Abstract
The bacterial flagellar motor is driven by an ion flux through a channel called MotAB in Escherichia coli or Salmonella and PomAB in Vibrio alginolyticus. PomAB is composed of two transmembrane (TM) components, PomA and PomB, and converts a sodium ion flux to rotation of the flagellum. Its homolog, MotAB, utilizes protons instead of sodium ions. PomB/MotB has a peptidoglycan (PG)-binding motif in the periplasmic domain, allowing it to function as the stator by being anchored to the PG layer. To generate torque, PomAB/MotAB is thought to undergo a conformational change triggered by the ion flux and to interact directly with FliG, a component of the rotor. Here, we present the first three-dimensional structure of this torque-generating stator unit analyzed by electron microscopy. The structure of PomAB revealed two arm domains, which contain the PG-binding site, connected to a large base made of the TM and cytoplasmic domains. The arms lean downward to the membrane surface, likely representing a “plugged” conformation, which would prevent ions leaking through the channel. We propose a model for how PomAB units are placed around the flagellar basal body to function as torque generators.
INTRODUCTION
There are many essential biological processes coupled to ion potentials across the membrane. These include ATP synthesis by the F1Fo-ATPase and rotation of the bacterial flagellar motor. In Escherichia coli or Salmonella, an inner membrane complex composed of two components, MotA and MotB (9, 50, 8, 62), converts proton flux into rotation of the flagellum to power bacterial motility (2, 25, 52). The MotAB complex also functions as a stator by being anchored to the peptidoglycan (PG) layer of the cell through a binding motif in the periplasmic domain of MotB (10, 23). Other channel complexes, ExbBD and TolQR, have weak sequence similarity to MotAB but utilize the proton motive force to power very different physiological functions (14, 24). ExbBD is involved in active transport of iron siderophores and vitamin B12 through the outer membrane (24), whereas TolQR is known to maintain the integrity of the outer membrane (14). Both complexes need other subunits, TonB for ExbBD or TolAB and Pal for TolQR, to bridge the periplasmic space.
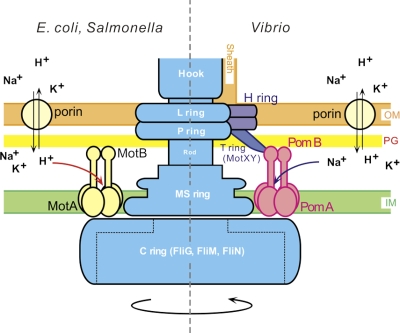
Some bacteria, such as alkalophilic Bacillus and Vibrio species, use an electrochemical potential of sodium ions to drive the flagellar motor (59, 39). In Vibrio species, which normally have only one polar flagellum, the four proteins PomA, PomB, MotX, and MotY are necessary to generate torque (59, 39). The maximum rotational speed of the sodium-driven motor in Vibrio alginolyticus is ∼1,700 Hz (37), whereas the maximum speed of the proton-driven motor in E. coli is ∼300 Hz (7, 35). PomA and PomB are closely related to MotA and MotB, respectively, whereas paralogs of MotX and MotY (40, 41, 44) are not found in E. coli. Although the functions of MotX and MotY are not yet clear, it has been shown that they form a ring structure, called the T ring, that protrudes into the periplasmic space (Fig. 1), where it could interact with PomAB (51, 20). Another ring structure, named the H ring, has recently been identified around the LP ring. The H ring is necessary for T ring formation (Fig. 1) (53).
Fig. 1.
A schematic diagram of two types of flagellar motors. A hypothetical model for the proton-driven motor of E. coli or Salmonella (left) and the sodium-driven motor of Vibrio (right). The sodium-driven polar flagellum of Vibrio is sheathed. The energy source for flagellar motor rotation is provided by an electrochemical potential gradient across the inner membrane. The functional units of the stator are thought to be (MotA)4(MotB)2 and (PomA)4(PomB)2 (3, 26, 49). IM, inner membrane; PG, peptidoglycan layer; OM, outer membrane.
PomA and PomB are thought to form a sodium channel complex. MotAB from E. coli can drive the polar flagellum of Vibrio cholerae (15) and V. alginolyticus (1) using the proton motive force. Also, a complex of PomA and a chimeric protein made of PomB and MotB segments can convert a proton-driven motor into a sodium-driven one (1). Hence, PomAB or MotAB determines the ion specificity of the motor, and the two types of motors most likely share similar mechanisms for ion-driven torque generation. Figure 1 shows a schematic drawing that compares the proton-type motor of E. coli and Salmonella with the sodium-type motor of Vibrio.
Despite the importance of torque generation, structural information for the intact torque-generating unit is limited although cross-linking experiments (3, 4) proposed an arrangement for the transmembrane (TM) helices. Recently, tomographic reconstructions of the spirochete flagella have reveled the motor structures in situ (30, 33, 43). However, these reconstructions, at 70- to 35-Å resolution, gave little information about the molecular boundary and shape of the torque-generating unit (30, 33, 43). Here, we report the first three-dimensional structure reconstructed from molecular images of isolated PomAB by electron microscopy (EM), single-particle analysis, and EM tomography.
MATERIALS AND METHODS
Sample preparation and electron microscopy.
