Abstract
Tetrapyrroles are ubiquitous molecules in nearly all living organisms. Heme, an iron-containing tetrapyrrole, is widely distributed in nature, including most characterized aerobic and facultative bacteria. A large majority of bacteria that contain heme possess the ability to synthesize it. Despite this capability and the fact that the biosynthetic pathway has been well studied, enzymes catalyzing at least three steps have remained “missing” in many bacteria. In the current work, we have employed comparative genomics via the SEED genomic platform, coupled with experimental verification utilizing Acinetobacter baylyi ADP1, to identify one of the missing enzymes, a new protoporphyrinogen oxidase, the penultimate enzyme in heme biosynthesis. COG1981 was identified by genomic analysis as a candidate protein family for the missing enzyme in bacteria that lacked HemG or HemY, two known protoporphyrinogen oxidases. The predicted amino acid sequence of COG1981 is unlike those of the known enzymes HemG and HemY, but in some genomes, the gene encoding it is found neighboring other heme biosynthetic genes. When the COG1981 gene was deleted from the genome of A. baylyi, a bacterium that lacks both hemG and hemY, the organism became auxotrophic for heme. Cultures accumulated porphyrin intermediates, and crude cell extracts lacked protoporphyrinogen oxidase activity. The heme auxotrophy was rescued by the presence of a plasmid-borne protoporphyrinogen oxidase gene from a number of different organisms, such as hemG from Escherichia coli, hemY from Myxococcus xanthus, or the human gene for protoporphyrinogen oxidase.
INTRODUCTION
With the availability of an increasing number of completed microbial genome sequences, metabolic pathways that were long considered to have been conserved across species have now been found to have missing pieces (enzymes, transporters, etc.) in some organisms (16, 36). Such is the case with microbial tetrapyrrole biosynthesis. By the start of 2011, over 500 sequenced microbial genomes were complete for organisms believed to possess functional heme biosynthetic pathways. Yet most bacteria lacked one or more identifiable genes for enzymes that are necessary to catalyze heme biosynthetic steps (40). The situation for Archaea is even less settled, although a recent bioinformatics-based approach has added significant clarity to the issue (47). This is important, given the fact that hemes are essential biological molecules in virtually all living organisms.
Heme, the iron-containing tetrapyrrole protoporphyrin IX, functions as an enzymatic cofactor with a diverse background, including electron transfer in cytochromes (4), catalysis in catalases and peroxidases (51), diatomic gas transport in hemoglobin (41), activation of oxygen in P450 enzymes (42), and signal molecule recognition (15). In prokaryotes, heme can also serve as an iron source (49), a regulator of pathogenicity and dormancy (18, 48), and a transcriptional regulator for heme-responsive global iron networks (46). The biosynthesis of protoheme from the first committed precursor, δ-aminolevulinic acid (ALA), is relatively well conserved. However, two distinct mechanisms exist for ALA synthesis, the so-called 5-C route from glutamyl tRNA, which is present in plants, most bacteria, and Archaea, and the C-4 route from glycine and succinyl coenzyme A, which is found in the metazoa and a few bacteria (21, 22). However, the remaining seven enzymatic steps are highly conserved in eukaryotes and the entire system is tightly regulated. The pathway is absent in only a few organisms, such as helminths and trypanosomes, where it appears that heme biosynthesis was lost in favor of heme uptake (25, 43). In some instances, eukaryotes lacking the ability to synthesize heme maintain a bacterial endosymbiont to synthesize and provide the necessary heme (12). In prokaryotes, this essential biological pathway is more diverse and less well characterized.
Examination of available bacterial genomes for heme biosynthetic enzymes reveals several noteworthy gaps where recognizable forms of characterized enzymes cannot be located (40). Interestingly, this phenomenon is not limited to one specific taxon but occurs across a diverse set of microorganisms. The first gap occurs during the cyclization of the linear tetrapyrrole, where uroporphyrinogen synthase cannot be identified in some Alphaproteobacteria and Chlamydiaceae. The next missing step is the antepenultimate step, where two distinct enzymes are known to catalyze this reaction (7), i.e., an oxygen-dependent coproporphyrinogen oxidase named HemF (3) and an oxygen-independent, radical S-adenosyl-l-methionine-utilizing coproporphyrinogen dehydrogenase named HemN (29). Some heme-synthesizing bacteria lack both of these proteins, so it is clear that there is an as-yet-undiscovered form of coproporphyrinogen oxidase.
Until recently, there was a third example, where an identified enzyme for the penultimate step, the six-electron oxidation of protoporphyrinogen IX to protoporphyrin IX, was lacking. Some bacteria, such as Myxococcus and Aquifex, possess an oxygen-dependent, membrane-associated enzyme, HemY, which is homologous to eukaryotic protoporphyrinogen oxidase (PPOX; the general abbreviation for protoporphyrinogen oxidase is PPO) (6, 7, 50). Firmicutes and Actinobacteria contain a soluble version of HemY that is different in that it lacks the membrane binding domain (8) and the enteric Gammaproteobacteria employ the oxygen-independent enzyme HemG (20, 44), which is a quinone-dependent flavodoxin with protoporphyrinogen dehydrogenase (PpdH) activity that can be coupled to aerobic or anaerobic respiration (2, 34). The Deltaproteobacteria, such as Desulfovibrio, lack typical PPO activity since they utilize a different pathway for the terminal steps (19, 31). Interestingly, a homolog of either HemG or HemY is absent from many cyanobacteria and nonenteric Gram-negative bacteria, including many pathogenic microbial genera, such as Bordetella, Brucella, Campylobacter, Fusobacterium, Helicobacter, Pseudomonas, and Rickettsia, for example.
