Abstract
High-throughput mRNA sequencing (RNA-Seq) holds the promise of simultaneous transcript discovery and abundance estimation1-3. We introduce an algorithm for transcript assembly coupled with a statistical model for RNA-Seq experiments that produces estimates of abundances. Our algorithms are implemented in an open source software program called Cufflinks. To test Cufflinks, we sequenced and analyzed more than 430 million paired 75bp RNA-Seq reads from a mouse myoblast cell line representing a differentiation time series. We detected 13,692 known transcripts and 3,724 previously unannotated ones, 62% of which are supported by independent expression data or by homologous genes in other species. Analysis of transcript expression over the time series revealed complete switches in the dominant transcription start site (TSS) or splice-isoform in 330 genes, along with more subtle shifts in a further 1,304 genes. These dynamics suggest substantial regulatory flexibility and complexity in this well-studied model of muscle development.
Recently, high-throughput sequencing of mRNA (RNA-Seq) has revealed tissue-specific alternative splicing4, novel genes and transcripts5, and genomic structural variations6. Deeply sampled RNA-Seq permits measurement of differential gene expression with greater sensitivity than expression7 and tiling8 microarrays. However, the analysis of RNA-Seq data presents major challenges in transcript assembly and abundance estimation arising from the ambiguous assignment of reads to isoforms8-10. In earlier RNA-Seq experiments conducted by some of us, we estimated the relative expression for each gene as the fraction of reads mapping to its exons after normalizing for gene length11. We did not attempt to allocate reads to specific alternate isoforms although we found ample evidence that multiple splice and promoter isoforms are often co-expressed in a given tissue 2. This raised biological questions about how the different forms are distributed across cell types and physiological states. In addition, our prior methods relied on annotated gene models that, even in mouse, are incomplete. Longer reads (here 75bp versus 25bp in our prior work) and pairs of reads from both ends of each RNA fragment can reduce uncertainty in assigning reads to alternative splice variants12. To produce useful transcript-level abundance estimates from paired-end RNA-Seq data, we developed a new algorithm that can identify complete novel transcripts and probabilistically assign reads to isoforms.
For our initial demonstration of Cufflinks, we performed a time course of paired-end 75bp RNA-Seq on a well-studied model of skeletal muscle development, the C2C12 mouse myoblast cell line13 (Methods). Regulated RNA expression of key transcription factors drives myogenesis and the execution of the differentiation process involves changes in expression of hundreds of genes14,15. Prior studies have not measured global transcript isoform expression, though there are well-documented expression changes at the whole gene level for a set of marker genes in this system. We aimed to establish the prevalence of differential promoter use and differential splicing, because such data could reveal much about the model's regulatory behavior. A gene with isoforms that code for the same protein may be subject to complex regulation in order to maintain a certain level of output in the face of changes in expression of its transcription factors. Alternatively, genes with isoforms that code for different proteins could be functionally specialized for different cell types or states. By analyzing changes in relative abundances of transcripts produced by the alternative splicing of a single primary transcript, we hoped to infer the impact of post-transcriptional processing (e.g. splicing) on RNA output separately from rates of primary transcription. Such analysis could identify key genes in the system and suggest experiments to establish how they are regulated.
We first mapped sequenced fragments to the mouse genome using an improved version of TopHat16, which can align reads across splice junctions without relying on gene annotation (Supplementary Methods Section 2). Out of 215 million fragments, 171 million (79%) mapped to the genome, and 46 million spanned at least one putative splice junction (Supplementary Table 1). Of the splice junctions spanned by fragment alignments, 70% were present in transcripts annotated by UCSC, Ensembl, or Vega.
To recover the minimal set of transcripts supported by our fragment alignments, we designed a comparative transcriptome assembly algorithm. EST assemblers such as PASA introduced the idea of collapsing alignments to transcripts based on splicing compatibility17, and Dilworth's Theorem18 has been used to assemble a parsimonious set of haplotypes from virus population sequencing reads19. Cufflinks extends these ideas, reducing the transcript assembly problem to finding a maximum matching in a weighted4 bipartite graph that represents compatibilities17 among fragments (Fig. 1a,b,c and Supplementary Methods Section 4). Non-coding RNAs20 and microRNAs21 have been reported to regulate cell differentiation and development, and coding genes are known to produce noncoding isoforms as a means of regulating protein levels through nonsense-mediated decay22. For these biologically motivated reasons, the assembler does not require that assembled transcripts contain an open reading frame. Since Cufflinks does not make use of existing gene annotations during assembly, we validated the transcripts by first comparing individual time point assemblies to existing annotations. We recovered a total of 13,692 known isoforms and 12,712 new isoforms of known genes. We estimate that 77% of the reads originated from previously known transcripts (Supplementary Table 2). Of the new isoforms, 7,395 (58%) contain novel splice junctions, with the remainder being novel combinations of known splicing outcomes. 11,712 (92%) have an open reading frame (ORF), 8,752 of which end at an annotated stop codon. Although we sequenced deeply by current standards, 73% of the moderately abundant (15-30 FPKM) transcripts detected at the 60 hour time point with three lanes of GAII transcriptome sequencing were fully recovered with just a single lane. Because distinguishing a full-length transcript from a partially assembled fragment is difficult, we conservatively excluded novel isoforms that were unique to a single time point from further analyses. Out of the new isoforms, 3,724 were present in multiple time points, and 581 were present at all time points. 6,518 (51%) of the new isoforms and 2,316 (62%) of the multiple time point novel isoforms were tiled by high-identity EST alignments or matched RefSeq isoforms from other organisms, and endpoint RT-PCR experiments confirmed new isoforms in genes of interest (Supplementary Table 3). We concluded that a majority of the unannotated transcripts we found are in the myogenic transcriptome, and that the mouse annotation remains incomplete.
