Abstract
Two-photon microscopy has substantially advanced our understanding of cellular dynamics in the immune system. Cell migration can now be imaged in real time in the living animal. Strikingly, the migration of naive lymphocytes in secondary lymphoid tissue appears predominantly random. It is unclear, however, whether directed migration may escape detection in this random background. Using a combination of mathematical modeling and experimental data, we investigate the extent to which modern two-photon imaging can rule out biologically relevant directed migration. For naive T cells migrating in uninfected lymph nodes (LNs) at average 3D speeds of around 18 μm/min, we rule out uniform directed migration of more than 1.7 μm/min at the 95% confidence level, confirming that T cell migration is indeed mostly random on a timescale of minutes. To investigate whether this finding still holds for longer timescales, we use a 3D simulation of the naive T cell LN transit. A pure random walk predicts a transit time of around 16 h, which is in good agreement with experimental results. A directional bias of only 0.5 μm/min—less than 3% of the cell speed—would already accelerate the transit twofold. These results jointly strengthen the random walk analogy for naive T cell migration in LNs, but they also emphasize that very small deviations from random migration can still be important. Our methods are applicable to cells of any type and can be used to reanalyze existing datasets.
Keywords: lymphocyte migration, statistical analysis, lymph node transit
Two-photon microscopy has fundamentally changed our view of immune cell migration. The first groups who imaged lymphocyte migration in intact organs (1, 2) reported that cells move in a run and tumble fashion and found no evidence for synchronization or directionality. This finding came as a surprise to many immunologists; previous research had emphasized the role of chemokines, “immunology's high impact factors” (3), and therefore, put forward a view of lymphocyte migration being nicely orchestrated. However, lack of evidence for cell synchronization does not necessarily rule out directed migration. Biased deviation from random motion can create an effect that, like directed migration, causes cells to displace gradually. In biology, such deviation is most often caused by an external stimulus, like a chemokine gradient or a flowing liquid, and is then called taxis. Specific ways in which cells could respond to a directed stimulus include faster migration (orthotaxis), preferential turning (topotaxis), and increased persistence (klinotaxis) to the stimulus (Fig. S1). These different kinds of taxis are called taxis modes (4, 5).
Strong taxis, which was found in B cells (6) and neutrophils (7), is obvious to the naked eye. However, detecting more subtle taxis can require sophisticated data analysis; for instance, Castellino et al. (8) used angle analysis to show that naive CD8+ T cells move to sites where naive CD4+ T cells and dendritic cells interact. These examples emphasize that detecting taxis is crucial, because it may point to important biological functions of lymphocytes (9). However, is it then appropriate at all to describe lymphocyte migration as random, or must we always assume that a substantial amount of taxis is hidden in the data? Without knowing the quantitative limits of two-photon cell tracking with regard to taxis detection, this question is hard to answer, and doors remain wide open for speculation. For example, although the migration of B cells in the germinal center was first described as random (10), it was then argued that two-photon data are consistent with both random and directed migration (11, 12), and very recently, a small directional bias to the light zone was revealed (13). In light of such controversy, we ask here the following two questions. (i) How strong is the random walk analogy—in other words, how and to what extent can we rule out taxis based on current two-photon data? (ii) Is the range of taxis that we cannot rule out still large enough to be biologically relevant?
Apparently, our ability to detect taxis is determined to a large extent by the data analysis method itself. Several methods are available and have been discussed in detail elsewhere (14, 15), but the need for a systematic study of their quantitative power (16) has so far not been addressed. Therefore, we benchmark both existing and new methods on experimental and computer-generated data. We discuss our approach and its biological implications in a simple, well-defined scenario: naive T cells migrating in the lymph nodes (LNs) in the absence of antigen. In this setting, the random walk turns out to be a surprisingly accurate model for T cell motility on a short timescale, and it also appears sufficient to explain T cell motility on a long timescale.
Results
Random Walk Accounts for >90% of T Cell Speed.
We imaged adoptively transferred polyclonal T cells in surgically exposed, uninfected LNs of two recipient mice, resulting in datasets of 1,132 and 1,355 tracks, respectively, in a 492 × 492 × 40-μm imaging region (Fig. 1A). The imaged cells migrated at a mean 3D speed of 17.9 ± 2.2 μm/min; no directed migration was evident from aligning the tracks to a common starting point (Fig. 1B). To analyze the data for directionality, we applied Hotelling's T2 test (17), a generalization of the well-known 1D T test, to a set of cell steps (15) extracted from the tracks (Fig. 1C, SI Text, and Fig. S2). Hotelling's test provides a confidence region for the mean cell step (Fig. 1D), and the taxis speed can be estimated by dividing the length of this mean step by the step duration, which in our case, is 60.9 s (three imaging intervals). For random migration, the mean step should be the null vector—cells go nowhere on average. If the null vector lies outside the confidence region, then this finding indicates significant taxis. Otherwise, the point in the confidence region that is farthest from the null vector gives an upper speed bound, which is the amount of taxis that we can safely rule out (Fig. 1D). For our naive T cell datasets, the mean step lengths are 0.62 and 0.73 μm, but the mean steps are not significantly different from random migration (P = 0.4 and P = 0.16). The upper 3D taxis speed bounds are 1.66 and 1.68 μm/min, respectively, at the 95% confidence level. Thus, at most, 10% of the cell speed can still contribute to uniform taxis. To quantify the random T cell migration, we estimated the T cell motility coefficient (14) using a method designed to work around the underestimation that is caused by the finite imaging volume (SI Text, Figs. S3 and S4). This method resulted in an estimate of 100 μm2/min.
