Abstract
Pacidamycins are a family of uridyl tetra/pentapeptide antibiotics with antipseudomonal activities through inhibition of the translocase MraY in bacterial cell wall assembly. The biosynthetic gene cluster for pacidamycins has recently been identified through genome mining of the producer Streptomyces coeruleorubidus, and the highly dissociated nonribosomal peptide assembly line for the uridyl tetrapeptide scaffold of pacidamycin has been characterized. In this work a hypothetical protein PacB, conserved in known uridyl peptide antibiotics gene clusters, has been characterized by both genetic deletion and enzymatic analysis of the purified protein. PacB catalyzes the transfer of the alanyl residue from alanyl-tRNA to the N terminus of the tetrapeptide intermediate yielding a pentapeptide on the thio-templated nonribosomal peptide synthetase (NRPS) assembly line protein PacH. PacB thus represents a new group of tRNA-dependent peptide bond-forming enzymes in secondary metabolite biosynthesis in addition to the recently identified cyclodipeptide synthases. The characterization of PacB completes the assembly line reconstitution of pacidamycin pentapeptide antibiotic scaffolds, bridging the primary and secondary metabolic pathways by hijacking an aminoacyl-tRNA to the antibiotic biosynthetic pathway.
Keywords: aminoacyltransferase, uridyl pentapeptide, peptidyl carrier protein
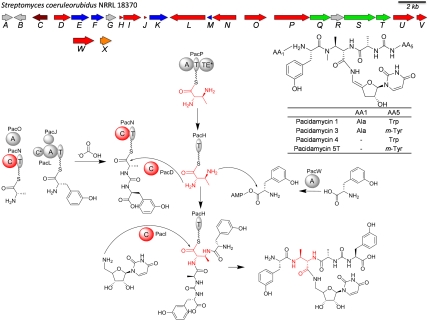
Pacidamycins are a family of uridyl tetra/pentapeptide antibiotics produced by Streptomyces coeruleorubidus (1). More than 10 related family members have been reported (1, 2): Their common scaffold contains a central Nβ-methyl 2S,3S-diaminoburytic acid (DABA) moiety which is α-amino-capped by a ureido dipeptide (C-terminal), β-amino-capped by a single amino acid or a dipeptide (N-terminal), and carboxy-linked to a 3′-deoxy-4′,5′-enaminouridine (Fig. 1). Pacidamycins were reported to exert antibiotic activities by virtue of functioning as a substrate analog of the UDP-MurNAc-pentapeptide of MraY in bacterial cell wall assembly of the pentapeptidyl-bactoprenol intermediate (3, 4). Pacidamycins are of interest due to their unusual peptidyl-nucleoside structure features, and for the development of next generation antibacterial drugs inhibiting the clinically underexplored cell wall enzyme target MraY. The biosynthetic gene clusters for pacidamycins and closely related napsamycins were identified in parallel recently by three research groups via genome mining (5–7). Bioinformatic analysis revealed the building blocks in pacidamycins are assembled using a nonribosomal peptide synthetase (NRPS) thiotemplate logic, whereas the encoded NRPS modules and domains are highly dissociated and predicted to work with each other in trans. We further characterized the functions of the separate NRPS protein domains in vitro to delineate catalytic steps for activation and insertion of each building block in the pacidamycin scaffold (8, 9). We have shown that nine NRPS enzymes encoded by the producing organism S. coeruleorubidus, when heterologously expressed and purified from Escherichia coli, allowed reconstitution of the 5′-aminouridyl-tetrapeptide framework of the pacidamycin family (Fig. 1). However, the biosynthetic route to pentapeptides from that set of tetrapeptide scaffolds remained unknown.
Fig. 1.
Structures of selected pacidamycins, map of pacidamycin gene cluster (6), and biosynthetic pathway for uridyl tetrapeptide using nine NRPS enzymes (9). Domain notation: T, thiolation; A, adenylation; C, condensation; TE, thioesterase. The diamine spacer DABA is shown in red.