Figure 1.

Overview of Cufflinks. The algorithm takes as input cDNA fragment sequences that have been (a) aligned to the genome by software capable of producing spliced alignments, such as TopHat. With paired-end RNA-Seq, Cufflinks treats each pair of fragment reads as a single alignment. The algorithm assembles overlapping ‘bundles’ of fragment alignments (b-c) separately, which reduces running time and memory use because each bundle typically contains the fragments from no more than a few genes. Cufflinks then estimates the abundances of the assembled transcripts (d-e). (b) The first step in fragment assembly is to identify pairs of ‘incompatible’ fragments that must have originated from distinct spliced mRNA isoforms. Fragments are connected in an ‘overlap graph’ when they are compatible and their alignments overlap in the genome. Each fragment has one node in the graph, and an edge, directed from left to right along the genome, is placed between each pair of compatible fragments. In this example, the yellow, blue, and red fragments must have originated from separate isoforms, but any other fragment could have come from the same transcript as one of these three. (c) Assembling isoforms from the overlap graph. Paths through the graph correspond to sets of mutually compatible fragments that could be merged into complete isoforms. The overlap graph here can be minimally ‘covered’ by three paths, each representing a different isoform. Dilworth's Theorem states that the number of mutually incompatible reads is the same as the minimum number of transcripts needed to “explain” all the fragments. Cufflinks implements a proof of Dilworth's Theorem that produces a minimal set of paths that cover all the fragments in the overlap graph by finding the largest set of reads with the property that no two could have originated from the same isoform. (d) Estimating transcript abundance. Fragments are matched (denoted here using color) to the transcripts from which they could have originated. The violet fragment could have originated from the blue or red isoform. Gray fragments could have come from any of the three shown. Cufflinks estimates transcript abundances using a statistical model in which the probability of observing each fragment is a linear function of the abundances of the transcripts from which it could have originated. Because only the ends of each fragment are sequenced, the length of each may be unknown. Assigning a fragment to different isoforms often implies a different length for it. Cufflinks can incorporate the distribution of fragment lengths to help assign fragments to isoforms. For example, the violet fragment would be much longer, and very improbable according to Cufflinks' model, if it were to come from the red isoform instead of the blue isoform. (e) The program then numerically maximizes a function that assigns a likelihood to all possible sets of relative abundances of the yellow, red and blue isoforms (γ1,γ2,γ3), producing the abundances that best explain the observed fragments, shown as a pie chart.
For the purposes of estimating transcript abundances, we first selected a set of 11,079 genes containing 17,416 high-confidence isoforms (Supplementary File 1). Of these, 13,692 (79%) were known and the remaining 3,724 (21%) were novel isoforms of known genes present in multiple time points. We then developed a statistical model of RNA-Seq parameterized by the abundances of these transcripts (Fig. 1d,e,f, Supplementary Methods Section. 3). Cufflinks' model allows for the probabilistic deconvolution of RNA-Seq fragment densities to account for cases where genome alignments of fragments do not uniquely correspond to source transcripts. The model incorporates minimal assumptions23 about the sequencing experiment, and extends the unpaired read model of Jiang and Wong8 to the paired-end case. Abundances were reported in expected Fragments Per Kilobase of transcript per Million fragments mapped (FPKM). A fragment corresponds to a single cDNA molecule, which can be represented by a pair of reads from each end. Confidence intervals for estimates were obtained using a Bayesian inference method based on importance sampling from the posterior distribution. Abundances of spiked control sequences (R2=0.99) and benchmarks with simulated data (R2=0.96) revealed that Cufflinks' abundance estimates are highly accurate. The inclusion of novel isoforms of known genes during abundance estimation had a dramatic impact on the estimates of known isoforms in many genes (R2 only 0.90), highlighting the importance of coupling transcript discovery together with abundance estimation.
