Abstract
During filamentous fungus development, multinucleated hyphae employ a system for long-range nuclear migration to maintain an equal nuclear density. A decade ago the microtubule motor dynein was shown to play a central role in this process. Previous studies with Ashbya gossypii revealed extensive bidirectional movements and bypassings of nuclei, an autonomous cytoplasmic microtubule (cMT) cytoskeleton emanating from each nucleus, and pulling of nuclei by sliding of cMTs along the cortex. Here, we show that dynein is the sole motor for bidirectional movements and bypassing because these movements are concomitantly decreased in mutants carrying truncations of the dynein heavy-chain DYN1 promoter. The dynactin component Jnm1, the accessory proteins Dyn2 and Ndl1, and the potential dynein cortical anchor Num1 are also involved in the dynamic distribution of nuclei. In their absence, nuclei aggregate to different degrees, whereby the mutants with dense nuclear clusters grow extremely long cMTs. As in budding yeast, we found that dynein is delivered to cMT plus ends, and its activity or processivity is probably controlled by dynactin and Num1. Together with its role in powering nuclear movements, we propose that dynein also plays (directly or indirectly) a role in the control of cMT length. Those combined dynein actions prevent nuclear clustering in A. gossypii and thus reveal a novel cellular role for dynein.
INTRODUCTION
Migration of nuclei in multinucleated hyphae of filamentous fungi is a fascinating but also a very complex dynamic process which is far from being understood. The efficient polar growth of hyphae continuously generates new cytoplasmic space. Hyphae maintain a relatively even distribution of nuclei by adjusting the migration and the division of nuclei to the hyphal growth speed. Important components of this control system were first identified as mutants exhibiting nuclear distribution defects and were termed nud in Aspergillus nidulans and ropy in Neurospora crassa. Most of the affected genes coded for components of the microtubule motor dynein and its activating complex, dynactin (27, 30, 42, 43). Dynein and dynactin components were found to form comet-like structures at the plus end (+end) of cytoplasmic microtubules (cMT) in N. crassa and A. nidulans, thereby indicating that cMTs and very likely the dynamics of cMTs are important for nuclear migration (10, 27, 45).
Dynein was also shown to be essential for functional distribution of nuclei in Nectria haematococca, Ashbya gossypii, and the dimorphic fungus Ustilago maydis (1, 13, 37). However, surprising phenotypic differences were observed, questioning the existence of a single mechanism controlling nuclear distribution in fungi. In the absence of dynein, nuclei remain in the germ bubble of A. nidulans, where they form clusters, whereas in A. gossypii, all nuclei move out of the germ bubble and form clusters at hyphal tips (1, 42). Hyphae lacking dynein generate more stable or longer cMTs in A. nidulans and A. gossypii, whereas fewer and shorter cMTs form in N. haematococca (1, 10, 13). An additional complication is the double role of dynein in filamentous fungi because this motor mediates nuclear migration as well as organelle transport (34, 35, 40). The only exception seems to be A. gossypii, because in this fungus, cMTs, as in budding yeast, are involved only in nuclear migration and not in transport of other organelles or vesicles (7, 17).
Our initial in vivo studies on long-range nuclear migration in A. gossypii had shown that nuclei exert autonomous forward and backward movements during all nuclear cycle stages, including mitosis, and that nuclei frequently bypass adjacent nuclei (1, 7). Thus, forces acting in opposite directions on nuclei result in a dynamic distribution, with an average distance between nuclei of 5 to 6 μm. Bidirectional nuclear migration was also demonstrated in A. nidulans, and it probably also exists in young hyphae of N. crassa, as inferred from the observed shorter and longer distances between nuclei, whereas nuclei in the much wider and very fast-growing mature hyphae of N. crassa migrate in one direction, i.e., with the cytoplasmic flow (27, 31, 32).
Further studies with A. gossypii revealed that the spindle pole body (SPB) of each nucleus initiates a microtubule cytoskeleton consisting of 4 to 6 short and long cMTs. The long cMTs emanate in opposite directions with respect to the growth axis and often extend beyond adjacent nuclei. Those studies also showed that mutants growing only short cMTs still perform bidirectional movements but lack nuclear bypassing events, that mutants with no or very short cMTs lack both bidirectional movements and bypassing of nuclei, and that mutants with slightly increased lengths of cMTs show higher frequencies of bidirectional movements and bypassing of nuclei (8, 18, 19). Knowing that 4 to 6 cMTs emanate from each nucleus in wild-type (WT) hyphae, it is difficult to understand how nuclei attached to short and long cMTs pointing to different directions can bypass each other without getting entangled in the narrow hyphal tube (the diameter of which is only twice the diameter of nuclei). It is possible that they sometimes do and that a rescue system operates to untangle those nuclei to prevent larger nuclear aggregates to form. We hypothesize that a combination of bidirectional forces alternately acting on nuclei and a length control for cMTs dissolves entangled nuclei, thus preventing the formation of nuclear aggregates in A. gossypii.
Since loss of dynein activity leads to formation of nuclear clusters in filamentous fungi, including A. gossypii, the dynein/dynactin complex is most likely an important component of such a rescue system. This huge multisubunit complex acts in all eukaryotes as a minus-end (–end)-directed MT motor. It consists of two heavy chains with motor activity and MT-binding capacity, several intermediate and light chains, and accessory proteins for its activation (14). In the yeast Saccharomyces cerevisiae, the dynein pathway has a very specific cellular function which is unlikely to be conserved in multinucleated hyphae. It acts together with the Kar9 pathway to position the nucleus at the bud neck and direct the pulling of one daughter nucleus into the bud (24, 29). The pulling function of dynein is restricted to anaphase in S. cerevisiae. According to the current model, which may also be valid for filamentous fungi, these pulling forces depend on the lateral contacts of cMTs with the cortex mediated by their interactions with the minus-end motor dynein, which is attached to the cell cortex via the anchor protein Num1 (6, 20, 29, 38).
In this paper, we first aimed at identifying the origin of the forces responsible for bidirectional movement and bypassing of nuclei in A. gossypii. Then, we tested whether hyphae lacking dynactin components or other dynein accessory proteins show the same or less severe degrees of nuclear aggregation than that originally observed in the dynein deletion mutant. Next, we monitored the localization of fluorescently labeled dynein expressed from its endogenous promoter to see potential alterations between the wild type and different mutants, including a mutant of the presumptive cortical anchor Num1. Finally, we compared the sizes of cMTs in mutants with decreased dynein expression and hyphae lacking dynein activity to try to differentiate between effects of overly long cMTs and a lowered level of dynein in formation of nuclear aggregates. The individual results will be compared mainly with the dynein/dynactin system of budding yeast because A. gossypii carries a budding yeast-like genome (4) and because we want to demonstrate the evolution of the dynein pathway, as it is known from studies in S. cerevisiae, to an essential cellular function in multinucleated hyphae.
MATERIALS AND METHODS
A. gossypii media and growth conditions.
A. gossypii media and culturing protocols are described in references 2 and 41. Strains were grown in Ashbya full medium (AFM; 1% Bacto peptone, 1% yeast extract, 2% glucose, and 0.1% myo-inositol) at 30°C. Transformants derived from A. gossypii reference strains were selected on AFM plates containing 200 mg/ml of G418/Geneticin (ForMedium Ltd., England) or 50 μg/ml ClonNAT (Werner BioAgents, Germany). To test the effect of benomyl on growth, AFM plates were prepared by adding 33 μM benomyl (Sigma-Aldrich) (dissolved in dimethyl sulfoxide [DMSO]) directly into the medium before pouring it onto the plates. A total of 15 μg/ml of nocodazole (Sigma-Aldrich) (dissolved in DMSO) was directly added to AFM liquid cultures, and cells were monitored on microscopy slide 15 to 30 min after treatment.
Strain construction.
A. gossypii transformation protocols are described in references 2 and 41. All strains constructed in this study were derived from reference strains expressing either a histone H4-green fluorescent protein (GFP) fusion (ASG46 strain, ADE2-HHF1-yeGFP) (7), an exogenous GFP-Tubulin1 fusion (ADE2-GFP-AgTUB1) (19), or the BIK1-Cherry fusion (8). A. gossypii deletion mutants were made by a PCR-based one-step gene targeting approach (41). PCR were performed using standard methods with Taq polymerase obtained from Roche, and oligonucleotides were synthesized at Microsynth (Balgach, Switzerland). Oligonucleotide primers are listed in Table S1 in the supplemental material.
