Abstract
A gene predicted to encode Trypanosoma brucei glucosamine 6-phosphate N-acetyltransferase (TbGNA1; EC 2.3.1.4) was cloned and expressed in Escherichia coli. The recombinant protein was enzymatically active, and its high-resolution crystal structure was obtained at 1.86 Å. Endogenous TbGNA1 protein was localized to the peroxisome-like microbody, the glycosome. A bloodstream-form T. brucei GNA1 conditional null mutant was constructed and shown to be unable to sustain growth in vitro under nonpermissive conditions, demonstrating that there are no metabolic or nutritional routes to UDP-GlcNAc other than via GlcNAc-6-phosphate. Analysis of the protein glycosylation phenotype of the TbGNA1 mutant under nonpermissive conditions revealed that poly-N-acetyllactosamine structures were greatly reduced in the parasite and that the glycosylation profile of the principal parasite surface coat component, the variant surface glycoprotein (VSG), was modified. The significance of results and the potential of TbGNA1 as a novel drug target for African sleeping sickness are discussed.
INTRODUCTION
The African trypanosomes are protozoan parasites that cause human sleeping sickness and nagana in cattle, both of which are fatal diseases if they are untreated. The current drugs are highly toxic; for example, melarsoprol, the front-line drug for sleeping sickness with central nervous system involvement, kills 5% of patients and is difficult to administer. With the exception of eflornithine, an unsuccessful anticancer drug that was registered for treatment of human African trypanosomiasis in 1990, no new drug has been registered for clinical use in over 50 years.
In the mammalian bloodstream form (BSF) of the parasite, a dense coat of 5 × 106 glycosylphosphatidylinositol (GPI)-anchored variant surface glycoprotein (VSG) homodimers decorates the plasma membrane. The parasite survives the immune attack of the vertebrate host due to antigenic variation, a process that involves replacement of the VSG coat by another composed of antigenically different VSG molecules (7). In addition to VSG, the parasite also expresses a number of less abundant glycoproteins, some of which are specific to and essential for its infectious bloodstream form. Examples include the ESAG6/ESAG7 heterodimeric transferrin receptor (45), the major lysosomal glycoprotein p67 (1, 38), and the Trypanosoma brucei membrane-bound histidine acid phosphatase MBAP1 (TbMBAP1) (13). All of these molecules contain mannose, galactose, and N-acetylglucosamine (GlcNAc) residues. The GlcNAc sugar is present in the N-acetylchitobiosyl cores of all glycoprotein N-linked glycans; and glucosamine (GlcN), derived from GlcNAc by de-N-acetylation (11), is present in protein-linked and free GPI structures. N-Acetylglucosamine is also found in N-acetyllactosamine (LacNAc) units of Galβ(1-4)GlcNAc that decorate conventional complex N-linked glycans (28) and appear in unusual giant poly-LacNAc-containing N-linked glycans throughout the flagellar pocket and endosomal/lysosomal system of the bloodstream form of the parasite (3). LacNAc repeats and lacto-N-biose repeats of Galβ(1-3)GlcNAc also appear as side chains of the procyclin GPI anchor and free GPIs in the procyclic form of the organism (35, 42, 48).
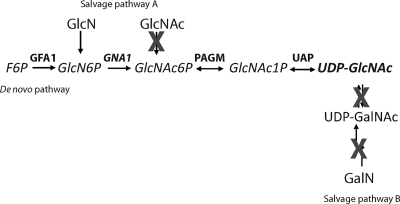
Sugar nucleotides are used by glycosyltransferases as donor substrates in glycosylation reactions (15). They are synthesized in the cytoplasm and used there and/or transported through specific transporters into the lumen of the Golgi apparatus (and/or endoplasmic reticulum [ER]). Sugar nucleotides may be formed by a de novo pathway or by salvage pathways, the latter involving the activation of the sugar using a kinase and a pyrophosphorylase. The source of GlcNAc in eukaryotic glycoconjugates is the sugar nucleotide UDP-GlcNAc. The sequence of reactions for the eukaryotic de novo pathway is represented in Fig. 1, starting from fructose-6-phosphate (F-6P). Two alternative salvage routes can also exist. In one case, GlcN or GlcNAc taken up by the cell from the medium (or from degradation of oligosaccharide moieties) is phosphorylated (19) to generate GlcN-6-phosphate (GlcN-6P) and GlcNAc-6-phosphate (GlcNAc-6P), respectively, and this then feeds the biosynthetic pathway. This salvage pathway has been confirmed by biosynthetic radiolabeling of T. brucei glycoproteins with [3H]GlcN (14), presumably via the action of hexokinase (GlcN → GlcN-6P). As T. brucei is unable to interconvert UDP-GalNAc to UDP-GlcNAc (42), the second salvage route, starting from galactosamine (GalN), can be discounted (Fig. 1). The de novo pathway from glucose is probably the most important in vivo, since T. brucei does not take up GlcNAc (4) and free GlcN is not an abundant sugar in either mammals or insects.
Fig. 1.
UDP-GlcNAc biosynthetic pathway in eukaryotes. Abbreviations: GFA1, glucosamine 6-phosphate synthetase (EC 2.6.1.16); PAGM, phosphoacetylglucosamine mutase (EC 5.4.2.3).
The biosynthesis of several sugar nucleotides has been shown to be essential for the survival and for infectivity of trypanosomatid parasites. Thus, UDP-galactose is essential in both bloodstream and procyclic forms T. brucei and also appears to be essential in T. cruzi (25, 42, 43), GDP-mannose is essential for infectivity in Leishmania major and survival in T. brucei (9, 10), and UDP-GlcNAc is essential for L. major pathogenesis in the mammalian host (34) and the growth and survival of bloodstream-form T. brucei (46).
The glucosamine 6-phosphate N-acetyltransferase (GNA1) enzymes (EC 2.3.1.4; Fig. 1) catalyze the acetylation of GlcN-6P using the cofactor acetyl coenzyme A (acetyl-CoA) and have been characterized in several eukaryotes, including Mus musculus and Saccharomyces cerevisiae (5, 32). The crystal structures of the S. cerevisiae, Aspergillus fumigatus, and human enzymes have been solved (20, 39, 53). GNA1 has also been considered a potential antifungal drug target since Candida albicans GNA1 mutants show reduced virulence in mice (31). Here, we present biochemical and structural properties, subcellular localization, and drug target genetic validation of T. brucei GNA1 (TbGNA1).
MATERIALS AND METHODS
Parasite culture.
Strain 427, variant MITat1.2 (also known as variant 221), T. brucei bloodstream-form parasites that express T7 polymerase and tetracycline repressor protein under G418 selection were cultured in HMI-9 medium (55) up to a density of ∼2 × 106 cells/ml at 37°C with 5% CO2.
Cloning and sequencing of TbGNA1.
The putative TbGNA1 open reading frame (ORF) identified in the T. brucei genome database (GeneDB accession number Tb11.01.2886) was amplified by PCR from genomic DNA with Platinum Taq High Fidelity DNA polymerase (Invitrogen) using a forward primer containing a BamHI site (underlined), 5′-CGCGGATCCATGACCGATATAGTGGACCTGGA-3′, and a reverse primer containing an XhoI site (boldface), 5′-CGCCTCGAGCTACAAATCCAGCCTCATTTGTCT-3′. The products of three separate PCRs were cloned into the BamHI/XhoI site of pGEX 6P-1 (GE Healthcare), and a representative clone from each PCR was sequenced in both directions. The TbGNA1 ORF sequence has been deposited in the EMBL nucleotide sequence database (accession number FR715998). Oligonucleotides were obtained from the University of Dundee oligonucleotide facility. All plasmids were verified by sequencing (DNA Sequencing Service, College of Life Sciences, University of Dundee).
Reverse transcription-PCR (RT-PCR).
RNA was extracted using RNeasy extraction kits with on-column DNase digestion (RNase-free DNase; Qiagen). RNA samples, 50 ng, were treated with Omniscript reverse transcriptase (Qiagen) to generate cDNA. The cDNAs were then amplified by PCR using Taq polymerase and TbGNA1 ORF primers (forward, 5′-ATGACCGATATAGTGGACCTGGA-3′; reverse, 5′-CTACAAATCCAGCCTCATTTGTCT-3′) and Dol-P-Man synthetase (DPMS) primers (forward, 5′-AATGGATGCGGACCTTCAGCACCCAC-3′; reverse, 5′-TAGAACCGTGAGCGCGGTGCCATAC-3′) as a constitutively expressed positive control.
Southern blotting.
Genomic DNA was prepared from 1 × 108 to 2 × 108 cells using DNAzol (Helena Biosciences). The hygromycin phosphotransferase resistance gene (HYG), puromycin acetyltransferase resistance gene (PAC), TbGNA1, and TbGNA1 5′ untranslated region (UTR) probes were amplified, purified, 32P labeled by an Amersham Rediprime II random prime labeling system (GE Healthcare), and used to probe the diverse digests of 3 μg of T. brucei genomic DNA.
Construction of a TbGNA1 conditional null mutant (TbGNA1cKO).