The purpose of the present study was to identify and characterize the protein(s) responsible for PPO activity in this broad class of prokaryotes by using a marriage of bioinformatics and experimental techniques. By employing the SEED platform (38), we identified a protein family, uncharacterized at the time, that we predicted may fulfill the “missing” function of PPOX in these taxa. Utilizing the naturally transformable bacterium Acinetobacter baylyi ADP1 (23), which lacks hemG and hemY, we determined that the COG1981 protein family possesses PPO activity in vivo and in vitro.
MATERIALS AND METHODS
Identification of genes for putative bacterial PPO.
The Signature tool allows homology-blind identification of candidate protein families based solely on their phyletic occurrence profile (as described at http://www.nmpdr.org/FIG/wiki/view.cgi/FIG/SigGenes). It requires as input an inclusion and an exclusion set of genomes. The latter is formed of diverse genomes, all of which harbor a clear homolog(s) of a known PPO gene(s) and hence are not expected to contain a novel nonhomologous form of PPO. The inclusion set consists of genomes that encode the majority of heme biosynthetic genes yet lack any known form of PPO and hence are expected to encode an as-yet-undiscovered nonhomologous form of the enzyme (see Table S1 in the supplemental material). The Signature tool identifies and returns protein families represented in the genomes of the inclusion set but absent from genomes of the exclusion set. In order to shorten and optimize the output list of candidate protein families, only the smallest genomes were chosen for the inclusion set, while larger genomes were selected for the exclusion set. After several iterations, an output file was obtained containing only 31 candidate protein families.
Cloning.
For the sequences of the PCR primers used in this study, see Table S2 in the supplemental material. A. baylyi ADP1 hemJ was obtained by PCR of genomic DNA from this organism. The DNA for the open reading frame (ORF) was ligated into the NheI and HindIII sites of pTrcHisA (Invitrogen). Escherichia coli hemG, Myxococcus xanthus hemY, and the human gene for PPOX were cloned as previously described (2, 7, 9). For expression in A. baylyi ADP1, these genes were inserted into the NheI and HindIII sites of pBBR1sp. pBBR1sp was made by inserting the following short sequences into the KpnI and XhoI sites of the broad-host-range cloning vector pBBR1MCS (26): 5′-CAAGGAGCAAGTAATGGCTAGCC-3′ and 3′-CATGGTTCCTCGTTCATTACCGATCGGAGCT-5′. These sequences contain an Acinetobacter ribosomal binding site (AGGA), an ATG start codon, and a unique NheI site (in bold). The unique ApaI site of pBBR1MCS between KpnI and XhoI was abolished in the process and was used for verification of the insertion. The plasmid was confirmed by sequencing by the Georgia Genomics Facility at the University of Georgia.
Construction of hemJ knockout.
The A. baylyi hemJ knockout strain, termed ACN1054, was constructed by a three-step gene-targeting method. First, a DNA sequence containing flanking regions ∼400 bp upstream and ∼800 bp downstream of hemJ was amplified by PCR and cloned into the EcoRI and HindIII sites of pUC19. This plasmid was named pBAC883. Next, a kanamycin resistance cassette was excised from pUI1637 with XbaI and inserted into the unique SpeI site within the hemJ sequence of pBAC883, and the resulting plasmid was named pBAC884. Finally, pBAC884 was introduced into the bacterial chromosome by allelic exchange as previously described (5). The wild-type strain was transformed with linear pBAC884, and recombination events were selected on LB agar containing 10 μg/ml hemin and 30 μg/ml kanamycin. Positive recombination was further screened by plating on medium without hemin. A lack of growth was used to indicate a phenotype auxotrophic for heme. ACN1054 was verified by sequencing of genomic DNA.
PPO assays.
Enzymatic assays were carried out using a continuous fluorimetric assay as previously described (45). Briefly, crude extracts were obtained by sonicating 15 g of cells three times for 30 s on ice in 60 ml solubilization buffer containing 100 mM Tris-morpholinepropanesulfonic acid (MOPS; pH 8.1), 100 mM KCl, and 0.1% Na cholate. The resulting extracts were diluted 1:100 in the same buffer and assayed for PPO activity in reaction mixtures consisting of 10 μl extract, 50 mM NaH2PO4 (pH 8.0), 0.2% (wt/vol) Tween 20, 2.5 mM glutathione, and 25 μM protoporphyrinogen IX. Extract protein concentrations were on the order of 0.1 mg/ml. For soluble and membrane fractions, crude extracts were centrifuged at 100,000 × g and the soluble supernatant fraction was collected. The pelleted membrane fraction was then resuspended in an equivalent volume of solubilization buffer containing 1% Triton X-100 and centrifuged again at 100,000 × g. The resulting supernatant was then collected as the membrane fraction. These extracts were diluted 1:10 before use in reaction mixtures. Protoporphyrinogen IX was generated by reducing protoporphyrin IX (Frontier Scientific, Logan, UT) with an amalgam of 3.5 g sodium and 75 g mercury and then adjusted to neutral pH with 1.0 M Na-MOPS. Product formation was monitored by accumulation of fluorescence at 545 nm using a Synergy HTI plate reader (BioTek, Winooski, VT).
