Abstract
The O-antigen component of the lipopolysaccharide (LPS) represents a population of polysaccharide molecules with nonrandom (modal) chain length distribution. The number of the repeat O units in each individual O-antigen polymer depends on the Wzz chain length regulator, an inner membrane protein belonging to the polysaccharide copolymerase (PCP) family. Different Wzz proteins confer vastly different ranges of modal lengths (4 to >100 repeat units), despite having remarkably conserved structural folds. The molecular mechanism responsible for the selective preference for a certain number of O units is unknown. Guided by the three-dimensional structures of PCPs, we constructed a panel of chimeric molecules containing parts of two closely related Wzz proteins from Salmonella enterica and Shigella flexneri which confer different O-antigen chain length distributions. Analysis of the O-antigen length distribution imparted by each chimera revealed the region spanning amino acids 67 to 95 (region 67 to 95), region 200 to 255, and region 269 to 274 as primarily affecting the length distribution. We also showed that there is no synergy between these regions. In particular, region 269 to 274 also influenced chain length distribution mediated by two distantly related PCPs, WzzB and FepE. Furthermore, from the 3 regions uncovered in this study, region 269 to 274 appeared to be critical for the stability of the oligomeric form of Wzz, as determined by cross-linking experiments. Together, our data suggest that chain length determination depends on regions that likely contribute to stabilize a supramolecular complex.
INTRODUCTION
Lipopolysaccharide (LPS) is a major component of the outer membrane in Gram-negative bacteria and consists of the following three regions: lipid A, core oligosaccharide (OS), and in some bacteria, the O-antigen polysaccharide (OAg) (30). The OAg is formed as an undecaprenyl-phosphate (Und-P-P)-linked intermediate and ligated to lipid A-core OS with the release of Und-P-P. While lipid A and core OS structures are relatively conserved, the OAg is highly diverse, giving rise to thousands of O-specific serotypes among Gram-negative bacteria. In certain bacteria, the OAg plays a role in pathogenesis by influencing macrophage recognition and resistance to the lytic action of the complement system (3, 25, 26, 32), epithelial cell invasion (7, 38), and intracellular survival (27).
Und-P-P-OAg polymers are made of multiple oligosaccharide-repeating units. The synthesis of the OAg starts at the cytosolic face of the plasma membrane by the formation of a sugar phosphoanhydride linkage with Und-P, a reaction catalyzed by two different families of integral membrane proteins (36). Following the initiation reaction, additional sugars are added to complete the OAg subunit through reactions catalyzed by specific glycosyltransferases. Polymeric OAg assembly occurs by mechanisms referred to as Wzy (polymerase)-dependent and ATP-binding cassette (ABC)-dependent pathways (36). In ABC-dependent pathways, the OAg polymer is formed on the cytoplasmic side of the inner membrane and subsequently exported across the inner membrane by an ABC transporter. In most systems, Wzm and Wzt are the permease and ATPase components of the ABC transporter (4), while in others, a single protein mediates the energy-dependent export (14). In the Wzy-dependent pathway, Und-P-P-linked OAg repeating units are synthesized at the cytosolic side of the inner membrane. Each unit is subsequently translocated across the membrane by an ATP hydrolysis-independent mechanism mediated by Wzx flippase (8, 20–22). On the periplasmic side of the plasma membrane, translocated subunits polymerize to a certain length, unique to each OAg, by the concerted functions of Wzy (OAg polymerase) and Wzz (OAg chain length regulator or copolymerase). The resulting polysaccharide is ligated “en bloc” to the lipid A-core OS by the WaaL ligase (30). LPS is then transported to the outer leaflet of the outer membrane by the recently discovered Lpt multiprotein complex (31, 33).
LPS on the bacterial cell surface consists of a population of molecules with OAg chains that have nonrandom (modal) chain length distribution (1). The chain length distribution of OAgs assembled by the Wzy-dependent pathway requires Wzz, a member of a protein family collectively referred to as polysaccharide copolymerases (PCPs) (24). Different Wzz proteins confer a wide range of modal lengths (4 to >100 repeat units). Gram-negative bacteria often have two different Wzz proteins that confer two distinct OAg modal chain lengths, one longer and one shorter (23). The Wzz proteins are 36- to 40-kDa inner membrane proteins with substantial variation in sequence identity (∼15 to ∼80%) but a conserved structural organization. They have two transmembrane regions (TM1 and TM2) located near the amino and carboxyl termini, a proline- and glycine-rich motif sequence located adjacent to and overlapping with TM2, and a large soluble region between TM1 and TM2 extending to the periplasm. A second protein, designated FepE or WzzFepE, is also present in several bacteria, including Escherichia coli and Salmonella enterica serovar Typhimurium (herein S. Typhimurium), Shigella flexneri, and Pseudomonas aeruginosa, and is responsible for the very-long-OAg-chain modality.
