Abstract
Alphaproteobacteria are common members of marine bacterioplankton assemblages, but are believed to be rare in lacustrine systems. However, uncultured Alphaproteobacteria of the freshwater LD12 lineage form a tight monophyletic sister group with the numerically dominant bacteria in marine epipelagic waters, the SAR11 clade or genus Pelagibacter. Comparative rRNA sequence analysis reveals a global occurrence of LD12 bacteria in freshwater systems. The association of genotypic subclades with single-study systems moreover suggests a regional diversification. LD12 bacteria exhibit distinct and annually recurring spatio-temporal distribution patterns in prealpine lakes, as assessed by seasonally resolved vertical profiling and high-throughput cell counting. During the summer months, these ultramicrobacteria can form cell densities in the surface (epilimnetic) water layers that are comparable to those of their marine counterparts (>5 × 108 cells per l). LD12 bacteria had a pronounced preference for glutamine and glutamate over 7 other amino acids in situ, and they exhibited substantially higher uptake of these two substrates (and glycine) than the microbial assemblage in general. In addition, members of LD12 were also able to exploit other monomeric sources of organic carbon such as glucose, fructose or acetate. LD12 seemed to follow an oligotrophic lifestyle with slow but efficient uptake already at low substrate concentrations. Thus, LD12 bacteria do not only share phenotypic and metabolic traits with Pelagibacter, but also seem to thrive in the analogous spatiotemporal niche in freshwaters. The two groups together form one of the rare monophyletic lineages of ultramicrobacteria that have successfully traversed the barrier between marine and freshwater habitats.
Keywords: bacterioplankton, freshwater lakes, LD12/SAR11
Introduction
The freshwater bacterioplankton is believed to be numerically dominated by small Actinobacteria, Betaproteobacteria and microbes related to the Flavo- and Sphingobacteriales (Glöckner et al., 1999, 2000; Zwart et al., 2002). By contrast, Alphaproteobacteria are regarded to be rare in the pelagic zones of most lacustrine ecosystems (as opposed to marine pelagic habitats), with only very few exceptions (Kirchman et al., 2004; Nishimura and Nagata, 2007). This view is mainly based on the results of quantitative rRNA-based methodology, for example, of whole-cell fluorescence in situ hybridization techniques (for review see Table VI in Klammer et al., 2002, and Appendix 3 in Nishimura and Nagata, 2007). However, the presumably most abundant microbial species on earth, the marine bacterium Pelagibacter ubique and relatives—also known as SAR11-cluster (Alphaproteobacteria) (Morris et al., 2002; Rappe et al., 2002; Carlson et al., 2008)—has both a brackish and a freshwater sister group (Bahr et al., 1996; Carlson et al., 2008). Members of the species-like (>98% sequence similarity) freshwater cluster, termed LD12 (Zwart et al., 1998), alpha V (Glöckner et al., 2000) or SAR11 IV (Carlson et al., 2008), are only known from environmental 16S rRNA gene sequences. A ubiquitous occurrence of these bacteria in lakes (Bahr et al., 1996; Zwart et al., 2002) has been suggested, albeit based on a limited number of complete sequences available in public databases. It is unknown whether members of the LD12 clade are only an exotic remnant lineage and form small populations in lake water, or whether they are as successful and ecologically relevant in freshwater pelagic habitats as their marine counterparts.
The success of SAR11 bacteria in marine waters is attributed to a number of features that render them well adapted to the nutrient and energy-limited conditions in offshore habitats such as the photic zone of the Sargasso Sea (Morris et al., 2002). Cultivated representatives (Pelagibacter ubique) are very small (ultramicrobacteria, cell volume <0.01 μm3; Rappe et al., 2002) with a streamlined genome dominated by transport functions (Giovannoni et al., 2005b; Sowell et al., 2009), which are driven by light-dependent proton pumps (proteorhodopsins; (Giovannoni et al., 2005a)). Moreover, they are centrally involved in the remineralization of low molecular weight organic matter such as amino acids or glucose (Malmstrom et al., 2004; Alonso and Pernthaler, 2006), but require exogenous-reduced sulfur compounds for growth (Tripp et al., 2008). Several ecotypes within the SAR11 group have been identified and linked to either spatio-temporal habitat preferences (Carlson et al., 2008) or to ocean productivity (Schwalbach et al., 2010).
We investigated the quantitative importance and potential role of bacteria of the LD12 lineage in freshwater habitats by studying (i) their occurrence and quantitative importance in several freshwater lakes with different hydrochemical and limnological properties, (ii) their seasonal and vertical distribution patterns at high spatio-temporal resolution and (iii) selected metabolic traits, that is, the incorporation of various low molecular weight organic compounds. Our results suggest that members of the LD12 clade are one of the most abundant ubiquitous lineages of freshwater bacteria, the quantitative importance of which so far has been largely overlooked. We furthermore could gain first insight into the ecological niche of LD12 bacteria both by identifying their spatio-temporal distribution maxima and by assessing their specific substrate preferences.
Materials and methods
Sampling
Lake Zurich is a large, deep, prealpine, mesotrophic lake, characterized by persistent annual blooms of the toxic cyanobacterium Planktothrix rubescens (Bossard et al., 2001; Van den Wyngaert et al., 2011). Water samples of 53 depth profiles were collected nearby the deepest point of the lake (136 m) in a biweekly interval between January 17th 2008 and December 21st 2009. Core sampling depths were 0, 5, 10, 20, 30, 40, 60, 80 and 100 m, and additional samples were taken from seemingly interesting depths (that is, depths of chlorophyll a maxima or maxima of P. rubescens), resulting in a total sample number of 490. A volume of 40 ml of water was fixed with formaldehyde (2% final concentration) for flow cytometry, and 5–10 ml were fixed with buffered paraformaldehyde (2% f.c.) for CARD–FISH analyses. Temperature profiles were recorded with an YSI multiprobe.
Piburger See, a small oligo-mesotrophic lake with a maximum depth of 24.6 m is situated in the Austrian Alps and was sampled in a monthly interval between February 2005 and February 2006 (Salcher et al., 2008). Water samples of 3, 9, 18 and 24 m depth were fixed as mentioned above, and unfixed water for autoradiographic assays (see below) was transported to the lab within 2 h. Water temperature was directly determined during sampling. Thalwiler Waldweiher is a small, shallow (5.5 m maximum depth), eutrophic pond, which was sampled in a monthly interval in a 1-m depth resolution during the ice-free period between May 2008 and July 2009. Temperature profiles were recorded with an YSI multiprobe. Lake Constance is a large, deep (250 m), oligotrophic lake connected to Gnadensee (shallow, mesotrophic) and Zellersee (shallow, mesotrophic) (Güde et al., 1998). Lake Constance, Gnadensee, Zeller See, Badsee (small, shallow, eutrophic) and Federsee (small, shallow, eutrophic) were sampled in July 2009. Samples from 0, 5, 10, 15, 20 and 30 m depth were analyzed from Lake Constance, and surface-water samples (Table 1) were taken from the other lakes. Egelsee is a shallow, meromictic, meso-eutrophic lake with a pronounced layer of green and purple sulfur bacteria (Chlorobium sp., Thiopedia sp. and Chromatium sp.) between 6–7 m depth. A depth profile of this lake was acquired in September 2007. Depth profiles of the large, deep and oligo-mesotrophic lakes Lago Maggiore (372 m depth) and oligotrophic Lake Thun (217 m depth) were gained in July 2010 and Gossenköllesee (small, ultraoligotrophic, high mountain lake (Pernthaler et al., 1998)) was sampled in August 2005. Horgener Waldweiher (small, shallow, eutrophic) was sampled in June 2009.