Porphyrin accumulation.
To induce porphyrin accumulation in vivo, ACN1054 cells were grown on LB agar containing 25 μg/ml ALA and 5 μg/ml hemin. After 72 h at 30°C in the dark, plates were examined under UV light and visible red fluorescence could be seen in cells positive for accumulation. For identification and quantification, 1.0 ml of a 5.0-ml 48-h culture of ACN1054 was grown under the same conditions as above and centrifuged at 10,000 rpm for 3 min. The resulting pellet was then suspended in a solution of a 4:1 (vol/vol) ethyl acetate-acetic acid solution for extraction of porphyrins. This suspension was centrifuged at 10,000 × g for 3 min, and the supernatant was collected. Porphyrins were then separated and quantified using high-performance liquid chromatography (HPLC) as previously described (13). The lowest level of detection for any porphyrin in this system is 1 pmol.
Complementation and growth analysis.
The plasmid pBBR1sp containing one of the PPOs listed was transformed into ACN1054 cells from a 5-ml overnight culture in LB broth to which 2.0 mM succinate had been added 30 min prior to the transformation procedure. For transformation, 2.0 μl of each respective vector and 2.0 μl of culture were mixed and spotted onto an LB plate. After 24 h, the resulting growth was transferred to a fresh LB plate containing 10 μg/ml hemin, 20 μg/ml chloramphenicol, and 30 μg/ml kanamycin. Selected colonies arising from this procedure were transferred to LB with both antibiotics but no hemin to verify complementation. Transformation of the isolated cultures was verified by extraction of the original plasmid and identification by restriction digestion.
For growth analysis, 1.0 ml of a culture with an optical density at 600 nm of 1.0 was used to inoculate 100 ml LB medium containing appropriate antibiotics. Flasks were shaken at 37°C, and the culture density was measured hourly for 10 h using a Klett-Summerson colorimeter bearing a number 66 filter.
RESULTS
Identification of HemJ.
Since protein-based searches identified no homologs of HemG or HemY in many genomes of heme-synthesizing prokaryotes, a non-homology-based technique was employed using the SEED genomic platform (http://www.theseed.org) (38). A subsystem encoding the heme and siroheme biosynthetic pathways was constructed as described by Osterman and Begley (37) for the nearly 690 eubacteria with complete and nearly complete genomes available in the SEED database at the time. An updated analysis can be viewed at http://theseed.uchicago.edu/FIG/seedviewer.cgi?page=Subsystems&subsystem=Heme_and_Siroheme _Biosynthesis. From this evaluation, 167 organisms were inferred to be incapable of heme biosynthesis based on their genomic data (with 123 of these incapable of any tetrapyrrole biosynthesis). The heme biosynthetic pathway could be asserted in the remaining 521 eubacteria with sequenced genomes. Four representative species are shown in Fig. S1 in the supplemental material, illustrating several common operational variants of this subsystem. Of the 521 potentially heme-synthesizing bacteria, approximately half (254) lacked an identifiable homolog of hemG or hemY. The NMPDR Signature tool (http://www.nmpdr.org/FIG/wiki/view.cgi/FIG/SigGenes) (33), with a similarity score cutoff of 1e−10 and default parameters, was then utilized to identify hypothetical protein families with the following desired occurrence profile: (i) present only in genomes encoding other enzymes of heme biosynthesis, specifically, the terminal enzymes surrounding PPO; (ii) not found in genomes with known forms of PPO; and (iii) present in nearly all candidate genomes missing hemG and hemY. The inclusion and exclusion sets of genomes required as input for the Signature tool were composed as described in Materials and Methods and shown in Table S1 in the supplemental material. Thirty-one hypothetical protein families with the desired occurrence profile were identified. The list of possible candidates was further shortened by examining the genomic context, as genes associated with the same metabolic pathway or functional complex tend to colocalize in prokaryotic genomes (14, 39). Using the Compare Regions tool of the SEED database, the presence of other known heme biosynthetic genes in the immediate vicinity of each of the 30 candidate gene families was assessed. Only one gene family, encoding COG1981, was found to cluster with either uroporphyrinogen decarboxylase and ferrochelatase genes or the ferrochelatase gene alone. This clustering was seen in 26% of the genomes examined.