Crystallographic studies of the periplasmic domains of WzzB from S. enterica (responsible for an OAg chain modal length of 16 to 35 repeat units) and FepE from E. coli O157:H7 (responsible for an OAg chain modal length of more than 80 repeat units) revealed that the two proteins adopt very similar three-dimensional folds at the protomer level (35). Despite their low sequence similarity, both proteins contain an α/β base domain and a long centrally located α-helix, about 100 Å in length. In the crystals, WzzB and FepE assembled into oligomers of various sizes, suggesting that the oligomerization state could be important to determine the modal length. Results obtained from cryoelectron microscopy upon reconstitutions of purified copolymerases in lipid vesicles challenge this notion (17). Therefore, the current structural information, while providing some clues with regard to the molecular basis of OAg modal distribution, has not yet led to a clear mechanistic view of this process.
Several attempts were made to identify the regions controlling the specificity toward a given O-unit repeat number. By analyzing WzzB homologs from closely related Shigella and E. coli species, Klee et al. (16) showed that minor variations at the primary sequence level give rise to different O-antigen modalities (16). Others (10) reported that several conserved substitutions of hydrophobic amino acids in the corresponding Wzz are responsible for the differences in modal lengths seen among several serotypes of E. coli (9). The differences in modalities imparted by WzzB from S. Typhimurium and S. flexneri were ascribed to their C-terminal regions comprising 134 residues (6). More recently, Papadopoulos and Morona (28) showed that the modal length could be modified by insertion of short sequences in various places in the copolymerase sequence.
Guided by structural information, we have probed regions of Wzz that are important for modal length determination. We used structural data on WzzB and FepE to construct a panel of chimeric and mutant proteins and examine their effects on OAg assembly in vivo in an E. coli K-12 strain reconstituted with an O-antigen synthesis pathway (8). We report the identification of several regions that dictate specificity toward a given modality.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Plasmids and bacterial strains used in this study are listed in Table 1. Strain EVV33 (E. coli W3110 ΔwzzB [18]) was used for in vivo complementation studies. Bacteria were cultured at 37°C in Luria-Bertani medium supplemented with ampicillin (100 μg/ml−1) and 0.2% (wt/vol) arabinose.
Table 1.
Strains and plasmids used in this study
| Strain/plasmid | Relevant property(ies) | Source/reference |
|---|---|---|
| Strains | ||
| DH5α | F− φ80lacZΔM15 endA recA hsdR (rK− mK−) supE thi gyrA relA Δ(lacZYA-argF)U169 | Laboratory stock |
| EVV33 | E. coli W3110 ΔwzzB | 18 |
| W3110 | rph-1 1N(rrnD-rrnE) | Laboratory stock |
| Plasmids | ||
| pBAD24 | Arabinose inducible, expression vector, Apr | 13 |
| pMF19 | wbbLEcO16 cloned into pEXT21, Spr | 8 |
| pWzzB | pEV6, parental wzzBEcK-12 cloned into pBAD24 | 37 |
| pWzzB-SF | parental wzzBSF cloned into pBAD24 | This study |
| pWzzB-ST | parental wzzBST cloned into pBAD24 | This study |
| pSK1 | wzzBST′(F95)-′wzzBSF(I96) encoding WzzB-ST-SF-T1 | This study |
| pSK2 | wzzBST′(L119)-′wzzBSF(T120) encoding WzzB-ST-SF-T2 | This study |
| pSK3 | wzzBST′(T139)-′wzzBSF(T140) encoding WzzB-ST-SF-T3 | This study |
| pSK4 | wzzBST′(Q199)-′wzzBSF(I200) encoding WzzB-ST-SF-T4 | This study |
| pSK5 | wzzBST′(Y255)-′wzzBSF(Q256) encoding WzzB-ST-SF-T5 | This study |
| pSK6 | wzzBST′(V274)-′wzzBSF(H275) encoding WzzB-ST-SF-T6 | This study |