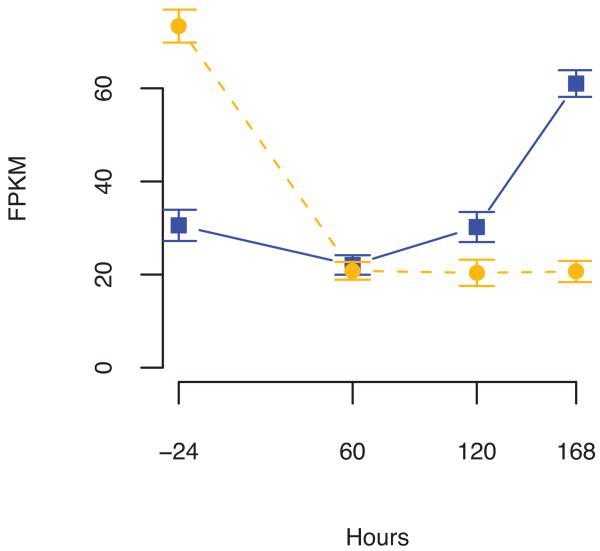
We identified 7,770 genes and 10,480 isoforms undergoing significant abundance changes between some successive pair of time points (FDR < 5%). Many genes display substantial transcript-level dynamics that are not reflected in their overall expression patterns (Supplementary File 2). For example, Myc, a proto-oncogene which is known to be transcriptionally and post-transcriptionally regulated during myogenesis24, is down-regulated overall during the time course, and while isoforms A and B follow this pattern, isoform C has a more complex expression pattern (Figure 2b). We noted that many genes displayed switching between major and minor transcripts, some containing isoforms with muscle-specific functions, such as tropomyosin I and II, which display a dramatic switch in isoform dominance upon differentiation (Supplementary Appendix B). However, many genes featured dynamics involving several isoforms with behavior too complex to be deemed “switching”. In light of these observations, we classified the patterns of expression dynamics for transcripts, assigning them one of four “trajectories” based on their expression curves being flat, increasing, decreasing or mixed (Methods). Based on trajectory classification, a total of 1,634 genes were found to have multiple isoforms with different trajectories in the time course, and we hypothesized that differential promoter preference and differential splicing were responsible for the divergent patterns.
Figure 2.
Distinction of transcriptional and post-transcriptional regulatory effects on overall transcript output. (a) When abundances of isoforms A, B, and C of Myc are grouped by TSS, changes in the relative abundances of the TSS groups indicate transcriptional regulation. Post-transcriptional effects are seen in changes in levels of isoforms of a single TSS group. (b) Isoforms of Myc have distinct expression dynamics. (c) Myc isoforms are downregulated as the time course proceeds. The width of the colored band is the measure of change in relative transcript abundance and the color is the log ratio of transcriptional and post-transcriptional contributions to change in relative abundances (plot construction detailed in Supplementary Method Section 5.3). Changes in relative abundances of Myc isoforms suggest that transcriptional effects immediately following differentiation at 0 hours give way to post-transcriptional effects later in the time course, as isoform A is eliminated.
To explore the impact of regulation on mRNA output and to check whether it could explain the variability of trajectories, we grouped transcripts by their start site (TSS) instead of just by gene. Changes in the relative abundances of mRNAs spliced from the same pre-mRNA transcript are by definition post-transcriptional, so this grouping effectively discriminated changes in mRNA output associated with differential transcription from changes associated with differential post-transcriptional processing. Of the 3,486 genes in our high confidence set with isoforms that shared a common TSS, 41% had TSS groups containing different isoform trajectories. Summing the expressions of isoforms sharing a TSS produces the trajectory for their primary transcript, and we identified 401 (48%) genes with multiple distinct primary transcript trajectories. However, trajectory classification was not precise enough to prioritize further investigation into individual genes and could not form the basis for statistical significance testing. We therefore formalized and quantified divergent expression patterns of isoforms within and between TSS groups with an information-theoretic metric derived from the Jensen-Shannon divergence. With this metric, relative transcript abundances move in time along a logarithmic spiral in a real Hilbert space25, and the distance moved measures the extent of change in relative expression. Quantification of expression change in this way revealed significant (FDR < 5%) differential transcriptional regulation and splicing in 882 of 3,486 (25%) and 273 of 843 (32%) candidate genes respectively, with 70 genes displaying both types of differential regulation (Supplementary Table 4). Myc (Fig. 2a,b) undergoes a shift in transcriptional regulation of transcript abundances to post-transcriptional control of abundances (Fig. 2c) between 60 and 90 hours, as myocytes are beginning to fuse into myotubes.
Focusing on the genes with significant promoter and isoform changes, we noted that in many cases changes in relative abundance reflected switch-like events in which there was an inversion of the dominant primary transcript. For example, in FHL3, a transcriptional regulator recently reported to inhibit myogenesis26, Cufflinks assembled the known isoform and another with a novel start site. We validated the 5′ exon of this isoform along with other novel start sites and splicing events by form-specific RT-PCR (Fig. 3a, Supplementary Methods Section 4). Limiting analysis to known isoforms would have produced an incorrect abundance estimate for the known isoform of FHL3. Moreover, the novel isoform is dominant prior to differentiation, so this potentially important differentiation-associated promoter switch would have been missed (Fig. 3b). In total, we tested and validated 153 of 185 putative novel transcription start sites by comparison against TAF1 and RNA polymerase II ChIP-Seq peaks. We also observed switches in the major isoform of alternatively spliced genes. In total, 10% of multi-promoter genes featured a switch in major primary transcript and 7% of alternatively spliced primary transcripts switched major isoforms. We concluded that not only is the impact of promoter-switching on mRNA output significant, many genes are also exhibiting evidence of post-transcriptionally induced expression changes, supporting a role for dynamic splicing regulation in myogenesis. A key question is whether genes that display divergent expression patterns of isoforms are differentially regulated in a particular system because they have isoforms that are functionally specialized for that system. Of the genes undergoing transcriptional or post-transcriptional isoform switches, 26%, respectively 24%, code for multiple distinct proteins according to annotation. Genes with novel isoforms were excluded from the coding sequence analysis, so this fraction likely underestimates the impact of differential regulation on coding potential. We thus speculate that differential RNA level isoform regulation, whether transcriptional, post-transcriptional, or mixed in underlying mechanism, suggests functional specialization of the isoforms in many genes.