The ORF for COG1981 occurs in nearly all eubacterial genomes where a known PPO is absent and is not present in organisms incapable of heme biosynthesis. The COG1981 homologs are found in cyanobacteria and all subdivisions of proteobacteria, except for Deltaproteobacteria. As stated previously, M. xanthus and its relatives contain eukaryotic-protein-like HemY, and others, such as D. gigas, employ a different route to heme that is based upon the siroheme pathway (19, 31). The enterics also lack COG1981, containing HemG instead (2, 20, 30, 44). In addition, COG1981 is present in a few subclasses of Bacteroides, though the majority of these contain HemY. These results led us to select COG1981 for characterization, and the gene was named hemJ (see Fig. S2 in the supplemental material). A previous publication had predicted that the COG1981 protein family was associated with heme synthesis, but no function was assigned by those researchers (17).
hemJ knockout in A. baylyi ADP1.
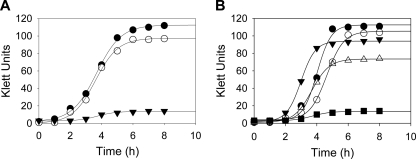
To elucidate the function of HemJ in vivo, we chose to knock out the gene in an organism that contains only hemJ and not hemG or hemY. A. baylyi ADP1 was selected since it possesses only hemJ (ACIAD0878), is naturally competent, and affords ease of genetic manipulation via homology-driven recombination (23). The hemJ gene was disrupted by the introduction of a linear plasmid containing a gene conferring kanamycin resistance. The resulting strain, named ACN1054, was selected for its ability to grow in the presence of kanamycin and hemin. ACN1054 cells do not grow in the absence of hemin, but wild-type growth characteristics are restored by hemin supplementation of the medium (Fig. 1A). This is consistent with the designation of this gene as essential in the complete A. baylyi knockout collection generated by de Berardinis et al. (11).
Fig. 1.
Comparison of A. baylyi ADP1 wild-type and ACN1054 cultures. (A) Growth characteristics of wild-type and ACN1054 mutant A. baylyi ADP1. Shown are data for ACN1054 cultures without added heme (▾) and supplemented with exogenous heme (○) and wild-type cells without heme supplementation (•). (B) Growth analysis of ACN1054 with expression of known PPO enzymes. Shown are data on the growth of ACN1054 cells alone (▪) and with plasmid-encoded expression of human PPOX (•), M. xanthus HemY (○), and E. coli HemG (▵). Data for wild-type A. baylyi ADP1 are shown for comparison (▾).
Complementation of ACN1054.
Functional complementation of heme auxotrophs has proven to be a valuable tool in identifying heme biosynthetic proteins (9, 10, 30, 35). To determine the role of HemJ, we cloned genes encoding several validated PPO enzymes into a newly created A. baylyi active expression vector. HemG from E. coli, HemY from M. xanthus, and human PPOX were selected as candidates for complementation of ACN1054 on the basis that each is a characterized, fully functioning PPO. In addition, HemJ from A. baylyi ADP1 was used as a control. Soluble HemY of the Firmicutes or Actinobacteria was not chosen, since it has been shown that it is not functional in E. coli unless supplied concomitantly with HemH and HemQ (8). Each individual PPO-encoding plasmid was transformed into ACN1054 and rescued heme auxotrophy. Growth analysis further revealed that each plasmid restored growth to wild-type status (Fig. 1B).
To further confirm that the gene under examination possessed PPO activity, two separate approaches were used: (i) examination of intracellular porphyrin accumulation, which is anticipated when PPO activity is not present, and (ii) in vitro assays of crude cell extracts. In other systems, such as E. coli, lack of PPO results in an increase in the protoporphyrin precursors uro-, copro-, and protoporphyrinogen (44). Accumulation of these intermediates is enhanced by the addition of ALA, the initial precursor compound in heme biosynthesis.
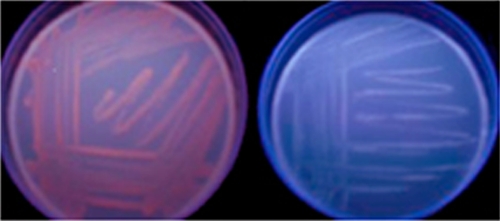
When supplemented with ALA, ACN1054 cells exhibited an orange-red fluorescence under UV illumination after 24 h of growth. This fluorescence increased up to approximately 72 h of incubation (Fig. 2). HPLC was used to separate and quantify oxidized samples obtained from cellular acid extracts. Data presented in Table 1 demonstrate an accumulation of protoporphyrin, coproporphyrin, uroporphyrin, and a heptacarboxyl porphyrin. The latter is an intermediate formed during the enzymatic decarboxylation of uroporphyrin. In comparison, quantifiable amounts of porphyrins were not found in extracts from wild-type cells supplemented with ALA in the same fashion.
Fig. 2.
Porphyrin accumulation in mutant and wild-type A. baylyi cells. After stimulation of the heme biosynthetic pathway, porphyrin accumulation is clearly seen in ACN1054 cells (left) but not in wild-type cells (right). Fluorescence is visible with exposure to long-wavelength UV light.
Table 1.
