Abstract
The retinoblastoma protein, pRb, is a key regulator of cell proliferation, differentiation, apoptosis, as well as checkpoint and stress responses. The function of Rb is often inactivated in many types of cancers, a feature that can potentially be used to target this specific subset of cancers. However little is known about how the loss of Rb function can be exploited in cancer therapies. In this review, we overview the functions of Rb, and discuss a genetic screen that led to the finding that inactivation of TSC2 and Rb induces synergistic cell death in both Drosophila developing tissues and human cancer cells. The mechanisms for synergistic cell death involve the accumulation of cellular stress, suggesting that inactivation of TSC2 and chemotherapeutic agents that result in induction of cellular stress can potentially be combined to treat cancers harboring inactivated Rb.
Keywords: Rb, E2F, TSC2, mTOR, synthetic lethality, cellular stress, ROS
Introduction
Isolation of the Rb gene [1, 2], encoding the first known tumor suppressor, was based on the observation that a subset of retinoblastomas exhibit deletions in the q14 region of chromosome 13 [3]. Some osteosarcomas, including those in patients without retinoblastoma, were also found to have alterations at this locus, which implicated Rb in the pathogenesis of cancer in general [4]. Rb was subsequently found to be lost at high frequency in small cell lung cancers (SCLCs) and, to a lesser degree, in bladder cancers but was actually present in the majority of cell lines derived from colon and breast carcinomas as well as melanomas [5]. Although these initial results showed that actual Rb deletion may not be involved in all cancers, the pRb gene product was nevertheless recognized as a critical regulator of cell proliferation and transformation. In the following decades of research, pRb inactivation has become established as a critical event for tumorigenesis [6].
Mammalian Rb and E2F family proteins
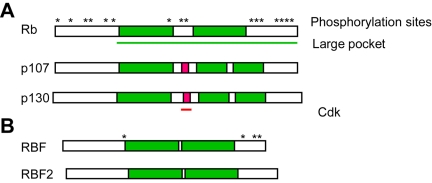
pRb, p107, and p130 are now known to comprise a family of “pocket proteins” that share structural features (Figure 1). The C-terminal domains of these proteins consist of conserved, bipartite hydrophobic pockets that are responsible for interaction with most endogenous binding partners and viral oncoproteins [7]. A spacer within the pockets is not as well conserved, and these regions contain binding sites for cyclin/ cdk complexes in p107 and p130 but not pRb [8, 9]. The activity of pocket proteins is regulated by phosphorylation, and pRb contains numerous sites that are targeted by Cyclin D/ Cdk4, Cyclin E/Cdk2, and Cyclin A/Cdk2 during the G1and S phases of the cell cycle [10-13]. In general, phoshorylation of pRb correlates with cell cycle progression and is reversed by phos-phatases during mitosis [14]. pRb is a stable protein that is present in both proliferating and non-proliferating cells but expression of the other pocket proteins is dynamic: p130 is most abundant in quiescent, differentiated cells and in early G1 [15] and p107 expression increases in mid to late G1 [15].
Figure 1.
Pocket protein structure. A. The general structure of mammalian Rb family proteins is conserved. The C-terminal pocket domains are divided by variable spacer regions. The spacers of p107 and p130 but not pRb harbor Cdk-binding sites. Asterisks denote known kinase target sites. B. Drosophila pocket proteins reflect the structure of mammalian counterparts.
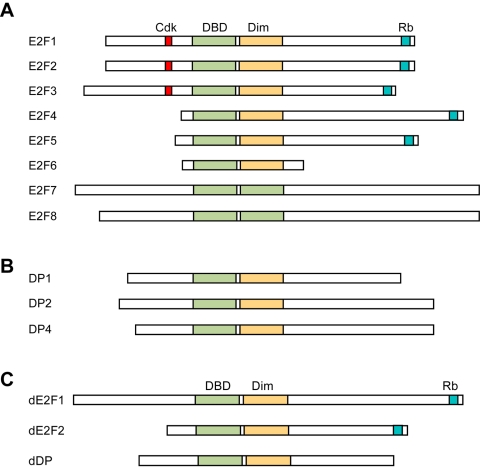
pRb has established functions as a tumor suppressor and cell cycle regulator, but also has important roles in differentiation and apoptosis. These functions are mediated by its interaction with different proteins, more than 100 of which have been identified [16], but the best-studied of these is the E2F family of transcription factors. There at least eight distinct E2F isoforms in mammals (Figure 2). E2F function generally requires dimerization with DP family proteins and DNA binding [17-19], although two atypical E2F family members, E2f7 and E2f8, function as transcriptional repressors independent of DP binding. Interestingly, a critical target of these atypical E2Fs is E2f1 during the S and G2 phases of the cell cycle [20].
Figure 2.
E2F and DP family structures. A. All E2F family members have a DNA binding domain (DBD). Activator-type E2Fs (E2F1-3) possess Cdk-binding sites near their N-termini, and both activator- and repressor-type E2Fs harbor C-terminal Rb family binding domains. Atypical E2Fs (E2F6-8) do not bind Rb family proteins, and all E2Fs except E2F7-8 have a dimerization domain (Dim) that mediates interaction with the DP family. B. DP family proteins all have domains for both DNA binding and E2F dimerization. C. Drosophila E2Fs consist of one activator, dE2F1, and one repressor, dE2F2. Both proteins bind DNA, heterodimerize with the lone dDP, and interact with RBF but only dE2F2 binds RBF2.
Binding of pRb to E2Fs inhibits the expression of target genes, notably those involved in cell cycle (e.g. Cyclin E1 and Cyclin A1) and DNA replication [21]. pRb is the main pocket protein that binds activator-type E2Fs (i.e. E2f1, E2f2, and E2f3), but pRb, p107, and p130 all interact with repressor-type E2Fs (E2f4 and E2f5) [22]. E2f6 and E2f7-8 are atypical E2Fs that do not interact with pocket proteins. The interactions of repressor-type E2Fs with pocket proteins are thought to be important for cell cycle inhibition during early G1, and activator-type E2Fs are observed to displace the repressors as cells approach the G1/S checkpoint [23].
Drosophila Rb and E2F
The Rb and E2F families are evolutionarily conserved. For example the fruitfly Drosophila melanogaster has two Rb isoforms, Rbf and Rbf2, and two E2Fs, dE2f1 and dE2f2 [24-28]. dE2f1 has been shown to functions primarily as a transcriptional activator [29, 30], while dE2f2 is thought to act as a repressor [30]. Additionally, Rbf can bind to either dE2f1 or dE2f2 [30] while Rbf2 has only been found to interact with dE2f2 [26]. Thus flies present a simpler yet well-conserved model system in which Rb/E2f biology can be studied using a genetic approach (see below). Additionally, there is functional conservation between fly and mammalian pocket proteins, E2Fs and cell cycle regulators: Rbf activity during G1 represses Drosophila E2F target genes important for DNA replication and cell cycle progression [31].
pRb at the center of cell cycle regulation
Early experiments using retroviral-mediated infection of tumor cells with exogenous Rb showed reductions of cell proliferation, growth in soft agar, and tumorigenic potential in mice [32]. Meanwhile pRb was found to associate with the large T antigen (TAg) of SV40 [33] and other polyoma viruses [34], the E7 protein of HPV [35], and the adenoviral E1A protein [36], showing that inhibition of pRb activity is the likely mechanism of transformation by these DNA tumor viruses. Numerous other studies showed that TAg competes with E2F and other endogenous proteins to bind pRb [37-39] and that pRb phosphorylation during late G1 [40] permits entrance into S-phase [41]. These studies established a foundation for much of our current understanding of pRb function and the other pocket proteins, both of which were identified on the basis of the conserved ability to bind the SV40 TAg and adenoviral E1A proteins [42, 43].
pRb function cooperates with cyclin-dependent kinase inhibitors (CKIs). p16Ink4a is one of two critical tumor suppressors encoded by the Cdkn2a gene, the other being p14Arf. p16 was originally identified based on its ability to physically interact with and inhibit Cdk4, thus promoting the activity of pRb [44], and the inhibitory function of p16 on the cell cycle was subsequently demonstrated to require Rb [45]. The p53-inducible gene Cdkn1a encodes another CKI, p21Cip1/Waf1, which binds to and inhibits the kinase activity of Cdk2. p21 thus also protects pRb from hyperphosphorylation by G1 cyclins [46, 47]. Additionally the Cdkn1b gene product p27Kip1, which is functionally and structurally related to p21, was shown to inhibit both Cdk4 and Cdk2 thereby promoting pRb activity [48, 49]. Importantly, the Rb protein family is essential for cell cycle arrest in response to a variety of signals including CKI activity [50].
Transcriptional repression by pRb
More recently pRb has been found to regulate transcriptional repression and cell cycle progression, at least in part, by affecting DNA methylation [51] and histone modification [52]. Recruitment of LXCXE motif-containing histone deacetylases (HDACs) by pRb to SWI/SNF complexes repress E2f1-mediated Cyclin E expression during G1, whereas HDACs are not important for the ability of these complexes to repress Cyclin A during S-phase [53]. Additional studies showed that histone acetylation is important for E2F target gene expression [23], and histone acetyltransferases (HATs) are recruited to target genes by activator E2F complexes at the G1/S transition [54]. These experiments suggest a model for pRb function that involves dynamic chromatin-assisted repression of specific E2F target genes.