His6-tagged PomA/PomB (PomA/PomB-His6) and PomA/PomB with a deletion of the C-terminal 120 residues of PomB (PomA/PomBΔC-His6) (56) were overproduced in E. coli BL21. The plasmids carrying these genes with a sequence for a hexahistidine tag fused to the C terminus were kindly provided by Toshiharu Yakushi, Yamaguchi University. The genes were expressed under the control of the tac promoter. We purified the PomAB complex as described previously (58), with some modifications. Here, Cymal-5 (Anatrace, Maumee, OH) was used instead of CHAPS (3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate) to extract proteins from the membrane fraction.
A few microliters of ∼0.03 mg/ml purified protein solution was applied onto a carbon-coated grid. It was washed with deionized water five times and negatively stained with 2% uranyl acetate. We examined the sample grids with an FEI Tecnai T20 electron microscope (FEI, Hillsboro, OR) with a LaB6 gun operated at an accelerating voltage of 120 kV. Images were recorded on a 4,000- by 4,000-pixel (4K-by-4K) slow-scan charge-coupled device (SSCCD) camera (UltraScan 4000; Gatan, Pleasanton, CA) at a final magnification of ∼×110,000 and at defocus settings of 5,700 to 21,000 Å. The magnification was calibrated from negatively stained catalase crystals. We collected a tilt series of negatively stained wild-type PomAB complexes on the SSCCD at the same magnification and at a defocus level of ∼30,000 Å by using UCSF Tomo software (61). A constant tilt increment of 3° or 4° or 5° was applied for each tilt series over a tilt range of ±60°. Tilt series were also acquired by SerialEM (38) with a JEM-2100 electron microscope (JEOL, Tokyo, Japan). We examined frozen-hydrated samples with a JEM-3200SFC electron microscope (JEOL, Tokyo, Japan) operated at 200 kV with a specimen temperature of ∼50 K. Zero-energy-loss images were recorded at defocus settings of 50,000 to 80,000 Å by selecting only electrons with an energy loss of less than 10 eV.
Image analysis.
For single-particle analysis of negatively stained particles, we first reduced image frames by a factor of 2. The phase reversal due to the contrast transfer function (CTF) was corrected by taking the astigmatism into account with a modified version of CTFCORRECT from the TOMOCTF package (11). The EMAN software suite (36) was used for the following single-particle analysis. We manually picked up individual molecular images with Boxer by applying a 9- by 9-pixel median filter to enhance the visibility of the particles. After a low-pass filter was applied, three-dimensional maps were constructed from those images by using startnrclasses and startAny with 2-fold symmetry enforced. The structures were refined by REFINE with PC clusters. For the wild-type PomAB complex, the three-dimensional map obtained was used as a starting model for higher-resolution refinement, with no filter applied. The total number of molecular images included in the three-dimensional reconstruction was 5,428 for the wild type, after a few dozen iterations starting from 7,018 images, and 2,254 for the deletion mutant (PomABΔC), starting from 2,838. The numbers of averaged molecules were doubled by 2-fold averaging. The Fourier shell correlation (FSC) was calculated between two volumes, each generated from half of the data set. The resolution was taken to be the spatial frequency at which the FSC drops below 0.5 and was measured to be 21.3 Å for the wild type and 30.1 Å for the deletion mutant (see Fig. S1 in the supplemental material). A full CTF correction produced a map with the identical structural features although the map appeared to be at lower resolution.
For electron tomography of negatively stained PomAB, each tilt series was aligned by using IMOD (31) after tilt angles were determined by using Priism software (6). Individual particles in the tilt series were cut out with BSHOW from the Bsoft suite (17), and molecular tomograms were reconstructed by weighted back-projection in a way similar to that described by Iwasaki et al. (21). Then, the tomograms were aligned without Fourier terms inside the missing wedge, using a modified version of BFIND (17). A total of 19 molecular tomograms were averaged (see Fig. S2B).
Map interpretation.
We extracted the conserved region of the PG-binding domain from the atomic model of the MotB C-terminal structure (MotBC) from Helicobacter pylori (Protein Data Bank [PDB] accession code 3CYP) (48). The model was manually docked into the density map with the graphics program O (22). To build a model in a conformation aligned to interact with the PG layer, we cut out the two arm domains with XDISPMSK (57) and made manual adjustments by using Chimera (46). The rotation and translation matrix thus obtained was converted using a handmade program (CHIMERA2OMAT) and applied to the density maps of the arm domain by MRCRTR. These programs are available on request. A two-dimensional average of the hook-basal body (HBB) complex (54) was kindly provided by David J. DeRosier, Brandeis University, and was cylindrically averaged around the rotational axis of the flagellum to produce a three-dimensional volume. We placed the modeled PomAB complexes manually around the HBB volume. Molecular structures presented in Fig. 3 to 5 and in Fig. S2 in the supplemental material were all prepared with Chimera (46).
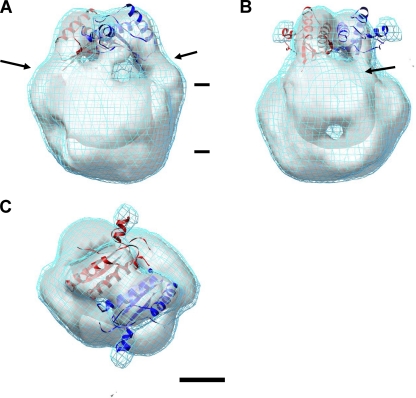
Fig. 3.