| pSK7 | wzzBSF′(L95)-′wzzBST(I96) encoding WzzB-SF-ST-F1 | This study |
| pSK8 | wzzBSF′(L119)-′wzzBST(T120) encoding WzzB-SF-ST-F2 | This study |
| pSK9 | wzzBSF′(Q139)-′wzzBST(T140) encoding WzzB-SF-ST-F3 | This study |
| pSK10 | wzzBSF′(Q199)-′wzzBST(I200) encoding WzzB-SF-ST-F4 | This study |
| pSK11 | wzzBSF′(Y255)-′wzzBST(Q256) encoding WzzB-SF-ST-F5 | This study |
| pSK12 | wzzBSF′(I274)-′wzzBST(H275) encoding WzzB-SF-ST-F6 | This study |
| pSK13 | wzzBSF(V67-L95 >A67-F95 wzzBST) encoding WzzB-SF-T7 | This study |
| pSK14 | wzzBSF(Q202-I274 >E202-V274 wzzBST) encoding WzzB-SF-T8 | This study |
| pSK15 | wzzBSF(Q202-A253 >E202-N253 wzzBST) encoding WzzB-SF-T9 | This study |
| pSK16 | wzzBSF(Q202-K214 >E202-Q214 wzzBST) encoding WzzB-SF-T10 | This study |
| pSK17 | wzzBSF(V217-E237 >I217-K237 wzzBST) encoding WzzB-SF-T11 | This study |
| pSK18 | wzzBSF(N260-I274 >T260-V274 wzzBST) encoding WzzB-SF-T12 | This study |
| pSK19 | wzzBSF(F269-I274 >V269-V274 wzzBST) encoding WzzB-SF-T13 | This study |
| pSK20 | wzzBST(A67-F95 >V67-L95 wzzBSF) encoding WzzB-ST-F7 | This study |
| pSK21 | wzzBST(E202-V274 >Q202-I274 wzzBSF) encoding WzzB-ST-F8 | This study |
| pSK22 | wzzBST(E202-N253 >Q202-A253 wzzBSF) encoding WzzB-ST-F9 | This study |
| pSK23 | wzzBST(E202-Q214 >Q202-K214 wzzBSF) encoding WzzB-ST-F10 | This study |
| pSK24 | wzzBST(I217-K237 >I217-E237 wzzBSF) encoding WzzB-ST-F11 | This study |
| pSK25 | wzzBST(T260-V274 >N260:I274 wzzBSF) encoding WzzB-ST-F12 | This study |
| pSK26 | wzzBST(V269-V274 >F269-I274 wzzBSF) encoding WzzB-ST-F13 | This study |
| pSK27 | wzzBST(E202-Q214 >Q202-K214; T260-V274 >N260-I274 wzzBSF) encoding WzzB-ST-F14 | This study |
| pSK28 | wzzBST(A67-F95 >V67-I95; E202-Q214 >Q202-K214 wzzBSF) encoding WzzB-ST-F15 | This study |
| pSK29 | wzzBST(E202-Q214 >Q202-K214; V269-V274 > F269-I274 wzzBSF) encoding WzzB-ST-F16 | This study |
| pSK30 | wzzBSF(V67-L95 >A67:F95; F269-I274 >V269-V274 wzzBST) encoding WzzB-SF-T15 | This study |
| pSK31 | wzzBSF(V67-L95 >A67-F95; Q202-K214 >E202-Q214 wzzBST) encoding WzzB-SF-T16 | This study |
| pSK32 | wzzBSF(Q202-K214 >E202-Q214; F269-I274 >V269-V274 wzzBST) encoding WzzB-SF-T14 | This study |
| pSK33 | wzzBSF(V67-L95 >A67:F95; Q202-K214 >E202-Q214; F269-I274 >V269-V274 wzzBST) encoding WzzB-SF-T17 | This study |
| pSK34 | Parental fepEEcO157 | This study |
| pSK35 | Parental wzzBEcO157 | This study |
| pSK36 | fepEEcO157(256-273 >231-250 wzzBEcO157) | This study |
| pSK37 | wzzBEcO157(64-295 >62-338 fepEEcO157) | This study |
| pSK38 | wzzBEcO157(32-317 >42-359 fepEEcO157) | This study |
| pSK39 | wzzBST(E202-Q214 >Q202-K214; V269-V274 >F269-I274 wzzBSF) encoding WzzB-ST-F17 | This study |
| pSK40 | fepEEcO157 with deletion of residues 256 to 273 | This study |
Chimera construction and mutagenesis.
Genes encoding the chimeric Wzz proteins were constructed using overlap PCR (10), with pBAD24 as an expression vector (11). PCR fragments and pBAD24 were digested with EcoRI and KpnI prior to ligation. The DNA sequences of the oligonucleotide primers used in this work can be found in Table S1 in the supplemental material. Site-directed mutagenesis was carried out using QuikChange (Stratagene), according to the manufacturer's instructions. Briefly, the corresponding oligonucleotides containing the desired substitutions were used for whole-plasmid amplification using Pfu polymerase for 16 cycles. The resulting products were subjected to DpnI treatment and were introduced into E. coli DH5α competent cells by transformation. Plasmids were recovered, and the DNA inserts were sequenced to verify the presence of the desired modifications.