Porphyrin content analysis of cell extracts from A. baylyi ADP1 wild-type and ACN1054 strains
| Strain | Avg porphyrin concn (nM) ± SDa |
||||
|---|---|---|---|---|---|
| Total | Uroporphyrin | Heptacarboxyl porphyrin | Coproporphyrin | Protoporphyrin | |
| ACN1054 | 4,802 ± 318 | 559 ± 81 | 81 ± 43 | 2,429 ± 149 | 1,734 ± 135 |
| Wild type | BDb | BD | BD | BD | BD |
Quantitation of porphyrins was done as described in Materials and Methods. Samples for analysis were obtained from 1 ml of 48-h A. baylyi ADP1 cultures grown in the presence of ALA.
BD, below detection level.
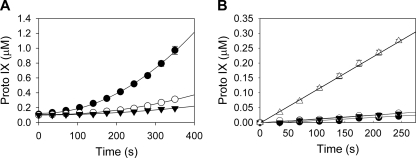
In vitro PPO activity was determined in assays performed with crude cell lysates of wild-type and ACN1054 cells (Fig. 3A). No PPO activity was present in the mutant. In vitro assays were also conducted using soluble and membrane fractions of wild-type A. baylyi extracts. Analysis of the HemJ amino acid sequence by the program TMHMM (27) yields a firm prediction for a membrane-bound protein. However, no measurable activity was found for isolated membrane or soluble fractions. Activity was seen only when the soluble and membrane fractions were present together (Fig. 3B).
Fig. 3.
PPO activity in Acinetobacter crude cell extracts as measured by continuous fluorescence spectroscopy. (A) In vitro PPO assay results are shown for wild-type (•) and ACN1054 (○) extracts, as well as an extract-free control (▾). (B) PPO activity in membrane and soluble fractions of Acinetobacter extracts. Activity levels in the soluble fraction (○), the membrane fraction (▾), an extract-free control (•), and the membrane and soluble fractions together (▵) are shown. Approximately 0.1 mg/ml protein was present in each extract. Proto, protoporphyrin.
DISCUSSION
Over the past decades, researchers have employed heme synthesis mutants of E. coli to identify enzymes of heme biosynthesis via complementation. This approach, while straightforward and robust, can work only if the sought-after enzyme is functional in E. coli. Another approach is to rely upon specific biochemical characteristics of the sought-after protein. Kato et al. recently identified HemJ in Synechocystis in this fashion by introducing plant-derived acifluorfen-sensitive PPO into cells and then screening mutants for acifluorfen sensitivity (24). This approach is suited for the identification of novel enzymes where a second, well-characterized enzyme exists that catalyzes the reaction of interest and for which a novel inhibitor also exists, but it does not easily lend itself to all enzyme systems. A third approach is to employ bioinformatics to initially identify genes of potential interest and then experimentally test these. This technique has a broad range of applications in biological systems for which significant genomic data exist. It requires little knowledge of actual function to identify novel components, relying instead on gene context and functional complementation. That was the approach we employed herein to identify hemJ.
We utilized the SEED (http://www.theseed.org) platform, in which a characterized biological system from one species can be accurately projected to other species within the database based on sequence similarity, nonhomologous contextual clues, and experimental evidence of enzymatic function (38). From this, “missing” steps become readily apparent as “gaps” for a given genome within the displayed subsystem. Once gaps are identified, the tools for comparative genome analysis within the SEED database facilitate the identification of candidate gene families that can then be experimentally verified (11, 15, 44, 45). As detailed above, this led to our identification of COG1981 as a strong candidate for the missing PPO, which was then experimentally verified as HemJ.
With HemG, HemY, and HemJ now identified, nearly all eubacterial genomes in the SEED database inferred to encode functional heme biosynthetic pathways have identifiable enzymes to catalyze the conversion of protoporphyrinogen IX to protoporphyrin IX (663 out of 671). The majority (46%) encode HemJ, with HemY or HemG present in 32% and 23%, respectively. The only two clear exceptions appear to be Tropheryma and Desulfovibrionaceae. For Desulfovibrionaceae, the explanation is simple. An alternative, ancient pathway for the biosynthesis of heme has been shown to be operational in the genus Desulfovibrio (19). Lobo et al. (31) presented evidence that protoheme biosynthesis in Desulfovibrio vulgaris occurs by branching off the common tetrapyrrole route not at the level of precorrin 2, as was hypothesized by Ishida et al. (19), but at the level of sirohydrochlorin. In this scenario, the activities of classic coproporphyrinogen oxidase, PPO, and possibly ferrochelatase are not involved in the terminal steps of protoheme synthesis. Instead, the D. vulgaris homologs of the NirJ and NirD enzymes participate in this novel transformation (31, 47).
Tropheryma whipplei, the causative agent of Whipple's disease, is an obligate human pathogen that has not been cultured in the absence of living eukaryotic cells (28). Compared to free-living species of the same order (Actinomycetales), this organism has undergone drastic genome reduction (to merely ∼930 kbp), resulting in a lack of key biosynthetic pathways and a reduced capacity for energy metabolism (1). The absence of a gene encoding any of the 3 known forms of PPO from the genome of this highly host-adapted organism might indicate a recent onset of pathway decay and the reliance on the host as a source of heme.
