Abstract
The genus Aeromonas has been described as comprising several species associated with the aquatic environment, which represents their principal reservoir. Aeromonas spp. are commonly isolated from diseased and healthy fish, but the involvement of such bacteria in human infection and gastroenteritis has frequently been reported. The primary challenge in establishing an unequivocal link between the Aeromonas genus and pathogenesis in humans is the extremely complicated taxonomy. With the aim of clarifying taxonomic relationships among the strains and phenotypes, a multilocus sequencing approach was developed and applied to characterize 23 type and reference strains of Aeromonas spp. and a collection of 77 field strains isolated from fish, crustaceans, and mollusks. All strains were also screened for putative determinants of virulence by PCR (ast, ahh1, act, asa1, eno, ascV, and aexT) and the production of acylated homoserine lactones (AHLs). In addition, the phenotypic fingerprinting obtained from 29 biochemical tests was submitted to the nonparametric combination (NPC) test methodology to define the statistical differences among the identified genetic clusters. Multilocus sequence typing (MLST) achieved precise strain genotyping, and the phylogenetic analysis of concatenated sequences delineated the relationship among the taxa belonging to the genus Aeromonas, providing a powerful tool for outbreak traceability, host range diffusion, and ecological studies. The NPC test showed the feasibility of phenotypic differentiation among the majority of the MLST clusters by using a selection of tests or the entire biochemical fingerprinting. A Web-based MLST sequence database (http://pubmlst.org/aeromonas) specific for the Aeromonas genus was developed and implemented with all the results.
INTRODUCTION
The strains ascribed to the genus Aeromonas are present in a wide range of habitats. These bacteria are usually associated with an aquatic environment, which represents their principal reservoir (30). Aeromonas spp. have been found in different sites in both freshwater and brackish water, and some strains seem to be resistant to the chlorination of drinking water (8, 58, 71). Moreover, this genus is usually isolated from different terrestrial ecosystems, such as food, invertebrates, vegetables, slurry, and fecal contents of farm animals but also as a digestive tract symbiont of fish, leeches, and bats (30). Some strains of motile and nonmotile aeromonads are involved in different fish diseases, such as septicemia, ulcerative disease, and furunculosis (2, 16, 75). The genus is also implicated in some infections of terrestrial vertebrates (46). Given the worldwide distribution of this genus, the occurrence of antibiotic resistance, and the ability of some strains to survive safety treatments, interest in this genus (including interest in its members as human pathogens) has grown over the past 2 decades (32).
Recently, several studies have investigated the role of Aeromonas species in human infections (34, 43, 64, 61) and the role of the involved virulence factors (6, 15, 49, 62). Several recent studies reported the involved virulence factors in fish infections (11, 17, 36). The primary challenge in establishing an unequivocal link between the Aeromonas genus and pathogenesis is the extremely complicated taxonomy. Furthermore, only a small subset of strains containing genes for potential virulence factors seems to cause infection or diarrhea (30).
Thus, considerable effort has been directed toward developing methods for correctly identifying and classifying the different species of the genus, especially those species that have been implicated in human diseases. The number of taxa ascribed to the genus Aeromonas has increased during the last decade; over 20 species have been described, but in some cases, the validity of the designation is not universally accepted (14). Phenotypic classification keys and numerical taxonomy have been proposed by some authors to describe the more frequently isolated species and include some new phenospecies (1, 13, 72). However, the chemotaxonomic methods that have worked with large numbers of tests need simplification for routine use. In addition, problems in the accuracy of discrimination between species developed when the variability of strain data sets was improved (1). The classification of microorganisms on the basis of traditional microbiological methods (morphological, physiological, and biochemical) is not actually a reliable designation of their taxonomic status. Thus, a more comprehensive and pragmatic approach is required to furnish convincing information to derive a complete characterization of the bacteria.
DNA-based molecular methods have become more popular and widely acceptable due to their reproducibility, simplicity, and high discriminatory power (54). Several molecular methods for discriminating Aeromonas species have been applied in the last decade, and these methods include DNA-DNA hybridization, 16S rRNA gene ribotyping, randomly amplified polymorphic DNA (RAPD) PCR, amplified fragment length polymorphism (AFLP), restriction fragment length polymorphism (RFLP), pulsed-field gel electrophoresis (PFGE), and multiplex PCR; however, the majority of these methods are very laborious, are not reproducible, and in most cases do not give discriminatory results (40, 59). The limited intragenomic heterogeneity reported for the 16S rRNA genes in the genus Aeromonas suggests that a single-gene-based identification approach may not be appropriate for characterizing this bacterial genus (45).
In 1998, multilocus sequence typing (MLST) was proposed as a portable and universal method for characterizing bacteria on the basis of sequence polymorphisms within internal fragments of housekeeping genes. Each gene fragment is translated into a distinct allele, and each isolate is classified as a sequence type (ST) by the combination of the alleles of the housekeeping loci (70). Therefore, this type of sequence analysis is effective for genomic species identification and is extremely useful for determining branching orders in evolution, which is difficult to achieve using other methods, such as DNA-DNA hybridization (76). Recently, MLST (often called multilocus sequence analysis [MLSA]) has been applied to different bacterial genera as a rapid and simple method for species delineation (18, 50).
In the present study, we applied the traditional microbiological tests for the identification of 100 strains preliminarily attributed to the Aeromonas genus (23 reference/type strains and 77 isolates), and at the same time, we developed a molecular method based on a comparative sequence analysis of six relevant markers. The two methods were compared to assess the congruence of the respective results. Furthermore, we have developed and implemented a Web-based MLST sequence database (http://pubmlst.org/aeromonas) specific for the Aeromonas genus (31). Derived phylogenetic analyses were inferred to investigate Aeromonas interspecies relationships, particularly between very close species, and to investigate internal genetic structures and recombination rates within the main Aeromonas groups.
To complete the characterization of the Aeromonas strains, a PCR approach was applied for a preliminary test to verify the presence of a selection of genes involved in virulence processes. In this way, the distribution of the virulence factors were related to the taxonomic position of the Aeromonas strains.
MATERIALS AND METHODS
Bacterial strains and genus phenotypic identification.
A total of 23 reference and type strains were selected to develop an MLST scheme comprising the 15 major taxa of the genus Aeromonas (A. hydrophila, A. bestiarum, A. salmonicida, A. caviae, A. media, A. eucrenophila, A. sobria [sensu stricto], A. veronii [two biotypes], A. jandaei, A. schubertii, A. enteropelogenes [A. trota], A. encheleia, A. allosaccharophila, A. popoffii, and A. sharmana [now proposed as Manjusharmella aquatica]) (41).
A collection of 77 field strains isolated between 1999 and 2009 was analyzed. Approximately 93.5% of the samples were derived from specimens of diseased freshwater and marine fish, and the remaining fraction was isolated from crustaceans (3.9%) and mollusks (2.6%) collected in the Veneto region in the northeast of Italy. The field isolates had previously been characterized to the genus level with the following phenotypic traits: they are Gram negative, oxidase positive, have facultative anaerobic metabolism, show resistance to O/129 (150 μg) (Oxoid discs), perform glucose fermentation on a Kligler iron agar (KIA) slant, and were presumptively confirmed by a miniaturized API-20E system (bioMérieux, Inc., Hazelwood, MO). In this trial, nonmotile strains were also included and assigned to an A. salmonicida group (A. hydrophila complex) that grows at lower temperatures (22°C) and produces a brownish pigment. The complete list of the 100 strains included in the study is presented in Table 1. Other designations regarding type and reference strains are reported in Table S1 in the supplemental material.
Table 1.
Origins and typing data of Aeromonas strains analyzed in this study
| Strain | Species/complexa | Source/organ | Additional informationc | STb | Alleleb |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| gyrB | groL | gltA | metG | ppsA | recA | |||||
| Reference/type strains | ||||||||||
| CECT 4199T | A. allosaccharophila/A. veronii | Anguilla anguilla (eel) | 13 | 14 | 13 | 14 | 13 | 13 | 13 | |
| NCIMB 1134 | A. bestiarum/A. hydrophila | Rainbow trout | 4 | 5 | 4 | 5 | 4 | 4 | 4 | |
| DSM 13956T | A. bestiarum | Infected fish | 8 | 9 | 8 | 9 | 8 | 8 | 8 | |
| CECT 838T | A. caviae/A. punctata subsp. caviae | Epizootic of young guinea pigs | 12 | 13 | 12 | 13 | 12 | 12 | 12 | |
| NCIMB 882 | A. caviae | Goldfish (Crassius auratus) | 3 | 4 | 3 | 4 | 3 | 3 | 3 | |
| DSM 11577T | A. encheleia | Healthy eel in freshwater | 7 | 8 | 7 | 8 | 7 | 7 | 7 | |
| CECT 4487T | A. enteropelogenes | Human feces | 18 | 20 | 19 | 20 | 19 | 17 | 19 | |
| CECT 4255T | A. enteropelogenes/A. trota | Human feces | 16 | 18 | 17 | 18 | 17 | 16 | 17 | |