One interesting feature observed during the reconstitution of pacidamycin scaffold is the mechanistic versatility in linking various building blocks. In total, three condensation (C) domain-containing NRPS Pac enzymes were identified in our biochemical studies: a C-T didomain protein PacN (T: thiolation), and two freestanding C domain proteins PacD and PacI. PacN is responsible for the ureido dipeptide formation, which is then ligated to the α-amino of DABA catalyzed by one of the freestanding C domain proteins PacD; a modified uridine moiety was incorporated at the carboxyl group of DABA via amide linkage catalyzed by the other freestanding C domain PacI. No putative C domain was found to be involved in the attachment of L-Ala/L-m-Tyr to the β-amino of DABA after activation by the dedicated adenylation (A) domains (PacU/PacW) (Fig. 1) (9). As the functions of all of the NRPS enzymes encoded by the gene cluster have been elucidated, the molecular mechanism of the activation and insertion of the N-terminal L-Ala/L-Gly in the pacidamycin pentapeptidyl scaffold may therefore involve an NRPS-unrelated mechanism. The pacidamycin gene cluster encodes three hypothetical proteins (PacA, PacB, and PacG) with no significant sequence homology to any characterized proteins in the NCBI database (6). In this work, the online program HHpred was used to detect structural homology of these hypothetical proteins and predict their functions (10). This led to the discovery of the role played by PacB in tRNA-dependent biosynthesis of the pacidamycin group of pentapeptidyl nucleoside antibiotics.
Results
Bioinformatic Analysis of PacA, PacB, and PacG.
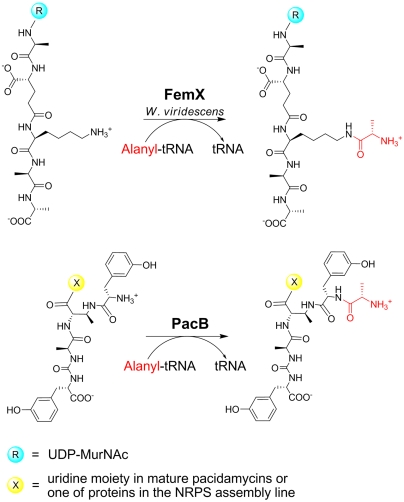
After BLASTP analysis failed to provide useful information on the putative functions of PacA, PacB, and PacG, structural homology search of these hypothetical proteins using the online program HHpred was performed. PacA was predicted to contain an N-terminal forkhead-associated domain related to phosphor-Ser/Thr binding, and a C-terminal helix-turn-helix DNA-binding motif. PacA was therefore postulated to play a regulatory role in the pacidaymcin biosynthesis. PacG showed high structural homology to iron(II)-dependent cysteine dioxygenase and thus could function as an oxidoreductase in the formation of N-terminal bicyclic moiety (6-hydroxy-tetrahydro-isoquinoline carboxylic acid) or the modification of uridine. HHpred analysis of PacB revealed structural homology to Weissella viridescens FemX (11) and Staphylococcus aureus FemA (12). These Fem enzymes catalyze the addition of amino acid (FemX: Ala; FemA: Gly) on the peptidoglycan precursor using an aminoacylated tRNA as a substrate for subsequent peptide bridge cross-linking to strengthen the cell wall (Fig. 2A) (13). We thus proposed that PacB may similarly utilize aminoacylated tRNAs as substrates for the installation of the N-terminal Ala/Gly in the uridyl pentapeptide scaffold of pacidamycins (Fig. 2B).
Fig. 2.
Schematic of the reaction catalyzed by a tRNA-dependent aminoacyltransferase FemX and proposed function of PacB. FemX catalyzes the addition of Ala to a peptidylglycan precursor; PacB is predicted to add Ala or Gly (Ala shown) to mTyr2-DABA3-Ala4-CO-mTyr5.
In Vivo Gene Disruption of pacB.
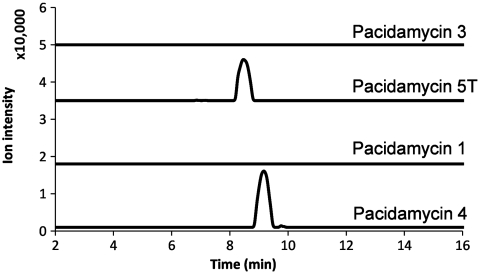
Both tetrapeptides and pentapeptides of pacidamycins were routinely detected from the culture of wild-type strain by liquid chromatography–high-resolution mass spectrometry (LC-HRMS) analysis following the standard growth condition and extraction technique (6). A pacB disruption experiment was carried out to probe its role in the biosynthesis of pacidamycins. The gene was deleted in-frame through double crossover according to standard methods (14), and the resulting mutants were confirmed by PCR (Fig. S1). The deletion of pacB abolished the production of all of the pentapeptide compounds, while uridyl tetrapeptides were produced, albeit at lower yields (Fig. 3). The pacB knockout result suggested that PacB may be specifically related to the incorporation of the N-terminal amino acid to make the uridyl pentapeptide framework.
Fig. 3.