Kato et al. (24) noted that they could not identify hemJ, hemG, or hemY in Archaea, Actinobacteria, Acidibacteria, Deinococcus, Fusobacteria, Spirochaetes, and Thermotogae and posited that these organisms may contain other, yet-to-be-identified, novel PPO enzymes. This is clearly true in Archaea, where, with limited exceptions, the organisms do not possess HemF/N, HemG/Y/J, or HemH as the terminal pathway enzymes essential for protoheme biosynthesis but instead utilize a different pathway from uroporphyrinogen to heme that involves S-adenosyl-L-methionine-dependent uroporphyrinogen III methyltransferase, precorrin-2 dehydrogenase, and the ahb-nir genes (47). While some Spirochaetes contain the siroheme biosynthetic pathway but not the terminal enzymes for protoheme synthesis, the remaining Spirochaetes and Thermotogae contain no tetrapyrrole biosynthetic enzymes. However, it should be noted that hemY is present in the genomes of Acidobacteria and Actinobacteria and has been expressed and characterized from Actinobacteria (8). The examination of the remaining groups using the SEED subsystem readily explains the absence of hemG, hemY, or hemJ in these organisms.
The present work does not identify whether HemJ is a stand-alone PPO enzyme or a subunit of a larger complex. Although it is clear that HemJ is an essential protein for the cellular oxidation of protoporphyrinogen to protoporphyrin, our observation that both the membrane fraction containing HemJ and the soluble cellular fractions are required for measurable activity in vitro argues for an additional component. Whether this additional component is an as-yet-unidentified protein that did not manifest itself in our genome screen or simply a redox cofactor is something that requires additional study. Interestingly, Kato et al. (24) reported that a partially purified HemJ-maltose binding protein fusion from Rhodobacter sphaeroides could catalyze the oxidation of protoporphyrinogen to protoporphyrin without any added cofactors. This led them to speculate that HemJ could utilize molecular oxygen for the reaction. However, no description or evidence of the level of protein purity was presented and no UV/visible spectrum was presented that would have identified chromophoric cofactors such as flavin mononucleotide or flavin adenine dinucleotide. During our studies, we cloned and attempted to express and purify hemJ from A. baylyi, Pseudomonas aeruginosa, and R. sphaeroides using a variety of different methods, including maltose binding protein fusions. However, in all our efforts, the membrane-associated enzyme was expressed poorly and was not purified to a level that would allow confident kinetic studies. Since neither Kato et al. nor we managed to obtain pure enzyme, a comparison of our results is not possible. However, the specific activity of our combined membrane and soluble fractions is greater than a factor of 10 higher than their published data for purified enzyme.
In addition to the use of SEED to identify target genes, the current work also has additional impact in that we have utilized A. baylyi ADP1 rather than E. coli in our gene target verification protocol. While our choice was in part driven by the fact that A. baylyi ADP1 lacks hemG and hemY, we were also attracted to the high natural transformability and ease with which one can produce gene mutations, deletions, and homologous recombinations (23). Interestingly, had we chosen E. coli ΔhemG for complementation with the sought-after HemJ protein, we would have not been successful, since we discovered that HemJ does not function in E. coli (data not shown).
HemJ represents a novel drug target for many human pathogens. Acifluoren, a potent herbicide, is already a widely used plant PPO inhibitor (32), and it is imaginable that HemJ could present a high-specificity target in those organisms that possess it. Many clinically significant pathogens and opportunists that have HemJ are among those organisms that have evolved multiantibiotic resistance, and for these, HemJ presents a new antimicrobial target. The prokaryotic heme biosynthetic pathway has previously been proposed as a drug target for symbiont-containing parasites (12), and HemJ's absence from most human flora suggests that it could be a prime candidate.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grant DK32303 (H.A.D.), NSF grant MCB-0920619 (E.L.N.), and NIAID contract HHSN266200400042C (S.G.).
We acknowledge D. Vavilin for generous sharing of unpublished data related to HemJ of Synechocystis and T. A. Dailey for editing of the manuscript.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 3 June 2011.