| DSM 17534T | A. eucrenophila | Freshwater fish | 9 | 10 | 9 | 10 | 9 | 9 | 9 | |
| CECT 4228T | A. jandaei | Feces from patient with diarrhea | 14 | 15 | 14 | 15 | 14 | 14 | 14 | |
| ATCC 7966T | A. hydrophila | Milk | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| CECT 398 | A. hydrophila | Human feces of a child with diarrhea | 11 | 12 | 11 | 12 | 11 | 11 | 11 | |
| DSM 4881T | A. media | Fish farm effluent | 6 | 7 | 6 | 7 | 6 | 6 | 6 | |
| DSM 19604T | A. popoffii | Drinking water | 10 | 11 | 10 | 11 | 10 | 10 | 10 | |
| NCIMB 1109 | A. salmonicida subsp. achromogenes | Diseased sea trout, Salmo trutta | 3 | 2 | 3 | 2 | 0 | 2 | ||
| NCIMB 2020 | A. salmonicida subsp. masoucida | Masou, Oncorhynchus sp. | 2 | 2 | 3 | 2 | 0 | 2 | ||
| NCIMB 1102T | A. salmonicida subsp. salmonicida | Atlantic salmon | 2 | 2 | 2 | 2 | 2 | 2 | 2 | |
| DSM 17445T | A. sharmana | Warm spring | 82 | |||||||
| CECT 4240T | A. schubertii | Forehead abscess | 15 | 16 | 15 | 16 | 15 | 15 | 15 | |
| CECT 4245T | A. sobria | Fish | 19 | 21 | 20 | 21 | 20 | 18 | 20 | |
| NCIMB 75 | A. sobria | Diseased freshwater fish | 5 | 6 | 5 | 6 | 5 | 5 | 5 | |
| CECT 4257T | A. veronii bv. veronii | Sputum of drowning victim | 17 | 19 | 18 | 19 | 18 | 16 | 18 | |
| CECT 4246 | A. veronii bv. sobria | Infected frog (red leg disease) | 17 | 16 | 17 | 16 | 0 | 16 | ||
| Field strains | ||||||||||
| Ae1 | A. sobria-A. veronii | European catfish (Ameiurus melas)/kidney | H1W2S1 | 20 | 22 | 21 | 22 | 21 | 19 | 20 |
| Ae2 | A. hydrophila | Rainbow trout (Oncorhynchus mykiss)/kidney | H1W1S1 | 21 | 23 | 22 | 23 | 22 | 20 | 21 |
| Ae3 | A. sobria-A. veronii | Rainbow trout (Oncorhynchus mykiss)/kidney | H1W1S2 | 22 | 24 | 23 | 24 | 23 | 21 | 22 |
| Ae4 | A. sobria-A. veronii | Sea bass (Dicentrarchus labrax)/kidney | H3W2S2 | 23 | 25 | 24 | 25 | 24 | 22 | 23 |
| Ae5 | A. sobria-A. veronii | Sturgeon (Acipenser sp.)/kidney | H1W2S2 | 24 | 26 | 25 | 26 | 25 | 23 | 24 |
| Ae6 | A. hydrophila | Northern pike (Esox lucius)/kidney | H1W2S3 | 25 | 27 | 26 | 27 | 26 | 24 | 25 |
| Ae7 | A. sobria-A. veronii | Trout (Salmo trutta)/kidney | H1W1S3 | 26 | 28 | 27 | 28 | 27 | 25 | 26 |
| Ae8 | A. sobria-A. veronii | European catfish (Ameiurus melas)/kidney | H1W2S3 | 27 | 29 | 28 | 17 | 21 | 26 | 27 |
| Ae9 | A. hydrophila | Crayfish (Procambarus clarkii)/hemolymph | H1W2S4 | 28 | 30 | 29 | 29 | 28 | 27 | 28 |
| Ae10 | A. sobria-A. veronii | Carp (Cyprinus carpio)/kidney | H1W2S3 | 29 | 31 | 30 | 30 | 29 | 28 | 29 |
| Ae11 | A. sobria-A. veronii | Carp (Cyprinus carpio)/kidney | H1W2S4 | 30 | 24 | 31 | 31 | 30 | 29 | 30 |
| Ae12 | A. sobria-A. veronii | Carp (Cyprinus carpio)/kidney | H1W2S4 | 31 | 25 | 32 | 32 | 31 | 3o | 31 |
| Ae13 | A. sobria-A. veronii | European grayling (Thymallus thymallus)/kidney | H1W1S4 | 32 | 26 | 33 | 33 | 32 | 31 | 33 |
| Ae14 | A. sobria-A. veronii | Arctic char (Salvelinus alpinus)/kidney | H1W1S4 | 33 | 32 | 34 | 34 | 33 | 32 | 34 |
| Ae15 | A. sobria-A. veronii | Arctic char (Salvelinus alpinus)/kidney | H1W1S4 | 34 | 33 | 35 | 35 | 34 | 33 | 35 |
| Ae16 | A. hydrophila | Rainbow trout (Oncorhynchus mikiss)/kidney | H1W1S4 | 35 | 34 | 36 | 36 | 35 | 34 | 36 |
| Ae17 | A. sobria-A. veronii | Trout (Salmo trutta)/kidney | H1W1S1 | 36 | 35 | 37 | 37 | 36 | 35 | 37 |
| Ae18 | A. sobria-A. veronii | Eel (Anguilla anguilla)/kidney | H2W2S4 | 37 | 36 | 38 | 38 | 37 | 36 | 38 |
| Ae19 | A. hydrophila | Sea bream (Sparus aurata)/kidney | H3W2S1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| Ae20 | A. sobria-A. veronii | Carp (Cyprinus carpio)/kidney | H1W2S1 | 38 | 37 | 39 | 39 | 38 | 37 | 39 |
| Ae21 | A. sobria-A. veronii | Goldfish (Carassius auratus)/spleen | H1W2S2 | 39 | 38 | 40 | 40 | 39 | 38 | 40 |
| Ae22 | A. caviae-A. media | Sturgeon (Acipenser sp.)/kidney | H1W2S2 | 40 | 39 | 41 | 41 | 40 | 39 | 41 |
| Ae23 | A. sobria-A. veronii | Goldfish (Carassius auratus)/kidney | H1W2S3 | 41 | 40 | 42 | 42 | 41 | 40 | 42 |
| Ae24 | A. sobria-A. veronii | Carp (Cyprinus carpio)/kidney | H1W2S3 | 42 | 41 | 43 | 43 | 42 | 41 | 43 |
| Ae25 | A. sobria-A. veronii | Goldfish (Carassius auratus)/spleen | H1W2S3 | 43 | 42 | 44 | 44 | 43 | 42 | 44 |
| Ae26 | A. sobria-A. veronii | Arctic char (Salvelinus alpinus)/liver | H1W1S3 | 44 | 43 | 45 | 45 | 27 | 43 | 45 |
| Ae27 | A. hydrophila | Arctic char (Salvelinus alpinus)/spleen | H1W1S3 | 45 | 44 | 46 | 46 | 44 | 44 | 46 |
| Ae28 | A. caviae-A. media | Goldfish (Carassius auratus)/spleen | H1W2S3 | 46 | 45 | 47 | 47 | 45 | 45 | 47 |
| Ae29 | A. sobria-A. veronii | Sturgeon (Acipenser sp.)/kidney | H1W2S3 | 47 | 46 | 48 | 48 | 46 | 46 | 48 |
| Ae30 | A. sobria-A. veronii | Channel catfish (Ictalurus punctatus)/kidney | H1W2S3 | 48 | 47 | 49 | 49 | 47 | 47 | 49 |
| Ae31 | A. hydrophila | Trout (Salmo trutta)/kidney | H1W1S3 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| Ae32 | A. sobria-A. veronii | European catfish (Ameiurus melas)/liver | H1W2S3 | 49 | 48 | 50 | 50 | 48 | 48 | 50 |
| Ae33 | A. sobria-A. veronii | European catfish (Ameiurus melas)/liver | H1W2S3 | 50 | 49 | 51 | 51 | 49 | 49 | 51 |
| Ae34 | A. sobria-A. veronii | Carp (Cyprinus carpio)/liver | H1W2S3 | 51 | 50 | 52 | 50 | 50 | 50 | 52 |
| Ae35 | A. sobria-A. veronii | European catfish (Ameiurus melas)/kidney | H1W2S4 | 52 | 51 | 53 | 52 | 51 | 51 | 53 |
| Ae36 | A. sobria/A. veronii | Rainbow trout (Oncorhynchus mikiss)/kidney | H1W1S4 | 53 | 52 | 54 | 31 | 52 | 52 | 54 |
| Ae37 | A. hydrophila | Carp (Cyprinus carpio)/kidney | H1W2S4 | 54 | 14 | 55 | 14 | 53 | 53 | 13 |
| Ae38 | A. sobria-A. veronii | Rainbow trout (Oncorhynchus mikiss)/kidney | H1W1S4 | 55 | 53 | 56 | 53 | 54 | 54 | 55 |
| Ae39 | A. sobria-A. veronii | Sea bream (Sparus aurata)/kidney | H3W2S1 | 56 | 54 | 57 | 54 | 55 | 55 | 56 |
| Ae40 | A. sobria-A. veronii | Trout (Salmo trutta)/kidney | H1W1S1 | 38 | 37 | 39 | 39 | 38 | 37 | 39 |
| Ae41 | A. hydrophila | Trout (Salmo trutta)/kidney | H1W1S1 | 57 | 9 | 58 | 55 | 8 | 56 | 57 |
| Ae42 | A. sobria-A. veronii | Goldfish (Carassius auratus)/kidney | H1W2S1 | 58 | 55 | 59 | 56 | 56 | 57 | 58 |
| Ae43 | A. hydrophila | Rainbow trout (Oncorhynchus mykiss)/kidney | H1W1S1 | 59 | 56 | 60 | 57 | 57 | 58 | 59 |
| Ae44 | A. sobria-A. veronii | Rainbow trout (Oncorhynchus mykiss)/kidney | H1W1S1 | 60 | 57 | 61 | 58 | 58 | 59 | 60 |
| Ae45 | A. hydrophila | Rainbow trout (Oncorhynchus mykiss)/kidney | H1W1S1 | 38 | 37 | 39 | 39 | 38 | 37 | 39 |
| Ae46 | A. hydrophila | Trout (Salmo trutta)/kidney | H1W1S1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| Ae47 | A. sobria-A. veronii | Rainbow trout (Oncorhynchus mykiss)/kidney | H1W1S1 | 61 | 58 | 62 | 59 | 59 | 60 | 61 |
| Ae48 | A. sobria-A. veronii | Carp (Cyprinus carpio)/kidney | H1W2S2 | 62 | 38 | 63 | 60 | 60 | 61 | 62 |
| Ae49 | A. sobria-A. veronii | European perch (Perca fluviatilis)/kidney | H1W2S2 | 63 | 59 | 64 | 61 | 61 | 62 | 63 |
| Ae50 | A. sobria-A. veronii | Carp (Cyprinus carpio)/kidney | H1W2S2 | 64 | 60 | 65 | 62 | 62 | 63 | 64 |
| Ae51 | A. hydrophila | Crayfish (Procambarus clarkii)/hemolymph | H1W2S3 | 65 | 61 | 66 | 63 | 63 | 64 | 65 |
| Ae52 | A. sobria-A. veronii | Crayfish (Procambarus clarkii)/hemolymph | H1W2S3 | 66 | 62 | 67 | 64 | 64 | 65 | 66 |
| Ae53 | A. sobria-A. veronii | Carp (Cyprinus carpio)/kidney | H1W2S3 | 67 | 38 | 40 | 40 | 39 | 66 | 40 |
| Ae54 | A. sobria-A. veronii | Crayfish (Procambarus clarkii)/hemolymph | H1W2S3 | 68 | 63 | 68 | 65 | 65 | 67 | 67 |
| Ae55 | A. sobria-A. veronii | Manila clam (Ruditapes philippinarum)/foot | H3W2S2 | 69 | 64 | 69 | 66 | 66 | 68 | 68 |
| Ae56 | A. sobria-A. veronii | Freshwater mussel (Anodonta sp.)/foot | H1W2S3 | 70 | 65 | 70 | 67 | 67 | 69 | 69 |
| Ae57 | A. sobria-A. veronii | Sea bass (Dicentrarchus labrax)/kidney | H3W2S1 | 71 | 66 | 71 | 68 | 68 | 70 | 70 |
| Ae58 | A. hydrophila | Carp (Cyprinus carpio)/kidney | H1W2S1 | 72 | 67 | 72 | 69 | 69 | 71 | 71 |
| Ae59 | A. sobria-A. veronii | Sea bass (Dicentrarchus labrax)/kidney | H3W2S3 | 23 | 25 | 24 | 25 | 24 | 22 | 24 |
| Ae60 | A. sobria-A. veronii | Eel (Anguilla anguilla)/kidney | H2W2S3 | 73 | 68 | 73 | 70 | 70 | 72 | 11 |
| Ae61 | A. hydrophila | Flathead gray mullet (Mugil cephalus)/kidney | H2W2S4 | 74 | 69 | 74 | 71 | 71 | 73 | 72 |
| Ae62 | A. hydrophila | Sea bream (Sparus aurata)/kidney | H3W2S1 | 75 | 70 | 75 | 72 | 72 | 74 | 73 |
| Ae63 | A. hydrophila | Rainbow trout (Oncorhynchus mykiss)/kidney | H1W1S2 | 76 | 71 | 76 | 73 | 73 | 75 | 74 |
| Ae64 | A. sobria-A. veronii | Chub (Leuciscus cephalus)/skin | H2W2S2 | 77 | 66 | 77 | 74 | 74 | 76 | 75 |
| Ae65 | A. hydrophila | Carp (Cyprinus carpio)/skin | H1W2S2 | 78 | 72 | 78 | 75 | 44 | 77 | 76 |
| Ae66 | A. sobria-A. veronii | Carp (Cyprinus carpio)/skin | H1W2S2 | 79 | 73 | 79 | 76 | 75 | 78 | 77 |
| Ae67 | A. sobria-A. veronii | Rainbow trout (Oncorhynchus mykiss)/brain | H1W1S2 | 80 | 14 | 80 | 14 | 13 | 79 | 78 |
| Ae68 | Atypical | Rainbow trout (Oncorhynchus mykiss)/brain | H1W1S3 | 81 | 74 | 81 | 77 | 76 | 80 | 79 |
| Ae69 | A. sobria-A. veronii | Trout (Salmo trutta)/kidney | H1W1S3 | 82 | 75 | 82 | 78 | 20 | 81 | 80 |
| Ae70 | A. sobria-A. veronii | Trout (Salmo trutta)/kidney | H1W1S3 | 83 | 76 | 83 | 39 | 77 | 82 | 81 |
| Ae71 | A. sobria-A. veronii | European perch (Perca fluviatilis)/kidney | H1W2S3 | 84 | 77 | 84 | 79 | 78 | 83 | 82 |
| Ae72 | A. hydrophila | European catfish (Ameiurus melas)/liver | H1W2S4 | 85 | 78 | 85 | 80 | 79 | 84 | 83 |
| Ae73 | A. sobria-A. veronii | Rainbow trout (Oncorhynchus mykiss)/skin | H1W1S4 | 86 | 79 | 86 | 81 | 80 | 85 | 84 |
| Ae74 | A. sobria-A. veronii | Sea bream (Sparus aurata)/brain | H3W2S3 | 87 | 66 | 87 | 82 | 81 | 86 | 85 |
| Ae75 | A. sobria-A. veronii | European perch (Perca fluviatilis)/kidney | H1W2S3 | 87 | 66 | 87 | 82 | 81 | 86 | 85 |
| Ae76 | A. sobria-A. veronii | Rainbow trout (Oncorhynchus mykiss)/eyes | H1W1S3 | 88 | 80 | 88 | 83 | 82 | 87 | 86 |
| Ae77 | A. sobria-A. veronii | Tench (Tinca tinca)/spleen | H1W2S4 | 89 | 81 | 89 | 84 | 83 | 88 | 87 |
The phenotypic group names (i.e., complexes, which are shown for field isolates) were assigned according to the proposed attribution provided by Khajanchi et al. (32), using the keys of identification previously described (1, 13).