LPS extraction and SDS-PAGE.
LPS was prepared from bacteria grown in LB with 0.2% (wt/vol) arabinose as previously described (19). Gel loading was normalized to the bacterial density so that each sample represented the same number of cells (∼1 × 108 CFU). Samples were separated on 14% (wt/vol) Tris-SDS-PAGE, and the gels stained with silver nitrate (19).
Chemical cross-linking.
Chemical cross-linking was performed by a modification of the method of Papadopoulos and Morona (28). Briefly, EVV33(pMF19) bacteria containing plasmids encoding selected Wzz chimeras were grown overnight, diluted 1:200 into fresh broth, and grown to an optical density at 600 nm (OD600) of 0.6. Wzz expression was induced with 0.2% l-arabinose, and the cultures were grown for 2 h. Bacterial cells (3.5 × 108) were harvested, washed with ice-cold buffer A (10 mM sodium potassium phosphate buffer, pH 7.0), and then incubated with 0.5% (vol/vol) formaldehyde (Sigma) in buffer A at room temperature for 1 h. Samples incubated without formaldehyde were used as controls. After washing with ice-cold buffer A, samples were heated to 60°C or 100°C for 10 min, separated on a 12% SDS-PAGE gel, and subjected to immunoblotting. The primary antibody used was a 1:500 dilution of anti-WzzB polyclonal antiserum. The secondary antibody used was goat anti-rabbit IgG antibody conjugated with IRDye 800CW (Rockland, PA). The reacting bands were detected by fluorescence with an Odyssey infrared imaging system (Li-COR Biosciences).
RESULTS
Two regions in WzzBSF and WzzBST are critical to determine modality.
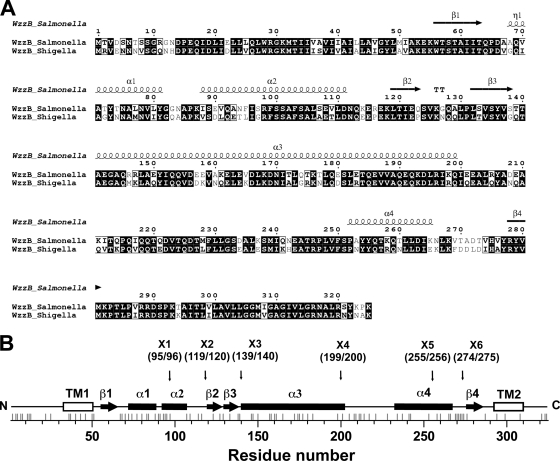
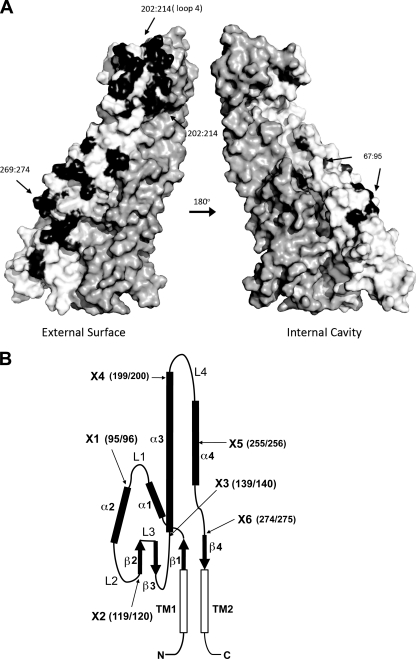
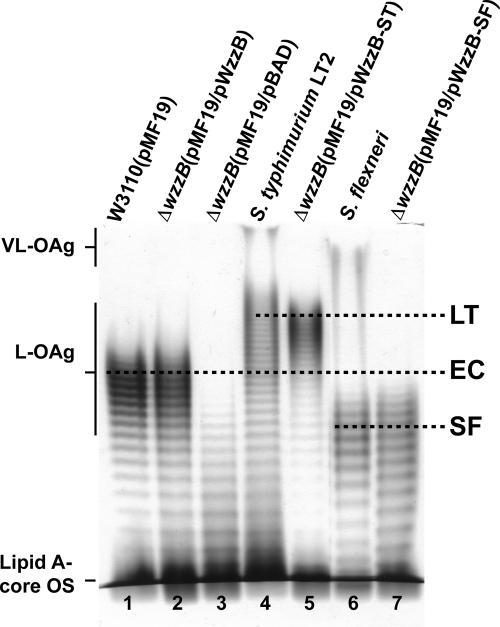
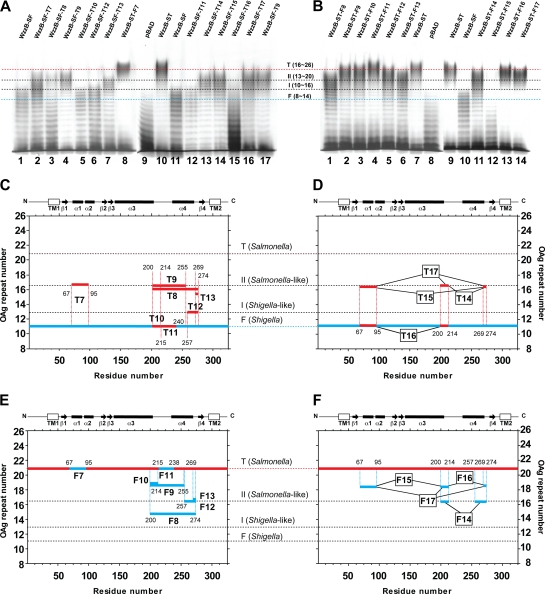
The strategy to identify regions of the copolymerase that are critical for chain length distribution (or modality) involved using a pair of proteins that confer readily discernible phenotypes. Such a pair consists of the highly similar WzzB copolymerases from S. enterica serovar Typhimurium (WzzBST) and S. flexneri (WzzBSF). Both proteins have ∼325 amino acids and share 72% amino acid sequence identity (Fig. 1). There are 68 amino acid differences between their periplasmic domains, and the majority of these residues map to the external surface in the context of the WzzB oligomer (Fig. 2 A). However, WzzBST gives rise to O-antigen polysaccharides with an average modal length of ∼26 repeats, while WzzBSF confers an average modal length of ∼14 repeats (6). Each of these proteins served as parent molecules to build a set of chimeras with junction points distributed along the length of the copolymerase. The choice of junction points was decided based on the known three-dimensional structure of the copolymerase protomer (35) and occurred either within the loops connecting the secondary structures or at regions of high sequence conservation, as guided by sequence alignment (Fig. 2B; Table 