell senescence in cancer treatment. Cancer Res. 2003;63:2705–2715. [PubMed] [Google Scholar]
- 7.Aguirre-Ghiso JA. The problem of cancer dormancy: Understanding the basic mechanisms and identifying therapeutic opportunities. Cell Cycle. 2006;5:1740–1743. doi: 10.4161/cc.5.16.3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sell S. Potential gene therapy strategies for cancer stem cells. Curr Gene Ther. 2006;6:579–591. doi: 10.2174/156652306778520674. [DOI] [PubMed] [Google Scholar]
- 9.Song CM, Lim SJ, Tong JC. Recent advances in computer-aided drug design. Brief Bioinform. 2009;10:579–591. doi: 10.1093/bib/bbp023. [DOI] [PubMed] [Google Scholar]
- 10.Deiss LP, Kimchi A. A genetic tool used to identify thioredoxin as a mediator of a growth inhibitory signal. Science. 1991;252:117–120. doi: 10.1126/science.1901424. [DOI] [PubMed] [Google Scholar]
- 11.Holzmayer TA, Pestov DG, Roninson IB. Isolation of dominant negative mutants and inhibitory antisense RNA sequences by expression selection of random DNA fragments. Nucleic Acids Res. 1992;20:711–717. doi: 10.1093/nar/20.4.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gudkov AV, et al. Cloning mammalian genes by expression selection of genetic suppressor elements: Association of kinesin with drug resistance and cell immortalization. Proc Natl Acad Sci USA. 1994;91:3744–3748. doi: 10.1073/pnas.91.9.3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berns K, et al. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature. 2004;428:431–437. doi: 10.1038/nature02371. [DOI] [PubMed] [Google Scholar]
- 14.Silva JM, et al. Second-generation shRNA libraries covering the mouse and human genomes. Nat Genet. 2005;37:1281–1288. doi: 10.1038/ng1650. [DOI] [PubMed] [Google Scholar]
- 15.Bernards R, Brummelkamp TR, Beijersbergen RL. shRNA libraries and their use in cancer genetics. Nat Methods. 2006;3:701–706. doi: 10.1038/nmeth921. [DOI] [PubMed] [Google Scholar]
- 16.Shtutman M, et al. Function-based gene identification using enzymatically generated normalized shRNA library and massive parallel sequencing. Proc Natl Acad Sci USA. 2010;107:7377–7382. doi: 10.1073/pnas.1003055107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beck R, Rawet M, Wieland FT, Cassel D. The COPI system: Molecular mechanisms and function. FEBS Lett. 2009;583:2701–2709. doi: 10.1016/j.febslet.2009.07.032. [DOI] [PubMed] [Google Scholar]
- 18.Razi M, Chan EY, Tooze SA. Early endosomes and endosomal coatomer are required for autophagy. J Cell Biol. 2009;185:305–321. doi: 10.1083/jcb.200810098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hahn WC, et al. Creation of human tumour cells with defined genetic elements. Nature. 1999;400:464–468. doi: 10.1038/22780. [DOI] [PubMed] [Google Scholar]
- 20.Styers ML, O'Connor AK, Grabski R, Cormet-Boyaka E, Sztul E. Depletion of beta-COP reveals a role for COP-I in compartmentalization of secretory compartments and in biosynthetic transport of caveolin-1. Am J Physiol Cell Physiol. 2008;294:C1485–C1498. doi: 10.1152/ajpcell.00010.2008. [DOI] [PubMed] [Google Scholar]
- 21.Fung C, Lock R, Gao S, Salas E, Debnath J. Induction of autophagy during extracellular matrix detachment promotes cell survival. Mol Biol Cell. 2008;19:797–806. doi: 10.1091/mbc.E07-10-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klionsky DJ, Elazar Z, Seglen PO, Rubinsztein DC. Does bafilomycin A1 block the fusion of autophagosomes with lysosomes? Autophagy. 2008;4:849–850. doi: 10.4161/auto.6845. [DOI] [PubMed] [Google Scholar]
- 23.Citterio C, et al. Unfolded protein response and cell death after depletion of brefeldin A-inhibited guanine nucleotide-exchange protein GBF1. Proc Natl Acad Sci USA. 2008;105:2877–2882. doi: 10.1073/pnas.0712224105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Platini F, Pérez-Tomás R, Ambrosio S, Tessitore L. Understanding autophagy in cell death control. Curr Pharm Des. 2010;16:101–113. doi: 10.2174/138161210789941810. [DOI] [PubMed] [Google Scholar]
- 25.Filimonenko M, et al. Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. J Cell Biol. 2007;179:485–500. doi: 10.1083/jcb.200702115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Broude EV, et al. Mitotic Catastrophe in Cancer Therapy. Beyond Apoptosis: Cellular Outcomes of Cancer Therapy. New York: Informa Healthcare; 2008. pp. 307–320. [Google Scholar]
