Abstract
The GU-rich element (GRE) was identified as a conserved sequence enriched in the 3′ UTR of human transcripts that exhibited rapid mRNA turnover. In mammalian cells, binding to GREs by the protein CELF1 coordinates mRNA decay of networks of transcripts involved in cell growth, migration, and apoptosis. Depending on the context, GREs and CELF1 also regulate pre-mRNA splicing and translation. GREs are highly conserved throughout evolution and play important roles in development of organisms ranging from worms to man. In humans, abnormal GRE-mediated regulation contributes to disease states and cancer. Thus, GREs and CELF proteins serve critical functions in gene expression regulation and define an important evolutionarily conserved posttranscriptional regulatory network.
Keywords: GU-rich element, CELF1, CUG-binding protein 1, RNA-binding proteins, CELF, mRNA degradation, posttranscriptional gene regulation
Introduction
The precise control of gene expression during cellular processes such as activation, proliferation, differentiation, and development requires multiple levels of regulation, including transcriptional and posttranscriptional mechanisms. Steady-state protein levels within a cell correlate poorly with steady-state levels of mRNA, suggesting that large numbers of transcripts undergo post-transcriptional regulation [1]. Cis-acting regulatory sequences found in coding regions and in 3′ and 5′ untranslated regions (UTRs) of mRNA allow selective recognition by RNA-binding proteins (RBPs) or microRNAs which direct the fate of the mRNA by controlling posttranscriptional processes such as translation and mRNA degradation (reviewed in references [2]•, [3], [4], [5]). Here, we review the GU-rich element (GRE) as an example of an evolutionarily conserved cis element in mRNA that controls posttranscriptional gene expression networks through its interaction with the protein CELF1.
Cis Elements in the Coordinate Regulation of mRNA Decay
Cis elements in mRNA function at posttranscriptional levels to coordinately regulate gene expression through their interactions with microRNAs or RBPs. microRNAs are small endogenous RNA molecules that bind to specific sequences in mRNA and regulate translation and/or mRNA degradation during growth and development [6]•, [7]•, [8]. Overexpression of a specific microRNA can lead to the down-regulation of hundreds of mRNAs suggesting that microRNAs play important roles in orchestrating mRNA degradation. In addition to microRNAs, certain RBPs bind to specific mRNA sequences and also coordinately regulate mRNA degradation and/or translation. The combinatorial interplay between various miRNAs and RBPs that bind to a given mRNA transcript control many developmental decisions in a variety of species [9], [10], [11].
Considerable insight into the mechanisms of coordinate mRNA degradation by cis elements has come from studies involving the AU-rich element (ARE) and ARE-binding proteins. The ARE is a well characterized cis-acting mRNA sequence that regulates mRNA decay by binding to a variety of RBPs depending on the cellular context (for reviews see [12],[13]). Several ARE-binding proteins, including AUF1 [14], BRF1 and TTP [15],[16],[17], and KSRP [18], [19], promote ARE-mediated mRNA decay, whereas other ARE-binding proteins such as HuR [20],[21], HuB, HuC, and HuD stabilize target mRNAs and stimulate their translation [22]. ARE-binding proteins rapidly modulate the stability and/or translation of mRNA during cell proliferation and development [23],[24],[25] •. For example, the destabilizing protein TTP is induced following T cell activation and functions to mediate the degradation of multiple ARE-containing transcripts that encode inflammatory mediators such as interleukin-2 and interferon-gamma [26],[27]. TTP mediates the decay of ARE-containing transcripts by recruiting components of the mRNA decay machinery to the transcript [28],[16],[29]. This mechanism allows for the coordinate down-regulation of multiple genes at the appropriate time following T cell activation. The characterization of the ARE as a cis-acting regulator of mRNA decay led to a more systematic classification of ARE-containing genes, including the construction of ARE databases [30],[31]•,[32] and examination of the mRNA decay rates of ARE-containing transcripts using microarray technology [33]. These methodologies enabled biologists to globally assess the physiological significance of ARE-mediated mRNA decay regulation and to identify coordinate gene expression networks regulated by AREs [34],[35],[13],[36]. This approach can be applied to identify and understand other posttranscriptional regulatory networks.
In this review, we describe how knowledge about coordinate gene regulation by conserved cis sequences in mRNA led to the identification of the GRE, which defines a posttranscriptional regulatory network that has been conserved through evolution. By using bioinformatic sequence motif discovery methods, in conjunction with gene expression clustering, the GRE was identified as a highly conserved sequence that was enriched in the 3′ UTR of mRNA transcripts with short half lives and was shown to function in human cells as a regulator of mRNA decay [37] ••. In human cells, the GRE is a target of CELF1, also known as CUG-binding protein 1 (CUGBP1), a member of the CELF (CUGBP and embryonically lethal abnormal vision-type RNA binding protein 3-like factors) family of RNA-binding proteins. CELF1 has been implicated as a regulator of alternative splicing [38],[39],[40], translation [41], deadenylation [42], and mRNA degradation [37]••,[43] ••. Together, the GRE and CELF1 define an evolutionarily conserved posttranscriptional regulatory network.
