Abstract
DNA polymerase zeta (Pol ζ) participates in translesion synthesis (TLS) of DNA adducts that stall replication fork progression. Previous studies have led to the suggestion that the primary role of Pol ζ in TLS is to extend primers created when another DNA polymerase inserts nucleotides opposite lesions. Here we test the non-exclusive possibility that Pol ζ can sometimes perform TLS in the absence of any other polymerase. To do so, we quantified the efficiency with which S. cerevisiae Pol ζ bypasses abasic sites, cis-syn cyclobutane pyrimidine dimers and (6-4) photoproducts. In reactions containing dNTP concentrations that mimic those induced by DNA damage, a Pol ζ derivative with phenylalanine substituted for leucine 979 at the polymerase active site bypasses all three lesions at efficiencies between 27–73%. Wild-type Pol ζ also bypasses these lesions, with efficiencies that are lower and depend on the sequence context in which the lesion resides. The results are consistent with the hypothesis that, in addition to extending aberrant termini created by other DNA polymerases, Pol ζ has the potential to be the sole DNA polymerase involved in TLS.
Keywords: DNA polymerase zeta, translesion synthesis, UV photoproducts, abasic sites
1. INTRODUCTION
When lesions in DNA impede DNA synthesis, the impediment can be alleviated by translesion DNA synthesis (TLS) catalyzed by specialized DNA polymerases. Included among several TLS polymerases is eukaryotic DNA polymerase ζ, an exonuclease-deficient, heterodimeric (Rev3–Rev7) DNA polymerase that participates in a variety of DNA transactions. These transactions contribute to spontaneous mutagenesis [1–4], mutagenesis induced by DNA damaging agents such as UV irradiation [5–8], mutagenesis associated with repair of double-strand DNA breaks [9–12], mutagenesis associated with high levels of transcription, cytosine deamination-dependent somatic hypermutation of immunoglobulin genes [13,14], and mutagenesis in cells defective in NER, BER, replication fork progression and post-replication repair [15–20]. That the cellular functions of Pol ζ are important is further revealed by the embryonic lethality resulting from loss of the mouse REV3L gene [21–23], the increased cancer susceptibility observed in a conditional knockout mouse model [24], and the increased cisplatin sensitivity of lung tumors in mice upon suppression of Rev3 [25].
Many of the phenotypes observed in genetic studies of Pol ζ are thought to reflect the ability of Pol ζ to participate in TLS, a subject that has been extensively investigated [reviewed by 1,26,27]. Several biochemical studies have reported that Pol ζ alone cannot efficiently bypass a UV-induced cis-syn cyclobutane pyrimidine dimer (CPD) [28,29], a (6-4) photoproduct [28–30] or an abasic site [29,31]. This limitation has been ascribed to inefficient insertion of a nucleotide opposite the first base of the dipyrimidine lesions or opposite the abasic site. However, Pol ζ is highly efficient at extending aberrant primer-templates, especially those that already contain a nucleotide present opposite a lesion [29–34]. These observations have led to a now widespread view that the primary role of Pol ζ in TLS is to extend primer-templates after a nucleotide has first been inserted opposite a lesion by another DNA polymerase. This is referred to as the two-polymerase model for TLS.
While there is substantial experimental support for the two-polymerase TLS model [1,26,27,35–37], an additional and non-exclusive possibility is that Pol ζ is sometimes the sole TLS polymerase involved in lesion bypass. This possibility is supported by several observations. In their seminal description of the discovery of yeast Pol ζ, Nelson et al. [38] reported that Pol ζ could bypass a T-T cis-syn CPD ten-fold more efficiently than could the catalytic subunit of yeast Pol α. Yeast Pol ζ was later reported to also perform bypass of thymine glycol [39], limited bypass of a (6-4) photoproduct [28], and to bypass photoproducts generated by UV irradiation of a poly(dT)29 template [40]. A study of the efficiency with which lesion-containing plasmids transform wild-type yeast strains versus strains deficient in different TLS polymerases led to the suggestion that Pol ζ is responsible not only for extension, but also for insertion opposite lesions, at least for bypass events other than those in which Pol η participates [41].
That Pol ζ might be the sole TLS polymerase involved in UV photoproduct bypass in vivo is further suggested by a genetic study utilizing a variant of yeast Pol ζ containing a phenylalanine substituted for leucine 979, a conserved residue at the active site in the catalytic Rev3 subunit of the Rev3–Rev7 heterodimeric polymerase. A yeast strain harboring the rev3-L979F allele has wild-type survival following UV irradiation [42], consistent with the fact that purified L979F Pol ζ has robust polymerase activity [43], as needed to enhance survival following UV irradiation [5–8]. The rev3-L979F strain also has an elevated UV-induced mutation frequency compared to the wild-type REV3 strain [42], consistent with the fact that L979F Pol ζ (L979F Rev3–Rev7) has lower fidelity than wild-type Pol ζ during DNA synthesis in vitro [43]. This indicates that, like wild-type Pol ζ, L979F Pol ζ also participates in mutagenic bypass of UV photoproducts in vivo. Moreover, UV-induced mutagenesis is further elevated when the RAD30 gene encoding Pol η is deleted from the rev3-L979F strain [42]. This demonstrates that L979F Pol ζ contributes to bypassing UV photoproducts in vivo even in the absence of Pol η, the major yeast TLS polymerase implicated in insertion opposite lesions in the two-polymerase TLS model. Thus, either a polymerase other than Pol η or Pol ζ performs the initial insertion, or yeast Pol ζ alone can perform TLS in vivo.
The observations discussed above raise the issue of how efficiently yeast L979F Pol ζ and wild-type yeast Pol ζ perform TLS without assistance from other DNA polymerases. Here we examine this issue by performing biochemical studies of yeast Pol ζ lesion bypass that are similar to earlier TLS studies yet take into account three additional parameters. Many studies have examined TLS using amounts of polymerase and incubation times that result in a large, but often unknown, number of cycles of polymerase binding, synthesis and termination, which Nelson et al. (1996) called “forcing conditions”. When bypass is observed under such conditions, it is not possible to quantify bypass efficiency per synthesis cycle, making it difficult to compare TLS efficiency from one study to another. Here, as in several of our earlier TLS studies [44–47], we determine the relative bypass efficiency of Pol ζ per cycle of polymerization, thus permitting direct comparisons to other polymerases when analyzed in the same manner [44]. We also take into account the fact that the concentrations of the four dNTPs are not equal in vivo and that yeast cells respond to exposure to DNA damaging agents by up-regulating dNTP pools [48,49]. As a consequence, yeast TLS polymerases may perform bypass in vivo using dNTP concentrations that are unequal and that may be higher than those used previously for bypass studies in vitro. Unequal dNTP concentrations could influence bypass efficiency in a sequence-dependent manner. High, damage-induced dNTP concentrations can also increase TLS efficiency, as evidenced by the increased efficiency with which DNA Pol ε bypasses 8-oxo-guanine at high dNTP concentrations [47], and the increased efficiency with which DNA polymerases δ and ε bypass rNMPs in template DNA at high dNTP concentrations [50].
Here we examine the ability of Pol ζ to bypass lesions using dNTP concentrations approximating those induced in yeast by exposure using dNTP concentrations approximating those induced upon exposure of yeast to UV light [49] or chronic exposure to 4-NQO [48], which has frequently been used as a UV mimetic. The results show that, without assistance from other DNA polymerases, L979F Pol ζ can bypass synthetic abasic sites, T-T cis-syn CPDs and T-T (6-4) photoproducts in two different sequence contexts. Bypass per cycle of polymerization is remarkably efficient, supporting a model wherein L979F Pol ζ is the only polymerase needed for UV photoproduct bypass in the rev3-L979F rad30Δ strain. The results show that wild-type Pol ζ can also bypass these lesions, albeit with lower efficiencies that vary depending on the lesion and the DNA sequence in which it is embedded. These data imply that, in addition to a prominent role in extending aberrant primers in a two-polymerase model, Pol ζ has the biochemical potential to function as the sole polymerase involved in TLS.
