Abstract
Cellulose is an abundant and renewable biopolymer that can be used for biofuel generation; however, structural entrapment with other cell wall components hinders enzyme-substrate interactions, a key bottleneck for ethanol production. Biomass is routinely subjected to treatments that facilitate cellulase-cellulose contacts. Cellulases and glucosidases act by hydrolyzing glycosidic bonds of linear glucose β-1,4-linked polymers, producing glucose. Here we describe eight high-temperature-operating cellulases (TCel enzymes) identified from a survey of thermobacterial and archaeal genomes. Three TCel enzymes preferentially hydrolyzed soluble cellulose, while two preferred insoluble cellulose such as cotton linters and filter paper. TCel enzymes had temperature optima ranging from 85°C to 102°C. TCel enzymes were stable, retaining 80% of initial activity after 120 h at 85°C. Two modes of cellulose breakdown, i.e., with endo- and exo-acting glucanases, were detected, and with two-enzyme combinations at 85°C, synergistic cellulase activity was observed for some enzyme combinations.
INTRODUCTION
Cellulose, an abundant and renewable biopolymer, constitutes one-third of all existing plant cell wall material (45). Cellulose fibers are composed of bundles of linear polymers of d-glucose linked exclusively by β-1,4-glycosyl bonds (10), generating solely glucose molecules upon complete hydrolysis (46). Glucose is the starting material in bioconversion (fermentation) processes that produce ethanol (31, 45), other biofuels (9), feedstock chemicals (44), and pharmaceuticals (9, 23, 39).
Hydrolysis of cellulose can be achieved by chemical disruption of glycosidic bonds with high temperature and a strong acid or by enzymatic methods with lower temperatures and weakly acidic conditions, which are ideal for coupled fermentation processes (1, 8, 12, 17, 24, 25, 35, 40, 43). Microorganisms contain cellulase genes, which translate into hydrolytic proteins capable of completely decomposing cellulose into glucose molecules (31, 46, 47). Cellulose degradation typically involves the concerted activities of at least three enzymes, endo-β-glucanase, exo-β-glucanase (in some cases termed cellobiohydrolase), and β-glucosidase, which interact synergistically in producing glucose (4, 18, 33). Endoglucanases randomly hydrolyze internal β-1,4-glycosidic bonds to decrease the length of the cellulose chain (28–30), exo-β-glucanases (and in fungal systems the so-called cellobiohydrolases) split off cellobiose from the shortened cellulose chain (2, 36, 48), and β-glucosidases hydrolyze cellobiose to render glucose (37, 42).
Microorganisms that live in elevated-temperature environments are capable of assimilating complex carbohydrates as a carbon source (3). Hyperthermophilic marine archaea contain genes for utilization of alpha- and beta-linked glucans, such as starch, barley glucan, laminarin, and chitin, while hyperthermophilic bacteria utilize the same glucans as well as hemicellulose, such as xylans and mannans (3). The majority of extremely thermophilic enzymes (genes) suggested to be implicated in the deconstruction of cellulosic materials have been identified only by bioinformatics (3) and have been vaguely associated in biochemical studies with enzymes produced under nonideal conditions. Biochemical characterization of these enzymes will require intensive effort but is likely to generate new opportunities for the use of renewable resources as biofuels (3).
A number of hyperthermophilic β-1,4-endoglucanases have been studied. EGPh from the hyperthermophilic archaeon Pyrococcus horikoshii shows crystalline cellulose-hydrolyzing activity (6, 19–22). Extensive mutational analysis showed that this enzyme contains a recognizable cellulase active center even though the enzyme lacks a true carbohydrate-binding domain (19, 20). Another cellulase, encoded by celA from Thermotoga maritima, hydrolyzes oligosaccharide substrates by an exoglucanase mode of action; however, it rapidly reduces the viscosity of carboxymethylcellulose (CMC), suggesting an endoglucanase mode of action (5, 11, 26). Finally, the thermoacidophilic archaeon Sulfolobus solfataricus P2 encodes three hypothetical endo-β-glucanases, SSO1354, SSO1949, and SSO2534. SSO1949 hydrolyzes carboxymethylcellulose as well as cello-oligomers, with cellobiose and cellotriose as the main reaction products and with a pH optimum of 1.8 and a temperature optimum of 80°C. The enzyme is thermostable, with a half-life of 8 h at 80°C and pH 1.8. The catalytic domain of SSO1949 is similar to those of other mesophilic, acidophilic, and neutral cellulases (16, 27).
Here we report on a set of six high-temperature cellulases and two glucosidases identified from a comprehensive survey of sequenced thermophilic bacterial and archaeal genomes. To determine their usefulness as additives in biomass degradation, we expressed the potential genes and produced the corresponding proteins in the tractable laboratory model Escherichia coli, purified proteins from crude cell extracts, and determined key enzymatic and biophysical properties. We recognized multiple functional forms of endoglucanases, exoprocessive glucanases (but not a typical fungal cellobiohydrolase), and β-d-glucosidases as well as synergistic and additive enzymatic interactions with combinations of two enzymes at 85°C.
MATERIALS AND METHODS
Materials.