1). To investigate the modal distribution of the O-antigen polysaccharides mediated by each chimera, the proteins were expressed in the wzz null strain EVV33 containing pMF19 (18). The presence of pMF19 allows us to reconstruct O-antigen expression in E. coli K-12, as previously described (8). This approach was feasible since the modality of the O-antigen length is dependent mainly on the cognate WzzB. As demonstrated in Fig. 3, the recombinant WzzBST and WzzBSF proteins confer a modality to the O16 antigen that is similar to the modality observed in the respective S. enterica serovar Typhimurium and S. flexneri strains (compare lanes 4 and 5 and lanes 6 and 7, respectively).
Fig. 1.
Sequences and secondary structure of WzzB proteins. (A) Sequence alignment of WzzBST and WzzBSF. The secondary structures for α-helices and β-strands in S. Typhimurium WzzB are indicated by Greek letters and numbers. The alignment was generated using the program ClustalW (2), and this figure was prepared by using ESPript (11) (B) Linear representation of WzzB, with secondary structure marked along the polypeptide chain.
Fig. 2.
Structural map of WzzB. (A) The differences in amino acids between WzzBST and WzzBSF were mapped onto the surface of the periplasmic domain of chimeric WzzB, composed of various portions of WzzBSF and WzzBST. One protomer is colored in white, the nonconserved residues are colored in black, and the other protomer is colored dark gray. The regions found to be functionally important are labeled, and their positions are indicated with arrows. This figure was prepared using PyMOL (http://pymol.org). (B) Topological diagram of WzzST indicating the most important structural regions of the Wzz protomer. α, α-helix; β, β-strand; L, loop; TM, transmembrane domain. The arrows point to the locations of junction points (indicated by amino acid number in parentheses) for the construction of chimeric WzzST/WzzSF and WzzSF/WzzST derivatives.
Fig. 3.
Modality of O-antigen polysaccharides expressed in E. coli K-12, S. enterica, and S. flexneri. Silver-stained polyacrylamide gel showing the O-antigen LPS profiles of the E. coli K-12 W3110 (lane 1) and EVV33 (ΔwzzB) isogenic strains (lanes 2, 3, 5, and 7) containing pMF19 to allow expression of O16 LPS. The LPS profiles of parental S. Typhimurium (lane 4) and S. flexneri (lane 6) are also shown. For the expression of recombinant WzzB, EVV33 also the contained pBAD vector control (lane 3) and WzzB expressing plasmids from E. coli K-12 (pWzzB; lane 2), S. Typhimurium (pWzzB-ST; lane5) and S. flexneri (pWzzB-SF; lane 7). The regions corresponding to the migration of the lipid A-core OS, long OAg (L-OAg), and very long OAg (VL-OAg) polysaccharide are indicated, as well as the average modalities of each strain (LT, S. Typhimurium; EC, E. coli K-12; and SF, S. flexneri). Note that VL-OAg is normally produced in S. Typhimurium and S. flexneri and imparted by FepE.