2. MATERIALS AND METHODS
2.1. Measurements of dNTP pools and cell cycle progression in yeast strains
The previously described wild-type and rev3-L979F strains [42] were grown in YPDA media (1% yeast extract, 2% bacto-peptone, 250mg/L adenine). Measurements of dNTP levels in extracts prepared from asynchronous, logarithmically growing yeast cells were performed by HPLC analysis as described [51]. Flow cytometry was performed as previously described [47].
2.2. Protein purification
S. cerevisiae Pol ζ (Rev3–Rev7) and L979F Pol ζ (L979F Rev3–Rev7) were over-expressed and purified from yeast as previously described [40]. SDS-PAGE gels of purified two-subunit wild-type and L979F Pol ζ were shown previously [43]. S. cerevisiae Pol δ was purified as previously described [52].
2.3. DNA substrates
All substrates are listed in Table 1. Substrates were prepared by annealing 32P-labelled primer oligonucleotides and template oligonucleotides in a 1:1.5 ratio, as described previously [45].
Table 1.
Substrates
| Substrate | Sequencea |
|---|---|
| 49-mer UV substrates | |
| 25-mer primer | 5’-AGCTATGACCATGATTACGAATTGC |
| Undamaged template | 3’-TCGATACTGGTACTAATGCTTAACGAATTAAGCACGTCCGTACCATCGA |
| CPD templateb | 3’-TCGATACTGGTACTAATGCTTAACGAATTAAGCACGTCCGTACCATCGA |
| (6-4) PP templateb | 3’-TCGATACTGGTACTAATGCTTAACGAATTAAGCACGTCCGTACCATCGA |
| 45-mer UV substrates | |
| 25-mer primer | 5’-AATTTCTGCAGGTCGACTCCAAAGG |
| Undamaged template | 3’-TTAAAGACGTCCAGCTGAGGTTTCCGATTGGGCCATGGCTCGACC |
| CPD templateb | 3’-TTAAAGACGTCCAGCTGAGGTTTCCGATTGGGCCATGGCTCGACC |
| (6-4) PP templateb | 3’-TTAAAGACGTCCAGCTGAGGTTTCCGATTGGGCCATGGCTCGACC |
| 45-mer AP substrates | |
| 23-mer primer | 5’-AATTTCTGCAGGTCGACTCCAAA |
| Undamaged template | 3’-TTAAAGACGTCCAGCTGAGGTTTCCGATTGGGCCATGGCTCGACC |
| TAF templatec | 3’-TTAAAGACGTCCAGCTGAGGTTTCCFATTGGGCCATGGCTCGACC |
| TFG templatec | 3’-TTAAAGACGTCCAGCTGAGGTTTCCGFTTGGGCCATGGCTCGACC |
2.4. Lesion bypass assay
Wild-type and L979F Pol ζ reactions (30 µl) contained 40 mM Tris-HCL (pH 7.8), 100 mM NaCl, 20 mg/ml BSA, 8 mM MgAc, 1 pmol DNA substrate, 120 fmol Pol ζ or L979F Pol ζ and dNTPs. The dNTP concentrations were either 16 µM dATP, 30 µM dTTP, 14 µM dCTP, and 12 µM dGTP (referred to as “normal”), or 10-fold higher concentrations of each dNTP (refered to as “high). Pol δ reactions (30 µl) contained 20 mM Tris-HCL (pH 7.8), 90 mM NaCl, 200 mg/ml BSA, 1 mM DTT, 8 mM MgAc, 160 µM dATP, 300 µM dTTP, 140 µM dCTP, and 120 µM dGTP, 1 pmol DNA substrate and the indicated amount of polymerase. Reactions were performed in duplicate. All components except polymerase were mixed on ice and incubated at 30° C for 2 min. Polymerase was added to start the reactions, which were incubated at 30° C. Aliquots were removed at the times specified and added to an equal volume of formamide loading buffer (95% deionized formamide, 25 mM EDTA, 0.01% bromophenol blue and 0.01% xylene cyanol) to stop the reaction. DNA products were separated by electrophoresis using 12% denaturing polyacrylamide gels and quantified using ImageQuant version 5.2 (Molecular Dynamics). As previously described [53,54], bypass probability is defined as the summed intensity of all product bands corresponding to synthesis after the lesion (e.g. in Fig. 2, positions +1 and greater) divided by the summed intensity of all bands corresponding to synthesis before the lesion is encountered (e.g. in Fig. 2., positions −1 and −2). Termination probability of a particular template position is calculated dividing the intensity of the band that corresponds to that position by the sum of the intensity of that position plus the intensity of all larger products [53,54]. All reactions using wild-type and L979F Pol ζ were considered to be “single hit”; enzyme to DNA ratios were sufficiently low so as to generate termination probabilities at each position along the template that remained constant (± 10%) over the short time course of the reaction, thus demonstrating that each DNA primer is extended only once. The depiction of certain overexposed lanes in gel images are shown simply to more clearly reveal the less abundant DNA products of lesion bypass.
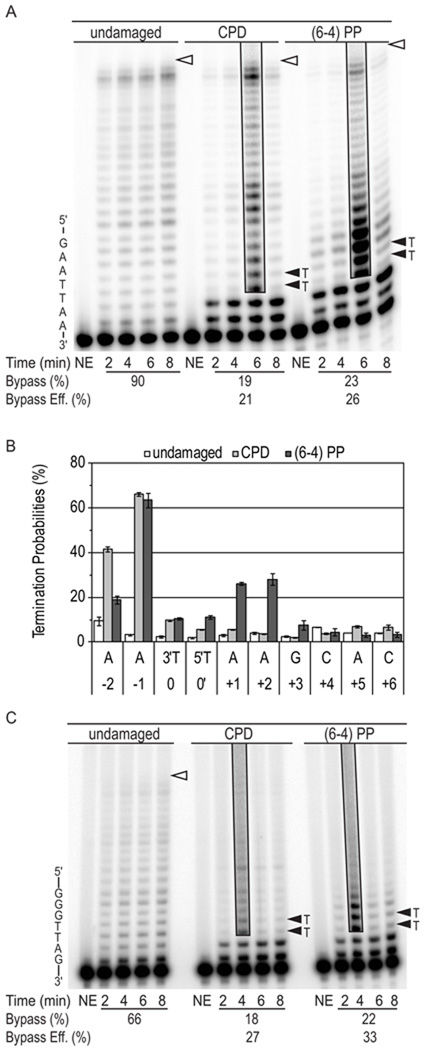
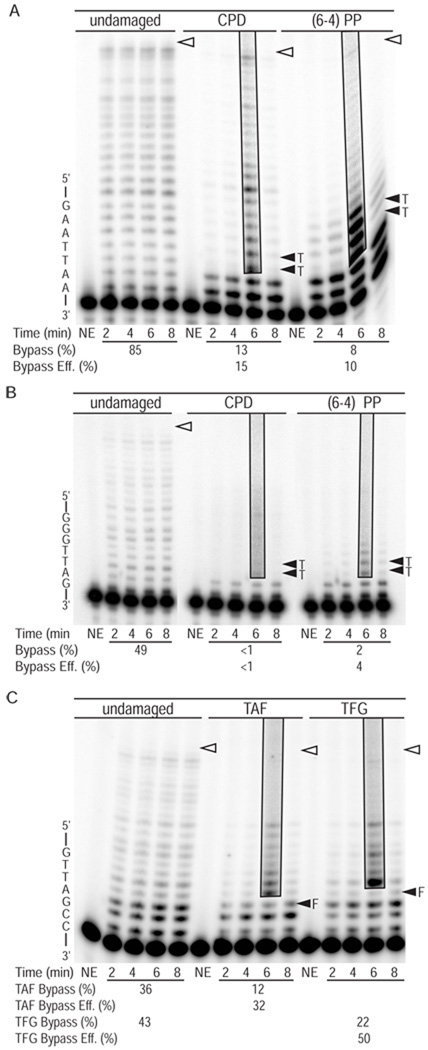
Figure 2. Bypass of UV photoproducts by L979F Pol ζ.