Three-dimensional structure of PomAB constructed from single-particle analysis. (A) Structure viewed parallel to the membrane plane. Top and bottom correspond to the periplasmic and cytoplasmic sides of the membrane, respectively. (B) The same structure as in panel A but rotated by 90° around the vertical (2-fold) axis. (C) Structure viewed from the periplasm. Subdomains are labeled 1 to 4. (D) Three-dimensional structure of a PomAB mutant with the PG-binding domain deleted (PomABΔC) (56) shown in the solid density in blue. The magenta net corresponds to the solid density of the wild-type PomAB in panels A to C. Arrows in A and C indicate the shoulder of one arm domain. The contour level of the solid densities in blue is ∼1.8 σ and that of the nets in cyan is ∼1.5 σ. The counter-level of the nets in panels A to C corresponds to ∼100% volume recovery based on a molecular mass of ∼180 kDa for the complex. Scale bar, 25 Å.
Fig. 5.
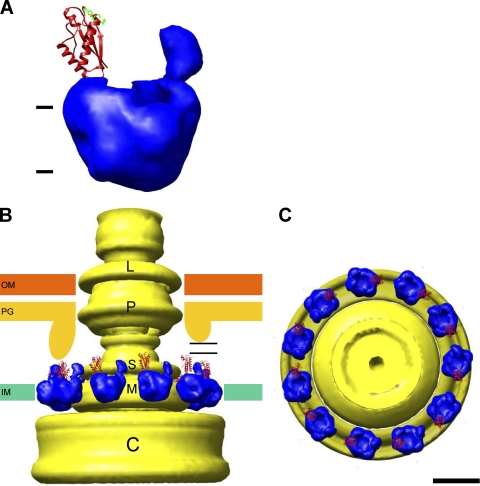
Models of PomAB anchored to the PG layer in position for torque generation. (A) The two arm domains are modeled to face the PG layer. For clarity, the counter-level of the map is the same as for the solid density in Fig. 3 and 4. The atomic model of the MotBC core region (48) is displayed for one of the arms. Residues involved in glycan binding (48) are displayed in green. (B and C) Eleven of the PomAB models shown in panel A are placed around a cylindrical average of the HBB complex (54). The structure is viewed parallel to the membrane in panel B and from the periplasm in panel C. Part of the PG layer supported by MotXY in Vibrio protrudes into the periplasmic space so that the arm of PomAB can be anchored to it (left side in panel B). When the plug region of PomB/MotB stands upright from the membrane, the PG-binding domains can reach an even smaller protrusion from the PG layer over a gap, indicated with horizontal lines (right side). IM, inner membrane; PG, peptidoglycan layer; OM, outer membrane. Scale bar, 100 Å.
RESULTS
Characterization of the PomAB complex.
We used tac-based expression plasmids in E. coli to overexpress the PomAB complex. PomAB was well represented in the membrane fraction and did not significantly slow cell growth after induction by isopropyl-β-d-thiogalactopyranoside (IPTG). Because previous samples in CHAPS detergent aggregated, based on observation by EM (58), we screened more than 20 different detergents to find one that minimized aggregation. Cymal-5 was found to solubilize the complex from the membrane without disassociation of PomA and PomB. A modified purification protocol using Cymal-5 reproducibly generated monodispersed particles.
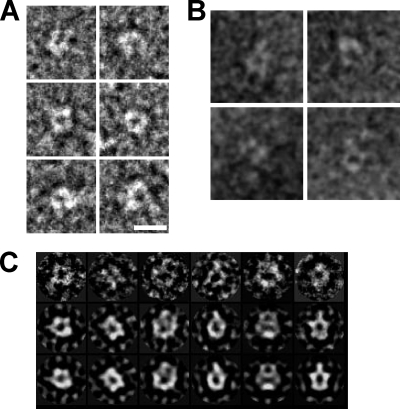
Several studies suggest that the stoichiometry of the flagellar stator unit is (PomA)4(PomB)2/(MotA)4(MotB)2 (3, 26, 49). The relative amount of PomA and PomB on an SDS-PAGE gel appeared to be consistent with this. The molecular mass of (PomA)4(PomB)2 is ∼180 kDa. After gel filtration of the solubilized complex, a single peak was seen at 200 to 250 kDa, as calibrated using soluble globular proteins as standards. This is less than half that observed with the complex solubilized by CHAPS (∼550 kDa) (58) or sucrose monocaprate (∼900 kDa) (55). Considering that detergent micelles are probably bound to the complex, the molecular mass calculated from gel filtration is consistent with the particles being complete PomAB complexes. The relatively small size of the complex makes it suitable for single-particle analysis of negatively stained images. Figure 2A shows typical EM images of negatively stained particles. We also examined frozen-hydrated samples by cryo-electron microscopy (cryo-EM). Most of the particles were absorbed onto carbon film on EM grids, but particles embedded in ice resemble negatively stained ones (compare Fig. 2A and B).
Fig. 2.
EM images of PomAB. (A) Images of negatively stained PomAB. (B) Frozen-hydrated molecules. The particles show similar dimensions and features to those shown in panel A. (C) Selected raw images of particles (upper row) aligned with their respective classes (middle row) in the same column and corresponding reprojections (lower row) of the three-dimensional volume shown in Fig. 3. Scale bar in panel A, 100 Å.