Extracted ion chromatograms showing the production of selected pacidamycins by the S. coeruleorubidus pacB mutant. The molecular structures of pacidamycins are shown in Fig. 1. The calculated mass with 10-ppm mass error tolerance was used.
In Vitro Production of Uridyl Pentapeptides Using Purified PacB.
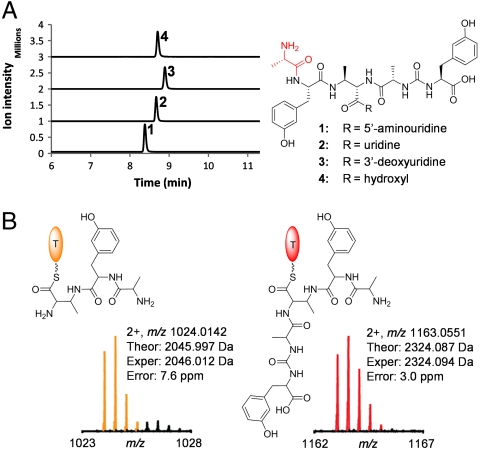
PacB was cloned as an N-terminal His6-tagged protein, expressed and purified from E. coli with a yield of 1.6 mg/L (Fig. S2). We have previously shown that uridyl tetrapeptides with N-terminal m-Tyr tethered to the β-amino of DABA could be formed using the nine NRPS and associated enzymes PacDHIJLNOPW in vitro (9). No uridyl pentapeptide could be detected from those enzymatic reactions. The purified PacB was then added to the reaction mixture of the nine NRPS enzymes, with the addition of E. coli aminoacyl-tRNA synthetase (aatRS, from Sigma) and tRNA (Sigma) to generate alanyl-tRNA in situ. Uridyl pentapeptides (1–3) were formed as the major products in vitro, with the newly added Ala1 attached to the amino group of m-Tyr2 at the N terminus as confirmed by MS/MS analysis (Fig. 4A and Figs. S3–S5). The in vitro reconstitution confirmed the essential role of PacB in the uridyl pentapeptide biosynthesis. It is notable that 1–3 were also produced in the absence of tRNA at slower apparent rates, indicating that activated L-Ala, probably through alanyl-AMP formed by aatRS or PacO, could also be transferred to the m-Tyr residue.
Fig. 4.
Characterization of PacB by LC-MS analysis. (A) Extracted ion chromatograms showing production of pacidamycin group of uridyl pentapeptides (1–3) and the hydrolytic intermediate 4. The calculated mass with a 10-ppm mass tolerance was used to create the extracted ion chromatograms. (B) Detection of PacH-bound biosynthetic intermediates with the incorporation of N-terminal Ala catalyzed by PacB. The intermediates were analyzed by FTMS after trypsin digestion of PacH.
PacB Catalyzes Transfer of the Alanyl1 Residue on NRPS Assembly Line.
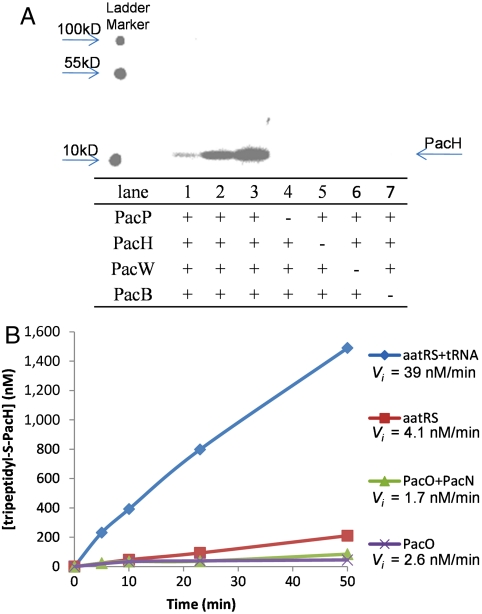
The timing of the aminoacylation during pacidamycin scaffold assembly was probed by LC-MS analysis. The enzyme-free uridyl tetrapeptide was prepared from the nine NRPS enzymatic reactions after dialysis through a 5,000 molecular weight cutoff membrane and incubated with PacB and alanyl-tRNA generated in situ. The free uridyl tetrapeptide was not converted to the uridyl pentapeptide by PacB. This result indicated that PacB might function to elongate intermediates on the NRPS assembly line. PacB assay was then performed using the tetrapeptide substrate still tethered on the pantetheinyl arm of PacH, a 10-kDa freestanding T domain. Pentapeptide product 4 (Ala1-mTyr2-DABA3-Ala4-CO-mTyr5) was formed and confirmed by LC-MS after hydrolytic release of the peptidyl intermediate from PacH with thioesterase TycF (Fig. 4A and Fig. S6) (15). The formation of pentapeptidyl-S-PacH was further confirmed by Fourier transform mass spectrometry (FTMS) after trypsin digestion (Fig. 4B and Fig. S7). We have previously shown that the aminoacylation of α- and β-amino of tethered DABA could take place independently (Fig. 1) (9); therefore the necessity of the C-terminal ureido dipeptide on the assembly line for PacB recognition