Figure 3.
Excluding isoforms discovered by Cufflinks from the transcript abundance estimation impacts the abundance estimates of known isoforms, in some cases by orders of magnitude. Four-and-a-half-LIM domains 3 (Fhl3) inhibits myogenesis by binding MyoD and attenuating its transcriptional activity. (a) The C2C12 transcriptome contains a novel isoform that is dominant during proliferation. The new TSS for Fhl3 is supported by proximal TAF1 and RNA polymerase II ChIP-Seq peaks. (b) The known isoform (solid line) is preferred at time points following differentiation.
Although Cufflinks was designed to investigate transcriptional and splicing regulation in this experiment, it is applicable to a broad range of RNA-Seq studies (Fig. 4). The open-source software runs on commonly available and inexpensive hardware, making it accessible to any researcher using RNA-Seq data. We are currently exploring the use of the Cufflinks assembler to annotate genomes of newly sequenced organisms, and to quantify the impact of various mechanisms of gene regulation on expression. When coupled with assays of upstream regulatory activity, such as chromatin state mapping or promoter occupancy, Cufflinks should help unveil the range of mechanisms governing RNA manufacture and processing.
Figure 4.
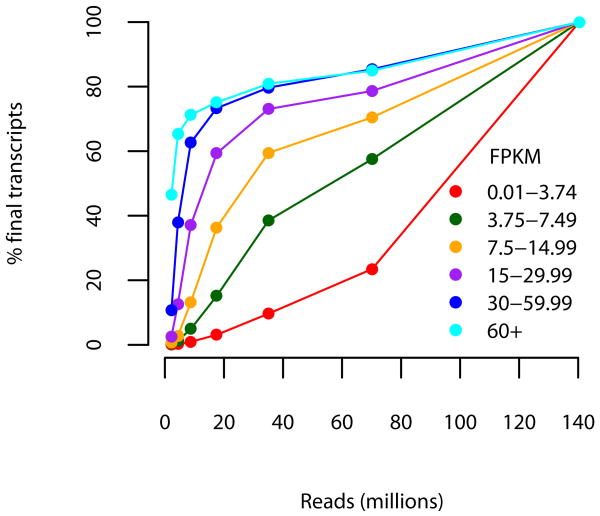
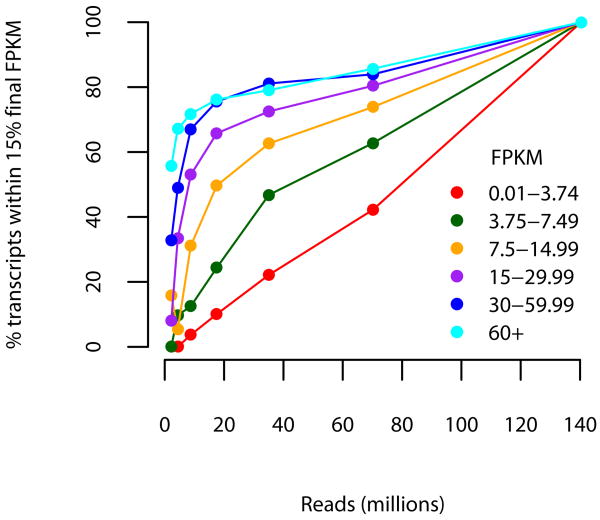
Robustness of assembly and abundance estimation as a function of expression level and depth of sequencing. Subsets of the full 60-hour read set were mapped and assembled with TopHat and Cufflinks and the resulting assemblies were compared for structural and abundance agreement with the full 60 hour assembly. Colored lines show the results obtained at different depths of sequencing in the full assembly; e.g., the light blue line tracks the performance for transcripts with FPKM greater than 60. (a) The fraction of transcript fragments fully recovered increases with additional sequencing data, though nearly 75% of moderately expressed (≥15 FPKM) are recovered with less than 40 million 75bp paired-end reads (20 million fragments), a fraction of the data generated by a single run of the sequencer used in this experiment. (b) Abundance estimates are similarly robust. At 40 million reads, transcripts determined to be moderately expressed using all 60 hour reads were estimated at within 15% of their final FPKM values.
Methods
RNA isolation
Mouse skeletal muscle C2C12 cells were initially plated on 15 cm plates in DMEM with 20% fetal bovine serum. At confluence, the cells were switched to low serum medium to initiate myogenic differentiation. For extraction of total RNA, cells were first rinsed in PBS and then lysed in Trizol reagent (Invitrogen catalog # 15596-026) either during exponential growth in high serum medium, or at 60 hrs, 5 days and 7 days after medium shift. Residual contaminating genomic DNA was removed from the total RNA fraction using Turbo DNA-free (Ambion catalog # AM1907M). mRNA was isolated from DNA-free total RNA using the Dynabeads mRNA Purification Kit (Invitrogen catalog # 610-06).