Genomic DNAs of P. horikoshii OT3 and Thermotoga petrophila RKU-1 were obtained from the American Type Culture Collection (ATCC 700860D-5 and ATCC BAA-488D-5, respectively). Codon-optimized synthetic DNAs were purchased from Gene Synthesis (Houston, TX) (GenBank accession numbers JF715060 [TCel1], JF715061 [TCel4], and JF715062 [TCel6]). Cellulosic and hemicellulosic substrates were purchased from the best sources possible, Sigma Aldrich (MO) and Megazyme (Ireland). Commercially purified cellobiohydrolase and endoglucanase from Trichoderma reesei were purchased from Megazyme (Ireland). APTS (8-aminopyrene-1,3,6-trisulfonic acid trisodium salt)-labeled cellopentaose was prepared from cellopentaose, β-d-Glc-(1→4)4–d-Glc (Sigma Aldrich, MO), and APTS as described by Naran et al. (34). For details, see Materials and Methods in the supplemental material.
Bioinformatics analysis was done with putative cellulases selected from the CAZy (carbohydrate-active enzymes database) website (13, 14) using BLASTp searches among predicted proteins from sequenced archaeal and thermophilic bacterial genomes deposited at the National Center for Biotechnology Information (NCBI) website (GenBank release 161.0). For details, see Materials and Methods in the supplemental material.
Putative cellulase (TCel1 through TCel6) and beta-glucosidase (TCel11 and TCel12) gene models were cloned in E. coli, and proteins were expressed, recovered and purified by immobilized metal chelate affinity chromatography (Qiagen, CA), validated for purity by SDS-PAGE (41), and used for biochemical studies. For details, see Materials and Methods in the supplemental material. Enzymatic activity, pH optima, and thermostability were determined by adding 10 μl of purified enzyme (1.16 ± 0.156 μg protein/μl) to 50 μl of 1% (wt/vol) substrate in 100 mM phosphate buffer, pH 6.0 (or as specified), and incubating with agitation at 85°C or as specified for 30 to 60 min. The reaction was terminated by addition of 60 μl of dinitrosalicyclic acid (DNS) reagent, and the mixture was incubated in a boiling (95°C) water bath for 5 min. The enzymatic release of reducing sugars was spectrophotometrically quantified at 575 nm and compared with glucose and cellobiose standard curves (32). The activity of β-glucosidase was determined spectrophotometrically by monitoring the release of p-nitrophenol from the substrate p-nitrophenol-β-d-glucoside (pNPG) (Sigma Aldrich, MO). Specific activity was defined as U per mg protein at 85°C, where the unit was the amount of enzyme that produced 1 μmol of reducing sugar (glucose or cellobiose) per minute. For further details on substrates and analytical details, see Materials and Methods in the supplemental material.
RESULTS
Discovery and selection of hyperthermophilic cellulases.
Bacterial carbohydrate-active enzyme protein models (glycosyl hydrolases, lyases, esterases, and carbohydrate-binding domain-containing enzymes) were derived from CAZy (7) and used in BLASTp surveys of sequenced archaeal and thermophilic bacterial genomes (GenBank release 161.0). The search resulted in 32 prokaryotic species with at least three hits in one of the four functional categories of carbohydrate-active enzymes defined by CAZy (data not shown); however, only nine genomes showed significant carbohydrate-hydrolyzing enzyme content.
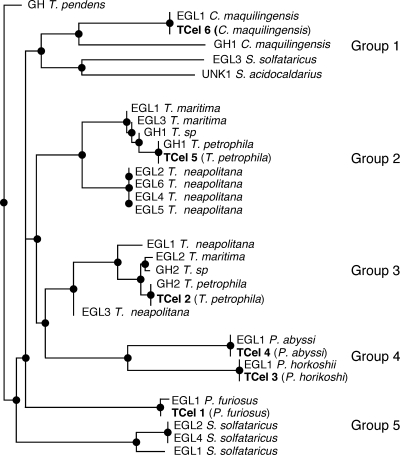
Figure 1 displays a neighbor-joining phylogenetic tree (38) of all BLASTp-identified archaeal and bacterial putative cellulases. Based on the relatedness of cellulases displayed in the tree, we chose six putative cellulases and two clearly identified β-glucosidases (not shown in the tree) for further biochemical studies. Genes noted as TCel1 through TCel6 correspond to putative cellulases, and TCel11 and TCel12 correspond to putative β-glucosidases.
Fig. 1.
Neighbor-joining phylogenetic tree of archaeal and thermophilic bacterial putative cellulases. Proteins selected for further characterization are noted as TCel1 through TCel6, and molecular properties are indicated in Table 1.
Thermophilic cellulase activity.
Cloned TCel genes were expressed and proteins produced in E. coli TOP 10F′ cells. The proteins were purified by His6 tag nickel-chelate affinity chromatography of boiled crude cell extracts. Recovered proteins were further analyzed for biochemical activity as well as other physical and biochemical properties (see Materials and Methods in the supplemental material).