was tested. PacB was able to catalyze the transfer of the alanyl residue from tRNA to mTyr2-DABA3-S-PacH, and the resulting Ala1-mTyr2-DABA3-S-PacH species was confirmed by FTMS (Fig. 4B and Figs. S8 and S9). To confirm the minimal NRPS components required for the PacB assays, the transfer of the alanyl residue to mTyr2-DABA3-S-PacH was further investigated using [14C]L-Ala as the substrate and visualized by SDS-PAGE autoradiography. Radioactivity migrated with PacH (10 kD) but not PacP (90 kD), and the labeling of PacH by [14C]L-Ala was dependent on the presence of PacBHPW (Fig. 5A). We have previously revealed that the central building block DABA was activated by and loaded onto holo-PacP (an A-T-TE* tridomain NRPS, TE*: thioesterase predicted to be inactive), and transferred to holo-PacH; the freestanding A domain PacW activated m-Tyr, which was then attached to the β-amino of DABA tethered on PacH (Fig. 1) (9). The autoradiography results confirmed that mTyr2-DABA3-S-PacH was a minimal biosynthetic intermediate for PacB recognition. It is notable that PacB could not catalyze the aminoacylation of the free amino acid m-Tyr, demonstrated by LC-MS analysis.
Fig. 5.
Characterization of PacB using the [14C]l-Ala substrate. (A) Autoradiograph of SDS-PAGE gel illustrating the covalent loading of [14C]l-Ala on dipeptidyl-S-PacH. ATP, m-Tyr, DABA, aatRS, and tRNA were present in all assays. Lanes 1–3 show reactions stopped at different time intervals (30, 60, and 120 min, respectively). Reactions in lane 4–7 were stopped after 120 min. (B) Time-course studies of the formation of [14C]Ala1-mTyr2-DABA3-S-PacH using various activated alanyl species.
PacB Prefers Alanyl-tRNA over Other Activated Alanyl Species.
No formation of Ala1-mTyr2-DABA3-S-PacH species was detected by FTMS when E. coli tRNA was omitted from the PacBHPW reaction (Fig. S10), strongly suggesting that PacB catalyzed the aminoacyltransfer reaction in a tRNA-dependent manner. However, the in vitro total synthesis of uridyl pentapeptides above indicated that PacB could probably take activated alanyl species other than alanyl-tRNA. The preference of PacB toward various activated alanyl species was then probed using time course studies on the labeling intensity of PacH by [14C]L-Ala. During the in vitro reconstitution of uridyl pentapeptides biosynthesis, the activated alanyl species included (i) alanyl-tRNA generated by aatRS and tRNA, (ii) Ala-S-PacN involved in the ureido dipeptide synthesis, and (iii) putative alanyl-AMP generated and released by aatRS or the freestanding A domain of PacO. The utilization of these alanyl species was studied in the [14C]Ala1-mTyr2-DABA3-S-PacH formation assays containing aatRS and tRNA (providing alanyl-tRNA), PacO and PacN (providing Ala-S-PacN), aatRS only (providing alanyl-AMP), and PacO only (providing alanyl-AMP), respectively. The apparent initial tripeptidyl-S-PacH formation rate (Vi) using alanyl-tRNA as a substrate was approximately 39 nM/ min, which was at least 10-fold higher than the rates using other activated alanyl species (Fig. 5B). This rate comparison demonstrated that PacB preferred alanyl-tRNA (from bulk E. coli tRNA) as a donor over other activated alanyl species in transferring the alanyl residue to the NRPS assembly line.