- 27.Chang BD, et al. Effects of p21Waf1/Cip1/Sdi1 on cellular gene expression: Implications for carcinogenesis, senescence, and age-related diseases. Proc Natl Acad Sci USA. 2000;97:4291–4296. doi: 10.1073/pnas.97.8.4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Satyanarayana A, Kaldis P. Mammalian cell-cycle regulation: Several Cdks, numerous cyclins and diverse compensatory mechanisms. Oncogene. 2009;28:2925–2939. doi: 10.1038/onc.2009.170. [DOI] [PubMed] [Google Scholar]
- 29.Kobayashi H, et al. Variant sublines of early-stage human melanomas selected for tumorigenicity in nude mice express a multicytokine-resistant phenotype. Am J Pathol. 1994;144:776–786. [PMC free article] [PubMed] [Google Scholar]
- 30.Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14(10A):1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lehmann U, et al. Epigenetic inactivation of microRNA gene hsa-mir-9-1 in human breast cancer. J Pathol. 2008;214:17–24. doi: 10.1002/path.2251. [DOI] [PubMed] [Google Scholar]
- 32.Hiroki E, et al. Changes in microRNA expression levels correlate with clinicopathological features and prognoses in endometrial serous adenocarcinomas. Cancer Sci. 2010;101:241–249. doi: 10.1111/j.1349-7006.2009.01385.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Braconi C, Huang N, Patel T. MicroRNA-dependent regulation of DNA methyltransferase-1 and tumor suppressor gene expression by interleukin-6 in human malignant cholangiocytes. Hepatology. 2010;51:881–890. doi: 10.1002/hep.23381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen LL, Carmichael GG. Decoding the function of nuclear long non-coding RNAs. Curr Opin Cell Biol. 2010;22:357–364. doi: 10.1016/j.ceb.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu XM, et al. Overexpression of miR-152 leads to reduced expression of human leukocyte antigen-G and increased natural killer cell mediated cytolysis in JEG-3 cells. Am J Obstet Gynecol. 2010;202:592–597. doi: 10.1016/j.ajog.2010.03.002. e1–e7. [DOI] [PubMed] [Google Scholar]
- 36.Stuart RO, et al. In silico dissection of cell-type-associated patterns of gene expression in prostate cancer. Proc Natl Acad Sci USA. 2004;101:615–620. doi: 10.1073/pnas.2536479100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu W, Lin J, Jin C, Xia B. Solution structure of human zeta-COP: Direct evidences for structural similarity between COP I and clathrin-adaptor coats. J Mol Biol. 2009;386:903–912. doi: 10.1016/j.jmb.2008.12.083. [DOI] [PubMed] [Google Scholar]
- 39.Sausville EA, et al. Antiproliferative effect in vitro and antitumor activity in vivo of brefeldin A. Cancer J Sci Am. 1996;2:52–58. [PubMed] [Google Scholar]
- 40.Chang BD, et al. A senescence-like phenotype distinguishes tumor cells that undergo terminal proliferation arrest after exposure to anticancer agents. Cancer Res. 1999;59:3761–3767. [PubMed] [Google Scholar]
- 41.Zhulidov PA, et al. Simple cDNA normalization using kamchatka crab duplex-specific nuclease. Nucleic Acids Res. 2004;32:e37. doi: 10.1093/nar/gnh031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wiznerowicz M, Trono D. Conditional suppression of cellular genes: Lentivirus vector-mediated drug-inducible RNA interference. J Virol. 2003;77:8957–8961. doi: 10.1128/JVI.77.16.8957-8961.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sanchez-Carbayo M, Socci ND, Lozano J, Saint F, Cordon-Cardo C. Defining molecular profiles of poor outcome in patients with invasive bladder cancer using oligonucleotide microarrays. J Clin Oncol. 2006;24:778–789. doi: 10.1200/JCO.2005.03.2375. [DOI] [PubMed] [Google Scholar]
- 44.Varambally S, et al. Integrative genomic and proteomic analysis of prostate cancer reveals signatures of metastatic progression. Cancer Cell. 2005;8:393–406. doi: 10.1016/j.ccr.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 45.Yu YP, et al. Gene expression alterations in prostate cancer predicting tumor aggression and preceding development of malignancy. J Clin Oncol. 2004;22:2790–2799. doi: 10.1200/JCO.2004.05.158. [DOI] [PubMed] [Google Scholar]
- 46.Sabates-Bellver J, et al. Transcriptome profile of human colorectal adenomas. Mol Cancer Res. 2007;5:1263–1275. doi: 10.1158/1541-7786.MCR-07-0267. [DOI] [PubMed] [Google Scholar]
- 47.Talantov D, et al. Novel genes associated with malignant melanoma but not benign melanocytic lesions. Clin Cancer Res. 2005;11:7234–7242. doi: 10.1158/1078-0432.CCR-05-0683. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.