The gene replacement cassettes were generated by PCR amplification of 500 bp of UTR immediately flanking the 5′ and 3′ ends of the TbGNA1 ORF with Platinum Taq High Fidelity DNA polymerase (Invitrogen) using forward primers 5′-CGTTGTCGACCATTTGGTCAAGGTGCTGGT-3′ (SalI site, boldface) and 5′-GTTTAAACTTACGGACCGTCAAGCTTTGTATAGCTTCACAACTTTCG-3′ and reverse primers 5′-gacggtccgtaagtttaaacGGATCCttttttttgtctgcattttcgat-3′ and 5′-ATAAGTAAGCGGCCGCTAGCATACAGCACCTGTTAC-3′ (NotI site, boldface). The two PCR products were then used in a separate PCR to produce a construct containing the 5′ UTR linked to the 3′ UTR by a short HindIII, PmeI (italics), or BamHI (capital letters) cloning site. The resulting DNA fragment was then ligated into the pGEM-5Zf(+) vector (Promega) using SalI/NotI sites. Antibiotic resistance markers were cloned into the HindIII/BamHI restriction sites between the two UTRs to produce two constructs, one containing PAC and one containing HYG. To generate the tetracycline-inducible ectopic copy of TbGNA1 ORF, the primers 5′-GTTTAAACTTACGGACCGTCAAGCTTATGACCGATATAGTGGACCTGGA-3′ and 5′-CGCGGATCCCTACAAATCCAGCCTCATTTGTCT-3′ were used to PCR amplify the ORF, which was cloned into the vector pLew100 using the HindIII and BamHI sites, respectively (italics). These constructs were purified using a Qiagen Maxiprep kit, double digested with SalI/NotI to linearize the DNA, precipitated, washed twice with 70% ethanol, and redissolved in sterile water. The linearized DNA was electroporated into T. brucei bloodstream cells (strain 427, variant 221) that were stably transformed to express T7 RNA polymerase and the tetracycline repressor protein under G418 selection. This cell line will be referred to as “wild type” from here on. Cell culture, transformation, and selection were carried out as previously described (55).
Protein expression and purification.
Expression was performed using Escherichia coli ArcticExpress RP(DE3) cells (Stratagene) transformed with the pGEX 6P-1 vector containing the TbGNA1 ORF. The cells were grown at 30°C until their optical density reached 1.2, and then isopropyl β-d-1-thiogalactopyranoside (IPTG) was added to a final concentration of 0.025 mM. The cell culture was then grown for 48 h at 12°C. Cells were harvested; washed in 50 mM Tris-HCl, pH 7.5–0.25 M NaCl (buffer A), 1 mg/ml lysozyme, and complete protease inhibitor cocktail tablets (Roche); and then lysed in a French press. The lysate was cleared by centrifugation (20,000 × g, 30 min, 4°C), and the supernatant containing N-terminally glutathione S-transferase (GST)-tagged TbGNA1 protein was incubated at 4°C for 2 h with glutathione-Sepharose 4B beads (GE Healthcare) previously equilibrated with buffer A. After washing with buffer A, the GST tag was cleaved in situ using PreScission protease (a kind gift of Daan Van Aalten, University of Dundee) in buffer A with 1 mM dithiothreitol (DTT) for 16 h at 4°C. The sample was further purified by gel filtration using a Superdex 200 HR30 high-performance liquid chromatography (HPLC) column (Amersham Biosciences). Proteins were eluted at 0.5 ml/min in buffer A and monitored by measurement of the absorbance at 280 nm. Fractions (2 ml) were collected, and the presence of TbGNA1 in peak fractions was verified by SDS-PAGE. The protein of interest was then concentrated using a Vivaspin concentrator (Vivascience) with a 3,500-kDa-molecular-mass cutoff at 4°C. TbGNA1 was stored at −80% in buffer A–10% glycerol. For raising antibodies (see below), GST-TbGNA1 was eluted from the glutathione-Sepharose 4B beads with buffer A containing 10 mM glutathione. Recombinant protein identity was performed by the Proteomics and Mass Spectrometry Facility, College of Life Sciences, University of Dundee.
Recombinant TbGNA1 (1 mg/ml in buffer A) was analyzed by sedimentation velocity centrifugation using a Beckman Optima XL-1 analytical ultracentrifuge with an AN50-Ti rotor at 32,000 rpm at 20°C. Absorbance data (72 scans at 280 nm) were collected and analyzed using the SEDFIT program (44). TbGNA1 was assumed to be globular, and its density was predicted from its amino acid composition.
For crystallization purposes, TbGNA1 was expressed in resistant strain E. coli BL21(DE3)-V2R-pRARE2 using a Lex bioreactor (Harbinger Biotechnology and Engineering, Markham, Ontario, Canada) and purified according to methods described previously (51). The protein sample identity and purity were evaluated by mass spectrometry and SDS-PAGE.
Crystallographic analysis, data processing, and refinement.
TbGNA1 with the His6 tag intact was crystallized by mixing 1.5 μl of protein (at a concentration of 10.9 mg/ml in a buffer of 10 mM HEPES, pH 7.5, 500 mM NaCl) with 1.5 μl of reservoir solution containing 19% polyethylene glycol 3350 and 0.2 M ammonium tartrate in a hanging-drop vapor diffusion setup at 18°C in VDX plates (Hampton Research, Aliso Viejo, CA). Crystals appeared in 2 to 3 days and were grown to 80 by 150 μm in 2 weeks. Crystals were briefly soaked in the reservoir solution supplemented with 15% ethylene glycol before they were flash frozen in liquid nitrogen for data collection. Single-wavelength data were collected with an FR-E Superbright rotating anode X-ray source on a Raxis IV++ image plate (Rigaku, The Woodlands, TX).
Diffraction data were reduced and scaled with the program XDS (22). Molecular replacement solution was obtained with the program balbes (24) using S. cerevisiae GNA1 as a searching model (Protein Data Bank [PDB] accession number 1I1D). Iterative cycles of model building and refinement were carried out using the Coot and refmac programs (12, 22, 33) The refinement statistics data for TbGNA1 (PDB accession number 3I3G) are listed in Table 1. All structural figures were produced using the PyMOL program (www.pymol.org).
Table 1.
Crystallographic parameters for TbGNA1a
| Parameter | Value(s) for TbGNA1b |
|---|---|
| Data collection statistics | |
| Space group | P212121 |
| Unit cell dimensions | |
| a (Å) | 50.86 |
| b (Å) | 64.44 |
| c (Å) | 82.77 |
| Alpha (°) | 90.00 |
| Beta (°) | 90.00 |
| Gamma (°) | 90.00 |
| Wavelength | 1.5418 |
| Resolution range (Å) | 40–1.86 |
| No. of unique reflections | 23,634 |
| No. of reflections observed | 23,625 |
| Rsym | 0.049 (0.237) |
| I/σ〈I〉 | 28.94 (7.5) |
| Completeness (%) | 99.9 (99.9) |
| Redundancy | 7.0 (6.9) |
| Refinement statistics | |
| Resolution range | 35.96–1.86 (1.91–1.86) |
| No. of reflections | 22,408 (1,628) |
| No. of reflections in test set | 1,217 (95) |
| Rwork/Rfree | 0.243/0.282 (0.309/0.331) |
| No. of atoms | |
| Protein | 2,265 |
| Water | 238 |
| Mean B factor (Å2) | 20.24 |
| Ramachandran (%) | |
| Favored | 98.15 |
| Allowed | 1.85 |
| Disallowed | 0 |
| RMSD | |
| Bond length (Å) | 0.002 |
| Bond angle (°) | 0.531 |
PDB accession number 3I3G.
Values in parentheses are for outer shell.
Glucosamine 6-phosphate N-acetyltransferase activity assay.
Acetyl coenzyme A, GlcN-6P, and coenzyme A were supplied by Sigma. All measurements were performed at room temperature in triplicate in a 96-well-plate format (Nunc). For activity measurement, 500 μM GlcN-6P and 500 μM acetyl-CoA were dissolved in 25 mM Tris-HCl–150 mM NaCl, pH 7.2, in a total volume of 50 μl. The reactions were initiated by adding 50 ng TbGNA1 protein and stopped with 50 μl of a solution containing 25 mM bis-Tris-propane, 250 mM NaCl, 2 mM EDTA, and 6.4 M guanidine chloride, pH 7.5. Fifty microliters of dithio-bis(2-nitrobenzoic acid) (DTNB) solution (1 mM DTNB in 0.1% dimethyl sulfoxide containing 25 mM Tris-HCl, 250 mM NaCl, and 2 mM EDTA, pH 7.5) was added. Reaction of DTNB with coenzyme A, a by-product of the acetylation of GlcN, generates 2-nitro-5-thiobenzoate anion, and its absorbance was measured at 412 nm. Absorbance was quantified using a Spectra Max 340 PC spectrometer (Molecular Devices), and data were analyzed with the Grafit (version 5) program. For the determination of kinetic constants, different concentrations up to 1.5 mM GlcN-6P and acetyl-CoA were used.
TbGNA1 localization.