Table 1. Relative LD12 abundance (% of all bacteria) in different European lakes determined by CARD–FISH with probes LD12–121 and EUB I–III.
| Lake | Coordinates | Trophic status | Sampling depths (max depth) | Sampling (n) | Mean % of bacteria | Range |
|---|---|---|---|---|---|---|
| Lake Zurich | 47.31 N, 8.58 E | M | 0–100 (136) | 2 years (490) | 9.2 | 1.9–35.6 |
| Thalwiler Waldweiher | 47.28 N, 8.56 E | E | 0–5.5 (5.5) | 2 years (64) | 14.3 | 1.9–42.9 |
| Piburger See | 47.20 N, 10.89 E | O-M | 3–24 (24.6) | 1 year (44) | 4.7 | 0.4–21.5 |
| Lake Constance | 47.65 N, 9.29 E | M | 0–30 (252) | Summer (6) | 12.8 | 7.5–17.9 |
| Egelsee | 47.40 N, 8.36 E | M-E | 0–9 (9) | Autumn (6) | 6.0 | 0.4–12.7 |
| Lago Maggiore | 45.96 N, 8.63 E | O-M | 0–350 (372) | Summer (14) | 9.1 | 1.9–20.3 |
| Lake Thun | 46.69 N, 7.72 E | O | 0–213 (217) | Summer (12) | 9.7 | 3.6–13.6 |
| Gossenköllesee | 47.23 N, 11.01 E | UO | 0–8 (8) | Summer (5) | 0.8 | 0.7–1.0 |
| Gnadensee | 47.70 N, 9.09 E | M | 5 | Summer (1) | 18.2 | — |
| Zeller See | 47.72 N, 8.97 E | M | 5 | Summer (1) | 19.4 | — |
| Badsee | 47.75 N, 10.0 E | E | 1 | Summer (1) | 21.7 | — |
| Federsee | 48.08 N, 9.63 E | E | 1 | Summer (1) | 6.3 | — |
| Horgener Weiher | 47.24 N, 8.60 E | E | 0.5 | Summer (1) | 17.8 | — |
Abbreviations: E, eutrophic; M, mesotrophic; M-E, meso-eutrophic; O, oligotrophic; O-M, oligo-mesotrophic; UO, ultraoligotrophic.
Cloning, sequence analysis of 16S rRNA genes, and probe design
A volume of 250 ml of unfixed water samples from Lake Zurich from 10, 12.5 and 15 m depth from October 1st 2007 were filtered onto white polycarbonate membranes and stored frozen until further processing. DNA was extracted with the powerbead soil isolation kit (MOBIO, Carlsbad, CA, USA) and 16S rRNA genes were amplified with primers GM3f and GM4r (Muyzer et al., 1995). PCR products were purified with the QIAquick PCR purification kit (QIAGEN, Hilden, Germany), inserted into TOPO vectors (TOPO TA cloning kit for sequencing; Invitrogen, Carlsbad, CA, USA) and cloned into competent E. coli according to the manufacturer's instructions. After screening of the clones for right-sized inserts, plasmid preparations were carried out with the QIAprep Spin Miniprep Kit (QIAGEN). The sequencing reactions were accomplished with the primers GM1f (Muyzer et al., 1993), the vector primers M13f and M13r (Messing et al., 1983) and the ABI BigDye chemistry on an ABI 3730 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA). Partial sequences were assembled with the DNA baser software (HeracleSoftware, Lilienthal, Germany) and checked for chimeric origin using Mallard and Pintail (Ashelford et al., 2005). These sequence data have been submitted to the EMBL database under accession No. FN665702–FN665785. Phylogenetic analyses were performed with the ARB software package (Ludwig et al., 2004) using the ARB reference database SILVA SSU100. All sequences were automatically aligned by the SINA web aligner (http://www.arb-silva.de) and alignments were subsequently manually optimized. Only sequences >1200 nucleotides were used for tree reconstruction, where first a maximum parsimony tree was built. All good quality sequences affiliated with SAR11 (2569) and LD12 (370) were selected for bootstrapped maximum likelihood analyses (100 repetitions) using the RAxML web server (Stamatakis et al., 2005). The obtained trees were manually compared, reduced by 251 almost identical LD12 sequences (mostly from Lake Gatun (Rusch et al., 2007)), and nodes with bootstrap values <50% were collapsed into multifurcations. Probe design for the LD12 cluster was carried out with the respective ARB tools probe_design and probe_check, and obtained probes were also checked for specificity in the ribosomal database project (http://rdp.cme.msu.edu). The resulting probe LD12–121 (5′-CACAAGGCAGATTCCCACAT-3′) was tested with different formamide concentrations until stringency was achieved at 35%.
Enumeration of microbes
Total bacterial abundances were counted by an inFlux V-GS cell sorter (Becton Dickinson, Franklin Lakes, NJ, USA) equipped with a UV laser (Lightwave Electronics, Milpitas, CA, USA, CY-PS, 60 mW, wavelength of 355 nm), a blue laser (Coherent, Saphire, 200 mW, wavelength of 488 nm), and detectors for two scatter and six fluorescence channels. All samples were stained with DAPI (4′,6-diamidino-2-phenylindole, 1 μg ml−1 final concentration), and scatter plots were analyzed with the software FlowJo 7.2.2. (Tree Star, Ashland, OR, USA).
CARD–FISH (fluorescence in situ hybridization followed by catalyzed reporter deposition) was carried out as previously described (Sekar et al., 2003) with slight modifications for probe LD12–121, such as longer signal amplification (45 min) and a higher concentration of Alexa488 labeled tyramides (4 μl ml−1 amplification buffer). CARD–FISH-stained filters were analyzed by fully automated high-throughput microscopy (Zeder and Pernthaler, 2009), where three images were recorded: UV excitation for DAPI stained cells, blue excitation for hybridized cells and green excitation for autofluorescent cells, which otherwise interfere with hybridization signals. All images were analyzed with an image analysis macro (M Zeder, unpublished), and interfering autofluorescent cyanobacteria or debris were individually subtracted from hybridized cells. A total of 10–36 high-quality images were analyzed for each sample. Replicated hybridization of 28 randomly chosen filters resulted in a mean s.d. of 0.2% of DAPI (CV=0.082).
Cell volume estimations of bacteria hybridized with probes LD12–121 and EUB I–III were carried out with the image analysis program LUCIA D (http://www.lim.cz) as described in Posch et al. (2009) for one sample of Lake Zurich (August 14th 2008, 0 m depth). In all, 400–500 cells were measured and significant differences between length, width and biovolume of bacteria hybridized with LD12 and EUB I–III were identified by paired t-tests computed in SPSS 16.0.
