Abstract
Many mitochondrial proteins are synthesized as preproteins carrying amino-terminal presequences in the cytosol. The preproteins are imported by the translocase of the outer mitochondrial membrane (TOM) and the presequence translocase of the inner membrane (TIM). Tim50 and Tim23 transfer preproteins through the intermembrane space to the inner membrane. We report the crystal structure of the intermembrane space domain of yeast Tim50 to 1.83 Å resolution. A protruding β-hairpin of Tim50 is crucial for interaction with Tim23, providing a molecular basis for the cooperation of Tim50 and Tim23 in preprotein translocation to the protein-conducting channel of the mitochondrial inner membrane.
Keywords: mitochondrial inner membrane, preprotein, protein sorting, Saccharomyces cerevisiae, Tim23
Mitochondria play central functions in cellular bioenergetics, metabolism, ion homeostasis and apoptosis.1-5 99% of the ~1,000 different mitochondrial proteins are synthesized in the cytosol and imported into mitochondria.1,2,6-8 More than half of the mitochondrial preproteins are synthesized with N-terminal presequences that form positively charged amphipathic α-helices and target the preproteins to mitochondria. The preproteins are imported by the general translocase of the outer membrane (TOM complex) and the presequence translocase of the inner membrane (TIM23 complex).2,9-14 After passing through the TOM complex, the preproteins are received by Tim50, an essential component of the TIM23 complex exposed to the intermembrane space (IMS).15-17 Tim50 interacts with the N-terminal IMS-domain of Tim23 to transport preproteins to the transmembrane channel formed by the C-terminal domain of Tim23.13,18-22 No structural information on Tim50 has been reported and thus the molecular mechanism of Tim50 and its mode of interaction with Tim23 are unknown.
In this study, we report the crystal structure of the IMS-domain of Tim50 that contains a large groove as putative binding site for presequences and an exposed β-hairpin. We show that the β-hairpin is crucial for the interaction of Tim50 with Tim23, suggesting a cooperative function of these two essential TIM proteins in preprotein import.
Structure of the IMS-domain of Tim50
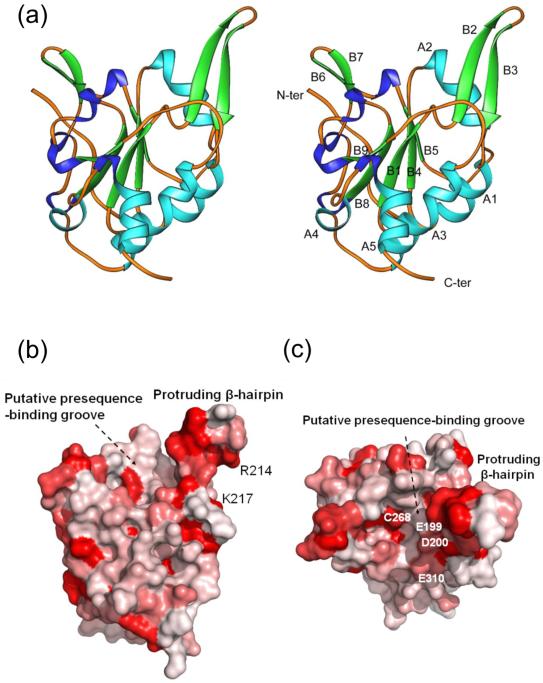
Tim50 consists of an N-terminal presequence, a transmembrane anchor and a large C-terminal IMS-domain (residues 133-476 in Saccharomyces cerevisiae).15-17 We expressed the C-terminal domain in E. coli and generated a trypsin-resistant fragment (residues 164-361). The crystal structure of the trypsin-resistant fragment was determined to 1.83 Å resolution using the molecular replacement method (Table 1). The final model of Tim50IMS contains residues 176-361, representing the conserved core of Tim50 (Fig. S1). Tim50IMS forms a monomer in the crystal structure and consists of five α-helices (A1-A5) and nine β-strands (B1-B9) (Fig. 1a). The core of the structure is constituted by a parallel β-sheet formed by B1, B4, B5, B8 and B9 with B1 in the middle. A proline-rich region forms an extended loop region at the N-terminus of the structure. Three α-helices A3, A4 and A5 constitute an anti-parallel bundle at the C-terminus of the structure. A β-hairpin protruding out of the Tim50 molecular surface by ~15 Å is formed by B2 and B3 and the short loop between B2 and B3 (Fig. 1a). The protruding β-hairpin represents the largest conserved area on the Tim50 molecular surface (Fig. 1b). Close to the protruding β-hairpin, Tim50IMS contains a large groove with a dimension of 15×10×5 Å (L×W×D) (Fig. 1b and c). The bottom of the groove is formed by the loops between B1/B2, B4/A2, B5/B6 and B8/B9.