Two BALB/c adult mice were used to raise polyclonal antibodies against GST-tagged TbGNA1. About 0.1 mg per mouse was used for immunization with Freund's complete adjuvant. Each mouse received two further immunizations with Freund's incomplete adjuvant over 2 months. Antibodies were then affinity purified on CNBr-Sepharose-immobilized TbGNA1 that had had its GST tag removed with PreScission protease. A similar protocol was used to immunize two New Zealand rabbits with 0.5 mg of purified TbGNA1 protein per animal. Wild-type bloodstream-form T. brucei cells were grown in HMI-9 medium to a density of 1 × 106 cells/ml, harvested by centrifugation, and resuspended in trypanosome dilution buffer (0.1 M Na2HPO4, 0.01 M NaH2PO4, 0.025 M KCl, 0.4 M NaCl, 5 mM MgSO4, 0.1 M glucose adjusted to pH 7.45 with HCl) to a density of 4 × 107 cells/ml. Aliquots (15 μl) were added to 13-mm coverslips, left at 4°C for 15 min, and fixed in 1 ml 4% paraformaldehyde in phosphate-buffered saline (PBS) for 30 min, followed by three 5-min washes in 2 ml PBS. Cells were permeabilized with 0.1% Triton X-100 in PBS for 10 min at room temperature. Samples were then blocked in 5% fish skin gelatin (FSG) in PBS containing 10% normal goat serum. The coverslips were incubated with immunopurified mouse anti-TbGNA1 (0.5 μg/ml) and rabbit anti-glyceraldehyde-3-phosphate dehydrogenase (anti-GAPDH) antiserum at a 1:2,000 dilution or rabbit antienolase antiserum at a 1:4,000 dilution in 1% FSG in PBS and 0.05% Triton X-100. Both anti-GAPDH and antienolase were kind gifts of Paul Michels (Catholic University of Louvain). Samples were then washed as described above in PBS and incubated with 50 μl of Alexa 594-conjugated anti-mouse IgG and Alexa 488-conjugated anti-rabbit IgG for 1 h. Coverslips were mounted on glass slides, sealed with Hydromount containing 2.5% 1,4-diazabicyclo[2.2.2]octane, and left to dry in the dark for 30 min. Microscopy was performed on a Zeiss LSM 510 META confocal microscope.
Sugar nucleotide analysis.
Sugar nucleotide analysis was performed as described previously (49). Briefly, cells were pelleted by centrifugation, washed in ice-cold PBS, and lysed in 70% ethanol in the presence of 20 pmol of GDP-glucose internal standard. Sugar nucleotides were extracted using EnviCarb columns (Supelco) and analyzed using multiple-reaction-monitoring (MRM) liquid chromatography-tandem mass spectroscopy (LC-MS/MS).
Lectin blotting.
T. brucei cells washed with trypanosome dilution buffer and hypotonically lysed in 300 μl of water containing 0.1 mM 1-chloro-3-tosylamido-7-amino-2-heptone (TLCK) and 1 μg/ml leupeptin. Cell ghosts were harvested by centrifugation (13,000 × g for 10 min), and the pellet was resuspended in SDS-sample buffer containing 8 M urea. The lysed extracts were subjected to electrophoresis under reducing conditions with 1.5 × 107 cell equivalents/lane on a NuPAGE 4 to 12% bis-Tris gradient (Invitrogen) using morpholinepropanesulfonic acid (MOPS)-SDS running buffer. Proteins were then transferred to a nitrocellulose membrane under normal Western blotting conditions.
Membranes were stained with Ponceau S solution to demonstrate equal loading; blocked with 5% bovine serum albumin, 0.05% Igepal detergent (Sigma), and 0.15 M NaCl in 50 mM Tris-HCl, pH 7.4; and then incubated with 0.33 μg/ml of biotinylated tomato lectin (Vector Labs), with or without 3 mg/ml chitin hydrolysate (Vector Labs), and then with 1:10,000-diluted ExtraAvidin-horseradish peroxidase conjugate (Sigma). All membranes were then developed by chemiluminescent detection (ECL-plus; GE Healthcare).
Purification of sVSG.
The VSG coat of trypanosomes can be conveniently released in a soluble form (sVSG) through osmotic cell lysis at 37°C (14). We used a modified version of this procedure of Cross without the cell lysis and membrane washing step at 0°C (26). Briefly, T. brucei cultures (100 ml) were washed in trypanosome dilution buffer, resuspended in 300 μl of lysis buffer (10 mM NaH2PO4-Na2HPO4, pH 8.0, 0.1 mM TLCK, 1 μg/ml leupeptin, and 1 μg/ml aprotinin), and incubated at 37°C for 10 min. The lysate was then cooled on ice for 2 min and centrifuged at 16,000 × g for 5 min. The supernatant was applied to 200 μl of DE52 (Whatman) preequilibrated in 10 mM NaH2PO4-Na2HPO4, pH 8.0, buffer and eluted 4 times with 200 μl of fresh lysis buffer. The eluates were pooled and concentrated to 100 μl using a YM-10 spin concentrator (Microcon), yielding about 50 μg of sVSG. The majority of the buffer salts were removed by diafiltration with three additions of 0.5 ml of water. This procedure yields sVSG preparations that are sufficiently pure (>90% by SDS-PAGE and Coomassie blue staining) for analysis by mass spectrometry (26, 46).
Electrospray mass spectrometry of sVSG.
Intact sVSG was diluted to 0.05 μg/μl in 50% methanol–1% formic acid and loaded into Micromass type F nanotips. The sVSG was analyzed by positive-ion electrospray tandem mass spectrometry using an Applied Biosystems Q-StarXL instrument (26, 46). The data were processed using the Bayesian protein reconstruction algorithm (ABI Analyst Software).
Accession numbers.
DNA sequence for T. brucei GNA1 has been deposited in the EMBL nucleotide sequence database, accession number FR715998. Coordinates for TbGNA1 determined by X-ray crystallography have been deposited in the Protein Data Bank, with accession number FR715998 for 1.86-Å resolution.
RESULTS
Identification, cloning, and expression of TbGNA1.
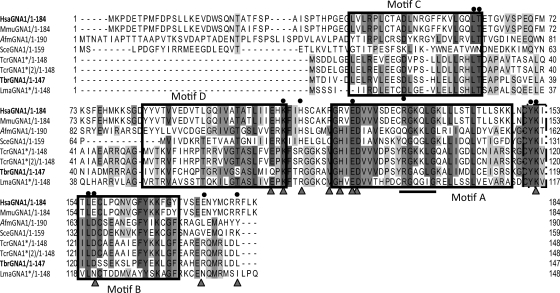
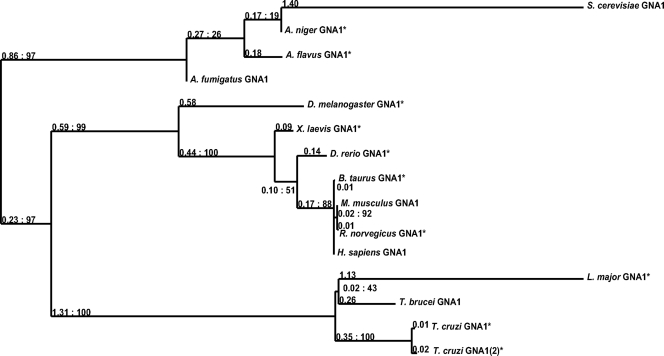
GNA1 is an amino sugar N-acetyltransferase and a member of the GCN5-related N-acetyltransferase (GNAT) superfamily (30, 32). A BLASTp search of the T. brucei predicted protein database with the Saccharomyces cerevisiae GNA1 amino acid sequence (NCBI accession number NP_116637.1) and human GNA1 (NCBI accession number NP_932332.1) revealed a putative TbGNA1 gene (GeneDB accession number Tb11.01.2886). The 444-bp TbGNA1 open reading frame (EMBL accession number FR715998) was amplified by PCR from genomic DNA prepared from T. brucei strain 427. There is a single-nucleotide difference, with a corresponding codon change, between the 427 sequence reported here and the 927 genome strain (G184 is replaced by C184). Sequence analysis suggests that TbGNA1 shares structural similarities with other members of the GNAT superfamily and, in particular, with GNA1 orthologues (Fig. 2). The closest relatives to TbGNA1 are putative GNA1s predicted from other kinetoplastid genome sequences. T. cruzi contains two putative genes for TcGNA1 with 63 and 62% identities to the T. brucei gene sequence, while the L. major gene is the least similar with 44% identity. In a maximum likelihood tree obtained using the PhyML program (2), kinetoplastid GNA1 sequences have a separate clade (Fig. 3). Compared to the sequence of its human orthologue (Fig. 2), the T. brucei GNA1 shares only 26% identity, although most of the residues involved in activity are conserved. Thus, only three conservative changes are observed in the 9 amino acids described to be essential for activity (23, 30). The charge distribution at the proposed GlcN-6P binding pocket has an important role not only for substrate affinity but also for catalytic activity. The consensus motif for GCN5-related N-acetyltransferases (R/Q)-XX-G-X-G (52), where X is any amino acid, is also conserved in T. brucei GNA1, corresponding to R-GA-G-L-G (amino acids 94 to 99). Nevertheless, the low sequence similarity between TbGNA1 and Homo sapiens GNA1 (HsaGNA1) prompted an investigation of whether differences also extended to the crystal structure.
Fig. 2.
Alignment of GNA1 predicted amino acid sequences. The sequences of Homo sapiens (Hsa), Mus musculus (Mmu), Aspergillus fumigatus (Afm), Saccharomyces cerevisiae (Sce), T. cruzi (Tcr), T. brucei, and L. major (Lma) were aligned using the Clustal W (http://www.ebi.ac.uk/) and Jalview (http://www.jalview.org/) programs. Residues required for activity (circles) and those involved in charge distribution (triangles) are highlighted. The classical four motifs for GNA1s (motifs A to D) are boxed, and the consensus motif [(R/Q)-X-X-Q-X-G] for GNC5-related N-acetyltransferase family is underlined in motif A.