STs and alleles were determined by MLST.
H, habitat (1, freshwater; 2, brackish water; 3, marine water); W, water (1, cold water; 2, warm water); S, season (1, winter; 2, spring; 3, summer; 4, autumn).
Phenotypic characterization and acylated homoserine lactone (AHL) production.
All strains were tested for 31 phenotypic traits. The incubation was conducted at 28°C (72) except for growth tests, which were conducted at 42°C and 4°C. The media used for biochemical analysis were inoculated from overnight tryptone soy broth (TSB) cultures. The following tests were applied in this study: motility, production of diffusible brown pigment on tryptic soy agar (TSA), catalase, gelatin salt (3%) liquefaction, Voges-Proskauer test, ornithine and lysine decarboxylase activity, arginine dihydrolase activity, requirement of salt (0 and 3% [wt/vol] NaCl in tubes), gas production from d-glucose in Durham tubes, indole production in tryptone tryptophan media (TTM), growth on TCBS plates (Oxoid), acid production from the carbohydrates d-mannitol, sucrose, and l- arabinose (1), hydrolysis of esculin and starch, lecithinase and phospholipase activities and proteolytic activity on egg yolk agar, citrate utilization, urease production, cephalothin and ampicillin susceptibility by the Bauer-Kirby method (13), beta-hemolysis in blood sheep agar, Kligler iron agar slant to detect lactose utilization, and gas and hydrogen sulfide production.
A qualitative screening for AHLs on agar plates was conducted according to the methods of Ravn and colleagues (55) with the Chromobacterium violaceum CV026 monitor strain. Most tests were recorded daily with a 48-h endpoint as suggested by Abbott and colleagues (1) for clinical laboratories. Antibiotic resistance, AHL production, and catalase were read at day 1, while growth at 42°C or 4°C was read at 7 days. A first biochemical classification of Aeromonas spp. as members of the A. hydrophila complex, A. caviae-A. media complex, and A. sobria-A. veronii complex was conducted according to previous literature (1, 13, 32) and is reported in Table 1.
Design of primers.
Six housekeeping genes (gyrB, groL, gltA, metG, ppsA, and recA) were chosen for the MLST analysis by using the following criteria: presence as a single copy in all strains, conservation of sequence, and wide distribution across the chromosome. Six genes (aexT, ascV, eno, ast, act-asa, and ahh1) were selected as potential markers of virulence. All of the available partial and full-length sequences of the six Aeromonas housekeeping genes and of the Aeromonas aexT, ascV, and eno virulence genes were downloaded from the GenBank database and aligned by the ClustalW program (http://www.ebi.ac.uk). Primers were designed from the most conserved regions by using Primer3 software (http://frodo.wi.mit.edu/primer3/), with a length of 19 to 25 nucleotides and, for MLST primers, with the constraint of displaying the same annealing temperature range. Primers for the amplification of ast, act-asa, and ahh1 were obtained from previous studies (33, 58, 74). The complete list of genes analyzed in this study and all primers used for PCR amplifications and sequencing is listed in Table 2.
Table 2.
Primers used for amplifications and sequencing
| Primer | Sequence (5′-3′) | Gene product | Size of PCR amplicon (bp) | Size of the target sequence (bp) | Annealing temp (°C) | Reference |
|---|---|---|---|---|---|---|
| gyrB_F | GGGGTCTACTGCTTCACCAA | DNA gyrase, β subunit | 669 | 477 | 59 | This study |
| gyrB_R | CTTGTCCGGGTTGTACTCGT | |||||
| groL_F | CAAGGAAGTTGCTTCCAAGG | Chaperonin GroEL | 782 | 510 | 56 | This study |
| groL_R | CATCGATGATGGTGGTGTTC | |||||
| gltA_F | TTCCGTCTGCTCTCCAAGAT | Citrate synthase I | 626 | 495 | 58 | This study |
| gltA_R | TTCATGATGATGCCGGAGTA | |||||
| metG_F | TGGCAACTGATCCTCGTACA | Methionyl-tRNA synthetase | 657 | 504 | 57 | This study |
| metG_R | TCTTGTTGGCCATCTCTTCC | |||||
| ppsA_F | AGTCCAACGAGTACGCCAAC | Phosphoenolpyruvate synthase | 619 | 537 | 60 | This study |
| ppsA_R | TCGGCCAGATAGAGCCAGGT | |||||
| recA_F | AGAACAAACAGAAGGCACTGG | Recombinase A | 640 | 561 | 57 | This study |
| recA_R | AACTTGAGCGCGTTACCAC | |||||
| ahh1_F | GCCGAGCGCCCAGAAGGTGAGTT | Extracellular hemolysin | 130 | 60 | 74 | |
| ahh1_R | GAGCGGCTGGATGCGGTTGT | |||||
| asa1_F | TAAAGGGAAATAATGACGGCG | Hemolysin | 249 | 56 | 74 | |
| asa1_R | GGCTGTAGGTATCGGTTTTCG | |||||
| act_F | AGAAGGTGACCACCAAGAACA | Cytotoxic enterotoxin | 232 | 56 | 33 | |
| act_R | AACTGACATCGGCCTTGAACTC | |||||
| ast_F | TCTCCATGCTTCCCTTCCACT | Heat-stable cytotonic enterotoxin | 331 | 60 | 58 | |
| ast_R | GTGTAGGGATTGAAGAAGCCG | |||||
| ascV_F | CTCGAACTGGAAGAGCAGAATG | Type III secretion system inner | 577 | 60 | This study | |
| ascV_R | GAACATCTGGCTCTCCTTCTCGATG | membrane component | ||||
| eno_F | CGCCGACAACAACGTCGACATC | Enolase | 598 | 60 | This study | |
| eno_R | CTTGATGGCAGCCAGAGTTTCG | |||||
| aexT_F | ATGCAGATTCAAGCAAACAC | ADP-ribosylating toxin | 226 | 54 | This study | |
| aexT_R | TTGCCGATCCACTCTTTGAT |
DNA extraction and PCR amplification.
For DNA extraction, a single colony from a fresh culture was resuspended in 100 μl nuclease-free water, vortexed at high speed for 5 s, and incubated at 94°C for 10 min. The tube was vortexed again and centrifuged for 2 min at 14,000 rpm. The supernatant was transferred to a fresh tube and stored at −20°C.
The PCR amplification was performed in a Euroclone One Advanced thermal cycler (Celbio, Milan, Italy). The PCRs were performed in a final volume of 20 μl of amplification mix containing 1 U of GoTaq polymerase (Promega, Madison, WI), 1× GoTaq buffer, 1.5 mM MgCl2, 0.2 mM each deoxynucleoside triphosphate (dNTP), 250 mM each primer, and 5 ng of genomic DNA as the template.
For the amplification of the six housekeeping genes, conditions for direct sequencing without any additional purification of templates were used (0.1 mM dNTPs, 0.02 mM both primers). The reaction mixture was subjected to a touchdown PCR as follows: an initial step at 94°C for 2 min, followed by 35 cycles each of denaturation at 94°C for 10 s, annealing at changing temperatures (i.e., the temperature changed from 65°C to 60°C in 0.5°C decrements during the first 10 cycles) for 30 s, and extension at 72°C for 2 min. Amplification conditions for virulence genes were comprised of an initial 2-min denaturation step at 94°C followed by 35 cycles of 20 s at 94°C, 30 s at different temperatures, depending on the amplified target, and 50 s at 72°C, with a final extension at 72°C for 7 min.
Amplified products were analyzed by electrophoresis on 1.8% agarose-Tris-acetate-EDTA (TAE) gels, stained with SYBR Safe (Invitrogen, Carlsbad, CA), and visualized on a UV transilluminator.
Bidirectional sequencing of the six target genes for the MLST analysis was performed using the respective primer pairs used for PCR amplifications as sense and antisense sequencing primers. The nucleotide sequences were determined using the BigDye Terminator cycle sequencing ready reaction kit with AmpliTaq DNA polymerase (Applied Biosystems, Foster City, CA), and the electrophoresis was performed on an ABI Prism 3100 Genetic Analyzer (Applied Biosystems, Foster City, CA) automated sequencer, according to the manufacturer's instructions. The sequences of the amplicons were verified by BLAST search (7) to indicate whether they had homology to the respective genes for which the primers were designed.
MLST data treatment and phylogenetic analyses.
Analysis, editing, and comparison of the 1,452 chromatograms and sequences obtained for the six genes from the 96 bacterial strains (4 strains are not included in the MLST analysis because of amplification problems) were performed using FinchTV software (Geospiza). The consensus sequence for each gene fragment was determined by alignment of the forward and reverse sequences by using the ClustalW program (http://www.ebi.ac.uk). The coding sequences used for the housekeeping genes were read in frame. Allele sequences that differed from each other by one or more polymorphisms were attributed to a unique allele number in the order of discovery. Each unique allelic profile, as defined by the allele numbers of the 6 loci, was assigned a sequence type (ST) number. The same ST was used for some strains if they shared the same allelic profile. Multiple alignments containing the concatenated sequences were straightforward and were performed according to the genomic gene order, gyrB, groL, gltA, metG, ppsA, and recA. All analyzed MLST sequences had the same length (3,084 nucleotides). Diversity indices, such as the G+C content of each locus, number of polymorphic sites, average numbers of synonymous and nonsynonymous sites, Tajima's D, nucleotide diversity per site (π), and the average number of nucleotide differences per site (θ), were calculated using DnaSP version 5.10 (37).
For phylogenetic analysis, concatenated sequences were aligned and analyzed by using MEGA v4.1 (69). Genetic distances were computed by the Kimura two-parameter model, and the phylogenetic tree was constructed using the neighbor-joining method (see Fig. 1). At the same time, a phylogenetic tree was also constructed for each gene to create a comparison between the six single-gene trees and the concatenated tree (see Fig. S1 in the supplemental material).