GREs as Regulators of mRNA Decay
The GRE consensus sequence, UGUUUGUUUGU, was identified as a sequence that was highly enriched in the 3′ UTR of short-lived transcripts expressed in primary human T cells [37] ••. The GRE is a bona fide mRNA decay element because it conferred instability upon reporter transcripts when it was inserted into their 3′ UTR. The CELF1 protein binds to GREs, and knockdown of CELF1 leads to stabilization of GRE-containing transcripts, indicating that CELF1 is essential for GRE-mediated mRNA decay. More recently, RNA-immunoprecipitation (RNA-IP) was performed in cytoplasmic extracts from HeLa cells using an anti-CELF1 antibody, and CELF1-associated transcripts were identified using oligonucleotide microarrays. A bioinformatics search for conserved sequences in immunoprecipitated transcripts, using the program BioProspector (and overrepresentation algorithm), found the previously described UGUUUGUUUGU sequences as well as the GU-repeat sequence UGUGUGUGUGU sequences to be overrepresented [43] ••. Interestingly, the GU-repeat sequence was previously identified through systemic evolution of ligands exponential enrichment (SELEX) as a CELF1-binding sequence [44], and CELF1 binds with high affinity to GU-repeat sequences [45],[46]. Insertion of a GU-repeat sequence into the 3′ UTR of a reporter transcript conferred instability to the reporter construct, demonstrating that this GU-repeat sequence functioned as a decay element [43]. Because the UGUUUGUUUGU sequence and the GU-repeat sequence both bound to CELF1 and functioned as decay elements, the GRE was redefined to contain both of these sequences ([43] ••, see Table 1). In myoblasts, a similar RNA-IP approach using an anti-CELF1 antibody identified GRE hexamers to be significantly overrepresented in short-lived transcripts that co-immunoprecipitated with CELF1 [47] ••. In this system, knockdown of CELF1 led to the stabilization of certain GRE-containing targets, confirming that CELF1 regulated the stability of those transcripts. In Xenopus, target transcripts identified by RNA-IP using an antibody against embryo deadenylation element binding protein (EDEN-BP), the CELF1 orthologue, were enriched in GU-rich sequences, very similar to GREs and the 15 nucleotide consensus motif (UGU/UG)n was predicted to be a target of CELF1 [48]••,[49]. Overall, these studies revealed that GU-rich sequences function as mRNA decay elements and serve as binding sites for CELF1 in a manner that has been conserved through evolution.
Table 1.
GU-rich motifs known to bind CELF1.
Evolutionary Conservation of GREs and CELF Proteins
Translation and mRNA decay are often coupled with one another to control of gene expression in response to environmental and developmental changes. In several organisms, translation is regulated by deadenylation, which is also an early step in the mRNA decay pathway. The deadenylation and translation of genes important in development are regulated by GU-rich sequences and CELF proteins across diverse species [50],[51]. In Xenopus, the CELF1 orthologue, EDEN-BP, binds to the GU-rich EDEN element, which functions as a deadenylation signal in Xenopus embryos after fertilization and regulates translational activation [52],[53]. In Drosophila, the CELF1 orthologue, Bru-3 (Bruno-3), binds specifically to (UG)15 repeats to regulate translation of proteins involved in embryogenesis and organogenesis [54], [55], [56]. The Zebrafish orthologue, Bru-l, also binds preferentially to GU-rich RNAs and regulates development [57]. NMR-based solution studies demonstrated that human CELF1 RNA recognition motifs bound specifically to RNA UGUU or UGUG sequences [58]•,[59] •.
CELF proteins are essential post-transcriptional regulators of development in lower organisms such as Xenopus where they regulate deadenylation and translation [60]. Whereas GRE-mediated deadenylation often regulates translation in lower organisms, the deadenylation is usually the first step leading to mRNA degradation in mammalian cells. The consequences of deadenylation differ in different organisms, although the mechanism of deadenylation appears to be evolutionarily conserved. For example, a GU-rich sequence from human c-jun mRNA substituted for the EDEN element as a deadenylation signal in Xenopus extracts [61]. Furthermore, human CELF1, which has 88% identity with EDEN-BP, was able to functionally substitute for EDEN-BP to mediate transcript deadenylation in Xenopus extracts [53], suggesting that the deadenylation function of GU-rich sequences and CELF proteins were conserved in diverse species. Human CELF1 was shown to associate with poly A ribonuclease (PARN) and to stimulate poly A tail shortening in a cell-free assay using S100 extracts from human cells, suggesting that CELF1 mediates mRNA decay through deadenylation [42]. Thus, the deadenylation function of GREs and CELF1 is conserved through evolution and may be responsible for coordinated mRNA decay in mammalian cells.
In addition to regulating deadenylation and translation, CELF proteins regulate alternative splicing in diverse species by binding to GU-rich or U-rich sequences (reviewed in [62] •, [4]). CELF-mediated regulation of alternative splicing is necessary for maintenance of normal muscle structure and function [63],[64],[65]. Recently, a RNA cross-linking immunoprecipitation (RNA-CLIP) approach was used to identify 315 CELF1 RNA targets in whole cell extracts from mouse hindbrain [66] ••. RNA binding targets for CELF1 were enriched in UG repeat sequences with 64% of target sequences found in introns and 25% found in 3′ UTR sequences. Thus, by binding to GU-rich sequences, CELF1 may function to regulate pre-mRNA splicing, translation, and/or deadenylation/decay, depending on the context.
Coordinate Regulation of the GRE/CELF1 Network in Cellular Activation and Differentiation
In primary human T cells, GREs and CELF1 appear to regulate rapid changes in gene expression following T cell receptor-mediated activation. Figure 1 shows a network of short-lived GRE-containing transcripts that are involved in T cell signaling. Many of these GRE-containing transcripts were expressed transiently following T cell activation and then rapidly disappeared [67], suggesting that GRE-mediated mRNA decay plays a central role in the coordinate down-regulation of these genes following T cell activation [37]••,[68] ••. Thus, GRE-mediated mRNA decay appears to be an important regulatory step in the early stages of T cell activation.