Role of pRb in maintaining genome stability
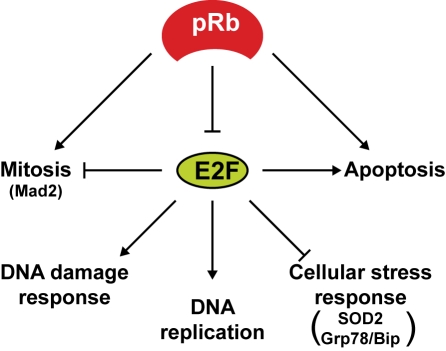
The ability of pRb to repress the expression of E2F target genes has implications for the cell cycle beyond DNA replication during S-phase (Figure 3). Various microarray analyses have identified E2F target genes that are known to be involved in DNA damage response, the spindle checkpoint, and mitosis [55, 56] suggesting that one of the key functions of pRb is to maintain genome fidelity prior to cell division. Indeed Rb inactivation in mouse or human fibroblasts resulted in E2f1-induced expression of Mad2, a checkpoint regulator that senses improper microtubule attachment to kinetochores during mitosis. This accumulation of Mad2 leads to mitotic defects, ultimately resulting in abnormal ploidy [57]. More recent experiments in Drosophila melanogaster showed improper chromatin condensation in the absence of the Rb gene ortholog Rbf, owing to reduced recruitment of the Condensin II complex component dCAP-D3 [58]. This group subsequently linked abnormal Condensin II function to chromosomal instability in human retinal cells [59]. Additionally, Condensin II dysfunction was independently implicated in experiments using cells derived from mice that were engineered to express an Rb knock-in allele that lacks its LXCXE-interacting motif. Importantly these Rb alleles were observed to enhance tumorigenesis in Trp53-/-mice, and lymphoma cells from these animals are aneuploid [60]. Another related study showed that Rb-/-, p107-/-, p130-/- triple-knockout MEFs accumulate double-strand breaks and undergo G2 arrest upon mitogen deprivation. Addition of mitogens allows the G2-arrested triple knockout cells to resume proliferation without fully repaired DNA, leading to aneuploidy [61]. These reports show that pRb is a critical regulator of different phase of the cell cycle, and its functions outside of the G1/S transition are also important for tumor suppression.
Figure 3.
E2F functions. The Rb and E2F families work together to ensure genome fidelity during multiple phases of the cell cycle. While some E2F target genes are directly involved in DNA replication, others are important for the orderly progression through mitosis. In the absence of pRb function E2F hyperactivity can induce cell death directly by affecting apoptotic target gene expression, or indirectly by promoting the response to DNA damage and inhibiting responses to oxidative and ER stress. Several checkpoint and stress response targets of E2F discussed in this review are indicated in parentheses.
Rb and cell death
Rb also has critical roles in a variety of cellular processes outside of the cell cycle. pRb has long been known to repress E2F-mediated cell death, as suggested by experiments using E2f1 knockout mice which were shown to develop tumors in a variety of tissues within 1-2 years [62]. A litany of subsequent experiments have demonstrated the ability of E2f1 to induce apoptosis by both p53-dependent and -independent mechanisms in different model systems [63]. Importantly p53 stabilization is induced by E2f1-mediated expression of p14Arf, which functions to inhibit the polyubiquitination and degradation of p53 by Mdm2 [64]. p53 accumulation induces the transcription of an array of pro-apoptotic genes [65]. Additionally, E2f1 has been shown to have roles in DNA damage response by promoting p53 phosphorylation and activation independent of p14Arf [66]. E2f1 can directly affect expression of the transcription factor p73 [67-69], which itself controls apoptotic gene targets shared by but not requiring its paralog p53 [70]. E2f1 has also been shown to directly induce the transcription of pro-apoptotic genes independent of the p53 family such as Apaf-1 [71], thereby promoting apical caspase activity via apoptosome assembly, and several effector caspases including Caspase 3 and Caspase 7 [72]. Finally, E2f1 affects target genes that antagonize mitochondrial membrane integrity such as Bad, Bid and Bak [73]. Thus E2F1 has multiple roles not only in proliferation and transformation, but also apoptosis in mammalian cells.
Interesting and unexpected functions have been attributed to pRb by recent studies, including the activation of transcription during apoptosis [74] as well as differentiation [75]. The ability of pRb to promote either arrest or apoptosis seems to be context dependent, with apoptosis being favored in proliferating cells. In proliferating cells treated with the DNA-damaging agent doxorubicin, hyperphosphorylated pRb was found to be in a complex with E2f1 and the histone acetyltransferase P/CAF bound to the transcriptionally-active Caspase 7 and p73 promoters. Intact Rb was shown to be important for this DNA damage-induced apoptotic phenotype both in vitro and in vivo [74]. While the ability of pRb to promote transcription of apoptotic target genes is a novel finding, conserved pRb and E2F functions during cell death are known to depend on developmental cues. Studies in Drosophila show that the activator E2F, dE2f1, can either promote or antagonize apoptosis through the Smac/Diablo ortholog Hid depending on both tissue type and the position of cells within tissues during fly development [76-79]. Though experiments in mammalian cells indicate that Smac/Diablo expression is generally induced by activator E2Fs [80, 81], E2f1-/-, E2f2-/-, E2f3-/-triple-knockout (E2f1-3 TKO) retinas still exhibit apoptosis during development due to decreased expression of the p53 deacetylase Sirt1 [81]. Thus the activator E2Fs may also have an important cell survival role in retinal progenitor cells. Another study found similar results using independently-derived, conditional E2f1-3 TKO mice. These experiments showed that progenitor cells, but not differentiated cells, in the intestine can proliferate in the absence of activator E2Fs but accumulate DNA damage and undergo apoptosis, although in this case the phenotype was observed to be p53-independent [82]. These experiments indicate that activator E2Fs are not required for the proliferation of some undifferentiated cell types, but rather are important for survival. Thus cellular context is a critical determinant of whether activator E2Fs have pro- or anti-apoptotic functions, a distinction that has important consequences for the role of Rb deficiency in tumorigenesis.
Rb inactivation in cancers
In addition to their well-documented roles in retinoblastoma, inherited Rb mutations are thought to cause a number of different tumors. Osteosarcoma, for example, is commonly observed in retinoblastoma survivors [83]. Susceptibility to small cell lung carcinoma (SCLC) has also been attributed to germline Rb mutation and retinoblastoma survivors are also at increased risk for this and several other cancers [84]. The frequency of Rb loss in SCLC is extremely high (>80%), but is much less common in non-small cell lung cancer (NSCLC) [85]. Other cancers display somatic loss of Rb less frequently than SCLC but still in significant proportions, including bladder, espophageal, liver, brain, breast, and prostate cancers, as well as chronic myelogenous leukemia (CML) but less so in other leukemias. Rb loss is perhaps involved in disease progression in some cancers rather than initiation, which has been suggested for retinoblastoma and SCLC [86], as the majority of these diseases are not associated with inheritance but rather have lost Rb sporadically and at lower frequency. Indeed a recent study found that Rb deletion is associated with more advanced forms of prostate cancer (CaP) due to E2f1-dependent expression of Androgen receptor (AR) [87].
Importantly the deletion of Rb is not necessary for tumorigenesis in many cancers, as pRb function is affected by other genetic alterations. Studies have shown activator E2Fs to be overexpressed or amplified in some tumor cells, such as E2f3 in CaP [88] and bladder cancer [89], which likely overwhelms the regulatory activity of pRb. Additionally, functional inactivation of pRb due to loss of cooperating tumor suppressors such as p16Ink4a is also common. The molecular alterations in SCLC vs. NSCLC illustrate the cooperation between these genes, as the great majority of NSCLC tumors display either p16 loss or overexpression of Cyclin D [85]. Further, the inverse correlation of p16 and Rb loss is observed in a wide range of other tumors and suggests that inactivation of the pRb pathway is nearly universal in cancer [90]. However this functional inactivation is unlikely to simply phenocopy the effect of Rb loss. For example, cooperating mutations in Trp53 are more frequent in SCLC than in NSCLC [91]. It is possible that cell types differ in the relative expression of pRb and p16, and these differences could affect how loss of function mutations in either tumor suppressor impact tumorigenesis. Consistent with this, it was shown that loss of Rb but not p16 results in the accumulation of DNA double strand breaks induced by deregulated E2f1 [92]. In addition, hyperphosphorylated Rb may have a function regulating apoptosis depending on the cellular context [74]. Nevertheless as loss of p53 activity is also generally required for tumorigenesis, functional pRb inactivation must also cooperate with other alterations that affect p53 such as concomitant loss of p14Arf due to deficiency at the Ink4a locus.