Fragmentation and reverse transcription
Preparation of cDNA followed the procedure described in Mortazavi et al.2, with minor modifications as described below. Prior to fragmentation, a 7 uL aliquot (∼ 500 pgs total mass) containing known concentrations of 7 “spiked in” control transcripts from A. thaliana and the lambda phage genome were added to a 100 ng aliquot of mRNA from each time point. This mixture was then fragmented to an average length of 200 nts by metal ion/heat catalyzed hydrolysis. The hydrolysis was performed in a 25 uL volume at 94°C for 90 seconds. The 5X hydrolyis buffer components are: 200 mM Tris acetate, pH 8.2, 500 mM potassium acetate and 150 mM magnesium acetate. After removal of hydrolysis ions by G50 Sephadex filtration (USA Scientific catalog # 1415-1602), the fragmented mRNA was random primed with hexamers and reverse-transcribed using the Super Script II cDNA synthesis kit (Invitrogen catalog # 11917010). After second strand synthesis, the cDNA went through end-repair and ligation reactions according to the Illumina ChIP-Seq genomic DNA preparation kit protocol (Illumina catalog # IP102-1001), using the paired end adapters and amplification primers (Illumina Catalog # PE102-1004). Ligation of the adapters adds 94 bases to the length of the cDNA molecules.
Size selection
The cDNA library was size-fractionated on a 2% TAE low melt agarose gel (Lonza catalog # 50080), with a 100 bp ladder (Roche catalog # 14703220) run in adjacent lanes. Prior to loading on the gel, the ligated cDNA library was taken over a G50 Sephadex column to remove excess salts that interfere with loading the sample in the wells. After post-staining the gel in ethidium bromide, a narrow slice (∼2mm) of the cDNA lane centered at the 300 bp marker was cut. The slice was extracted using the QiaEx II kit (Qiagen catalog # 20021), and the extract was filtered over a Microcon YM-100 microconcentrator (Millipore catalog # 42409) to remove DNA fragments shorter than 100 bps. Filtration was performed by pipeting the extract into the upper chamber of a microconcentrator, and adding ultra pure water (Gibco catalog # 10977) to a volume of 500 uLs. The filter was spun at 500 X g until only 50 uLs remained in the upper chamber (about 20 minutes per spin) and then the upper chamber volume was replenished to 500 uLs. This procedure was repeated 6 times. The filtered sample was then recovered from the filter chamber according to the manufacturer's protocol. Fragment length distributions obtained after size selection were estimated from the spike-in sequences and are show in Supplementary Fig. 1.
Amplification
One-sixth of the filtered sample volume was used as template for 15 cycles of amplification using the paired-end primers and amplification reagents supplied with the Illumina ChIP-Seq genomic DNA prep kit. The amplified product was then cleaned up over a Qiaquick PCR column (Qiagen catalog # 28104), and then the filtration procedure using the Microcon YM-100 microconcentrators described above was repeated, to remove both amplification primers and amplification products shorter than 100 bps. A final pass over a G50 Sephadex column was performed, and the library was quantified using the Qubit fluorometer and PicoGreen quantification reagents (Invitrogen catalog # Q32853). The library was then used to build clusters on the Illumina flow cell according to protocol.
Mapping cDNA fragments to the genome
Fragments were mapped to build 37.1 of the mouse genome using TopHat version 1.0.13. We extended our previous algorithms to exploit the longer paired reads used in the study. TopHat version 1.0.7 and later splits a read 75bp or longer in three or more segments of approximately equal size (25bp), and maps them independently. Reads with segments that can be mapped to the genome only non-contiguously are marked as possible intron-spanning reads. These “contiguously unmappable” reads are used to build a set of possible introns in the transcriptome. TopHat accumulates an index of potential splice junctions by examining segment mapping for all contiguously unmappable reads. For each junction the program then concatenates kbp upstream of the donor to kbp downstream of the acceptor to form a synthetic spliced sequence around the junction. The segments of the contiguously unmappable reads are then aligned against these synthetic sequences with Bowtie. The resulting contiguous and spliced segment alignments for these reads are merged to form complete alignments to the genome, each spanning one or more splice junctions. Further details of how version 1.0.13 of TopHat differs from the published algorithm are provided in Section 2 of the Supplementary Methods.
Transcript abundance estimation
We estimated transcript abundances using a generative statistical model of RNA-Seq experiments. The model was parameterized by the relative abundances of the set of all transcripts in a sample. For computational convenience, abundances of non-overlapping transcripts in disjoint genomic loci were calculated independently. The parameters of the model were the non-negative abundances ρt. Denoting the fragment distribution by F, we defined the effective length of a transcript to be
where l(t) is the length of a transcript. The likelihood function for our model was then given by:
where the products were over all fragment alignments R and transcripts T in the transcriptome, and It(r) was the implied length of a fragment determined by a pair of reads assuming it originated from transcript t (Supplementary Fig. 2). This is the likelihood function for a linear model, and therefore, assuming the model was identifiable, the likelihood function had a unique maximum, which our implementation calculated via a numerical optimization procedure. Rather than reporting this estimate, we instead found the MAP estimate using a Bayesian inference procedure based on importance sampling from the posterior distribution. The proposal distribution we used was multivariate normal with mean given by the maximum likelihood estimate discussed above, and variance-covariance matrix given by the inverse of the observed Fisher information matrix. The samples were also used to compute 95% confidence intervals for the maximum a posteriori (MAP) estimates. The MAP estimates and (and associated confidence intervals) were used for differential expression testing.