(A) Image of denaturing polyacrylamide gels showing products of synthesis by L979F Pol ζ using undamaged, CPD and (6-4) photoproduct (PP) 49-mer substrates. Primer extension reactions were performed in the presence of high dNTP concentrations intended to simulate damage-induced dNTP concentrations. Each reaction contained an 8-fold excess of substrate over polymerase. Part of the template sequence is shown on the left. The position of the 3′ and 5′ T of the CPDs and (6-4) PPs are indicated by closed arrowheads. Open arrowheads indicate the positions of full-length reaction products. Boxed areas of the gel show reaction products at an overexposed level of contrast to more clearly reveal less abundant DNA products of lesion bypass. NE indicates control lanes for reaction mixtures to which no enzyme was added. The average bypass probability for 2, 4, 6 and 8 min time points of each reaction and the relative bypass efficiency (bypass of damaged substrate relative to bypass of undamaged substrate) are shown under each gel. Duplicate reactions were performed with similar results. (B) Graph showing average probability of termination of processive synthesis at template nucleotides for 49-mer substrates, with error bars indicating standard deviation. (C) Image of denaturing polyacrylamide gels showing products of synthesis by L979F Pol ζ using undamaged, CPD and (6-4) PP 45-mer substrates.
3. RESULTS
3.1. dNTP pools and cell cycle progression
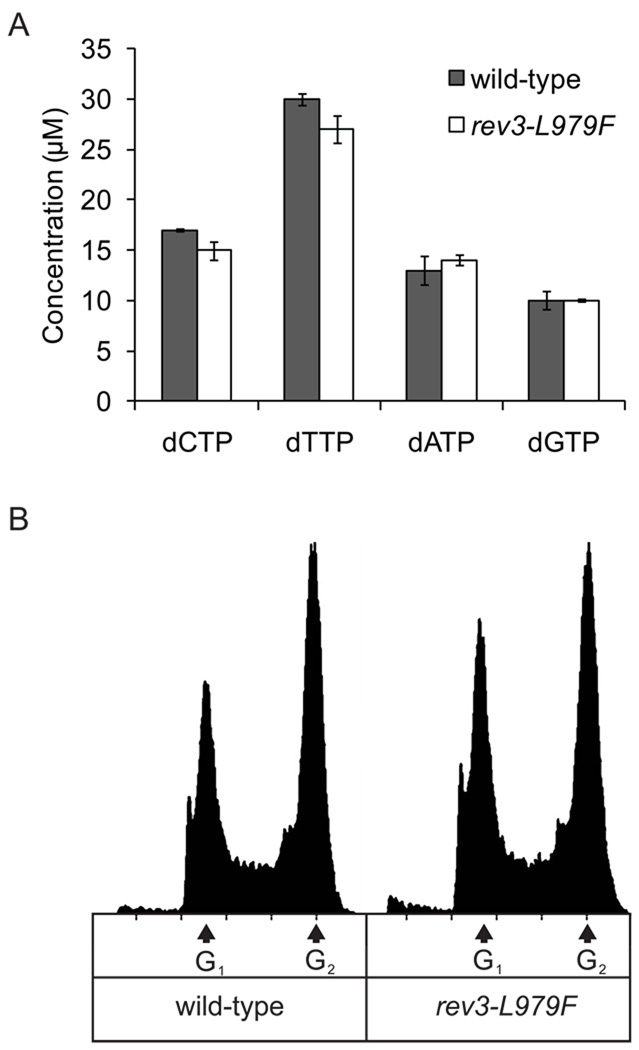
We began by measuring the dNTP concentrations in extracts of asynchronously growing wild-type (REV3) and rev3-L979F yeast cells in log phase. The dNTP concentrations in extracts of each strain (Fig.1A) were similar to those previously measured in wild-type strains [51]. The DNA content of asynchronous cultures indicated that both strains progressed normally through S-phase (Fig. 1B). Thus, dNTP pools in both strains are naturally slightly biased, and the rev3-L979F mutation does not alter cellular dNTP pools or cell cycle progression. Based on the values that had been previously described [51], DNA synthesis reactions described below (sections 3.2 through 3.6) contain either 16 µM dATP, 30 µM dTTP, 14 µM dCTP, and 12 µM dGTP, hereafter referred to as “normal”, or 160 µM dATP, 300 µM dTTP, 140 µM dCTP, and 120 µM dGTP, hereafter referred to as “high”. The high dNTP concentrations used here are slightly higher than those observed in yeast after treatment with UV, 4-NQO or MMS [48,49] and are intended to approximate the dNTP levels that would be observed during stress induced by multiple sources of DNA damage.
Figure 1. Characteristics of wild-type and rev3-L979F strains of S. cerevisiae.
(A) dNTP concentrations in yeast strains, determined as described [51]. Values are the average of two experiments and error bars represent the standard deviation. (B) Flow cytometry histograms for wild-type and rev3-L979F strains, obtained as described [47]. Arrows indicate the DNA content of cells in the G1 and G2 phases of the cell cycle.
3.2. Pol ζ synthesis with undamaged templates
When copying undamaged DNA under single-hit conditions, L979F Pol ζ generates products ranging in length from 1–25 nucleotides (Fig. 2A/C). Thus, like wild-type Pol ζ [38,40,43 and Fig. 5A–D], L979F Pol ζ is moderately processive. The probability of termination of processive synthesis varies along the template (see open bars in Fig. 2B and Fig. 3B), but tends to be highest after inserting the first nucleotide. Similar termination probabilities were obtained when dNTP concentrations were high (Figs. 2 and 3) or normal (Fig. 4). Likewise, the processivity of wild-type Pol ζ is unaffected by dNTP concentrations (Fig. 5), and is not in Section 3.3 increased in the presence of RPA, RFC and PCNA [43].
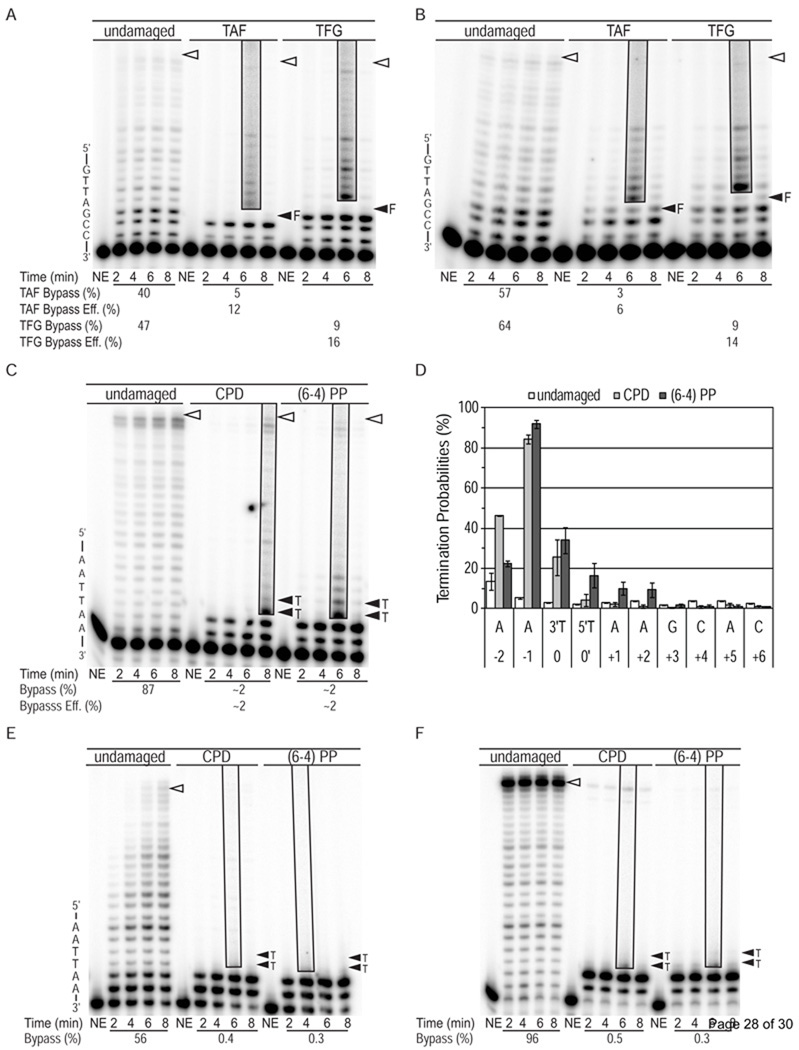
Figure 5. Primer extension reactions using wild-type Pol ζ and Pol δ.