REFERENCES
- 1. Bentley S. D., et al. 2003. Sequencing and analysis of the genome of the Whipple's disease bacterium Tropheryma whipplei. Lancet 361:637–644 [DOI] [PubMed] [Google Scholar]
- 2. Boynton T. O., Daugherty L. E., Dailey T. A., Dailey H. A. 2009. Identification of Escherichia coli HemG as a novel, menadione-dependent flavodoxin with protoporphyrinogen oxidase activity. Biochemistry 48:6705–6711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Breckau D., Mahlitz E., Sauerwald A., Layer G., Jahn D. 2003. Oxygen-dependent coproporphyrinogen III oxidase (HemF) from Escherichia coli is stimulated by manganese. J. Biol. Chem. 278:46625–46631 [DOI] [PubMed] [Google Scholar]
- 4. Chance B. 1972. The nature of electron transfer and energy coupling reactions. FEBS Lett. 23:3–20 [DOI] [PubMed] [Google Scholar]
- 5. Collier L. S., Gaines III G. L., Neidle E. L. 1998. Regulation of benzoate degradation in Acinetobacter sp. strain ADP1 by BenM, a LysR-type transcriptional activator. J. Bacteriol. 180:2493–2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Corradi H. R., et al. 2006. Crystal structure of protoporphyrinogen oxidase from Myxococcus xanthus and its complex with the inhibitor acifluorfen. J. Biol. Chem. 281:38625–38633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dailey H. A., Dailey T. A. 1996. Protoporphyrinogen oxidase of Myxococcus xanthus. Expression, purification, and characterization of the cloned enzyme. J. Biol. Chem. 271:8714–8718 [DOI] [PubMed] [Google Scholar]
- 8. Dailey T. A., et al. 2010. Discovery and characterization of HemQ: an essential heme biosynthetic pathway component. J. Biol. Chem. 285:25978–25986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dailey T. A., Dailey H. A. 1996. Human protoporphyrinogen oxidase: expression, purification, and characterization of the cloned enzyme. Protein Sci. 5:98–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dailey T. A., Dailey H. A., Meissner P., Prasad A. R. 1995. Cloning, sequence, and expression of mouse protoporphyrinogen oxidase. Arch. Biochem. Biophys. 324:379–384 [DOI] [PubMed] [Google Scholar]
- 11. de Berardinis V., et al. 2008. A complete collection of single-gene deletion mutants of Acinetobacter baylyi ADP1. Mol. Syst. Biol. 4:174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Foster J., et al. 2005. The Wolbachia genome of Brugia malayi: endosymbiont evolution within a human pathogenic nematode. PLoS Biol. 3:e121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Franklin M. R., Phillips J. D., Kushner J. P. 1997. Cytochrome P450 induction, uroporphyrinogen decarboxylase depression, porphyrin accumulation and excretion, and gender influence in a 3-week rat model of porphyria cutanea tarda. Toxicol. Appl. Pharmacol. 147:289–299 [DOI] [PubMed] [Google Scholar]
- 14. Galperin M. Y., Koonin E. V. 2000. Who's your neighbor? New computational approaches for functional genomics. Nat. Biotechnol. 18:609–613 [DOI] [PubMed] [Google Scholar]
- 15. Gilles-Gonzalez M. A. 2001. Oxygen signal transduction. IUBMB Life 51:165–173 [DOI] [PubMed] [Google Scholar]
- 16. Hanson A. D., Pribat A., Waller J. C., de Crecy-Lagard V. 2010. ‘Unknown’ proteins and ‘orphan’ enzymes: the missing half of the engineering parts list—and how to find it. Biochem. J. 425:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Harrington E. D., et al. 2007. Quantitative assessment of protein function prediction from metagenomics shotgun sequences. Proc. Natl. Acad. Sci. U. S. A. 104:13913–13918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ioanoviciu A., Meharenna Y. T., Poulos T. L., Ortiz de Montellano P. R. 2009. DevS oxy complex stability identifies this heme protein as a gas sensor in Mycobacterium tuberculosis dormancy. Biochemistry 48:5839–5848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ishida T., et al. 1998. A primitive pathway of porphyrin biosynthesis and enzymology in Desulfovibrio vulgaris. Proc. Natl. Acad. Sci. U. S. A. 95:4853–4858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jacobs N. J., Jacobs J. M. 1977. Evidence for involvement of the electron transport system at a late step of anaerobic microbial heme synthesis. Biochim. Biophys. Acta 459:141–144 [DOI] [PubMed] [Google Scholar]
- 21. Jahn D., Heinz D. W. 2009. Biosynthesis of 5-aminolevulinic acid, p. 29–42 In Warren M. J., Smith A. G. (ed.), Tetrapyrroles: birth, life, and death. Landes Bioscience, Austin, TX [Google Scholar]
- 22. Jordan P. M. 1990. The biosynthesis of 5-aminolevulinic acid and its transformation in coproporphyrinogen in animals and bacteria, p. 55–121 In Dailey H. A. (ed.), Biosynthesis of heme and chlorophylls. McGraw-Hill, New York, NY [Google Scholar]
- 23. Juni E., Janik A. 1969. Transformation of Acinetobacter calco-aceticus (Bacterium anitratum). J. Bacteriol. 98:281–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kato K., Tanaka R., Sano S., Tanaka A., Hosaka H. 2010. Identification of a gene essential for protoporphyrinogen IX oxidase activity in the cyanobacterium Synechocystis sp. PCC6803. Proc. Natl. Acad. Sci. U. S. A. 107:16649–16654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Korený L., Lukes J., Obornik M. 2010. Evolution of the haem synthetic pathway in kinetoplastid flagellates: an essential pathway that is not essential after all? Int. J. Parasitol. 40:149–156 [DOI] [PubMed] [Google Scholar]