For gene deletions, A. gossypii cells were transformed with PCR products amplified with the pAG140 (GEN3) or pAG100 (NAT) templates and the “gene name”-del5/del3 oligonucleotide pairs, which contained 45-bp homology upstream and downstream of the open reading frames (ORFs). The primary transformation produces heterokaryon cells, which have a mixture of “wild-type” and transformed nuclei. Transformed heterokaryons were verified with oligonucleotide pairs “gene name”-VER5/G2.2 and “gene name”-VER3/G3.3 for GEN3 cassettes (“gene name”-VER5/V2*NAT1 and “gene name”-VER3/V3*NAT1 for NAT cassettes) (see Table S1 in the supplemental material). For the DYN1 promoter truncations, cells were transformed with PCR products obtained with the pAG140 plasmid and the oligonucleotide pairs DYN1-180-NS1/DYN1PR-DELF2 (only the 180 bp upstream of the ATG were retained; the sequence from −673 to −181 bp was replaced by the GEN3 cassette) and DYN1-130-NS1/DYN1PR-DELF2 (only the 130 bp upstream of the ATG were retained; the sequence from −673 to −131 bp was replaced by GEN3). The correct integration of the two cassettes was then checked by PCR with the same primer pairs, DYN1-PROM-I2/DYN1-PROM-G4 and DYN1-PROM-I1/DYN1-PROM-G1 (for the absence of a WT promoter) and G2.1/DYN1-PROM-G4 and G3.3/DYN1-PROM-G1 (for the presence of the deletion cassette). Three homokaryons (obtained after sporulation of three independent verified transformants) were characterized for each mutant.
For the Dyn1-TandemTomato (tdTomato) fusion, a tdTomato-NAT-tagging cassette was produced by PCR using, as a template, the pAGT213 plasmid (15) and the primers DYN1-TAG3 and DYN1-TAG5 (homologous to the 45 bp upstream and downstream of the AgDyn1 stop codon, respectively). The resulting PCR product was cotransformed into yeast with the pAG1011 plasmid (encoding the C-terminal part of Dyn1). After verification of the fusion by sequencing and digestion with ClaI/BglII, the fragment of DNA showing long regions of homology with the 3′ end of Dyn1 was transformed into GFP-Tub1 A. gossypii cells. Verifications of the heterokaryons and homokaryons were performed as described above with the primer pairs DYN1-VER3/V3*NAT1.
Immunofluorescence and time-lapse microscopy.
We used an Axioplan 2 imaging microscope equipped with a Plan-Apochromat 100× objective/1.40-numerical-aperture oil differential interference contrast (DIC), a Plan-Apochromat 63× objective/1.40-numerical-aperture oil DIC (Carl Zeiss AG, Feldbach, Switzerland), and appropriate filters (Zeiss and Chroma Technology, Brattleboro, VT). The light source used for fluorescence microscopy was a Polychrome V monochromator (TILL Photonics GmbH, Gräfelfing, Germany). Images were acquired at room temperature using a CoolSNAP HQ cooled charge-coupled-device camera (Photometrics, Tucson, AZ) with MetaMorph 6.2r6 software (Molecular Devices Corp., Downingtown, PA). The distance between two planes in stack acquisitions was set to 1 μm for H4-GFP movies and 0.3 μm for anti-tubulin and actin stainings. Brightness and contrast were adjusted using MetaMorph's “scale image” command. Stacks were deconvolved with MetaMorph's “2-D deconvolution” module and flattened by maximum projection with the “stack arithmetic” function. The Bik1-Cherry GFP-Tub1 and Dyn1-TdTomato GFP-Tub1 movies were one Z-plane movies treated with Flatten Background/Kernel/Equalize light functions. Images were colored and overlaid using MetaMorph's “overlay images” command. Time-lapse picture series were processed as described above and converted into QuickTime MPEG-4 movies (QuickTime Player Pro; Apple Inc.). Immunofluorescence stainings were performed as described previously (2, 7). Rat anti-alpha-tubulin (YOL1/34; Serotec, Oxford, United Kingdom) was used at a 1:50 dilution, and Alexa Fluor 568 goat anti-rat IgG (Invitrogen, Carlsbad, CA) was used at a 1:200 dilution. For time-lapse acquisition, small pieces of 2-day-old mycelium were cultured on agarose slides as previously described (19).
RESULTS
Hyphae lacking Kar9 and Bim1 show even nuclear distributions.
A strong nuclear clustering phenotype was observed in hyphae lacking the dynein heavy chain Dyn1. Since all nuclei in germlings of this mutant clustered in the tip of the first emerging hypha, it was anticipated that dynein is the motor responsible for retrograde movements, while another motor was responsible for the tip-directed movements (1). In an attempt to identify this motor, all six kinesin genes of A. gossypii were deleted, but none of the deletions showed nuclear clustering (C. Alberti-Segui, unpublished results).
We then asked whether the A. gossypii Kar9 (AgKar9) protein could be responsible for the tip-directed nuclear movements because of the function of its S. cerevisiae homolog. Indeed, in budding yeast, Kar9 specifically accumulates at the bud-proximal SPB and at cMTs emerging from it to ensure that only this SPB will move into the bud later, during anaphase. This bud tip-directed guidance of cMTs also depends on Bim1, the myosin motor Myo2, and actin cables. In the current model, Kar9 binds to cMT +ends via Bim1. The Kar9-loaded MTs are then pulled toward the bud tip along actin cables due to the interaction of Kar9 with Myo2 (12, 24). In A. gossypii, actin cables emerge at hyphal tips and are oriented along the polarity axis to allow directed transport of secretory vesicles (16). We hypothesized that Kar9 and Bim1 may be involved in tip-directed (forward) nuclear movements via a mechanism similar to that in budding yeast. AgKar9 is 31.6% identical to S. cerevisiae Kar9 (ScKar9) and contains an additional 158-amino-acid (aa) C-terminal extension. A. gossypii Bim1 (AgBim1) shares 55.4% identity with its budding yeast orthologue. We constructed Agkar9Δ and Agbim1Δ deletions in a histone H4-GFP background and studied the effect of the deletions on nuclear dynamics and distribution.
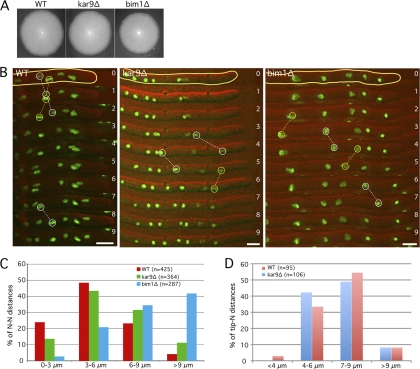
As shown in Fig. 1A, the deletions of KAR9 and BIM1 only slightly reduced the radial growth rate (97% and 90% of that of the WT [H4-GFP], respectively). We performed 1-min-interval time-lapse movies with WT and mutant hyphae to quantify the ability of nuclei to oscillate and bypass each other. These cMT-dependent movements are superimposed on the overall nuclear migration with the cytoplasmic stream, which, for nuclei close to the hyphal tip, is similar to the growth speed of the hyphae (19). For our measurements, we defined “forward events” (faster than the cytoplasmic stream) and “backward events” (against the cytoplasmic stream) as follows. Forward movements during 1 min with at least twice the speed of the hyphal tip were counted as forward events. Backward movements during 1 min with at least half the speed of the hyphal tip were counted as backward events. This strict definition was also previously employed (8). The 5 nuclei closest to the hyphal tips were tracked for 30 min in 7 different WT and mutant hyphae (Fig. 1B and Table 1). We also measured the distances between neighboring nuclei in the WT, kar9Δ, and bim1Δ strains, which are presented as histograms of four size ranges (Fig. 1C).
Fig. 1.
The absence of Kar9 and Bim1 does not significantly affect maximal hyphal growth and nuclear movements. (A) Radial growth of wild-type, kar9Δ, and bim1Δ strains after 5 days at 30°C on full medium (AFM). (B) Nuclear dynamics in wild-type, kar9Δ, and bim1Δ hyphae with H4-GFP-labeled nuclei. Images of selected time-lapse movies are presented (green, H4-GFP; red, DIC). Yellow circles show examples of backward nuclear movements, whereas white circles represent forward events. Time is indicated in minutes. Bars, 5 μm. (C) Dynamic distributions of nuclei. Distances between neighboring nuclei were measured in the wild type and the promoter mutants and plotted in four different size ranges. (D) Distances between the hyphal tip and the first nucleus (N) were measured in the wild type and the kar9Δ strain.
Table 1.