Reaction products are shown for synthesis by wild-type Pol ζ for (A) undamaged, TAF and TFG substrates in the presence of low dNTP concentrations, (B) TAF and TFG substrates in the presence of high dNTP concentrations and (C) synthesis of undamaged, CPD and (6-4) PP 49-mer substrates in the presence of high dNTP concentrations. These primer extension reactions contained an 8-fold excess of substrate over polymerase. The average bypass probability and the relative bypass efficiency (bypass of damaged substrate relative to bypass of undamaged substrate) for each reaction are shown under each gel. (D) Graph showing termination probabilities at template nucleotides for synthesis of undamaged, CPD and (6-4) PP 49-mer substrates by wild-type Pol ζ. (E and F) Images of denaturing polyacrylamide gels show control reactions using yeast Pol δ to synthesize undamaged, CPD and (6-4) PP 49-mer substrates in the presence of high dNTP concentrations. Reactions contained (E) 100-fold and (F) 10-fold excess substrate over polymerase. All markings are as in Figure 1. Note that in the boxed, overexposed areas of the Pol δ gels, the pixels at the bottom are from the band at the −1 position relative to the lesions, i.e., there is no evidence that Pol δ can insert a nucleotide opposite the 3′ T of either a CPD or (6-4) PP.
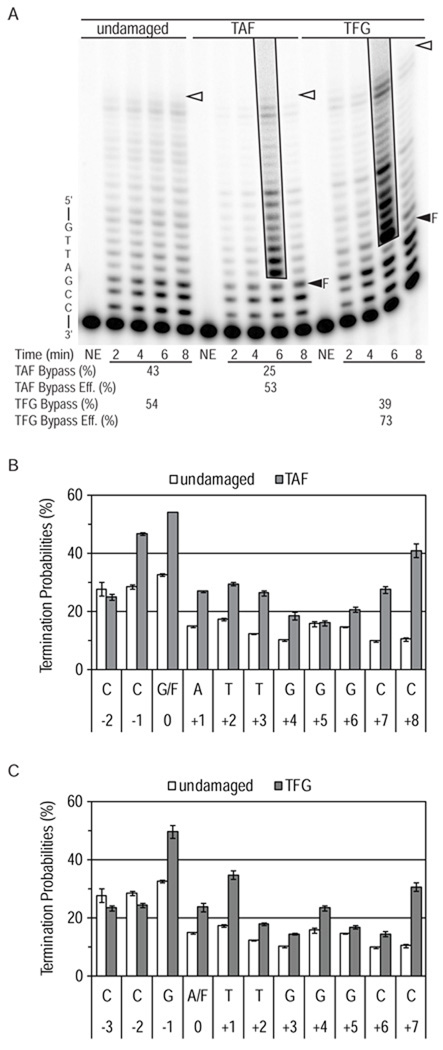
Figure 3. Bypass of AP sites L979F Pol ζ.
(A) Image of denaturing polyacrylamide gels showing products of synthesis by L979F Pol ζ using undamaged, TAF and TFG substrates. TAF and TFG templates contain a tetrahydrofuran (F) whose location is marked with a closed arrowhead. Other markings are as in Figure 1. Each reaction contained 8-fold excess of substrate over polymerase and high dNTP concentrations. The average bypass probability and the relative bypass efficiency (bypass of damaged substrate relative to bypass of undamaged substrate) for each reaction are shown under the gel. Graphs show average probability of termination of processive synthesis for positions along (B) the TAF template and (C) the TFG template with error bars indicating standard deviation.
Figure 4. Bypass of UV photoproducts and AP sites by L979F Pol ζ when dNTP concentrations are at normal levels.
Images of denaturing polyacrylamide gels showing products of synthesis by L979F Pol ζ with (A) undamaged, CPD and (6-4) PP 49-mer substrates, (B) CPD and (6-4) PP 45-mer substrates, and (C) undamaged, TAF and TAG substrates. Each primer extension reaction contained an 8-fold excess of substrate over polymerase and normal dNTPs concentrations. The average bypass probability and the relative bypass efficiency (bypass of damaged substrate relative to bypass of undamaged substrate) for each reaction are shown under the gel. Markings are as in Figure 1.
3.3. UV photoproducts bypass by L979F Pol ζ with high dNTP concentrations
We first examined the efficiency with which L979F Pol ζ bypasses template-strand UV photoproducts located two bases downstream from the 3′ terminus of a primer hybridized to a 49-mer template (Table 1). For each template position we determined termination probability, which is likelihood with which Pol ζ will stop synthesis at a given position rather than insert the next nucleotide. In reactions containing high dNTP concentrations (Fig. 2A/B), when L979F Pol ζ attempts to bypass a T-T cis-syn CPD (respectively designated 0 and 0´ in Fig. 2B), increased termination of processive synthesis is seen at positions −2 through +1 when compared to synthesis of the undamaged substrate (compare light gray and open bars in Fig. 2B). The termination probability for a particular position is inversely proportional to the probability of insertion at the subsequent position. For example, the termination probability for the −1 position of the CPD-containing template is 66% (Fig. 2B) and the insertion probability for the subsequent 3´ T position is 34% (i.e., 100% – 66% = 34%; Fig. 6). Thus, insertion by L979F Pol ζ is least efficient opposite the 3´ T of the CPD (Fig. 2B), which is consistent with published studies with wild-type Pol ζ [28,29]. Insertions opposite the 5´ T of the CPD and opposite the following two template bases are also inefficient, after which the CPD has no effect (e.g., at +2). When these DNA product distributions are used to calculate bypass for a single cycle of synthesis, L979F Pol ζ bypassed CPDs with 21% of the relative efficiency with which it bypassed the corresponding undamaged T-Ts (Fig. 2A). We also examined bypass with a second substrate in which a T-T cis-syn CPD was embedded in a different sequence context (45-mer UV substrates in Table 1). L979F Pol ζ bypassed this CPD with 27% of the efficiency with which it bypassed the undamaged T-Ts (Fig. 2C).