- 26. Kovach M. E., Phillips R. W., Elzer P. H., Roop II R. M., Peterson K. M. 1994. pBBR1MCS: a broad-host-range cloning vector. Biotechniques 16:800–802 [PubMed] [Google Scholar]
- 27. Krogh A., Larsson B., von Heijne G., Sonnhammer E. L. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567–580 [DOI] [PubMed] [Google Scholar]
- 28. La Scola B., et al. 2001. Description of Tropheryma whipplei gen. nov., sp. nov., the Whipple's disease bacillus. Int. J. Syst. Evol. Microbiol. 51:1471–1479 [DOI] [PubMed] [Google Scholar]
- 29. Layer G., et al. 2006. The substrate radical of Escherichia coli oxygen-independent coproporphyrinogen III oxidase HemN. J. Biol. Chem. 281:15727–15734 [DOI] [PubMed] [Google Scholar]
- 30. Lermontova I., Kruse E., Mock H. P., Grimm B. 1997. Cloning and characterization of a plastidal and a mitochondrial isoform of tobacco protoporphyrinogen IX oxidase. Proc. Natl. Acad. Sci. U. S. A. 94:8895–8900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lobo S. A., Brindley A., Warren M. J., Saraiva L. M. 2009. Functional characterization of the early steps of tetrapyrrole biosynthesis and modification in Desulfovibrio vulgaris Hildenborough. Biochem. J. 420:317–325 [DOI] [PubMed] [Google Scholar]
- 32. Matringe M., Camadro J. M., Labbe P., Scalla R. 1989. Protoporphyrinogen oxidase as a molecular target for diphenyl ether herbicides. Biochem. J. 260:231–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. McNeil L. K., et al. 2007. The National Microbial Pathogen Database Resource (NMPDR): a genomics platform based on subsystem annotation. Nucleic Acids Res. 35:D347–D353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Möbius K., et al. 2010. Heme biosynthesis is coupled to electron transport chains for energy generation. Proc. Natl. Acad. Sci. U. S. A. 107:10436–10441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Narita S., et al. 1996. Molecular cloning and characterization of a cDNA that encodes protoporphyrinogen oxidase of Arabidopsis thaliana. Gene 182:169–175 [DOI] [PubMed] [Google Scholar]
- 36. Osterman A., Overbeek R. 2003. Missing genes in metabolic pathways: a comparative genomics approach. Curr. Opin. Chem. Biol. 7:238–251 [DOI] [PubMed] [Google Scholar]
- 37. Osterman A. L., Begley T. P. 2007. A subsystems-based approach to the identification of drug targets in bacterial pathogens. Prog. Drug Res. 64:131, 133–170. [DOI] [PubMed] [Google Scholar]
- 38. Overbeek R., et al. 2005. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 33:5691–5702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Overbeek R., Fonstein M., D'Souza M., Pusch G. D., Maltsev N. 1999. The use of gene clusters to infer functional coupling. Proc. Natl. Acad. Sci. U. S. A. 96:2896–2901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Panek H., O'Brian M. R. 2002. A whole genome view of prokaryotic haem biosynthesis. Microbiology 148:2273–2282 [DOI] [PubMed] [Google Scholar]
- 41. Perutz M. 1942. X-ray analysis of haemoglobin. Nature 149:491–494 [DOI] [PubMed] [Google Scholar]
- 42. Raikhman L. M., Annaev B., Mamedniiazov O. N., Rozantsev E. G. 1973. Molecular structure and mechanism of functioning of cytochrome P-450. Biofizika 18:228–234 [PubMed] [Google Scholar]
- 43. Rao A. U., Carta L. K., Lesuisse E., Hamza I. 2005. Lack of heme synthesis in a free-living eukaryote. Proc. Natl. Acad. Sci. U. S. A. 102:4270–4275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Săsărman A., et al. 1979. Mapping of a new hem gene in Escherichia coli K12. J. Gen. Microbiol. 113:297–303 [DOI] [PubMed] [Google Scholar]
- 45. Shepherd M., Dailey H. A. 2005. A continuous fluorimetric assay for protoporphyrinogen oxidase by monitoring porphyrin accumulation. Anal. Biochem. 344:115–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Singleton C., et al. 2010. Heme-responsive DNA binding by the global iron regulator Irr from Rhizobium leguminosarum. J. Biol. Chem. 285:16023–16031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Storbeck S., et al. 2010. A novel pathway for the biosynthesis of heme in Archaea: genome-based bioinformatic predictions and experimental evidence. Archaea 2010:175050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Torres V. J., et al. 2007. A Staphylococcus aureus regulatory system that responds to host heme and modulates virulence. Cell Host Microbe 1:109–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wandersman C., Delepelaire P. 2004. Bacterial iron sources: from siderophores to hemophores. Annu. Rev. Microbiol. 58:611–647 [DOI] [PubMed] [Google Scholar]
- 50. Wang K. F., Dailey T. A., Dailey H. A. 2001. Expression and characterization of the terminal heme synthetic enzymes from the hyperthermophile Aquifex aeolicus. FEMS Microbiol. Lett. 202:115–119 [DOI] [PubMed] [Google Scholar]
- 51. Zámocký M., Furtmuller P. G., Obinger C. 2010. Evolution of structure and function of class I peroxidases. Arch. Biochem. Biophys. 500:45–57 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.