Fig. 1.
Quantitative bounds for taxis in two-photon data. (A) x and y projections of 1,355 naive T cell tracks. (Inset) A 40 × 40 × 40 μm subvolume for comparison. (B) The tracks from A with aligned starting positions. No preferential direction is apparent by eye. The small square corresponds to the coordinate system in D. (C) Scatter plot of cell steps (duration = 60.9 s) extracted from the tracks in B. (D) Hotelling's T2 test gives a confidence region (ellipsoid) for the mean step (υmean). Because the null vector (cross) is inside the confidence region, data are consistent with random motion. The point in the confidence region that is farthest from the null vector (υmax) gives an upper speed bound for taxis that may escape this analysis. Data are shown for the x and y dimensions, and 3D values are in the text.
Benchmarking Taxis Detection Methods.
Hotelling's test has, to our knowledge, not been applied previously to lymphocyte migration data. Thus, we next investigated how the sensitivity of this method compares with that of more established ones using computer generated cell tracks with and without taxis. We limit our discussion here to two important alternative methods: displacement analysis, which is widely used (14, 15, 18), and angle analysis, because it has recently received much attention (13, 15). For displacement analysis, we generated mean square displacement (MSD) plots, which are expected to be linear for random migration and curve up in case of directed migration (14). For angle analysis, we calculated the mean 3D angle of the cell steps (Fig. 1C) to the taxis direction. This angle should be 90° for random walk, and a lower angle indicates taxis (15). Note that this approach was only applicable to our simulated data for which we knew the taxis direction. Because our movies did not allow us to clearly identify medulla or sinuses, we had no hypothetical taxis direction for our naive T cell data. Still, we were interested in whether angle analysis outperforms Hotelling's test if this information is available.
To generate the tracks, we used a simple simulation model proposed by Beauchemin et al. (19). This model captures a key feature of short-term lymphocyte motility (1): relatively straight runs alternate with pauses, during which the cell acquires a new orientation. We analyzed this model mathematically (SI Text), enabling us to calculate its motility coefficient and extend it to simulate taxis. Beauchemin et al. (19) extensively validated the short-term migration characteristics of their model against experimental data; by matching its motility coefficient to our estimate, we ensured that the long-term migration is also reasonably modeled.
We simulated three scenarios (Fig. 2): pure random walk, random walk with taxis, and mixed populations containing 50% taxing and 50% randomly walking cells. The taxing cells performed orthotaxis at 3 μm/min (roughly two times our upper bound) for the uniform population and 5.1 μm/min (the maximum possible in our simulation) for the heterogeneous population along the z axis. Crucially, we only tracked the simulated cells in a volume having the size of our imaging region (Materials and Methods). This method ensured that the bias caused by the finite region was present in our simulated tracks as well. The track snapshots were then aligned (Fig. 2A) to generate MSD plots (Fig. 2B). From the aligned tracks, we extracted cell steps (Fig. 2C) for angle analysis (Fig. 2D) and Hotelling's test (Fig. 2E). The results revealed two issues that are discussed below in more detail.
Fig. 2.
Benchmarking two-photon data analysis methods. The power of two-photon data analysis methods can be evaluated systematically using computer-generated data (Materials and Methods) as shown here for the three methods discussed in this paper. (A) Naive T cell data (red; 1,132 tracks) are displayed for comparison alongside three simulated datasets: random walk (black; 842 tracks), random walk with taxis (blue; 3 μm/min along z axis, 897 tracks), and a mixed population with 50% taxing cells and 50% randomly walking cells (green; 5.1 μm/min along z axis, 893 tracks). Simulated cells were tracked in a finite volume having the size of our two-photon imaging region. (B) Mean square displacement of the data in A plotted as a function of time. (C) Cell steps extracted from the tracks in A. Crosses indicate the null vector (red = 703 steps; black = 693 steps; blue = 773 steps; green = 749 steps). (D) Analysis of the mean angle between the cell steps (C) and the taxis direction, which is known only for the simulated data. A mean angle of less than 90° indicates taxis (mean angles and 95% confidence intervals are shown; asterisk indicates significance). (E) Hotelling's T2 test applied to the y and z dimensions (compare with Fig. 1D).