Table 1 describes the physical and biochemical properties derived from the predicted amino acid sequence of all eight proteins as well as the temperature and pH optima found experimentally. Two clear groups of cellulases appear in Table 1, i.e., CAZy family GH12, with molecular masses of 31 to 38 kDa and pI 4.8 to 5.7, and CAZy family GH5, with molecular masses of 52 to 60 kDa and pI 6.5 to 7. The two β-glucosidases TCel11 and TCel12 belong to CAZy families GH3 and GH1, respectively.
Table 1.
Physical and functional properties of high-temperature-operating cellulases
| Sp actb |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Proteina | CAZy family | Accession no. | Mol wt | pI | Charge at pH 7 | Optimum temp (°C) | Optimum pH | mU/mg protein |
U/mg protein, pNPG | |||
| CMC | PASC | CL | FP | |||||||||
| TCel1 | GH12 | JF715060 | 34,005 | 4.80 | −13.10 | 102 | 7.50 | 47.92 | 44.03 | 9.44 | 7.29 | |
| TCel2 | GH12 | YP001244857 | 31,816 | 4.77 | −13.30 | 98 | 6.50 | 162.65 | 5.63 | 1.53 | 2.01 | |
| TCel3 | GH5 | NP143072 | 51,930 | 6.47 | −3.60 | 94 | 6.00 | 47.68 | 33.68 | 5.83 | 5.97 | |
| TCel4 | GH5 | JF715061 | 59,980 | 7.05 | 0.30 | 95 | 5.50 | 37.33 | 4.72 | 1.53 | 2.85 | |
| TCel5 | GH12 | YP001244858 | 38,226 | 5.58 | −6.60 | 96 | 5.50 | 91.17 | 23.68 | 4.72 | 5.56 | |
| TCel6 | GH12 | JF715062 | 31,818 | 5.66 | −5.00 | 85 | 5.50 | 116.17 | 14.31 | 3.54 | 3.54 | |
| TCel11 | GH3 | YP001244492 | 81,243 | 5.38 | −16.90 | 98 | 5.00 | 8.00 | 8.00 | 8.00 | 694 | |
| TCel12 | GH1 | YP001244546 | 51,509 | 5.84 | −9.10 | 92 | 6.50 | 13.00 | 17.00 | 15.00 | 609 | |
| CbhI | E-CBHI | 65,000 | 70 | 4.50 | 78.4 | 27.08 | 27.29 | 0.01 | ||||
| EG II | E-CELTR | 57,250 | 4.70 | 70 | 4.50 | 221.94 | 33.47 | 38.89 | 1.87 | |||
Cellobiohydrolase (CBHI) (EC 3.2.1.91) and endocellulase (EG II) (EC 3.2.1.4) were from Megazyme.
Activity of cellulase, μM reducing glucose/min at 85°C and pH 6; activity of beta-glucosidase, μM reducing pNPG/min at 85°C and pH 6. CMC, carboxymethylcellulose; PASC, phosphoric acid-swollen Avicel PH-101; CL, cotton linters (Sigma Cell50 microcrystalline cellulose); FP, Whatman no. 3 filter paper; pNPG, p-nitrophenylglycoside.
Table 1 describes cellulase specific activities for our enzyme collection on carboxymethylcellulose (CMC) (a soluble substituted cellulose substrate) or more crystalline (insoluble) forms of cellulose, such as cotton linters (CL), filter paper (FP), and phosphoric acid-swollen Avicel (PASC). TCel2, TCel5, and TCel6 efficiently hydrolyzed soluble carboxymethylcellulose (162.64, 91.11, and 116.18 mU/mg protein, respectively) but were much less active on insoluble substrates such as PASC (5.62, 23.68, and 14.3 mU/mg protein, respectively), cotton linters (1.53, 4.72, and 3.54 mU/mg protein, respectively), and filter paper (2.01, 5.55, and 3.54 mU/mg protein, respectively). On the other hand, TCel1 and TCel3 efficiently hydrolyzed PASC (44.03 and 33.68 mU/mg protein, respectively), cotton linters (9.44 and 5.83 mU/mg protein, respectively), and filter paper (7.29 and 5.97 mU/mg protein, respectively) and were less active on CMC than the other enzymes (TCel2, TCel5, and TCel6). TCel4 overlapped both groups and showed reduced activities across all substrates tested (Table 1).
Table 1 compares the specific enzyme activities of our TCel enzymes on various cellulosic substrates with those of benchmark commercial enzymes, namely, Megazyme's cellobiohydrolase (E.C. 3.2.1.91) and endoglucanase (E.C. 3.2.1.4). As expected, cellobiohydrolase showed low activity on CMC (78.4 mU/mg protein) and comparable activities on insoluble substrates (27.08, 27.29, and 0.01 mU/mg protein with PASC, CL, and FP, respectively). On the other hand, endoglucanase showed high specific activity toward CMC (221.94 mU/mg protein). Thus, TCel enzymes function on insoluble cellulosic substrates with specific activities equivalent to those of fungal cellobiohydrolases.