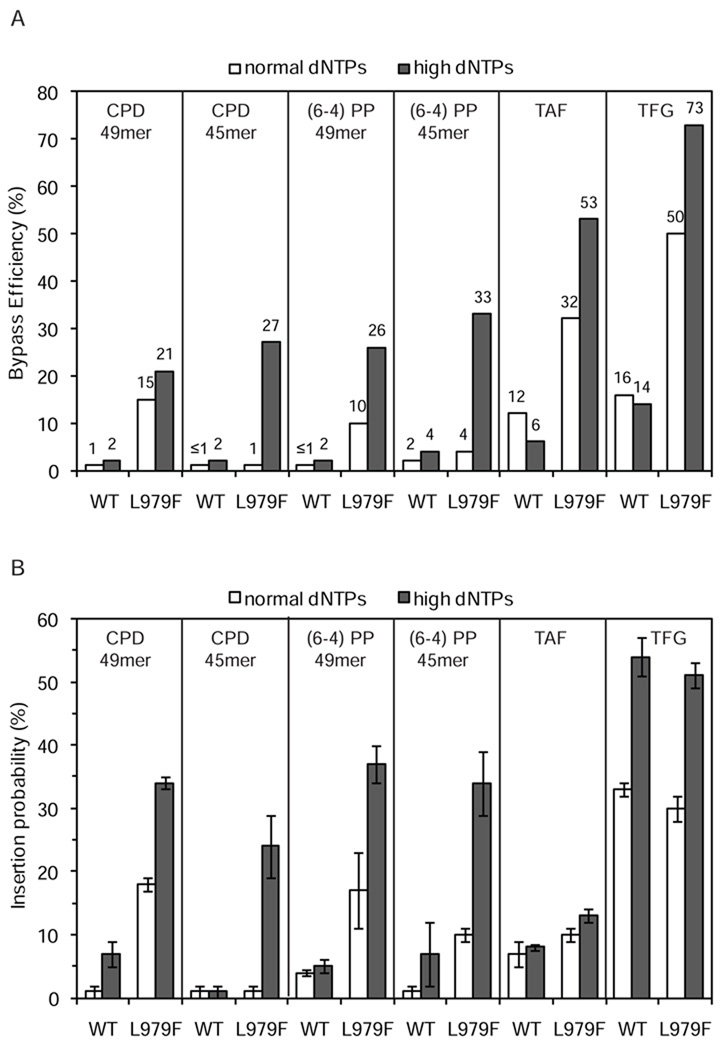
Figure 6. Bypass efficiencies and insertion probabilities.
(A) Graph showing average bypass efficiency (bypass of damaged substrate relative to bypass of undamaged substrate) for each lesion tested in the presence of normal or high dNTP concentrations. WT and L979F indicate synthesis by wild-type Pol ζ and L979F Pol ζ, respectively. (B) Graph showing average probability of inserting nucleotides opposite each lesion with error bars indicating standard deviation. For CPDs and (6-4) photoproducts, insertion probability is shown for insertion opposite the 3′ T.
A similar experiment was performed with a (6-4) photoproduct located in the same position as the CPD in the 49-mer template. In this case, insertion by L979F Pol ζ was reduced opposite both bases of the photoproduct and opposite four subsequent nucleotides as synthesis proceeded (dark gray bars in Fig. 2B). Thus, the more helix-distorting (6-4) photoproduct affects insertion over a greater distance than does a CPD. Despite this, L979F Pol ζ bypassed the (6-4) photoproducts with 26% of the efficiency with which it bypassed the corresponding undamaged T-Ts (Fig. 2A). When bypass of a (6-4) photoproduct in the second sequence context (45-mer substrates in Table 1) was examined, L979F Pol ζ bypassed this lesion with 33% of the efficiency with which it bypassed the undamaged T-Ts (Fig. 2C).
3.4. Synthetic abasic site bypass by L979F Pol ζ with high dNTP concentrations
We then examined the ability of L979F Pol ζ to bypass a tetrahydrofuran (designated F), used as a stable surrogate for a natural abasic site, when present in either of two different locations (designated TAF and TFG in Table 1 and Fig. 3). In reactions containing high dNTP concentrations, the results reveal that relative bypass efficiencies were high for each substrate, 53% for TAF and 73% for TFG (Fig. 3). These high efficiencies reflect the fact that insertions opposite the lesions, and insertions opposite subsequent template bases, were only slightly reduced compared to those occurring opposite the equivalent template bases in the undamaged control (Fig. 3B/C).
3.5. Lesion bypass by L979F Pol ζ with normal dNTP concentrations
Next we examined the ability of L979F Pol ζ to bypass CPDs, (6-4) photoproducts and abasic sites in reactions containing normal dNTP concentrations. Under these conditions, the relative bypass efficiency of L979F Pol ζ for the CPD in the 49-mer template was 15% (Fig. 4A), only slightly lower than the 21% bypass observed using high dNTP concentrations (Fig. 2A). In contrast, the relative bypass efficiency of L979F Pol ζ for the CPD in the 45-mer template was less than 1% (Fig. 4B), much lower than the 27% relative bypass efficiency observed with high dNTP concentrations (Fig. 2C). Using normal dNTP concentrations, L979F Pol ζ bypassed the (6-4) photoproduct in the 49-mer with 10% efficiency (Fig. 4A), as compared to 26% efficiency with high dNTPs (Fig. 2A). Again in contrast, the relative bypass efficiency of L979F Pol ζ for the (6-4) photoproduct in the 45-mer was approximately 4% (Fig.4B), as compared to the 33% observed with high dNTP concentrations (Fig. 2C). Finally, L979F Pol ζ bypassed the abasic sites with 32% (TAF) and 50% (TFG) efficiency in reactions with normal dNTP concentrations (Fig. 4C), compared to 53% (TAF) and 73% efficiency (TFG) with high dNTP concentrations (Fig. 3A). Thus high dNTP concentrations improve the efficiency with which L979F Pol ζ bypasses CPDs, (6-4) photoproducts and synthetic abasic sites, but to variable extents depending on the lesion and sequence context.
3.6. Lesion bypass by wild-type Pol ζ
The above measurements demonstrate that L979F Pol ζ performs TLS without assistance from another polymerase. To determine if this is also the case for wild-type Pol ζ, we first measured bypass of synthetic abasic sites. Relative bypass efficiencies were 12% (TAF) and 16% (TFG) with normal dNTP concentrations (Fig. 5A), and 6% (TAF) and 14% (TFG) with high dNTP concentrations (Fig. 5B). Thus unlike L979F Pol ζ, the efficiency with which wild-type Pol ζ bypasses abasic sites was not increased by high dNTP concentrations. We then examined the ability of wild-type Pol ζ to bypass the two UV photoproducts in reactions containing high dNTP concentrations to simulate the effects of treating yeast with UV [49]. The relative bypass efficiency of wild-type Pol ζ during a single cycle of synthesis was ~2% for both photoproducts (Fig. 5C). These values are approximately 10-fold lower than the corresponding values for L979F Pol ζ (Fig. 6). Nonetheless, wild-type Pol ζ appears to be performing authentic photoproduct bypass, as indicated by two observations. First, wild-type and L979F Pol ζ both respond similarly to the UV-induced lesions (compare Fig. 5D to 2B). Insertion efficiency is reduced opposite both the 3´ and 5´ Ts of either photoproduct compared to the undamaged T-Ts. There is also a greater effect on subsequent insertions observed with the (6-4) photoproduct as compared to the CPD. Second, little if any bypass of either photoproduct was observed in single-hit reactions with a major replicative polymerase, Pol δ, even when multiple cycles of polymerization were allowed (Fig. 5E; similar results were obtained for the 45-mer substrates, data not shown). In fact, when the Pol δ concentration was increased by 10-fold, such that all initial primers were extended even at the earliest time point, only trace amounts of long DNA products were observed (Fig. 5F). Under such extreme “forcing conditions” [38], these long products could result from photoproduct bypass achieved by a very large number of polymerization cycles, or they could represent copying of a trace amount of template that lacks the photoproduct. Regardless, the amount of photoproduct bypass achieved by wild-type Pol ζ during a single cycle of synthesis (Fig. 5C) is clearly substantially higher than by Pol δ (Fig. 5E/F).