Microautoradiography coupled with CARD–FISH (MAR–FISH)
Autoradiography assays were carried out with the following radioactive substrates (all purchased from Amersham, GE Healthcare, Chalfont St Giles, UK): [3H]-thymidine (77 Ci mmol−1 specific activity), [3H]-glucose (29 Ci mmol−1 s.a.), [14C]-fructose (0.316 Ci mmol−1 s.a.), [14C]-acetate (56 Ci mmol−1 s.a.), [3H]-amino acids (45 Ci mmol−1 s.a.), [3H]-arginine (49 Ci mmol−1 s.a.), [3H]-lysine (73 Ci mmol−1 s.a.), [3H]-glutamine (53 Ci mmol−1 s.a.), [3H]-glutamate (45 Ci mmol−1 s.a.), [14C]-alanine (0.154 Ci mmol−1 s.a.), [14C]-aspartate (0.207 Ci mmol−1 s.a.), [14C]-serine (0.255 Ci mmol−1 s.a.), [14C]-glycine (0.101 Ci mmol−1 s.a.) and [3H]-leucine (61 Ci mmol−1 s.a.). All substrates were added at 10 nmol l−1 final concentrations and incubated for 2 h at in situ temperature for Lake Zurich samples from 5 m depth (September 16th, 2009), whereas monthly samples from Piburger See were spiked with 5 nmol l−1 [3H]-amino acids (48 Ci mmol−1 s.a.) and incubated for 1 h at in situ temperature. Additionally, a ‘substrate affinity' experiment was carried out with [3H]-Glutamine (60 Ci mmol−1 s.a.), for Lake Zurich samples (5 m depth, September 28th, 2010). Four different concentrations (0.1, 1, 10 and 100 nmol l−1, 120-min incubation each) and five different incubation times (15, 30, 60, 120 and 240 min, 10 nmol l−1 concentration each) were used to gain more information on the affinity of LD12 to the selected tracer and on incorporation speed. Triplicates plus two prefixed controls were processed as outlined in Salcher et al. (2008) with the modification that filter sections were glued (1% agarose) onto cover slips with bacteria facing down, carefully peeled off, and cover slips were mounted onto microscopic slides for autoradiography. Therefore, the very small LD12 cells were not covered by silver grains, but lying above the photo emulsion. Cover slips were thereafter embedded in a mounting medium containing DAPI and samples were manually evaluated with a Zeiss Microscope (Carl Zeiss, Oberkochen, Germany) with bright field illumination and blue excitation. At least 100 hybridized cells were inspected per sample.
Results and Discussion
Phylogenetic relationship of freshwater LD12 and marine SAR11 bacteria
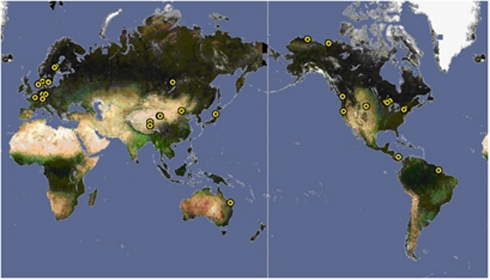
Bacterial 16S rRNA gene sequences affiliated with LD12 (n=370) originated from habitats distributed over several continents and from both hemispheres (Figure 1), supporting the notion of their ubiquitous distribution (Glöckner et al., 2000; Zwart et al., 2002). Sequences from a single lake (Lake Gatun) in the GOS data set (Rusch et al., 2007) contributed almost two thirds of all LD12 sequences (n=202). Our own collection of bacterial 16S rRNA gene sequences from the large mesotrophic Lake Zurich, Switzerland, also yielded a surprisingly high proportion of LD12-related genotypes (84 of 442 sequences). Although the majority of LD12 sequences have been obtained from freshwater habitats, some have also been found in estuarine habitats such as the Chesapeake Bay (n=3) or the Delaware estuary (n=20).
Figure 1.
Ubiquitous distribution of LD12 bacteria detected either by rRNA gene sequences (33 locations) or CARD–FISH (13 locations).
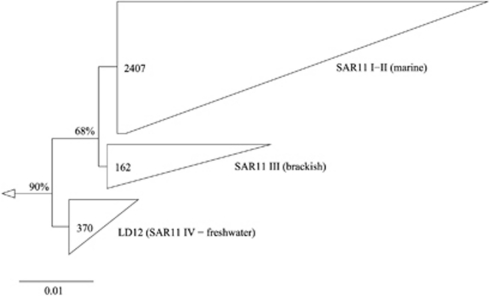
Phylogenetic analyses gave significant support for the separation of the marine SAR11 and the freshwater LD12 group (90% bootstrap support, Figure 2). The mean distance between the LD12 cluster and a cultivated SAR11 representative, Pelagibacter ubique HTCC1062 (88.9%), indicates that the two lineages are different genera. By contrast, the high sequence similarity within the LD12 cluster (mean similarity: 99.5%) suggests that these bacteria might be classified as a single species. Nevertheless, several distinct subclusters within the LD12 lineage were supported by bootstrap values of >50% (Supplementary Figure S1). Most of these subclusters contained sequences that have been obtained from a single habitat (that is, Wang Yang River, Lake Gatun, or Lake Zurich), and they typically featured rather low branch lengths. However, a more deeply branching subclade of 10 sequences from Lake Zurich (53% bootstrap support) even contained another well-separated subcluster (63% bootstrap support). This microdiversification points to the existence of specific ecotypes in different lakes and possibly even in a single lake. Distinct ecotypes of the marine SAR11 cluster have been reported to differ in their spatio-temporal distribution patterns in the Sargasso Sea (Carlson et al., 2008).
Figure 2.
Unrooted maximum likelihood tree of 16S rDNA sequences affiliated with SAR11 I–II, SAR11 III and LD12. The scale bar at the bottom refers to 0.01% of sequence divergence.
Occurrence and habitat preference of LD12 bacteria
We identified and quantified LD12 bacteria in several freshwater habitats by CARD–FISH with a newly designed specific rRNA-targeted oligonucleotide probe. The available probe that targets the majority of Alphaproteobacteria (ALF968, Amann and Fuchs, 2008) is not suitable for the detection of these microbes, as it features several mismatches with genotypes from the LD12 cluster. Therefore, LD12 bacteria have been overlooked in the analysis of freshwater microbial communities by FISH until now.
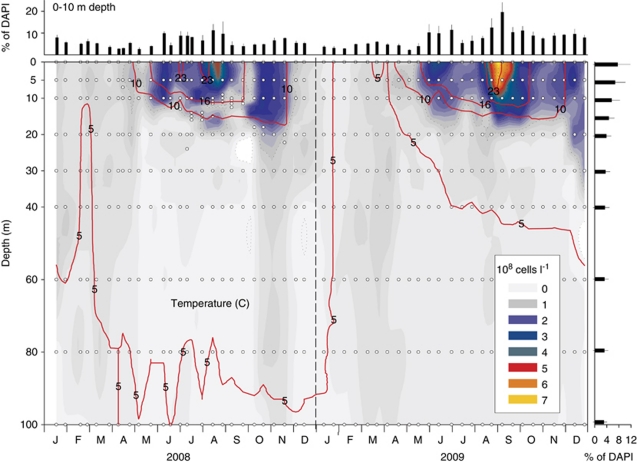
A high-resolution sampling campaign (n=490 samples) over two consecutive years unveiled the spatio-temporal population dynamics of LD12 bacteria in Lake Zurich. These bacteria were always present in lake water, but they also exhibited pronounced depth-distribution patterns and seasonal fluctuations (Figure 3). The highest numbers were found in the warm epilimnetic water strata during summer (up to 8.0 × 108 cells per l). Three distinct abundance maxima were discernable in both successive years: (i) shortly after water temperatures reached >16 °C (May–June), (ii) during the warmest period at temperatures >22 °C (August–September) and (iii) in autumn (November), immediately before or at the onset of the epilimnetic mixis. Only low proportions of LD12 bacteria were present during winter or in deep hypolimnetic water (<5% of all Bacteria). Thus, the seasonal population dynamics of freshwater LD12 bacteria are to some extent comparable to those of their marine sister group: members of the SAR11 clade have been shown to peak during summer stratification in the euphotic zones of various marine habitats such as the oligotrophic North Pacific Ocean (Eiler et al., 2009), the northwestern Sargasso Sea (Carlson et al., 2008) and the Mediterranean Sea (Alonso-Saez et al., 2007).