Finally, we also assayed beta-glucosidases (TCel11 and TCel12) for the ability to hydrolyze p-nitrophenol-β-d-glucoside (pNPG), a colorimetric substrate commonly used to determine specific activity for beta-glucosidases, and found that both were active, at 694 and 609 U/mg protein, respectively (Table 1). Both TCel11 and TCel12 were true β-glucosidases even though they have distinct physical properties: molecular masses of 81.3 and 51.5 kDa, pIs of 5.38, 5.84, and net charges at pH 7 of −16.90 and −9.10, respectively (Table 1).
Temperature optimum and stability.
Table 2 shows the optimum temperatures (also shown in Table 1) for catalytic activity for all six cellulases (TCel1 to -6) and two beta-glucosidases (TCel11 and TCel12) along with percentages of maximum activity at 60, 45, and 20°C. Temperature optima were 102, 96, 98, 85, 94, and 95°C for TCel1, TCel2, Tcel3, TCel4, TCel5, and TCel6, respectively. Interestingly, at 60°C, all enzymes except TCel4 showed below 60% of their optimum activity, and at 20°C, all enzymes retained less than 20% of optimal activity.
Table 2.
High-temperature cellulases are inactive at room temperature
| Substrate and cellulasea | Activity (mU)b | Optimum temp (°C) | % Retaining activity at temp (°C): |
||
|---|---|---|---|---|---|
| 60 | 45 | 20 | |||
| PASC | |||||
| TCel1 | 44.03 | 102 | 56.0 | 40.0 | 20.0 |
| TCel2 | 5.62 | 96.0 | 53.6 | 34.1 | 14.6 |
| TCel3 | 33.68 | 98.0 | 42.2 | 25.3 | 14.1 |
| TCel4 | 4.72 | 85.0 | 71.4 | 46.4 | 14.3 |
| TCel5 | 23.68 | 94.0 | 44.7 | 21.1 | 10.5 |
| TCel6 | 14.31 | 95.0 | 26.0 | 11.6 | 4.3 |
| pNPG | |||||
| TCel11 | 69.4 | 98.0 | 4.4 | 1.1 | 1.0 |
| TCel12 | 60.9 | 92.0 | 25.0 | 12.5 | 2.5 |
PASC, phosphoric acid-swollen Avicel; pNPG, p-nitrophenylglycoside.
Cellulase specific activity, μM reducing glucose/min/mg protein at 85°C and pH 6; beta-glucosidase specific activity, μM pNP/min/mg protein at 85°C and pH 6.
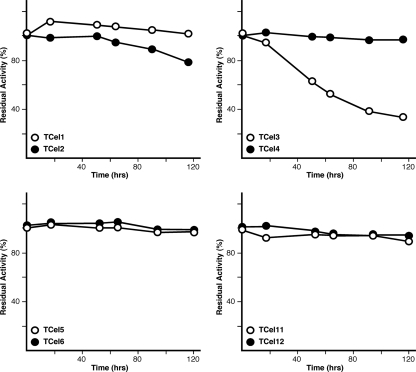
We investigated the thermostability of our TCel collection of enzymes over a 5-day period (120 h). One microgram (1.16 ± 0.156 μg protein/assay) of enzyme was incubated in phosphate-citrate buffer at 90°C for various time periods (Fig. 2) and then assayed for activity on CMC or pNPG (see Materials and Methods in the supplemental material for details of the assay). Figure 2 shows that all enzymes except TCel3 retained at least 80% of the optimal activity after a 5-day period (120 h), indicating robust activity at high temperatures over the period studied.
Fig. 2.
TCel thermostability. TCel enzymes (1.16 ± 0.156 μg protein/assay) were incubated at 90°C in phosphate-citrate buffer for the indicated number of hours, a sample was drawn from the master mix, and cellulase (CMC) or β-glucosidase (pNPG) activity was determined. The amount of residual activity is shown. With the exception of TCel3, all cellulases retained >80% of activity after 120 h at 90°C.
Catalytic mode of operation.
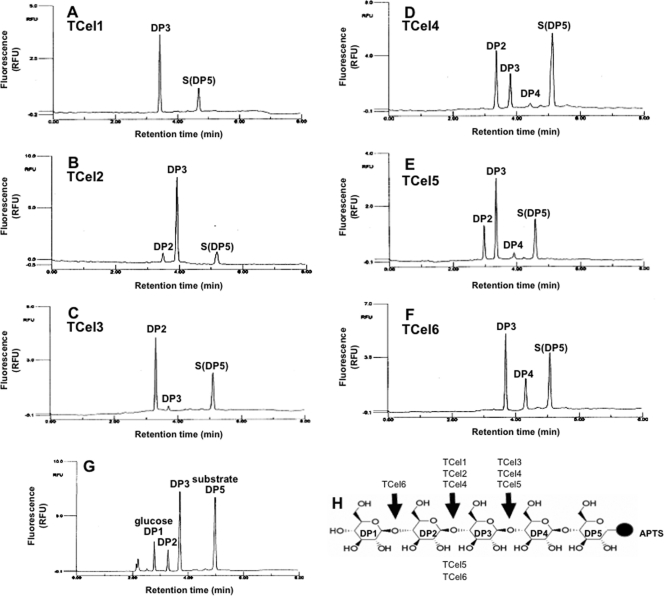
Figure 3 describes our attempt to define the mode of operation of TCel cellulases using fluorescently 1-aminopyrene-3,6,8-trisulfonate (APTS)-reducing-end-labeled substrate (cellopentaose) combined with capillary zone electrophoresis (CZE). After hydrolysis by TCel1 and TCel2, the main fluorescent breakdown products detected by CZE were APTS-labeled trimers (Fig. 3A and B), and TCel3 produced almost exclusively dimers (Fig. 3C). TCel4 (Fig. 3D) and TCel5 (Fig. 3E) produced dimers and trimers, and TCel6 (Fig. 3F) produced trimers and tetramers.