The cooperation of Rb and p53 in tumor suppression is observed in mouse models of cancer. Although Rb-/-mice die during embryogenesis [93-95], Rb+/- animals develop neuroendocrine tumors displaying loss-of-heterozygosity with high penetrance and have shortened life-spans [96], which prevents the assessment of Rb involvement in other cancers. However Rb+/-, Trp53+/- mice exhibit a slightly broader tumor spectrum and further shortened lifespans. Although Rb+/- mice do not develop retinoblastoma, the incidence of retinal dysplasia in Rb+/-, Trp53+/- mice was observed to be dramatically increased [97]. Interestingly subsequent studies showed that Rb mutation cooperates with loss of either p107 or p130 in the development of retinoblastoma in mice [98]. A multitude of later studies have shown that conditional deletion of Rb in a range of tissues leads to apoptotic as well as hyperplastic phenotypes, but that concomitant Trp53 deletion often results in actual tumor growth [99].
Rb-/-, E2f1-/-double-knockout mice exhibit significant suppression of apoptosis in multiple tissues and survive somewhat longer than Rb-/-alone [100]. Additionally E2f1-/- extends life-span and reduces tumor incidence in Rb+/-mice and these animals exhibit a phenotype similar to E2f1-/- on its own [101]. These studies demonstrated that E2f1 is important for tumorigenesis downstream of Rb loss, most likely due to expression of target genes involved in cell proliferation because E2f1-/-mice also exhibit defective apoptosis in different tissues and develop tumors themselves [62]. Therefore inhibition of E2f1-induced cell death is a critical function of pRb during mouse development, and loss of this function is correlated with tumorigenesis when cooperating mutations such as Trp53 abrogate cell death. While many apoptotic E2f1 target genes have been established, an understanding of how intersecting signaling pathways affect E2F-mediated apoptosis could provide novel therapeutic targets for cancers in which pRb is inactivated.
Strategies for targeting Rb-deficient cancers
Rb status as a marker of drug sensitivity
Given the range of mechanisms by which the pRb pathway is deregulated in different cancers, e.g. Rb locus alteration/deletion, p16 loss or Cyclin D amplification, and activator E2F overexpression, it is likely that exploiting the dependence of tumors on different modes of pRb inactivation will require a multitude of therapeutic options. One such strategy is to use Rb status as a prognostic indicator or predictor of therapy outcomes. In addition to the aforementioned association of advanced CaP with loss of Rb [87], reduced pRb expression was correlated with more aggressive forms of breast carcinoma, including p53- and ER-negative tumors [102]. Experiments using p16-negative breast cancer cells showed that knockdown of Rb, while promoting the proliferation of cells in culture and xenografted tumors, confers sensitivity to DNA-damage induced by cisplatin or ionizing radiation (IR). However Rb knockdown was seen to desensitize cells to Tamoxifen, and a gene expression signature used to indicate disrupted pRb function in a cohort of ER-positive tumor samples correlated with recurrence after Tamoxifen treatment [103]. Later studies also found that loss of pRb expression correlated with patient response to adjuvant antimetabolite treatment with 5-fluorouracil (5-FU) and methotrexate, and knockdown of Rb breast cancer cells enhanced their sensitivity to these same drugs [104]. Therefore deciphering Rb status or pRb pathway disruption is potentially useful in predicting responses of breast cancer patients to different forms of therapy. However given that actual loss of Rb expression is not frequently observed in these tumors more sophisticated methods for detecting reduced pRb activity are likely to be required, some of which have already been approved for use in the clinical setting as diagnostic tools [105].
Targeting Rb loss in cancers by synthetic lethality
A concept that could be useful in developing novel therapies for Rb-deficient cancers is synthetic lethality, which generally occurs when tumor cells with particular genetic alterations become reliant on the functions of distinct, cooperating genes that confer protection against cell death. Identification of such genes could provide targets whose selective inactivation would induce death specifically in cells that harbor the relevant alteration but not in normal cells. This kind of intervention would theoretically have less severe side effects than cytotoxic agents that are currently employed [106]. However, identifying relevant targets is a major challenge in developing this therapeutic strategy.
Identification of Rb synthetic lethal mutations through genetic screens
A method for identifying potential genes whose inactivation is synthetically lethal in combination with Rb deficiency is to use genetic screens in animal models. We have taken advantage of the Drosophila melanogaster system in our lab, which combines a well-conserved Rb/E2F signaling pathway with established genetic tools that allow for relatively efficient and unbiased screening of large numbers of genes in vivo. One such “forward” genetic screening approach [107] utilizes Ethyl-methanesulfonate (EMS)-induced mutagenesis and mosaic clone analysis to identify novel genes that cooperate with Rb during fly development [77, 78, 108]. An interesting gene that came out of our screen is the fly ortholog of Tsc2, an important regulator of mTOR.
Inactivation of TSC2 and Rb lead to synergistic cell death in both flies and human cancer cells
Tsc2 is one of two tumor suppressors, the other being Tsc1, whose loss of function causes tuberous sclerosis complex (TSC) [109, 110], a syndrome characterized by the presence of benign tumors called hamartomas in different tissues [111]. The TSC1 and TSC2 proteins form a heterodimer that stimulates GTP hydrolysis by the small G protein Rheb, a crucial activator of mTOR in a rapamycin-sensitive complex termed TORC1 [112-115]. TORC1 affects a variety of cellular processes including ribosome biogenesis and cap-dependent mRNA translation, lipid synthesis, and autophagy [116].
We showed that simultaneous inactivation of the fly Rb and Tsc2 orthologs causes synergistic cell death during larval eye development, leading to adults with significantly reduced double-mutant eye tissue. This synergistic cell death is dependent on the deregulation of both the TORC1 effector dS6k and dE2f1-mediated transcriptional activation, as well as increased stress signaling [78]. Importantly, this synergistic lethality observed in flies is conserved in mammals, as inactivation of Tsc2 using short hairpin RNA (shTSC2) specifically kills Rb-deficient cancer cells under stress conditions and inhibits cancer cell growth in both soft agar and mouse xenografts. Cell death induction by shTSC2 depends on Rb deficiency because re-expression of wildtype Rb suppresses shTSC2-induced death in Rb-mutant cancer cells while knockdown of both Rb and Tsc2 is required for synergistic death in Rb-wildtype cancer cells.
Mechanisms by which Rb and TSC2 inactivation leads to synergistic cell death induction
What are the mechanisms that lead to synergistic cell death when both Rb and Tsc2 are inactivated? TSC tumor suppressors are known to be central mediators of energy- and nutrient-sensing signaling pathways that are generally altered in cancers. AMPK, which is activated by low cellular ATP levels [117], phosphorylates and activates Tsc2 so that cells can respond to energy deficiency by inhibiting the TORC1 pathway. In addition, Tsc2 is activated in response to low oxygen levels to regulate TORC1 activity [118-120] and Tsc2 is required for the survival of cells cultured in the absence of glucose [117]. Therefore regulation of TORC1 activity by the TSC proteins is required for cell survival under adverse conditions such as hypoxia and nutrient deprivation that are expected to be encountered by cells during different stages of tumorigenesis [116].
We found that inactivation of Tsc2 leads to increased cellular stress in Rb-mutant cancer cells, including the accumulation of reactive oxygen species (ROS) and an induction of the unfolded protein response (UPR) [78]. It is possible that the increased ROS is due to deregulated mTOR activity, which increases the rates of protein synthesis and mitochondrial oxidative phosphorylation [121]. In support of this, inhibition of mTOR activity by rapamycin or inhibition of protein synthesis by G418 significantly decreases the accumulation of ROS and decreased the level of cell death induction [78]. Importantly, reducing the level of ROS, either by antioxidant treatment or expression of ROS scavenger enzymes, significantly decreases cell death and restores growth in soft agar. These observations provide strong support for the notion that ROS induction contributes to the synergistic cell death phenotype.
In addition to ROS induction, inactivation of Tsc2 also has a conserved function of reducing the activation of Akt, a strong survival factor, by mTOR in a complex referred to as TORC2 [122, 123]. Aside from the potential titration of mTOR, TORC1 signaling has also been shown to inhibit Akt activation through p70/S6k-mediated phos-phoryation of both the TORC2 component Rictor [124, 125] and the PI3K-activator IRS-1 [126, 127]. Despite decreased Akt survival signaling, enforced expression of activated forms of Akt are unable to rescue the cell death phenotype induced by Rb and TSC2 co-inactivation in either cultured cells or flies. Therefore, although PI3K/Akt survival signaling has been shown to antagonize an E2f1-mediated cell death program [128], our experiments suggest that cell death induced by Rb and TSC2 inactivation is not the result of decreased Akt signaling. These observations are consistent with the notion that increased ROS contributes to cell death due to Rb and TSC2 inactivation. Independent studies indicate that although Akt signaling can suppress cell death induced by a variety of signaling pathways, ROS-mediated cell death is an exception because Akt inhibits the transcription factor FoxO, which results in reduced expression of vital ROS scavengers and increased sensitivity to oxidative stress [129]. However Akt has also been shown to promote NF-κB activity, which also directs the expression of ROS scavenging enzymes [130, 131]. As the induction of NF-κB by Akt may only be important in certain contexts such as TNFa treatment [130], it is possible that reduced Akt activity could contribute to Rb and Tsc2 inactivation-induced synthetic lethality in some cell types.