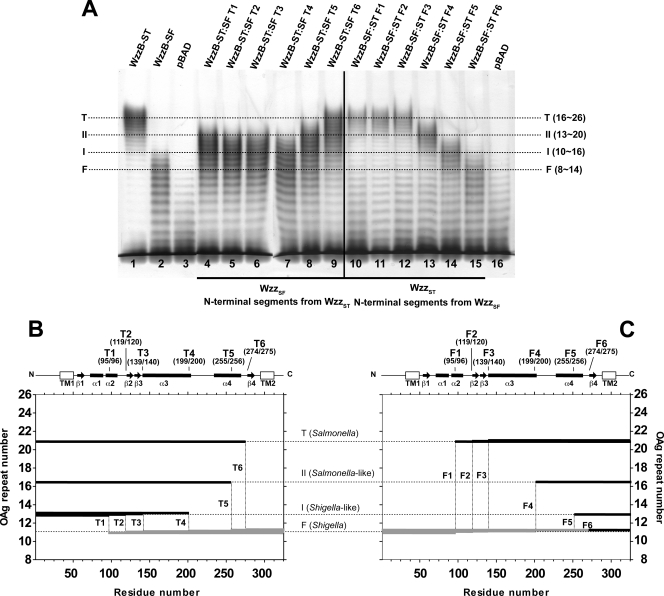
We constructed six chimeras containing progressively increasing N-terminal segments from WzzBST and correspondingly decreasing C-terminal parts from WzzBSF (Table 1; Fig. 4). The introduction of the WzzBST N-terminal sequence (first 95 residues) in the chimeras led to an increase of O-antigen chain length distribution from 8 to 14 repeat units in WzzBSF to 10 to 16 (Fig. 4A, lane 4). This pattern remained largely unchanged in the chimeras containing 119, 139, and 199 N-terminal residues from WzzBST (Fig. 4A, lanes 5 to 7). The chimera, including 255 N-terminal residues from WzzBST, resulted in a further increase of modality of 13 to 23 repeats (Fig. 4A, lane 8), while a small extension of the WzzBST component to include 274 residues resulted in modality that was comparable to that of intact WzzBST (Fig. 4A, compare lanes 1 and 9). These observations suggest that the region spanning amino acids 1 to 95 (region 1 to 95) (spanning the TM, helix α1, and part of helix α2) and region 199 to 274 (spanning loop 4 [L4], helix α4, and loop L5 in the three-dimensional structure of the copolymerase protomer) are important for increasing the modal length (Fig. 3A and B).
Fig. 4.
Effect of reciprocal domain swapping of large regions of WzzB from S. flexneri and S. Typhimurium. (A) Silver-stained polyacrylamide gel showing the O-antigen LPS profile of E. coli EVV33(pMF19) expressing parental and swapped versions of WzzB (Table 1). The designations T1 to T6 indicate that the chimeras contain N-terminal segments from WzzBST in the WzzBSF background. The designations F1 to F6 indicate that the chimeras contain N-terminal segments from WzzBSF in the WzzBST background. Lane 1, parental WzzBST; lane 2, parental WzzBSF; lanes 3 and 16, pBAD vector control; lane 4, WzzBST(F95)-WzzBSF(I96) (WzzB ST-SF-T1); lane 5, WzzBST(L119)-WzzBSF(T120) (WzzB ST-SF-T2); lane 6, WzzBST(T139)-WzzBSF(T140) (WzzB ST-SF-T3); lane 7, WzzBST(Q199)-WzzBSF(I200) (WzzB ST-SF-T4); lane 8, WzzBST(Y255)-WzzBSF(Q256) (WzzB ST-SF-T5); lane 9, WzzBST(V274)-WzzBSF(H275) (WzzB ST-SF-T6); lane 10, WzzBSF(L95)-WzzBST(I96) (WzzB SF-ST-F1); lane 11, WzzBSF(L119)-WzzBST(T120) (WzzB SF-ST-F2); lane 12, WzzBSF(Q139)-WzzBST(T140) (WzzB SF-ST-F3); lane 13, WzzBSF(Q199)-WzzBST(I200) (WzzB SF-ST-F4); lane 14, WzzBSF(Y255)-WzzBST(Q256) (WzzB SF-ST-F5); and lane 15, WzzBSF(I274)-WzzBST(H275) (WzzB SF-ST-F6). Dotted lines indicate the average modality for the E. coli K-12 (O16) LPS OAg molecules, as determined by WzzBST (T), WzzBST-like (II), WzzBSF-like (I), and WzzBSF (F) proteins. (B and C) Graphical representation of the changes in modality observed for hybrids shown in panel A, lanes 4 to 9 and lanes 10 to 15, respectively. Hybrid polypeptides are plotted according to residue number (on the x axis) and average modality (OAg repeat number; on the y axis). Thick black segments indicate the regions derived from WzzBST, while thick gray segments indicate WzzBSF regions. A linear representation of the structural map of WzzB and the location of the hybrid endpoints are also indicated.