Fig. 3.
(A to F) Capillary zone electrophoresis of breakdown products of APTS-labeled cellopentaose incubated with TCel1 (A), TCel2 (B), TCel3 (C), TCel4 (D), TCel5 (E), or TCel6 (F). (G) CZE retention times of purified monomer (DP1), dimer (DP2), trimer (DP3), tetramer (DP4), and the substrate cellopentaose (DP5). (H) Predicted cleavage patterns between DP2, DP3, and DP4. Assay conditions were as follows: substrate, APTS-cellopentaose; buffer, 50 mM sodium phosphate-50 mM citrate, incubation at 95°C and pH 6. CZE retention times vary slightly among electrophoresis runs. Since we used a defined substrate, retention time variations were not corrected.
Two-enzyme combinations.
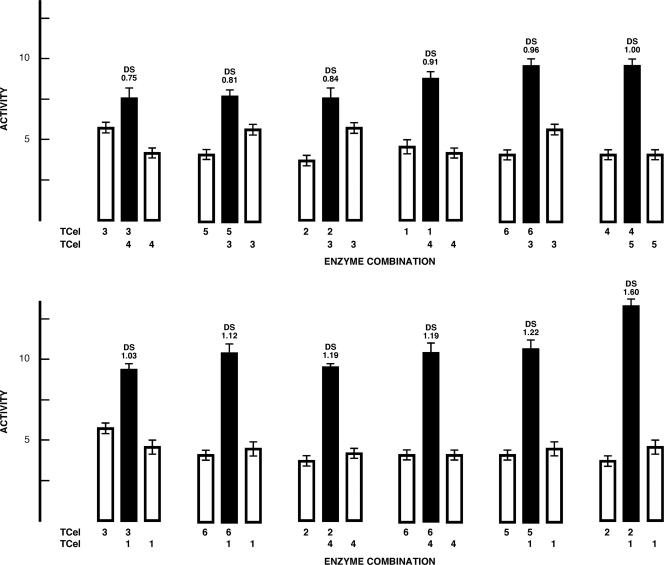
Figure 4 compares the cellulase activities of enzymes working solely (open bars) or interactively (closed bars) in a two-enzyme cellulose breakdown system. The degree of synergism (DS) was determined by comparing single-enzyme (a + b) breakdown activities with two-enzyme (ab) combinations as defined in reference 33, where DS values above 1.0 were deemed significant (closed bars). For two-enzyme interactions, a positive gain of cellulose breakdown activity (DS) was observed for five of the 12 combinations tested: TCel1 with TCel6 (DS = 1.12), TCel4 with TCel2 (DS = 1.19) or TCel6 (DS = 1.19), and TCel1 with TCel5 (DS = 1.22) or TCel2 (DS = 1.60). The remaining five enzyme combinations did not indicate interactive enzyme action.
Fig. 4.
Interactive two-enzyme cellulose breakdown system. One (open bars) or two (closed bars) TCel enzymes (1.16 ± 0.156 μg protein/system) were incubated at 85°C in the presence of PASC and activity measured as the release of reducing sugars after a 60-min incubation period. The two-enzyme system degree of synergism (DS) was calculated as ab/(a + b) (33).
DISCUSSION
In this study, we surveyed thermophilic archaeal and bacterial genomes for the occurrence of glycosyl hydrolases (cellulases) and found a limited set of genes that are consistent among archaeal and bacterial genomes (Fig. 1). Among archaeal hydrolases, only three showed significant glycosyl hydrolase content, i.e., those from Pyrococcus furiosus, Thermophilum pendens, and Picrophilus torridus, with 15, 15, and 13 gene models, respectively.
The temperature optimum could not be directly correlated to the organism source (e.g., bacterial or archaeal); while bacterial proteins had temperature optima of 98°C (TCel2) and 96°C (TCel6), the archaeal protein TCel6 had a temperature optimum of 85°C. Figure 2 shows that all tested cellulases and β-glucosidases were stable at high temperatures (5 days at 85°C), with the exception of TCel4, which was less active after the second day.
All of the tested (six) glycosyl hydrolase proteins showed cellulose degradation activity (Table 1; Fig. 3); however, they showed differential specificity and affinity toward the soluble and insoluble substrates. According to the specific activities on different substrates (Table 1), five cellulases fall into two groups: (i) TCel2, TCel5, and TCel6, with preferential activity on CMC and little to no activity on crystalline cellulose substrates, and (ii) TCel1 and TCel3, which are less active on CMC but active on insoluble substrates. TCel4, the sixth enzyme, showed low activity with carboxymethylcellulose, PASC, Avicel, and filter paper.