Discussion
In this work we have characterized PacB as a tRNA-dependent aminoacyltransferase involved in peptide bond formation in secondary metabolism, specifically in the addition of an N-terminal alanyl1 residue to a tethered di- to tetrapeptidyl intermediate in the highly dissociated NRPS assembly line in pacidamycin biosynthesis. In the absence of PacB action, a tetrapeptidyl-S-pantetheinyl-PacH species is ammonylized by a 5′-aminouridine moiety for enzymatic-mediated chain release. When PacB has acted, a full-length Ala1-mTyr2-DABA3-Ala4-CO-mTyr5 thioester forms on PacH and is released by the aminouridine to generate the pentapeptidyl nucleoside antibiotic scaffold. Uridyl pentapeptidyl forms of pacidamycins were reported to be isolated as major products from the fermentation studies (1). We have noticed that in early stages of pacidamycin production the pentapeptidyl forms predominated, whereas tetrapeptidyl forms came up later, perhaps due to some combination of proteolysis, transport, or limiting metabolite concentration in stationary phase cultures of S. coeruleorubidus. We therefore assign PacB a chain elongation role on the NRPS assembly line. The deletion of the pacB gene confirms this assignment, given the selective loss of pentapeptidyl nucleoside but not tetrapeptidyl nucleoside forms of this antibiotic family.
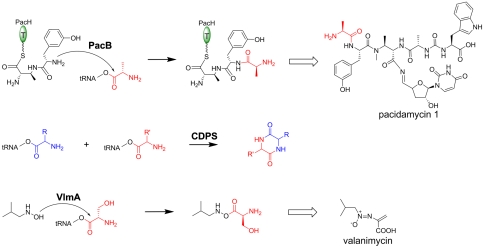
In terms of amino acid addition to a peptidyl chain, PacB has functional homologies to the Fem ligases involved in the modification of peptidylglycans (13, 16). FemX of W. viridescens adds the first alanyl residue from alanyl-tRNA to the ϵ-amino group of Lys at the third position of the pentapeptide stem to initiate an interpeptide cross-bridge (Fig. 2A); FemXAB enzymes of S. aureus deliver glycyl moieties from the glycyl-tRNA to build a Gly5 cross-bridge at the same Lys3 position. Similarly, PacB delivers an alanyl moiety from alanyl-tRNA to the m-Tyr2 residue of the growing pacidamycin scaffold attached to the peptidyl carrier protein PacH (Fig. 6).
Fig. 6.
Summary of three characterized tRNA-dependent enzymes for aminoacylation of aminoacyl/peptidyl scaffolds in secondary metabolism. CDPS: cyclodipeptide synthase.
Recently another family of enzymes, cyclodipeptide synthase (CDPS), has been identified to be tRNA-dependent in peptide bond formation in secondary metabolism (Fig. 6) (17). CDPSs utilize two aminoacylated tRNAs as their substrates to catalyze the formation of cyclic dipeptides, and structural studies suggest that they share a common class I aatRS-like architecture without the amino acid activation activity (18, 19). Distinct from CDPSs, PacB is not predicted by HHpred to harness the aatRS-like architecture. The structural model of PacB shown in Fig. S11 was generated based on the highest scoring template FemX (PDB ID code 3GKR), in which PacB is presumed to have two domains separated by a cleft. One domain is predicted to have a three-dimensional fold similar to the fold of the GCN5-like N-acetyltransferase superfamily (20). The residues interacting with the UDP-MurNAc-pentapeptide substrate of FemX are not conserved in PacB, consistent with the different substrate specificities of the two enzymes. It is notable that PacB has only 11% sequence identity and 26% sequence similarity to FemX demonstrated by ClustalW analysis, so it is not picked up in sequence-based search comparisons (Fig. S12).
Other enzymes that catalyze the transfer of an amino acid in a tRNA-dependent manner include the protein argininyl and phenylalanyl/leucyl transferases (21), and the phosphatidylglycerol lysyl/alanyl transferase MprF (22). These enzymes function in protein or lipid modifications, respectively, in primary metabolism. Recently, a homolog of MprF, VlmA, was identified from Streptomyces viridifaciens to promote the ester bond formation between L-Ser and the hydroxyl group of isobutylhydroxylamine in the biosynthesis of the antibiotic valanimycin (Fig. 6) (23). VlmA functions with a distinct class II seryl-tRNA synthetase paralog VlmL encoded in the valanimycin gene cluster, which provides seryl-tRNA as a substrate for VlmA (24). In contrast, no dedicated alanyl/glycyl-tRNA synthetase was identified in the pacidamycin gene cluster. Two putative “housekeeping” alanyl-tRNA synthetases (amino acid size: 390 and 890) are identifiable in the producer S. coeruleorubidus genome by BLASTP analysis, both of which are conserved in the published genomes of Streptomyces coelicolor, Streptomyces lividans, Streptomyces griseus subsp. griseus, Streptomyces sviceus, etc. In addition, the online program tRNAscan-SE (25) predicts the absence of a putative Ala specific tRNA near the pacidamycin gene cluster. Therefore, PacB presumably hijacks alanyl/glycyl-tRNAs from primary metabolism, bridging the primary and secondary metabolic pathways in pacidamycin biosynthesis. As both uridyl tetrapeptides and uridyl pentapeptides exhibited similar antipseudomonal activities in vitro (26), the rationale to produce both versions of pacidamycins can be deduced only by further study of timing and maturation processes in the producer S. coeruleorubidus.