Displacement Analysis Can Be Misleading.
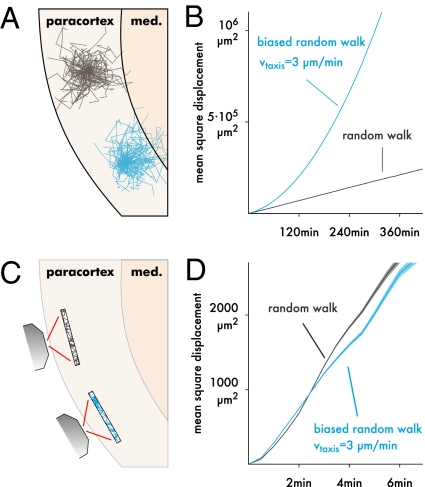
None of the MSD plots in Fig. 2B indicate directed migration. We suspected the reason to be that a curved MSD plot can only arise if taxing cells are followed for a very long time (Fig. 3 A and B). Because we are imaging within a limited region (Fig. 3C), we quickly lose track of fast cells, and long tracks should, thus, predominantly represent slower than average cells, leading to an artificial impression of cell confinement (15). To test this hypothesis, we generated MSD plots for n = 10,000 simulated cells with and without taxis along the z axis (Fig. 3D). Surprisingly, the MSD of the taxing cells is significantly lower from 3 to 4 min on, and the MSD plot is slightly more linear (R2 = 0.998 for t ≤ 5 min) compared with the randomly walking cells (R2 = 0.994 for t ≤ 5 min). From these MSD plots, one would conclude that the taxing cells are less motile than the randomly walking ones, when in fact, the opposite is true.
Fig. 3.
Displacement analysis can lead to wrong conclusions. (A) For example, assume that one subset of T cells (black) migrates purely randomly, whereas another subset (blue) performs taxis to the medulla. (B) If one could follow the cell populations for a very long time, the difference could be detected using a mean square displacement (MSD) plot, which is shown here for two simulated cell populations tracked for 6 h without placing bounds on the trajectories. Pure random walk (black; M = 100 μm2/min) leads to a linear MSD, and taxis (3 μm/min) gives a curved MSD. Data are averaged over 10,000 simulated cells, giving a negligibly small SEM. (C) Two-photon imaging is limited to a finite region in which faster cells are underrepresented, because they exit more quickly. (D) This effect can be reproduced by truncating the simulated data from B to a finite region (shading ± SEM for 10,000 simulated cells; region size = 492 × 492 × 40 μm as in our experimental setup). If we compared the two populations based on this plot alone, we would conclude that the blue population has lower motility (slope) and persistence (curvedness) than the black one, when in fact, the opposite is true.
This seemingly counterintuitive result can be explained by considering the effect of the finite imaging volume in more detail. Because our imaging region is thinnest along the z axis, cells performing taxis in that direction leave the imaging region more quickly than those cells migrating randomly, which amplifies the aforementioned artificial confinement. This artifact would be much weaker if taxis were along the x or y direction, but at least for certain combinations of imaging region shape and taxis direction, it can apparently be strong enough to give misleading MSD plots.
Orthotaxis Can Escape from Angle Analysis.
Because we assumed that the taxis direction is exactly known for applying angle analysis, it may be surprising to observe (Fig. 2 D and E) that angle analysis detected only the uniformly biased population but not the partially biased one, whereas Hotelling's test detected both without knowing the taxis direction. More detailed analysis revealed that this finding was caused by the taxis mode; our simulations use orthotaxis, which hardly affects the migration angle distribution. Klinotaxis and topotaxis were more easily detected; however, the sensitivity of angle analysis to these taxis modes was only slightly better than that of Hotelling's test (Fig. S5). This finding emphasizes that angle analysis is a parametric test whose power depends on the taxis mode. We conjecture that this problem can be addressed by combining migration angles with movement speeds, giving a version of Hotelling's test that checks only for taxis in a single direction rather than in all directions simultaneously.
In summary, our results indicate that the power of Hotelling's test is superior to displacement analysis and similar to angle analysis with known taxis direction. Thus, it gives the best currently possible upper bounds for ruling out taxis of unknown direction and mode.
Impact of Taxis on the T Cell LN Transit.