Figure 1. T-cell receptor signaling pathways regulated by GREs.
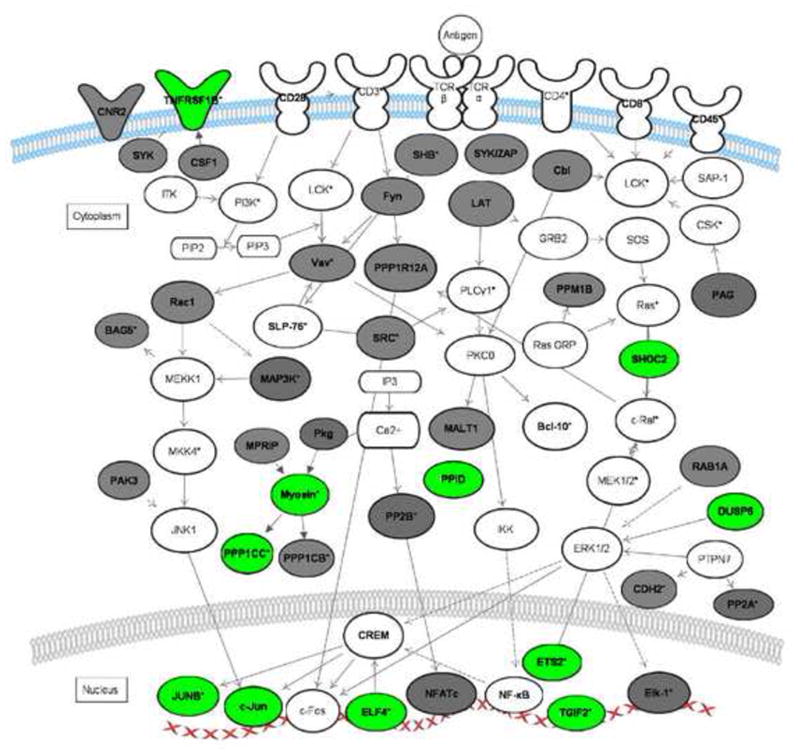
The network diagram depicts the coordinate regulation of GRE-containing transcripts involved in T cell receptor signaling. Transcripts in bold are GRE-containing. Transcripts in grey were identified as CELF1 targets in HeLa cells by RNA-IP [43].
Transcripts in green represent short-lived GRE-containing transcripts expressed in primary human T cells [33], [37]. Transcripts labeled with an asterisk exhibited changes in steady state levels following T-cell receptor stimulation [33]. This network diagram was built using Ingenuity Pathway Assistant Software.
In mouse myoblasts, RNA-IP followed by microarray analysis identified a variety of CELF1 target transcripts that contained GU-rich sequences, including networks of transcripts that regulate cell cycle and intracellular signaling cascades involved in intracellular transport and cell survival (Figure 2) [47] ••. Many of these CELF1 target transcripts were found to be significantly stabilized in CELF1 knockout myoblasts [47] ••, suggesting CELF1 mediates the decay of a network of transcripts that may be involved in myoblast growth and differentiation. Interestingly, many of the CELF1 target transcripts in mouse myobasts were also found to be target transcripts of EDEN-BP in Xenopus tropicalis extracts (Figure 2), providing further evidence that GRE/CELF1 posttranscriptional networks were conserved through evolution.
Figure 2. A posttranscriptional network of CELF1 target transcripts in mouse myoblasts.
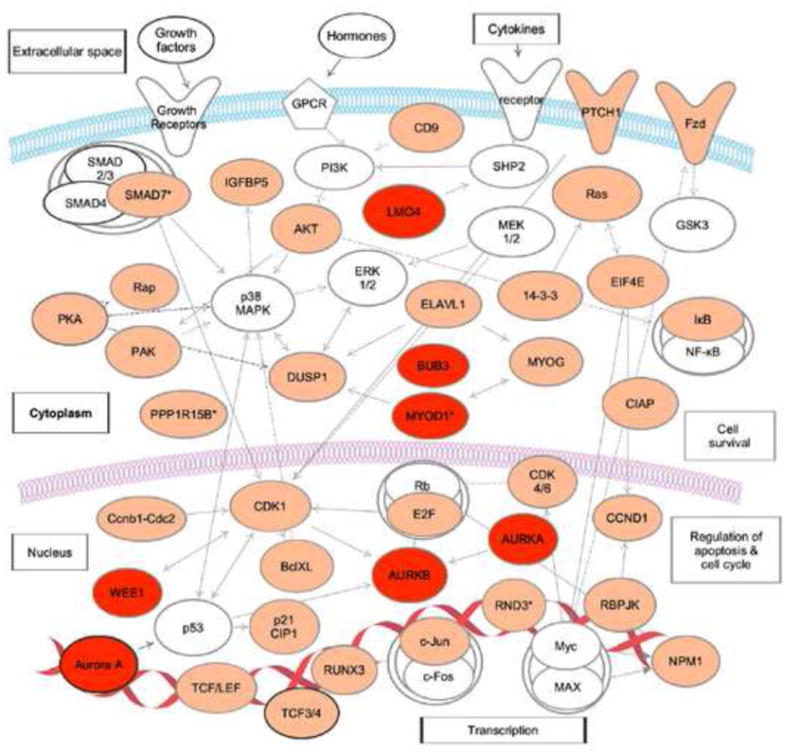
Transcripts shown in orange are CELF1 targets in mouse myoblasts [47]. Transcripts shown in red are CELF1 targets in mouse myoblasts and are also targets of EDEN-BP in Xenopus tropicalis extracts [48]. Transcripts, marked with asterisk (*), were stabilized in CELF1 knockout myoblasts. This network diagram was built using Ingenuity Pathway Assistant Software.