Figure 3.
Spatio-temporal distribution of LD12 bacteria in Lake Zurich, Switzerland. The red lines indicate temperature isopleths and open circles refer to sampling points. Mean proportions and s.d. of LD12 from 0 to 10 m depths are shown on top and mean seasonal values for all depths on the right (in % of DAPI-stained particles).
SAR11 bacteria usually account for one quarter of marine bacterioplankton or more, and their abundances range between 0.3–6.3 × 108 cells per l in the North Pacific Ocean (Eiler et al., 2009), 0.2–3.2 × 108 cells per l in the Sargasso Sea (Morris et al., 2002; Carlson et al., 2008) and 0.5–3 × 108 cells per l in the Mediterranean (Alonso-Saez et al., 2007). The abundances of LD12 bacteria in the epilimnion of Lake Zurich during summer stratification (2.1±1.2 × 108 cells per l, range 0.3–8.0 × 108 cells per l, n=109) were comparable to those of their marine counterparts in the euphotic zone (mean, 2 × 108 cells per l (Morris et al., 2002)). LD12 cell densities in the lower strata (hypolimnion) of the lake and during winter (0.5±0.3 × 108 cells per l, range 0.05–2.0 × 108 cells per l, n=381) exceeded typical values for SAR11 in the aphotic zone, for example, those in the mesopelagic realms of the Sargasso Sea (0.2 × 108 cells per l (Morris et al., 2002; Carlson et al., 2008)).
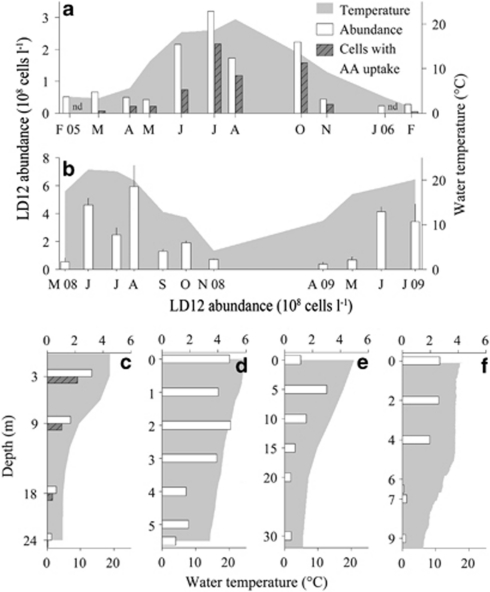
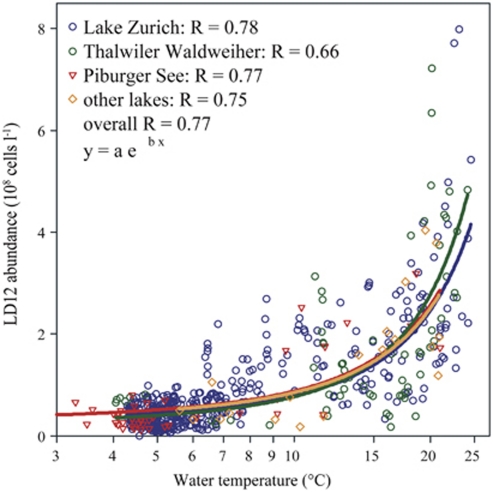
Seasonal population dynamics of LD12 comparable to those in Lake Zurich were also found in surface waters of two other systems with contrasting limnological properties, an oligo-mesotrophic mountain lake (Piburger See, Austria) and a eutrophic pond with only 40 days of water renewal time (Thalwiler Waldweiher, Switzerland) (Figures 4a and b). Although the sampling resolution in these habitats was lower (monthly), two distinct summer maxima were nevertheless discernable in both cases. Depth profiles from these two lakes, from a large mesotrophic lake (Lake Constance, Germany), and from a shallow, eutrophic lake (Egelsee, Switzerland) furthermore supported the conclusion that LD12 bacteria may generally prefer the upper epilimnetic water layers (Figures 4c–f). Genome analyses of P. ubique revealed the presence of a light-driven proton pump (proteorhodopsin (Giovannoni et al., 2005a)), and proteorhodopsin genes related to SAR11 have been obtained from several lakes (Atamna-Ismaeel et al., 2008). This suggests that LD12 bacteria might possess such light harvesting pigments, and it could help to explain their high densities in the light-penetrated uppermost layers (Supplementary Figures S2 and S3). Moreover, the close relationship between water temperature and the abundances of LD12 bacteria (Figure 5, Pearson's correlation coefficient=0.81, P>0.001, n=621) also suggests that these microbes in some aspects share habitat preferences with their marine counterparts SAR11 (Morris et al., 2002; Alonso-Saez et al., 2007; Carlson et al., 2008).
Figure 4.
Seasonal and depth distribution of LD12 abundances (white bars) in relation to water temperature (gray area) in temperate lakes. (a) Seasonal development of LD12 bacteria in Piburger See, Austria, in 3 m depth and corresponding abundances of LD12 bacteria with active amino acid uptake (gray bars). (b) Mean values and s.d. (n=3) in Thalwiler Waldweiher, Switzerland, in 0–2 m depth. (c) Depth distribution in Piburger See in July 2005 and corresponding abundances of LD12 bacteria with active amino acid uptake. (d) Depth distribution in Thalwiler Waldweiher in June 2008, (e) in Lake Constance, Germany, in July 2009 and (f) in Egelsee, Switzerland, in September 2007. nd, amino acid uptake assay not conducted.
Figure 5.
Relationship between LD12 abundance and water temperature in Lake Zurich, Switzerland (blue circles, n=490), Piburger See, Austria (red triangles, n=44), Thalwiler Waldweiher, Switzerland (green circles, n=64) and six other lakes (orange diamonds, n=19).
We also assessed the occurrence of LD12 bacteria in the epilimnion of eight more lakes with different limnological characteristics, ranging from ultra-oligotrophic to eutrophic (Table 1). LD12 bacteria were also abundant in highly productive systems, which is in contrast with the adaptation to nutrient poor habitats of their marine counterparts. Interestingly, LD12 bacteria were extremely rare in samples from a cold, ultra-oligotrophic high mountain lake (Gossenköllesee, Austria) that is usually covered by ice for 7–8 months and features water temperatures around 10 °C during summer (Pernthaler et al., 1998).
Finally, LD12 and SAR11 bacteria also seem to share a conspicuously small cell size (mean LD12 biovolume, 0.017 μm3, Table 2) (Rappe et al., 2002; Nicastro et al., 2006). The miniature size of Pelagibacter ubique has mainly been attributed to an effective adaptation to nutrient-limited environments, as their optimized surface-to-volume ratio points to advantages in nutrient and substrate uptake (Giovannoni et al., 2005b). In view of the apparent success of LD12 bacteria in eutrophic ponds and lakes, other advantages of the ultramicrobacterial morphotype might also be of relevance, for example, the protection against size-selective protistan grazing (Pernthaler, 2005).