Quantification of numbers and amplitudes of oscillation events in WT and mutant A. gossypii strains
| Strain | No. of eventsa |
Event P valueb |
Maximal amplitudec |
Avg growth speedd | ||||
|---|---|---|---|---|---|---|---|---|
| Bypassing | Forward | Backward | Forward | Backward | Forward | Backward | ||
| WT | 9 | 164 | 98 | 3.75 | 3.06 | 0.75 | ||
| kar9Δ | 19 | 153 | 149 | 0.54 | <0.0001 | 9.07 | 4.48 | 0.77 |
| bim1Δ | 3 | 168 | 119 | 0.85 | 0.15 | 5.22 | 4.87 | 0.66 |
| prom180-DYN1 | 6 | 89 | 40 | <0.0001 | <0.0001 | 3.36 | 1.94 | 0.759 |
| prom130-DYN1 | 5 | 44 | 7 | <0.0001 | <0.0001 | 2.57 | 1.05 | 0.734 |
| num1ΔCt | 4 | 87 | 78 | <0.0001 | 0.13 | 2.4 | 2.71 | 0.68 |
Total number of events observed during 30 min for 35 nuclei. The 5 front nuclei of 7 different hyphae were followed for 30 min (see examples of movies shown in Fig. 1B). The time interval used was 1 min. Definitions for forward and backward are given in Results.
P values derived from two-tailed Fisher's exact test (R software).
Amplitudes (in μm/min) indicate the maximal distance covered by a nucleus within a 1-min time interval.
This speed is an average of the growth speeds of the 7 hyphae used for the quantification.
As evident from Fig. 1B, nuclei in kar9Δ and bim1Δ hyphae did not form clusters and were still able to move back and forth and bypass each other. There was no significant difference in the number of forward events between the WT and the mutant hyphae (Table 1). Interestingly, we noticed significantly more backward events and twice as many bypassing events in the kar9Δ strain, indicating an overall increase in nuclear dynamics compared to that in the WT (Table 1). Hyphae lacking Bim1 reveal slightly more backward events and a strong decrease in bypassing events (3 in the bim1Δ strain, 9 in the WT, and 19 in the kar9Δ strain). This low number is probably related to the overall increase in nuclear distances in bim1Δ hyphae (nucleus-to-nucleus [N-N] distances > 6 μm: 27.5% in the WT; 76.3% in the bim1Δ strain) (Fig. 1C). Budding yeast Bim1 is known to control the dynamics of kinetochore MTs during anaphase (46). The A. gossypii homolog most likely plays a similar role, and it is therefore conceivable that the increased N-N distances in hyphae lacking Bim1 result from a lower mitotic index. Since budding yeast Kar9 is important to direct the nucleus toward the bud tip, we tested whether hyphae lacking Kar9 would show an increased distance between the first nucleus and the hyphal tip. No such increase was found (Fig. 1D).
All together, our results strongly suggest that forward nuclear movements do not depend on the potential ability of Kar9/Bim1 to guide the cMTs/SPB along polarized actin cables and to pull nuclei (especially the nucleus closest to the tip) in the direction of the hyphal tips. The overall higher nuclear mobility in the kar9Δ mutant is partly reminiscent of the kar9Δ phenotype in S. cerevisiae, with its increased dynein-dependent oscillations of the nucleus through the bud neck (44). The increased frequency in backward events in this mutant suggests a role of Kar9, even if minor, in transiently anchoring cMT +ends on actin cables to counterbalance dynein-dependent backward events or in controlling the number of cMTs directed toward the hyphal tips.
Dynein drives backward and forward nuclear movements.
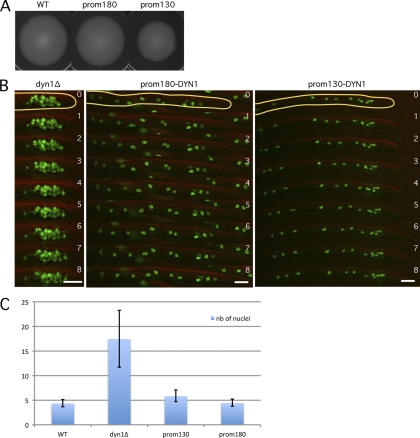
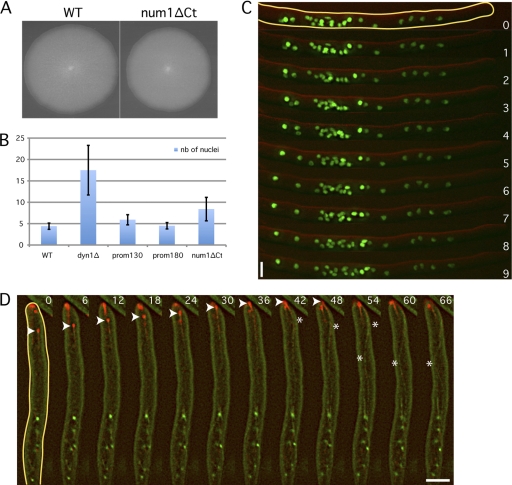
Since the Kar9 homolog of A. gossypii does not play a role in forward movements, we hypothesized that dynein, in addition to its obvious role in backward movements, could also be responsible for forward movements. It was already recently suggested that dynein could be the motor which induces cMT sliding along the cell cortex and actively pulls on nuclei, not only in the opposite direction of the hyphal tip but also toward it (8). It is not possible to test this hypothesis by directly measuring nuclear movements in a dyn1Δ strain because, already in germlings, nuclei move to the tip of emerging hyphae, where they form clusters (1). Nuclei in these clusters are so close to each other that oscillatory movements cannot be analyzed by 1-min-interval movies (Fig. 2B). To overcome this difficulty, we decided to observe the effects of reduced expression of the motor. Several attempts to exchange the strong promoter of the dynein gene with a weak A. gossypii promoter failed. We then decided to construct and test truncations of the DYN1 promoter. The following two shortened DYN1 promoter mutants were created: the prom180-DYN1 strain carries only 180-bp original sequences upstream of the start codon, and the prom130-DYN1 strain carries only 130-bp original sequences upstream of the start codon. The prom180-DYN1 and prom130-DYN1 strains grew at close to 100% and 80% of the WT radial growth rate, respectively (Fig. 2A). They showed only a weak tendency of nuclear clustering compared with that of the dynein deletion mutant, which allowed us to monitor the mobility of individual nuclei with 1-min-interval movies (Fig. 2B).
Fig. 2.
Dynein is the motor for forward and backward nuclear movements. (A) Radial growth of the wild type and strains with shortened DYN1 promoters on full medium (AFM) for 4 days at 30°C. prom180 and prom130 indicate the lengths (in base pairs) of the remaining DNA sequence upstream of the start codon. (B) Nuclear distribution in the dyn1Δ, prom180-DYN1, and prom130-DYN1 mutants. Images of time-lapse movies are shown (green, H4-GFP; red, DIC). Time is indicated in minutes. Note the dense nuclear cluster in the tip region of the dyn1Δ hyphae and the loose nuclear aggregate in the tip region of the prom130-DYN1 hyphae. Bars, 5 μm. (C) Nuclear clustering index of the dyn1Δ and the DYN1 promoter mutants. The average number (nb) of nuclei within 15 hyphal segments, each 15 μm long, showing a higher nuclear content was determined in the different mutants and compared with the average density of nuclei in randomly picked 15-μm-long regions in wild-type hyphae. Analyses were performed with images as described for panel B. Bars indicate standard deviations.
In both strains, the ability of nuclei to oscillate and bypass each other is impaired (Table 1). Compared to the WT, the number of both forward and backward events was decreased by approximately 50% in the prom180-DYN1 mutant. The prom130-DYN1 strain displayed an even stronger decrease in oscillation events (27% of WT forward events and 7% of WT backward events). Indeed, fewer and less intense dynein foci were observed when a DYN1-tdTomato fusion was expressed with the 130-bp promoter than with the WT or 180-bp promoter (data not shown). In order to compare different degrees of clustering, we counted the number of nuclei within 15-μm-long hyphal segments containing nuclear clusters and defined the average number as the clustering index. The highest density was measured in dyn1Δ hyphae, with an average of 17.5 nuclei (maximum of 25 nuclei), compared to an average of 4.5 nuclei in 15-μm-long WT hyphal segments. The loose nuclear clusters visible in the prom130-DYN1 mutant have a clustering index of 6, whereas the prom180-DYN1 mutant, which retains 50% of the bidirectional nuclear movements, has a clustering index like that of the wild type.
All together, these results strongly indicate that dynein is the key motor for both forward and backward nuclear movements in A. gossypii and that its decreased expression in the prom130-DYN1 strain leads to a pronounced reduction of bidirectional mobility, concomitant with the formation of loose nuclear aggregates.
Dynactin plays a key role in preventing aggregation of nuclei.
Because dynein is essential for the dynamic homogeneous distribution of nuclei and the prevention of nuclear cluster formation in A. gossypii hyphae, we asked whether dynactin, the complex necessary for activating dynein, also plays a similar role. Hyphae lacking dynactin components may show different degrees of nuclear distribution defects depending on their importance for dynein activation. A complete loss of dynein activation could cause dense clusters of nuclei and an almost complete absence of single nuclei, as observed in hyphae lacking the dynein heavy chain (1).