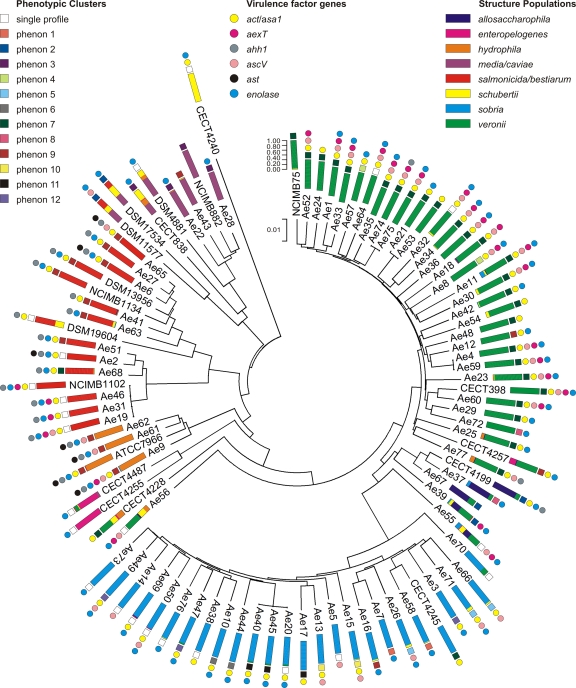
Fig. 1.
Neighbor-joining phylogenetic tree of 96 Aeromonas strains constructed from the concatenated sequences of the six genes included in this study. Colored rectangles represent the eight ancestral populations identified with Structure analysis. For each strain, the length of the colored segments indicates the proportion of nucleotides from each of the eight ancestral populations. Colored circles indicate, for each strain, the presence of the virulence factor genes analyzed in this study. Colored squares represent the phenotypic clusters obtained with Jaccard's coefficient. The scale bar length correlates with the length of the concatenated sequence, expressed as a percentage.
Recombination analyses and horizontal gene transfer detection.
Evidence for recombination between STs of each allele was investigated by using different approaches. Split-decomposition trees were constructed with 1,000 bootstrap replicates based on parsimony splits as implemented in SplitsTree 4.0 (28). The resulting trees, for individual loci and for the concatenated sequences, were analyzed using the pairwise homoplasy index (PHI) test (12) to identify alleles with significant evidence of recombination.
Recombination was also investigated by analyzing all STs with five algorithms implemented in the RDP3 program (RDP, Chimaera, GENCONV, MaxChi, and 3Seq) (39). Evidence for recombination was accepted if significant (P < 0.001) and obtained with at least three tests implemented in the RDP3 software.
The linkage model was used to identify groups with distinct allele frequencies in Structure software (21). This procedure assigns a probability of ancestry for each polymorphic nucleotide for a given number of groups, K, and it estimates q, the combined probability of ancestry from each of the K groups for each individual isolate. Eight groups were chosen for this report because repeated analyses (5 iterations, following a burn-in period of 100,000 iterations; Markov chain Monte Carlo [MCMC] = 50,000) with a K between 1 and 10 showed that the model probability was best at a K value of 8.
eBURST and ClonalFrame analysis.
Strain relationships were analyzed using the eBURST program (http://eburst.mlst.net/default.asp) to identify potential clonal complexes and founders (22). eBURST analysis was performed using the default parameters, in which STs are assigned to the same group only if five out of six alleles in the MLST loci are identical. ClonalFrame (19) was also used to investigate the population structure. ClonalFrame is a method for using multilocus sequence data to infer the clonal relationship of bacteria and assumes that recombination events were introduced at a constant rate of substitution to a contiguous region of sequence. This model is reported to have advantages over other methods, including bootstrapping and eBURST, for subdividing recombinogenic bacteria (73).
The recombination to mutation (r/m) values were calculated as reported by Vos and Didelot (73) for the main represented Aeromonas groups (A. sobria, A. veronii, and A. salmonicida-A. bestiarum) and for the entire population analyzed.
Statistical analysis of phenotypic traits, virulence factors, and environmental information.
As suggested by Valera and Esteve (72), the individual test error (Si2) was evaluated by examining 15 reference strains in duplicate (15% of the total strains), and the estimation of the average error probability (S2) was calculated according to the method proposed by Sneath and Johnson (65).
A hierarchical cluster analysis was performed on the phenotypic data set; the matrix of the results was scored with values 1 (positive) and 0 (negative) and analyzed with the program R (http://www.r-project.org). The dissimilarity distance matrixes for the variables were based on Gower's coefficient (25) and Jaccard's coefficient; the method applied for clustering was unweighted-pair group mathematical averaging (UPGMA) (72). The cophenetic correlation coefficient was applied to evaluate the distortion of the obtained dendrograms (66), and the identification of the phenotypic clusters was conducted (24).
The obtained phenotypic data were also submitted to the nonparametric combination (NPC) test methodology to define the statistical differences between the identified genetic clusters. As a general rule, considering a k-dimensional hypothesis-testing problem, the NPC solution was processed in two steps. First, a suitable set of k one-dimensional permutation tests, called partial tests, was defined. Each partial test examined the marginal contribution of any single response variable (e.g., phenotypic test) in the comparison between groups (51). Second, the nonparametric combination of dependent tests into a second-order combined test, which was suitable for testing possible global differences between the multivariate distributions of groups (all phenotypic profiles), was performed. NPC test analysis was conducted with the free software NPC Test R10 (http://www.gest.unipd.it/∼salmaso/NPC_TEST.htm), using 10,000 iterations. Partial P values were corrected for multiplicity and the global P values were obtained using the Tippet combining function. NPC permits a more flexible analysis in terms of both specification of the multivariate hypotheses and the nature of the involved variables; this approach is also useful when the number of variables is larger than the sample data set. Moreover, the NPC test methodology is proposed to solve some multivariate problems, as in the case of different variable types (i.e., categorical and numeral variables) (52). The same NPC test procedure was adopted for AHL production and for virulence factor patterns according to the Structure clustering.
Multinomial logistic regression (MLR) analysis was also applied to study the association between Structure population groups and environmental information used as a set of independent categorical variables (SPSS 17.0; SPSS Inc., Chicago, IL). The predictors used were three categorical variables (habitat [3 levels], water [2 levels], and season [4 levels]). The additional information codes and the categorical levels for each variable are reported in Table 1.
Nucleotide sequence accession numbers.
All DNA sequences were deposited in the Aeromonas MLST database (http://pubmlst.org/aeromonas) (31) and in GenBank with the accession numbers JF323072 to JF32357.
RESULTS
MLST scheme and genetic diversity.
The portions of the six housekeeping genes selected for the study were successfully amplified and sequenced in all 100 strains, except for the ppsA locus, which was not amplified in the Aeromonas type strains NCIMB 1409, NCIMB 2020, and CECT 4246. In addition, amplification in A. sharmana was not successful for any locus except for the gyrB gene. Therefore, these samples were not included in the MLST analysis. Examination of the obtained sequences revealed 11 times more synonymous substitutions than nonsynonymous substitutions, indicating that the selected six genes are appropriate for population studies. The mean G+C content of these genes varied from 57.6% (metG) to 63.7% (ppsA), with little interstrain variation; the mean G+C content of the whole A. hydrophila genome is 61%. The nucleotide diversity (the average number of nucleotide differences per site from two randomly selected sequences) was high in all genes (ranging from 0.057 for gyrB to 0.098 for ppsA). The genetic equilibrium of alleles was analyzed by using the Tajima's D neutrality test (68). All of the obtained D values were less than zero, supporting a diversifying selection of the alleles of these genes (Table 3) . Following the MLST approach, the allelic profiles of the 96 strains with no missing genes were determined (Table 1). The sequence similarity between all Aeromonas strains was 66%, which corresponded to 1,073 polymorphic sites (nucleotide diversity of 0.078) in the concatenated sequence. The genotypic diversity was high, and 89 distinct STs were identified. This high number of different alleles was expected because distinct species/taxa were processed. No ST was observed with high frequency, and only a few STs comprised more than one isolate.
Table 3.
Nucleotide diversity observed within the Aeromonas strains characterized in this studya
| Locus or concatenated sequence for cluster | Fragment size (bp) | No. of alleles | G+C content | No. (%) of polymorphic sites | No. of parsimony informative sites | Synonymous changes | Nonsynonymous changes | Ks | Ka | Tajima's D test | θ | π |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Locus (avg values) | ||||||||||||
| gyrB | 477 | 81 | 0.596 | 140 (29.3) | 96 | 174 | 9 | 0.23873 | 0.00474 | −1.10866 | 0.057 | 0.053 |
| groL | 510 | 89 | 0.584 | 199 (39) | 145 | 176 | 21 | 0.40048 | 0.01206 | −0.58984 | 0.094 | 0.083 |
| gltA | 495 | 84 | 0.603 | 150 (30.3) | 126 | 156 | 15 | 0.31618 | 0.01663 | −0.33598 | 0.080 | 0.072 |
| metG | 504 | 83 | 0.576 | 178 (35.3) | 143 | 137 | 15 | 0.35801 | 0.01965 | −0.61507 | 0.084 | 0.075 |
| ppsA | 537 | 88 | 0.637 | 233 (43.3) | 171 | 176 | 25 | 0.40523 | 0.01801 | −0.94769 | 0.098 | 0.086 |
| recA | 561 | 87 | 0.595 | 176 (31.3) | 136 | 194 | 9 | 0.24533 | 0.00443 | −1.01709 | 0.058 | 0.054 |
| Concatenated sequence | 3,084 | 89 | 0.599 | 1,073 (34.7) | 807 | 1,013 | 91 | 0.25229 | 0.01233 | −0.84311 | 0.078 | 0.070 |
| Concatenated sequence for: | ||||||||||||
| A. salmonicida-A. bestiarumb | 3,084 | 12 | 0.601 | 395 (12.8) | 250 | 382 | 38 | 0.16307 | 0.00709 | −0.09657 | 0.048 | 0.045 |
| A. veroniic | 3,084 | 35 | 0.598 | 571 (18.5) | 337 | 572 | 73 | 0.12883 | 0.00275 | −1.38050 | 0.035 | 0.033 |
| A. allosaccharophila | 3,084 | 3 | 0.598 | 68 (2.2) | 0 | 59 | 10 | 0.05092 | 0.00315 | 0.015 | 0.015 | |
| A. schubertiid | 3,084 | 2 | 0.619 | 399 (13) | 0 | 307 | 92 | 0.38629 | 0.04596 | 0.156 | 0.129 | |
| A. enteropelogenes | 3,084 | 2 | 0.607 | 99 (3.2) | 0 | 99 | 3 | 0.12679 | 0.00171 | 0.033 | 0.032 | |
| A. sobria | 3,084 | 27 | 0.589 | 394 (12.7) | 258 | 377 | 28 | 0.10795 | 0.00219 | −0.92751 | 0.029 | 0.028 |
| A. hydrophila | 3,084 | 4 | 0.617 | 115 (3.7) | 24 | 114 | 2 | 0.08056 | 0.00043 | −0.23817 | 0.020 | 0.020 |
| A. media-A. caviaee | 3,084 | 7 | 0.619 | 414 (13.4) | 145 | 384 | 48 | 0.18365 | 0.00951 | −0.86238 | 0.056 | 0.052 |
π, nucleotide diversity per site; θ, average number of nucleotide differences per site; Ks, number of synonymous changes per synonymous site; Ka, number of nonsynonymous changes per nonsynonymous site.
This group also includes DSM 19604, type strain of A. popoffii.
This group also includes CECT 4228, type strain of A. jandaei.
This group also includes DSM 17534, type strain of A. eucrenophila.
This group also includes DSM 11577, type strain of A. encheleia.
Phylogeny based on MLST data.