Table 1.
Data collection and refinement statistics for Tim50IMS structure
| Tim50IMS | |
|---|---|
| Data collection | |
| Space group | P6122 |
| Cell dimensions | |
| a, c (Å) | 84.109, 116.549 |
| Wavelength (Å) | 1.000 |
| Resolution (Å) | 1.83 |
| Rsym | 0.0613 (0.266) |
| I / sigmaI | 42.1 (5.2) |
| Completeness (%) | 98.8 (91.9) |
| Redundancy | 8.2 (7.1) |
| Refinement | |
| No. reflections | 20436 |
| Rfactor / Rfree | 0.193/0.224 |
| No. atoms (Non- water) |
1532 |
| No. waters | 132 |
| R.m.s deviations | |
| Bond lengths (Å) | 0.015 |
| Bond angles (°) | 1.578 |
The coding region of S. cerevisiae Tim50 (residues 132-476) was cloned into the vector pET28b for expression in E. coli. A trypsin-resistant fragment (residues 164-361), termed Tim50IMS, was identified by using limited proteolysis. The coding region of Tim50IMS was cloned into the vector pET28b for expression in E. coli. Tim50IMS was concentrated to 20 mg ml−1 in 10 mM Tris buffer (pH 7.5), 150 mM NaCl and subjected to crystallization trials. Large rod-shaped crystals (0.5 × 0.2 × 0.2 mm) were obtained by the hanging-drop vapor-diffusion method using Linbro plates at room temperature. The well solution consisted of 1 ml 100 mM MES buffer (pH 6.0), 30% (w/v) PEG4K, 100 mM calcium acetate and 200 mM ammonium acetate. The native Tim50 crystals diffracted X-ray to 1.83 Å in the beamline SER-CAT at APS. The crystals were flash frozen at 100 K in a Nitrogen gas stream in the cryoprotectant consisting of 100 mM MES buffer (pH 6.0), 30% (w/v) PEG4K, 200 mM ammonium acetate and 20% (v/v) ethylene glycol. The atomic coordinates of Scp1 (PDB code: 2GHQ) were used to search for molecular replacement solutions for the crystals of Tim50 by the program PHASER.26 The initial model was built by use of WARP/ARP,27 followed by manual model building using COOT.28 Residues 176-361 of Tim50 were modeled into the resultant electron density map. The subsequent structure refinement was performed by using the program CNS and Refmac5 against the native data set of 1.83 Å resolution.26,29 After each cycle of refinement, the model was manually rebuilt based on the resultant 2Fo-Fc and Fo-Fc maps. The Tim50 structure was refined with an Rfactor of 19.3% and Rfree of 22.4%. The refinement gave reasonable rms derivations from the ideal geometry at this resolution. A Ramachandran plot of the final model by use of the program Probity (http://kinemage.biochem.duke.edu) revealed that 98.3% of the residues were in favored regions.30 The final model contains Tim50IMS, two calcium atoms, one acetate molecule and one penta-ethylene glycol (PEG400) molecule. Highest-resolution shell is shown in parentheses in the table.