We also measured the ability of wild-type Pol ζ to bypass the photoproducts in the 49-mer templates in reactions containing normal dNTP concentrations. The bypass efficiencies for the CPD and (6-4) photoproduct were each about 1% (Fig 6A). Finally, we measured the ability of wild-type Pol ζ to bypass a CPD and a (6-4) photoproduct located in a second sequence context by using the 45-mer template. Bypass efficiencies for CPDs and (6-4) photoproducts were ≤1% and 2%, respectively, in reactions containing normal dNTPs and 2% and 4%, respectively, in reactions containing high dNTPs (Fig 6A). In each of these reactions with wild-type Pol ζ, the inefficiency of bypass is at least partially due to inefficient insertion opposite the 3′T of each photoproduct (Fig. 6B).
4. DISCUSSION
Pol ζ has been implicated as the second polymerase to operate in a two-polymerase model for TLS partly based on reports of inefficient insertion of a nucleotide opposite the 3´T of UV photoproducts [28–30] or an abasic site [29,31]. In the present study, one third (4 of 12) of the TLS reactions performed with wild-type yeast Pol ζ similarly show inefficient insertion opposite a lesion. Little to no insertion was observed opposite the 3´ T of the CPD in the 49-mer with normal dNTP concentrations, the CPD in the 45-mer with either normal or high dNTP concentrations, or the (6-4) photoproduct in the 45-mer with normal dNTP concentrations (Fig. 6B). In addition, neither insertion nor bypass was observed when L979F Pol ζ attempted to bypass the CPD in the 45-mer with normal dNTP concentrations. As a result, the bypass efficiency for these five reactions was less than 2% (Fig, 6A). These results are therefore consistent with published observations [28–31,38] and the proposal that the role of Pol ζ in TLS may sometimes be limited to extension following insertion opposite a lesion by another polymerase.
For the majority of the 24 TLS reactions performed here, wild-type and L979F Pol ζ were able to insert a nucleotide opposite the lesion (Fig. 6B) and bypass it (Fig. 6A) when given only a single opportunity for bypass. These data clearly demonstrate that Pol ζ has the biochemical potential to function as the sole polymerase involved in TLS. This potential is most apparent for L979F Pol ζ (gray bars in Fig. 6), which inserts a dNTP opposite, and ultimately bypasses, both abasic sites and UV photoproducts. The latter data support a model wherein L979F Pol ζ is the only polymerase needed for bypass of UV photoproducts to promote the survival of a rev3-L979F rad30Δ strain [42]. However, the fact that wild type and L979F Pol ζ do not bypass all replication-blocking and potentially lethal lesions equally well may account for the fact that a rev3-L679F rad30Δ double mutant strain does not survive UV irradiation any better than does a rad30 strain expressing wild type Pol ζ. An effect of rev3-L979F on survival in the rad30Δ background might be seen upon inactivation of template switching, which competes with TLS to tolerate lesions by copying the same sequence from an undamaged template.
The capacity to act as the sole TLS polymerase is not exclusive to L979F Pol ζ, because even wild-type Pol ζ can insert nucleotides opposite these lesions and bypass them. As mentioned in the Introduction, some evidence for this conclusion preceded the present study. The original biochemical study of yeast Pol ζ [38] reported that it could bypass a CPD ten-fold more efficiently than could yeast Pol α. In subsequent studies involving an unreported number of polymerization cycles, yeast Pol ζ was also reported to perform limited bypass of a (6-4) photoproduct [28], to bypass an abasic site [40], and to bypass photoproducts generated by UV irradiation of a poly(dT)29 template [40]. In our studies involving a single cycle of synthesis, bypass by wild-type Pol ζ is most obvious with the two abasic sites, which were bypassed with efficiencies of 6–16%. Wild-type Pol ζ also inefficiently bypassed CPDs and the (6-4) photoproducts in the 49-mer templates in reactions containing high dNTP concentrations (Fig. 5C) that approximate those present in yeast treated with UV [49]. Although insertion opposite the 3´ T of both photodimers is inefficient (5–7%, Fig. 6B), it is nonetheless readily observable. This contrasts with Pol δ, which did not insert a dNTP opposite these lesions or perform bypass even when given many attempts to do so (Fig. 5 E/F). The results with Pol δ confirm our earlier study indicating that neither Pol δ nor Pol ε, the enzymes that perform the bulk of replication of the nuclear genome, has the ability to bypass or insert a dNTP opposite the 3´ T of a CPD [55]. Thus the bypass observed with wild-type Pol ζ again supports the hypothesis that Pol ζ can be both the inserter and the extender, and thus can act as the sole polymerase involved in TLS. As previously proposed based on genetic data [41], this situation could arise when Pol η is unavailable for CPD bypass, for example in a rad30 strain or an XPV patient, or for bypass of lesions that Pol η reportedly cannot bypass, such as a (6-4) photoproduct [30,41].
The results presented here also support the idea that Pol ζ may be better able to act as the sole TLS polymerase when dNTP concentrations are increased in response to DNA damage. It is also possible that other proteins, such as Rev1, RPA and RFC/PCNA, may enhance the TLS capacity of Pol ζ. Indeed, both Rev1 and monoubiquitylated PCNA are strongly implicated in recruiting TLS polymerases to lesion sites [56–58]. A previous study has shown stimulation of TLS by these accessory proteins in reactions involving multiple cycles of synthesis and bypass of a site-specific abasic site and UV photoproducts randomly introduced into a poly(dT)29 template [40]. We have not yet examined the effects of accessory proteins on Pol ζ bypass efficiency using the current quantitative approach, but if stimulation is observed, that would only strengthen the interpretation that Pol ζ can operate in a one-TLS polymerase pathway. Finally, it is worth noting that even for the same lesion, the decision between a two-polymerase and a one-polymerase TLS reaction may depend on the sequence context in which a lesion resides, as suggested here by the fact that L979F Pol ζ bypass of the CPD and the (6-4) photoproduct was more efficient in the 49-mer template (Fig. 4A) as compared to the 45-mer template (Fig. 4B).
ACKNOWLEDGEMENTS
We thank Mercedes Arana, Katarzyna Bebenek and Danielle Watt for helpful comments on the manuscript. This work was supported in part by the Division of Intramural Research of the National Institute of Environmental Health Sciences, National Institutes of Health [Project Z01 ES065070 to TAK], by the Swedish Foundation for Strategic Research, the Swedish Research Council and the Swedish Cancer Society [to AC], by National Institutes of Health [grant GM032431 to PMB], and by Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology, Japan [to SI].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONFLICT OF INTEREST STATEMENT
The authors declare that there is no conflict of interest.