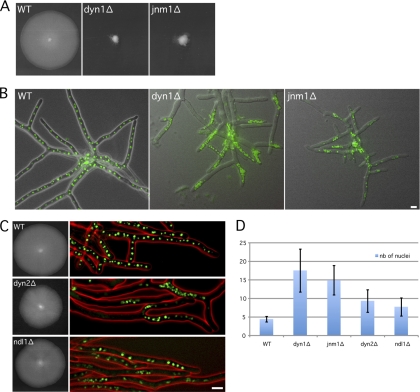
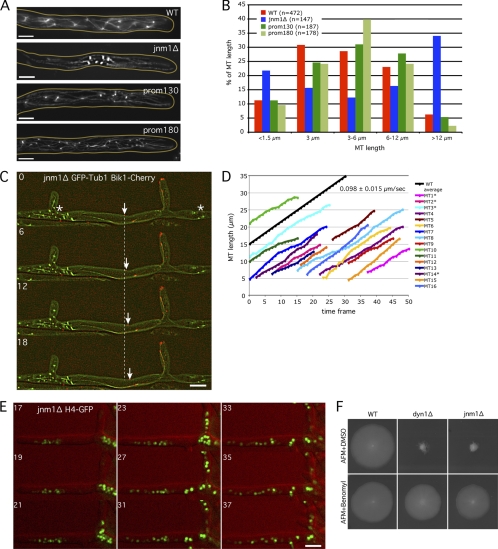
We first confirmed the dynein deletion phenotypes by deleting the ORF for the dynein heavy chain in a strain expressing a histone H4-GFP fusion. As expected, this dyn1Δ strain showed a strong growth defect and dense nuclear clustering mainly at the tips of young hyphae and later also at branching sites of mature hyphae (Fig. 2B and 3A and B). We also noticed that the size of the dyn1Δ nuclei was reduced in the clusters. We then deleted the complete open reading frame of JNM1, a gene encoding the homolog of a subunit of the dynactin complex, in the H4-GFP strain. As mentioned above, the dynactin complex is an essential dynein adaptor found to be involved in most of dynein functions, and its inhibition is phenotypically similar to a complete loss of dynein function (14). Even though A. gossypii Jnm1 (AgJnm1) is only weakly related to S. cerevisiae Jnm1 (ScJnm1) (28.2% identity), its deletion induced phenotypes also observed in dyn1Δ hyphae, i.e., a strong growth defect, severe nuclear clustering, and an almost complete absence of single nuclei (Fig. 3A and B). The only apparent difference between both mutants is seen during germination. All nuclei in germinating spores carrying the dyn1Δ allele move to the tip of the first emerging hyphae, which leads to tip-located nuclear clusters, and only later, clusters also form throughout the hyphae (Fig. 2B and 3B) (1). Nuclei in germinating spores carrying the jnm1Δ allele distribute similarly to the wild type, most likely because these spores still carry some Jnm1 protein. Clusters form only later throughout the branching hyphae and less frequently in tip regions (Fig. 3B). In jnm1Δ hyphae, nuclear clusters contained, on average, 14.9 nuclei (maximum of 19 nuclei), which is almost as dense as the nuclear clusters in dyn1Δ hyphae (Fig. 3D).
Fig. 3.
Radial growth and nuclear distribution in the dyn1Δ, jnm1Δ, dyn2Δ, and ndl1Δ mutants. (A) Size of wild-type, dyn1Δ, and jnm1Δ mycelia after 5 days at 30°C on full medium (AFM). (B) Nuclear distribution in branched mycelia of the wild type, dyn1Δ, and jnm1Δ strains. Hyphae were grown at 30°C in AFM for 18 h (green, H4-GFP; gray, DIC). Nuclei strongly cluster in dyn1Δ and jnm1Δ hyphae. (C) Radial growth and nuclear distribution in wild type, dyn2Δ, and ndl1Δ strains. (Left) Size of mycelia after 5 days at 30°C on full medium (AFM); (right) nuclear distribution in 17-h-old mycelia (green, H4-GFP; red, DIC). Note the loose nuclear aggregates in the mutant hyphae. (D) Nuclear clustering index of indicated mutants. The average number of nuclei within 15-μm-long regions with a higher nuclear content was determined in mutant hyphae and compared with the average nuclear density in the wild type. Bars indicate standard deviations. Scale bars, 5 μm.
Together, these results show that the dynactin component Jnm1 is also essential for nuclear distribution and for preventing the formation of nuclear aggregates.
Dyn2 and Ndl1 contribute to the dynamic distribution of nuclei.
The dynein heavy chain Dyn1 interacts with several light and intermediate chains within the dynein complex. In addition, several proteins that do not belong to the dynein complex itself are important for adapting the motor to its cellular function. We tested the deletion phenotypes of two of these factors, Dyn2 and Ndl1. In budding yeast, the role of the dynein light chain Dyn2 for dynein activity is not critical since a dyn2Δ mutant exhibits only partial defects in certain spindle position assays (29). The A. gossypii Dyn2 shows 71% identity with S. cerevisiae Dyn2 (ScDyn2). In contrast to dyn1Δ, the dyn2Δ mutant only has a slightly decreased radial growth rate (85% of that of the WT) (Fig. 3C). However, nuclear distribution is significantly impaired in dyn2Δ: nuclear clusters form with an average of 9.3 nuclei in 15-μm-long segments (4.5 in the WT), which is a lower nuclear density than those in clusters of dyn1Δ and jnm1Δ strains (Fig. 3D).
We also deleted the gene encoding A. gossypii Ndl1 (AgNdl1), the homolog of which is involved in the accumulation of dynein at cMT +ends in budding yeast, but its role in dynein function is not clearly understood (20, 23). The radial growth rate of the ndl1Δ mutant is only slightly decreased (94% of that of the WT), and nuclei only rarely form loose clusters, with an average density of 7.7 nuclei in 15-μm-long segments (Fig. 3C and D). Radial growth in these two mutants is much less affected by nuclear aggregates than that in the dyn1Δ and jnm1Δ mutants. Possible reasons for these growth differences may be the insufficient distribution of mRNAs and vesicles in the tightly clustered mutants (the fraction of nonaggregated nuclei is less than 10% in dyn1Δ and jnm1Δ mycelia, 29% in dyn2Δ mycelia, and 51% in ndl1Δ mycelia) or a severe impairment of organelle and secretory vesicle transport by dense nuclear aggregates.
All together, the dynein adaptors A. gossypii Dyn2 (AgDyn2) and AgNdl1 support nuclear distribution and play a role in preventing nuclear cluster formation in A. gossypii.
Dynein localizes to +ends of cMTs emanating from SPBs in both directions of the growth axis, to cMT –ends, and along cMTs.
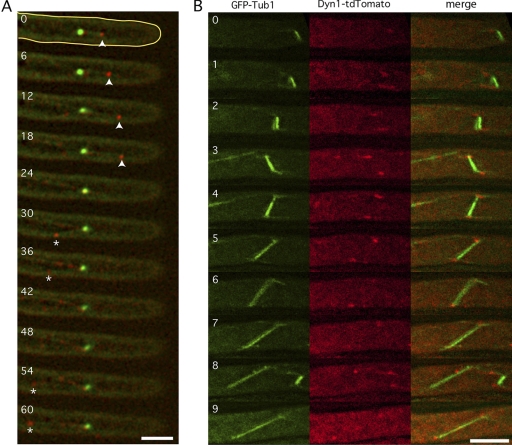
To begin to understand the underlying mechanisms of nuclear movements in A. gossypii, we looked at dynein localization. We fused the TandemTomato (tdTomato) fluorescent protein to the C-terminal end of dynein in hyphae expressing the GFP-Tubulin1 fusion. The Dyn1-tdTomato fusion was expressed at an endogenous level and was fully functional, since the strain expressing this fusion was growing at a wild-type rate (data not shown). Using time-lapse video microscopy, we first tested whether the dynein motor, in accordance with its role in the forward and backward movements of nuclei, could be observed at the +ends of cMTs growing in the direction of the hyphal tip and opposite to it. One example with 11 consecutive images taken in 6-s intervals is shown in Fig. 4A. Dynein (red) is seen at the +end of a cMT emanating from an SPB (green) and growing 2.7 μm toward the hyphal tip during the first four images. Starting in the sixth image, dynein is seen at the +end of a second cMT emanating from the same SPB but in the opposite direction and growing 4.5 μm during the next five images. Dynein can also clearly be observed at the SPB in all images and, during six images, along a third cMT, which may or may not emanate from the same SPB. Visual inspection of many images revealed that dynein localizes mainly at SPBs (cMT –ends) during all nuclear cycle stages and, as less intense foci, along cMTs and at cMT +ends (Fig. 5A and data not shown). Rarely, dynein foci were detectable at hyphal tips, probably when +ends of growing cMTs reached the tip cortex (see also below). With our label, we did not detect dynein foci along the hyphal cortex. Cortically located dynein could be seen in budding yeast but only using Dyn1 fused to 3 GFPs, and dynein localization at cMT +ends in budding yeast is more predominant compared to that in A. gossypii (20, 21).