ER stress is also induced upon inactivation of Tsc2 in Rb-mutant cancer cells. Interestingly, overexpression of Bip/Grp78, an ER resident and molecular chaperone involved in the unfolded protein response, decreases ER stress and inhibits shTSC2-induced death in Rb mutant cancer cells. Conversely, knockdown of Grp78 significantly increased the sensitivity of Rb-wildtype cancer cells to shTSC2-induced death (Li et al., unpublished results). These observations suggest that ER stress also plays an important role in the synergistic cell death induced by inactivation of Rb and TSC2. Further studies will be needed to establish the relationship between the induction of ROS and ER stress.
There is an abundance of evidence that shows that Tsc2 inactivation leads to the induction of multiple types of cellular stresses, including ROS, ER stress, and energy stress. So why is loss of Tsc2 on its own generally insufficient to cause cell death, and how does the loss of Rb sensitize cells to Tsc2 inactivation? It appears that the stress-response mechanisms such as induction of ROS scavenging enzymes and Grp78 provide some protective effects. Interestingly, previous studies have implicated E2f1 as a key regulator of genes involved in cellular stress responses, including the ROS scavenger SOD2 [132], the ER chaperone Grp78 [133], and the cellular energy regulator AMPKα2 [128]. Indeed, we found that SOD2 levels are regulated by pRb and the inability to upregulate SOD2 expression in response to shTSC2 contributes to the increased sensitivity of Rb-mutant cancer cells to synergistic death induction [78]. Furthermore, we found that upregulation of Grp78 in response to cellular stress induced by Tsc2 knockdown also requires Rb (Li et al., unpublished results), which also contributes to the sensitivity of Rb-mutant cells to shTSC2-induced cell death. These results indicate that pRb regulates the expression of targets that normally function to control the level of cellular stresses and are therefore critical for the ability of cells to adapt to a variety of cellular stresses. The inability to induce such genes renders Rb-mutant tumors susceptible to shTSC2-induced cell death.
It should be mentioned that TSC2 inactivation also inhibits autophagy, which is a dynamic cellular bulk-degradation process that is activated in response to environmental stress in order to preserve vital nutrients, conserve energy production, or remove damaged organelles [134]. The ULK1-Atg13-FIP200 complex is involved in the early stages of autophagosome formation, and is inhibited by TORC1-mediated phosphorylation of ULK1 and Atg13. Inhibition of TORC1 activity by amino acid starvation or rapamycin treatment results in the dissociation of TORC1 from and dephosphorylation of this complex [135, 136]. Although its role in tumorigenesis is controversial several studies suggest that autophagy is an important mechanism of survival for cells in some aggressive tumors [137-139]. As autophagy is thought to allow cells to cope with ER stress, ROS, and damaged organelles to prevent genomic instability and tumorigenesis [139], it is possible that the impairment of autophagy contributes to the sensitivity of Rb-deficient tumor cells to Tsc2 inactivation.
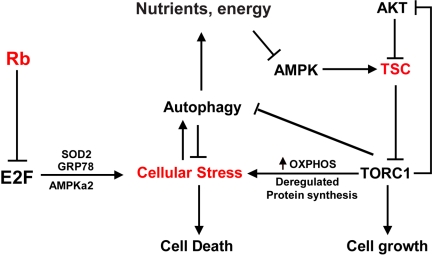
In summary, synthetic lethality induced by inactivation of both Rb and Tsc2 involves a combination of the induction of cellular stresses in conjunction with an impairment of the adaptive mechanisms that are critical for cell survival (Figure 4). It is possible that agents or treatments that increase cellular stress or decrease responses to stress can further enhance the sensitivity of Rb-mutant cells to Tsc2 inactivation. In this regard, it is interesting to note that current cancer therapies, which often cause DNA damage by inhibiting DNA replication or mitosis, also induce high levels of other forms of cellular stress. It will be interesting to test if these conventional cancer treatments can be combined with Tsc2 inactivation to increase the killing of Rb-mutant cancers. Indeed, Tsc2- or Tsc1-null cells, which exhibit reduced PI3K signaling through Akt [126, 127], were shown to be sensitive to apoptosis induced by the DNA damaging agents camptothecin and etoposide, and this effect was blocked by rapamycin [127]. Thus Tsc2 inactivation does appear to sensitize cells to some existing chemotherapeutic drugs.
Figure 4.
A model for cell death induced by Rb and TSC2 inactivation. Rb-deficient cancer cells are sensitive to death in part due to impairments in the responses to various forms of stress, including the accumulation of ROS and misfolded protein. These cells therefore are particularly sensitive to deregulation of TORC1 activity, which is an inherently energy-intensive and stress-inducing signaling pathway. Because the TSC proteins play a central role in mediating TORC1 inhibition by various signaling cues, their inactivation can cause catastrophic metabolic deregulation in the absence of compensatory stress responses.
Conclution
The inherent genomic instability of tumor cells presents a variety of challenges to developing effective therapies for treating cancers of different tissue origin and underlying molecular etiology, thus it stands to reason that current and future treatment regimens should be multifaceted. It is imperative for preclinical research to continually provide both insight into the genetic context of drug sensitivity, as well as novel targets for rational drug discovery, such that oncology in the future will have an arsenal of tools to deploy in the ongoing war against cancer. We propose that synthetic lethality will have an important role to play as therapeutic intervention becomes more sophisticated and personalized medicine advances the notion of tailoring specific treatments for individual patients. Further studies into the cooperative functions of particular genes has the potential to not only identify promising new targets for exploitation but to elucidate mechanisms underlying treatment efficacy, or lack thereof.
Acknowledgments
This work is supported in part by grants from the National Institute of Health [R01CA149275, R01 GM074197, P01AT004418] and a grant from DOD [W81XWH-10-1-0077] to WD.
Conflict of interest
Authors have no conflict of interest.
References
- 1.Friend SH, Bernards R, Rogelj S, Weinberg RA, Rapaport JM, Albert DM, Dryja TP. A human DNA segment with properties of the gene that predisposes to retinoblastoma and osteosarcoma. Nature. 1986;323:643–646. doi: 10.1038/323643a0. [DOI] [PubMed] [Google Scholar]
- 2.Lee WH, Bookstein R, Hong F, Young LJ, Shew JY, Lee EYHP. Human Retinoblastoma Susceptibility Gene - Cloning, Identification, and Sequence. Science. 1987;235:1394–1399. doi: 10.1126/science.3823889. [DOI] [PubMed] [Google Scholar]
- 3.Dryja TP, Rapaport JM, Joyce JM, Petersen RA. Molecular detection of deletions involving band q14 of chromosome 13 in retinoblastomas. Proc Natl Acad Sci U S A. 1986;83:7391–7394. doi: 10.1073/pnas.83.19.7391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dryja TP, Rapaport JM, Epstein J, Goorin AM, Weichselbaum R, Koufos A, Cavenee WK. Chromosome-13 Homozygosity in Osteosarcoma without Retinoblastoma. American Journal of Human Genetics. 1986;38:59–66. [PMC free article] [PubMed] [Google Scholar]
- 5.Horowitz JM, Park SH, Bogenmann E, Cheng JC, Yandell DW, Kaye FJ, Minna JD, Dryja TP, Weinberg RA. Frequent Inactivation of the Retinoblastoma Antioncogene Is Restricted to a Subset of Human Tumor-Cells. Proc Natl Acad Sci U S A. 1990;87:2775–2779. doi: 10.1073/pnas.87.7.2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 7.Classon M, Dyson N. p107 and p130: versatile proteins with interesting pockets. Exp Cell Res. 2001;264:135–147. doi: 10.1006/excr.2000.5135. [DOI] [PubMed] [Google Scholar]
- 8.Zhu L, Harlow E, Dynlacht BD. p107 uses a p21CIP1-related domain to bind cyclin/cdk2 and regulate interactions with E2F. Genes Dev. 1995;9:1740–1752. doi: 10.1101/gad.9.14.1740. [DOI] [PubMed] [Google Scholar]