To investigate the long-term impact of small taxis on T cell migration, we constructed a partial differential equation-based model of the T cell transit from paracortex to sinuses in a rat LN (SI Text, Figs. S6 and S7). The kinetics of lymphocyte recirculation in rats were thoroughly studied during the 1980s and 1990s by cannulation experiments, which reached recovery rates of up to >90% (20, 21). Because the data available for mice are scarce in comparison, we preferred to model the transit through a rat LN at the cost of assuming that microscopic T cell migration in rats and mice is similar. In a careful metaanalysis of several rat experiments, Stekel et al. (22) estimated that LN transit times in rats range between 7 and 20 h. For our simulation, we used a mesenteric LN (Fig. 4), which at a diameter of 4–6 mm, is among the largest LNs; therefore, the transit time should lie within the upper end of the range estimated by Stekel et al. (22). Our model (Fig. 4 A and B) distinguishes between paracortex (defined as containing few macrophages and few B cells), sinuses (many macrophages and B cells), and follicles (few macrophages and many B cells). If cells are released uniformly in the paracortex and assumed to reach the sinuses by pure random walk with M = 100 μm2/min, the model predicts a median transit time of 16.1 h (Fig. 5 A and B).
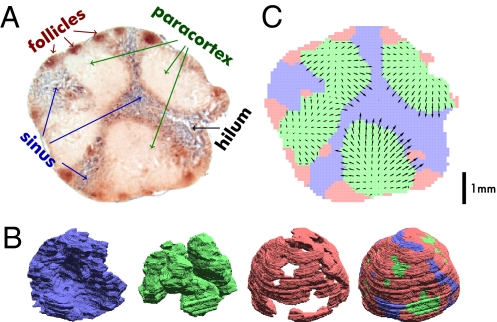
Fig. 4.
Modeling the naive T cell LN transit. (A) Microtome slice from a mesenteric rat LN with B cells stained in brown and macrophages stained in blue. (B) Using image processing algorithms (SI Text), the lymph node volume was divided into three compartments: sinus (blue; abundant macrophages), paracortex (green; few macrophages and B cells), and follicles (red; few macrophages and abundant B cells). 3D renderings of the reconstructed compartments are shown. (C) The 3D reconstruction was used to simulate the transit of naive T cells from paracortex to sinus (a 2D projection of the central LN slice is shown). The model assumes that naive T cells are released uniformly in the paracortex (green), transit to a nearby sinus region (blue), and then, exit after around 1 h. For simulating taxis, a hypothetical chemokine gradient (arrows) pointing from paracortex to sinus was created. Arrows indicate direction of the gradient and taxis speed per minute, with arrow lengths magnified 250-fold based on a mean taxis speed of 0.5 μm/min.
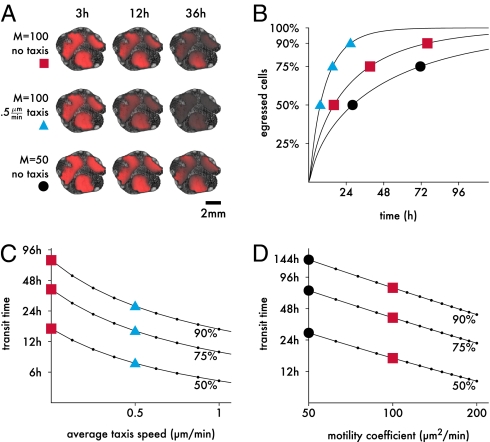
Fig. 5.
Impact of taxis on the LN transit. (A) 2D snapshots of T cell concentrations from the central slice of the reconstructed LN (Fig. 4C) superimposed on grayscale images of the slice. Intensity of the red color is proportional to T cell concentration. Top corresponds to random T cell motility as estimated from our data. In the biased population (Middle), cells perform taxis to the sinuses with 0.5 μm/min on average. Bottom shows the effect of a 50% lower motility coefficient. (B) Detailed exit kinetics of the three simulated populations (symbols on the curve indicate the matching populations in A). For random T cell migration (squares), our simulation gives a realistic median transit time of 16.1 h, which taxis accelerates roughly twofold (7.33 h). (C) Transit time quantiles as a function of the taxis speed with M = 100 μm2/min. Squares and triangles mark the same values as in A. (D) Transit time as a function of the random walk motility coefficient in the absence of taxis. Squares and circles mark the same values in A.
To determine the impact of taxis on this transit time, we augmented our model by a hypothetical long-range chemokine gradient pointing to the sinuses (Fig. 4C and SI Text) and performed simulations with varying cell responsiveness to this gradient, giving average paracortical taxis speeds of, at most, 2 μm/min. The impact of taxis on transit time was dramatic (Fig. 5 A and C). Merely 0.5 μm/min accelerated the median transit time 2.2-fold, and the time at which 90% of the T cells had egressed decreased 2.84-fold. For comparison, we also analyzed the impact of varying the motility coefficient on the transit time (Fig. 5 A and D). Here, we found, consistent with the definition of the motility coefficient, a roughly antiproportional relationship (i.e., to reduce the median transit time by 50%, the motility coefficient would need to increase twofold). Thus, the impact of over- or underestimating the motility coefficient by 20% would be much less than that of 0.5 μm/min of taxis.