The GRE/CELF1 Posttranscriptional Network in Human Diseases
CELF1 and its GRE-containing target transcripts define posttranscriptional regulatory networks that functions to control cellular growth, activation, and differentiation. Disruptions in GRE-mediated mRNA regulation may play a role in developmental pathology [69],[62] or cancer. CELF1 was found in a transposon-based genetic screen in mice to be one of the top ten genes to drive tumorigenesis if mutated and/or dysregulated [70] •, suggesting that CELF1/GRE networks may be regulated abnormally in cancer. RNA-IP followed by microarray analysis to identify CELF 1 target transcripts in human Hela cells (carcinoma cell line), revealed that numerous target transcripts play roles in processes important for cancer development including cell growth, apoptosis, and cell migration [43] ••. For example, CELF1 targets in HeLa cells included numerous transcripts encoding regulators of G-protein signaling pathway and G-protein coupled receptor ligands (Figure 3). These pathway activate/or repress cell-cell interaction, cell migration and invasion, and thereby play important roles in cancer development and metastasis. Thus, CELF1/GRE networks may be aberrantly regulated in malignant cells.
Figure 3. A posttranscriptional network of CELF1 target transcripts in malignant cells.
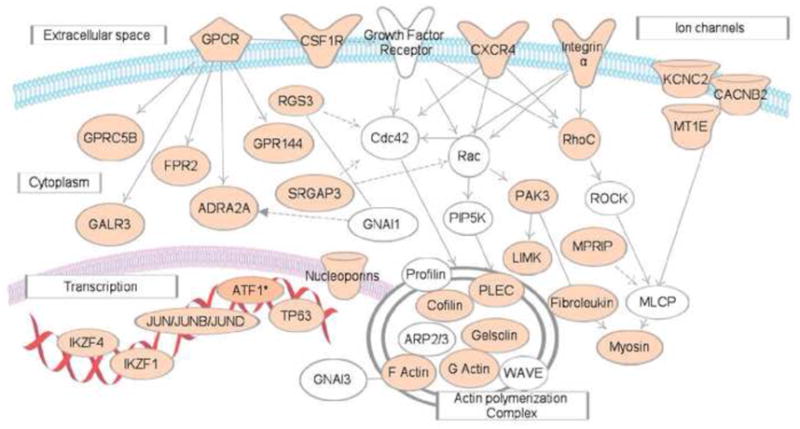
This network represents transcripts that involved in G protein coupled receptor signaling pathways. Transcripts depicted in tan represent GRE-containing CELF1 target transcripts in HeLa cells [43]. This network diagram was built using Ingenuity Pathway Assistant Software.
Conclusions
Posttranscriptional regulation of gene expression is controlled through a highly dynamic and combinatorial interaction of RBPs, microRNAs, and mRNA that forms complex ribonucleoprotein particles. Sequences and structures within a given mRNA species may interact with numerous regulatory proteins and microRNAs that function together to determine the fate of the transcript. Networks of transcripts may share regulatory sequences, such as the GRE, that allow for coordinated expression during cellular activation or development. Coordination of mRNA degradation by the GRE in mammalian cells depends on the CELF1 protein, but further work is needed to understand the mechanisms by which CELF1 mediates mRNA decay and how this process responds to environmental signals during cellular activation and differentiation. A better understanding of the molecular mechanisms through which GREs and CELF1 regulate mRNA decay and how this process is disrupted in disease states such as malignancy may provide new avenues for therapeutic modalities.
Acknowledgments
This work was supported by grant 1R01AI072068 from the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- •1.Mansfield KD, Keene JD. The ribonome: a dominant force in co-ordinating gene expression. Biol Cell. 2009;101(3):169–181. doi: 10.1042/BC20080055. This review discusses the post-transcriptional RNA operon theory of co-regulated gene expression, whereby the coordinated dynamics of RNA-binding proteins allows for the recombination and remodelling of the RNPs (ribonucleoproteins) to generate new combinations of functionally related proteins and keep the global mRNA environment in balance. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Keene JD. Biological clocks and the coordination theory of RNA operons and regulons. Cold Spring Harb Symp Quant Biol. 2007;72:157–165. doi: 10.1101/sqb.2007.72.013. [DOI] [PubMed] [Google Scholar]
- 3.Moroy T, Heyd F. The impact of alternative splicing in vivo: mouse models show the way. RNA. 2007;13(8):1155–1171. doi: 10.1261/rna.554607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee JE, Cooper TA. Pathogenic mechanisms of myotonic dystrophy. Biochem Soc Trans. 2009;37(Pt 6):1281–1286. doi: 10.1042/BST0371281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jin J, Wang GL, Timchenko L, Timchenko NA. GSK3beta and aging liver. Aging (Albany NY) 2009;1(6):582–585. doi: 10.18632/aging.100060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •6.Chekulaeva M, Filipowicz W. Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Curr Opin Cell Biol. 2009;21(3):452–460. doi: 10.1016/j.ceb.2009.04.009. This article reviews understanding how microRNAs function in animal cells and discusses controversies regarding different modes of microRNA function to affect both the translation and stability of mRNAs. [DOI] [PubMed] [Google Scholar]
- •7.Belver L, Papavasiliou FN, Ramiro AR. MicroRNA control of lymphocyte differentiation and function. Curr Opin Immunol. 2011 doi: 10.1016/j.coi.2011.02.001. This review outlines our current understanding of microRNA function in lymphocytes as it impacts expression of mRNA in the context of proper development. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Agami R. microRNAs, RNA binding proteins and cancer. Eur J Clin Invest. 2010;40(4):370–374. doi: 10.1111/j.1365-2362.2010.02279.x. [DOI] [PubMed] [Google Scholar]