The phylogeny of the 96 Aeromonas strains was analyzed by constructing a neighbor-joining tree from the 3,084-bp concatenated sequences of the six loci (Fig. 1). The tree revealed two major phylogroups, one of which contained only the A. schubertii reference strain (CECT 4240T), while all other strains belonged to the second group in which different branches are easily distinguishable. The majority of the branches contained reference/type strains corresponding to named species (A. veronii, A. allosaccharophila, A. sobria, A. jandaei, A. enteropelogenes, and A. hydrophila), except for two groups containing reference strains of different species (A. salmonicida-A. bestiarum and A. media-A. caviae) that are located in different branches but are genotypically related. The phylogenetic tree that resulted from the concatenated sequence analysis was compared to the topologies of the six trees constructed independently from each gene to verify if there were important differences and to determine whether one of the six genes influenced this tree topology. The general sample classification of the single-locus trees was very similar to that of the concatenated one, even if there were differences in the distributions of some reference strains, but the main cluster divisions were maintained. The only exception was given by the trees derived from groL, metG, and recA, in which A. caviae, A. media, A. eucrenophila, and A. encheleia species clustered together with A. schubertii (see Fig. S1 in the supplemental material). However, the distribution of the concatenated phylogeny, also supported by three single-locus trees (for gyrB, gltA, and ppsA), was more reliable and demonstrated that the distribution of Aeromonas species into two groups did not result from the allelic diversity of a single gene but more likely from a general tendency of the whole genome.
The concatenated phylogeny demonstrates that two bona fide reference strains previously assigned to the A. hydrophila group (NCIMB 1434 and CECT 398) clustered into different phylogenetic groups, A. bestiarum and A. veronii, respectively. The reference strain NCIMB 75 was purchased as A. sobria, but our analyses characterized it as A. veronii bv. sobria.
Evidence of recombination in Aeromonas spp. and strain relationships.
Microevolutionary relationships among closely related genotypes may be best disclosed by analysis of allelic profiles rather than sequences because the former approach is less affected by the disturbing effect of homologous recombination (38). By use of MLST data, clonal families are typically defined as groups of strains linked by a single allelic mismatch (in our case, five common alleles out of six). Relationships between Aeromonas species were analyzed by using the eBURST algorithm (22), which focuses on allelic profiles and identifies clonal complexes (CCs) by linking single (or double)-locus variants. The eBURST analysis revealed the rarity of closely related genotypes, with the presence of only one CC formed by two A. veronii strains (ST 39 and ST 67). ClonalFrame analysis of the concatenated sequences (see Fig. S4 in the supplemental material) confirmed the association identified in the eBURST analysis. The majority of STs occurred as doublets, and some occurred as singlets with no apparent clonal relationship to each other. In other words, most strains were distantly related, and the populations of these species do not appear to be structured, based on the present sampling, into highly prevalent clonal families. The r/m ratio was calculated for the entire population and for the three most represented groups identified with Structure analysis (A. veronii, A. sobria, and A. salmonicida-A. bestiarum) to evaluate whether the high genotypic diversity could be due to recombination events. The r/m value for the entire population was found to be 0.15, while a lower value was found for the three populations, ranging between 0.07 and 0.13.
Evidence for recombination in the MLST loci was also investigated with the SplitsTree program, which used the split-decomposition method separately on each locus and on the concatenated sequences of all STs (see Fig. S2 and S3 in the supplemental material). Most of the genes were not significantly affected by intragenic recombination, but in all cases, parallelogram formation (indicative of some recombination events) was evident. Furthermore, only recA exhibited significant evidence of recombination (P < 0.05). When the concatenated sequences of all STs were investigated, evidence of significant recombination was found (P < 0.0001). The split-decomposition analysis (28) showed a “rectangular” network structure in which A. schubertii and all other Aeromonas species were clearly distant. As a confirmation of the neighbor-joining method, the distribution of the clusters previously identified was visible, and most of them corresponded to a different species. However, a separation between A. salmonicida, A. popoffii, and A. bestiarum, which was not clearly highlighted in the phylogenetic analysis, resulted in the split graph. When the three most represented populations identified with Structure analysis were investigated (A. sobria, A. veronii, and A. salmonicida-A. bestiarum), the trees showed limited parallelogram formation (see Fig. S3 in the supplemental material). To detect the sites of recombination, we searched the MLST data set by using five algorithms implemented in the RDP3 package (39). RDP3 disclosed 51 possible intergenic events, among which 21 were supported by more than 3 algorithms.
Structure software was used to identify the main groups (which differed in terms of their allele frequencies) and more subtle recombination events to also detect strains carrying foreign DNA. The software identified eight distinct ancestral sources of nucleotides (corresponding to eight colors in Fig. 1). Within the same species, most strains were homogeneous, even if some strains presented gene sequences typical of other species. In fact, some strains presented mixed colors in the corresponding column, demonstrating the import of gene sequences from other species. These isolates seemed to have a partially mixed origin. The A. schubertii strain formed a unique population, even though some regions typical of this group were found in isolates of other species (A. popoffii DSM 19604T, Ae56, A. jandaei CECT 4228T, A. eucrenophila DSM 17534T, A. encheleia DSN 14577T, and A. caviae CECT 838T). The A. encheleia and A. eucrenophila strains presented almost the same structure pattern, as supported by the phylogenetic analysis, and the entire distribution of all of the strains in the tree was clearly supported by Structure analysis.
Phenotypic traits and sources.
The phenotypic traits considered in this study were selected from the most useful tests applied in routine laboratory assessment (1, 13).
Considering all of the 31 phenotypic traits tested, the average error probability was 1.5% (S2 = 0.015). The tests with nonzero Si2 values were the following: beta-hemolysis, lactose utilization in Kligler iron agar, hydrogen sulfide production in Kligler iron agar, gelatin liquefaction, acid from sucrose and d-mannitol, growth on TCBS and at 42°C (3.3%), lysine decarboxylase, production of gas in Kligler iron agar, and citrate (6.6%) tests. Growth on 0% salt and urease production were not considered in the NPC test, due to uniform results from all strains.
Biochemical characteristics were used to build two dendrograms according to Jaccard's and Gower's indexes. The cophenetic correlations indicated that Jaccard's index provided a better description of clusters than Gower's index (0.89 versus 0.80); furthermore, both specified a good adjustment of the original distance matrixes (66). Moreover, Gower's dendrogram failed in the differentiation between the two A. veronii biovars (A. veronii bv. veronii and A. veronii bv. sobria) that were clustered together. According to these observations, UPGMA hierarchical clustering based on Jaccard's distance seems to be more reliable than the Gower's data. Therefore, only the phenotypic clusters obtained by Jaccard's dendrogram (see Fig. S5 in the supplemental material) were reported in Fig. 1 to highlight the position of each strain according to the genetic and biochemical analyses. Considering a cutoff value of 0.26 in the Jaccard's index dendrogram, 12 clusters and 24 single strain profiles were assigned.
The widest cluster, named phenon 7, was almost totally constituted by strains ascribed to the A. sobria complex (A. veronii bv. sobria, A. jandaei, and A. sobria) proposed by Abbott and colleagues (1). Phenotypically closely related species, such as A. encheleia (DSM 14577T) and A. eucrenophila (DSM 17534T), were grouped together with A. allosaccharophila (phenon 2); these species can be easily differentiated with the esculin hydrolysis test. The type strain of A. caviae (CECT 838) was located near the NCIMB 882 reference strain. The number of reference and type strains did not allow a clustering for some species, which was suggested by the presence of single profiles (A. popoffii, A. enteropelogenes, A. schubertii, A. enteropelogenes [A. trota], A. media, and A. salmonicida).
According to source information, strains showing the same ST were not always isolated from the same host species. The four isolates typed as ST 2 (A. salmonicida subsp. salmonicida NCIMB 1402T, Ae46, Ae31, and Ae19) arose from different species of salmonids and from one marine fish. The two isolates typed as ST 38 (Ae40 and Ae20) were derived from a cold-freshwater species and warm-water species and did not present the same phenotype (Fig. 1). However, the distribution of genetic groups seems to reflect the host environmental range and seasonality of sampling. The putative isolates ascribed to the cluster A. sobria were entirely isolated from freshwater fish, most of which were cold-water species (about 70%), during autumn and winter. The A. veronii isolates showed a heterogeneous host range with particular preferences for warm-water species (87%) in different habitats (freshwater, brackish water, and marine water). A reduction in the number of clusters considered in the MLR analysis was necessary to understand the influences of each environmental predictor. The build model with three structure clusters (A. veronii, A. sobria, and A. salmonicida-A. bestiarum) as a categorical dependent outcome showed that 71% of strains were overall correctly classified by using only the warm- and cold-water attribution (Nagelkerke pseudo-R-square of 0.47; P < 0.001). To solve the quasicomplete separation of variables, the number of categories was reduced. The marine and brackish habitat categories were combined into a single dummy variable, and the seasons were also divided into two categories, combining spring-winter and summer-fall (cold and warm seasons). The MLR model considering all new categorical variables showed a significant contribution of each variable (Nagelkerke pseudo-R-square of 0.61; habitat P = 0.008; water P = 0.0001; season P = 0.009) and an increase in the overall percentage of correctly classified samples (74%). However, the prediction of the A. salmonicida-A. bestiarum group failed.
Statistical analyses through the NPC test.
The NPC test of phenotypic traits was applied to the clusters identified by Structure analysis. Fifteen contrasts were generated only between taxa containing more than three strains. Among the eight identified groups, only A. schubertii and A. enteropelogenes were excluded. The results of the NPC test are reported in Tables 4 and 5 . The global P value showed that, among the identified Structure clusters, 14 contrasts were significantly different (significance α level equals 5%). In particular, Structure populations were not differentiated by the examined phenotypic characteristics for the A. salmonicida-A. bestiarum group compared to the A. allosaccharophila group, the A. hydrophila group, or the A. media-A. caviae group. Moreover, the contrast between the A. allosaccharophila group and the A. media-A. caviae group was not significantly different. The global multivariate difference can be explained by 14 of the 29 considered phenotypic variables, which have been denoted by an individual significant P value. The following tests were selected for group identification: cephalothin resistance, motility, esculin, beta-hemolysis, Voges-Proskaeur, gas from glucose (both Kligler and Durham methods), citrate, sucrose, l-arabinose, lipase on egg yolk agar, gelatin hydrolysis, and growth at 42°C and on 3% NaCl.
Table 4.
Multivariate analysis of phenotypic traits through NPC test of the identified Structure populations (group comparisons)
| Cluster comparisona | Global P valueb | Partial P value for test/parameterb |
Partial P value for test/parameterb |
|||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Catalase | Cephalothin resistance | Ampicillin resistance | Motility | Indole | Esculin | β-Hemolysis | Voges-Proskauer | LDCc | ODCd | ADH | Kligler iron agar |
Citrate | Acid from: |
Gas from glucose | Egg yolk agar |
Gelatin | Starch | Growth |
Brown pigment | |||||||||||
| Lactose | Gas | H2S | Sucrose | d-Mannitol | l-Arabinose | Protease | Lecithinase | Lipase | TCBS | 4°C | 42°C | NaCl (3%) | ||||||||||||||||||
| C1 vs C2 | ** | ** | ** | ** | ** | ** | ||||||||||||||||||||||||
| C1 vs C3 | ||||||||||||||||||||||||||||||
| C1 vs C6 | ** | * | * | ** | ** | ** | ** | |||||||||||||||||||||||
| C1 vs C7 | ||||||||||||||||||||||||||||||
| C1 vs C8 | ||||||||||||||||||||||||||||||
| C2 vs C3 | ** | * | * | ** | ** | |||||||||||||||||||||||||
| C2 vs C6 | ** | ** | ** | ** | ** | ** | ** | |||||||||||||||||||||||
| C2 vs C7 | *** | * | *** | ** | ||||||||||||||||||||||||||
| C2 vs C8 | ** | ** | ** | * | ** | ** | * | |||||||||||||||||||||||
| C3 vs C6 | * | * | * | * | ||||||||||||||||||||||||||
| C3 vs C7 | * | * | * | |||||||||||||||||||||||||||
| C3 vs C8 | ||||||||||||||||||||||||||||||
| C6 vs C7 | *** | * | *** | * | ||||||||||||||||||||||||||
| C6 vs C8 | ** | * | ** | * | ** | * | ||||||||||||||||||||||||
| C7 vs C8 | * | * | ||||||||||||||||||||||||||||
C1, A. salmonicida-A. bestiarum; C2, A. veronii; C3, A. allosaccharophila; C4, A. schubertii; C5, A. enteropelogenes; C6, A. sobria; C7, A. hydrophila; C8, A. media-A. caviae.