Our simple model assumes that T cells freely transmigrate between sinuses and paracortex. The precise mechanisms of T cell egress into the sinus have received much attention (23–27), and these studies proposed the following multistep model. T cells enter branched cortical sinusoid structures, which are mostly located in a narrow transitional region between paracortex and sinuses (27), in an S1P-dependent manner. From there, they flow to the macrophage-rich regions within the sinus and ultimately, reach the medulla from where they exit through efferent lymphatic vessels connected to the hilum. In a recent study, Grigorova et al. (27) simulated this multistep egress using a 3D reconstruction of the cortical sinusoid structure of a mouse LN. In their model, cells can exit only through the sinusoids rather than across the whole paracortex–sinus interface. To approximate the effect of a transitional region that partially obstructs transzonal migration in our model, we lowered cell motility near the paracortex–sinus boundary (SI Text and Fig. S8). On the one hand, this modification increases the time that cells need to reach the sinuses; on the other hand, after reaching the boundary region, cells will more likely remain near it, which should decrease the transit time. Simulations of the modified model show cell crowding near the boundary (Fig. S8C), consistent with the observations by Wei et al. (28) that cells apparently queue to exit near the sinuses. However, because cells still spend most of their time in the deep paracortex, 0.5 μm/min taxis still decrease the median transit time 1.74-fold from 16.15 to 9.3 h. Thus, our qualitative conclusions are unaffected by this more realistic transmigration model.
Discussion
We have revisited a fundamental question. Is naive T cell migration in LNs in the absence of antigen directed or random? Or in the words of Bajénoff et al. (16), how many cells would need to migrate nonrandomly for it to be detected by two-photon microscopy? Using Hotelling's T2 test, we can rule out uniform taxis of 1.67 μm/min 3D speed with a confidence of 95%. This upper bound is less than 10% of the mean cell speed and less than one cell diameter in 10 min. The result extends to populations only partially affected by taxis (e.g., a 50% subpopulation with taxis of 3.34 μm/min is also excluded). Hence, the pure random walk (with persistence) is a surprisingly accurate model of naive T cell migration at the two-photon imaging timescale. Hotelling's test is applicable to any cell type and can be used to reanalyze existing datasets.
Our benchmark results for two-photon data analysis methods indicate that, for klinotaxis and topotaxis, Hotelling's test and angle analysis are similarly powerful provided that the taxis direction for angle analysis is exactly known. Even then, angle analysis misses orthotaxis of up to 4 μm/min in simulated T cell data. This issue should be addressable by analyzing angles together with speeds, which Beltman et al. (13) did most recently. Both angle analysis and Hotelling's test outperformed displacement analysis, presumably because both are based on cell steps (Fig. 1C) instead of entire tracks (Fig. 1B). Our imaging volume is axially thin to increase image acquisition speed and avoid phototoxicity. In such a volume, long tracks mostly represent slower than average cells moving in the x and y plane. Beltman et al. (15) argued that this effect may compromise cell-based methods such as displacement analysis but not step-based methods. Our analysis confirms that downward taxis is not missed by step-based methods, whereas displacement analysis gives confusing results; the downward taxis causes the observed displacement to decrease rather than increase as expected, which would lead to a lower motility coefficient estimate instead of a larger one. Extreme care should, thus, be taken when using motility coefficients to compare different cell populations, which is common practice (14). The results should be supported by additional quantitative analysis methods, and simulations such as ours can be used to investigate whether this artifact could arise under the given experimental conditions.
There is recent evidence that lymphocyte migration in secondary lymphoid organs is guided by stromal cell networks (29). T cells in LNs were shown to crawl on fibroblastic reticular cells (30). Because the taxis mode can be relevant for data analysis, it is instructive to consider how different taxis modes could be realized by cells crawling on a network. For simplicity, let us assume that cells can change directions only at network intersections. Then, orthotaxis could be caused by a fluid flow that influences cell speeds, but it is hard to envisage how such a flow could be maintained with the turbulent T cell motion. Alternatively, T cells might have preferred turning directions at network intersections, but in the densely packed space, they would have to compete for these directions. Cells moving in their preferred direction would try to keep going but could be pushed away by other cells. This scenario would likely lead to a mix of slow klinotaxis and topotaxis.
Slow taxis, which we cannot safely rule out, can still have a huge long-term impact. In our mesenteric LN model, merely 0.5 μm/min of taxis (less than 3% of the cell speed) accelerate the naive T cell transit time roughly two times. A simple calculation can illustrate why this occurs. If we fill a ball with a radius of 800 μm with cells that migrate to the surface at 0.5 μm/min, then all cells within 90 μm from the surface would leave the ball within 3 h. Although 90 μm seems small compared with the radius, only 100·(710/800)3 = 70% of the cells would remain. However, the transit times predicted by our pure diffusion model are more realistic. Consistent with our results, Grigorova et al. (27) predict a transit time of 4–5 h for a small (0.5–1 mm) inguinal LN using a random walk model, which even without taxis, is already very fast. Chemokinesis alone might, thus, explain naive T cell motility. This view is supported by findings that chemical factors promote T cell motility but not directionality; the main chemical cues driving T cell motility are the CCR7 ligands CCL19 and CCL21, which are present in the LN paracortex (31). In CCR7-deficient mice, T cell motility within LNs is markedly reduced, but the directionality seems unaffected (32).