Fig. 1.
The Tim50 IMS-domain structure. (a) Ribbons drawing of the Tim50IMS monomer structure in side-by-side stereo mode. α-helices (A1 to A5) and β-strands (B1 to B9) are shown in light blue and green, respectively. Turns are shown in dark blue. (b) Sequence conservation drawing of the Tim50IMS molecular surface. The Tim50 sequences from ten species (S. cerevisiae, C. elegans, H. sapiens, D. melanogaster, N. crassa, D. rerio, M. musculus, B. taurus, P. pastoris and D. hansenii) were put into the ClustalW program for sequence alignment. The aligned sequences in multiple sequence alignment formats were converted into a property file by use of the program ProtSkin (http://www.mcgnmr.mcgill.ca/ProtSkin/). The property file was then visualized by using the program PyMol (http://www.pymol.org). Red color denotes conserved regions. The orientation is similar to that in A. (c) Tim50IMS of b is rotated along the horizontal axis by 90° and the protruding β-hairpin is facing the reader.
Interaction of Tim50 with Tim23 in organello
To map the region of Tim50 that interacts with Tim23, we established an in organello assay by importing modified forms of Tim50 into mitochondria that carried a protein A-tag at Tim23. It was shown that the N-terminal presequence and transmembrane domain of Tim50 are not essential for its function.21 However, the crystallized Tim50IMS also lacks 115 C-terminal residues. To test if this C-terminal region of Tim50 was required for its biogenesis or binding to Tim23, we synthesized and radiolabeled a C-terminally truncated Tim50. Tim501-361 was efficiently imported into yeast mitochondria (Fig. S2a) and co-precipitated by protein A-tagged Tim23 after lysis of the mitochondria with a non-ionic detergent (Fig. S2b). Control proteins like Atp19 of the mitochondrial ATP synthase and Tim10 of the carrier translocase were not co-precipitated. Thus, the C-terminal region of Tim50 (residues 362-476) is neither essential for biogenesis of Tim50 nor binding to Tim23. The exact function of the C-terminal region of Tim50 is unknown; it may potentially contribute to the interaction with preproteins or the cooperation of Tim50 with the TOM complex or further TIM subunits.
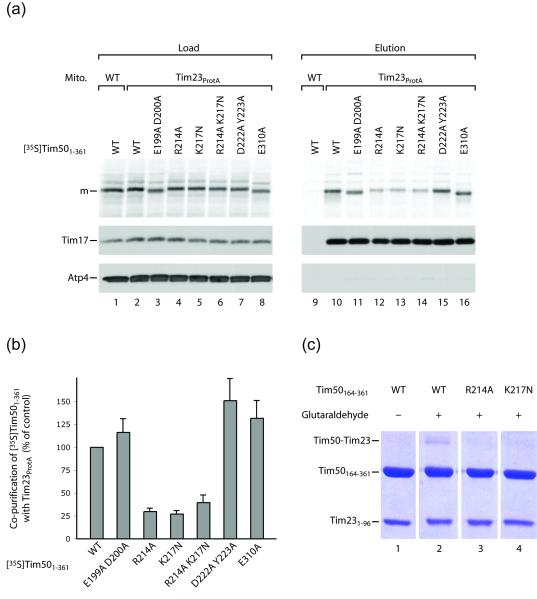
Using a chemical crosslinking reagent, the only cysteine of yeast Tim50 (C268) was reported to be in proximity to Tim23.23 C268 is located at the bottom of the large groove and near the protruding β-hairpin (Fig. 1c). We performed a structure-based mutagenesis of Tim50 residues located at the protruding β-hairpin or the groove. The resulting mutant forms of Tim501-361 were efficiently imported into mitochondria (Fig. 2a, lanes 3-8), but differed significantly in their interaction with Tim23, as analyzed by pull-down with tagged Tim23 (Fig. 2a, lanes 11-16). Replacement of R214 and K217 of the β-hairpin strongly reduced the ability of Tim50 to bind to Tim23. In contrast, replacement of the residues E199 and D200 did not impair the Tim50-Tim23 interaction, whereas replacement of D222, Y223 and E310 slightly enhanced the interaction (Fig. 2a and b). In the crystal structure, residues R214 and K217 are located at the lateral surface of the protruding β-hairpin; E199, D200 and E310 are located in the large groove; and residues D222 and Y223 are positioned at the side of the Tim50 molecule below the β-hairpin (Fig. 1b and c; Fig. S3). In addition to the in organello interaction studies, we also analyzed the interaction of recombinant Tim50164-361 with Tim23IMS (residues 1-96; pI 4.12) by crosslinking. Replacement of R214 or K217 inhibited the Tim50-Tim23 interaction in vitro (Fig. 2c) comparable to the findings in organello. Taken together, these results indicate that the protruding β-hairpin of Tim50IMS, including the positively charged residues 214 and 217, is critical for binding to the acidic IMS-domain of Tim23.