To test whether the same regions are also involved in the O-antigen length control when the order of the components making chimeric WzzB is reversed, we constructed a reciprocal set of hybrids containing increasing N-terminal regions from WzzBSF and decreasing C-terminal regions of WzzBST with the same junction points as used previously (Fig. 1 and 2). The modality conferred by the chimeras containing N-terminal sequences from WzzBSF to positions 95, 119, and 139 was very similar to that conferred by parental WzzBST (Fig. 4A, compare lane 1 with lanes 10 to 12). The chimeras with fusion junction points at positions 199 and 255 showed progressively smaller modal lengths (Fig. 4A, lanes 13 and 14), while the chimera containing 274 residues of WzzBSF conferred modality comparable to that conferred by the parental WzzBSF (Fig. 4A, compare lanes 2 and 15). The reciprocal experiment showed that the N-terminal region did not much affect the modality and confirmed that region 200 to 274 spanning L4, α4, and L5 in the three-dimensional structure of the copolymerase protomer is important to mediating modality in WzzBSF (Fig. 4C).
Structural segments that individually affect modality exhibit no cooperativity.
To narrow down the regions determining modality, we created a second set of chimeras by replacing shorter internal segments of WzzBSF with the corresponding segments from WzzBST. We first swapped residues 67 to 95 spanning helix α1 and part of helix α2. We chose this region because in the first series of chimeras, the crossover at position 95 gave an intermediate modality change compared to that imparted by parental WzzBSF (Fig. 4A, lane 4). This construct also conferred an intermediate phenotype, as indicated by an average modal length of ∼16 repeat units, with fewer longer chains but also with a noticeably larger amount of shorter chain polymers (Fig. 5 A, lane 2). Swapping residues 200 to 274, 200 to 255, and 269 to 274 (Fig. 5C, T8, T9, and T13) also resulted in increased modal lengths, but these were still shorter on average than those of the parental WzzBST (Fig. 5A, lanes 3, 4, and 7). In contrast, substitution of smaller segments within this range showed that regions 200 to 214, 215 to 240, and 257 to 274 (Fig. 5A, lanes 5, 6, and 12, and Fig. 5C, T10 to T12, respectively) did not alter much modal length distribution and retained a WzzBSF-like phenotype.
Fig. 5.
Effect of reciprocal single- and multiple-domain swapping of small regions of WzzB from S. flexneri and S. Typhimurium. (A) silver-stained polyacrylamide gel showing the O-antigen LPS profile of E. coli EVV33 (pMF19) expressing different forms of WzzB (Table 1). The designations T7 to T13 indicate that the chimera contains one segment from WzzBST in the WzzBSF background, while T14 to T17 denote two or more segments of WzzBST in the WzzBSF background. The designations F7 to F13 indicate that the chimera contains one segment from WzzBSF in the WzzBST background. F14 to F17 denotes two or more segments of WzzBSF in the WzzBST background. Lanes 1 and 11, parental WzzBSF; lane 2, WzzBSFV67-L95>A67-F95WzzBST (WzzB-SF-T7); lane 3, WzzBSFQ202-I274>E202-V274WzzBST (WzzB-SF-T8); lanes 4 and 17, WzzBSFQ202-A253>E202-N253WzzBST (WzzB-SF-T9); lane 5, WzzBSFQ202-K214>E202-Q214WzzBST (WzzB-SF-T10); lane 6, WzzBSFN260-I274>T260-V274WzzBST (WzzB-SF-T12); lane 7, WzzBSFF269-I274>V269-V274WzzBST (WzzB-SF-T13); lane 8, WzzBSTA67-F95>V67-L95WzzBSF (WzzB-ST-F7); lane 9, pBAD vector control; lane 10, parental WzzBST; lane 12, WzzBSFV217-E237>I217-K237WzzBST (WzzB-SF-T11); lane 13, WzzBSFQ202-K214>E202-Q214/F269-I274>V269-V274WzzBST (WzzB-SF-T14); lane 14, WzzBSFV67-L95>A67:F95/F269-I274>V269-V274 WzzBST (WzzB-SF-T15); lane 