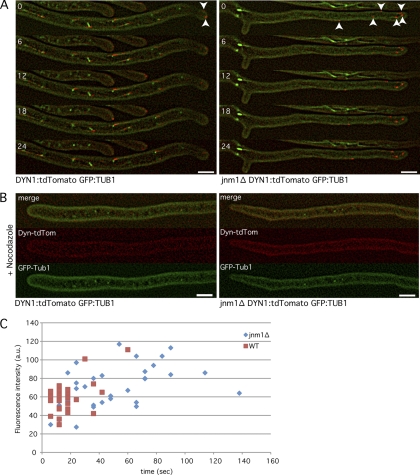
Fig. 4.
A. gossypii dynein localizes at cMT +ends and –ends and along cMTs. Hyphae expressing Dyn1-tdTomato (red) at endogenous levels and GPF-Tub1 (green) were monitored by time-lapse microscopy. (A) Dynein interacts with cMTs directed toward the hyphal tip (arrowheads) or backward (*). Numbers indicate time in seconds. Pictures represent 1 Z plane. (B) Pictures are stacks of 5 Z planes with 0.75-μm intervals. Dynein localizes symmetrically on SPBs during both metaphase and anaphase. Numbers indicate time in minutes. Bars, 5 μm.
Fig. 5.
Dynein strongly accumulates at cMT plus-end tips (+tips) in the absence of Jnm1. (A) One-Z-plane movies of dynein-tdTomato GFP-Tub1 wild-type and jnm1Δ cells. Time is shown in seconds. Arrowheads indicate +tips of cMTs. (B) Dynein-tdTomato GFP-Tub1 wild-type and jnm1Δ cells treated with 15 μg/ml nocodazole for 15 min (5-Z-plane pictures). (C) Quantification of the lifetime and fluorescence intensity (in arbitrary units [a.u.]) of dynein-tdTomato dots in wild-type (Dyn1-tdTomato GFP-Tub1) and jnm1Δ cells. Quantifications were made with movies as shown in the legend to panel A. Bars, 5 μm.
Another very important difference between budding yeast and A. gossypii is the functional distinction between the two SPBs in metaphase. In yeast, dynein loading is restricted in metaphase to the SPB/cMT, which enters the bud in order to help the spindle to align along the polarity axis and to control the pulling of only one SPB through the bud neck (9). In A. gossypii, nuclear division and cytokinesis are not strictly coordinated, and nuclei are in continuous motion. Thus, a requirement for an asymmetric distribution of dynein at metaphase SPBs is very unlikely. Indeed, metaphase spindles often changed orientations with respect to the growth axis prior to anaphase, and a dynein signal was always visible on both SPBs (Fig. 4B).
In conclusion, dynein is seen at all SPBs, independently of the nuclear cycle stage. It is transiently visible at +ends of cMTs, emanating in both directions from SPBs with respect to the growth axis, which is consistent with the role of this motor in bidirectional nuclear movements.
Jnm1 is important for transient localization of dynein at cMT +ends.
Since A. gossypii dynein accumulation at cMT +ends is transient, it is possible that dynein gets regularly “off-loaded” from the cMT +ends to the cell cortex, as it has been proposed in budding yeast (20). The “dynein off-loading model” requires that first an inactive form of dynein is transported to cMT +ends. The dynein complex then detaches from the +end when it reaches the cell cortex. This leads to a stable cortex association of dynein. At the same time, the dynactin complex and some cortical factors (Num1 for example) may stimulate the –end motor activity of the anchored dynein (see review by Moore and Cooper [28]).
To test whether this model is relevant in A. gossypii, we determined the amount of time that dynein was visible at +ends of cMTs in the WT and the jnm1Δ dynactin mutant and, in addition, measured the intensity of the dynein signals at cMT +ends. Our time-lapse video microscopy (1 Z plane, 6-s intervals) (Fig. 4A and 5A) revealed that the motile dynein dots (most probably associated with cMT +ends) are visible only for short periods in the WT (17.5 s on average) (Fig. 5C). For comparison, the MT +end tracking protein Bik1 can be visible for more than 3 min at cMT +ends in A. gossypii (8). Compared to the WT, dynein localization in jnm1Δ significantly increased at cMT +ends and concomitantly decreased at SPBs, indicating that less dynein migrates to the cMT –ends (Fig. 5A). Moreover, the average time dynein spent at the cMT +ends in jnm1Δ significantly increased compared to that in the WT (17.5 s for the WT and 47.7 s for jnm1Δ; n = 35) (Fig. 5C). When WT and jnm1Δ hyphae were treated with the MT-destabilizing drug nocodazole (NZ), dynein localization at SPBs was completely lost (Fig. 5B).
Together, these results indicate that dynein is most likely not directly loaded on SPBs but travels along cMTs toward the –ends/SPBs. Our data also strongly suggest that dynein accumulation at SPBs is dynactin dependent and that dynactin is necessary to keep the accumulation of dynein at cMT +ends very transient, most probably because dynactin activates the –end motor activity of dynein. The off-loading model presented in S. cerevisiae may therefore also be valid in A. gossypii.
AgNum1 is involved in nuclear distribution and in the control of dynein accumulation at cMT +ends.
Num1 has been shown in budding yeast to be necessary for the cortical attachment of dynein. Num1 associates with dynein and with the plasma membrane as discrete nonmotile patches (5, 6, 11). Recent work has shown that S. cerevisiae Num1 (ScNum1) is targeted to the plasma membrane via its C-terminal Pleckstrin homology (PH) domain (aa 2563 to 2692) (29, 38). The A. gossypii Num1 (AgNum1) homolog (3,645 aa) shares 29.4% identity with ScNum1 (2,748 aa). Interestingly, a stretch of 178 aa in the C terminus of AgNum1 shares 67% identity with the C terminus of the budding yeast ortholog and also contains a PH domain. We therefore supposed that AgNum1 is also targeted to the plasma membrane and tested whether its role in nuclear migration is conserved in A. gossypii. So far, our attempts to delete the complete 10,938-bp NUM1 ORF or its 3′-end half have not been successful. However, we were able to delete the last 515 bp of the ORF (encoding the PH domain) in the H4-GFP background.
This carboxy-terminal deletion, named num1ΔCt, showed a mild growth defect (91% of the WT growth rate) but frequently formed loose nuclear clusters with, on average, 8.4 nuclei in 15-μm segments (Fig. 6A, B, and C). These clusters showed densities similar to those in the dyn2Δ and ndl1Δ strains (averages of 9.3 and 7.7 nuclei, respectively), and were less dense than clusters in dyn1Δ (average of 17.5 nuclei). We also looked at the ability for individual nuclei to oscillate and bypass each other. The results compiled in Table 1 reveals that the numbers of bypassing, forward, and backward events are overall reduced in the num1ΔCt mutant compared to those in the WT strain but to a lesser extent than in the prom130-DYN1 mutant. Interestingly, this indicates that dynein can still exert pulling forces, although reduced, on nuclei in hyphae expressing the carboxy-terminal truncation of Num1. This remaining dynein activity exceeds the dynein activity in the low-level-expressing prom130-DYN1 mutant.
Fig. 6.
Num1 is involved in the control of nuclear distribution and dynein localization. (A) Radial growth of wild-type and num1ΔCt strains after 5 days at 30°C on full medium (AFM). (B) Average numbers of nuclei within 15 clusters. Regions (15 μm long) with higher nuclear content for the different strains were analyzed (see example in panel C). (C) Nuclear dynamics in H4-GFP num1ΔCt cells (green, H4-GFP; red, DIC). Some nuclei move individually toward the hyphal tip, whereas others form clusters. Time is indicated in minutes. (D) Dynein (in red) only weakly localizes at SPBs (green dots) but concentrates mainly at cMT +ends in the num1ΔNt mutant. The arrowheads focus on the +end of a growing cMT. The asterisks show the curvatures of the cMT after its +end has reached the hyphal tip. Time is indicated in seconds. Bars, 5 μm.