TCel1 and TCel2 produced APTS-labeled trimers (Fig. 3A and B), indicating that they require at least two glucose residues at either side of the hydrolysis site in the glucan substrate. TCel3 produced almost exclusively dimers, indicating that it preferred three glucoses toward the nonreducing end from the hydrolysis site (Fig. 3C), and TCel4 (Fig. 3D) and TCel5 (Fig. 3E) produced both APTS-labeled dimers and trimers, suggesting that they had a shorter substrate binding site needing only one intact glucose residue toward the reducing side of the hydrolysis site. TCel6 (Fig. 3F) produced both APTS-labeled trimers and tetramers, showing that it needed only one glucose residue toward the nonreducing end from the hydrolysis site but needed at least two toward the reducing end.
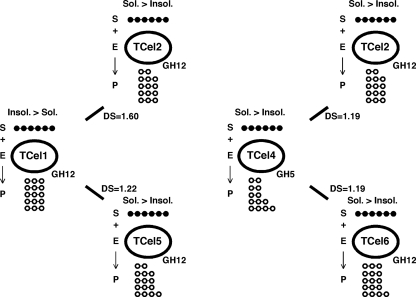
Figure 5 diagrams the characteristics of two-enzyme combinations with enhanced cellulase activity: TCel1 with TCel2 (DS = 1.60) or TCel5 and TCel4 with TCel2 (DS = 1.19) or TCel6 (DS = 1.19). TCel1, TCel2, and TCel5 are CAZy GH12 enzymes (Table 1), TCel2 and TCel5 preferentially hydrolyzed soluble forms of cellulose (CMC), and TCel1 preferred insoluble cellulosic substrates (Table 1). TCel1 cleaved only dimers off a pentameric substrate, while TCel2 cleaved dimers and trimers and TCel5 cleaved dimers, trimers, and to a small extent monomers (Fig. 3). TCel4 is a CAZy GH5 enzyme with low cellulase activity on soluble and/or insoluble substrates (Table 1); however, it sequentially cleaved dimers, trimers, and tetramers off our pentameric substrate (Fig. 3), and interactions with TCel2 and TCel6 (Fig. 4) resulted in enhanced cellulase activity (DS = 1.19).
Fig. 5.
Diagram describing the two-enzyme interactive system. S, substrate (cellopentaose); E, TCel enzyme; P, hydrolysis products (dimers, trimers, or tetramers).
Thus, we conclude that five enzymes, TCel1, TCel2, TCel4, TCel5, and TCel6 work well alone or in specific combinations to degrade PASC, a semicrystalline form of cellulosic substrate (10, 15). At 90°C, all five enzymes are catalytically active at their peak performance and are thermostable for over a 2-day period (Table 2 and Fig. 2).
Supplementary Material
ACKNOWLEDGEMENTS
We are indebted to Mike Himmel, Stephen Decker, and friends at the National Renewable Energy Laboratory (NREL) for the insightful discussion.
This work was funded by Department of Energy awards 06103-OKL and ZDJ-7-77608-01.
TCel enzymes are a substantial portion of patent applications 61/079,208 and 61/145,665 licensed to Edenspace Corporation.
Footnotes
Present address: Laboratório de Ciência e Tecnologia do Bioetanol (CTBE) do Centro Nacional de Pesquisa em Energia e Materiais, Campinas/SP, Brazil.
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 17 June 2011.
REFERENCES
- 1. Abe S., Takagi M. 1991. Simultaneous saccharification and fermentation of cellulose to lactic acid. Biotechnol. Bioeng. 37:93–96 [DOI] [PubMed] [Google Scholar]
- 2. Baker J. O., Ehrman C. I., Adney W. S., Thomas S. R., Himmel M. E. 1998. Hydrolysis of cellulose using ternary mixtures of purified celluloses. Appl. Biochem. Biotechnol. 70-72:395–403 [DOI] [PubMed] [Google Scholar]
- 3. Blumer-Schuette S. E., Kataeva I., Westpheling J., Adams M. W., Kelly R. M. 2008. Extremely thermophilic microorganisms for biomass conversion: status and prospects. Curr. Opin. Biotechnol. 19:210–217 [DOI] [PubMed] [Google Scholar]
- 4. Boisset C., Fraschini C., Schulein M., Henrissat B., Chanzy H. 2000. Imaging the enzymatic digestion of bacterial cellulose ribbons reveals the endo character of the cellobiohydrolase Cel6A from Humicola insolens and its mode of synergy with cellobiohydrolase Cel7A. Appl. Environ. Microbiol. 66:1444–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bronnenmeier K., Kern A., Liebl W., Staudenbauer W. L. 1995. Purification of Thermotoga maritima enzymes for the degradation of cellulosic materials. Appl. Environ. Microbiol. 61:1399–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cady S. G., et al. 2001. Beta-endoglucanase from Pyrococcus furiosus. Methods Enzymol. 330:346–354 [DOI] [PubMed] [Google Scholar]