Abundances were reported in FPKM (expected fragments per kilobase of transcript per million fragments sequenced). This unit is a scalar multiple of the parameters ρt. FPKM is conceptually analogous to the reads per kilobase per million reads sequenced (RPKM) measure, but it explicitly accommodates sequencing data with one, two, or – if needed for future sequencing platforms – higher numbers of reads from single source molecules.
Abundance estimates were validated using spike-in sequences (Supplementary Fig. 3) and simulations (Supplementary Fig. 4). In order to confirm that all transcripts of a gene are necessary for accurate abundance estimation, novel transcripts were removed from the analysis (Supplementary Fig. 5) showing that resulting estimates may be biased.
Transcript assembly
Transcripts were assembled from the mapped fragments sorted by reference position. Fragments were first divided into non-overlapping loci, and each locus was assembled independently of the others using the Cufflinks assembler. The assembler was designed to find the minimal number of transcripts that “explain” the reads (i.e. every read should be contained in some transcript). First erroneous spliced alignments or reads from incompletely spliced RNAs were filtered out. The algorithm for assembly was based on a constructive proof of Dilworth's Theorem (see Supplementary Methods, Appendix A, Theorem 17). Each fragment alignment was assigned a node in an “overlap graph” G. A directed edge (x,y) was placed between nodes x and y when the alignment for x started at a lower coordinate than y, the alignments overlapped in the genome, and the fragments were “compatible” (Supplementary Fig. 6). Compatibility was defined for overlapping fragments for which every implied intron in one fragment matched an identical implied intron in the other fragment. The resulting directed, acyclic graph was transitively reduced to produce G, to avoid including redundant path information. Cufflinks then found a minimum path cover of G, meaning that every fragment node was contained in some path in the cover, and the cover contained as few paths as possible. Each path in the cover corresponded to a set of mutually compatible fragments overlapping each other on the left and right (except initial and terminal fragments on the path). Dilworth's theorem implied that this path cover could be constructed by first finding the largest set of fragments with the property that no two are compatible. This set was determined by finding a maximum matching in a bipartite graph constructed from the transitive closure of G. The bipartite “reachability graph” had a node in each partition for all fragments in G, and nodes were connected if there was a path between them in G. Given a maximum cardinality matching M, any fragment without an incident edge in M was a member of an antichain. Each member of this antichain could be extended to a path, and this extension was a minimum path cover of G.
The minimum cardinality chain decomposition computed using the approach above was not guaranteed to be unique. In order to “phase” distant exons, we leveraged the fact that abundance inhomogeneities could link distant exons via their coverage. We therefore weighted the edges of the bipartite reachability graph based on the percent-spliced-in metric introduced by Wang et al.4 Cufflinks arbitrated between multiple parsimonious assemblies by choosing the minimum-cost maximum matching in the reachability graph. In our setting, the percent- spliced-in ψx for an alignment x was computed by counting the alignments overlapping x in the genome that were compatible with x and dividing by the total number of alignments that overlap x, and normalizing for the length of the x. The cost C(y, z) assigned to an edge between alignments y and z reflected the belief that they originated from different transcripts:
A useful feature of the Cufflinks assemblies is that they resulted in provably identifiable models. Complete details of the Cufflinks assembler are provided in the Supplementary Material (Section 4), along with proofs of several key theorems.
Structural comparison of time point assemblies
To validate Cufflinks transfrags (assembled transcript fragments) against annotated transcriptomes, and also to find transfrags common to multiple assemblies, we developed a tool called “Cuffcompare” that builds structural equivalence classes of transcripts. We ran Cuffcompare on the assembly from each time point against the combined annotated transcriptomes of UCSC, Ensembl, and Vega (Supplementary Fig. 7). Because of the stochastic nature of sequencing, assembly of the same transcript in two different samples may result in transfrags of slightly different lengths. A Cufflinks transfrag was considered a complete match when there was a transcript with an identical chain of introns in the combined annotation. When no complete match was found between a Cufflinks transfrag and the transcripts in the combined annotation, Cuffcompare determined and reported if another potentially significant relationship existed with any of the annotation transcripts that could be found in or around the same genomic locus.
Assembly and abundance robustness analysis
A total of 61,787,833 cDNA fragments were sequenced at 60 hours. We mapped and assembled subsets of these fragments (at fractions 1/64, 1/32, 1/16, 1/8, 1/4, and 1/2 of the total) using TopHat and Cufflinks.
Each assembly of parts of the data was compared to the assembly obtained with the full fragment set using Cuffcompare. We counted transcripts recovered in assemblies from partial data that structurally matched some transcripts in the assembly using all the reads. We assessed robustness of abundance estimation by counting the fraction of assembled transcripts that were assigned abundances within 15% of the FPKM value reported for the full fragment set transcript.