In summary, we have predicted the function of a hypothetical protein PacB in the biosynthesis of the pacidamycin group of pentapeptidyl nucleoside antibiotics and validated the function of PacB as a tRNA-dependent alanyl1 transferase by both genetic and enzymatic studies. PacB has high sequence identity to homologs encoded by other uridyl peptide biosynthetic gene clusters, including NpsN (67% identity) encoded by the napsamycin gene cluster from Streptomyces sp. DSM 5940 (7), and a hypothetical protein SrosN15_15085 (68% identity) encoded by a putative uridyl peptide gene cluster from Streptomyces roseosporus. A gene encoding a hypothetical protein with 44% sequence identity to PacB is also present in the genome of S. sviceus and is located in a putative secondary metabolite gene cluster that also encodes nonribosomal peptide synthetases. The functions of these hypothetical proteins are yet to be established. PacB utilized a tRNA-loaded amino acid as donor and a T domain-loaded nonribosomal dipeptide as acceptor (Fig. 6) and might therefore represent a unique group of transferases linking ribosomal and nonribosomal peptide synthesis.
Materials and Methods
pacB Disruptions in S. coeruleorubidus and Mutant Analysis.
Streptomyces coeruleorubidus NRRL 18370 obtained from USDA ARS Culture Collection was maintained on ISP4 agar or in tryptic soy broth medium at 30 °C. The pacB knockout cassette was constructed using the ReDirect technology (27). A 5-kb fragment containing pacB flanked by 2-kb arms was PCR amplified from genomic DNA and inserted into PCRBlunt vector (Invitrogen) (primers listed in Table S1). This plasmid was introduced into E. coli BW25113/pIJ790 by electroporation. The acc(3)IV-oriT cassette amplified by PCR from pIJ773 was then introduced to replace the entire pacB using PCR targeting and λ-Red-mediated recombination. The resulting knockout cassette was transformed into E. coli WM6026 (a diaminopimelic acid auxotroph) for conjugation with S. coeruleorubidus. The double-crossover strain was obtained from antibiotic selection (ApraRKanS) and confirmed by PCR using one internal primer from acc(3)IV-oriT cassette and one external primer from an adjacent gene (Fig. S1). Mutant analysis was carried out following the reported procedure (6) and is detailed in SI Appendix. HRMS analysis was performed on an Agilent Technologies 6520 Accurate-Mass QTOF LC-MS instrument with a Luna 3u C18 column (4.6 × 75 mm) from Phenomenex. A linear gradient of 2 to 95% CH3CN (vol/vol) over 15 min in H2O supplemented with 0.1% (vol/vol) formic acid at a flow rate of 0.5 mL/ min was used.
LC-HRMS Product Assays.
All proteins were expressed and purified with His6 tag following the general protocol (see SI Appendix). A typical LC-HRMS product assay was performed in 100 μL of 50 mM Hepes (pH 8.0) containing 5 mM ATP, 2 mM MgCl2, 1 mM TCEP [tris(2-carboxyethyl)phosphine], 5 mM amino acids and indicated uridine substrate, 10 μM purified forms of the eight enzymes PacDIJLNOPW (9), 50 μM PacH, 385 units E. coli aatRS (Sigma), 100 μg E. coli tRNA (Sigma), and 1 μM PacB. Reactions were incubated at 25 °C and 25-μL samples were quenched at different time points by adding 25 μL of 10% trichloroacetic acid (TCA). Protein precipitate was pelleted by centrifugation and the supernatant was subjected to LC-HRMS and MS/MS analysis. A linear gradient of 2 to 80% CH3CN (vol/vol) over 15 min in H2O supplemented with 0.1% (vol/vol) formic acid at a flow rate of 0.5 mL/ min was used. In the assay without PacI, TycF (5 μM) was added to the reaction mixture and further incubated for 30 min to release the product from PacH before TCA precipitation.
LC-FTMS Analysis of PacH-Bound Products.