- 7. Cantarel B. L., et al. 2009. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37:D233–D238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Corredor D. Y., et al. 2009. Evaluation and characterization of forage sorghum as feedstock for fermentable sugar production. Appl. Biochem. Biotechnol. 158:164–179 [DOI] [PubMed] [Google Scholar]
- 9. Detroy R. W., St. Julian G. 1983. Biomass conversion: fermentation chemicals and fuels. Crit. Rev. Microbiol. 10:203–228 [DOI] [PubMed] [Google Scholar]
- 10. Ding S. Y., Himmel M. E. 2006. The maize primary cell wall microfibril: a new model derived from direct visualization. J. Agric. Food Chem. 54:597–606 [DOI] [PubMed] [Google Scholar]
- 11. Evans B. R., Gilman A. K., Cordray K., Woodward J. 2000. Mechanism of substrate hydrolysis by a thermophilic endoglucanase from Thermotoga maritima. Biotechnol. Lett. 22:735–740 [Google Scholar]
- 12. Geddes C. C., et al. 2010. Optimizing the saccharification of sugar cane bagasse using dilute phosphoric acid followed by fungal cellulases. Bioresour. Technol. 101:1851–1857 [DOI] [PubMed] [Google Scholar]
- 13. Henrissat B. 1991. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 280:309–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Henrissat B., Bairoch A. 1993. New families in the classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 293:781–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Himmel M. E., et al. 2007. Biomass recalcitrance: engineering plants and enzymes for biofuels production. Science 315:804–807 [DOI] [PubMed] [Google Scholar]
- 16. Huang Y., Krauss G., Cottaz S., Driguez H., Lipps G. 2005. A highly acid-stable and thermostable endo-beta-glucanase from the thermoacidophilic archaeon Sulfolobus solfataricus. Biochem. J. 385:581–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jacobsen S. E., Wyman C. E. 2000. Cellulose and hemicellulose hydrolysis models for application to current and novel pretreatment processes. Appl. Biochem. Biotechnol. 84-86:81–96 [DOI] [PubMed] [Google Scholar]
- 18. Jeoh T., Wilson D. B., Walker L. P. 2002. Cooperative and competitive binding in synergistic mixtures of Thermobifida fusca cellulases Cel5A, Cel6B, and Cel9A. Biotechnol. Prog. 18:760–769 [DOI] [PubMed] [Google Scholar]
- 19. Kang H. J., Ishikawa K. 2007. Analysis of active center in hyperthermophilic cellulase from Pyrococcus horikoshii. J. Microbiol. Biotechnol. 17:1249–1253 [PubMed] [Google Scholar]
- 20. Kang H. J., Uegaki K., Fukada H., Ishikawa K. 2007. Improvement of the enzymatic activity of the hyperthermophilic cellulase from Pyrococcus horikoshii. Extremophiles 11:251–256 [DOI] [PubMed] [Google Scholar]
- 21. Kashima Y., Udaka S. 2004. High-level production of hyperthermophilic cellulase in the Bacillus brevis expression and secretion system. Biosci. Biotechnol. Biochem. 68:235–237 [DOI] [PubMed] [Google Scholar]
- 22. Kim H. W., Takagi Y., Hagihara Y., Ishikawa K. 2007. Analysis of the putative substrate binding region of hyperthermophilic endoglucanase from Pyrococcus horikoshii. Biosci. Biotechnol. Biochem. 71:2585–2587 [DOI] [PubMed] [Google Scholar]
- 23. Kundu S., Mahapatra A. C., KumarNigam V., Kundu K. 2003. Continuous production of cephalosporin-C by immobilized microbial cells using symbiotic mode in a packed bed bioreactor. Artif. Cells Blood Substit. Immobil. Biotechnol. 31:313–327 [DOI] [PubMed] [Google Scholar]
- 24. Lee K. C., Bulls M., Holmes J., Barrier J. W. 1997. Hybrid process for the conversion of lignocellulosic materials. Appl. Biochem. Biotechnol. 66:1–23 [DOI] [PubMed] [Google Scholar]
- 25. Li H., Kim N. J., Jiang M., Kang J. W., Chang H. N. 2009. Simultaneous saccharification and fermentation of lignocellulosic residues pretreated with phosphoric acid-acetone for bioethanol production. Bioresour. Technol. 100:3245–3251 [DOI] [PubMed] [Google Scholar]
- 26. Liebl W., et al. 1996. Analysis of a Thermotoga maritima DNA fragment encoding two similar thermostable cellulases, CelA and CelB, and characterization of the recombinant enzymes. Microbiology 142:2533–2542 [DOI] [PubMed] [Google Scholar]
- 27. Limauro D., Cannio R., Fiorentino G., Rossi M., Bartolucci S. 2001. Identification and molecular characterization of an endoglucanase gene, celS, from the extremely thermophilic archaeon Sulfolobus solfataricus. Extremophiles 5:213–219 [DOI] [PubMed] [Google Scholar]