Fig. 3.
Phylogenetic analyses of glucosamine 6-phosphate N-acetyltransferases. The amino acid sequences of the GNA1s from mouse (NCBI accession number 54342), rat (NCBI accession number 498486), bovine (NCBI accession number 512299), Homo sapiens (NCBI accession number 64841), Saccharomyces cerevisiae (NCBI accession number 850529), Aspergillus flavus (NCBI accession number 7920275), Aspergillus niger (NCBI accession number 4986135), Aspergillus fumigatus (NCBI accession number 3505194), Danio rerio (NCBI accession number 554143), Xenopus laevis (NCBI accession number 379524), Trypanosoma brucei (NCBI accession number 3665679), T. cruzi (NCBI accession numbers 3551278/3544900), and L. major (NCBI accession number 5653516) were aligned, and the phylogenetic relationships were analyzed using the Jalview (version 2.6) (54) and PhyML (2) programs. The tree was rooted midpoint after calculation, and the numbers after the colon correspond to bootstrap support for each branch. The branch lengths given before the colon were calculated under the WAG model. Asterisks indicate putative GNA1s identified by sequence homology.
The TbGNA1 ORF was cloned into a pGEX 6P-1 expression vector with a GST tag at the N terminus and a PreScission protease site between the protein and the tag. The plasmid was transformed into E. coli DH5α cells and purified as described in Materials and Methods. Analysis of the untagged recombinant TbGNA1 protein by gel filtration and analytical ultracentrifugation at 0.5 mg/ml and 0.75 mg/ml indicated that the majority of TbGNA1 is dimeric at these concentrations (see Fig. S1 in the supplemental material).
Crystal structure of TbGNA1.
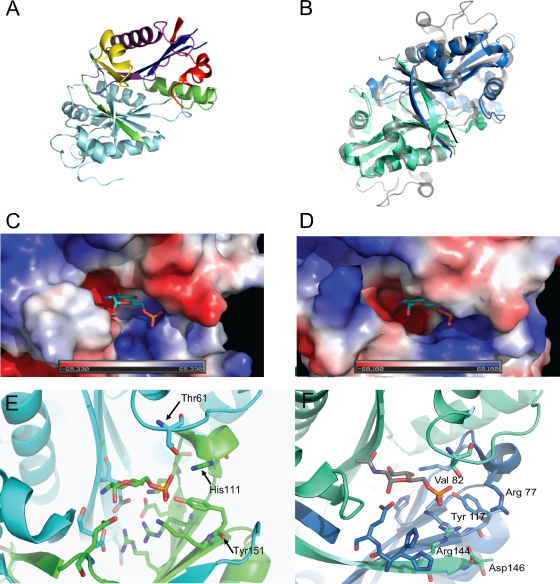
Several TbGNA1 constructs were prepared in order to achieve crystallization; only a construct without the first 4 N-terminal residues was successful. The refined structure contains a homodimer, with each monomer (named chains A and B in the crystal structure) composed of 146 residues and with only the amino-terminal valine missing for chain A in the electron density map. Chain B includes all residues from the construct, including 6 residues from the amino-terminal His tag. Its structure resembles a classic GNAT fold (39, 52), containing all four motifs (A to D; Fig. 2) conserved among the GNATs. Unfortunately, no structure with the natural ligands (GlcN-6P, acetyl-CoA, GlcNAc-6P, CoA) or glucose-6-phosphate (21) could be obtained. Similar to other GNATs, including the Saccharomyces cerevisiae (PDB accession number 1I1D) and the human (PDB accession number 2HUZ) homologues, TbGNA1 (PDB accession number 3I3G) features a dimeric structure created by the intertwining of the C-terminal beta strands (Fig. 4 B). The TbGNA1 dimer is present not only in the crystal lattice but also in solution, as demonstrated from gel filtration and analytical centrifugation results (see Fig. S1 in the supplemental material). The GNAT superfamily signature is clearly apparent: an N-terminal strand (β1), followed by three α helices (α3-α4-α5), three antiparallel β strands, a signature central helix (α8), a fifth β strand, two α helices (α9 and α10), and a sixth and final β strand (Fig. 4A). TbGNA1 adopts the same overall fold as its human orthologue and shows the typical intertwining C-terminal beta strands, but it maintains a longer hydrogen-bonding network in this area and in this respect more resembles the S. cerevisiae GNA1 structure (PDB accession number 1I1D). The HsaGNA1 structure has 35 extra residues at the N terminus that are not found in the T. brucei gene sequence. Overall, the root mean square difference (RMSD) between TbGNA1 main chains and S. cerevisiae GNA1 is 1.41 Å, while compared to the HsaGNA1 sequence, the RMSD is 1.55 Å, suggesting no major conformational differences but not exact secondary structure. However, when the RMSD is narrowed to just the C terminus (residues 63 to 134), the results for S. cerevisiae GNA1 (SceGNA1) are extremely close, with 0.804 Å compared to 1.411 Å for humans. An Aspergillus fumigatus GNA1 (AfmGNA1) structure (PDB accession number 2VEZ) was recently reported as well (20) and also adopts a C-terminal fold similar to that of TbGNA1. As a common feature, TbGNA1, AfmGNA1, and SceGNA1 all adopt a long beta strand that lines the bottom of the sugar binding pocket, whereas the human structure shows an interruption, placing several residues outside the pocket lining, pointing away from the active-site entrance (Fig. 4B, arrow).
Fig. 4.
Crystal structure of TbGNA1. (A) Representation of the two domains present in the TbGNA1 monomer from N-terminal motif C (red), motif D (blue), motif A (violet), and motif B (yellow). Light blue, the second chain that forms the dimer. (B) Overall structure comparison of TbGNA1 (cyan and blue, for each domain in the dimeric structure) and HsaGNA1 (gray). Arrows point to the C-terminal discrepancies between H. sapiens and T. brucei at the entrance to the sugar binding site. (C) Close-up of the HsaGNA1 active site, representing the surface charge and shape of the pocket. The ribbon cartoon shows the specific interactions with the substrates. (D) TbGNA1 active site, representing the surface charge and shape of the pocket. (E) Ribbon cartoon with sticks showing the specific interactions with the substrates in H. sapiens GNA1 structure (green and blue, corresponding to different domains). (F) Ribbon cartoon with sticks showing the specific interactions with the substrates in T. brucei GNA1 structure. As TbGNA1 could not be crystallized in the presence of substrates, the figure is created by modeling in an N-acetyl-d-glucosamine-6-phosphate from the human structure.
As noted before with the AfmGNA1 structure, the acetyl-CoA pocket is relatively unchanged between HsaGNA1 and TbGNA1, but the sugar binding pocket contains some significant differences. Several residues essential for the interactions with the substrates, such as K118 and Y117 (involved in positioning the phosphate group) and D87 (which positions the carbohydrate residue in the back of the pocket), are highly conserved between the three structures. In spite of the high conservation observed for the residues involved in activity, a significantly different environment for the pocket lining can also be found in the parasite enzyme compared to the HsGNA1. In the parasite enzyme, V82 replaces R116 and I110 replaces T76, changing the shape and charge of the pocket. In addition, TbGNA1 H139 creates a smaller entrance to the pocket (compared to E174 in HsaGNA1 or A151 in SceGNA1), so that overall the pocket shape is narrower and deeper than the pocket of the human protein (Fig. 4D). The narrowness of the TbGNA1 pocket could possibly be attributed to the fact that this structure is without bound substrate. However, neither the SceGNA1 nor AfmGNA1 pockets are much affected by substrate binding, making this unlikely. Lastly, replacement of H111 (Fig. 4E; conserved in S. cerevisiae, H. sapiens, and A. fumigatus and involved in phosphate positioning) with R77 is also interesting, as this residue is positioned out of the pocket. The longer residue could possibly link to a phosphate via a linking water from this outside position, but it could also potentially crowd out correct phosphate positioning if it moved into a similar position as the histidines in all other structures (Fig. 4F).
Enzyme kinetics.
To address the question whether TbGNA1 encodes an active acetyltransferase, enzymatic assays on the recombinant protein were performed as described in Materials and Methods. Analysis of the kinetic data demonstrated that it obeyed simple Michaelis-Menten kinetics for both substrates with linear Lineweaver-Burke plots (see Fig. S2 in the supplemental material). The apparent Km values for GlcN-6P (144 ± 11 μM) and acetyl-CoA (234 ± 44 μM) are similar to those reported for other GNA1s (23, 32). The activities of the recombinant proteins were not affected by the presence of His6 or GST tags (data not shown). Recombinant TbGNA1 can be stored at −80°C for several months in 10% glycerol with no significant loss of activity.
Subcellular localization of TbGNA1.
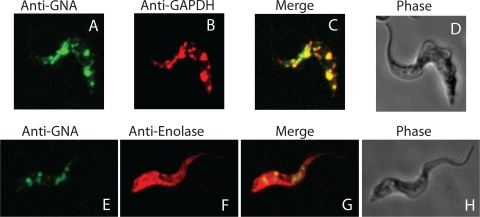
Mouse anti-TbGNA1 affinity-purified antibodies were used together with rabbit anti-GAPDH antibody as a glycosomal marker. The secondary antibodies were antimouse Alexa 488 (red) and antirabbit Alexa 594 (green). The fluorescence micrographs of wild-type cells show that the anti-GAPDH colocalized with the anti-TbGNA1, indicating that TbGNA1 is located predominantly in glycosome microbodies in bloodstream-form T. brucei (Fig. 5 A to C). A similar experiment was performed with rabbit antienolase antibodies as a cytosolic marker (Fig. 5E to G), where no colocalization was found. Additional images showing more cells are shown in Fig. S3 in the supplemental material.