- 9.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2011;11(9):597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 10.Kedde M, Agami R. Interplay between microRNAs and RNA-binding proteins determines developmental processes. Cell Cycle. 2008;7(7):899–903. doi: 10.4161/cc.7.7.5644. [DOI] [PubMed] [Google Scholar]
- 11.Bitel CL, Perrone-Bizzozero NI, Frederikse PH. HuB/C/D, nPTB, REST4, and miR-124 regulators of neuronal cell identity are also utilized in the lens. Mol Vis. 2011;16:2301–2316. [PMC free article] [PubMed] [Google Scholar]
- 12.Clark A, Dean J, Tudor C, Saklatvala J. Post-transcriptional gene regulation by MAP kinases via AU-rich elements. Front Biosci. 2009;14:847–871. doi: 10.2741/3282. [DOI] [PubMed] [Google Scholar]
- •13.Khabar KS. Post-transcriptional control during chronic inflammation and cancer: a focus on AU-rich elements. Cell Mol Life Sci. 2010;67(17):2937–2955. doi: 10.1007/s00018-010-0383-x. This review addresses the cellular and molecular mechanisms that are common between inflammation and cancer and that also govern ARE-mediated post-transcriptional control. It examines the role of the ARE-genes in inflammation and cancer and sequence characteristics of AU-rich elements and addresses the common signaling pathways in inflammation and cancer that regulate the ARE-mediated pathways and how their deregulations affect ARE-gene regulation and disease outcome. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ishimaru D, Zuraw L, Ramalingam S, Sengupta TK, Bandyopadhyay S, Reuben A, Fernandes DJ, Spicer EK. Mechanism of regulation of bcl-2 mRNA by nucleolin and A+U-rich element-binding factor 1 (AUF1) J Biol Chem. 2011;285(35):27182–27191. doi: 10.1074/jbc.M109.098830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stoecklin G, Colombi M, Raineri I, Leuenberger S, Mallaun M, Schmidlin M, Gross B, Lu M, Kitamura T, Moroni C. Functional cloning of BRF1, a regulator of ARE-dependent mRNA turnover. EMBO J. 2002;21(17):4709–4718. doi: 10.1093/emboj/cdf444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stoecklin G, Ming XF, Looser R, Moroni C. Somatic mRNA turnover mutants implicate tristetraprolin in the interleukin-3 mRNA degradation pathway. Mol Cell Biol. 2000;20(11):3753–3763. doi: 10.1128/mcb.20.11.3753-3763.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanduja S, Blanco FF, Dixon DA. The roles of TTP and BRF proteins in regulated mRNA decay. WIREs RNA. 2011;2(1):42–57. doi: 10.1002/wrna.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gherzi R, Lee KY, Briata P, Wegmuller D, Moroni C, Karin M, Chen CY. A KH domain RNA binding protein, KSRP, promotes ARE-directed mRNA turnover by recruiting the degradation machinery. Mol Cell. 2004;14(5):571–583. doi: 10.1016/j.molcel.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 19.Diaz-Moreno I, Hollingworth D, Kelly G, Martin S, Garcia-Mayoral M, Briata P, Gherzi R, Ramos A. Orientation of the central domains of KSRP and its implications for the interaction with the RNA targets. Nucleic Acids Res. 2011;38(15):5193–5205. doi: 10.1093/nar/gkq216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peng SS, Chen CY, Xu N, Shyu AB. RNA stabilization by the AU-rich element binding protein, HuR, an ELAV protein. EMBO J. 1998;17(12):3461–3470. doi: 10.1093/emboj/17.12.3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kurosu T, Ohga N, Hida Y, Maishi N, Akiyama K, Kakuguchi W, Kuroshima T, Kondo M, Akino T, Totsuka Y, Shindoh M, et al. HuR keeps an angiogenic switch on by stabilising mRNA of VEGF and COX-2 in tumour endothelium. Br J Cancer. 2011;104(5):819–829. doi: 10.1038/bjc.2011.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiao L, Wang JY. Posttranscriptional regulation of gene expression in epithelial cells by polyamines. Methods Mol Biol. 2011;720:67–79. doi: 10.1007/978-1-61779-034-8_4. [DOI] [PubMed] [Google Scholar]
- 23.Hambardzumyan D, Sergent-Tanguy S, Thinard R, Bonnamain V, Masip M, Fabre A, Boudin H, Neveu I, Naveilhan P. AUF1 and Hu proteins in the developing rat brain: implication in the proliferation and differentiation of neural progenitors. J Neurosci Res. 2009;87(6):1296–1309. doi: 10.1002/jnr.21957. [DOI] [PubMed] [Google Scholar]
- 24.Hasegawa H, Kakuguchi W, Kuroshima T, Kitamura T, Tanaka S, Kitagawa Y, Totsuka Y, Shindoh M, Higashino F. HuR is exported to the cytoplasm in oral cancer cells in a different manner from that of normal cells. Br J Cancer. 2009;100(12):1943–1948. doi: 10.1038/sj.bjc.6605084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •25.Papadaki O, Milatos S, Grammenoudi S, Mukherjee N, Keene JD, Kontoyiannis DL. Control of thymic T cell maturation, deletion and egress by the RNA-binding protein HuR. J Immunol. 2009;182(11):6779–6788. doi: 10.4049/jimmunol.0900377. The authors assess HuR’s role in the staged progression of thymic T cell differentiation by means of its genetic ablation. Mice with an early deletion of HuR in thymocytes possess enlarged thymi but display a substantial loss of peripheral T cells. [DOI] [PubMed] [Google Scholar]