Assays were set up as for the LC-HRMS product assays without the addition of uridine substrate and PacI. After approximately 4 h of incubation at 25 °C, 0.1 M Tris and trypsin (1∶5 wt∶wt trypsin∶total protein) were added and further incubated at 30 °C for 15 min. The reactions were quenched with 25% formic acid and analyzed by nano-capillary LC-MS using a 100 mm × 75 μm C18 column in-line with a LTQ-FT (7 Tesla). Refer to ref. 9 for MS methods. All data were analyzed using QualBrowser, part of the Xcalibur software packaged with the ThermoFisher LTQ-FT. All mass values reported are for the neutral monoisotopic peaks.
Loading Assays with [14C]L-Ala.
A typical assay contained, in a total volume of 50 μL, 5 mM ATP, 2 mM MgCl2, 1 mM TCEP, 5 mM L-m-Tyr and DABA, 50 μM [14C]L-Ala (0.32 μCi), 10 μM PacPW, 50 μM PacH, 192 units aatRS, 50 μg tRNA, 1 μM PacB, and 50 mM Hepes, pH 8.0. For rate analysis using various activated alanyl species, 192 units aatRS and 50 μg tRNA, 192 units aatRS, 40 μM PacNO, 40 μM PacO were included in the reactions, respectively. All reaction components except PacB were preincubated at 25 °C for 30 min to build up substrates mTyr2-DABA3-S-PacH and activated alanyl species. After adding PacB, reactions were incubated at 25 °C and 10-μL samples were quenched at different time points by adding 1× SDS sample buffer. Following SDS-PAGE, radiolabeled proteins were detected using a BAS-III imaging plate (Fuji Film, 48–96 h of exposure) and a Typhoon 9400 phosphorimager (GE Healthcare). The amounts of radioactive products were determined by counting the radioactive intensity and compared to the standard curve generated by [14C]L-Ala in the same image.
Supplementary Material
Acknowledgments.
We thank Dr. Chris Neumann (Harvard Medical School, Boston, MA) for providing the purified TycF, Dr. Steven J. Malcolmson (Harvard Medical School, Boston, MA) for providing 5′-aminouridine, and Prof. Daniel Kahne (Harvard University, Cambridge, MA) for providing DABA. This work was supported by NIH Grant GM49338 (to C.T.W.) and GM067725-08 (to N.L.K).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1109539108/-/DCSupplemental.
References
- 1.Chen RH, Buko AM, Whittern DN, McAlpine JB. Pacidamycins, a novel series of antibiotics with anti-Pseudomonas aeruginosa activity. II. Isolation and structural elucidation. J Antibiot (Tokyo) 1989;42:512–520. doi: 10.7164/antibiotics.42.512. [DOI] [PubMed] [Google Scholar]
- 2.Fronko RM, et al. New pacidamycins produced by Streptomyces coeruleorubidus, NRRL 18370. J Antibiot (Tokyo) 2000;53:1405–1410. doi: 10.7164/antibiotics.53.1405. [DOI] [PubMed] [Google Scholar]
- 3.Boojamra CG, et al. Stereochemical elucidation and total synthesis of dihydropacidamycin D, a semisynthetic pacidamycin. J Am Chem Soc. 2001;123:870–874. doi: 10.1021/ja003292c. [DOI] [PubMed] [Google Scholar]
- 4.Winn M, Goss RJ, Kimura K, Bugg TD. Antimicrobial nucleoside antibiotics targeting cell wall assembly: Recent advances in structure-function studies and nucleoside biosynthesis. Nat Prod Rep. 2010;27:279–304. doi: 10.1039/b816215h. [DOI] [PubMed] [Google Scholar]
- 5.Rackham EJ, Gruschow S, Ragab AE, Dickens S, Goss RJ. Pacidamycin biosynthesis: Identification and heterologous expression of the first uridyl peptide antibiotic gene cluster. Chembiochem. 2010;11:1700–1709. doi: 10.1002/cbic.201000200. [DOI] [PubMed] [Google Scholar]
- 6.Zhang W, Ostash B, Walsh CT. Identification of the biosynthetic gene cluster for the pacidamycin group of peptidyl nucleoside antibiotics. Proc Natl Acad Sci USA. 2010;107:16828–16833. doi: 10.1073/pnas.1011557107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaysser L, et al. Identification of a napsamycin biosynthesis gene cluster by genome mining. Chembiochem. 2011;12:477–487. doi: 10.1002/cbic.201000460. [DOI] [PubMed] [Google Scholar]