Fig. 2.
Interaction of mutant forms of Tim50 with Tim23. (a) Radiolabeled Tim501-361 and mutant forms were imported into yeast mitochondria carrying protein A-tagged Tim23 or into wild-type (WT) mitochondria. After lysis with digitonin and affinity-purification, mature-sized (m) Tim501-361 was analyzed by SDS-PAGE and autoradiography (see Supplementary Material). Control proteins, Tim17 and Atp4, were detected by Western blotting. Load, 10%; elution, 100%. (b) Quantification of co-purification of wild-type (WT) and mutant Tim501-361 forms with Tim23, normalized to input/load. The yield of co-purification of imported WT Tim501-361 was set to 100%. Shown are averages ± SEM (n=4) (range with n=2 for the R214A K217N double mutant form). (c) In vitro crosslinking of purified Tim50164-361 forms and Tim231-96. Crosslinking reactions with glutaraldehyde were carried out at room temperature for 20 min. 10 μM Tim231-96 were incubated with equimolar amounts of Tim50164-361 or mutant forms in 20 mM HEPES, 0.1 M NaCl, pH 8 buffer. 0.005% glutaraldehyde was added. The crosslinking reaction was quenched by adding SDS-PAGE sample buffer, and products were analyzed by 12% SDS polyacrylamide gel electrophoresis.
Model for cooperation of Tim50 and Tim23 in protein import
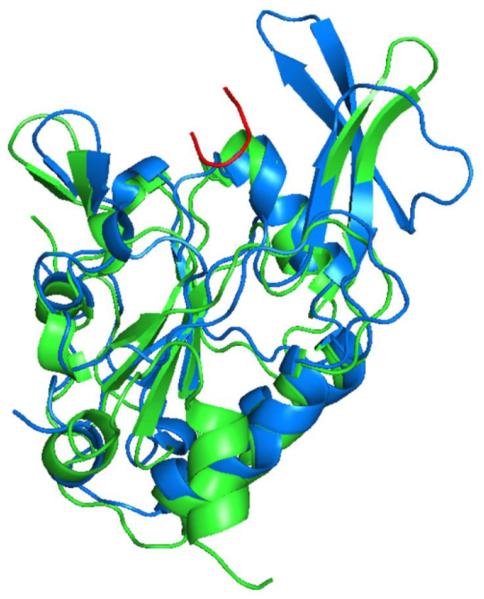
The large groove in Tim50IMS contains several exposed negatively charged residues (Fig. 1c). We speculate that this groove is ideally suited as binding site for positively charged presequences/preproteins (additionally, hydrophobic residues located around the rim of the groove may contribute to an interaction with the hydrophobic surfaces of the amphipathic presequences), though an involvement of the groove in the cooperation of Tim50 with the TOM complex or further TIM subunits cannot be excluded. The crystal structure of Tim50IMS was determined by molecular replacement using the phosphatase Scp1 that is specific for the C-terminal domain of RNA polymerase II.24 Scp1 is the top structural homologue of Tim50IMS (Z-score of 22.8), exhibiting similar protein folding with rms derivation of 2.2 Å for the main chain atoms (Fig. 3) (the sequence identity between human Scp1 and yeast Tim50IMS is 33.5% [calculated by ClustalW]). Interestingly, the crystal structure of Scp1 was solved in complex with the C-terminal phosphorylated peptide of RNA Polymerase II.24 The peptide forms a U-turn in a groove that corresponds to the putative presequence-binding groove in Tim50IMS (Fig. 3). In Tim50IMS, the protruding β-hairpin swings away from the groove, providing more space for the presequence-binding groove to likely accommodate an α-helix formed by the presequence. The bottom of the peptide-binding groove of Scp1 contains a DXDX(T/V) phosphatase motif24 that is not present in Tim50 (and Tim50 does not show phosphatase activity).15 The bottom of the putative presequence-binding groove of Tim50IMS is only weakly conserved (Fig. 1c), which may reflect that Tim50 has to interact with a large variety of mitochondrial presequences.