It is important to point out that our results do not imply that taxis of less than 10% the cell speed is impossible to detect—ruling out taxis is not the same as showing taxis. For the latter problem, assumptions like a potential target region or a specific affected cell subtype can be exploited to increase sensitivity and detect even highly nonuniform or localized taxis that could escape our generic analysis. For instance, if the putative taxis direction is known, one can normalize tracks from multiple experiments accordingly and combine them in a single test. This method was recently used by a study by Beltman et al. (13), which revealed taxis of germinal center B cells to the light zone at less than 5% of the cell speed. If taxis is suspected to act only within a region that can be clearly identified, one can focus the analysis on the tracks within that region. Similarly, suppose there were several gradients pointing in opposite directions, and each affected equal shares of the observed population. If we have a hypothesis identifying the affected subpopulations, we can apply Hotelling's test to each one separately and therefore, reveal the taxis.
In summary, we have presented a method to rule out uniform taxis in two-photon data and showed that the displacement plots used in the past to provide evidence for random migration can miss large amounts of such taxis. For naive T cells migrating in the LN paracortex in the absence of antigen, the remaining gap for taxis is narrow, and it is on the same quantitative range as subtle imaging artifacts like tissue drift (15). To narrow the gap even more, we used a simulation that extrapolates subtle motility features from the two-photon timescale to a larger one, where they accumulate to substantial, testable changes. Still, it is not yet possible to rule out (relevant) taxis completely, because even very small taxis can have a big long-term impact. How can the remaining gap be closed? One option would be increasing the imaging volume, because the width of the confidence region is roughly antiproportional to the square root of the number of cells visible at the same time (e.g., a 16 times larger volume would yield a fourfold lower bound). Independently, data analysis and modeling techniques could be further improved. One limitation of our and similar modeling approaches (13, 19, 27) is the assumption that cells are massless particles. Although such models can faithfully reproduce individual cell trajectories, they cannot account for some effects of dense cell packing. For example, our diffusion model predicts a cell concentration gradient across the paracortex at equilibrium. Spatially explicit approaches like the cellular Potts model (33) or cellular automata (34) better reflect the effects of cell crowding, but they do not always faithfully reproduce trajectory properties like turning angles or persistence (33). Future work is needed to devise modeling techniques that can account for both cell- and population-level properties of cell migration in densely crowded lymphoid organs.
Materials and Methods
Intravital Two-Photon Microscopy.
Magnetic bead negatively selected polyclonal CD4+ and CD8+ T cells (5 × 106, >95% purity; Miltenyi) were fluorescently labeled with 4 μM 5-chloromethylfluorescein diacetate (Cell Tracker Green; Molecular Probes) and adoptively transferred to sex-matched 6-wk-old C57BL/6 recipient mice. After 24 h, mice were anesthetized by an initial i.p. injection of ketamine (50 mg/kg) and xylazine (10 mg/kg). The right popliteal LN was prepared microsurgically for intravital microscopy and positioned on a custom-built microscope stage. Care was taken to spare blood vessels and afferent lymph vessels. The prepared LN was submerged in normal saline and covered with a glass coverslip. A thermocouple was placed next to the LN to monitor local temperature, which was maintained at 37 °C. Two-photon imaging was performed with an Olympus BX50WI fluorescence microscope equipped with a 20×, 0.95 numerical aperture objective (Olympus) using a Prairie Technologies Ultima Two-Photon Microscope. For two-photon excitation and second harmonic generation, a Tsunami Ti:sapphire laser with a 10-W MilleniaXs pump laser (Spectra-Physics) with Deepsee module was tuned to 800 nm. For 4D analysis of cell migration, stacks of 10 square x and y sections with 4 μm z spacing were acquired every 20.3 s with electronic zooming up to two times to provide image volumes 40 μm in depth. Cell tracks were extracted using Volocity software. All of the above experiments were in accordance with National Institutes of Health guidelines and were approved by the Committees on Animal Care and Use of both Harvard Medical School and the Immune Disease Institute.
Statistical Analysis of Cell-Tracking Data.
Analyses of migration angles relative to a known taxis direction were performed as described by Beltman et al. (15). All other analyses were performed according to the step by step protocols given in SI Text.
Simulating Cell Tracks with Taxis.