*, P value < 0.05; **, P value < 0.01; ***, P value < 0.001.
LDC, lysine decarboxylase.
ODC, ornithine decarboxylase.
Table 5.
Multivariate analysis of phenotypic traits through NPC test of the identified Structure populations (cluster values)
| Clustera | No. of strains | % positive for the phenotypic characteristic/test/parameter |
% positive for the phenotypic characteristic/test/parameter |
|||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Catalase | Cephalothin resistance | Ampicillin resistance | Motility | Indole | Esculin | β-Hemolysis | Voges-Proskauer | LDCe | ODCf | ADH | Kligler iron agar |
Citrate | Acid from: |
Gas from glucose | Egg yolk agar |
Gelatin | Starch | Growth |
Brown pigment | |||||||||||
| Lactose | Gas | H2S | Sucrose | d-Mannitol | l-Arabinose | Protease | Lecithinase | Lipase | TCBS | 4°C | 42°C | NaCl (3%) | ||||||||||||||||||
| C1b | 15 | 93 | 87 | 87 | 60 | 60 | 73 | 80 | 60 | 27 | 33 | 80 | 7 | 60 | 0 | 73 | 67 | 93 | 73 | 80 | 87 | 0 | 80 | 73 | 87 | 0 | 87 | 27 | 93 | 13 |
| C2 | 37 | 97 | 13 | 97 | 100 | 92 | 11 | 92 | 86 | 35 | 49 | 97 | 0 | 84 | 0 | 89 | 92 | 95 | 5 | 97 | 89 | 5 | 81 | 70 | 78 | 10 | 73 | 95 | 97 | 3 |
| C3 | 3 | 100 | 100 | 100 | 100 | 100 | 33 | 0 | 33 | 0 | 0 | 67 | 0 | 67 | 0 | 0 | 100 | 100 | 100 | 100 | 100 | 0 | 100 | 100 | 33 | 0 | 100 | 100 | 100 | 0 |
| C4c | 2 | 100 | 50 | 100 | 100 | 50 | 50 | 50 | 50 | 50 | 0 | 100 | 0 | 100 | 0 | 50 | 0 | 50 | 50 | 50 | 100 | 0 | 100 | 100 | 100 | 0 | 50 | 100 | 100 | 0 |
| C5 | 2 | 100 | 50 | 50 | 100 | 100 | 0 | 50 | 50 | 0 | 0 | 100 | 0 | 100 | 0 | 50 | 50 | 100 | 50 | 100 | 100 | 0 | 0 | 0 | 50 | 50 | 0 | 100 | 50 | 0 |
| C6 | 26 | 84 | 35 | 100 | 96 | 92 | 39 | 23 | 58 | 35 | 39 | 92 | 0 | 65 | 4 | 19 | 50 | 81 | 8 | 100 | 100 | 0 | 96 | 15 | 85 | 0 | 92 | 23 | 61 | 0 |
| C7 | 4 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 0 | 25 | 100 | 0 | 75 | 25 | 100 | 100 | 100 | 100 | 100 | 100 | 25 | 75 | 100 | 100 | 0 | 75 | 100 | 100 | 0 |
| C8d | 7 | 100 | 100 | 100 | 86 | 100 | 86 | 29 | 14 | 0 | 0 | 100 | 14 | 29 | 14 | 57 | 100 | 100 | 86 | 57 | 86 | 0 | 29 | 57 | 100 | 0 | 71 | 86 | 100 | 14 |
C1, A. salmonicida-A. bestiarum; C2, A. veronii; C3, A. allosaccharophila; C4, A. schubertii; C5, A. enteropelogenes; C6, A. sobria; C7, A. hydrophila; C8, A. media-A. caviae.
The A. salmonicida-A. bestiarum cluster also included A. popoffii DSM 19604T.
The A. schubertii cluster was formed by A. schubertii CECT 4240T and A. eucrenophila DSM 17534T.
The A. media-A. caviae cluster also included A. encheleia DSM 11577T.
LDC, lysine decarboxylase.
ODC, ornithine decarboxylase.
Distribution of virulence factors and AHL production.
In the present study, the presence of six genomic markers potentially linked to a virulence phenotype was investigated by a PCR approach. The distribution of the six virulence genes in the Aeromonas strains is reported in Fig. 1. Almost all strains contained the enolase gene, and positive reactions for act or asa1 were found in all populations with the exception of the A. media-A. caviae group. The ast gene seems to be rarely represented in the studied data set, with distributions of 20% and 100% in A. salmonicida-A. bestiarum and A. hydrophila groups, respectively (see Table S2 in the supplemental material). The genes ascV and aexT were present in 23 (24%) strains, namely, the 21 strains belonging to the A. veronii group and only one strain of the A. hydrophila and A. salmonicida-A. bestiarum groups.
In some cases, strains with the same ST showed identical prevalences of virulence genes (Ae40, Ae45, A20, Ae4, and Ae59); however, some discrepancies were also found (i.e., Ae74 and Ae75).
The fingerprinting of virulence genes was evaluated through the NPC test for each Structure group (see Table S2 in the supplemental material). The A. allosaccharophila group was not differentiable from the majority of the other clusters, while the other groups showed statistical differences in the prevalence of all virulence genes. The A. salmonicida-A. bestiarum cluster presented a higher number of strains positive for ahh1 than the others but was similar to the A. hydrophila population. These two groups were distinguished by the prevalence of the ast gene, which was present in all strains of A. hydrophila. The data related to the A. media-A. caviae cluster did not prove the presence of ast, ahh1, act or asa1, and aexT genes, and this virulence profile was statistically different from those of the others (see Table S2). Discrepancies existed in the 3 levels of AHLs among A. hydrophila, A. salmonicida-A. bestiarum, and other clusters, including A. veronii, A. sobria, and A. media-A. caviae. Moreover, the majority of the strains presented a high level of AHL production (68.8%).
DISCUSSION
Aeromonas is a genus of growing interest due to its pathogenicity for aquatic organisms, its potentially pathogenic effects in humans (30, 56), and its spoilage action in food. The knowledge of the main characteristics of Aeromonas species and strains, such as ecological, environmental, and host distributions, is currently hampered by the lack of precise delineation of genetic clusters at the species, subspecies, and clone levels. Presently, MLST is considered to be one of the most promising methods for bacterial species delineation (10, 26, 27). The main objective of this study was to apply the MLST approach to a collection of strains (reference/type and field strains) belonging to the Aeromonas genus that have been well defined from the phenotypic point of view. Among the 96 strains, the developed MLST scheme identified a large number of STs (89) and a considerable divergence among the sequence of the six concatenated alleles, considering both strains contained in the same branch (intracluster) and strains in different branches (intercluster) of the phylogenetic tree. The majority of STs occurred as singletons, which confirms the high level of sequence diversity detected, evident in π and θ values. The analysis of the concatenated gene phylogeny clearly separated the major species, and strain grouping was consistent with recently published phylogenetic studies on Aeromonas spp., with the exclusion of A. sharmana DSM 17445T from the genus (44) and the clustering of A. schubertii at the deepest branch (35, 67, 77).
However, the resolution and the discrimination power on intracluster strains achieved in this study with the application of an MLST scheme showed higher sensitivity than in previous studies. The study of six gene sequences increased the resolution of the analysis by joining the combined capacities of all molecular clocks. In fact, the reliability of differentiating closely related taxa was significantly improved, as attested by the comparison of the concatenated sequence tree with the single-gene trees (Fig. 1; see also Fig. S1 in the supplemental material). The Structure analysis of the MLST data revealed the primary genomic populations and allowed the investigation of the potential presence of foreign DNA and of gene transfer. The eight populations clearly showed genomic relationships between the Aeromonas strains, giving results similar to those of the phylogenetic analysis. All of the MLST data were also processed to evaluate potential clonal relationships and to detect the presence of recombination events. The results suggested that the emergence of clonal descents among the analyzed Aeromonas species was limited. This result could be due to inability of the six-locus MLST data to provide enough information on longer timescales, and the interrelationships among the lineages corresponding to clonal complexes remain unresolved. Moreover, as recently reported for Neisseria meningitis, an increase in the number of analyzed loci from 7 to 20 did not solve the clonal relationships among the strains. In this case, the impact of recombination events could be much more important, producing many strains with remote genotypes; this effect appears to be different in different bacterial lineages (19). However, the real effect of recombination is not easy to evaluate (20). In the case of Aeromonas, the impact of the recombination may not be relevant, resulting from the very similar topologies of the phylogenetic tree (see Fig. S1) and the dendrogram produced with Clonal Frame (see Fig. S4) and according to the low r/m value obtained from the global population. However, the split-tree analysis reported significant evidence of recombination (see Fig. S3). The r/m values calculated for single groups, such as A. sobria, A. veronii, and A. salmonicida-A. bestiarum (the only three groups that are represented by more than 10 STs), were low as well, suggesting a reduced intragroup rate of recombination.
Similar results could be visualized by the single-group split trees that, despite a significant value of recombination, present a reduced network structure (see Fig. S3 in the supplemental material). These results were compared with those obtained by Silver et al. (63) for A. veronii, in which a relevant effect of recombination was reported and a more reticulated structure was evident. This discrepancy could be partially due to the different habitats of A. veronii strains and to the physical separation, in time or space. In fact, different ecologic conditions for growth and spread or, on the contrary, the sharing of the same ecological niche could influence the horizontal gene transfer among bacteria. In conclusion, as discussed in detail by Didelot and Maiden in 2010 (20), the estimation of the recombination rate in bacteria remains a problematic task due to differences in sampling schemes and analytical methodologies across studies. The Structure analysis demonstrates a clear separation of eight populations with only a few groups (such as A. allosaccharophila and A. media-A. caviae) or a single isolate (such as Ae56 and Ae55), in which the genotype results were mixed; however, such strains are probably not well defined, due to the absence of enough isolates representing one individual group.
The comparison of the MLST data with the phenotypic results revealed several discrepancies between phenotypes and STs. Phenotypic characteristics were strongly related to habitat and were submitted to a selective pressure. Moreover, some biochemical tests showed an individual test variance (Si2) that suggests the possibility of erroneous results due to discrepancies between replicates. In any case, the S2 value was acceptable and similar to previous results reported for Aeromonas spp. (65, 72). The NPC test permitted the selection of the most useful phenotypic characteristics to differentiate the species in our data set. The tests selected by this statistical approach were a subset of those previously proposed for routine uses in the clinical laboratory (1, 13, 72).
Recently, through an MLST approach using sequencing of five genes, several new species have been described (4, 5, 23); however, the numbers of isolates available for each of these new species are very limited. Coverage of all groups with a satisfactory number of isolates will be interesting and will allow a better definition of the genetic taxonomy (30). In our data set, including the eight branches corresponding to the taxa A. veronii, A. allosaccharophila, A. sobria, A. jandaei, A. enteropelogenes, A. hydrophila, A. salmonicida-A. bestiarum, and A. media-A. caviae, several controversial phylogenetic and taxonomic positions were clarified with MLST data, particularly for groups represented by a sufficient number of isolates. Similar results were obtained in a recently published taxonomic study in which a phylogenetic analysis of the sequences of seven genes was performed (42).
A. veronii biovars and A. allosaccharophila.
The phylogenetic and taxonomic statuses of A. veronii are controversial; other studies have differentiated this species in the two biovars A. veronii bv. veronii and A. veronii bv. sobria, but the genotype divergence is very low, even if they represent two heterogeneous phenotypes (67). In contrast, our data demonstrate that, despite the high percentage of nucleotide variations, the A. veronii population is phenotypically homogeneous. Seventy-five percent of the strains were ascribed to or found to be closely related to phenon 7 (Fig. 1), 5% could be ascribed to A. veronii bv. veronii (cephalothin sensitive, esculin positive), and the other 20% presented atypical or unclustered profiles. Moreover, the A. veronii cluster could be phenotypically characterized by 12 tests (Tables 4 and 5) that give statistically different results in comparison to the other genetic clusters.