We also wanted to check dynein localization in a NUM1 deletion mutant. However, as mentioned above, our attempts for deletion of this long gene failed. We could delete only the first 1,500 bp of the NUM1 ORF in the Dyn1-tdTomato GFP-Tub1 background. Since the GEN3 deletion cassette replaced the coding sequence of the first 500 amino acids of Num1, we assumed that the protein was not expressed in this N-terminal deletion, named num1ΔNt. Hyphae of this strain also exhibit nuclear clustering, visualized by regions with densely populated GFP-labeled SPBs, and grow at a rate similar to that of num1ΔCt (data not shown). In this mutant, we also clearly observed very long cMTs. Some long-growing cMTs were curving while reaching hyphal tips (Fig. 6D, asterisk). These curvatures were probably induced by physical constrictions and not lateral cortex interactions characteristic of a sliding event. In contrast to WT cells but similar to jnm1Δ, the concentration of dynein at SPBs in the num1ΔNt mutant was weak and significantly increased at cMT +ends (compare Fig. 5A and Fig. 6D).
Together, these results indicate that Num1, like Jnm1/dynactin, plays an important role in the dynamic localization of dynein at cMT +ends in A. gossypii,and that Num1 very likely has to bind to the plasma membrane to fulfill its function. Thus, major control components for dynein localization and activity are conserved in S. cerevisiae and A. gossypii, even though the specific cellular roles of dynein in both organisms have changed during evolution.
Hyphae lacking Jnm1 exhibit very long cMTs which maintain nuclei in clusters.
Previous work has shown that the lack of Dyn1 is associated with the formation of unduly long cMTs in A. gossypii (1). MT immunostainings of jnm1Δ hyphae also show that very long cMTs were spreading in the anucleated regions and that very short cMTs are visible only in nuclear clusters (Fig. 7A). On average, cMT length was 9.6 μm in jnm1Δ (n = 147; maximum length, 40 μm) and 5.6 μm in the WT (n = 165; maximum length, 26 μm) (Fig. 7B). Only 9% of the WT cMTs are longer than 12 μm. This category reaches 34% in jnm1Δ cells. Figure 7B also shows a WT-like cMT length distribution in hyphae expressing less dynein (prom180-DYN1 and prom130-DYN1 strains).
Fig. 7.
The very long cMTs observed in dynein/dynactin mutants are responsible for nuclear clustering. (A) Anti-alpha-tubulin immunostaining of jnm1Δ, prom130-DYN1, prom180-DYN1, and wild-type strains. Pictures correspond to stacks of 5 Z planes with 0.75-μm distance. (B) Quantification of cMT length in WT and mutant cells using anti-Tub1 immunostaining pictures, as shown in panel A. (C) Localization of Bik1-Cherry (red) at cMTs +ends (GFP-Tub1 in green) in jnm1Δ cell. Nuclei/SPBs cluster at some places (*), while long cMTs investigate the nuclei-free lateral branch. The arrow points to one growing cMT +end. The dashed line represents the position of this +end at time point 0. Time is indicated in seconds. (D) Quantification of the cMT polymerization rate using 1-Z-plane movies as shown in panel C. cMTs were measured from their –end (SPB observed with GFP-Tub1) to their +tip (Bik1-Cherry signal). Due to increased cMT length in the jnm1Δ mutant, SPBs and +tips were often in different focal planes (*), corresponding to the real length of the cMTs (SPB to +tip), whereas the other cMTs have an underestimated length (visible part of the cMT to the +tip). One frame corresponds to 6 s. The average cMT polymerization rate in the wild type is shown in black as a reference (see reference 8). (E) Nuclear clusters disappear and nuclei redistribute after treatment of jnm1Δ H4-GFP cells with 15 μg/ml nocodazole (red, DIC; green, H4-GFP). Time indicates minutes after addition of nocodazole. (F) Growth defects of jnm1Δ and dyn1Δ cells are partially rescued by the MT-destabilizing drug benomyl. Bars, 5 μm.
We then looked at cMT's behavior and polymerization rate in jnm1Δ hyphae in vivo (in a Bik1-Cherry GFP-Tubulin1 background). Bik1 fused to the fluorescent protein Cherry is a good marker to follow growth at cMT +ends (8). Using this marker, we have previously shown that in A. gossypii WT hyphae, cMTs do not undergo dynamic instability (consecutive phases of growth and shrinkage) but spend most of their time growing. As shown in Fig. 7C and D, cMTs in jnm1Δ cells are also mainly in growing phase. The average MT polymerization rates in the jnm1Δ mutant and in the WT were very similar, as follows: 0.098 ± 0.015 μm/s for jnm1Δ and 0.108 ± 0.035 μm/s for the WT (see reference 8). Therefore, cMTs in jnm1Δ strain hyphae are much longer than cMTs in WT hyphae because of a prolonged growing phase but not because of a faster MT polymerization rate.
We hypothesized that by being more stable and/or constantly growing, long jnm1Δ cMTs (with an approximately polymerization rate of 6 μm/min) prevent nuclear progression toward the growing hyphal tips (maximal growth speed of jnm1Δ and dyn1Δ hyphae, 0.4 μm/min; that of WT hyphae, 3.0 μm/min) and thus maintain nuclei in clusters. As shown in Fig. 7C, cMTs can also enter into nuclei-free lateral branches. Since sliding of those cMTs along the cell cortex does not occur, nuclei are not pulled into the lateral branches, and their progression is blocked within the main hyphae. The ability of jnm1Δ long cMTs to enter lateral branches and stably interact with them can also, at least partially, explain how nuclear clusters form in mutants. The conclusion that nuclei are maintained in dense clusters by the presence of extra-long cMTs is confirmed by the following experiment. Treatment of jnm1Δ cells with the MT-destabilizing drug nocodazole induces dissolution of nuclear clusters. After 25 to 30 min of incubation, jnm1Δ nuclei redistributed and migrated into nuclei-free spaces and lateral branches (Fig. 7E). In addition, as it had been already demonstrated for the dyn1Δ mutant, the severe growth defect of the jnm1Δ mutant can be rescued by the addition of the MT-destabilizing drug benomyl (Fig. 7F) (1). This suggests that dense nuclear clusters caused by unrestricted growth of cMTs and the absence of oscillatory nuclear movements act as a physical barrier for normal distribution of organelles and sufficient tip-directed transport of secretory vesicles, thereby causing the severe growth defect.
DISCUSSION
The goals in this work were, first, to identify the motor(s) responsible for the bidirectional movements of nuclei in the filamentous fungus A. gossypii, second, to find the underlying cellular function for this continuous nuclear motion, and third, to gain insights into the mechanism directing nuclear movements. As a result, we could demonstrate that the dynein network, from its rather restricted and nonessential role during anaphase in budding yeast, has evolved in A. gossypii into an essential system for maintaining a dynamic distribution of nuclei and preventing deleterious formations of nuclear clusters. When we realized that dynein is the motor exerting bidirectional forces on nuclei, we wanted to know to what degree the partner proteins of dynein, known from work in yeast and other organisms, contribute to dynein activity and localization and bidirectional movements.
Nuclear oscillations prevent the formation of nuclear clusters in multinucleated hyphae.
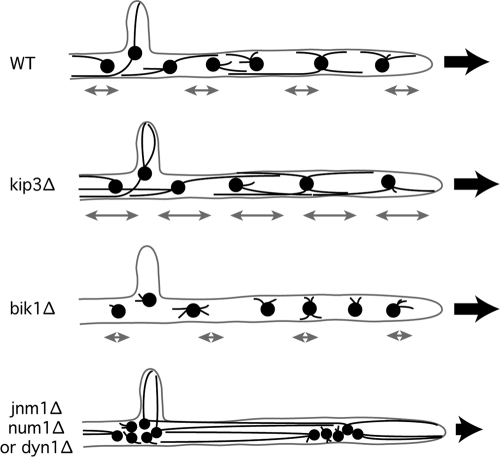
Ten years ago, it was noted that nuclei in A. gossypii hyphae are in continuous motion and that the distribution of nuclei is dynamic, resulting in an average distance between adjacent nuclei of 5 to 6 μm (1). We tried several approaches to identify a cellular function of oscillatory nuclear movements. So far, it was not possible only to eliminate or decrease these movements without affecting at the same time the microtubule cytoskeleton. First, it was shown that mutants (bik1Δ and kip2Δ) with shortened cMTs due to decreased cMT +end stability have decreased back and forth movements and do not show any nuclear bypassing events (8). Those mutants do not show any clustering phenotype, grow almost like the WT, and do not reveal clues about a potential cellular role of nuclear oscillations. Second, when cMTs are slightly longer than those in the WT (kip3Δ mutant), the frequency of nuclear oscillation increases, but no nuclear clusters or cell growth defects are observed (8). Third, loss of dynein/dynactin causes very long cMTs (much longer than those in the kip3Δ mutant), concomitant with the formation of tight nuclear cluster and a strong growth defect. WT-like oscillations events and bypassing are also abolished in this mutant (Fig. 8). Here, we analyzed a dynein promoter truncation (prom130-DYN1) which showed decreased oscillations and bypassing events and triggered formation of loose nuclear aggregates without observably altered cMT growth control. We propose that nuclei, each nucleating up to six cMTs in different directions, can entangle in the narrow hyphal tube, e.g., during bypassing attempts, and that dynein-driven oscillatory movements in WT frequencies can dissolve blocked pairs of nuclei (e.g., first two nuclei of the WT in Fig. 1B). Nuclei in mutants with short cMTs do not entangle since they do not bypass each other. However, in mutants with WT-like cMTs (such as the prom130-DYN1 mutant), the absence of sufficient oscillatory movements does not dissolve loose nuclear clusters.