Fig. 5.
Subcellular localization of TbGNA1. Wild-type bloodstream-form T. brucei cells were stained with affinity-purified mouse anti-TbGNA1 and Alexa 488-conjugated antimouse antibody (green channel) (A and E) and with rabbit anti-GAPDH and Alexa 594-conjugated antirabbit antibody (red channel) (B) to mark the glycosomes or rabbit antienolase and Alexa 594-conjugated anti-rabbit antibody (red channel) (F) to mark the cytosol. Merged images are shown (C and G), and corresponding phase-contrast images are shown (D and H).
Construction of a TbGNA1 conditional null mutant.
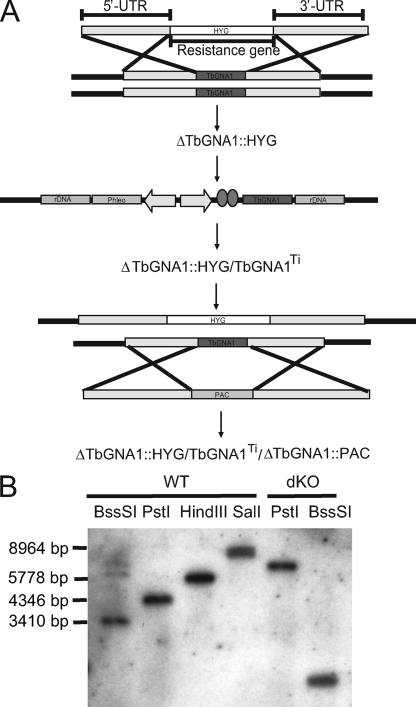
RT-PCR with the primers described in Materials and Methods for TbGNA1 ORF expression showed that this gene is transcribed in both the procyclic and bloodstream forms of the parasite (data not shown). The conditional null mutant construction strategy summarized in Fig. 6 A was made possible because genome assembly clearly indicates that TbGNA1 is present as a single copy per haploid genome. A Southern blot analysis using a TbGNA1 ORF probe was also consistent with that conclusion (Fig. 6B).
Fig. 6.
Creation of the bloodstream-form T. brucei TbGNA1 conditional null mutant. (A) Schematic representation of the genetic transformations. The first endogenous allele of TbGNA1 was replaced with HYG, creating ΔTbGNA1::HYG. The ectopic copy (pLew100-TbGNA1) was then inserted into the rDNA region, creating ΔTbGNA1::HYG/TbGNA1Ti. The second allele of TbGNA1 was then replaced with PAC, creating ΔTbGNA1::PAC/TbGNA1Ti/ΔTbGNA1::HYG. (B) Southern blots. Genomic DNA from the wild-type (WT) T. brucei 427 was digested with BssSI, PstI, HindIII, and SalI and probed with the TbGNA1 ORF probe, showing that TbGNA1 is a single-copy gene. Genomic DNA from TbGNA1 conditional null mutant (dKO) was digested with PstI and BssSI and probed with the TbGNA1 open reading frame, confirming that the only copy present in these mutants is the ectopic gene.
The first TbGNA1 allele was replaced by homologous recombination following electroporation of the parasites in the presence of linear DNA containing HYG flanked by about 500 bp of the TbGNA1 5′ and 3′ UTRs. Following selection with hygromycin, a ΔTbGNA1::HYG clone was selected and transformed with an ectopic, tetracycline-inducible copy of TbGNA1 introduced into the ribosomal DNA (rDNA) locus under phleomycin selection (55). After tetracycline induction, the second endogenous allele was replaced by a PAC gene to yield the desired ΔTbGNA1::HYG/TbGNA1Ti/ΔTbGNA1::PAC clone. After each round of transformation, selected clones were analyzed by Southern blotting using a TbGNA1 ORF probe. The blot obtained after the last transfection (Fig. 6B) shows the successful introduction of the ectopic copy and replacement of both endogenous alleles in the ΔTbGNA1::HYG/TbGNA1Ti/ΔTbGNA1::PAC clone used for further studies. This TbGNA1 conditional null mutant cell line is referred to as TbGNA1cKO.
The TbGNA1 gene is essential to bloodstream-form T. brucei in vitro.
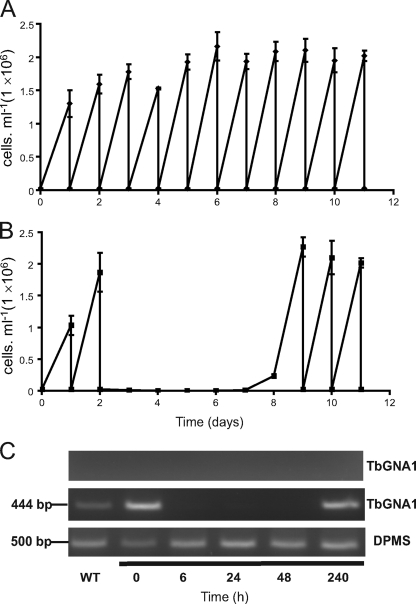
Triplicate cultures of wild-type and TbGNA1 conditional null mutant cells were inoculated under permissive and nonpermissive conditions (i.e., with and without tetracycline, respectively) at 2.5 × 105 cells/ml and subcultured every day. The TbGNA1 conditional null mutant cultures under permissive conditions had growth rates similar to the rate for the wild type (Fig. 7 A). Under nonpermissive conditions, the cells grew for 2 days but upon subculturing failed to grow for 5 days (Fig. 7B). Nevertheless, these cultures resumed growth on day 8. Analysis by reverse transcription-PCR confirmed that TbGNA1 mRNA was undetectable by 6 h of tetracycline removal but that the cells that resumed growth 10 days after tetracycline removal were reexpressing TbGNA1 mRNA (Fig. 7C). This phenomenon, where conditional null mutants become tetracycline independent, is well known in trypanosomatids and is further evidence for gene essentiality (43).
Fig. 7.
T. brucei TbGNA1 conditional null mutant: confirmation of gene essentiality in culture. (A) Growth curve for TbGNA1 conditional null mutant cells, subcultured every day, grown under permissive (plus tetracycline) conditions. (B) Growth curve for TbGNA1 conditional null mutant cells, subcultured every day, grown under nonpermissive (minus tetracycline) conditions. (C) Ethidium bromide-stained agarose gel of reverse transcription-PCR products from RNA extracted from TbGNA1 conditional null mutant cells after 0, 6, 24, and 48 h and 10 days without tetracycline, as indicated. The middle panel shows reverse transcription-PCR products using TbGNA1 primers, the upper panel is a control without reverse transcriptase, and the lower panel is a control using DPMS primers to show equal RNA input.
In order to check if addition of GlcN was sufficient for the rescue of the GNA1 knockout, medium with added GlcN (1 mM) was used in parallel cultures. However, the phenotype was not changed and the parasites died after 2 days (data not shown), showing that the de novo pathway of UDP-GlcNAc biosynthesis is the main source of this sugar nucleotide in T. brucei.
Phenotypic characterization of TbGNA1 conditional null mutant.
To analyze the effect of the selective removal of TbGNA1 gene expression on parasite UDP-GlcNAc levels, sugar nucleotides were extracted from the TbGNA1 conditional null mutant under permissive and nonpermissive conditions and quantified by the ion-pair HPLC-electrospray ionization-MS/MS method described previously (49). Briefly, sugar nucleotides were extracted from T. brucei, separated by HPLC, and quantitated by MRM tandem mass spectrometry using an internal standard, GDP-glucose, a sugar nucleotide that is not found in trypanosomes. The levels of UDP-GlcNAc, UDP-Glc, UDP-Gal, and GDP-Man in the TbGNA1 conditional null cell line under permissive conditions agree reasonably well with the wild-type levels (46, 49). However, a dramatic reduction in UDP-GlcNAc levels, to about 6%, occurs by 48 h of tetracycline removal (Table 2). The same cell cultures stopped dividing at between 48 and 72 h and died, by cell lysis, by about 72 h.
Table 2.
Quantification of sugar nucleotides by liquid chromatography-tandem mass spectrometrya
| Sugar nucleotide | Sugar nucleotide concn (pmol/107 cells) |
|||
|---|---|---|---|---|
| Wild type | 0 h | 24 h | 48 h | |
| UDP-glucose | 182 ± 15 | 210 ± 5 | 281 ± 50 | 267 ± 40 |
| UDP-galactose | 44 ± 9 | 68 ± 15 | 78 ± 8 | 94 ± 6 |
| UDP-GlcNAc | 209 ± 12 | 270 ± 14 | 224 ± 40 | 17 ± 2 |
| GDP-mannose | 12 ± 4 | 19 ± 3 | 26 ± 5 | 39 ± 12 |
| GDP-fucose | 0.18 ± 0.03 | 0.2 ± 0.04 | 0.15 ± 0.07 | 0.1 ± 0.03 |
Sugar nucleotides extracted from the TbGNA1 conditional null mutant at 0, 24, and 48 h after withdrawal of tetracycline. The experiments were performed in triplicate.