- 9.Dynlacht BD, Flores O, Lees JA, Harlow E. Differential regulation of E2F transactivation by cyclin/cdk2 complexes. Genes Dev. 1994;8:1772–1786. doi: 10.1101/gad.8.15.1772. [DOI] [PubMed] [Google Scholar]
- 10.Hinds PW, Mittnacht S, Dulic V, Arnold A, Reed SI, Weinberg RA. Regulation of retinoblastoma protein functions by ectopic expression of human cyclins. Cell. 1992;70:993–1006. doi: 10.1016/0092-8674(92)90249-c. [DOI] [PubMed] [Google Scholar]
- 11.Ewen ME, Sluss HK, Sherr CJ, Matsushime H, Kato J, Livingston DM. Functional interactions of the retinoblastoma protein with mammalian D-type cyclins. Cell. 1993;73:487–497. doi: 10.1016/0092-8674(93)90136-e. [DOI] [PubMed] [Google Scholar]
- 12.Kato J, Matsushime H, Hiebert SW, Ewen ME, Sherr CJ. Direct binding of cyclin D to the retinoblastoma gene product (pRb) and pRb phosphorylation by the cyclin D-dependent kinase CDK4. Genes Dev. 1993;7:331–342. doi: 10.1101/gad.7.3.331. [DOI] [PubMed] [Google Scholar]
- 13.Resnitzky D, Hengst L, Reed SI. Cyclin Aassociated kinase activity is rate limiting for entrance into S phase and is negatively regulated in G1 by p27Kip1. Mol Cell Biol. 1995;15:4347–4352. doi: 10.1128/mcb.15.8.4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ludlow JW, Glendening CL, Livingston DM, DeCarprio JA. Specific enzymatic dephosphorylation of the retinoblastoma protein. Mol Cell Biol. 1993;13:367–372. doi: 10.1128/mcb.13.1.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Claudio PP, Zamparelli A, Garcia FU, Claudio L, Ammirati G, Farina A, Bovicelli A, Russo G, Giordano GG, McGinnis DE, Giordano A, Cardi G. Expression of cell-cycle-regulated proteins pRb2/p130, p107, p27(kip1), p53, mdm-2, and Ki-67 (MIB-1) in prostatic gland adenocarcinoma. Clin Cancer Res. 2002;8:1808–1815. [PubMed] [Google Scholar]
- 16.Morris EJ, Dyson NJ. Retinoblastoma protein partners. Adv Cancer Res. 2001;82:1–54. doi: 10.1016/s0065-230x(01)82001-7. [DOI] [PubMed] [Google Scholar]
- 17.Dyson N. The regulation of E2F by pRB-family proteins. Genes Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 18.Attwooll C, Lazzerini Denchi E, Helin K. The E2F family: specific functions and overlapping interests. Embo J. 2004;23:4709–4716. doi: 10.1038/sj.emboj.7600481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Du W, Pogoriler J. Retinoblastoma family genes. Oncogene. 2006;25:5190–5200. doi: 10.1038/sj.onc.1209651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li J, Ran C, Li E, Gordon F, Comstock G, Siddiqui H, Cleghorn W, Chen HZ, Kornacker K, Liu CG, Pandit SK, Khanizadeh M, Weinstein M, Leone G, de Bruin A. Synergistic function of E2F7 and E2F8 is essential for cell survival and embryonic development. Dev Cell. 2008;14:62–75. doi: 10.1016/j.devcel.2007.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DeGregori J, Kowalik T, Nevins JR. Cellular targets for activation by the E2F1 transcription factor include DNA synthesis- and G1/Sregulatory genes. Mol Cell Biol. 1995;15:4215–4224. doi: 10.1128/mcb.15.8.4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rowland BD, Bernards R. Re-evaluating cell-cycle regulation by E2Fs. Cell. 2006;127:871–874. doi: 10.1016/j.cell.2006.11.019. [DOI] [PubMed] [Google Scholar]
- 23.Takahashi Y, Rayman JB, Dynlacht BD. Analysis of promoter binding by the E2F and pRB families in vivo: distinct E2F proteins mediate activation and repression. Genes Dev. 2000;14:804–816. [PMC free article] [PubMed] [Google Scholar]
- 24.Dynlacht BD, Brook A, Dembski M, Yenush L, Dyson N. DNA-binding and transactivation properties of Drosophila E2F and DP proteins. Proc Natl Acad Sci U S A. 1994;91:6359–6363. doi: 10.1073/pnas.91.14.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Du W, Vidal M, Xie JE, Dyson N. RBF, a novel RB-related gene that regulates E2F activity and interacts with cyclin E in Drosophila. Genes Dev. 1996;10:1206–1218. doi: 10.1101/gad.10.10.1206. [DOI] [PubMed] [Google Scholar]
- 26.Stevaux O, Dimova D, Frolov MV, Taylor-Harding B, Morris E, Dyson N. Distinct mechanisms of E2F regulation by Drosophila RBF1 and RBF2. Embo J. 2002;21:4927–4937. doi: 10.1093/emboj/cdf501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sawado T, Yamaguchi M, Nishimoto Y, Ohno K, Sakaguchi K, Matsukage A. dE2F2, a novel E2F-family transcription factor in Drosophila melanogaster. Biochem Biophys Res Commun. 1998;251:409–415. doi: 10.1006/bbrc.1998.9407. [DOI] [PubMed] [Google Scholar]
- 28.Ohtani K, Nevins JR. Functional properties of a Drosophila homolog of the E2F1 gene. Mol Cell Biol. 1994;14:1603–1612. doi: 10.1128/mcb.14.3.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Du W. Suppression of the rbf null mutants by a de2f1 allele that lacks transactivation domain. Development. 2000;127:367–379. doi: 10.1242/dev.127.2.367. [DOI] [PubMed] [Google Scholar]
- 30.Frolov MV, Huen DS, Stevaux O, Dimova D, Balczarek-Strang K, Elsdon M, Dyson NJ. Functional antagonism between E2F family members. Genes Dev. 2001;15:2146–2160. doi: 10.1101/gad.903901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van den Heuvel S, Dyson NJ. Conserved functions of the pRB and E2F families. Nat Rev Mol Cell Biol. 2008;9:713–724. doi: 10.1038/nrm2469. [DOI] [PubMed] [Google Scholar]
- 32.Huang HJS, Yee JK, Shew JY, Chen PL, Bookstein R, Friedmann T, Lee EYHP, Lee WH. Suppression of the Neoplastic Phenotype by Replacement of the Rb Gene in Human Cancer-Cells. Science. 1988;242:1563–1566. doi: 10.1126/science.3201247. [DOI] [PubMed] [Google Scholar]
- 33.Decaprio JA, Ludlow JW, Figge J, Shew JY, Huang CM, Lee WH, Marsilio E, Paucha E, Livingston DM. Sv40 Large Tumor-Antigen Forms a Specific Complex with the Product of the Retinoblastoma Susceptibility Gene. Cell. 1988;54:275–283. doi: 10.1016/0092-8674(88)90559-4. [DOI] [PubMed] [Google Scholar]
- 34.Dyson N, Bernards R, Friend SH, Gooding LR, Hassell JA, Major EO, Pipas JM, Vandyke T, Harlow E. Large T-Antigens of Many Polyomaviruses Are Able to Form Complexes with the Retinoblastoma Protein. Journal of Virology. 1990;64:1353–1356. doi: 10.1128/jvi.64.3.1353-1356.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dyson N, Howley PM, Munger K, Harlow E. The Human Papilloma Virus-16 E7-Oncoprotein Is Able to Bind to the Retinoblastoma Gene-Product. Science. 1989;243:934–937. doi: 10.1126/science.2537532. [DOI] [PubMed] [Google Scholar]
- 36.Whyte P, Buchkovich KJ, Horowitz JM, Friend SH, Raybuck M, Weinberg RA, Harlow E. Association between an Oncogene and an Anti -Oncogene - the Adenovirus E1a Proteins Bind to the Retinoblastoma Gene-Product. Nature. 1988;334:124–129. doi: 10.1038/334124a0. [DOI] [PubMed] [Google Scholar]
- 37.Kaelin WG, Jr, Pallas DC, DeCaprio JA, Kaye FJ, Livingston DM. Identification of cellular proteins that can interact specifically with the T/E1A-binding region of the retinoblastoma gene product. Cell. 1991;64:521–532. doi: 10.1016/0092-8674(91)90236-r. [DOI] [PubMed] [Google Scholar]
- 38.Bagchi S, Weinmann R, Raychaudhuri P. The retinoblastoma protein copurifies with E2F-I, an E1A-regulated inhibitor of the transcription factor E2F. Cell. 1991;65:1063–1072. doi: 10.1016/0092-8674(91)90558-g. [DOI] [PubMed] [Google Scholar]
- 39.Chellappan SP, Hiebert S, Mudryj M, Horowitz JM, Nevins JR. The E2F transcription factor is a cellular target for the RB protein. Cell. 1991;65:1053–1061. doi: 10.1016/0092-8674(91)90557-f. [DOI] [PubMed] [Google Scholar]
- 40.Pardee AB. G1 Events and Regulation of Cell-Proliferation. Science. 1989;246:603–608. doi: 10.1126/science.2683075. [DOI] [PubMed] [Google Scholar]
- 41.Goodrich DW, Wang NP, Qian YW, Lee EYHP, Lee WH. The Retinoblastoma Gene-Product Regulates Progression through the G1 Phase of the Cell-Cycle. Cell. 1991;67:293–302. doi: 10.1016/0092-8674(91)90181-w. [DOI] [PubMed] [Google Scholar]