Fig. 8.
The absence of oscillation and the presence of very long cMTs observed in dynein/dynactin mutants are responsible for nuclear clustering. In the kip3Δ mutant, cMTs are slightly longer than those in the WT and induce more frequent nuclear oscillations and higher oscillation amplitudes (see gray double arrows). In bik1Δ, cMTs are much shorter than those in the wild type. Nuclear dynamics is significantly decreased in this mutant (8). In the jnm1Δ, num1Δ, or dyn1Δ mutant, MTs are also longer than those in the WT, but since dynein activity is abolished, no oscillations are observed. MTs push nuclei apart and prevent their progression within the hyphae. The black arrows illustrate the direction of cell growth and of the cytoplasmic stream responsible for the passive forward nuclear movements. A reduced number of cMTs emerging from each SPB is shown in this figure for reasons of clarity.
Nuclei in dynein and dynactin mutants form tight clusters and do not oscillate (Fig. 8). Those clusters dissolve in the presence of MT-destabilizing drugs such as nocodazole (NZ), suggesting that the increased stability and length of cMTs lead to tight nuclear clustering. We showed here that the dissolution of nuclear clusters by NZ takes about 30 min, a relatively long period of time. This is probably dependent on the cytoplasmic stream since nuclear oscillations are abolished in NZ-treated cells (19).
Dyn1, dynactin, and Num1 are involved in the control of cMT dynamics.
In budding yeast, mutants lacking Num1 or dynein/dynactin display longer MTs than those in WT cells (3, 5, 26). We show here that this “longer MT” phenotype is also visible in A. gossypii dynactin and num1 mutants.
In previous work, we have shown that cMTs in A. gossypii do not exhibit the dynamic instability characteristic of cMTs in other organisms. Therefore, even if A. gossypii cMTs do not go through consecutive phases of growth and shrinkage, it seems that their overall maximal length is controlled in WT cells. This control seems to be particularly affected in the dynein, dynactin, and num1 mutants. The fact that a complete loss of dynein or an abnormal accumulation of dynein at cMT +ends (in jnm1Δ or num1Δ hyphae) induces the same “long cMT” phenotype is still not understood. One hypothesis could be that the absence of dynein activity directly or indirectly induces cMT bundling. This bundling could affect cMT dynamics by increasing cMTs stability. Our time-lapse microscopy data on jnm1Δ Bik1-Cherry GFP-Tub1 cells go in favor of this hypothesis since only a few very long cMTs were visible in the nuclei-free regions (compared to the overall number of WT cMTs) and those very long cMTs had an increased GFP fluorescence intensity (see an example in Fig. 7C). The establishment of the whole set of interactions at cMT +ends in A. gossypii will definitely help understanding of the contribution of dynein and dynein accessory proteins to MT dynamics in filamentous fungi but also in other organisms.
Control of dynein loading on cMT +ends.
In filamentous fungi like A. nidulans and U. maydis, dynein accumulation at cMT +ends is essential for the retrograde movement of early endosomes. Recent publications have shown that in U. maydis, approximately 55 dynein motors concentrate at cMTs +ends. The mechanism by which dynein concentrates there is a combination of stochastic accumulation and active retention by the dynactin/EB1 complex (22, 33, 34). In A. gossypii, dynein is not only involved in cell growth and endosome movements but also accumulates at cMT +ends. However, the control of dynein loading on cMT +ends seems to be different from that in U. maydis. Indeed, in contrast to U. maydis, the dynactin complex prevents dynein accumulation at cMT +ends.
In budding yeast and A. gossypii, dynein is involved mainly in nuclear movements. As already mentioned previously, yeast dynein is first maintained in an inactive form at cMT +ends (with maximal retention at anaphase). It gets activated only (and released from the +end) after its interaction with the cell cortex and especially Num1 (20, 25, 36). Since dynein is a highly processive –end MT motor, it must be in an inactive form to be able to stably interact with cMT +ends. This high dynein concentration then induces a strong pulling of the nucleus through the bud neck during anaphase.
Our results show that A. gossypii dynein requires similar mechanisms (and especially Num1 and dynactin) to be activated and therefore released from the cMT +ends. However, our time-lapse microscopy of GFP-Tub1 Dyn1-tdTomato hyphae clearly showed that dynein accumulation at cMT +ends is weaker and more transient than that in S. cerevisiae. This suggests that dynein loading and/or retention at +ends is regulated slightly differently in the filamentous fungus. In budding yeast, it seems that dynein +end accumulation and activation are cell cycle regulated (36). In the multinucleated A. gossypii hyphae, mitoses are asynchronous, and little is known about the mode of actions of cell cycle regulators (7). However, the fact that nuclei constantly oscillate throughout A. gossypii hyphae and that dynein localizes to SPBs during the entire nuclear cycle suggest that dynein activation is not dependent on cell cycle regulators.
In the future, it will be necessary to test whether kinesins (such as Kip2, for example) are involved in dynein loading on cMT +ends. It will also be interesting to test whether AgNum1 spreads all over the hyphal cortex like its budding yeast orthologue (25, 29, 38).
Another important question to solve in the future: how do nuclei coordinate the (opposite) pulling forces applied on their SPBs? One attractive hypothesis, in agreement with our dynein localization data, is that the loading of dynein on cMTs occurs only on specific cMTs. It has been shown in the fission yeast Schizosaccharomyces pombe that dynein can quickly relocate from one MT to another during meiotic nuclear oscillations. In their model, Vogel et al. proposed that in response to load forces, dynein motors detach from the trailing cMTs (39). Dyneins first detach from the cell cortex and then from the cMTs. After redistribution via the cytoplasm, they attach along the leading/sliding MTs, thereby producing asymmetric forces necessary for oscillations. We hope to be able to generate in the future new variants of fluorescent tubulin, allowing a better visualization of cMTs dynamics and sliding, and to create strong fluorescent variants of dynein. These tools will be necessary to better characterize the relationship between the dynamic distribution of dynein and cortical pulling forces.
Supplementary Material
ACKNOWLEDGMENTS
We are very grateful to Sue Jaspersen for many helpful comments, to Kerry Bloom for suggesting the experiment with decreased dynein expression, to Mark Finlayson for valuable discussions, and to Doug Phanstiel for help in statistical analyses. Major parts of this publication were written during a sabbatical of P.P. with Mike Snyder, Stanford University.
This work was supported by a grant from the Swiss National Science Foundation (grant 31003A-112688).
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 3 June 2011.