- 26.Ogilvie RL, Abelson M, Hau HH, Vlasova I, Blackshear PJ, Bohjanen PR. Tristetraprolin down-regulates IL-2 gene expression through AU-rich element-mediated mRNA decay. J Immunol. 2005;174(2):953–961. doi: 10.4049/jimmunol.174.2.953. [DOI] [PubMed] [Google Scholar]
- 27.Ogilvie RL, Sternjohn JR, Rattenbacher B, Vlasova IA, Williams DA, Hau HH, Blackshear PJ, Bohjanen PR. Tristetraprolin mediates interferon-gamma mRNA decay. J Biol Chem. 2009;284(17):11216–11223. doi: 10.1074/jbc.M901229200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lykke-Andersen J, Wagner E. Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains in the proteins TTP and BRF-1. Genes Dev. 2005;19(3):351–361. doi: 10.1101/gad.1282305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hau HH, Walsh RJ, Ogilvie RL, Williams DA, Reilly CS, Bohjanen PR. Tristetraprolin recruits functional mRNA decay complexes to ARE sequences. J Cell Biochem. 2007;100(6):1477–1492. doi: 10.1002/jcb.21130. [DOI] [PubMed] [Google Scholar]
- 30.Bakheet T, Williams BR, Khabar KS. ARED 3.0: the large and diverse AU-rich transcriptome. Nucleic Acids Res. 2006;34(Database issue):D111–114. doi: 10.1093/nar/gkj052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •31.Halees AS, El-Badrawi R, Khabar KS. ARED Organism: expansion of ARED reveals AU-rich element cluster variations between human and mouse. Nucleic Acids Res. 2008;36(Database issue):D137–140. doi: 10.1093/nar/gkm959. The authors performed quantitative assessment of ARE conservation in human, mouse and rat transcripts. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gruber AR, Fallmann J, Kratochvill F, Kovarik P, Hofacker IL. AREsite: a database for the comprehensive investigation of AU-rich elements. Nucleic Acids Res. 39(Database issue):D66–69. doi: 10.1093/nar/gkq990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raghavan A, Bohjanen PR. Microarray-based analyses of mRNA decay in the regulation of mammalian gene expression. Brief Funct Genomic Proteomic. 2004;3(2):112–124. doi: 10.1093/bfgp/3.2.112. [DOI] [PubMed] [Google Scholar]
- 34.Raghavan A, Dhalla M, Bakheet T, Ogilvie RL, Vlasova IA, Khabar KS, Williams BR, Bohjanen PR. Patterns of coordinate down-regulation of ARE-containing transcripts following immune cell activation. Genomics. 2004;84(6):1002–1013. doi: 10.1016/j.ygeno.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 35.Al-Souhibani N, Al-Ahmadi W, Hesketh JE, Blackshear PJ, Khabar KS. The RNA-binding zinc-finger protein tristetraprolin regulates AU-rich mRNAs involved in breast cancer-related processes. Oncogene. 2011;29(29):4205–4215. doi: 10.1038/onc.2010.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mukherjee N, Lager PJ, Friedersdorf MB, Thompson MA, Keene JD. Coordinated posttranscriptional mRNA population dynamics during T-cell activation. Mol Syst Biol. 2009;5:288. doi: 10.1038/msb.2009.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••37.Vlasova IA, Tahoe NM, Fan D, Larsson O, Rattenbacher B, Sternjohn JR, Vasdewani J, Karypis G, Reilly CS, Bitterman PB, Bohjanen PR. Conserved GU-rich elements mediate mRNA decay by binding to CUG-binding protein 1. Mol Cell. 2008;29(2):263–270. doi: 10.1016/j.molcel.2007.11.024. The authors identified GU-rich element consensus sequence UGUUUGUUUGU using computational algorithms in the 3′ UTR of transcripts that exhibited rapid decay in primary human T cells. Authors demonstrate that the GRE mediates coordinated mRNA decay by binding to CELF1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Charlet BN, Savkur RS, Singh G, Philips AV, Grice EA, Cooper TA. Loss of the muscle-specific chloride channel in type 1 myotonic dystrophy due to misregulated alternative splicing. Mol Cell. 2002;10(1):45–53. doi: 10.1016/s1097-2765(02)00572-5. [DOI] [PubMed] [Google Scholar]
- 39.Sureau A, Sauliere J, Expert-Bezancon A, Marie J. CELF and PTB proteins modulate the inclusion of the beta-tropomyosin exon 6B during myogenic differentiation. Exp Cell Res. 2011;317(1):94–106. doi: 10.1016/j.yexcr.2010.09.010. [DOI] [PubMed] [Google Scholar]