- 42.Ewen ME, Xing YG, Lawrence JB, Livingston DM. Molecular cloning, chromosomal mapping, and expression of the cDNA for p107, a retinoblastoma gene product-related protein. Cell. 1991;66:1155–1164. doi: 10.1016/0092-8674(91)90038-z. [DOI] [PubMed] [Google Scholar]
- 43.Mayol X, Grana X, Baldi A, Sang N, Hu Q, Giordano A. Cloning of a new member of the retinoblastoma gene family (pRb2) which binds to the E1A transforming domain. Oncogene. 1993;8:2561–2566. [PubMed] [Google Scholar]
- 44.Serrano M, Hannon GJ, Beach D. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature. 1993;366:704–707. doi: 10.1038/366704a0. [DOI] [PubMed] [Google Scholar]
- 45.Lukas J, Parry D, Aagaard L, Mann DJ, Bartkova J, Strauss M, Peters G, Bartek J. Retinoblastoma-protein-dependent cell-cycle inhibition by the tumour suppressor p16. Nature. 1995;375:503–506. doi: 10.1038/375503a0. [DOI] [PubMed] [Google Scholar]
- 46.Harper JW, Adami GR, Wei N, Keyomarsi K, Elledge SJ. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclindependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 47.el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler KW, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 48.Polyak K, Lee MH, Erdjument-Bromage H, Koff A, Roberts JM, Tempst P, Massague J. Cloning of p27Kip1, a cyclin-dependent kinase inhibitor and a potential mediator of extracellular antimitogenic signals. Cell. 1994;78:59–66. doi: 10.1016/0092-8674(94)90572-x. [DOI] [PubMed] [Google Scholar]
- 49.Toyoshima H, Hunter T. p27, a novel inhibitor of G1 cyclin-Cdk protein kinase activity, is related to p21. Cell. 1994;78:67–74. doi: 10.1016/0092-8674(94)90573-8. [DOI] [PubMed] [Google Scholar]
- 50.Sage J, Mulligan GJ, Attardi LD, Miller A, Chen S, Williams B, Theodorou E, Jacks T. Targeted disruption of the three Rb-related genes leads to loss of G(1) control and immortalization. Genes Dev. 2000;14:3037–3050. doi: 10.1101/gad.843200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Robertson KD, Ait-Si-Ali S, Yokochi T, Wade PA, Jones PL, Wolffe AP. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat Genet. 2000;25:338–342. doi: 10.1038/77124. [DOI] [PubMed] [Google Scholar]
- 52.Magnaghi-Jaulin L, Groisman R, Naguibneva I, Robin P, Lorain S, Le Villain JP, Troalen F, Trouche D, Harel-Bellan A. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature. 1998;391:601–605. doi: 10.1038/35410. [DOI] [PubMed] [Google Scholar]
- 53.Zhang HS, Gavin M, Dahiya A, Postigo AA, Ma D, Luo RX, Harbour JW, Dean DC. Exit from G1 and S phase of the cell cycle is regulated by repressor complexes containing HDAC-RbhSWI/SNF and Rb-hSWI/SNF. Cell. 2000;101:79–89. doi: 10.1016/S0092-8674(00)80625-X. [DOI] [PubMed] [Google Scholar]
- 54.Taubert S, Gorrini C, Frank SR, Parisi T, Fuchs M, Chan HM, Livingston DM, Amati B. E2Fdependent histone acetylation and recruitment of the Tip60 acetyltransferase complex to chromatin in late G1. Mol Cell Biol. 2004;24:4546–4556. doi: 10.1128/MCB.24.10.4546-4556.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ren B, Cam H, Takahashi Y, Volkert T, Terragni J, Young RA, Dynlacht BD. E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes Dev. 2002;16:245–256. doi: 10.1101/gad.949802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ishida S, Huang E, Zuzan H, Spang R, Leone G, West M, Nevins JR. Role for E2F in control of both DNA replication and mitotic functions as revealed from DNA microarray analysis. Mol Cell Biol. 2001;21:4684–4699. doi: 10.1128/MCB.21.14.4684-4699.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hernando E, Nahle Z, Juan G, Diaz-Rodriguez E, Alaminos M, Hemann M, Michel L, Mittal V, Gerald W, Benezra R, Lowe SW, Cordon-Cardo C. Rb inactivation promotes genomic instability by uncoupling cell cycle progression from mitotic control. Nature. 2004;430:797–802. doi: 10.1038/nature02820. [DOI] [PubMed] [Google Scholar]
- 58.Longworth MS, Herr A, Ji JY, Dyson NJ. RBF1 promotes chromatin condensation through a conserved interaction with the Condensin II protein dCAP-D3. Genes Dev. 2008;22:1011–1024. doi: 10.1101/gad.1631508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Manning AL, Longworth MS, Dyson NJ. Loss of pRB causes centromere dysfunction and chromosomal instability. Genes Dev. 2010;24:1364–1376. doi: 10.1101/gad.1917310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Coschi CH, Martens AL, Ritchie K, Francis SM, Chakrabarti S, Berube NG, Dick FA. Mitotic chromosome condensation mediated by the retinoblastoma protein is tumor-suppressive. Genes Dev. 2010;24:1351–1363. doi: 10.1101/gad.1917610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van Harn T, Foijer F, van Vugt M, Banerjee R, Yang F, Oostra A, Joenje H, te Riele H. Loss of Rb proteins causes genomic instability in the absence of mitogenic signaling. Genes Dev. 2010;24:1377–1388. doi: 10.1101/gad.580710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yamasaki L, Jacks T, Bronson R, Goillot E, Harlow E, Dyson NJ. Tumor induction and tissue atrophy in mice lacking E2F-1. Cell. 1996;85:537–548. doi: 10.1016/s0092-8674(00)81254-4. [DOI] [PubMed] [Google Scholar]
- 63.Ginsberg D. E2F1 pathways to apoptosis. FEBS Lett. 2002;529:122–125. doi: 10.1016/s0014-5793(02)03270-2. [DOI] [PubMed] [Google Scholar]
- 64.Bates S, Phillips AC, Clark PA, Stott F, Peters G, Ludwig RL, Vousden KH. p14ARF links the tumour suppressors RB and p53. Nature. 1998;395:124–125. doi: 10.1038/25867. [DOI] [PubMed] [Google Scholar]
- 65.Vousden KH. p53: death star. Cell. 2000;103:691–694. doi: 10.1016/s0092-8674(00)00171-9. [DOI] [PubMed] [Google Scholar]
- 66.Rogoff HA, Pickering MT, Debatis ME, Jones S, Kowalik TF. E2F1 induces phosphorylation of p53 that is coincident with p53 accumulation and apoptosis. Mol Cell Biol. 2002;22:5308–5318. doi: 10.1128/MCB.22.15.5308-5318.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Irwin M, Marin MC, Phillips AC, Seelan RS, Smith DI, Liu W, Flores ER, Tsai KY, Jacks T, Vousden KH, Kaelin WG., Jr Role for the p53 homologue p73 in E2F-1-induced apoptosis. Nature. 2000;407:645–648. doi: 10.1038/35036614. [DOI] [PubMed] [Google Scholar]
- 68.Lissy NA, Davis PK, Irwin M, Kaelin WG, Dowdy SF. A common E2F-1 and p73 pathway mediates cell death induced by TCR activation. Nature. 2000;407:642–645. doi: 10.1038/35036608. [DOI] [PubMed] [Google Scholar]
- 69.Stiewe T, Putzer BM. Role of the p53-homologue p73 in E2F1-induced apoptosis. Nat Genet. 2000;26:464–469. doi: 10.1038/82617. [DOI] [PubMed] [Google Scholar]
- 70.Marin MC, Jost CA, Brooks LA, Irwin MS, O'Nions J, Tidy JA, James N, McGregor JM, Harwood CA, Yulug IG, Vousden KH, Allday MJ, Gusterson B, Ikawa S, Hinds PW, Crook T, Kaelin WG., Jr A common polymorphism acts as an intragenic modifier of mutant p53 behaviour. Nat Genet. 2000;25:47–54. doi: 10.1038/75586. [DOI] [PubMed] [Google Scholar]
- 71.Moroni MC, Hickman ES, Lazzerini Denchi E, Caprara G, Colli E, Cecconi F, Muller H, Helin K. Apaf-1 is a transcriptional target for E2F and p53. Nat Cell Biol. 2001;3:552–558. doi: 10.1038/35078527. [DOI] [PubMed] [Google Scholar]
- 72.Muller H, Bracken AP, Vernell R, Moroni MC, Christians F, Grassilli E, Prosperini E, Vigo E, Oliner JD, Helin K. E2Fs regulate the expression of genes involved in differentiation, development, proliferation, and apoptosis. Genes Dev. 2001;15:267–285. doi: 10.1101/gad.864201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stanelle J, Stiewe T, Theseling CC, Peter M, Putzer BM. Gene expression changes in response to E2F1 activation. Nucleic Acids Res. 2002;30:1859–1867. doi: 10.1093/nar/30.8.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ianari A, Natale T, Calo E, Ferretti E, Alesse E, Screpanti I, Haigis K, Gulino A, Lees JA. Proapoptotic function of the retinoblastoma tumor suppressor protein. Cancer Cell. 2009;15:184–194. doi: 10.1016/j.ccr.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Thomas DM, Carty SA, Piscopo DM, Lee JS, Wang WF, Forrester WC, Hinds PW. The retinoblastoma protein acts as a transcriptional coactivator required for osteogenic differentiation. Mol Cell. 2001;8:303–316. doi: 10.1016/s1097-2765(01)00327-6. [DOI] [PubMed] [Google Scholar]