- 8.Zhang W, Heemstra JR, Jr, Walsh CT, Imker HJ. Activation of the pacidamycin PacL adenylation domain by MbtH-like proteins. Biochemistry. 2010;49:9946–9947. doi: 10.1021/bi101539b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang W, et al. Nine enzymes are required for assembly of the pacidamycin group of peptidyl nucleoside antibiotics. J Am Chem Soc. 2011;133:5240–5243. doi: 10.1021/ja2011109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Biarrotte-Sorin S, et al. Crystal structures of Weissella viridescens FemX and its complex with UDP-MurNAc-pentapeptide: Insights into FemABX family substrates recognition. Structure. 2004;12:257–267. doi: 10.1016/j.str.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 12.Benson TE, et al. X-ray crystal structure of Staphylococcus aureus FemA. Structure. 2002;10:1107–1115. doi: 10.1016/s0969-2126(02)00807-9. [DOI] [PubMed] [Google Scholar]
- 13.Hegde SS, Shrader TE. FemABX family members are novel nonribosomal peptidyltransferases and important pathogen-specific drug targets. J Biol Chem. 2001;276:6998–7003. doi: 10.1074/jbc.M008591200. [DOI] [PubMed] [Google Scholar]
- 14.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical Streptomyces Genetics. Norwich, UK: John Innes Foundation; 2000. [Google Scholar]
- 15.Yeh E, Kohli RM, Bruner SD, Walsh CT. Type II thioesterase restores activity of a NRPS module stalled with an aminoacyl-S-enzyme that cannot be elongated. Chembiochem. 2004;5:1290–1293. doi: 10.1002/cbic.200400077. [DOI] [PubMed] [Google Scholar]
- 16.Fonvielle M, et al. Aminoacyl-tRNA recognition by the FemXWv transferase for bacterial cell wall synthesis. Nucleic Acids Res. 2009;37:1589–1601. doi: 10.1093/nar/gkn1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gondry M, et al. Cyclodipeptide synthases are a family of tRNA-dependent peptide bond-forming enzymes. Nat Chem Biol. 2009;5:414–420. doi: 10.1038/nchembio.175. [DOI] [PubMed] [Google Scholar]
- 18.Vetting MW, Hegde SS, Blanchard JS. The structure and mechanism of the Mycobacterium tuberculosis cyclodityrosine synthetase. Nat Chem Biol. 2010;6:797–799. doi: 10.1038/nchembio.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bonnefond L, et al. Structural basis for nonribosomal peptide synthesis by an aminoacyl-tRNA synthetase paralog. Proc Natl Acad Sci USA. 2011;108:3912–3917. doi: 10.1073/pnas.1019480108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dyda F, Klein DC, Hickman AB. GCN5-related N-acetyltransferases: A structural overview. Annu Rev Biophys Biomol Struct. 2000;29:81–103. doi: 10.1146/annurev.biophys.29.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Watanabe K, et al. Protein-based peptide-bond formation by aminoacyl-tRNA protein transferase. Nature. 2007;449:867–871. doi: 10.1038/nature06167. [DOI] [PubMed] [Google Scholar]
- 22.Roy H, Ibba M. RNA-dependent lipid remodeling by bacterial multiple peptide resistance factors. Proc Natl Acad Sci USA. 2008;105:4667–4672. doi: 10.1073/pnas.0800006105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garg RP, Qian XL, Alemany LB, Moran S, Parry RJ. Investigations of valanimycin biosynthesis: Elucidation of the role of seryl-tRNA. Proc Natl Acad Sci USA. 2008;105:6543–6547. doi: 10.1073/pnas.0708957105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garg RP, Gonzalez JM, Parry RJ. Biochemical characterization of VlmL, a Seryl-tRNA synthetase encoded by the valanimycin biosynthetic gene cluster. J Biol Chem. 2006;281:26785–26791. doi: 10.1074/jbc.M603675200. [DOI] [PubMed] [Google Scholar]
- 25.Schattner P, Brooks AN, Lowe TM. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 2005;33:W686–689. doi: 10.1093/nar/gki366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fernandes PB, et al. Pacidamycins, a novel series of antibiotics with anti-Pseudomonas aeruginosa activity. III. Microbiologic profile. J Antibiot (Tokyo) 1989;42:521–526. doi: 10.7164/antibiotics.42.521. [DOI] [PubMed] [Google Scholar]
- 27.Gust B, Challis GL, Fowler K, Kieser T, Chater KF. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc Natl Acad Sci USA. 2003;100:1541–1546. doi: 10.1073/pnas.0337542100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.