- 28. Macarron R., et al. 1993. Mode of action of endoglucanase III from Trichoderma reesei. Biochem. J. 289:867–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Macarron R., van Beeumen J., Henrissat B., de la Mata I., Claeyssens M. 1993. Identification of an essential glutamate residue in the active site of endoglucanase III from Trichoderma reesei. FEBS Lett. 316:137–140 [DOI] [PubMed] [Google Scholar]
- 30. McCarthy T., Hanniffy O., Savage A. V., Tuohy M. G. 2003. Catalytic properties and mode of action of three endo-beta-glucanases from Talaromyces emersonii on soluble beta-1,4- and beta-1,3;1,4-linked glucans. Int. J. Biol. Macromol. 33:141–148 [DOI] [PubMed] [Google Scholar]
- 31. Merino S. T., Cherry J. 2007. Progress and challenges in enzyme development for biomass utilization. Adv. Biochem. Eng. Biotechnol. 108:95–120 [DOI] [PubMed] [Google Scholar]
- 32. Miller G. L. 1959. Use of dintirosalicilic acid reagent for determination of reducing sugar. Anal. Chem. 31:426–428 [Google Scholar]
- 33. Mishra C., Rao M. 1988. Mode of action and synergism of cellulases from Penicillium funiculosum. Appl. Biochem. Biotechnol. 19:139–150 [DOI] [PubMed] [Google Scholar]
- 34. Naran R., Pierce M. L., Mort A. J. 2007. Detection and identification of rhamnogalacturonan lyase activity in intercellular spaces of expanding cotton cotyledons. Plant J. 50:95–107 [DOI] [PubMed] [Google Scholar]
- 35. Neureiter M., Danner H., Thomasser C., Saidi B., Braun R. 2002. Dilute-acid hydrolysis of sugarcane bagasse at varying conditions. Appl. Biochem. Biotechnol. 98-100:49–58 [DOI] [PubMed] [Google Scholar]
- 36. Nimlos M. R., et al. 2007. Molecular modeling suggests induced fit of family I carbohydrate-binding modules with a broken-chain cellulose surface. Protein Eng. Des. Sel. 20:179–187 [DOI] [PubMed] [Google Scholar]
- 37. Nunoura N., Ohdan K., Yano T., Yamamoto K., Kumagai H. 1996. Purification and characterization of beta-d-glucosidase (beta-d-fucosidase) from Bifidobacterium breve clb acclimated to cellobiose. Biosci. Biotechnol. Biochem. 60:188–193 [DOI] [PubMed] [Google Scholar]
- 38. Papadopoulos J. S., Agarwala R. 2007. COBALT: constraint-based alignment tool for multiple protein sequences. Bioinformatics 23:1073–1079 [DOI] [PubMed] [Google Scholar]
- 39. Papas A., Wu S. H. 1997. Rumen-stable delivery systems. Adv. Drug Deliv. Rev. 28:323–334 [DOI] [PubMed] [Google Scholar]
- 40. Saha B. C., Iten L. B., Cotta M. A., Wu Y. V. 2005. Dilute acid pretreatment, enzymatic saccharification, and fermentation of rice hulls to ethanol. Biotechnol. Prog. 21:816–822 [DOI] [PubMed] [Google Scholar]
- 41. Shapiro A. L., Vinuela E., Maizel J. V., Jr 1967. Molecular weight estimation of polypeptide chains by electrophoresis in SDS-polyacrylamide gels. Biochem. Biophys. Res. Commun. 28:815–820 [DOI] [PubMed] [Google Scholar]
- 42. Sigurskjold B. W., Duus B., Bock K. 1991. Hydrolysis of substrate analogues catalysed by beta-d-glucosidase from Aspergillus niger. II. Deoxy and deoxyhalo derivatives of cellobiose. Acta Chem. Scand. 45:1032–1041 [DOI] [PubMed] [Google Scholar]
- 43. Soderstrom J., Pilcher L., Galbe M., Zacchi G. 2003. Combined use of H2SO4 and SO2 impregnation for steam pretreatment of spruce in ethanol production. Appl. Biochem. Biotechnol. 105-108:127–140 [DOI] [PubMed] [Google Scholar]
- 44. Varel V. H., Yen J. T. 1997. Microbial perspective on fiber utilization by swine. J. Anim. Sci. 75:2715–2722 [DOI] [PubMed] [Google Scholar]
- 45. Weng J. K., Li X., Bonawitz N. D., Chapple C. 2008. Emerging strategies of lignin engineering and degradation for cellulosic biofuel production. Curr. Opin. Biotechnol. 19:166–172 [DOI] [PubMed] [Google Scholar]
- 46. White A. R., Brown R. M. 1981. Enzymatic hydrolysis of cellulose: visual characterization of the process. Proc. Natl. Acad. Sci. U. S. A. 78:1047–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wood T. M. 1975. Properties and mode of action of cellulases. Biotechnol. Bioeng. Symp. 1975:111–133 [PubMed] [Google Scholar]
- 48. Yaoi K., et al. 2007. The structural basis for the exo-mode of action in GH74 oligoxyloglucan reducing end-specific cellobiohydrolase. J. Mol. Biol. 370:53–62 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.