Table 2. Size of bacteria affiliated with bacteria (probe EUB I–III), LD12 (probe LD12–121), SAR11 and Pelagibacter ubique.
| Length (μm) | Width (μm) | Biovolume (μm3) | Reference | |
|---|---|---|---|---|
| EUB I–III | 0.55 (0.21–4.21) | 0.28 (0.13–0.80) | 0.038 (0.002–0.569) | This study |
| LD12–121 | 0.38 (0.24–0.69) | 0.27 (0.19–0.38) | 0.017 (0.005–0.041) | This study |
| Marine SAR11 | — | — | 0.045 (0.033–0.060) | (Malmstrom et al., 2005) |
| Pelagibacter ubique HTCC1062 | 0.4 (0.37–0.89) | 0.2 (0.12–0.20) | 0.01 | (Rappe et al., 2002) |
| Pelagibacter ubique HTCC1062 | — | — | 0.025–0.045 | (Nicastro et al., 2006) |
Mean values (and ranges) of 400–500 measured cells are given, and significant differences between bacteria hybridized with probes LD12 and EUB are highlighted in bold (paired t-test, df=461, P>0.001).
It should be noted that the biomass contribution of LD12 might not be as high as its contribution to total cell numbers due to its small cell size. However, most pelagic bacteria in freshwater systems are of very small cell size (that is, Actinobacteria (Jezbera et al., 2009; Salcher et al., 2010), Polynucleobacter sp. (Wu and Hahn, 2006; Hahn et al., 2009, 2010) or Betaproteobacteria affiliated with the LD28 cluster (M Salcher, unpublished data)). The fraction of the typically small, so-called low nucleic acid-containing bacteria (LNA bacteria) was 77±5% in Lake Zurich (n=490), 83±8% in Piburger See (n=108) and 78±10% in Thalwiler Waldweiher (n=126) during the study period. More than 50% of these LNA bacteria were Actinobacteria and LD12 bacteria, whereas <5% of cells from these two groups were present in fractions of bacteria with high nucleic acid content (M Salcher, unpublished data).
Physiological traits of LD12 bacteria
Short-time incorporation assays with a tritiated mixture of amino acids over the course of 1 year in an oligo-mesotrophic lake (Piburger See) revealed high uptake of this important component of the labile dissolved organic matter pool (Weiss and Simon, 1999) by LD12 bacteria (Figures 4a and c). This agrees with reports that their marine relatives can account for 50% of total amino acid assimilation in the North Atlantic (Malmstrom et al., 2004) and around 25% in the Mediterranean, albeit with high seasonal variations (Alonso-Saez and Gasol, 2007). In Piburger See, highest amino acid uptake (up to 80% of all LD12 cells, or >2 × 108 active cells per l) coincided with the highest abundances of LD12 bacteria, which was observed in the epilimnion during the summer months. By contrast, only around 30% of LD12 microbes incorporated the radiolabelled amino acids during winter and in the deeper water layers (mean number of active cells in the hypolimnion: 0.09 × 108 cells per l). Thus, LD12 cells were most active when phosphorus concentrations were seasonally low (negative correlation between dissolved phosphorus and the proportion of active LD12 bacteria (r=−0.645, P<0.001)). Phosphorus can be the growth-limiting factor of freshwater planktonic bacteria (Carlsson and Caron, 2001), and members of the LD12 clade seem to be particularly well adapted to thrive in nutrient-limited situations (Supplementary Figures S2 and S3). Genomic and metaproteomic evidence suggests that their marine sister group may indeed be specialized for phosphorus transport and uptake (Giovannoni et al., 2005b; Sowell et al., 2009). Marine SAR11 bacteria are also centrally involved in phosphate turnover in the sea, for example, in the nutrient-depleted North Atlantic subtropical gyre (Zubkov et al., 2007). Besides phosphorus, also nitrogen is a limiting factor for bacterial growth, especially during summer at times of nitrate and ammonia depletion (Morris and Lewis, 1992; Carlsson and Caron, 2001). We found a significant negative relationship between LD12 cell densities and nitrate (Supplementary Figures S1 and S2) and ammonia concentrations (r=−0.54, P<0.001).
Analyses of the genome of P. ubique has revealed that despite its extreme reduction it nevertheless features a surprisingly high number of genes encoding for transport functions (Giovannoni et al., 2005b), in particular also transporters for nitrogenous compounds (for example, ammonia or amino acids). Several genes encode for ABC transporters with very narrow solute uptake specificity (Hosie and Poole, 2001) like glutamine or glycine/proline transport, others for general -amino acid transport with very broad solute range, and high-affinity transporters for branched amino acids like leucine have been detected in the genome of P. ubique HTCC1062 (Giovannoni et al., 2005b). Proteomic analyses of P. ubique and metaproteomic marine data also point to a dominance of transport functions in SAR11 bacteria (Sowell et al., 2008, 2009), with up to 67% of all spectra matching transport proteins, especially for polar amino acids (Sowell et al., 2009).
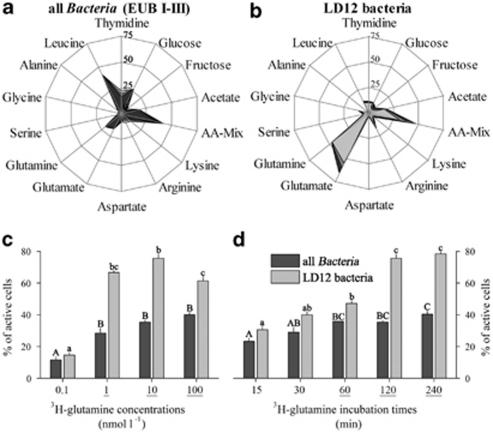
An ecophysiological assay with 14 different radiolabelled low molecular-weight substrates conducted in Lake Zurich in late summer 2009 (Figures 6a and b) revealed a pronounced potential preference of LD12 bacteria for glutamate and glutamine (60 and 46% of LD12 cells, respectively). These amino acids were 3.7 and 2.3 times more frequently incorporated by LD12 cells than by the bacterial community on average, with LD12 representing 48 and 30% of total glutamate or glutamine-assimilating bacterial cells, respectively. In addition, the proportion of LD12 cells capable of arginine incorporation was also more than twice as high than of other bacterial cells in general, with LD12 representing 30% of total arginine-incorporating bacterial cells. These three amino acids (together with proline) belong to the glutamate family, and arginine can also be de novo synthesized from glutamate (Philosof et al., 2009). Amino acids of the glutamate family are important as the main nitrogen carriers during the biosynthesis of other amino acids (Lengeler et al., 1999). Glutamate is the universal donor for α-amino groups in amino acids, which accounts for about 80% of the cellular nitrogen, and glutamine transfers another 15% of nitrogen. Although marine SAR11 are poised to assimilate inorganic nitrogen (ammonia (Sowell et al., 2009)), it is conceivable that their freshwater relatives cover their cellular nitrogen demand at least partially by a high assimilation of amino acids of the glutamate family (Figure 6b).
Figure 6.