- 76.Moon NS, Frolov MV, Kwon EJ, Di Stefano L, Dimova DK, Morris EJ, Taylor-Harding B, White K, Dyson NJ. Drosophila E2F1 has contextspecific pro- and antiapoptotic properties during development. Dev Cell. 2005;9:463–475. doi: 10.1016/j.devcel.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 77.Tanaka-Matakatsu M, Xu J, Cheng L, Du W. Regulation of apoptosis of rbf mutant cells during Drosophila development. Dev Biol. 2009;326:347–356. doi: 10.1016/j.ydbio.2008.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li B, Gordon GM, Du CH, Xu J, Du W. Specific killing of Rb mutant cancer cells by inactivating TSC2. Cancer Cell. 2010;17:469–480. doi: 10.1016/j.ccr.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Moon NS, Di Stefano L, Dyson N. A gradient of epidermal growth factor receptor signaling determines the sensitivity of rbf1 mutant cells to E2F-dependent apoptosis. Mol Cell Biol. 2006;26:7601–7615. doi: 10.1128/MCB.00836-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xie W, Jiang P, Miao L, Zhao Y, Zhimin Z, Qing L, Zhu WG, Wu M. Novel link between E2F1 and Smac/DIABLO: proapoptotic Smac/DIABLO is transcriptionally upregulated by E2F1. Nucleic Acids Res. 2006;34:2046–2055. doi: 10.1093/nar/gkl150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chen D, Pacal M, Wenzel P, Knoepfler PS, Leone G, Bremner R. Division and apoptosis of E2f-deficient retinal progenitors. Nature. 2009;462:925–929. doi: 10.1038/nature08544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chong JL, Wenzel PL, Saenz-Robles MT, Nair V, Ferrey A, Hagan JP, Gomez YM, Sharma N, Chen HZ, Ouseph M, Wang SH, Trikha P, Culp B, Mezache L, Winton DJ, Sansom OJ, Chen D, Bremner R, Cantalupo PG, Robinson ML, Pipas JM, Leone G. E2f1-3 switch from activators in progenitor cells to repressors in differentiating cells. Nature. 2009;462:930–934. doi: 10.1038/nature08677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chauveinc L, Mosseri V, Quintana E, Desjardins L, Schlienger P, Doz F, Dutrillaux B. Osteosarcoma following retinoblastoma: age at onset and latency period. Ophthalmic Genet. 2001;22:77–88. doi: 10.1076/opge.22.2.77.2228. [DOI] [PubMed] [Google Scholar]
- 84.Kaye FJ, Harbour JW. For whom the bell tolls: susceptibility to common adult cancers in retinoblastoma survivors. J Natl Cancer Inst. 2004;96:342–343. doi: 10.1093/jnci/djh080. [DOI] [PubMed] [Google Scholar]
- 85.Wikenheiser-Brokamp KA. Retinoblastoma regulatory pathway in lung cancer. Curr Mol Med. 2006;6:783–793. doi: 10.2174/1566524010606070783. [DOI] [PubMed] [Google Scholar]
- 86.Burkhart DL, Sage J. Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nat Rev Cancer. 2008;8:671–682. doi: 10.1038/nrc2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sharma A, Yeow WS, Ertel A, Coleman I, Clegg N, Thangavel C, Morrissey C, Zhang X, Comstock CE, Witkiewicz AK, Gomella L, Knudsen ES, Nelson PS, Knudsen KE. The retinoblastoma tumor suppressor controls androgen signaling and human prostate cancer progression. J Clin Invest. 2010;120:4478–4492. doi: 10.1172/JCI44239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Foster CS, Falconer A, Dodson AR, Norman AR, Dennis N, Fletcher A, Southgate C, Dowe A, Dearnaley D, Jhavar S, Eeles R, Feber A, Cooper CS. Transcription factor E2F3 overexpressed in prostate cancer independently predicts clinical outcome. Oncogene. 2004;23:5871–5879. doi: 10.1038/sj.onc.1207800. [DOI] [PubMed] [Google Scholar]
- 89.Feber A, Clark J, Goodwin G, Dodson AR, Smith PH, Fletcher A, Edwards S, Flohr P, Falconer A, Roe T, Kovacs G, Dennis N, Fisher C, Wooster R, Huddart R, Foster CS, Cooper CS. Amplification and overexpression of E2F3 in human bladder cancer. Oncogene. 2004;23:1627–1630. doi: 10.1038/sj.onc.1207274. [DOI] [PubMed] [Google Scholar]
- 90.Sherr CJ, McCormick F. The RB and p53 pathways in cancer. Cancer Cell. 2002;2:103–112. doi: 10.1016/s1535-6108(02)00102-2. [DOI] [PubMed] [Google Scholar]
- 91.Miller CW, Simon K, Aslo A, Kok K, Yokota J, Buys CH, Terada M, Koeffler HP. p53 mutations in human lung tumors. Cancer Res. 1992;52:1695–1698. [PubMed] [Google Scholar]
- 92.Pickering MT, Kowalik TF. Rb inactivation leads to E2F1-mediated DNA double-strand break accumulation. Oncogene. 2006;25:746–755. doi: 10.1038/sj.onc.1209103. [DOI] [PubMed] [Google Scholar]
- 93.Clarke AR, Maandag ER, van Roon M, van der Lugt NM, van der Valk M, Hooper ML, Berns A, te Riele H. Requirement for a functional Rb-1 gene in murine development. Nature. 1992;359:328–330. doi: 10.1038/359328a0. [DOI] [PubMed] [Google Scholar]
- 94.Jacks T, Fazeli A, Schmitt EM, Bronson RT, Goodell MA, Weinberg RA. Effects of an Rb mutation in the mouse. Nature. 1992;359:295–300. doi: 10.1038/359295a0. [DOI] [PubMed] [Google Scholar]
- 95.Lee EY, Chang CY, Hu N, Wang YC, Lai CC, Herrup K, Lee WH, Bradley A. Mice deficient for Rb are nonviable and show defects in neurogenesis and haematopoiesis. Nature. 1992;359:288–294. doi: 10.1038/359288a0. [DOI] [PubMed] [Google Scholar]
- 96.Hu N, Gutsmann A, Herbert DC, Bradley A, Lee WH, Lee EY. Heterozygous Rb-1 delta 20/ +mice are predisposed to tumors of the pituitary gland with a nearly complete penetrance. Oncogene. 1994;9:1021–1027. [PubMed] [Google Scholar]
- 97.Williams BO, Remington L, Albert DM, Mukai S, Bronson RT, Jacks T. Cooperative tumorigenic effects of germline mutations in Rb and p53. Nat Genet. 1994;7:480–484. doi: 10.1038/ng0894-480. [DOI] [PubMed] [Google Scholar]
- 98.Dannenberg JH, Schuijff L, Dekker M, van der Valk M, te Riele H. Tissue-specific tumor suppressor activity of retinoblastoma gene homologs p107 and p130. Genes Dev. 2004;18:2952–2962. doi: 10.1101/gad.322004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wikenheiser-Brokamp KA. Retinoblastoma family proteins: insights gained through genetic manipulation of mice. Cell Mol Life Sci. 2006;63:767–780. doi: 10.1007/s00018-005-5487-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tsai KY, Hu Y, Macleod KF, Crowley D, Yamasaki L, Jacks T. Mutation of E2f-1 suppresses apoptosis and inappropriate S phase entry and extends survival of Rb-deficient mouse embryos. Mol Cell. 1998;2:293–304. doi: 10.1016/s1097-2765(00)80274-9. [DOI] [PubMed] [Google Scholar]
- 101.Yamasaki L, Bronson R, Williams BO, Dyson NJ, Harlow E, Jacks T. Loss of E2F-1 reduces tumorigenesis and extends the lifespan of Rb1(+/-)mice. Nat Genet. 1998;18:360–364. doi: 10.1038/ng0498-360. [DOI] [PubMed] [Google Scholar]
- 102.Ceccarelli C, Santini D, Chieco P, Taffurelli M, Gamberini M, Pileri SA, Marrano D. Retinoblastoma (RB1) gene product expression in breast carcinoma. Correlation with Ki-67 growth fraction and biopathological profile. J Clin Pathol. 1998;51:818–824. doi: 10.1136/jcp.51.11.818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bosco EE, Wang Y, Xu H, Zilfou JT, Knudsen KE, Aronow BJ, Lowe SW, Knudsen ES. The retinoblastoma tumor suppressor modifies the therapeutic response of breast cancer. J Clin Invest. 2007;117:218–228. doi: 10.1172/JCI28803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Derenzini M, Donati G, Mazzini G, Montanaro L, Vici M, Ceccarelli C, Santini D, Taffurelli M, Trere D. Loss of retinoblastoma tumor suppressor protein makes human breast cancer cells more sensitive to antimetabolite exposure. Clin Cancer Res. 2008;14:2199–2209. doi: 10.1158/1078-0432.CCR-07-2065. [DOI] [PubMed] [Google Scholar]
- 105.Sparano JA, Paik S. Development of the 21-gene assay and its application in clinical practice and clinical trials. J Clin Oncol. 2008;26:721–728. doi: 10.1200/JCO.2007.15.1068. [DOI] [PubMed] [Google Scholar]
- 106.Kaelin WG., Jr The concept of synthetic lethality in the context of anticancer therapy. Nat Rev Cancer. 2005;5:689–698. doi: 10.1038/nrc1691. [DOI] [PubMed] [Google Scholar]
- 107.Bier E. Drosophila, the golden bug, emerges as a tool for human genetics. Nat Rev Genet. 2005;6:9–23. doi: 10.1038/nrg1503. [DOI] [PubMed] [Google Scholar]