The parameters used for the model by Beauchemin et al. (19) were tfree = 2 min, tpause = 0.5 min, and vfree = 19.1 μm/min; vfree was adjusted to match the desired motility coefficient. The model was extended for simulating taxis with a predefined speed as described in SI Text. For generating the datasets shown in Figs. 2 and 3D, 1,000 cells were set to random initial positions in a 550 × 550 × 550 μm3 torus and simulated for 60 min; their trajectories were tracked inside in a 492 × 492 × 40 μm3 subvolume (the imaging region size in our 2P experiments) of the torus. The cells in Fig. 3B were tracked in an unlimited space.
3D Reconstruction of a Rat LN.
Adult male Lewis rats were obtained from Charles River GmbH and were housed in the central animal facility of the University of Lübeck. A single mesenteric LN was harvested from a rat and fixated. The organ was cut in a microtome into 12-μm-thick slices. T cells and macrophages were stained on the sections using anti-IgM (brown) and anti-ED12 (blue) antibodies. The slices were photographed using a digital camera mounted on a microscope (Zeiss). 3D reconstruction of the LN and partial differential equation-based simulation of the T cell LN transit were then performed as described in SI Text. All experiments were in accordance with the German Animal Protection Law and were approved by the Animal Research Ethics Board of the Ministry of Environment.
Supplementary Material
Acknowledgments
J.T. thanks Rüdiger Reischuk, Vitaly Ganusov, and Rob de Boer for helpful discussions, Till Tantau and Alan Perelson for comments on earlier versions of the manuscript, and Joost Beltman for comments on an early version of this paper and sharing his data. J.W. was supported by Deutsche Forschungsgemeinschaft Grant SFB654, C4. U.H.v.A. was supported by National Institutes of Health Grants HL56949, AI069259, AI072252, CA71932, and AI078897. J.T. was supported by a Doktorandenstipendium from the Deutscher Akademischer Austauschdienst (DAAD).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. R.N.G. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1102288108/-/DCSupplemental.
References
- 1.Miller MJ, Wei SH, Parker I, Cahalan MD. Two-photon imaging of lymphocyte motility and antigen response in intact lymph node. Science. 2002;296:1869–1873. doi: 10.1126/science.1070051. [DOI] [PubMed] [Google Scholar]
- 2.Miller MJ, Wei SH, Cahalan MD, Parker I. Autonomous T cell trafficking examined in vivo with intravital two-photon microscopy. Proc Natl Acad Sci USA. 2003;100:2604–2609. doi: 10.1073/pnas.2628040100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mackay CR. Chemokines: Immunology's high impact factors. Nat Immunol. 2001;2:95–101. doi: 10.1038/84298. [DOI] [PubMed] [Google Scholar]
- 4.Dickinson RB, Tranquillo RT. Transport equations and indices for random and biased cell migration based on single cell properties. SIAM J Appl Math. 1995;55:1419–1454. [Google Scholar]
- 5.Ionides EL, Fang KS, Isseroff RR, Oster GF. Stochastic models for cell motion and taxis. J Math Biol. 2004;48:23–37. doi: 10.1007/s00285-003-0220-z. [DOI] [PubMed] [Google Scholar]
- 6.Okada T, et al. Antigen-engaged B cells undergo chemotaxis toward the T zone and form motile conjugates with helper T cells. PLoS Biol. 2005;3:e150. doi: 10.1371/journal.pbio.0030150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McDonald B, et al. Intravascular danger signals guide neutrophils to sites of sterile inflammation. Science. 2010;330:362–366. doi: 10.1126/science.1195491. [DOI] [PubMed] [Google Scholar]
- 8.Castellino F, et al. Chemokines enhance immunity by guiding naive CD8+ T cells to sites of CD4+ T cell-dendritic cell interaction. Nature. 2006;440:890–895. doi: 10.1038/nature04651. [DOI] [PubMed] [Google Scholar]
- 9.von Andrian UH, Mackay CR. T-cell function and migration. Two sides of the same coin. N Engl J Med. 2000;343:1020–1034. doi: 10.1056/NEJM200010053431407. [DOI] [PubMed] [Google Scholar]
- 10.Schwickert TA, et al. In vivo imaging of germinal centres reveals a dynamic open structure. Nature. 2007;446:83–87. doi: 10.1038/nature05573. [DOI] [PubMed] [Google Scholar]