As highlighted by other studies (44), A. allosaccharophila appeared in close proximity to the A. veronii group in the phylogenetic tree. Other genomic approaches, such as AFLP genotyping and dnaJ sequencing (29, 48), have suggested that A. allosaccharophila occupies a taxonomically uncertain position with respect to A. veronii, but it is considered to be a different species (77). In our phylogenetic tree obtained from the concatenated sequence, A. allosaccharophila strains are near A. veronii but are located in different phylogenetic lines. Therefore, our method was able to separate A. veronii from A. allosaccharophila and reported a high value of nucleotide diversity between these two groups (0.033). A similar result was reported by Martinez-Murcia et al. from the analysis of the sequence of seven genes (42). The problematic taxonomic position of the A. allosaccharophila group could be explained by the mixed genotypic situation resulting from the Structure analysis. According to genetic data, the A. allosaccharophila cluster could be phenotypically differentiated from A. veronii by using cephalothin resistance, beta-hemolysis, citrate, and l-arabinose tests (Tables 4 and 5).
A. sobria.
The A. sobria genogroup contains only CECT 4245T (A. sobria) as a reference strain and 25 field strains. The current taxonomical status of A. sobria is controversial (30). Some authors have included A. sobria and A. veronii bv. sobria in the same taxa (1, 53), while Valera and Esteve have found different results (72). Moreover, some authors have considered A. sobria to be synonymous with A. veronii bv. sobria (32). To clarify this point, the CECT 4246 type strain (A. veronii bv. sobria) was selected to be included in the MLST study. Unfortunately, despite several attempts with alternative primers for the ppsA gene, the DNA extracted from this strain was not amplified and was therefore excluded from the MLST analysis. However, to clarify the phylogenetic location of this type strain, the analysis was repeated using the concatenated sequence of five genes (excluding the ppsA sequence; data not shown) and the strains were maintained in the single-gene trees (see Fig. S1). All of these analyses clearly demonstrate the position of A. veronii bv. sobria in the A. veronii genogroup.
In the present study, the A. sobria population was composed of several phenogroups (phenons 1, 3, 5, 7, 8, 13, 14, and 15) and several single profiles. This heterogeneous distribution of phenons has also been observed by other authors (72). Moreover, CECT 4245T (A. sobria) showed the same phenotypic profile as A. veronii bv. sobria, but other clusters displayed higher variability on test reactions useful for the description of species (such as indole and acid from sucrose and d-mannitol). Furthermore, the A. sobria population can be phenotypically distinguished from other groups by using 14 alternative tests (Tables 4 and 5).
From the MLST and phylogenetic analysis, a definite division between A. sobria and A. veronii is clearly visible. Moreover, the A. veronii and A. sobria groups seemed to fit in different water environments, seasons, and host ranges, as reported by MLR analysis.
A. salmonicida-A. bestiarum and A. popoffii.
In previous studies, the interrelationship between A. salmonicida and A. bestiarum has been reported as difficult to define, due to a low level of nucleotide variation, analyzing both the 16S rRNA gene (40) and gyrB (77) sequences. In contrast, MLST analysis clearly discriminated the two groups with high nucleotide diversity (0.043), and the neighbor-joining tree clearly showed two distinct subbranches. Strains of A. popoffii were confirmed to be closely related to A. bestiarum, as previously reported (67), but were clearly separated with a nucleotide diversity value of 0.030. Unlike MLST data, the aggregation into phenoclusters failed to differentiate half of the strains of these species, ascribing them into the unique A. salmonicida-A. bestiarum group. These single phenotypic profiles were represented almost entirely by the A. salmonicida strains. In addition, as reported by other authors (1, 72), A. bestiarum and A. hydrophila fit into the same phenogroup, described as the A. hydrophila complex.
A. caviae-A. media and related species.
In agreement with previous studies (44), A. caviae, A. media, A. eucrenophila, and A. encheleia displayed related but different phylogenetic lines (Fig. 1) with 0.063 nucleotide diversity. In particular, an example of controversy within the genus Aeromonas is represented by A. encheleia and A. eucrenophila (77). In our phylogenetic analysis derived from the concatenated sequence, the two reference strains (DSM 14577T and DSM 17534T) clustered together but showed a very high nucleotide diversity value (0.054). From Structure analysis, A. eucrenophila belongs to the A. schubertii population, while A. encheleia clustered in the A. media/caviae group. Despite this division, which was not visible from phylogenetic analysis, the genomic compositions of these two strains are almost identical (represented by colors in Fig. 1). The only difference is that A. eucrenophila presents as a predominant source the A. schubertii population, while A. encheleia is composed of genomic regions more similar to the A. media-A. caviae group. Furthermore, the type strains of A. caviae (CECT 838) and A. media (DSM 4881) also exhibited a variety of putative ancestor populations (Fig. 1). Nevertheless, in this study, only one strain from each species was analyzed; therefore, further investigations using a considerable number of strains belonging to both species could give more reliable information. The phenotypic traits of A. media, A. caviae, A. encheleia, and A. eucrenophila type strains were in substantial agreement with previous literature (3, 29, 72). The subpartition of clusters in two related taxa (A. caviae-A. media and A. eucrenophila-A. encheleia) permitted a more reliable phenotypic identification of the A. caviae-A. media species through the NPC test methodology (data not shown).
To complete the description of the Aeromonas strains, a PCR approach was applied to investigate the presence of six virulence gene markers. As clearly demonstrated in Fig. 1, the majority of the virulence factors investigated appears to be present first in the A. hydrophila strains and in several A. veronii strains (the two species mostly indicated as pathogenic); the virulence factor distribution was in substantial agreement with previous studies on Aeromonas spp. isolated from the water environment (58). However, the high genetic diversity, evidenced by the MLST study among and inside the taxonomic/species groups in the Aeromonas genus, suggests that the PCR approach may not be appropriate to assess with certainty the presence of virulence genes as previously demonstrated also by Silver et al. (62). In fact, in addition to the frequent involvement of virulence genes in horizontal gene transfer, the gene sequence variations between different strains may prevent the amplification of PCR products. For this reason, other methods need to be applied for a deep study of virulence profiles. At the moment, the complete genome sequences for A. hydrophila ATCC 4966 (60), A. salmonicida subsp. salmonicida A449 (57), and (only recently) A. caviae Ae398 (9) are available. The increase of genomic information could allow a more extensive approach such as comparative genome hybridization (CGH) to investigate the presence of virulence genes as proposed by Nash and colleagues for A. salmonicida strains (47).
In conclusion, the MLST method developed in this study is broadly applicable for characterizing and identifying Aeromonas spp. and for strain typing. The chosen genes have proven to be excellent molecular markers for assessing phylogeny in the genus Aeromonas and for clarifying the controversial relationships between some species. Moreover, despite the variability observed in the present data set, the multivariate analysis from the NPC test provided a set of useful phenotypic characteristics to differentiate between the more numerous populations. The simultaneous use of phenotypic and genotypic approaches was extremely valuable and appropriate for the characterization of the Aeromonas strains, but in some cases, phenotypic studies can identify only a macrogroup level, while genotypic approaches are able to also characterize the strain level.
In particular, the results clearly indicate that the genus Aeromonas comprises several (at least eight, maybe more if additional sampling allows the implementation of less-represented groups) well-separated groups of strains, but each strain is highly divergent from the others. This result explains the taxonomic confusion and suggests that forcing Aeromonas isolates into a species scheme could delineate a pragmatic but not realistic scenario of the strain diversity.
Our results revealed clearly demarcated clusters and provide novel insights into the phylogenetic distinctions between the Aeromonas groups. Knowledge of the genetic structure of Aeromonas strains will provide a useful method to explore the phylogenetic distribution of relevant strain-dependent features and to understand potential spoilage and/or pathogenic properties.
Supplementary Material
ACKNOWLEDGMENT
The study was supported by the Department of Public Health, Comparative Pathology, and Veterinary Hygiene, University of Padova, financial fund for young researchers.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 3 June 2011.
REFERENCES
- 1. Abbott S. L., Cheung W. K. W., Janda J. M. 2003. The genus Aeromonas: biochemical characteristics, atypical reactions, and phenotypic identification schemes. J. Clin. Microbiol. 41:2348–2357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aberoum A., Jooyandeh H. 2010. A review on occurrence and characterization of the Aeromonas species from marine fishes. World J. Fish Mar. Sci. 2:519–523 [Google Scholar]
- 3. Allen D. A., Austin B., Colwell R. R. 1983. Aeromonas media, a new species isolated from river water. Int. J. Syst. Bacteriol. 33:599–604 [Google Scholar]
- 4. Alperi A., et al. 2010. Aeromonas fluvialis sp. nov., isolated from a Spanish river. Int. J. Syst. Evol. Microbiol. 60:72–77 [DOI] [PubMed] [Google Scholar]
- 5. Alperi A., et al. 2010. Aeromonas taiwanensis sp. nov. and Aeromonas sanarellii sp. nov., clinical species from Taiwan. Int. J. Syst. Evol. Microbiol. 60:2048–2055 [DOI] [PubMed] [Google Scholar]
- 6. Alperi A., Figueras M. J. 2010. Human isolates of Aeromonas possess Shiga toxin genes (stx1 and stx2) highly similar to the most virulent gene variants of Escherichia coli. Clin. Microbiol. Infect. 16:1564–1567 [DOI] [PubMed] [Google Scholar]
- 7. Altschul S. F., et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Alvandi S. V., Anathan S. 2003. Biochemical characteristics, serogroups, and virulence factors of Aeromonas species isolated from cases of diarrhea and domestic water samples in Chennai. Ind. J. Med. Microbiol. 21:233–238 [PubMed] [Google Scholar]
- 9. Beatson S. A., et al. 2011. Genome sequence of the emerging pathogen Aeromonas caviae. J. Bacteriol. 193:1286–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bishop C. J., et al. 2009. Assigning strains to bacterial species via the internet. BMC Biol. 7:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Boyd J. M., et al. 2008. Contribution of type IV pili to the virulence of Aeromonas salmonicida subsp. salmonicida in Atlantic salmon (Salmo salar L.). Infect. Immun. 76:1445–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bruen T. C., Philippe H., Bryant D. 2006. A simple and robust statistical test for detecting the presence of recombination. Genetics 172:2665–2681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Carnahan A. M., Behram S., Joseph S. W. 1991. Aerokey II: a flexible key for identifying clinical Aeromonas species. J. Clin. Microbiol. 29:2843–2849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Carnahan A. M., Joseph S. W. 2005. Genus I. Aeromonas Stanier 1943, 213AL, p. 557–578 In Brenner D. J., Krieg N. R., Staley J. T., Garrity G. M. (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 2. The proteobacteria. Part B. The gammaproteobacteria Springer, New York, NY [Google Scholar]