Three-dimensional structure.
We analyzed the three-dimensional structure of PomAB by electron tomography and single-particle analysis of negatively stained particles. We collected tilt series of molecules and carried out tomographic reconstructions (see Fig. S2 in the supplemental material). Individual molecular tomograms showed dimeric structures (see Fig. S2A). Hence, 2-fold averaging was applied for single-particle analysis. We reconstructed an initial three-dimensional map from two-dimensional class averages by the common-line method implemented in the EMAN suite (36) and used it as the reference for structure refinement. Figure 3 shows the three-dimensional structure calculated from ∼5,500 images after iterative refinement. Representative raw images show characteristic shapes similar to corresponding class averages and reprojections of the three-dimensional map (Fig. 2C). The density map looks similar to the average of individual tomograms (compare Fig. S2B in the supplemental material and Fig. 3A). The resemblance of these two independent reconstructions by tomography and single-particle analysis gives confidence in the reconstruction shown in Fig. 3. The resolution of the single-particle reconstruction was measured to be ∼21 Å by the Fourier shell correlation (see Fig. S1).
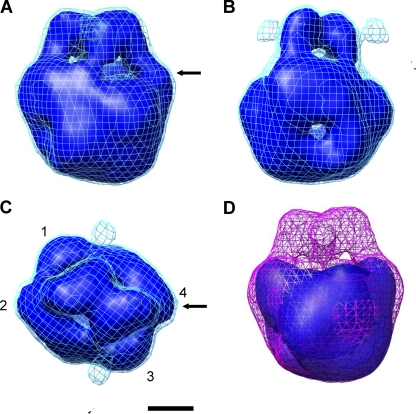
The structure is composed of two arm-like domains and an inverted pyramidal base domain (Fig. 3). The base domain is ∼60 Å tall and could be divided into four subdomains (Fig. 3C). The arm domains contact each other in the middle of the arms and lean back toward the base domain (Fig. 3).
Identification of the peptidoglycan-binding domain.
We then examined a mutant stator complex lacking the C-terminal 120 residues of PomB, which includes the PG-binding motif (PomABΔC) (56). This mutant complex was purified by the same procedure used for the wild-type complex, and similarly monodispersed particles were observed. We reconstructed the three-dimensional map from ∼2,250 molecular images. Although the structure of the mutant complex retains the same base domain as the wild type, it lacks the arm domains (Fig. 3D), indicating that the arms contain the PG-binding domain.
Interpretation of the map.
The PG-binding domain shares sequence similarity with other PG-associated proteins (16, 27, 29, 45, 48). X-ray crystallography and nuclear magnetic resonance (NMR) spectroscopy revealed that the core structure of the PG domain is composed of three α-helices, a four-stranded β-sheet, and loops connecting these elements (16, 27, 29, 45, 48). We docked an atomic model of the PG-binding domain from the H. pylori MotB C-terminal structure (MotBC) (48) into the arm domain of the EM density map (Fig. 4). The docked model consists of ∼110 amino acids, which is nearly equal to the deleted region in the PomB mutant protein (56). The arm domain is the same size as the atomic model of the PG-binding domain, which fits well into the EM density except for slight mismatches in loop regions. H. pylori MotBC forms a dimer in the crystal. Although the arm domains in the EM density make a dimer, the crystal structure requires shifting the two subunits to achieve the best fit to the EM density (Fig. 4).
Fig. 4.
Docking of atomic models into the PomAB map shown in Fig. 3A to C. (A) Structure viewed from the same direction as in Fig. 3A. (B) Structure rotated by 90°, as in Fig. 3B. (C) Structure viewed as in Fig. 3C. Two atomic models of the core of the MotBC PG-binding domain from H. pylori (PDB accession code 3CYP) (48) are shown in red and blue. They were docked into the arm domains. Arrows in panels A and B indicate the stalk and shoulder of the arm (see text), and the horizontal bars in panel A refer to the TM region. The scale bar in panel C represents 25 Å.
The base domain should contain the TM region and the cytoplasmic domain. Each of the four subdomains in the TM region should include one PomA monomer (Fig. 3C), indicating that the stoichiometry of PomA and PomB in the density map is consistent with the previous reports (3, 26, 49). Unfortunately, the resolution of the map is insufficient to interpret more structural details in the base domain. Bound detergent might impair the resolution of the base domain to some degree. There appears to be a cavity in the TM region. In general, it is difficult to analyze the TM region from negatively stained particles due to uneven staining, flattening, and other complicating factors. We cannot rule out that artifacts arising from negative staining and solubilization with detergent may affect the structure although the cavity seen in the base domain is also visible in cryo-EM images (Fig. 2B).
DISCUSSION
Here, we report the first three-dimensional structure for the flagellar torque generation unit. The structure was determined by single-particle analysis and corroborated by electron tomography. The structure can be summarized as a pyramidal base with four visible domains topped with two extended arm-like structures.
The orientation of the arm domains, which contain the PG-binding domains of PomB, relative to the cytoplasmic membrane surface is incompatible with binding to the PG layer. Thus, the isolated stator complex is likely to be somewhat different from the stator when it is bound to PG. The periplasmic domain of MotB is thought to reach back toward the membrane surface and plug the channel before the stator complex assumes its correct position relative to the MS ring and C ring (Fig. 1) (19). The structure of PomAB described here is probably in this plugged-channel conformation.