Fig. 3.
Superimposition of Tim50IMS (green) and Scp1 structures (blue). The orientation of Tim50 is similar to that in Fig. 1a. The bound peptide substrate of Scp1 is shown in red.
The close proximity between the putative preprotein-binding groove and the Tim23-interacting β-hairpin of Tim50 can provide a molecular explanation for the observation that binding of preproteins to Tim50 depends on the interaction of Tim50 with Tim23.21 Though NMR spectroscopy analysis indicated that Tim23IMS is intrinsically disordered,25 recent studies provided evidence that the Tim23 sites for preprotein binding and interaction with Tim50 are in close proximity. Crosslinking studies mapped a region of Tim23 including residues 70 and 71 interacting with Tim50.20,22 NMR spectroscopy suggested that a Tim23 region from residues 71–84 is involved in presequence binding.25 We propose that for Tim50 as well as Tim23, the site of preprotein recognition is in close proximity to the Tim50-Tim23 interaction site, leading to a working model that the receptor module of the presequence translocase is formed by a composite presequence binding pocket that involves both Tim50 and Tim23.
In summary, our structural and functional analysis of Tim50 revealed that the protruding β-hairpin is important for recruiting Tim23, supporting the hypothesis that Tim50 and Tim23 function cooperatively to direct preproteins to the transmembrane channel formed by the C-terminal domain of Tim23.
Supplementary Material
Acknowledgements
We are grateful to the staff scientists in APS beamline SER-CAT for their help in data collection. We thank I. Perschil, C. Prinz and A. Schulze-Specking for expert technical assistance. This work was supported by grants from NIH (R01 GM65959) and Army Research Office (51894LS) (to B.D.S), Deutsche Forschungsgemeinschaft, Sonderforschungsbereich 746, Excellence Initiative of the German Federal & State Governments (EXC 294 BIOSS; GSC-4 Spemann Graduate School), Bundesministerium für Bildung und Forschung, Landesforschungspreis Baden-Württemberg, Gottfried Wilhelm Leibniz Program and the Fonds der Chemischen Industrie.
Abbreviations used
- IMS
intermembrane space
- TIM23 complex
presequence translocase of the inner mitochondrial membrane
- Tim50
presequence translocase of inner membrane subunit of ~50 kDa
- TOM complex
translocase of the outer mitochondrial membrane
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession code Atomic coordinates have been deposited in the Protein Data Bank under accession code 3QLE.
Supplementary material Supplementary material associated with this article can be found online at doi: xxx.