Effects of UDP-GlcNAc starvation on protein glycosylation.
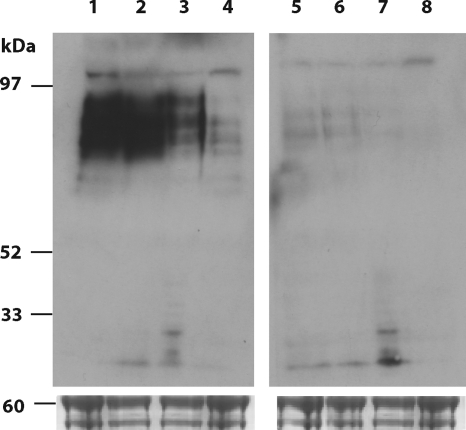
The poly-N-acetyllactosamine-specific lectin from Lycopersicon esculentum (tomato lectin) has been used for detection of glycoproteins in the flagellar pocket and endosomal/lysosomal system in T. brucei (36). The tomato lectin binding oligosaccharide structures have been shown to include a family of unusually large N-linked poly-N-acetyllactosamine-containing glycans with an average of 54 N-acetyllactosamine repeats per glycan (3). The effect of UDP-GlcNAc starvation on these structures was assessed using Western blots of whole-cell lysates from wild-type and TbGNA1 conditional null mutant cell lines probed with tomato lectin. A large smear was detected in the wild-type cell lysate, which decreased progressively from 0 to 48 h in the absence of tetracycline in the TbGNA1 conditional null mutant (Fig. 8, lanes 1 to 4). The decreased intensity of the tomato lectin binding to high-molecular-mass glycoproteins indicated a significant reduction in total poly-N-acetyllactosamine synthesis as the cellular levels of UDP-GlcNAc fall. The specificity of the tomato lectin blot for carbohydrate was confirmed by inhibition with chitin hydrolysate (Fig. 8, lanes 5 to 8).
Fig. 8.
Phenotypic characterization of TbGNA1 conditional null mutant. Tomato lectin blotting of wild-type and TbGNA1 conditional null mutant extracts. Extracts of wild type (lane 1) and TbGNA1 conditional null mutant cells 0, 24, and 48 h after tetracycline removal (lanes 2, 3, and 4, respectively) were subjected to SDS-PAGE, transferred to nitrocellulose, and probed with biotinylated tomato lectin, followed by horseradish peroxidase-streptavidin. Lanes 5 to 8, as in lanes 1 to 4, respectively, except that the tomato lectin inhibitor (chitin hydrolysate) was included with the lectin.
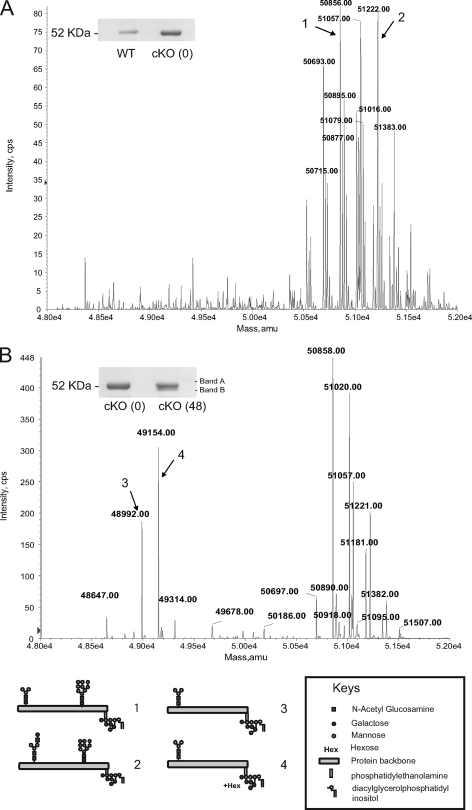
The effects of UDP-GlcNAc starvation on VSG glycosylation were also studied. sVSG was purified from wild-type and TbGNA1 conditional null mutant cells grown in the presence and absence of tetracycline for 48 h. Analysis by SDS-PAGE and Coomassie blue staining revealed that sVSGs from wild-type and TbGNA1 conditional null cells grown under permissive conditions appear as single bands, whereas sVSG from the TbGNA1 conditional null mutant grown for 48 h in the absence of tetracycline appeared as a doublet (Fig. 9 A and B). The VSG doublet present in the mutant represents VSG molecules that are similar to those found in the wild type (i.e., the upper band) and VSG molecules that have selectively lost their N-linked glycan from the C-terminal Asn428 site (i.e., the lower band), as previously described in detail for the T. brucei UDP-GlcNAc pyrophosphorylase (TbUAP; EC 2.7.7.23) conditional null mutant (46).
Fig. 9.
Electrospray mass spectrometry of sVSG from the TbGNA1 conditional null mutant. Cells grown in the presence of tetracycline (A) or absence of tetracycline (B) for 48 h were analyzed by positive-ion electrospray mass spectrometry, and the data were processed by Bayesian protein reconstruction to produce mass graphs of isobaric glycoforms. The SDS-PAGE bands for sVSG were detected by Coomassie blue staining. Models of some of the principal glycoforms similar to those observed for a TbUAP mutant are indicated. amu, atomic mass units.
DISCUSSION
In this paper, we have identified Tb11.01.2886 to be the gene encoding all GNA1 activity in T. brucei. This organism appears to have a typical eukaryotic UDP-GlcNAc biosynthetic pathway (46), and indeed, the dimeric nature and Km of TbGNA1 for GlcN-6P are similar to those of its human homologue. However, the parasite enzyme has an approximately 10-fold higher Km for acetyl-CoA than the human homologue. TbGNA1 and HsaGNA1 have only 26% amino acid sequence identity but share the four GNAT motifs, and like those of S. cerevisiae and Aspergillus, their crystal structures are quite similar (20, 21, 39, 53). Although we have only the apo structure for TbGNA1 (despite efforts to obtain complexes with acetyl-CoA, CoA, GlcN-6P, GlcNAc-6P, and Glc-6P), we can make useful comparisons between the parasite and human active-site pockets: in the human GNA1 active site, GlcN6P interacts closely with R116 and R181 (20); these two residues also directly contact the reaction product. Similar to the Aspergillus fumigatus enzyme, in TbGNA1, R116 is replaced by V82, a smaller, neutral residue, presumably reducing its ability to interact with the phosphate group. Noticeably, and in contrast to HsaGNA1, AfmGNA1, and SceGNA1, TbGNA1 replaces R181 with D146, a complete change in charge and function. A neighboring arginine (R144; Fig. 4F) is able to take over the phosphate positioning duties by poking into the pocket at the correct position and forming hydrogen bonds with the anomalous aspartate. As can be seen in Fig. 4D, in TbGNA1 there is a large positive patch that would still accommodate the phosphate moiety and a negative patch in the back, similar to the HsaGNA1. However, the size of the pocket created by the dimer is actually longer and narrower than in the human orthologue, providing potentially exploitable differences for the design or discovery of specific inhibitors.
GNA1 enzymes, like the other components of hexosamine biosynthesis, are typically cytosolic in eukaryotes, although murine GNA1 has been found in association with the cytoplasmic face of Golgi apparatus and other intracellular vesicle membranes (5). This association is based on a hydrophobic annexin V-like domain present in the mammalian GNA1s, which is absent in TbGNA1. In contrast, TbGNA1 localizes to the glycosomes. Glycosomes of trypanosomatids belong to the microbody family of organelles, containing the glyoxysomes of plants and peroxisomes of other eukaryotes. These organelles resemble each other with respect to morphology and contain both common and species-specific metabolic functions (29, 37). A unique feature of trypanosomatid glycosomes is their essential role in carbohydrate metabolism, i.e., glycolysis, gluconeogenesis, glycerol metabolism, and the pentose-phosphate pathway (18, 29). Glycosomal proteins are typically targeted to these organelles via a peroxisomal targeting sequence (PTS), which can be located in the C terminus (PTS-1) or N terminus (PTS-2) of the protein. Internal targeting sequences have also been proposed to be another targeting mechanism, with T. cruzi phosphoglucomutase and T. brucei triosephosphate isomerase being examples (16, 40). However, proteins may also traffic to the glycosome by piggybacking on other glycosome-targeted proteins (47). Since conventional trypanosome PTSs are not apparent for TbGNA1 or, indeed, for the T. cruzi or L. major orthologues (37), it seems likely that TbGNA1 uses either the piggybacking mechanism or some unidentified internal targeting sequence. In any case, TbGNA1 joins a list of enzymes of de novo sugar nucleotide biosynthesis that have been experimentally identified to be glycosomal components. These include the UDP-GlcNAc pyrophosphorylase (46), GDP-mannose 4,6-deydratase of GDP-fucose biosynthesis (50), UDP-glucose 4′-epimerase of UDP-galactose biosynthesis (42), and UDP-glucose pyrophosphorylase of UDP-glucose biosynthesis (27). In addition, hexokinase, necessary for all the above pathways, and phosphoglucose isomerase, necessary for GDP-mannose, GDP-fucose, and UDP-GlcNAc biosynthesis, are also known glycosomal components. Finally, phosphomannose isomerase, which contains a PTS1 signal and is necessary for GDP-mannose and GDP-fucose biosynthesis, has been found in the glycosome proteome (6). It is therefore possible that all or most of the sugar nucleotide biosynthesis machinery is localized to the glycosome in T. brucei and that the rate of synthesis of sugar nucleotides is controlled, to some degree, by compartmentalization. Such a mechanism has already been proposed to be an alternative to allosteric regulation for glycolysis in T. brucei (18).