- 40.Barron VA, Zhu H, Hinman MN, Ladd AN, Lou H. The neurofibromatosis type I pre-mRNA is a novel target of CELF protein-mediated splicing regulation. Nucleic Acids Res. 2011;38(1):253–264. doi: 10.1093/nar/gkp766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iakova P, Wang GL, Timchenko L, Michalak M, Pereira-Smith OM, Smith JR, Timchenko NA. Competition of CUGBP1 and calreticulin for the regulation of p21 translation determines cell fate. Embo J. 2004;23(2):406–417. doi: 10.1038/sj.emboj.7600052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moraes KC, Wilusz CJ, Wilusz J. CUG-BP binds to RNA substrates and recruits PARN deadenylase. Rna. 2006;12(6):1084–1091. doi: 10.1261/rna.59606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••43.Rattenbacher B, Beisang D, Wiesner DL, Jeschke JC, von Hohenberg M, St Louis-Vlasova IA, Bohjanen PR. Analysis of CUGBP1 targets identifies GU-repeat sequences that mediate rapid mRNA decay. Mol Cell Biol. 2010;30(16):3970–3980. doi: 10.1128/MCB.00624-10. The authors found the consensus GRE sequence and a GU-repeat sequence were both highly enriched in the 3′ UTRs of CELF1 targets identified using RNA-IP. Based on these results, they redefined the GRE to include this GU-repeat sequence. CELF1 coordinately regulates the mRNA decay of a network of transcripts involved in cell growth, cell motility, and apoptosis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marquis J, Paillard L, Audic Y, Cosson B, Danos O, Le Bec C, Osborne HB. CUG-BP1/CELF1 requires UGU-rich sequences for high-affinity binding. Biochem J. 2006;400(2):291–301. doi: 10.1042/BJ20060490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takahashi N, Sasagawa N, Suzuki K, Ishiura S. The CUG-binding protein binds specifically to UG dinucleotide repeats in a yeast three-hybrid system. Biochem Biophys Res Commun. 2000;277(2):518–523. doi: 10.1006/bbrc.2000.3694. [DOI] [PubMed] [Google Scholar]
- 46.Mori D, Sasagawa N, Kino Y, Ishiura S. Quantitative analysis of CUG-BP1 binding to RNA repeats. J Biochem. 2008;143(3):377–383. doi: 10.1093/jb/mvm230. [DOI] [PubMed] [Google Scholar]
- ••47.Lee JE, Lee JY, Wilusz J, Tian B, Wilusz CJ. Systematic analysis of cis-elements in unstable mRNAs demonstrates that CUGBP1 is a key regulator of mRNA decay in muscle cells. PLoS One. 2010;5(6):e11201. doi: 10.1371/journal.pone.0011201. The authors found that GU-rich and AU-rich elements are over-represented in the 3′ UTRs of short-lived mRNAs expressed in mouse myoblasts. RNA immunoprecipitation followed by microarray assay identified CELF1, HuR and PUM-associated transcripts. Finally, they showed that GRE-containing transcripts were stabilized in cells depleted of CELF1, consistent with the role of CELF1 as a destabilizing factor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••48.Graindorge A, Le Tonqueze O, Thuret R, Pollet N, Osborne HB, Audic Y. Identification of CUG-BP1/EDEN-BP target mRNAs in Xenopus tropicalis. Nucleic Acids Res. 2008;36(6):1861–1870. doi: 10.1093/nar/gkn031. The authors identified a number EDEN-BP target mRNAs complexes in Xenopus tropicalis egg extracts. GRE-containing transcripts were specifically targeted for EDEN-BP-dependent deadenylation after fertilization. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Le Tonqueze O, Gschloessl B, Namanda-Vanderbeken A, Legagneux V, Paillard L, Audic Y. Chromosome wide analysis of CUGBP1 binding sites identifies the tetraspanin CD9 mRNA as a target for CUGBP1-mediated down-regulation. Biochem Biophys Res Commun. 2010;394(4):884–889. doi: 10.1016/j.bbrc.2010.03.020. [DOI] [PubMed] [Google Scholar]
- 50.Ezzeddine N, Paillard L, Capri M, Maniey D, Bassez T, Ait-Ahmed O, Osborne HB. EDEN-dependent translational repression of maternal mRNAs is conserved between Xenopus and Drosophila. Proc Natl Acad Sci U S A. 2002;99(1):257–262. doi: 10.1073/pnas.012555499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Osborne HB, Gautier-Courteille C, Graindorge A, Barreau C, Audic Y, Thuret R, Pollet N, Paillard L. Post-transcriptional regulation in Xenopus embryos: role and targets of EDEN-BP. Biochem Soc Trans. 2005;33(Pt 6):1541–1543. doi: 10.1042/BST0331541. [DOI] [PubMed] [Google Scholar]
- 52.Paillard L, Omilli F, Legagneux V, Bassez T, Maniey D, Osborne HB. EDEN and EDEN-BP, a cis element and an associated factor that mediate sequence-specific mRNA deadenylation in Xenopus embryos. EMBO J. 1998;17(1):278–287. doi: 10.1093/emboj/17.1.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Paillard L, Legagneux V, Beverley Osborne H. A functional deadenylation assay identifies human CUG-BP as a deadenylation factor. Biol Cell. 2003;95(2):107–113. doi: 10.1016/s0248-4900(03)00010-8. [DOI] [PubMed] [Google Scholar]
- 54.Kim-Ha J, Kerr K, Macdonald PM. Translational regulation of oskar mRNA by bruno, an ovarian RNA-binding protein, is essential. Cell. 1995;81(3):403–412. doi: 10.1016/0092-8674(95)90393-3. [DOI] [PubMed] [Google Scholar]
- 55.Delaunay J, Le Mee G, Ezzeddine N, Labesse G, Terzian C, Capri M, Ait-Ahmed O. The Drosophila Bruno paralogue Bru-3 specifically binds the EDEN translational repression element. Nucleic Acids Res. 2004;32(10):3070–3082. doi: 10.1093/nar/gkh627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Horb LD, Horb ME. BrunoL1 regulates endoderm proliferation through translational enhancement of cyclin A2 mRNA. Dev Biol. 2011;345(2):156–169. doi: 10.1016/j.ydbio.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Suzuki H, Jin Y, Otani H, Yasuda K, Inoue K. Regulation of alternative splicing of alpha-actinin transcript by Bruno-like proteins. Genes Cells. 2002;7(2):133–141. doi: 10.1046/j.1356-9597.2001.00506.x. [DOI] [PubMed] [Google Scholar]