References
- 1.Dolezal P, Likic V, Tachezy J, Lithgow T. Evolution of the molecular machines for protein import into mitochondria. Science. 2006;313:314–318. doi: 10.1126/science.1127895. [DOI] [PubMed] [Google Scholar]
- 2.Neupert W, Herrmann JM. Translocation of proteins into mitochondria. Annu. Rev. Biochem. 2007;76:723–749. doi: 10.1146/annurev.biochem.76.052705.163409. [DOI] [PubMed] [Google Scholar]
- 3.Dimmer KS, Rapaport D. Proteomic view of mitochondrial function. Genome Biol. 2008;9:209. doi: 10.1186/gb-2008-9-2-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lill R. Function and biogenesis of iron-sulphur proteins. Nature. 2009;460:831–838. doi: 10.1038/nature08301. [DOI] [PubMed] [Google Scholar]
- 5.Schmidt O, Pfanner N, Meisinger C. Mitochondrial protein import: from proteomics to functional mechanisms. Nat. Rev. Mol. Cell. Biol. 2010;11:655–667. doi: 10.1038/nrm2959. [DOI] [PubMed] [Google Scholar]
- 6.Koehler CM. New developments in mitochondrial assembly. Annu. Rev. Cell Dev. Biol. 2004;20:309–335. doi: 10.1146/annurev.cellbio.20.010403.105057. [DOI] [PubMed] [Google Scholar]
- 7.Baker MJ, Frazier AE, Gulbis JM, Ryan MT. Mitochondrial protein-import machinery: correlating structure with function. Trends Cell Biol. 2007;17:456–464. doi: 10.1016/j.tcb.2007.07.010. [DOI] [PubMed] [Google Scholar]
- 8.Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N. Importing mitochondrial proteins: machineries and mechanisms. Cell. 2009;138:628–644. doi: 10.1016/j.cell.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abe Y, Shodai T, Muto T, Mihara K, Torii H, Nishikawa S, Endo T, Kohda D. Structural basis of presequence recognition by the mitochondrial protein import receptor Tom20. Cell. 2000;100:551–560. doi: 10.1016/s0092-8674(00)80691-1. [DOI] [PubMed] [Google Scholar]
- 10.Chacinska A, Lind M, Frazier AE, Dudek J, Meisinger C, Geissler A, Sickmann A, Meyer HE, Truscott KN, Guiard B, Pfanner N, Rehling P. Mitochondrial presequence translocase: switching between TOM tethering and motor recruitment involves Tim21 and Tim17. Cell. 2005;120:817–829. doi: 10.1016/j.cell.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 11.Oka T, Mihara K. A railroad switch in mitochondrial protein import. Mol. Cell. 2005;18:145–146. doi: 10.1016/j.molcel.2005.03.022. [DOI] [PubMed] [Google Scholar]
- 12.Wu Y, Sha B. Crystal structure of yeast mitochondrial outer membrane translocon member Tom70p. Nat. Struct. Mol. Biol. 2006;13:589–593. doi: 10.1038/nsmb1106. [DOI] [PubMed] [Google Scholar]
- 13.Alder NN, Jensen RE, Johnson AE. Fluorescence mapping of mitochondrial TIM23 complex reveals a water-facing, substrate-interacting helix surface. Cell. 2008;134:439–450. doi: 10.1016/j.cell.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 14.Vögtle FN, Wortelkamp S, Zahedi RP, Becker D, Leidhold C, Gevaert K, Kellermann J, Voos W, Sickmann A, Pfanner N, Meisinger C. Global analysis of the mitochondrial N-proteome identifies a processing peptidase critical for protein stability. Cell. 2009;139:428–439. doi: 10.1016/j.cell.2009.07.045. [DOI] [PubMed] [Google Scholar]
- 15.Geissler A, Chacinska A, Truscott KN, Wiedemann N, Brandner K, Sickmann A, Meyer HE, Meisinger C, Pfanner N, Rehling P. The mitochondrial presequence translocase: an essential role of Tim50 in directing preproteins to the import channel. Cell. 2002;111:507–518. doi: 10.1016/s0092-8674(02)01073-5. [DOI] [PubMed] [Google Scholar]