(a,b) Ecophysiological MAR–FISH assay with 14 radiolabeled substrates. Means and standard errors (n=3) of the percentage of all bacteria (a, probe EUB I–III) and LD12 bacteria (b) with active tracer incorporation are shown. AA-Mix; amino acid mixture. (c) Concentration and (d) incubation time-dependent incorporation of radiolabelled glutamine of all bacteria and LD12 bacteria. Significant differences between all bacteria and LD12 bacteria are indicated by underlined x-axis labels. Identical letters in uppercase (EUB) or lowercase (LD12 bacteria) indicate treatments that were statistically indistinguishable.
In additional experiments conducted in autumn 2010, we added 3H-glutamine at 0.1–100 nmol l−1 concentrations (2 h incubation) and incubated samples pulse-labeled with 10 nmol l−1 3H-glutamine for periods of 15–240 min. This revealed a typical adaptation of LD12 bacteria to an oligotrophic life style. High proportions of actively incorporating cells were already detected at very low tracer concentrations (Figure 6c): more LD12 bacteria incorporated glutamine at concentrations of only 1 nmol l−1, than at concentrations of 100 nmol l−1 (67 vs 61% MAR-positive cells). Moerover, although bacteria in general (as hybridized with probe EUB I–III) showed highest tracer uptake at the highest offered concentration (40% MAR-positive cells), LD12 bacteria had a distinct incorporation maximum at 10 nmol l−1 (76% MAR-positive cells), where they constituted 58% of all active bacteria. A similar adaptation to oligotrophic conditions was described for samples from the North Sea, where SAR11 bacteria showed a distinct affinity for glucose at concentrations of 1 nmol l−1 (Alonso and Pernthaler, 2006). On the other hand, LD12 bacteria showed slower uptake ‘kinetics' of radiolabeled glutamine (10 nmol l−1, Figure 6d). All bacteria showed a saturation of the numbers of MAR-positive cells already after 1 h of incubation (36% MAR-positive cells). By contrast LD12 bacteria reached a comparable saturation plateau only after 2–4 h of incubation (76% MAR-positive cells). Thus, we draw the prelimninary conclusion that LD12 seem to follow an oligotrophic lifestyle with slow but efficient uptake already at low substrate concentrations.
Glycine was also preferably incorporated by LD12 (7.5-fold higher number of MAR-positive cells than the bacterial average, 62% of the total bacterial glycine assimilation, Figures 6a and b). This is in accordance with the observation that P. ubique is auxotrophic for this amino acid (Tripp et al., 2009). On the other hand, the branched amino acid leucine, which is mostly applied for assessing activity and biomass synthesis of aquatic bacteria (Kirchman et al., 1985), was significantly selected against by LD12 (12% MAR-positive LD12 cells vs 41% MAR-positive bacteria, Figures 6a and b). By contrast, marine SAR11 bacteria readily incorporate leucine at frequencies comparable to the community average (Malmstrom et al., 2004; Alonso and Pernthaler, 2006) and the gene encoding its transporter has been identified in their genome (Giovannoni et al., 2005b).
Glucose was incorporated by only approximately one tenth of LD12 cells, accounting for only 5% of the total number of bacteria with glucose uptake. This is also in contrast to the high glucose uptake observed in their marine counterparts (that is, between 15–50% of total glucose uptake in the Delaware Estuary (Elifantz et al., 2005), Sargasso Sea (Malmstrom et al., 2005), North Sea (Alonso and Pernthaler, 2006), Mediterranean (Alonso-Saez and Gasol, 2007) and Arctic Ocean (Alonso-Saez et al., 2008)). However, Pelagibacter strains were found to differ in their ability to use glucose; P. ubique isolated from the open ocean entirely lack glycolysis-related genes (Giovannoni et al., 2005b), whereas alternative glucose oxidation pathways are present in isolates from the Oregon Coast (Schwalbach et al., 2010). Moreover, the total variants in the Entner–Doudoroff operon were highly correlated with chlorophyll a concentrations, suggesting diversification into glucose-using coastal ecotypes and open-ocean ecotypes incapable of glucose oxidation (Schwalbach et al., 2010). Similarly, variants with or without the ability for glucose assimilation might exist within the freshwater LD12 lineage.
Altogether, ultramicrobacteria affiliated with LD12 seem to be one of the most abundant single species of microbes in freshwater lakes with average to high primary productivity (for example, more than 30% of all bacteria in Lake Zurich). LD12 bacteria mainly thrive in the upper euphotic water layers during summer, and they have an important role in microbial assimilation of particular low molecular weight organic compounds. Generally, LD12 seem to follow an oligotrophic lifestyle with slow but efficient uptake at low substrate concentrations. They thus share basic features such as habitat preference and ecophysiological traits with their closely related marine sister group SAR11 (Morris et al., 2002; Alonso-Saez et al., 2007; Carlson et al., 2008; Eiler et al., 2009). The two groups together form one of the rare monophyletic lineages of ultramicrobacteria that have successfully traversed the barrier between marine and freshwater habitats (Urbach et al., 2001; Logares et al., 2010).
Acknowledgments
We thank E Loher and J Franzoi for help during sampling, M Steinkellner for help in the lab, and M Zeder for support with automated microscopy. H Güde is acknowledged for providing samples of Lake Constance, Gnadensee, Zeller See, Badsee, and Federsee, T Shabarova for providing samples of Lake Thun, and the water supply company Zurich is thanked for the provision of limnochemical data. We moreover thank the helpful suggestions of three anonymous reviewers. This study was supported by the Swiss National Science Foundation (SNF, Project 31003A-117765) and, as part of the European Science Foundation EUROCORES Programme EuroEEFG, was also supported by the SNF Project 31EE30-132771.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Alonso C, Pernthaler J. Roseobacter and SAR11 dominate microbial glucose uptake in coastal North Sea waters. Environ Microbiol. 2006;8:2022–2030. doi: 10.1111/j.1462-2920.2006.01082.x. [DOI] [PubMed] [Google Scholar]
- Alonso-Saez L, Balague V, Sa EL, Sanchez O, Gonzalez JM, Pinhassi J, et al. Seasonality in bacterial diversity in north-west Mediterranean coastal waters: assessment through clone libraries, fingerprinting and FISH. FEMS Microbiol Ecol. 2007;60:98–112. doi: 10.1111/j.1574-6941.2006.00276.x. [DOI] [PubMed] [Google Scholar]