REFERENCES
- 1.Lawrence CW. Cellular functions of DNA polymerase zeta and Rev1 protein. Adv. Protein Chem. 2004;69:167–203. doi: 10.1016/S0065-3233(04)69006-1. [DOI] [PubMed] [Google Scholar]
- 2.Minesinger BK, Abdulovic AL, Ou TM, Jinks-Robertson S. The effect of oxidative metabolism on spontaneous Pol zeta-dependent translesion synthesis in Saccharomyces cerevisiae. DNA Repair (Amst) 2006;5:226–234. doi: 10.1016/j.dnarep.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 3.Quah SK, von Borstel RC, Hastings PJ. The origin of spontaneous mutation in Saccharomyces cerevisiae. Genetics. 1980;96:819–839. doi: 10.1093/genetics/96.4.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sabbioneda S, Minesinger BK, Giannattasio M, Plevani P, Muzi-Falconi M, Jinks-Robertson S. The 9-1-1 checkpoint clamp physically interacts with pol zeta and is partially required for spontaneous pol zeta-dependent mutagenesis in Saccharomyces cerevisiae. J. Biol. Chem. 2005;280:38657–38665. doi: 10.1074/jbc.M507638200. [DOI] [PubMed] [Google Scholar]
- 5.Lawrence CW, Das G, Christensen RB. REV7, a new gene concerned with UV mutagenesis in yeast. Mol. Gen. Genet. 1985;200:80–85. doi: 10.1007/BF00383316. [DOI] [PubMed] [Google Scholar]
- 6.Abdulovic AL, Jinks-Robertson S. The in vivo characterization of translesion synthesis across UV-induced lesions in Saccharomyces cerevisiae: insights into Pol zeta-and Pol eta-dependent frameshift mutagenesis. Genetics. 2006;172:1487–1498. doi: 10.1534/genetics.105.052480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lawrence CW, Nisson PE, Christensen RB. UV and chemical mutagenesis in rev7 mutants of yeast. Mol. Gen. Genet. 1985;200:86–91. doi: 10.1007/BF00383317. [DOI] [PubMed] [Google Scholar]
- 8.Lemontt JF. Mutants of Yeast Defective in Mutation Induced by Ultraviolet Light. Genetics. 1971;68:21–33. doi: 10.1093/genetics/68.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rattray AJ, Shafer BK, McGill CB, Strathern JN. The roles of REV3 and RAD57 in double-strand-break-repair-induced mutagenesis of Saccharomyces cerevisiae. Genetics. 2002;162:1063–1077. doi: 10.1093/genetics/162.3.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hicks WM, Kim M, Haber JE. Increased mutagenesis and unique mutation signature associated with mitotic gene conversion. Science. 2010;329:82–85. doi: 10.1126/science.1191125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holbeck SL, Strathern JN. A role for REV3 in mutagenesis during double-strand break repair in Saccharomyces cerevisiae. Genetics. 1997;147:1017–1024. doi: 10.1093/genetics/147.3.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang Y, Sterling J, Storici F, Resnick MA, Gordenin DA. Hypermutability of damaged single-strand DNA formed at double-strand breaks and uncapped telomeres in yeast Saccharomyces cerevisiae. PLoS Genet. 2008;4:e1000264. doi: 10.1371/journal.pgen.1000264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diaz M, Lawrence C. An update on the role of translesion synthesis DNA polymerases in Ig hypermutation. Trends Immunol. 2005;26:215–220. doi: 10.1016/j.it.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 14.Diaz M, Verkoczy LK, Flajnik MF, Klinman NR. Decreased frequency of somatic hypermutation and impaired affinity maturation but intact germinal center formation in mice expressing antisense RNA to DNA polymerase zeta. J. Immunol. 2001;167:327–335. doi: 10.4049/jimmunol.167.1.327. [DOI] [PubMed] [Google Scholar]
- 15.Johnson RE, Torres-Ramos CA, Izumi T, Mitra S, Prakash S, Prakash L. Identification of APN2, the Saccharomyces cerevisiae homolog of the major human AP endonuclease HAP1, and its role in the repair of abasic sites. Genes Dev. 1998;12:3137–3143. doi: 10.1101/gad.12.19.3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Swanson RL, Morey NJ, Doetsch PW, Jinks-Robertson S. Overlapping specificities of base excision repair, nucleotide excision repair, recombination, and translesion synthesis pathways for DNA base damage in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:2929–2935. doi: 10.1128/mcb.19.4.2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hendricks CA, Razlog M, Matsuguchi T, Goyal A, Brock AL, Engelward BP. The S. cerevisiae Mag1 3-methyladenine DNA glycosylase modulates susceptibility to homologous recombination. DNA Repair (Amst) 2002;1:645–659. doi: 10.1016/s1568-7864(02)00072-1. [DOI] [PubMed] [Google Scholar]
- 18.Barbour L, Ball LG, Zhang K, Xiao W. DNA damage checkpoints are involved in postreplication repair. Genetics. 2006;174:1789–1800. doi: 10.1534/genetics.106.056283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Minesinger BK, Jinks-Robertson S. Roles of RAD6 epistasis group members in spontaneous pol zeta-dependent translesion synthesis in Saccharomyces cerevisiae. Genetics. 2005;169:1939–1955. doi: 10.1534/genetics.104.033894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harfe BD, Jinks-Robertson S. DNA polymerase zeta introduces multiple mutations when bypassing spontaneous DNA damage in Saccharomyces cerevisiae. Mol. Cell. 2000;6:1491–1499. doi: 10.1016/s1097-2765(00)00145-3. [DOI] [PubMed] [Google Scholar]
- 21.Bemark M, Khamlichi AA, Davies SL, Neuberger MS. Disruption of mouse polymerase zeta (Rev3) leads to embryonic lethality and impairs blastocyst development in vitro. Curr. Biol. 2000;10:1213–1216. doi: 10.1016/s0960-9822(00)00724-7. [DOI] [PubMed] [Google Scholar]
- 22.Esposito G, Godindagger I, Klein U, Yaspo ML, Cumano A, Rajewsky K. Disruption of the Rev3l-encoded catalytic subunit of polymerase zeta in mice results in early embryonic lethality. Curr. Biol. 2000;10:1221–1224. doi: 10.1016/s0960-9822(00)00726-0. [DOI] [PubMed] [Google Scholar]
- 23.Wittschieben J, Shivji MK, Lalani E, Jacobs MA, Marini F, Gearhart PJ, Rosewell I, Stamp G, Wood RD. Disruption of the developmentally regulated Rev3l gene causes embryonic lethality. Curr. Biol. 2000;10:1217–1220. doi: 10.1016/s0960-9822(00)00725-9. [DOI] [PubMed] [Google Scholar]
- 24.Wittschieben JP, Patil V, Glushets V, Robinson LJ, Kusewitt DF, Wood RD. Loss of DNA polymerase zeta enhances spontaneous tumorigenesis. Cancer Res. 2010;70:2770–2778. doi: 10.1158/0008-5472.CAN-09-4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doles J, Oliver TG, Cameron ER, Hsu G, Jacks T, Walker GC, Hemann MT. Suppression of Rev3, the catalytic subunit of Pol{zeta}, sensitizes drug-resistant lung tumors to chemotherapy. Proc Natl Acad Sci U S A. 2010;107:20786–20791. doi: 10.1073/pnas.1011409107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gan GN, Wittschieben JP, Wittschieben BO, Wood RD. DNA polymerase zeta (pol zeta) in higher eukaryotes. Cell Res. 2008;18:174–183. doi: 10.1038/cr.2007.117. [DOI] [PubMed] [Google Scholar]
- 27.Prakash S, Prakash L. Translesion DNA synthesis in eukaryotes: a one- or two-polymerase affair. Genes Dev. 2002;16:1872–1883. doi: 10.1101/gad.1009802. [DOI] [PubMed] [Google Scholar]
- 28.Guo D, Wu X, Rajpal DK, Taylor JS, Wang Z. Translesion synthesis by yeast DNA polymerase zeta from templates containing lesions of ultraviolet radiation and acetylaminofluorene. Nucleic Acids Res. 2001;29:2875–2883. doi: 10.1093/nar/29.13.2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson RE, Washington MT, Haracska L, Prakash S, Prakash L. Eukaryotic polymerases iota and zeta act sequentially to bypass DNA lesions. Nature. 2000;406:1015–1019. doi: 10.1038/35023030. [DOI] [PubMed] [Google Scholar]