- 108.Steele L, Sukhanova MJ, Xu J, Gordon GM, Huang Y, Yu L, Du W. Retinoblastoma family protein promotes normal R8-photoreceptor differentiation in the absence of rhinoceros by inhibiting dE2F1 activity. Dev Biol. 2009;335:228–236. doi: 10.1016/j.ydbio.2009.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Identification and characterization of the tuberous sclerosis gene on chromosome 16. Cell. 1993;75:1305–1315. doi: 10.1016/0092-8674(93)90618-z. [DOI] [PubMed] [Google Scholar]
- 110.van Slegtenhorst M, de Hoogt R, Hermans C, Nellist M, Janssen B, Verhoef S, Lindhout D, van den Ouweland A, Halley D, Young J, Burley M, Jeremiah S, Woodward K, Nahmias J, Fox M, Ekong R, Osborne J, Wolfe J, Povey S, Snell RG, Cheadle JP, Jones AC, Tachataki M, Ravine D, Sampson JR, Reeve MP, Richardson P, Wilmer F, Munro C, Hawkins TL, Sepp T, Ali JB, Ward S, Green AJ, Yates JR, Kwiatkowska J, Henske EP, Short MP, Haines JH, Jozwiak S, Kwiatkowski DJ. Identification of the tuberous sclerosis gene TSC1 on chromosome 9q34. Science. 1997;277:805–808. doi: 10.1126/science.277.5327.805. [DOI] [PubMed] [Google Scholar]
- 111.Crino PB, Nathanson KL, Henske EP. The tuberous sclerosis complex. N Engl J Med. 2006;355:1345–1356. doi: 10.1056/NEJMra055323. [DOI] [PubMed] [Google Scholar]
- 112.Garami A, Zwartkruis FJ, Nobukuni T, Joaquin M, Roccio M, Stocker H, Kozma SC, Hafen E, Bos JL, Thomas G. Insulin activation of Rheb, a mediator of mTOR/S6K/4E-BP signaling, is inhibited by TSC1 and 2. Mol Cell. 2003;11:1457–1466. doi: 10.1016/s1097-2765(03)00220-x. [DOI] [PubMed] [Google Scholar]
- 113.Castro AF, Rebhun JF, Clark GJ, Quilliam LA. Rheb binds tuberous sclerosis complex 2 (TSC2) and promotes S6 kinase activation in a rapamycin- and farnesylation-dependent manner. J Biol Chem. 2003;278:32493–32496. doi: 10.1074/jbc.C300226200. [DOI] [PubMed] [Google Scholar]
- 114.Inoki K, Li Y, Xu T, Guan KL. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 2003;17:1829–1834. doi: 10.1101/gad.1110003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tee AR, Manning BD, Roux PP, Cantley LC, Blenis J. Tuberous sclerosis complex gene products, Tuberin and Hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Curr Biol. 2003;13:1259–1268. doi: 10.1016/s0960-9822(03)00506-2. [DOI] [PubMed] [Google Scholar]
- 116.Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer Cell. 2007;12:9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 117.Inoki K, Zhu T, Guan KL. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115:577–590. doi: 10.1016/s0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- 118.Brugarolas J, Lei K, Hurley RL, Manning BD, Reiling JH, Hafen E, Witters LA, Ellisen LW, Kaelin WG., Jr Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes Dev. 2004;18:2893–2904. doi: 10.1101/gad.1256804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Reiling JH, Hafen E. The hypoxia-induced paralogs Scylla and Charybdis inhibit growth by down-regulating S6K activity upstream of TSC in Drosophila. Genes Dev. 2004;18:2879–2892. doi: 10.1101/gad.322704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.DeYoung MP, Horak P, Sofer A, Sgroi D, Ellisen LW. Hypoxia regulates TSC1/2-mTOR signaling and tumor suppression through REDD1-mediated 14-3-3 shuttling. Genes Dev. 2008;22:239–251. doi: 10.1101/gad.1617608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Schieke SM, Phillips D, McCoy JP, Jr, Aponte AM, Shen RF, Balaban RS, Finkel T. The mammalian target of rapamycin (mTOR) pathway regulates mitochondrial oxygen consumption and oxidative capacity. J Biol Chem. 2006;281:27643–27652. doi: 10.1074/jbc.M603536200. [DOI] [PubMed] [Google Scholar]
- 122.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 123.Yang Q, Inoki K, Kim E, Guan KL. TSC1/TSC2 and Rheb have different effects on TORC1 and TORC2 activity. Proc Natl Acad Sci U S A. 2006;103:6811–6816. doi: 10.1073/pnas.0602282103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Julien LA, Carriere A, Moreau J, Roux PP. mTORC1-activated S6K1 phosphorylates Rictor on threonine 1135 and regulates mTORC2 signaling. Mol Cell Biol. 2010;30:908–921. doi: 10.1128/MCB.00601-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Treins C, Warne PH, Magnuson MA, Pende M, Downward J. Rictor is a novel target of p70 S6 kinase-1. Oncogene. 2010;29:1003–1016. doi: 10.1038/onc.2009.401. [DOI] [PubMed] [Google Scholar]
- 126.Harrington LS, Findlay GM, Gray A, Tolkacheva T, Wigfield S, Rebholz H, Barnett J, Leslie NR, Cheng S, Shepherd PR, Gout I, Downes CP, Lamb RF. The TSC1-2 tumor suppressor controls insulin-PI3K signaling via regulation of IRS proteins. J Cell Biol. 2004;166:213–223. doi: 10.1083/jcb.200403069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Shah OJ, Wang Z, Hunter T. Inappropriate activation of the TSC/Rheb/mTOR/S6K cassette induces IRS1/2 depletion, insulin resistance, and cell survival deficiencies. Curr Biol. 2004;14:1650–1656. doi: 10.1016/j.cub.2004.08.026. [DOI] [PubMed] [Google Scholar]
- 128.Hallstrom TC, Mori S, Nevins JR. An E2F1-dependent gene expression program that determines the balance between proliferation and cell death. Cancer Cell. 2008;13:11–22. doi: 10.1016/j.ccr.2007.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Nogueira V, Park Y, Chen CC, Xu PZ, Chen ML, Tonic I, Unterman T, Hay N. Akt determines replicative senescence and oxidative or oncogenic premature senescence and sensitizes cells to oxidative apoptosis. Cancer Cell. 2008;14:458–470. doi: 10.1016/j.ccr.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ozes ON, Mayo LD, Gustin JA, Pfeffer SR, Pfeffer LM, Donner DB. NF-kappaB activation by tumour necrosis factor requires the Akt serine-threonine kinase. Nature. 1999;401:82–85. doi: 10.1038/43466. [DOI] [PubMed] [Google Scholar]
- 131.Romashkova JA, Makarov SS. NF-kappaB is a target of AKT in anti-apoptotic PDGF signalling. Nature. 1999;401:86–90. doi: 10.1038/43474. [DOI] [PubMed] [Google Scholar]
- 132.Tanaka H, Matsumura I, Ezoe S, Satoh Y, Sakamaki T, Albanese C, Machii T, Pestell RG, Kanakura Y. E2F1 and c-Myc potentiate apoptosis through inhibition of NF-kappaB activity that facilitates MnSOD-mediated ROS elimination. Mol Cell. 2002;9:1017–1029. doi: 10.1016/s1097-2765(02)00522-1. [DOI] [PubMed] [Google Scholar]
- 133.Racek T, Buhlmann S, Rust F, Knoll S, Alla V, Putzer BM. Transcriptional repression of the prosurvival endoplasmic reticulum chaperone GRP78/BIP by E2F1. J Biol Chem. 2008;283:34305–34314. doi: 10.1074/jbc.M803925200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ravikumar B, Vacher C, Berger Z, Davies JE, Luo S, Oroz LG, Scaravilli F, Easton DF, Duden R, O'Kane CJ, Rubinsztein DC. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36:585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 135.Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, Iemura S, Natsume T, Takehana K, Yamada N, Guan JL, Oshiro N, Mizushima N. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell. 2009;20:1981–1991. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Jung CH, Jun CB, Ro SH, Kim YM, Otto NM, Cao J, Kundu M, Kim DH. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20:1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, Mukherjee C, Shi Y, Gelinas C, Fan Y, Nelson DA, Jin S, White E. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Amaravadi RK, Yu D, Lum JJ, Bui T, Christophorou MA, Evan GI, Thomas-Tikhonenko A, Thompson CB. Autophagy inhibition enhances therapy-induced apoptosis in a Mycinduced model of lymphoma. J Clin Invest. 2007;117:326–336. doi: 10.1172/JCI28833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Guo JY, Chen HY, Mathew R, Fan J, Strohecker AM, Karsli-Uzunbas G, Kamphorst JJ, Chen G, Lemons JM, Karantza V, Coller HA, Dipaola RS, Gelinas C, Rabinowitz JD, White E. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 2011;25:460–470. doi: 10.1101/gad.2016311. [DOI] [PMC free article] [PubMed] [Google Scholar]