- Alonso-Saez L, Gasol JM. Seasonal variations in the contributions of different bacterial groups to the uptake of low-molecular-weight compounds in northwestern Mediterranean coastal waters. Appl Environ Microbiol. 2007;73:3528–3535. doi: 10.1128/AEM.02627-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Saez L, Sanchez O, Gasol JM, Balague V, Pedros-Alio C. Winter-to-summer changes in the composition and single-cell activity of near-surface Arctic prokaryotes. Environ Microbiol. 2008;10:2444–2454. doi: 10.1111/j.1462-2920.2008.01674.x. [DOI] [PubMed] [Google Scholar]
- Amann R, Fuchs BM. Single-cell identification in microbial communities by improved fluorescence in situ hybridization techniques. Nat Rev Microbiol. 2008;6:339–348. doi: 10.1038/nrmicro1888. [DOI] [PubMed] [Google Scholar]
- Ashelford K, Chuzhanova N, Fry J, Jones A, Weightman A. At least 1 in 20 rRNA sequence record currently held in public repositories is estimated to contain substantial anomalies. Appl Environ Microbiol. 2005;71:7724–7736. doi: 10.1128/AEM.71.12.7724-7736.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atamna-Ismaeel N, Sabehi G, Sharon I, Witzel K-P, Labrenz M, Jurgens K, et al. Widespread distribution of proteorhodopsins in freshwater and brackish ecosystems. ISME J. 2008;2:656–662. doi: 10.1038/ismej.2008.27. [DOI] [PubMed] [Google Scholar]
- Bahr M, Hobbie JE, Sogin ML. Bacterial diversity in an arctic lake: a freshwater SAR11 cluster. Aquat Microb Ecol. 1996;11:271–277. [Google Scholar]
- Bossard P, Gammeter S, Lehmann C, Schanz F, Bachofen R, Bürgi H-R, et al. Limnological description of the Lakes Zürich, Lucerne, and Cadagno. Aquat Sci. 2001;63:225–249. [Google Scholar]
- Carlson CA, Morris R, Parsons R, Treusch AH, Giovannoni SJ, Vergin K. Seasonal dynamics of SAR11 populations in the euphotic and mesopelagic zones of the northwestern Sargasso Sea. ISME J. 2008;3:283–295. doi: 10.1038/ismej.2008.117. [DOI] [PubMed] [Google Scholar]
- Carlsson P, Caron D. Seasonal variation of phosphorus limitation of bacterial growth in a small lake. Limnol Oceanogr. 2001;46:108–120. [Google Scholar]
- Eiler A, Hayakawa DH, Church MJ, Karl DM, Rappé MS. Dynamics of the SAR11 bacterioplankton lineage in relation to environmental conditions in the oligotrophic North Pacific subtropical gyre. Environ Microbiol. 2009;11:2291–2300. doi: 10.1111/j.1462-2920.2009.01954.x. [DOI] [PubMed] [Google Scholar]
- Elifantz H, Malmstrom R, Cottrell M, Kirchman D. Assimilation of polysaccharides and glucose by major bacterial groups in the Delaware Estuary. Appl Environ Microbiol. 2005;71:7799–7805. doi: 10.1128/AEM.71.12.7799-7805.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni SJ, Bibbs L, Cho J-C, Stapels MD, Desiderio R, Vergin KL, et al. Proteorhodopsin in the ubiquitous marine bacterium SAR11. Nature. 2005a;438:82–85. doi: 10.1038/nature04032. [DOI] [PubMed] [Google Scholar]
- Giovannoni SJ, Tripp HJ, Givan S, Podar M, Vergin KL, Baptista D, et al. Genome streamlining in a cosmopolitan oceanic bacterium. Science. 2005b;309:1242–1245. doi: 10.1126/science.1114057. [DOI] [PubMed] [Google Scholar]
- Glöckner F, Fuchs B, Amann R. Bacterioplankton compositions of lakes and oceans: a first comparison based on fluorescence in situ hybridization. Appl Environ Microbiol. 1999;65:3721–3726. doi: 10.1128/aem.65.8.3721-3726.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glöckner FO, Zaichikov E, Belkova N, Denissova L, Pernthaler J, Pernthaler A, et al. Comparative 16S rRNA analysis of lake bacterioplankton reveals globally distributed phylogenetic clusters including an abundant group of Actinobacteria. Appl Environ Microbiol. 2000;66:5053–5065. doi: 10.1128/aem.66.11.5053-5065.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Güde H, Rossknecht H, Wagner G. Anthropogenic impacts on the trophic state of Lake Constance during the 20th century. Arch Hydrobiol. 1998;53:85–108. [Google Scholar]
- Hahn MW, Lang E, Brandt U, Sproer C.2010Polynucleobacter acidiphobus sp. nov., a representative of an abundant group of planktonic freshwater bacteria Int J Syst Evol MicrobiolE-pub ahead of print, doi: 10.1099/ijs0.017350-0 [DOI] [PMC free article] [PubMed]
- Hahn MW, Lang E, Brandt U, Wu QL, Scheuerl T. Emended description of the genus Polynucleobacter and the species Polynucleobacter necessarius and proposal of two subspecies, P. necessarius subsp. necessarius subsp. nov. and P. necessarius subsp. asymbioticus subsp. nov. Int J Syst Evol Microbiol. 2009;59:2002–2009. doi: 10.1099/ijs.0.005801-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosie A, Poole P. Bacterial ABC transporters of amino acids. Res Microbiol. 2001;152:259–270. doi: 10.1016/s0923-2508(01)01197-4. [DOI] [PubMed] [Google Scholar]
- Jezbera J, Sharma AK, Brandt U, Doolittle WF, Hahn MW. ‘Candidatus Planktophila limnetica', an actinobacterium representing one of the most numerically important taxa in freshwater bacterioplankton. Int J Syst Evol Microbiol. 2009;59:2864–2869. doi: 10.1099/ijs.0.010199-0. [DOI] [PubMed] [Google Scholar]
- Kirchman D, K'nees E, Hodson R. Leucine incorporation and its potential as a measure of protein synthesis by bacteria in natural aquatic systems. Appl Environ Microbiol. 1985;49:599–607. doi: 10.1128/aem.49.3.599-607.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchman DL, Dittel AI, Findlay SEG, Fischer D. Changes in bacterial activity and community structure in response to dissolved organic matter in the Hudson River, New York. Aquat Microb Ecol. 2004;35:243–257. [Google Scholar]
- Klammer S, Posch T, Sonntag B, Griebler C, Mindl B, Psenner R. Dynamics of bacterial abundance, biomass, activity, and community composition in the oligotrophic Traunsee and the Traun river (Austria) Water Air Soil Poll Focus. 2002;2:137–163. [Google Scholar]
- Lengeler J, Drews G, Schlegel HG. Biology of the Prokaryotes. Georg Thieme Verlag: Stuttgart, Germany; 1999. [Google Scholar]
- Logares R, Brate J, Heinrich F, Shalchian-Tabrizi K, Bertilsson S. Infrequent Transitions between Saline and Fresh Waters in One of the Most Abundant Microbial Lineages (SAR11) Mol Biol Evol. 2010;27:347–357. doi: 10.1093/molbev/msp239. [DOI] [PubMed] [Google Scholar]
- Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, et al. ARB: a software environment for sequence data. Nucleic Acid Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmstrom R, Cottrell M, Elifantz H, Kirchman D. Biomass production and assimilation of dissolved organic matter by SAR11 bacteria in the northwest atlantic ocean. Appl Environ Microbiol. 2005;71:2979–2986. doi: 10.1128/AEM.71.6.2979-2986.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmstrom R, Kiene R, Cottrell M, Kirchman D. Contribution of SAR11 bacteria to dissolved dimethylsufoniopropionate and amino acid uptake in the North Atlantic Ocean. Appl Environ Microbiol. 2004;70:4129–4135. doi: 10.1128/AEM.70.7.4129-4135.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J, Ray Wu LG, Kivie M. Methods in Enzymology. Academic Press: Amsterdam, The Netherlands; 1983. New M13 vectors for cloning; pp. 20–78. [DOI] [PubMed] [Google Scholar]