The construction of a conditional null mutant for the TbGNA1 gene enabled us to test for its essentiality and to confirm that there are no alternative routes to GlcNAc-6P. Essentiality was the expected result, since TbUAP is known to be essential (46). However, because almost half of the genes in T. brucei encode hypothetical proteins of unknown function, that fewer still are experimentally annotated, and that the GNAT enzyme superfamily is one of the largest known, the possibility of redundancy certainly could not be excluded prior to experimental verification. A recent and relevant case in point is the unexpected nonessentiality of UDP-Glc pyrophosphorylase in L. major, where it transpires that its function can be compensated for by a relatively nonspecific UDP-sugar pyrophosphorylase (8) that is also present in T. cruzi (57).
The phenotype of the TbGNA1 conditional null mutant under nonpermissive conditions was similar to that of the TbUAP conditional null mutant, i.e., rapid cessation of growth and death by cell lysis 48 to 72 h after tetracycline removal (Fig. 7B). Also, like the TbUAP mutant, it took about 48 h for UDP-GlcNAc to fall to about 5% of wild-type levels. However, whereas UDP-glucose and UDP-galactose levels also dropped to about 60% of wild-type levels by 48 h in the TbUAP mutant, the levels of these sugar nucleotides slightly increased in the TbGNA1 mutant (Table 2). Intuitively, one would expect the latter, as a lack of UDP-GlcNAc should reduce the rate of UDP-Gal (and its UDP-Glc precursor) consumption for both the GPI anchor side chain and N-glycan processing. The reasons for the anomalous behavior in the TbUAP mutant are currently under investigation. The consequences of UDP-GlcNAc starvation, other than death by 72 h, were studied by isolating parasites 48 h after the removal of tetracycline. In these cells, two discrete sets of VSG glycoforms corresponding to the two bands seen by SDS-PAGE were observed by electrospray MS (Fig. 9): one set is similar to that of wild-type VSG, except that it is lacking in the higher-molecular-mass glycoforms, while the other set has masses consistent with the absence of oligomannose structures at the C-terminal (Asn428) N-glycosylation site. This is very similar to the situation described for the TbUAP and TbALG3 mutants (26, 46), and it reemphasizes the relative importance of N-glycosylation of the Asn263 site over that of the Asn428 site. A similar conclusion was also recently drawn for a conditional null mutant of T. brucei GDP-Man pyrophosphorylase (10).
In summary, in this paper we have extended our knowledge of the enzymology and location of hexosamine biosynthesis in T. brucei and determined that TbGNA1 is an essential gene for bloodstream-form T. brucei and, therefore, that it is a potential target of drugs with activity against the parasite. However, its therapeutic potential will obviously depend on our ability to find drug-like parasite-specific TbGNA1 inhibitors, as this enzyme is generally essential for eukaryotes (5, 31, 32). Nevertheless, inhibitors of other GNAT family members have been developed (17, 41, 56), and although the structure of TbGNA1 shows a similar dimerization mode as the human protein and both have similar activities and specificities, the active sites are dissimilar, in that several key residues are replaced. These differences could be exploited during the drug discovery process.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a Wellcome Trust Programme Grant (085622) to M.A.J.F. We are grateful to the University of Dundee Proteomics Facility and to Mark Agacan, supported by Wellcome Strategic Award 083481, for performing the proteomics and ultracentrifugation experiments. The Structural Genomics Consortium is a registered charity (number 1097737) that receives funds from the Canadian Institutes for Health Research, the Canadian Foundation for Innovation and Genome Canada, GlaxoSmithKline, Karolinska Institutet, the Knut and Alice Wallenberg Foundation, the Ontario Innovation Trust, the Ontario Ministry for Research and Innovation, Merck, the Novartis Research Foundation, the Swedish Agency for Innovation Systems, the Swedish Foundation for Strategic Research, and the Wellcome Trust.
We are grateful to Alan R. Prescott (Division of Cell Signaling and Immunology, University of Dundee) for valuable help with the microscopy. We also thank Paul Michels for providing anti-GAPDH and antienolase antibodies, Daan van Aalten for providing vectors, and Jim Procter and Geoff Barton for helpful advice on sequence comparisons.
We have no conflicts of interest to declare.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 29 April 2011.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1. Alexander D. L., Schwartz K. J., Balber A. E., Bangs J. D. 2002. Developmentally regulated trafficking of the lysosomal membrane protein p67 in Trypanosoma brucei. J. Cell Sci. 115:3253–3263 [DOI] [PubMed] [Google Scholar]
- 2. Anisimova M., Gascuel O. 2006. Approximate likelihood ratio test for branchs: a fast, accurate and powerful alternative. Syst. Biol. 55:539–552 [DOI] [PubMed] [Google Scholar]
- 3. Atrih A., Richardson J. M., Prescott A. R., Ferguson M. A. 2005. Trypanosoma brucei glycoproteins contain novel giant poly-N-acetyllactosamine carbohydrate chains. J. Biol. Chem. 280:865–871 [DOI] [PubMed] [Google Scholar]
- 4. Azema L., et al. 2004. Interaction of substituted hexose analogues with the Trypanosoma brucei hexose transporter. Biochem. Pharmacol. 67:459–467 [DOI] [PubMed] [Google Scholar]
- 5. Boehmelt G., et al. 2000. Cloning and characterization of the murine glucosamine-6-phosphate acetyltransferase EMeg32. Differential expression and intracellular membrane association. J. Biol. Chem. 275:12821–12832 [DOI] [PubMed] [Google Scholar]
- 6. Colasante C., Ellis M., Ruppert T., Voncken F. 2006. Comparative proteomics of glycosomes from bloodstream form and procyclic culture form Trypanosoma brucei brucei. Proteomics 6:3275–3293 [DOI] [PubMed] [Google Scholar]
- 7. Cross G. A. 1996. Antigenic variation in trypanosomes: secrets surface slowly. Bioessays 18:283–291 [DOI] [PubMed] [Google Scholar]
- 8. Damerow S., et al. 2010. Leishmania UDP-sugar pyrophosphorylase. J. Biol. Chem. 285:878–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Davis A. J., et al. 2004. Properties of GDP-mannose pyrophosphorylase, a critical enzyme and drug target in Leishmania mexicana. J. Biol. Chem. 279:12462–12468 [DOI] [PubMed] [Google Scholar]
- 10. Denton H., Fyffe S., Smith T. K. 2010. GDP-mannose pyrophosphorylase is essential in the bloodstream form of Trypanosoma brucei. Biochem. J. 425:603–614 [DOI] [PubMed] [Google Scholar]
- 11. Doering T. L., Masterson W. J., Englund P. T., Hart G. W. 1989. Biosynthesis of the glycosyl phosphatidylinositol membrane anchor of the trypanosome variant surface glycoprotein: origin of the non-acetylated glucosamine. J. Biol. Chem. 264:11168–11173 [PubMed] [Google Scholar]
- 12. Emsley P., Lohkamp B., Scott W. G., Cowtan K. 2010. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66:486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Engstler M., et al. 2005. The membrane-bound histidine acid phosphatase TbMBAP1 is essential for endocytosis and membrane recycling in Trypanosoma brucei. J. Cell Sci. 118:2105–2118 [DOI] [PubMed] [Google Scholar]
- 14. Ferguson M. A., Duszenko M., Lamont G. S., Overath P., Cross G. A. 1986. Biosynthesis of Trypanosoma brucei variant surface glycoproteins. N-Glycosylation and addition of a phosphatidylinositol membrane anchor. J. Biol. Chem. 261:356–362 [PubMed] [Google Scholar]
- 15. Freeze H. H., Elbein A. B. 2008. Glycosylation precursors, p. 47–61 In Varki A., Cummings R. D., Esko J. D., Freeze H. H., Stanley P., Bertozzi C. R., Hart G. W., Etzler M. E. (ed.). Essentials of glycobiology, 2nd ed. Cold Spring Harbor Laboratory Press, New York, NY [Google Scholar]
- 16. Galland N., de Walque S., Voncken F. G., Verlinde C. L., Michels P. A. 2010. An internal sequence targets Trypanosoma brucei triosephosphate isomerase to glycosomes. Mol. Biochem. Parasitol. 171:45–49 [DOI] [PubMed] [Google Scholar]
- 17. Gao F., et al. 2006. Synthesis and structure-activity relationships of truncated bisubstrate inhibitors of aminoglycoside 6-N-acetyltransferases. J. Med. Chem. 49:5273–5281 [DOI] [PubMed] [Google Scholar]