- 11.Figge MT, et al. Deriving a germinal center lymphocyte migration model from two-photon data. J Exp Med. 2008;205:3019–3029. doi: 10.1084/jem.20081160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meyer-Hermann M, Figge MT, Toellner KM. Germinal centres seen through the mathematical eye: B-cell models on the catwalk. Trends Immunol. 2009;30:157–164. doi: 10.1016/j.it.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 13.Beltman JB, Allen CD, Cyster JG, de Boer RJ. B cells within germinal centers migrate preferentially from dark to light zone. Proc Natl Acad Sci USA. 2011;108:8755–8760. doi: 10.1073/pnas.1101554108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cahalan MD, Parker I. Choreography of cell motility and interaction dynamics imaged by two-photon microscopy in lymphoid organs. Annu Rev Immunol. 2008;26:585–626. doi: 10.1146/annurev.immunol.24.021605.090620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beltman JB, Marée AFM, de Boer RJ. Analysing immune cell migration. Nat Rev Immunol. 2009;9:789–798. doi: 10.1038/nri2638. [DOI] [PubMed] [Google Scholar]
- 16.Bajénoff M, et al. Highways, byways and breadcrumbs: Directing lymphocyte traffic in the lymph node. Trends Immunol. 2007;28:346–352. doi: 10.1016/j.it.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 17.Oja H, Randles RH. Multivariate nonparametric tests. Statist Sci. 2004;19:598–605. [Google Scholar]
- 18.Sumen C, Mempel TR, Mazo IB, von Andrian UH. Intravital microscopy: Visualizing immunity in context. Immunity. 2004;21:315–329. doi: 10.1016/j.immuni.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 19.Beauchemin C, Dixit NM, Perelson AS. Characterizing T cell movement within lymph nodes in the absence of antigen. J Immunol. 2007;178:5505–5512. doi: 10.4049/jimmunol.178.9.5505. [DOI] [PubMed] [Google Scholar]
- 20.Westermann J, Puskas Z, Pabst R. Blood transit and recirculation kinetics of lymphocyte subsets in normal rats. Scand J Immunol. 1988;28:203–210. doi: 10.1111/j.1365-3083.1988.tb02432.x. [DOI] [PubMed] [Google Scholar]
- 21.Westermann J, Persin S, Matyas J, van der Meide P, Pabst R. IFN-γ influences the migration of thoracic duct B and T lymphocyte subsets in vivo. Random increase in disappearance from the blood and differential decrease in reappearance in the lymph. J Immunol. 1993;150:3843–3852. [PubMed] [Google Scholar]
- 22.Stekel DJ, Parker CE, Nowak MA. A model of lymphocyte recirculation. Immunol Today. 1997;18:216–221. doi: 10.1016/s0167-5699(97)01036-0. [DOI] [PubMed] [Google Scholar]
- 23.Matloubian M, et al. Lymphocyte egress from thymus and peripheral lymphoid organs is dependent on S1P receptor 1. Nature. 2004;427:355–360. doi: 10.1038/nature02284. [DOI] [PubMed] [Google Scholar]
- 24.Schwab SR, et al. Lymphocyte sequestration through S1P lyase inhibition and disruption of S1P gradients. Science. 2005;309:1735–1739. doi: 10.1126/science.1113640. [DOI] [PubMed] [Google Scholar]
- 25.Schwab SR, Cyster JG. Finding a way out: Lymphocyte egress from lymphoid organs. Nat Immunol. 2007;8:1295–1301. doi: 10.1038/ni1545. [DOI] [PubMed] [Google Scholar]
- 26.Grigorova IL, et al. Cortical sinus probing, S1P1-dependent entry and flow-based capture of egressing T cells. Nat Immunol. 2009;10:58–65. doi: 10.1038/ni.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grigorova IL, Panteleev M, Cyster JG. Lymph node cortical sinus organization and relationship to lymphocyte egress dynamics and antigen exposure. Proc Natl Acad Sci USA. 2010;107:20447–20452. doi: 10.1073/pnas.1009968107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wei SH, et al. Sphingosine 1-phosphate type 1 receptor agonism inhibits transendothelial migration of medullary T cells to lymphatic sinuses. Nat Immunol. 2005;6:1228–1235. doi: 10.1038/ni1269. [DOI] [PubMed] [Google Scholar]
- 29.Mueller SN, Germain RN. Stromal cell contributions to the homeostasis and functionality of the immune system. Nat Rev Immunol. 2009;9:618–629. doi: 10.1038/nri2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bajénoff M, et al. Stromal cell networks regulate lymphocyte entry, migration, and territoriality in lymph nodes. Immunity. 2006;25:989–1001. doi: 10.1016/j.immuni.2006.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mrass P, Petravic J, Davenport MP, Weninger W. Cell-autonomous and environmental contributions to the interstitial migration of T cells. Semin Immunopathol. 2010;32:257–274. doi: 10.1007/s00281-010-0212-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Worbs T, Mempel TR, Bölter J, von Andrian UH, Förster R. CCR7 ligands stimulate the intranodal motility of T lymphocytes in vivo. J Exp Med. 2007;204:489–495. doi: 10.1084/jem.20061706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beltman JB, Marée AF, Lynch JN, Miller MJ, de Boer RJ. Lymph node topology dictates T cell migration behavior. J Exp Med. 2007;204:771–780. doi: 10.1084/jem.20061278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Linderman JJ, et al. Characterizing the dynamics of CD4+ T cell priming within a lymph node. J Immunol. 2010;184:2873–2885. doi: 10.4049/jimmunol.0903117. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.