When the stator is correctly placed around the rotor, Hosking et al. (19) proposed that the plug domain of MotB should stand upright, perpendicular to the membrane, to achieve the ion-conducting state. They found that forming a disulfide cross-link between short segments just after the MotB TM helix in E. coli locks the stator in an unplugged, proton-conducting form (19). We previously observed that PomAB reconstituted into liposomes exposed a 60- to 70-Å-long domain that protruded from the liposome surface (58). To modify our structure to simulate the PG-bound state, we separated the arm domains from the base structure and rotated each of the arm domains so that the PG-binding site, which is located in a loop between α-helix 2 and β-strand 2 (48) (green residues in Fig. 5A), faces the PG layer (up in Fig. 5A). The positions of residues responsible for PG binding are well conserved relative to the periplasmic domain structure of Pal with a bound PG precursor (45). In our modified model, the arm domain extends ∼60 Å from the cytoplasmic membrane surface. Our previous cryo-EM observations showed a much smaller number of images in this conformation than were expected from the concentration of PomAB in the dialysis button (58). They probably represent the unplugged-channel conformation or an intermediate conformation extending toward the PG layer (see below), which may appear stochastically even when PomAB is not assembled around the rotor.
Single-molecule fluorescence microscopy demonstrated a quick turnover of motor-associated MotAB with its membrane pool (32), indicating that the binding of MotAB/PomAB to the PG layer is not tight. Also, a PG-binding assay found no MotBC dimer in the PG-associated fraction (28), suggesting that anchoring to the PG layer requires conformational changes in the MotB dimer. The disposition of the arm in our model is at least consistent with the possibility of dynamic association and dissociation of PomAB/MotAB with the PG layer around the flagellar rotor (28, 32).
The inner surface of the PG layer is more than 100 Å from the outer surface of the cytoplasmic membrane (12, 54). The arms in our model (Fig. 5A and B) are obviously not long enough to reach the PG layer. In addition to the PG-binding domain, there are another ∼160 amino acids in the periplasmic portion of PomB. The stalk and shoulder of the arm (arrows in Fig. 4A and B) probably correspond to those residues, and they might be able to stretch to the PG layer. If so, our model (Fig. 5A) and the complexes previously observed with liposomes (58) would represent an intermediate conformation between the plugged and the PG-bound states. In this scenario, most of the residues forming the stalk and shoulder would need to be extended to reach the PG layer. However, deletion of 50 to 90 amino acids in the MotB periplasmic region following the TM segment did not fully eliminate motility (42), implying that it is unlikely that this entire region must stretch out to contact the PG layer.
A recent study used disulfide cross-linking to show that MotB comes into at least occasional direct contact with FlgI, which forms the P ring (18). The P ring and the L ring form a bushing that allows the flagellar rod to pass through the PG layer and the outer membrane, respectively (Fig. 1). The efficiency of MotB-FlgI cross-linking was affected by the protonophore carbonyl m-chlorophenylhydrazone (CCCP) (18), which may indicate that the structure of the periplasmic domain of MotB changes in response to the proton motive force. It has been speculated that drastic structural changes in the periplasmic region of MotB are required to reach the PG layer and also to activate the proton channel (29). The sodium motive force may change the structure of the PomB periplasmic domain, and it has been observed that sodium ions are essential for assembly of the PomAB stator complex into the flagellar motor (13).
The other possibility is that the PG layer may be distorted around the flagellar motor in a way that makes contact with the PomAB/MotAB PG-binding domain possible. The formation of the P ring may push the PG layer aside and down toward the cytoplasmic membrane and make the periplasmic space smaller (Fig. 5B, left side). When the structures of the spirochete flagella were analyzed in situ (30, 33, 43), the density maps resolved a collar bridging the inner membrane and the PG layer. The mass of the collar was too large to contain only the stator complex, and most of the density may consist of unknown proteins (33) and/or PG. We do not know if a corresponding collar exists in the E. coli or Salmonella flagella because these bacteria are too thick for cryo-electron tomography in situ. This scenario, however, would match nicely with the idea that the plug region stands upright (Fig. 5B, right half). Indeed Vibrio species possess MotX and MotY, which protrude just below the H and P rings (Fig. 1) (20, 51, 53) into the periplasmic space, where they are suggested to interact with the PomAB complex. MotY has a PG-binding motif (27). We propose that MotY holds the PG layer close to PomB so that PomB is able to reach the PG. In E. coli or Salmonella, part of FlgI may project to the periplasmic space and interact with MotB (18).
The switch complex is made of three proteins, FliM, FliN, and the C-terminal domain of FliG, and these three form the C ring (Fig. 1). The switch complex is required for torque generation and switching of direction of the motor rotation (34). The cytoplasmic loop of PomA contains critical amino acids that are thought to interact with FliG (60). This loop, which connects TM helices 2 and 3, would lie below the base domain in a position to interact with the C-terminal domain of FliG. The loop consists of about 100 residues and constitutes the major mass of the stator complex in the cytoplasm. Four copies of the loop could be fitted onto the bottom of the base domain if they are tightly packed.