- 15. Chopra A. K., et al. 2009. Virulence factor-activity relationships (VFAR) with specific emphasis on Aeromonas species (spp.). J. Water Health 7(Suppl 1):S29–S54 [DOI] [PubMed] [Google Scholar]
- 16. Cristi L., Galindo A., Chopra K. 2007. Aeromonas and Plesiomonas species, p. 381–400 In Doyle M. P., Beuchat L. R. (ed.), Food microbiology fundamentals and frontiers, 3rd ed. ASM Press, Washington, DC [Google Scholar]
- 17. Dacanay A., et al. 2010. Aeromonas salmonicida type I pilus system contributes to host colonization but not invasion. Dis. Aquat. Organ. 88:199–206 [DOI] [PubMed] [Google Scholar]
- 18. Delétoile A., et al. 2010. Species delineation and clonal diversity in four Bifidobacterium species as revealed by multilocus sequencing. Res. Microbiol. 161:82–90 [DOI] [PubMed] [Google Scholar]
- 19. Didelot X., Falush D. 2007. Inference of bacterial microevolution using multilocus sequence data. Genetics 175:1251–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Didelot X., Maiden M. C. 2010. Impact of recombination on bacterial evolution. Trends Microbiol. 18:315–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Falush D., Stephens M., Pritchard J. K. 2003. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Feil E. J., et al. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186:1518–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Figueras M. J., et al. 2011. Aeromonas rivuli sp. nov., isolated from the upstream region of a karst water rivulet. Int. J. Syst. Evol. Microbiol. 61:242–248 [DOI] [PubMed] [Google Scholar]
- 24. Gonçalves L. S. A., et al. 2008. Comparison of multivariate statistical algorithms to cluster tomato heirloom accessions. Genet. Mol. Res. 7:1289–1297 [DOI] [PubMed] [Google Scholar]
- 25. Gower J. C. 1971. A general coefficient of similarity and some of its properties. Biometrics 27:623–637 [Google Scholar]
- 26. Hanage W. P., Fraser C., Spratt B. G. 2005. Fuzzy species among recombinogenic bacteria. BMC Biol. 3:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hanage W. P., Fraser C., Spratt B. G. 2006. Sequences, sequence clusters and bacterial species. Philos. Trans. R. Soc. Lond. B Biol. Sci. 361:1917–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Huson D. H., Bryant D. 2006. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23:254–267 [DOI] [PubMed] [Google Scholar]
- 29. Huys G., et al. 1997. Inclusion of Aeromonas DNA hybridization group 11 in Aeromonas encheleia and extended descriptions of the species Aeromonas eucrenophila and A. encheleia. Int. J. Syst. Bacteriol. 47:1157–1164 [DOI] [PubMed] [Google Scholar]
- 30. Janda J. M., Abbott S. L. 2010. The genus Aeromonas: taxonomy, pathogenicity, and infection. Clin. Microbiol. Rev. 23:35–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jolley K. A., Maiden M. 2010. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11:595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Khajanchi B. K., et al. 2010. Distribution of virulence factors and molecular fingerprinting of Aeromonas species isolates from water and clinical samples: suggestive evidence of water-to-human transmission. Appl. Environ. Microbiol. 76:2313–2325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kingombe C. E., et al. 1999. PCR detection, characterization, and distribution of virulence genes in Aeromonas spp. Appl. Environ. Microbiol. 65:5293–5302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kobayashi H., et al. 2009. Structural basis for the kexin-like serine protease from Aeromonas sobria as sepsis-causing factor. J. Biol. Chem. 284:27655–27663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Küpfer M., et al. 2006. Genetic relationships of Aeromonas strains inferred from 16S rRNA, gyrB and rpoB gene sequences. Int. J. Syst. Evol. Microbiol. 56:2743–2751 [DOI] [PubMed] [Google Scholar]
- 36. Li J., et al. 2011. Detection of three virulence genes alt, ahp and aerA in Aeromonas hydrophila and their relationship with actual virulence to zebrafish. J. Appl. Microbiol. 110:823–830 [DOI] [PubMed] [Google Scholar]
- 37. Librado P., Rozas J. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452 [DOI] [PubMed] [Google Scholar]
- 38. Maiden M. C. 2006. Multilocus sequence typing of bacteria. Annu. Rev. Microbiol. 60:561–588 [DOI] [PubMed] [Google Scholar]
- 39. Martin D. P., Williamson C., Posada D. 2005. RDP2: recombination detection and analysis from sequence alignments. Bioinformatics 21:260–262 [DOI] [PubMed] [Google Scholar]
- 40. Martinez-Murcia A. J., Benlloch S., Collins M. D. 1992. Phylogenetic interrelationships of members of the genera Aeromonas and Plesiomonas as determined by 16S ribosomal DNA sequencing: lack of congruence with results of DNA-DNA hybridizations. Int. J. Syst. Bacteriol. 42:412–421 [DOI] [PubMed] [Google Scholar]
- 41. Martínez-Murcia A. J., et al. 2007. The recently proposed species Aeromonas sharmana sp. nov., isolate GPTSA-6T, is not a member of the genus Aeromonas. Int. Microbiol. 10:61–64 [PubMed] [Google Scholar]
- 42. Martinez-Murcia A. J., et al. 2011. Multilocus phylogenetic analysis of the genus Aeromonas. Syst. Appl. Microbiol. 34:189–199 [DOI] [PubMed] [Google Scholar]
- 43. McCoy A. J., et al. 2010. Cytotoxins of the human pathogen Aeromonas hydrophila trigger, via the NLRP3 inflammasome, caspase-1 activation in macrophages. Eur. J. Immunol. 40:2797–2803 [DOI] [PubMed] [Google Scholar]
- 44. Miñana-Galbis D., et al. 2009. Phylogenetic analysis and identification of Aeromonas species based on sequencing of the cpn60 universal target. Int. J. Syst. Evol. Microbiol. 59:1976–1983 [DOI] [PubMed] [Google Scholar]
- 45. Morandi A., et al. 2005. Evolutionary and diagnostic implications of intragenomic heterogeneity in the 16S rRNA gene in Aeromonas strains. J. Bacteriol. 187:6561–6564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Moro E. M. P., et al. 1999. Aeromonas hydrophila isolated from cases of bovine seminal vesiculitis in south Brazil. J. Vet. Diagn. Invest. 11:189–191 [DOI] [PubMed] [Google Scholar]
- 47. Nash J. H., et al. 2006. Comparative genomics profiling of clinical isolates of Aeromonas salmonicida using DNA microarrays. BMC Genomics 7:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nhung P. H., et al. 2007. Use of the novel phylogenetic marker dnaJ and DNA-DNA hybridization to clarify interrelationships within the genus Aeromonas. Int. J. Syst. Evol. Microbiol. 57:1232–1237 [DOI] [PubMed] [Google Scholar]
- 49. Pablos M., et al. 2010. Identity, virulence genes, and clonal relatedness of Aeromonas isolates from patients with diarrhea and drinking water. Eur. J. Clin. Microbiol. Infect. Dis. 29:1163–1172 [DOI] [PubMed] [Google Scholar]
- 50. Pascual J., et al. 2010. Multilocus sequence analysis of the central clade of the genus Vibrio by using 16S rRNA, recA, pyrH, rpoD, gyrB, rctB, and toxR genes. Int. J. Syst. Evol. Microbiol. 60:154–165 [DOI] [PubMed] [Google Scholar]
- 51. Pesarin F. 2001. Multivariate permutation test: with application in biostatistics. Wiley, Chichester, United Kingdom [Google Scholar]
- 52. Pesarin F., Salmaso L. 2010. Permutation tests for complex data: theory, applications and software. Wiley, Chichester, United Kingdom [Google Scholar]
- 53. Popoff M. 1984. Genus III. Aeromonas Kluyver and Van Niel 1936, 398AL, p. 545–547 In Krieg N. R., Holt J. J. (ed.), Bergey's manual of systematic bacteriology, vol. 1, section 5a, 9th ed. Williams & Wilkins, Baltimore, MD [Google Scholar]
- 54. Prakash O., et al. 2007. Polyphasic approach of bacterial classification—an overview of recent advances. Indian J. Microbiol. 47:98–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ravn L., et al. 2001. Methods for detecting acylated homoserine lactones produced by Gram-negative bacteria and their application in studies of AHL-production kinetics. J. Microbiol. Methods 44:239–251 [DOI] [PubMed] [Google Scholar]
- 56. Rahman M., et al. 2007. Persistence, transmission, and virulence characteristics of Aeromonas strains in a duckweed aquaculture-based hospital sewage water recycling plant in Bangladesh. Appl. Environ. Microbiol. 73:1444–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Reith M. E., et al. 2008. The genome of Aeromonas salmonicida subsp. salmonicida A449: insights into the evolution of a fish pathogen. BMC Genomics 9:427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sen K., Rodgers M. 2004. Distribution of six virulence factors in Aeromonas species isolated from US drinking water utilities: a PCR identification. J. Appl. Microbiol. 97:1077–1086 [DOI] [PubMed] [Google Scholar]
- 59. Sen K. 2005. Development of a rapid identification method for Aeromonas species by multiplex-PCR. Can. J. Microbiol. 51:957–966 [DOI] [PubMed] [Google Scholar]
- 60. Seshadri R., et al. 2006. Genome sequence of Aeromonas hydrophila ATCC 7966T: jack of all trades. J. Bacteriol. 188:8272–8282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sha J., et al. 2009. Surface-expressed enolase contributes to the pathogenesis of clinical isolate SSU of Aeromonas hydrophila. J. Bacteriol. 191:3095–3107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Silver A. C., Graf J. 2009. Prevalence of genes encoding the type three secretion system and the effectors Aext and AexU in the Aeromonas veronii group. DNA Cell Biol. 8:383–388 [DOI] [PubMed] [Google Scholar]
- 63. Silver A. C., et al. 2011. Complex evolutionary history of the Aeromonas veronii group revealed by host interaction and DNA sequence data. PLoS One 6:e16751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sierra J. C., et al. 2010. Unraveling the mechanism of action of a new type III secretion system effector AexU from Aeromonas hydrophila. Microb. Pathog. 49:122–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sneath P. H. A., Johnson R. 1972. The influence on numerical taxonomic similarities of errors in microbiological tests. J. Gen. Microbiol. 72:377–392 [DOI] [PubMed] [Google Scholar]
- 66. Sokal R. R., Rohlf F. J. 1962. The comparison of dendrograms by objective methods. Taxon 11:33–40 [Google Scholar]
- 67. Soler L., et al. 2004. Phylogenetic analysis of the genus Aeromonas based on two housekeeping genes. Int. J. Syst. Evol. Microbiol. 54:1511–1519 [DOI] [PubMed] [Google Scholar]
- 68. Tajima F. 1989. Statistical methods to test for nucleotide mutation hypothesis by DNA polymorphism. Genetics 123:585–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Tamura K., et al. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 70. Urwin R., Maiden M. C. 2003. Multi-locus sequence typing: a tool for global epidemiology. Trends Microbiol. 11:479–487 [DOI] [PubMed] [Google Scholar]
- 71. Uyttendaele M., et al. 2004. Control of Aeromonas on minimally processed vegetables by decontamination with lactic acid, chlorinated water, or thyme essential oil solution. Int. J. Food Microbiol. 90:263–271 [DOI] [PubMed] [Google Scholar]
- 72. Valera L., Esteve C. 2002. Phenotypic study by numerical taxonomy of strains belonging to the genus Aeromonas. J. Appl. Microbiol. 93:77–95 [DOI] [PubMed] [Google Scholar]
- 73. Vos M., Didelot X. 2009. A comparison of homologous recombination rates in bacteria and archaea. ISME J. 3:199–208 [DOI] [PubMed] [Google Scholar]
- 74. Wang G., et al. 2003. Detection and characterization of the hemolysin genes in Aeromonas hydrophila and Aeromonas sobria by multiplex PCR. J. Clin. Microbiol. 41:1048–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Wiklund T., Dalsgaard I. 1998. Occurrence and significance of atypical Aeromonas salmonicida in non-salmonid and salmonid fish species: a review. Dis. Aquat. Organ. 32:49–69 [DOI] [PubMed] [Google Scholar]
- 76. Yamamoto S., Bouvet P. J. M., Harayama S. 1999. Phylogenetic structures of the genus Acinetobacter based on gyrB sequences: comparison with the grouping by DNA-DNA hybridization. Int. J. Syst. Bacteriol. 49:87–95 [DOI] [PubMed] [Google Scholar]
- 77. Yáñez M. A., et al. 2003. Phylogenetic analysis of members of the genus Aeromonas based on gyrB gene sequences. Int. J. Syst. Evol. Microbiol. 53:875–883 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.