- 16.Yamamoto H, Esaki M, Kanamori T, Tamura Y, Nishikawa S, Endo T. Tim50 is a subunit of the TIM23 complex that links protein translocation across the outer and inner mitochondrial membranes. Cell. 2002;111:519–528. doi: 10.1016/s0092-8674(02)01053-x. [DOI] [PubMed] [Google Scholar]
- 17.Mokranjac D, Paschen SA, Kozany C, Prokisch H, Hoppins SC, Nargang FE, Neupert W, Hell K. Tim50, a novel component of the TIM23 preprotein translocase of mitochondria. EMBO J. 2003;22:816–825. doi: 10.1093/emboj/cdg090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meinecke M, Wagner R, Kovermann P, Guiard B, Mick DU, Hutu DP, Voos W, Truscott KN, Chacinska A, Pfanner N, Rehling P. Tim50 maintains the permeability barrier of the mitochondrial inner membrane. Science. 2006;312:1523–1526. doi: 10.1126/science.1127628. [DOI] [PubMed] [Google Scholar]
- 19.van der Laan M, Meinecke M, Dudek J, Hutu DP, Lind M, Perschil I, Guiard B, Wagner R, Pfanner N, Rehling P. Motor-free mitochondrial presequence translocase drives membrane integration of preproteins. Nat. Cell Biol. 2007;9:1152–1159. doi: 10.1038/ncb1635. [DOI] [PubMed] [Google Scholar]
- 20.Gevorkyan-Airapetov L, Zohary K, Popov-Celeketic D, Mapa K, Hell K, Neupert W, Azem A, Mokranjac D. Interaction of Tim23 with Tim50 is essential for protein translocation by the mitochondrial TIM23 complex. J. Biol. Chem. 2009;284:4865–4872. doi: 10.1074/jbc.M807041200. [DOI] [PubMed] [Google Scholar]
- 21.Mokranjac D, Sichting M, Popov-Celeketic D, Mapa K, Gevorkyan-Airapetov L, Zohary K, Hell K, Azem A, Neupert W. Role of Tim50 in the transfer of precursor proteins from the outer to the inner membrane of mitochondria. Mol. Biol. Cell. 2009;20:1400–1407. doi: 10.1091/mbc.E08-09-0934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tamura Y, Harada Y, Shiota T, Yamano K, Watanabe K, Yokota M, Yamamoto H, Sesaki H, Endo T. Tim23-Tim50 pair coordinates functions of translocators and motor proteins in mitochondrial protein import. J. Cell Biol. 2009;184:129–141. doi: 10.1083/jcb.200808068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alder NN, Sutherland J, Buhring AI, Jensen RE, Johnson AE. Quaternary structure of the mitochondrial TIM23 complex reveals dynamic association between Tim23p and other subunits. Mol. Biol. Cell. 2008;19:159–170. doi: 10.1091/mbc.E07-07-0669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y, Kim Y, Genoud N, Gao J, Kelly JW, Pfaff SL, Gill GN, Dixon JE, Noel JP. Determinants for dephosphorylation of the RNA polymerase II C-terminal domain by Scp1. Mol. Cell. 2006;24:759–770. doi: 10.1016/j.molcel.2006.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de la Cruz L, Bajaj R, Becker S, Zweckstetter M. The intermembrane space domain of Tim23 is intrinsically disordered with a distinct binding region for presequences. Protein Sci. 2010;19:2045–2054. doi: 10.1002/pro.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.CCP4 The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 27.Langer G, Cohen SX, Lamzin VS, Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat. Protoc. 2008;3:1171–1179. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 29.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 30.Lovell SC, Davis IW, Arendall WB, III, de Bakker PI, Word JM, Prisant MG, Richardson JS, Richardson DC. Structure validation by Cα geometry: ϕ,ψ and Cβ deviation. Proteins. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.