- Morris D, Lewis W. Nutrient limitation of bacterioplankton growth in Lake Dillon, Colorado. Limnol Oceanogr. 1992;37:1179–1192. [Google Scholar]
- Morris RM, Rappe MS, Connon SA, Vergin KL, Siebold WA, Carlson CA, et al. SAR11 clade dominates ocean surface bacterioplankton communities. Nature. 2002;420:806–810. doi: 10.1038/nature01240. [DOI] [PubMed] [Google Scholar]
- Muyzer G, de Waal EC, Uitterlinden A. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyzer G, Teske A, Wirsen C, Jannasch H. Phylogenetic relationship of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturing gel electrophoresis of 16S rDNA fragments. Arch Microbiol. 1995;164:165–172. doi: 10.1007/BF02529967. [DOI] [PubMed] [Google Scholar]
- Nicastro D, Schwartz C, Pierson J, Cho J-C, Giovannoni S, McIntosh J. Three-dimensional Structure of the Tiny Bacterium Pelagibacter ubique Studied by Cryo-electron Tomography. Microsc Microanal. 2006;12:180–181. [Google Scholar]
- Nishimura Y, Nagata T. Alphaproteobacterial dominance in a large mesotrophic lake (Lake Biwa, Japan) Aquat Microb Ecol. 2007;48:231–240. [Google Scholar]
- Pernthaler J. Predation on prokaryotes in the water column and its ecological implications. Nat Rev Microbiol. 2005;3:537–546. doi: 10.1038/nrmicro1180. [DOI] [PubMed] [Google Scholar]
- Pernthaler J, Glockner FO, Unterholzner S, Alfreider A, Psenner R, Amann R. Seasonal community and population dynamics of pelagic bacteria and archaea in a high mountain lake. Appl Environ Microbiol. 1998;64:4299–4306. doi: 10.1128/aem.64.11.4299-4306.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philosof A, Sabehi G, Béjá O. Comparative analyses of actinobacterial genomic fragments from Lake Kinneret. Environ Microbiol. 2009;11:3189–3200. doi: 10.1111/j.1462-2920.2009.02024.x. [DOI] [PubMed] [Google Scholar]
- Posch T, Franzoi J, Prader M, Salcher MM. New image analysis tool to study biomass and morphotypes of three major bacterioplankton groups in an alpine lake. Aquat Microb Ecol. 2009;54:113–126. [Google Scholar]
- Rappe MS, Connon SA, Vergin KL, Giovannoni SJ. Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature. 2002;418:630–633. doi: 10.1038/nature00917. [DOI] [PubMed] [Google Scholar]
- Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S, et al. The Sorcerer II Global Ocean Sampling Expedition: Northwest Atlantic through Eastern Tropical Pacific. PLoS Biol. 2007;5:e77. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salcher M, Pernthaler J, Posch T. Spatiotemporal distribution and activity patterns of bacteria from three phylogenetic groups in an oligomesotrophic lake. Limnol Oceanogr. 2010;55:846–856. [Google Scholar]
- Salcher MM, Pernthaler J, Zeder M, Psenner R, Posch T. Spatio-temporal niche separation of planktonic Betaproteobacteria in an oligo-mesotrophic lake. Environ Microbiol. 2008;10:2074–2086. doi: 10.1111/j.1462-2920.2008.01628.x. [DOI] [PubMed] [Google Scholar]
- Schwalbach MS, Tripp HJ, Steindler L, Smith DP, Giovannoni SJ. The presence of the glycolysis operon in SAR11 genomes is positively correlated with ocean productivity. Environ Microbiol. 2010;12:490–500. doi: 10.1111/j.1462-2920.2009.02092.x. [DOI] [PubMed] [Google Scholar]
- Sekar R, Pernthaler A, Pernthaler J, Warnecke F, Posch T, Amann R. An improved protocol for quantification of freshwater Actinobacteria by fluorescence in situ hybridization. Appl Environ Microbiol. 2003;69:2928–2935. doi: 10.1128/AEM.69.5.2928-2935.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowell SM, Norbeck AD, Lipton MS, Nicora CD, Callister SJ, Smith RD, et al. Proteomic Analysis of Stationary Phase in the Marine Bacterium ‘Candidatus Pelagibacter ubique'. Appl Environ Microbiol. 2008;74:4091–4100. doi: 10.1128/AEM.00599-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowell SM, Wilhelm LJ, Norbeck AD, Lipton MS, Nicora CD, Barofsky DF, et al. Transport functions dominate the SAR11 metaproteome at low-nutrient extremes in the Sargasso Sea. ISME J. 2009;3:93–105. doi: 10.1038/ismej.2008.83. [DOI] [PubMed] [Google Scholar]
- Stamatakis A, Ludwig T, Meier H. RAxML-II: a program for sequential, parallel and distributed inference of large phylogenetic. Concurr Comput-Pract Exp. 2005;17:1705–1723. [Google Scholar]
- Tripp HJ, Kitner JB, Schwalbach MS, Dacey JWH, Wilhelm LJ, Giovannoni SJ. SAR11 marine bacteria require exogenous reduced sulphur for growth. Nature. 2008;452:741–744. doi: 10.1038/nature06776. [DOI] [PubMed] [Google Scholar]
- Tripp HJ, Schwalbach MS, Meyer MM, Kitner JB, Breaker RR, Giovannoni SJ. Unique glycine-activated riboswitch linked to glycine-serine auxotrophy in SAR11. Environ Microbiol. 2009;11:230–238. doi: 10.1111/j.1462-2920.2008.01758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbach E, Vergin K, Young L, Morse A, Larson G, Giovannoni S. Unusual bacterioplankton community structure in ultra-oligotrophic Crater Lake. Limnol Oceanogr. 2001;46:557–572. [Google Scholar]
- Van den Wyngaert S, Salcher MM, Pernthaler J, Zeder M, Posch T. Quantitative dominance of seasonally persistent filamentous cyanobacteria (Planktothrix rubescens) in the microbial assemblages of a temperate lake. Limnol Oceanogr. 2011;56:97–109. [Google Scholar]
- Weiss M, Simon M. Consumption of labile dissolved organic matter by limnetic bacterioplankton: the relative significance of amino acids and carbohydrates. Aquat Microb Ecol. 1999;17:1–12. [Google Scholar]
- Wu Q, Hahn M. Differences in structure and dynamics of Polynucleobacter communities in a temperate and a subtropical lake, revealed at three phylogenetic levels. FEMS Microbiol Ecol. 2006;57:67–79. doi: 10.1111/j.1574-6941.2006.00105.x. [DOI] [PubMed] [Google Scholar]
- Zeder M, Pernthaler J. Multispot live-image autofocusing for high-throughput microscopy of fluorescently stained bacteria. Cytometry Part A. 2009;75A:781–788. doi: 10.1002/cyto.a.20770. [DOI] [PubMed] [Google Scholar]
- Zubkov MV, Mary I, Woodward EMS, Warwick PE, Fuchs BM, Scanlan DJ, et al. Microbial control of phosphate in the nutrient-depleted North Atlantic subtropical gyre. Environ Microbiol. 2007;9:2079–2089. doi: 10.1111/j.1462-2920.2007.01324.x. [DOI] [PubMed] [Google Scholar]
- Zwart G, Crump B, Kamst-van Agterveld MP, Hagen F, Han S. Typical freshwater bacteria: an analysis of available 16S rRNA gene sequences from plankton of lakes and rivers. Aquat Microb Ecol. 2002;28:141–155. [Google Scholar]
- Zwart G, Hiorns W, Methé B, van Agterveld MP, Huismans R, Nold S, et al. Nearly identical 16S rRNA sequences recovered from lakes in North America and Europe indicate the existance of clades of globally distributed freshwater bacteria. Syst Appl Microbiol. 1998;21:546–556. doi: 10.1016/S0723-2020(98)80067-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.