Simulation-based validation
To assess the accuracy of Cufflinks' estimates, we simulated an RNA-Seq experiment using the FluxSimulator27, a freely available software package that models whole transcriptome sequencing experiments with the Illumina Genome Analyzer. The software works by first randomly assigning expression values to the transcripts provided by the user, constructing an amplified, size-selected library, and sequencing it. Mouse UCSC transcripts were supplied to the software, along with build 37.1 of the genome. FluxSimulator then randomly assigned expression levels to 18,935 UCSC transcripts. From these relative expression levels, the software constructed an in silico RNA-Seq sample, with each transcript assigned a number of library molecules according to its abundance. FluxSimulator produced 13,203,516 75bp paired-end RNA-Seq reads from 6,601,805 library fragments, which were mapped with TopHat to the mouse genome using identical parameters to those used to map the C2C12 reads. A total of 6,176,961 fragments were mapped (93% of the library). These alignments were supplied along with the exact set of expressed transcripts to Cufflinks, to measure Cufflinks' abundance estimation accuracy when working with a “perfect” assembly.
Validation of novel transcription start sites
Transcripts with 5′ exons not in UCSC, Ensembl, or VEGA were selected for validation. We excluded transcripts with estimated abundances less than 5.0 FPKM at all time points, as well as transcripts with a 5′ exon within 200bp of an annotated exon. To validate our novel observed 5′ exons, we conducted ChIP-Seq experiments as previously described28 at -24 and 60 hour time points using an antibody to the unphosphorylated CTD-repeat of RNA polymerase II (8WG16, Covance) as well as an antibody to TAF1 (SC-735, Santa Cruz) which marks promoters. For each candidate 5′end, we took the region +/- 200 bp and measured the normalized read density (RPKM) of each ChIP-Seq, requiring at least 1.5 RPKM of ChIP-Seq signal for both polymerase and TAF1 at either time point.
Endpoint RT-PCR validation of novel isoforms
Six genes with multiple assembled splice isoforms were chosen as cases for endpoint PCR validation, including three with novel isoforms (Supplementary Figs. 8,9). Amplification primers that cross the Cufflinks predicted spliced-exon junctions were purchased from Integrated DNA Technologies, Inc. (San Diego, CA). 5 ugs of total RNA from each time point was primed with oligodT(20) (Invitrogen catalog# 18418020), and reverse-transcribed at 50C using SuperScript III reverse transcriptase, (Invitrogen catalog # 18080044) according to the manufacturer's protocol. One tenth of the cDNA reaction was used as template for 35 rounds of PCR amplification with each pair of junction-crossing primers. The PCR reactions were cleaned up using the Qiaquick PCR cleanup kit (Qiagen catalog# 28104), and quantified using a Nanodrop spectrophotometer. An equal mass of DNA from each reaction (50 ngs) was then loaded in each lane of a 2.0% agarose gel, post-stained with Sybr Gold (Invitrogen Catalog # S11494) and visualized on a UV transilluminator.
Analysis of gene expression and regulation dynamics
In order to test for divergent expression dynamics among isoforms, we tested all high-confidence isoforms for significant changes between each time point using the abundance variance estimates produced by our statistical model (FDR < 5%). Trajectories were assigned to transcript expression curves based on significant (FDR<5%) increases or decreases in expression between consecutive time points. To be deemed significant, expression between consecutive time points also had to change by at least 25%. The possible trajectories were therefore reduced to 81 combinatorial possibilities (increasing, decreasing or flat between any of the three pairs of consecutive time points). Trajectories were then classified into 4 groups: increasing (3 consecutive increases), decreasing (3 consecutive decreases), flat (no changes) and mixed (presence of both increases and decreases in expression along the time course). To test for significant changes in relative abundance a group of transcripts, we calculated the square root of the Jensen-Shannon divergence on the relative abundances in each of two time points. The variance of this metric under the null hypothesis of no change in relative abundance can be estimated using the delta method from the variance-covariance matrix on abundances estimates. Using the estimated variance of the JS metric, we applied a one-sided t-test for significant changes in relative abundance of transcripts grouped by TSS and also primary transcripts grouped by gene. Type I errors were controlled with the Benjamini-Hochberg correction for multiple testing of differential expression, splicing, and promoter preference throughout the analysis. Supplementary Figs. 10,11 show examples of genes with significant changes in relative transcript abundances during the time-course.
Software availability
TopHat is freely available as source code at http://tophat.cbcb.umd.edu. It takes a reference genome (as a Bowtie29 index) and RNA-Seq reads as FASTA or FASTQ and produces alignments in SAM30 format. TopHat is distributed under the Artistic License and runs on Linux and Mac OS X.
The Cufflinks assembler and abundance estimation algorithms are open-source C++ programs and are freely available in both source and binary at http://cufflinks.cbcb.umd.edu/. The package includes the assembler along with utilities to structurally compare Cufflinks output between samples (Cuffcompare) and to perform differential expression testing (Cuffdiff). Cufflinks is distributed under the Boost License and runs on Linux and Mac OS X. The source code for Cufflinks version 0.8.0 is provided in Supplementary File 3.
Supplementary Material
Acknowledgments
This work was supported in part by NIH grants R01-LM006845 and ENCODE U54-HG004576, as well as the Beckman Foundation, Bren Foundation, Moore Foundation (Cell Center Program) and the Miller Research Institute. We thank Dr. Igor Antosechken and Lorian Schaeffer of the Caltech Jacobs Genome Center for DNA sequencing, and Diane Trout, Brandon King and Henry Amrhein for data pipeline and database design, operation, and display. We are grateful to Robert K. Bradley, Kiril Datchev, Ingileif Hallgrímsdóttir, Jane Landolin, Ben Langmead, Adam Roberts, Michael Schatz, and David Sturgill, for helpful discussions.