The flagellar hook-basal body (HBB) complex has previously been isolated from Salmonella and analyzed by cryo-EM (12, 54). A three-dimensional volume of the HBB was calculated by cylindrically averaging a two-dimensional image (54) around the flagellar axis of the HBB (see Materials and Methods). This reconstruction includes the C ring but not the stator complex. Based on the HBB reconstruction and the crystal structures of the Thermotoga maritima FliG C-terminal and middle domains, Brown et al. (5) proposed a model in which important charged residues of FliG are aligned on the membrane-facing surface (the upper side in Fig. 5B) of the C ring so as to interact with the cytoplasmic loop of MotA (or PomA). We placed our modeled PomAB stator complexes around the MS ring of the HBB and above the C ring (Fig. 5B and C). The radius of the C ring at the membrane-facing side is ∼200 Å and is ∼70 Å larger than that of the MS ring. The deck of the C ring has sufficient width to accommodate one PomAB unit (Fig. 5C). The maximum number of MotAB/PomAB stators around one flagellar motor in E. coli has been estimated to be at least 11 (47). The model shown in Fig. 5B and C can accommodate about this number of complexes around the MS ring, but little space remains for additional complexes unless some stators are tilted relative to the MS ring.
Supplementary Material
ACKNOWLEDGMENTS
We thank Toshiharu Yakushi for the plasmids, Hideyuki Matsunami and Tatsuo Atsumi for technical advice on protein production and purification, and Justin M. Kollman and Seiji Kojima for critically reading and improving the manuscript. We are also grateful to Michael B. Braunfeld, Shawn Q. Zheng, Eric Branlund, Jaap Brink, and David N. Mastronarde for technical assistance with electron tomography and Takayuki Kato and Tomoko Miyata with cryo-EM, David A. Agard, John W. Sedat, and Keiichi Namba for support, and David J. DeRosier for the two-dimensional image of the HBB.
This work was supported by funds from the W. M. Keck Advanced Microscopy Laboratory at UCSF and by Grand-in-Aid number 20370064 by the Ministry of Education, Culture, Sports, Science and Technology of JAPAN to K.Y.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 3 June 2011.
REFERENCES
- 1. Asai Y., Yakushi T., Kawagishi I., Homma M. 2003. Ion-coupling determinants of Na+-driven and H+-driven flagellar motors. J. Mol. Biol. 327:453–463 [DOI] [PubMed] [Google Scholar]
- 2. Berg H. C. 2003. The rotary motor of bacterial flagella. Annu. Rev. Biochem. 72:19–54 [DOI] [PubMed] [Google Scholar]
- 3. Braun T. F., Blair D. F. 2001. Targeted disulfide cross-linking of the MotB protein of Escherichia coli: evidence for two H+ channels in the stator complex. Biochemistry 40:13051–13059 [DOI] [PubMed] [Google Scholar]
- 4. Braun T. F., Al-Mawsawi L. Q., Kojima S., Blair D. F. 2004. Arrangement of core membrane segments in the MotA/MotB proton-channel complex of Escherichia coli. Biochemistry 43:35–45 [DOI] [PubMed] [Google Scholar]
- 5. Brown P. N., Hill C. P., Blair D. F. 2002. Crystal structure of the middle and C-terminal domains of the flagellar rotor protein FliG. EMBO J. 21:3225–3234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen H., Hughes D. D., Chan T.-A., Sedat J. W., Agard D. A. 1996. IVE (image visualization environment): a software platform for all three-dimensional microscopy applications. J. Struct. Biol. 116:56–60 [DOI] [PubMed] [Google Scholar]
- 7. Chen X., Berg H. C. 2000. Torque-speed relationship of the flagellar rotary motor of Escherichia coli. Biophys. J. 78:1036–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chun S. Y., Parkinson J. S. 1988. Bacterial motility: membrane topology of the Escherichia coli MotB protein. Science 239:276–278 [DOI] [PubMed] [Google Scholar]
- 9. Dean G. E., Macnab R. M., Stader J., Matsumura P., Burks C. 1984. Gene sequence and predicted amino acid sequence of the motA protein, a membrane-associated protein required for flagellar rotation in Escherichia coli. J. Bacteriol. 159:991–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. De Mot R., Vanderleyden J. 1994. The C-terminal sequence conservation between OmpA-related outer membrane proteins and MotB suggests a common function in both Gram-positive and Gram-negative bacteria, possibly in the interaction of these domains with peptidoglycan. Mol. Microbiol. 12:333–334 [DOI] [PubMed] [Google Scholar]
- 11. Fernández J. J., Li S., Crowther R. A. 2006. CTF determination and correction in electron cryotomography. Ultramicroscopy 106:587–596 [DOI] [PubMed] [Google Scholar]
- 12. Francis N. R., Sosinsky G. E., Thomas D., DeRosier D. J. 1994. Isolation, characterization and structure of bacterial flagellar motors containing the switch complex. J. Mol. Biol. 235:1261–1270 [DOI] [PubMed] [Google Scholar]
- 13. Fukuoka H., Wada T., Kojima S., Ishijima A., Homma M. 2009. Sodium-dependent dynamic assembly of membrane complexes in sodium-driven flagellar motors. Mol. Microbiol. 71:825–835 [DOI] [PubMed] [Google Scholar]