- •58.Teplova M, Song J, Gaw HY, Teplov A, Patel DJ. Structural insights into RNA recognition by the alternate-splicing regulator CUG-binding protein 1. Structure. 2010;18(10):1364–1377. doi: 10.1016/j.str.2010.06.018. Authors described crystal structures of CELF1 RRM1 and tandem RRM1/2 domains bound to RNAs containing tandem UGU(U/G) elements. Both RRMs use similar principles to target UGU(U/G) elements. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •59.Tsuda K, Kuwasako K, Takahashi M, Someya T, Inoue M, Terada T, Kobayashi N, Shirouzu M, Kigawa T, Tanaka A, Sugano S, et al. Structural basis for the sequence-specific RNA-recognition mechanism of human CUG-BP1 RRM3. Nucleic Acids Res. 2009;37(15):5151–5166. doi: 10.1093/nar/gkp546. The solution structure of the CELF1 RRM3 in the complex with (UG) RNA, and discovered that the UGU trinucleotide is specifically recognized by RRM3 to discriminate the short RNA segment from other sequences. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu J, Li C, Zhao S, Mao B. Differential expression of the Brunol/CELF family genes during Xenopus laevis early development. Int J Dev Biol. 2011;54(1):209–214. doi: 10.1387/ijdb.082685jw. [DOI] [PubMed] [Google Scholar]
- 61.Paillard L, Legagneux V, Maniey D, Osborne HB. c-Jun ARE targets mRNA deadenylation by an EDEN-BP (embryo deadenylation element-binding protein)-dependent pathway. J Biol Chem. 2002;277(5):3232–3235. doi: 10.1074/jbc.M109362200. [DOI] [PubMed] [Google Scholar]
- •62.Gallo JM, Spickett C. The role of CELF proteins in neurological disorders. RNA Biol. 2011;7(4):474–479. doi: 10.4161/rna.7.4.12345. The role of CELF1 and functionally related muscleblind-like 1 protein in a number of neurological conditions. The involvement of CELF proteins suggest that individual pathogenic pathways in a number of neurological conditions overlap at the level of RNA processing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ward AJ, Rimer M, Killian JM, Dowling JJ, Cooper TA. CUGBP1 overexpression in mouse skeletal muscle reproduces features of myotonic dystrophy type 1. Hum Mol Genet. 2011;19(18):3614–3622. doi: 10.1093/hmg/ddq277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ho TH, Bundman D, Armstrong DL, Cooper TA. Transgenic mice expressing CUG-BP1 reproduce splicing mis-regulation observed in myotonic dystrophy. Hum Mol Genet. 2005;14(11):1539–1547. doi: 10.1093/hmg/ddi162. [DOI] [PubMed] [Google Scholar]
- 65.Kress C, Gautier-Courteille C, Osborne HB, Babinet C, Paillard L. Inactivation of CUG-BP1/CELF1 causes growth, viability, and spermatogenesis defects in mice. Mol Cell Biol. 2007;27(3):1146–1157. doi: 10.1128/MCB.01009-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••66.Daughters RS, Tuttle DL, Gao W, Ikeda Y, Moseley ML, Ebner TJ, Swanson MS, Ranum LP. RNA gain-of-function in spinocerebellar ataxia type 8. PLoS Genet. 2009;5(8):e1000600. doi: 10.1371/journal.pgen.1000600. Data demonstrate novel RNA splicing targets of CELF1 in spinocerebellar ataxia type 8 brain cells. CUG(expanded) transcripts dysregulate MBNL/CELF pathways in the brain and provide mechanistic insight into the CNS effects of other CUG-expansion disorders. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Raghavan A, Ogilvie RL, Reilly C, Abelson ML, Raghavan S, Vasdewani J, Krathwohl M, Bohjanen PR. Genome-wide analysis of mRNA decay in resting and activated primary human T lymphocytes. Nucleic Acids Res. 2002;30(24):5529–5538. doi: 10.1093/nar/gkf682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vlasova IA, Bohjanen PR. Posttranscriptional regulation of gene networks by GU-rich elements and CELF proteins. RNA Biol. 2008;5(4):201–207. doi: 10.4161/rna.7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cooper TA, Wan L, Dreyfuss G. RNA and disease. Cell. 2009;136(4):777–793. doi: 10.1016/j.cell.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •70.Starr TK, Allaei R, Silverstein KA, Staggs RA, Sarver AL, Bergemann TL, Gupta M, O’Sullivan MG, Matise I, Dupuy AJ, Collier LS, et al. A transposon-based genetic screen in mice identifies genes altered in colorectal cancer. Science. 2009;323(5922):1747–1750. doi: 10.1126/science.1163040. The authors used a transposon-based genetic screen in mice to identify candidate cancer-causing genes for colorectal cancer. Mice harboring mutagenic Sleeping Beauty transposons were crossed with mice expressing sleeping beauty transposase in gastrointestinal tract epithelium. CELF1 was identified as a candidate which if mutated and/or dysregulated was likely to drive tumorigenesis. [DOI] [PMC free article] [PubMed] [Google Scholar]