REFERENCES
- 1. Alberti-Segui C., Dietrich F., Altmann-Johl R., Hoepfner D., Philippsen P. 2001. Cytoplasmic dynein is required to oppose the force that moves nuclei towards the hyphal tip in the filamentous ascomycete Ashbya gossypii. J. Cell Sci. 114:975–986 [DOI] [PubMed] [Google Scholar]
- 2. Ayad-Durieux Y., Knechtle P., Goff S., Dietrich F., Philippsen P. 2000. A PAK-like protein kinase is required for maturation of young hyphae and septation in the filamentous ascomycete Ashbya gossypii. J. Cell Sci. 113(Pt. 24):4563–4575 [DOI] [PubMed] [Google Scholar]
- 3. Carminati J. L., Stearns T. 1997. Microtubules orient the mitotic spindle in yeast through dynein-dependent interactions with the cell cortex. J. Cell Biol. 138:629–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dietrich F. S., et al. 2004. The Ashbya gossypii genome as a tool for mapping the ancient Saccharomyces cerevisiae genome. Science 304:304–307 [DOI] [PubMed] [Google Scholar]
- 5. Farkasovsky M., Kuntzel H. 1995. Yeast Num1p associates with the mother cell cortex during S/G2 phase and affects microtubular functions. J. Cell Biol. 131:1003–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Farkasovsky M., Kuntzel H. 2001. Cortical Num1p interacts with the dynein intermediate chain Pac11p and cytoplasmic microtubules in budding yeast. J. Cell Biol. 152:251–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gladfelter A. S., Hungerbuehler A. K., Philippsen P. 2006. Asynchronous nuclear division cycles in multinucleated cells. J. Cell Biol. 172:347–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Grava S., Philippsen P. 2010. Dynamics of multiple nuclei in Ashbya gossypii hyphae depend on the control of cytoplasmic microtubules length by Bik1, Kip2, Kip3, and not on a capture/shrinkage mechanism. Mol. Biol. Cell 21:3680–3692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Grava S., Schaerer F., Faty M., Philippsen P., Barral Y. 2006. Asymmetric recruitment of dynein to spindle poles and microtubules promotes proper spindle orientation in yeast. Dev. Cell 10:425–439 [DOI] [PubMed] [Google Scholar]
- 10. Han G., et al. 2001. The Aspergillus cytoplasmic dynein heavy chain and NUDF localize to microtubule ends and affect microtubule dynamics. Curr. Biol. 11:719–724 [DOI] [PubMed] [Google Scholar]
- 11. Heil-Chapdelaine R. A., Oberle J. R., Cooper J. A. 2000. The cortical protein Num1p is essential for dynein-dependent interactions of microtubules with the cortex. J. Cell Biol. 151:1337–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hwang E., Kusch J., Barral Y., Huffaker T. C. 2003. Spindle orientation in Saccharomyces cerevisiae depends on the transport of microtubule ends along polarized actin cables. J. Cell Biol. 161:483–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Inoue S., Turgeon B. G., Yoder O. C., Aist J. R. 1998. Role of fungal dynein in hyphal growth, microtubule organization, spindle pole body motility and nuclear migration. J. Cell Sci. 111(Pt. 11):1555–1566 [DOI] [PubMed] [Google Scholar]
- 14. Kardon J. R., Vale R. D. 2009. Regulators of the cytoplasmic dynein motor. Nat. Rev. Mol. Cell Biol. 10:854–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kaufmann A. 2009. A plasmid collection for PCR-based gene targeting in the filamentous ascomycete Ashbya gossypii. Fungal Genet. Biol. 46:595–603 [DOI] [PubMed] [Google Scholar]
- 16. Knechtle P., Dietrich F., Philippsen P. 2003. Maximal polar growth potential depends on the polarisome component AgSpa2 in the filamentous fungus Ashbya gossypii. Mol. Biol. Cell 14:4140–4154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kohli M., Galati V., Boudier K., Roberson R. W., Philippsen P. 2008. Growth-speed-correlated localization of exocyst and polarisome components in growth zones of Ashbya gossypii hyphal tips. J. Cell Sci. 121:3878–3889 [DOI] [PubMed] [Google Scholar]
- 18. Lang C., et al. 2010. Structural mutants of the spindle pole body cause distinct alteration of cytoplasmic microtubules and nuclear dynamics in multinucleated hyphae. Mol. Biol. Cell 21:753–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lang C., et al. 2010. Mobility, microtubule nucleation and structure of microtubule-organizing centers in multinucleated hyphae of Ashbya gossypii. Mol. Biol. Cell 21:18–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lee W. L., Kaiser M. A., Cooper J. A. 2005. The offloading model for dynein function: differential function of motor subunits. J. Cell Biol. 168:201–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lee W. L., Oberle J. R., Cooper J. A. 2003. The role of the lissencephaly protein PacI during nuclear migration in budding yeast. J. Cell Biol. 160:355–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lenz J. H., Schuchardt I., Straube A., Steinberg G. 2006. A dynein loading zone for retrograde endosome motility at microtubule plus-ends. EMBO J. 25:2275–2286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li J., Lee W. L., Cooper J. A. 2005. NudEL targets dynein to microtubule ends through LIS1. Nat. Cell Biol. 7:686–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liakopoulos D., Kusch J., Grava S., Vogel J., Barral Y. 2003. Asymmetric loading of Kar9 onto spindle poles and microtubules ensures proper spindle alignment. Cell 112:561–574 [DOI] [PubMed] [Google Scholar]
- 25. Markus S. M., Punch J. J., Lee W. L. 2009. Motor- and tail-dependent targeting of dynein to microtubule plus ends and the cell cortex. Curr. Biol. 19:196–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. McMillan J. N., Tatchell K. 1994. The JNM1 gene in the yeast Saccharomyces cerevisiae is required for nuclear migration and spindle orientation during the mitotic cell cycle. J. Cell Biol. 125:143–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Minke P. F., Lee I. H., Tinsley J. H., Bruno K. S., Plamann M. 1999. Neurospora crassa ro-10 and ro-11 genes encode novel proteins required for nuclear distribution. Mol. Microbiol. 32:1065–1076 [DOI] [PubMed] [Google Scholar]
- 28. Moore J. K., Cooper J. A. 2010. Coordinating mitosis with cell polarity: Molecular motors at the cell cortex. Semin. Cell Dev. Biol. 21:283–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Moore J. K., Stuchell-Brereton M. D., Cooper J. A. 2009. Function of dynein in budding yeast: mitotic spindle positioning in a polarized cell. Cell Motil. Cytoskeleton 66:546–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Morris N. R. 2000. Nuclear migration. From fungi to the mammalian brain. J. Cell Biol. 148:1097–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ramos-Garcia S. L., Roberson R. W., Freitag M., Bartnicki-Garcia S., Mourino-Perez R. R. 2009. Cytoplasmic bulk flow propels nuclei in mature hyphae of Neurospora crassa. Eukaryot. Cell 8:1880–1890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Requena N., et al. 2001. Genetic evidence for a microtubule-destabilizing effect of conventional kinesin and analysis of its consequences for the control of nuclear distribution in Aspergillus nidulans. Mol. Microbiol. 42:121–132 [DOI] [PubMed] [Google Scholar]
- 33. Schuster M., et al. 2011. Controlled and stochastic retention concentrates dynein at microtubule ends to keep endosomes on track. EMBO J. 30:652–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schuster M., Lipowsky R., Assmann M. A., Lenz P., Steinberg G. 2011. Transient binding of dynein controls bidirectional long-range motility of early endosomes. Proc. Natl. Acad. Sci. U. S. A. 108:3618–3623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Seiler S., Plamann M., Schliwa M. 1999. Kinesin and dynein mutants provide novel insights into the roles of vesicle traffic during cell morphogenesis in Neurospora. Curr. Biol. 9:779–785 [DOI] [PubMed] [Google Scholar]
- 36. Sheeman B., et al. 2003. Determinants of S. cerevisiae dynein localization and activation: implications for the mechanism of spindle positioning. Curr. Biol. 13:364–372 [DOI] [PubMed] [Google Scholar]
- 37. Straube A., et al. 2001. A split motor domain in a cytoplasmic dynein. EMBO J. 20:5091–5100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tang X., Punch J. J., Lee W. L. 2009. A CAAX motif can compensate for the PH domain of Num1 for cortical dynein attachment. Cell Cycle 8:3182–3190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vogel S. K., Pavin N., Maghelli N., Julicher F., Tolic-Norrelykke I. M. 2009. Self-organization of dynein motors generates meiotic nuclear oscillations. PLoS Biol. 7:e1000087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wedlich-Soldner R., Schulz I., Straube A., Steinberg G. 2002. Dynein supports motility of endoplasmic reticulum in the fungus Ustilago maydis. Mol. Biol. Cell 13:965–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wendland J., Ayad-Durieux Y., Knechtle P., Rebischung C., Philippsen P. 2000. PCR-based gene targeting in the filamentous fungus Ashbya gossypii. Gene 242:381–391 [DOI] [PubMed] [Google Scholar]
- 42. Xiang X., Beckwith S. M., Morris N. R. 1994. Cytoplasmic dynein is involved in nuclear migration in Aspergillus nidulans. Proc. Natl. Acad. Sci. U. S. A. 91:2100–2104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Xiang X., Fischer R. 2004. Nuclear migration and positioning in filamentous fungi. Fungal Genet. Biol. 41:411–419 [DOI] [PubMed] [Google Scholar]
- 44. Yeh E., et al. 2000. Dynamic positioning of mitotic spindles in yeast: role of microtubule motors and cortical determinants. Mol. Biol. Cell 11:3949–3961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhang J., Li S., Fischer R., Xiang X. 2003. Accumulation of cytoplasmic dynein and dynactin at microtubule plus ends in Aspergillus nidulans is kinesin dependent. Mol. Biol. Cell 14:1479–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zimniak T., Stengl K., Mechtler K., Westermann S. 2009. Phosphoregulation of the budding yeast EB1 homologue Bim1p by Aurora/Ipl1p. J. Cell Biol. 186:379–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.