- 14. Godlewska R., Wiśniewska K., Pietras Z., Jagusztyn-Krynicka E. K. 2009. Peptidoglycan-associated lipoprotein (Pal) of Gram-negative bacteria: function, structure, role in pathogenesis and potential application in immunoprophylaxis. FEMS Microbiol. Lett. 298:1–11 [DOI] [PubMed] [Google Scholar]
- 15. Gosink K. K., Häse C. C. 2000. Requirements for conversion of the Na+-driven flagellar motor of Vibrio cholerae to the H+-driven motor of Escherichia coli. J. Bacteriol. 182:4234–4240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Grizot S., Buchanan S. K. 2004. Structure of the OmpA-like domain of RmpM from Neisseria meningitidis. Mol. Microbiol. 51:1027–1037 [DOI] [PubMed] [Google Scholar]
- 17. Heymann J. B. 2001. Bsoft: image and molecular processing in electron microscopy. J. Struct. Biol. 133:156–169 [DOI] [PubMed] [Google Scholar]
- 18. Hizukuri Y., Kojima S., Homma M. 2010. Disulfide cross-linking between the stator and the bearing components in the bacterial flagellar motor. J. Biochem. 148:309–318 [DOI] [PubMed] [Google Scholar]
- 19. Hosking E. R., Vogt C., Bakker E. P., Manson M. D. 2006. The Escherichia coli MotAB proton channel unplugged. J. Mol. Biol. 364:921–937 [DOI] [PubMed] [Google Scholar]
- 20. Hosogi N., Shigematsu H., Terashima H., Homma M., Nagayama K. 2011. Zernike phase contrast cryo-electron tomography of sodium-driven flagellar hook-basal bodies from Vibrio alginolyticus. J. Struct. Biol. 173:67–76 [DOI] [PubMed] [Google Scholar]
- 21. Iwasaki K., et al. 2005. Electron tomography reveals diverse conformations of integrin αIIbβ3 in the active state. J. Struct. Biol. 150:259–267 [DOI] [PubMed] [Google Scholar]
- 22. Jones T. A., Zhou J. Y., Cowan S. W., Kjeldgaard M. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47:110–119 [DOI] [PubMed] [Google Scholar]
- 23. Koebnik R. 1995. Proposal for a peptidoglycan-associating alpha-helical motif in the C-terminal regions of some bacterial cell-surface proteins. Mol. Microbiol. 16:1269–1270 [DOI] [PubMed] [Google Scholar]
- 24. Koebnik R. 2005. TonB-dependent trans-envelope signalling: the exception or the rule? Trends. Microbiol. 13:343–347 [DOI] [PubMed] [Google Scholar]
- 25. Kojima S., Blair D. F. 2004. The bacterial flagellar motor: structure and function of a complex molecular machine. Int. Rev. Cytol. 233:93–134 [DOI] [PubMed] [Google Scholar]
- 26. Kojima S., Blair D. F. 2004. Solubilization and purification of the MotA/MotB complex of Escherichia coli. Biochemistry 43:26–34 [DOI] [PubMed] [Google Scholar]
- 27. Kojima S., et al. 2008. Insights into the stator assembly of the Vibrio flagellar motor from the crystal structure of MotY. Proc. Natl. Acad. Sci. U. S. A. 105:7696–7701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kojima S., Furukawa Y., Matsunami H., Minamino T., Namba K. 2008. Characterization of the periplasmic domain of MotB and implications for its role in the stator assembly of the bacterial flagellar motor. J. Bacteriol. 190:3314–3322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kojima S., et al. 2009. Stator assembly and activation mechanism of the flagellar motor by the periplasmic region of MotB. Mol. Microbiol. 73:710–718 [DOI] [PubMed] [Google Scholar]
- 30. Kudryashev M., Cyrklaff M., Wallich R., Baumeister W., Frischknecht F. 2010. Distinct in situ structures of the Borrelia flagellar motor. J. Struct. Biol. 169:54–61 [DOI] [PubMed] [Google Scholar]
- 31. Kremer J. R., Mastronarde D. N., McIntosh J. R. 1996. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116:71–76 [DOI] [PubMed] [Google Scholar]
- 32. Leake M. C., et al. 2006. Stoichiometry and turnover in single, functioning membrane protein complexes. Nature 443:355–358 [DOI] [PubMed] [Google Scholar]
- 33. Liu J., et al. 2009. Intact flagellar motor of Borrelia burgdorferi revealed by cryo-electron tomography: evidence for stator ring curvature and rotor/C-ring assembly flexion. J. Bacteriol. 191:5026–5036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lloyd S. A., Tang H., Wang X., Billings S., Blair D. F. 1996. Torque generation in the flagellar motor of Escherichia coli: evidence of a direct role for FliG but not for FliM or FliN. J. Bacteriol. 178:223–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lowe G., Meister M., Berg H. C. 1987. Rapid rotation of flagellar bundles in swimming bacteria. Nature 325:637–640 [Google Scholar]
- 36. Ludtke S. J., Baldwin P. R., Chiu W. 1999. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128:82–97 [DOI] [PubMed] [Google Scholar]
- 37. Magariyama Y., et al. 1994. Very fast flagellar rotation. Nature 371:752. [DOI] [PubMed] [Google Scholar]
- 38. Mastronarde D. N. 2005. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152:36–51 [DOI] [PubMed] [Google Scholar]