- 30.Johnson RE, Haracska L, Prakash S, Prakash L. Role of DNA polymerase eta in the bypass of a (6-4) TT photoproduct. Mol. Cell. Biol. 2001;21:3558–3563. doi: 10.1128/MCB.21.10.3558-3563.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haracska L, Unk I, Johnson RE, Johansson E, Burgers PM, Prakash S, Prakash L. Roles of yeast DNA polymerases delta and zeta and of Rev1 in the bypass of abasic sites. Genes Dev. 2001;15:945–954. doi: 10.1101/gad.882301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lawrence CW, Hinkle DC. DNA polymerase zeta and the control of DNA damage induced mutagenesis in eukaryotes. In: Lindahl T, editor. Cancer Surveys: Genetic Instability in Cancer. Vol. 20. 1996. pp. 21–31. [PubMed] [Google Scholar]
- 33.Lawrence CW, Gibbs PE, Murante RS, Wang XD, Li Z, McManus TP, McGregor WG, Nelson JR, Hinkle DC, Maher VM. Roles of DNA polymerase zeta and Rev1 protein in eukaryotic mutagenesis and translesion replication; Cold Spring Harb. Symp. Quant. Biol; 2000. pp. 61–69. [DOI] [PubMed] [Google Scholar]
- 34.Haracska L, Prakash S, Prakash L. Yeast DNA polymerase zeta is an efficient extender of primer ends opposite from 7,8-dihydro-8-Oxoguanine and O6-methylguanine. Mol Cell Biol. 2003;23:1453–1459. doi: 10.1128/MCB.23.4.1453-1459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoon JH, Prakash L, Prakash S. Error-free replicative bypass of (6-4) photoproducts by DNA polymerase zeta in mouse and human cells. Genes Dev. 2010;24:123–128. doi: 10.1101/gad.1872810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shachar S, Ziv O, Avkin S, Adar S, Wittschieben J, Reissner T, Chaney S, Friedberg EC, Wang Z, Carell T, Geacintov N, Livneh Z. Two-polymerase mechanisms dictate error-free and error-prone translesion DNA synthesis in mammals. Embo J. 2009;28:383–393. doi: 10.1038/emboj.2008.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu. Rev. Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 38.Nelson JR, Lawrence CW, Hinkle DC. Thymine-thymine dimer bypass by yeast DNA polymerase zeta. Science. 1996;272:1646–1649. doi: 10.1126/science.272.5268.1646. [DOI] [PubMed] [Google Scholar]
- 39.Johnson RE, Yu SL, Prakash S, Prakash L. Yeast DNA polymerase zeta (zeta) is essential for error-free replication past thymine glycol. Genes Dev. 2003;17:77–87. doi: 10.1101/gad.1048303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Garg P, Stith CM, Majka J, Burgers PM. Proliferating cell nuclear antigen promotes translesion synthesis by DNA polymerase zeta. J. Biol. Chem. 2005;280:23446–23450. doi: 10.1074/jbc.C500173200. [DOI] [PubMed] [Google Scholar]
- 41.Gibbs PE, McDonald J, Woodgate R, Lawrence CW. The relative roles in vivo of Saccharomyces cerevisiae Pol eta, Pol zeta, Rev1 protein and Pol32 in the bypass and mutation induction of an abasic site, T-T (6-4) photoadduct and T-T cis-syn cyclobutane dimer. Genetics. 2005;169:575–582. doi: 10.1534/genetics.104.034611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sakamoto AN, Stone JE, Kissling GE, McCulloch SD, Pavlov YI, Kunkel TA. Mutator alleles of yeast DNA polymerase zeta. DNA Repair (Amst) 2007;6:1829–1838. doi: 10.1016/j.dnarep.2007.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stone JE, Kissling GE, Lujan SA, Rogozin IB, Stith CM, Burgers PM, Kunkel TA. Low-fidelity DNA synthesis by the L979F mutator derivative of Saccharomyces cerevisiae DNA polymerase zeta. Nucleic Acids Res. 2009;37:3774–3787. doi: 10.1093/nar/gkp238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kokoska RJ, Bebenek K, Boudsocq F, Woodgate R, Kunkel TA. Low fidelity DNA synthesis by a y family DNA polymerase due to misalignment in the active site. J. Biol. Chem. 2002;277:19633–19638. doi: 10.1074/jbc.M202021200. [DOI] [PubMed] [Google Scholar]
- 45.McCulloch SD, Kokoska RJ, Masutani C, Iwai S, Hanaoka F, Kunkel TA. Preferential cis-syn thymine dimer bypass by DNA polymerase eta occurs with biased fidelity. Nature. 2004;428:97–100. doi: 10.1038/nature02352. [DOI] [PubMed] [Google Scholar]
- 46.McCulloch SD, Kokoska RJ, Garg P, Burgers PM, Kunkel TA. The efficiency and fidelity of 8-oxo-guanine bypass by DNA polymerases delta and eta. Nucleic Acids Res. 2009;37:2830–2840. doi: 10.1093/nar/gkp103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sabouri N, Viberg J, Goyal DK, Johansson E, Chabes A. Evidence for lesion bypass by yeast replicative DNA polymerases during DNA damage. Nucleic Acids Res. 2008;36:5660–5667. doi: 10.1093/nar/gkn555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chabes A, Georgieva B, Domkin V, Zhao X, Rothstein R, Thelander L. Survival of DNA damage in yeast directly depends on increased dNTP levels allowed by relaxed feedback inhibition of ribonucleotide reductase. Cell. 2003;112:391–401. doi: 10.1016/s0092-8674(03)00075-8. [DOI] [PubMed] [Google Scholar]
- 49.Lis ET, O'Neill BM, Gil-Lamaignere C, Chin JK, Romesberg FE. Identification of pathways controlling DNA damage induced mutation in Saccharomyces cerevisiae. DNA Repair (Amst) 2008;7:801–810. doi: 10.1016/j.dnarep.2008.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Watt DL, Arana ME, Johansson E, Burgers P, Kunkel TA. Bypass of rNMPs in DNA Templates by Yeast Replicative Polymerases. manuscript in submission. [Google Scholar]
- 51.Nick McElhinny SA, Watts BE, Kumar D, Watt DL, Lundstrom EB, Burgers PM, Johansson E, Chabes A, Kunkel TA. Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proc Natl Acad Sci U S A. 2010 doi: 10.1073/pnas.0914857107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fortune JM, Stith CM, Kissling GE, Burgers PM, Kunkel TA. RPA and PCNA suppress formation of large deletion errors by yeast DNA polymerase delta. Nucleic Acids Res. 2006;34:4335–4341. doi: 10.1093/nar/gkl403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kokoska RJ, McCulloch SD, Kunkel TA. The efficiency and specificity of apurinic/apyrimidinic site bypass by human DNA polymerase eta and Sulfolobus solfataricus Dpo4. J. Biol. Chem. 2003;278:50537–50545. doi: 10.1074/jbc.M308515200. [DOI] [PubMed] [Google Scholar]
- 54.McCulloch SD, Wood A, Garg P, Burgers PM, Kunkel TA. Effects of accessory proteins on the bypass of a cis-syn thymine-thymine dimer by Saccharomyces cerevisiae DNA polymerase eta. Biochemistry. 2007;46:8888–8896. doi: 10.1021/bi700234t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McCulloch SD, Kokoska RJ, Chilkova O, Welch CM, Johansson E, Burgers PM, Kunkel TA. Enzymatic switching for efficient and accurate translesion DNA replication. Nucleic Acids Res. 2004;32:4665–4675. doi: 10.1093/nar/gkh777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lehmann AR. Translesion synthesis in mammalian cells. Exp. Cell Res. 2006;312:2673–2676. doi: 10.1016/j.yexcr.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 57.Waters LS, Minesinger BK, Wiltrout ME, D'Souza S, Woodruff RV, Walker GC. Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiol Mol Biol Rev. 2009;73:134–154. doi: 10.1128/MMBR.00034-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guo C, Kosarek-Stancel JN, Tang TS, Friedberg EC. Y-family DNA polymerases in mammalian cells. Cell Mol Life Sci. 2009;66:2363–2381. doi: 10.1007/s00018-009-0024-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Murata T, Iwai S, Ohtsuka E. Synthesis and characterization of a substrate for T4 endonuclease V containing a phosphorodithioate linkage at the thymine dimer site. Nucleic Acids Res. 1990;18:7279–7286. doi: 10.1093/nar/18.24.7279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Iwai S, Shimizu M, Kamiya H, Ohtsuka E. Synthesis of a phosphoramidite coupling unit of the pyrimidine (6-4) pyrimidone photoproduct and its incorporation into oligodeoxynucleotides. Journal of the American Chemical Society. 1996;118:7642–7643. [Google Scholar]