- 18. Haanstra J. R., et al. 2008. Compartmentation prevents a lethal turbo-explosion of glycolysis in trypanosomes. Proc. Natl. Acad. Sci. U. S. A. 105:17718–17723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hinderlich S., et al. 1998. Purification and characterization of N-acetylglucosamine kinase from rat liver—comparison with UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase. Eur. J. Biochem. 252:133–139 [DOI] [PubMed] [Google Scholar]
- 20. Hurtado-Guerrero R., et al. 2008. Structural and kinetic differences between human and Aspergillus fumigatus d-glucosamine-6-phosphate N-acetyltransferase. Biochem. J. 415:217–223 [DOI] [PubMed] [Google Scholar]
- 21. Hurtado-Guerrero R., Raimi O., Shepherd S., van Aalten D. D. 2007. Glucose-6-phosphate as a probe for the glucosamine-6-phosphate N-acetyltransferase Michaelis complex. FEBS Lett. 581:5597–5600 [DOI] [PubMed] [Google Scholar]
- 22. Kabsch W. 1993. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 26:795–800 [Google Scholar]
- 23. Kato N., Mueller C. R., Wessely V., Lan Q., Christensen B. M. 2005. Mosquito glucosamine-6-phosphate N-acetyltransferase: cDNA, gene structure and enzyme kinetics. Insect Biochem. Mol. Biol. 35:637–646 [DOI] [PubMed] [Google Scholar]
- 24. Long F., Vagin A. A., Young P., Murshudov G. N. 2008. BALBES: a molecular-replacement pipeline. Acta Crystallogr. D Biol. Crystallogr. 64:125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. MacRae J. I., et al. 2006. The suppression of galactose metabolism in Trypanosoma cruzi epimastigotes causes changes in cell surface molecular architecture and cell morphology. Mol. Biochem. Parasitol. 147:126–136 [DOI] [PubMed] [Google Scholar]
- 26. Manthri S., Güther M. L. S., Izquierdo L., Acosta Serrano A., Ferguson M. A. 2008. Deletion of the TbALG3 gene demonstrates site-specific N-glycosylation and N-glycan processing in Trypanosoma brucei. Glycobiology 18:367–383 [DOI] [PubMed] [Google Scholar]
- 27. Mariño K., et al. 2010. Identification, subcellular localization, biochemical properties and high-resolution crystal structure of Trypanosoma brucei UDP-glucose pyrophosphorylase. Glycobiology 20:1619–1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mehlert A., Zitzmann N., Richardson J. M., Treumann A., Ferguson M. A. J. 1998. The glycosylation of the variant surface glycoproteins and procyclic acidic repetitive proteins of Trypanosoma brucei. Mol. Biochem. Parasitol. 91:145–152 [DOI] [PubMed] [Google Scholar]
- 29. Michels P. A., Bringaud F., Herman M., Hannaert V. 2006. Metabolic functions of glycosomes in trypanosomatids. Biochim. Biophys. Acta 1763:1463–1477 [DOI] [PubMed] [Google Scholar]
- 30. Milewski S., Gabriel I., Olchowy J. 2006. Enzymes of UDP-GlcNAc biosynthesis in yeast. Yeast 23:1–14 [DOI] [PubMed] [Google Scholar]
- 31. Mio T., Kokado M., Arisawa M., Yamada-Okabe H. 2000. Reduced virulence of Candida albicans mutants lacking the GNA1 gene encoding glucosamine-6-phosphate acetyltransferase. Microbiology 146:1753–1758 [DOI] [PubMed] [Google Scholar]
- 32. Mio T., Yamada-Okabe T., Arisawa M., Yamada-Okabe H. 1999. Saccharomyces cerevisiae GNA1, an essential gene encoding a novel acetyltransferase involved in UDP-N-acetylglucosamine synthesis. J. Biol. Chem. 274:424–429 [DOI] [PubMed] [Google Scholar]
- 33. Murshudov G. N., Vagin A. A., Dodson E. J. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53:240–255 [DOI] [PubMed] [Google Scholar]
- 34. Naderer T., Heng J., McConville M. J. 2010. Evidence that intracellular stages of Leishmania major utilize amino sugars as a major carbon source. PLoS Pathog. 6:e1001245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nagamune K., et al. 2004. Surface sialic acids taken from the host allow trypanosome survival in tsetse fly vectors. J. Exp. Med. 199:1445–1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nolan D. P., Geuskens M., Pays E. 1999. N-Linked glycans containing linear poly-N-acetyllactosamine as sorting signals in endocytosis in Trypanosoma brucei. Curr. Biol. 9:1169–1172 [DOI] [PubMed] [Google Scholar]
- 37. Opperdoes F. R., Szikora J. P. 2006. In silico prediction of the glycosomal enzymes of Leishmania major and trypanosomes. Mol. Biochem. Parasitol. 147:193–206 [DOI] [PubMed] [Google Scholar]
- 38. Peck R. F., et al. 2008. The LAMP-like protein p67 plays an essential role in the lysosome of African trypanosomes. Mol. Microbiol. 68:933–946 [DOI] [PubMed] [Google Scholar]
- 39. Peneff C., Mengin-Lecreulx D., Bourne Y. 2001. The crystal structures of apo and complexed Saccharomyces cerevisiae GNA1 shed light on the catalytic mechanism of an amino-sugar N-acetyltransferase. J. Biol. Chem. 276:16328–16334 [DOI] [PubMed] [Google Scholar]
- 40. Penha L. L., Sant'Anna C. B., Mendonca-Previato L., Cunha-e-Silva N. L., Previato J. O., Lima A. P. 2009. Sorting of phosphoglucomutase to glycosomes in Trypanosoma cruzi is mediated by an internal domain. Glycobiology 19:1462–1472 [DOI] [PubMed] [Google Scholar]
- 41. Poux A. N., Cebrat M., Kim C. M., Cole P. A., Marmorstein R. 2002. Structure of the GCN5 histone acetyltransferase bound to a bisubstrate inhibitor. Proc. Natl. Acad. Sci. U. S. A. 99:14065–14070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Roper J. R., et al. 2005. The suppression of galactose metabolism in procylic form Trypanosoma brucei causes cessation of cell growth and alters procyclin glycoprotein structure and copy number. J. Biol. Chem. 280:19728–19736 [DOI] [PubMed] [Google Scholar]
- 43. Roper J. R., Guther M. L., Milne K., Ferguson M. A. 2002. Galactose metabolism is essential for the African sleeping sickness parasite Trypanosoma brucei. Proc. Natl. Acad. Sci. U. S. A. 99:5884–5889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schuck P. 2004. A model for sedimentation in inhomogeneous media. I. Dynamic density gradients from sedimenting co-solutes. Biophys. Chem. 108:187–200 [DOI] [PubMed] [Google Scholar]
- 45. Steverding D., Kremp K. 1998. An improved method for the purification of Trypanosoma brucei variant surface glycoprotein (VSG). Parasitol. Res. 84:524–525 [DOI] [PubMed] [Google Scholar]
- 46. Stokes M. J., et al. 2008. The synthesis of UDP-N-acetylglucosamine is essential for bloodstream form Trypanosoma brucei in vitro and in vivo and UDP-N-acetylglucosamine starvation reveals a hierarchy in parasite protein glycosylation. J. Biol. Chem. 283:16147–16161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Titorenko V. I., Nicaud J. M., Wang H., Chan H., Rachubinski R. A. 2002. Acyl-CoA oxidase is imported as a heteropentameric, cofactor-containing complex into peroxisomes of Yarrowia lipolytica. J. Cell Biol. 156:481–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Treumann A., et al. 1997. Structural characterisation of two forms of procyclic acidic repetitive protein expressed by procyclic forms of Trypanosoma brucei. J. Mol. Biol. 269:529–547 [DOI] [PubMed] [Google Scholar]
- 49. Turnock D. C., Ferguson M. A. J. 2007. Sugar nucleotide pools of Trypanosoma brucei, Trypanosoma cruzi, and Leishmania major. Eukaryot. Cell 6:1450–1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Turnock D. C., Izquierdo L., Ferguson M. A. 2007. The de novo synthesis of GDP-fucose is essential for flagellar adhesion and cell growth in Trypanosoma brucei. J. Biol. Chem. 282:28853–28863 [DOI] [PubMed] [Google Scholar]
- 51. Vedadi M., et al. 2007. Genome-scale protein expression and structural biology of Plasmodium falciparum and related apicomplexan organisms. Mol. Biochem. Parasitol. 151:100–110 [DOI] [PubMed] [Google Scholar]
- 52. Vetting M. W., et al. 2005. Structure and functions of the GNAT superfamily of acetyltransferases. Arch. Biochem. Biophys. 433:212–226 [DOI] [PubMed] [Google Scholar]
- 53. Wang J., Liu X., Liang Y. H., Li L. F., Su X. D. 2008. Acceptor substrate binding revealed by crystal structure of human glucosamine-6-phosphate N-acetyltransferase 1. FEBS Lett. 582:2973–2978 [DOI] [PubMed] [Google Scholar]
- 54. Waterhouse A. M., Procter J. B., Martin D. M. A., Clamp M., Barton G. J. 2009. Jalview version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wirtz E., Leal S., Ochatt C., Cross G. A. 1999. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 99:89–101 [DOI] [PubMed] [Google Scholar]
- 56. Wolf E., De Angelis J., Khalil E. M., Cole P. A., Burley S. K. 2002. X-ray crystallographic studies of serotonin N-acetyltransferase catalysis and inhibition. J. Mol. Biol. 317:215–224 [DOI] [PubMed] [Google Scholar]
- 57. Yang T., Bar-Peled M. 2010. Identification of a novel UDP-sugar pyrophosphorylase with a broad substrate specificity in Trypanosoma cruzi. Biochem. J. 429:533–543 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.