- 39. McCarter L. L. 2001. Polar flagellar motility of the Vibrionaceae. Microbiol. Mol. Biol. Rev. 65:445–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. McCarter L. L. 1994. MotY, a component of the sodium-type flagellar motor. J. Bacteriol. 176:4219–4225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. McCarter L. L. 1994. MotX, the channel component of the sodium-type flagellar motor. J. Bacteriol. 176:5988–5998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Muramoto K., Macnab R. M. 1998. Deletion analysis of MotA and MotB, components of the force-generating unit in the flagellar motor of Salmonella. Mol. Microbiol. 29:1191–1202 [DOI] [PubMed] [Google Scholar]
- 43. Murphy G. E., Leadbetter J. R., Jensen G. J. 2006. In situ structure of the complete Treponema primitia flagellar motor. Nature 442:1062–1064 [DOI] [PubMed] [Google Scholar]
- 44. Okabe M., Yakushi T., Asai Y., Homma M. 2001. Cloning and characterization of motX, a Vibrio alginolyticus sodium-driven flagellar motor gene. J. Biochem. 130:879–884 [DOI] [PubMed] [Google Scholar]
- 45. Parsons L. M., Lin F., Orban J. 2006. Peptidoglycan recognition by Pal, an outer membrane lipoprotein. Biochemistry 45:2122–2128 [DOI] [PubMed] [Google Scholar]
- 46. Pettersen E. F., et al. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605–1612 [DOI] [PubMed] [Google Scholar]
- 47. Reid S. W., et al. 2006. The maximum number of torque-generating units in the flagellar motor of Escherichia coli is at least 11. Proc. Natl. Acad. Sci. U. S. A. 103:8066–8071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Roujeinikova A. 2008. Crystal structure of the cell wall anchor domain of MotB, a stator component of the bacterial flagellar motor: Implications for peptidoglycan recognition. Proc. Natl. Acad. Sci. U. S. A. 105:10348–10353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sato K., Homma M. 2000. Functional reconstitution of the Na+-driven polar flagellar motor component of Vibrio alginolyticus. J. Biol. Chem. 275:5718–5722 [DOI] [PubMed] [Google Scholar]
- 50. Stader J., Matsumura P., Vacante D., Dean G. E., Macnab R. M. 1986. Nucleotide sequence of the Escherichia coli motB gene and site-limited incorporation of its product into the cytoplasmic membrane. J. Bacteriol. 166:244–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Terashima H., Fukuoka H., Yakushi T., Kojima S., Homma M. 2006. The Vibrio motor proteins, MotX and MotY, are associated with the basal body of Na-driven flagella and required for stator formation. Mol. Microbiol. 62:1170–1180 [DOI] [PubMed] [Google Scholar]
- 52. Terashima H., Kojima S., Homma M. 2008. Flagellar motility in bacteria structure and function of flagellar motor. Int. Rev. Cell Mol. Biol. 270:39–85 [DOI] [PubMed] [Google Scholar]
- 53. Terashima H., Koike M., Kojima S., Homma M. 2010. The flagellar basal body-associated protein FlgT is essential for a novel ring structure in the sodium-driven Vibrio motor. J. Bacteriol. 192:5609–5615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Thomas D. R., Francis N. R., Xu C., DeRosier D. J. 2006. The three-dimensional structure of the flagellar rotor from a clockwise-locked mutant of Salmonella enterica serovar Typhimurium. J. Bacteriol. 188:7039–7048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yakushi T., Kojima M., Homma M. 2004. Isolation of Vibrio Na+-driven flagellar motor complex composed of PomA and PomB solubilized by sucrose monocaprate. Microbiology 150:911–920 [DOI] [PubMed] [Google Scholar]
- 56. Yakushi T., Hattori N., Homma M. 2005. Deletion analysis of the carboxyl-terminal region of the PomB component of the vibrio alginolyticus polar flagellar motor. J. Bacteriol. 187:778–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yonekura K., Toyoshima C. 2000. Structure determination of tubular crystals of membrane proteins. II. Averaging of tubular crystals of different helical classes. Ultramicroscopy 84:15–28 [DOI] [PubMed] [Google Scholar]
- 58. Yonekura K., et al. 2006. Electron cryomicroscopic visualization of PomA/B stator units of the sodium-driven flagellar motor in liposomes. J. Mol. Biol. 357:73–81 [DOI] [PubMed] [Google Scholar]
- 59. Yorimitsu T., Homma M. 2001. Na+-driven flagellar motor of Vibrio. Biochim. Biophys. Acta 1505:82–93 [DOI] [PubMed] [Google Scholar]
- 60. Yorimitsu T., Sowa Y., Ishijima A., Yakushi T., Homma M. 2002. The systematic substitutions around the conserved charged residues of the cytoplasmic loop of Na+-driven flagellar motor component PomA. J. Mol. Biol. 320:403–413 [DOI] [PubMed] [Google Scholar]
- 61. Zheng Q. S., Braunfeld M. B., Sedat J. W., Agard D. A. 2004. An improved strategy for automated electron microscopic tomography. J. Struct. Biol. 147:91–101 [DOI] [PubMed] [Google Scholar]
- 62. Zhou J., Fazzio R. T., Blair D. F. 1995. Membrane topology of the MotA protein of Escherichia coli. J. Mol. Biol. 251:237–242 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.