Footnotes
Author contributions: C.T. and L.P. developed the mathematics and statistics and designed the algorithms. B.A.W. and G.K. performed the RNA-Seq and B.A.W. designed and executed experimental validations. C.T. implemented Cufflinks and Cuffdiff. G.P. implemented Cuffcompare. M.v.B. and A.M tested the software. C.T., G.P. and A.M. performed the analysis. L.P., A.M. and B.J.W., conceived the project. C.T., L.P, A.M., B. J. W, and S.L.S. wrote the manuscript.
Accession: GEO, GSE20846
References
- 1.Cloonan N, et al. Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nature Methods. 2008;5:613–619. doi: 10.1038/nmeth.1223. [DOI] [PubMed] [Google Scholar]
- 2.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 3.Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D. The Transcriptional Landscape of the Yeast Genome Defined by RNA Sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang E, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Denoeud F, et al. Annotating genomes with massive-scale RNA sequencing. Genome Biology. 2008;9:R175. doi: 10.1186/gb-2008-9-12-r175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maher C, et al. Transcriptome sequencing to detect gene fusions in cancer. Nature. 2009;458:97–101. doi: 10.1038/nature07638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marioni J, Mason C, Mane S, Stephens M, Gilad Y. RNA-seq: An assessment of technical reproducibility and comparison with gene expression arrays. Genome Research. 2008;10:1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hiller D, Jiang H, Xu W, Wong W. Identifiability of isoform deconvolution from junction arrays and RNA-Seq. Bioinformatics. 2009;25:3056–3059. doi: 10.1093/bioinformatics/btp544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang H, Wong WH. Statistical Inferences for Isoform Expression in RNA-Seq. Bioinformatics. 2009;25:1026–1032. doi: 10.1093/bioinformatics/btp113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li B, Ruotti V, Stewart RM, Thomson JA, Dewey CN. RNA-Seq gene expression estimation with read mapping uncertainty. Bioinformatics. 2009;26:493–500. doi: 10.1093/bioinformatics/btp692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mortazavi A, Williams B, Mccue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 12.Pepke S, Wold B, Mortazavi A. Computation for ChIP-Seq and RNA-Seq studies. Nature Methods. 2009;6:S22–32. doi: 10.1038/nmeth.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yaffe D, Saxel O. A myogenic cell line with altered serum requirements for differentiation. Differentiation. 1977;7:159–166. doi: 10.1111/j.1432-0436.1977.tb01507.x. [DOI] [PubMed] [Google Scholar]
- 14.Yun K, Wold B. Skeletal muscle determination and differentiation: story of a core regulatory network and its context. Current opinion in cell biology. 1996;8:877–889. doi: 10.1016/s0955-0674(96)80091-3. [DOI] [PubMed] [Google Scholar]
- 15.Tapscott SJ. The circuitry of a master switch: Myod and the regulation of skeletal muscle gene transcription. Development. 2005;132:2685–2695. doi: 10.1242/dev.01874. [DOI] [PubMed] [Google Scholar]
- 16.Trapnell C, Pachter L, Salzberg S. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haas BJ, et al. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Research. 2003;31:5654–5666. doi: 10.1093/nar/gkg770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dilworth R. A decomposition theorem for partially ordered sets. Annals of Mathematics. 1950;51:161–166. [Google Scholar]
- 19.Eriksson N, et al. Viral Population Estimation Using Pyrosequencing. PLoS Computational Biology. 2008;4:e1000074. doi: 10.1371/journal.pcbi.1000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guttman M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;457:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cordes KR, et al. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature. 2009;460:705–710. doi: 10.1038/nature08195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lareau LF, Inada M, Green RE, Wengrod JC, Brenner SE. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature. 2007;446:926–929. doi: 10.1038/nature05676. [DOI] [PubMed] [Google Scholar]
- 23.Bullard J, Purdom E, Hansen K, Durinck S, Dudoit S. Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments. BMC Bioinformatics. 2010;11:94. doi: 10.1186/1471-2105-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Endo T, Nadal-Ginard B. Transcriptional and posttranscriptional control of c-myc during myogenesis: its mRNA remains inducible in differentiated cells and does not suppress the differentiated phenotype. Mol Cell Biol. 1986;6:1412–1421. doi: 10.1128/mcb.6.5.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fuglede B, Topsøe F. Proceedings of the IEEE International Symposium on Information Theory. 2004;3 [Google Scholar]
- 26.Cottle DL, McGrath MJ, Cowling BS, Coghill ID. FHL3 binds MyoD and negatively regulates myotube formation. Journal of Cell Science. 2007;120:1423–1435. doi: 10.1242/jcs.004739. [DOI] [PubMed] [Google Scholar]
- 27.Sammeth M, Lacroix V, Ribeca P, Guigó R. The FLUX Simulator. doi: 10.1093/nar/gks666. http://flux.sammeth.net. [DOI] [PMC free article] [PubMed]
- 28.Johnson D, Mortazavi A, Myers R, Wold B. Genome-Wide Mapping of in Vivo Protein-DNA Interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 29.Langmead B, Trapnell C, Pop M, Salzberg S. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biology. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.