15, WzzBSFV67-L95>A67-F95/Q202-K214>E202-Q214WzzBST (WzzB-SF-T16); and lane 16, WzzBSFV67-L95>A67:F95/Q202-K214>E202-Q214/F269-I274>V269-V274WzzBST (WzzB-SF-T17). (B) Silver-stained polyacrylamide gel showing the O-antigen LPS profile of E. coli EVV33 (pMF19) expressing various recombinant WzzB forms (Table 1). Lane 1, WzzBSTE202-V274>Q202-I274WzzBSF (WzzB-ST-F8); lane 2, WzzBSTE202-N253>Q202-A253 WzzBSF (WzzB-ST-F9); lane 3, WzzBSTE202-Q214>Q202-K214WzzBSF (WzzB-ST-F10); lane 4, WzzBSTI217-K237>I217-E237WzzBSF (WzzB-ST-F11); lane 5, WzzBSTT260-V274>N260:I274WzzBSF (WzzB-ST-F12); lane 6, WzzBSTV269-V274>F269-I274WzzBSF (WzzB-ST-F13); lanes 7 and 9, parental WzzBST; lane 8, pBAD vector control; lane 10, parental WzzBSF; 11, WzzBSTE202-Q214>Q202-K214/T260-V274>N260-I274WzzBSF (WzzB-ST-F14); lane 12, WzzBSTA67-F95>V67-I95/E202-Q214>Q202-K214WzzBSF (WzzB-ST-F15); lane 13, WzzBSTE202-Q214>Q202-K214/V269-V274>F269-I274WzzBSF (WzzB-ST-F16); and lane 14, WzzBSTA67-F95>V67-I95/V269-V274>F269-I274WzzBSF (WzzB-ST-F17. Dotted lines indicate the average modality for the E. coli K-12 (O16) LPS OAg molecules, as determined by WzzBST (T), WzzBST-like (II), WzzBSF-like (I), and WzzBSF (F) proteins. (C and D) Graphical representation of the changes in modality observed for hybrids in panel A, lanes 2 to 7 and 12 as well as lanes 13 to 16, respectively. (E and F) Graphical representation of the changes in modality observed for hybrids in panels A (lane 8) and B (lanes 1 to 6) as well as in panel B, lanes 11 to 14, respectively. Hybrid polypeptides are plotted according to the residue number (on the x axis) and the average modality (OAg repeat number; on the y axis). Thick red segments indicate the regions derived from WzzBST, while thick blue segments indicate WzzBSF regions. A linear representation of the structural map of WzzB and the location of the hybrid endpoints are also indicated in panels C through F.
As done previously, we also conducted reciprocal experiments by investigating whether the introduction of smaller regions from WzzBSF into the WzzBST background would be sufficient to change the specificity of WzzBST toward the shorter modality. In this series of chimeras, the changes in modal lengths were less pronounced. Swapping residues 67 to 95 of WzzBST with similar residues from WzzBSF did not have any noticeable effect on the WzzBST modality (Fig. 5A, lane 8, and Fig. 5E, F7). Swapping the regions 200 to 255, 200 to 215, and 215 to 238 decreased modal length only minimally compared to the parental WzzBST (Fig. 5B, lanes 2 to 4, and Fig. 5E, F9 to F11, respectively). However, swapping residues 257 to 274 and 269 to 274 (Fig. 5B, lanes 5 and 6, and Fig. 5E, F12 and F13) caused a somewhat more pronounced reduction in the modality imparted by the recombinant copolymerase, but the pattern of the O-polysaccharide chain length remained similar to that of WzzBST. Finally, the replacement of a longer region encompassing residues 200 to 274 (Fig. 5B, lane 1, and Fig. 5E, F8) resulted in a reduction of modality that was closer to that of WzzBSF.
Since we identified several short segments that individually affected the modality of the O antigen, we investigated whether combining these segments would cause any further changes in modal length. Thus, we constructed several additional chimeras containing various combinations of the three segments from WzzBST within the WzzBSF host molecule (Fig. 5D) and the reciprocal constructs within WzzBST (Fig. 5F). Chimeras containing any two or all three regions within either WzzBSF or WzzBST as the host protein led to no additional changes in modal length distribution compared to that with the replacement of single regions (Fig. 5A, lanes 13 to 16, and Fig. 5B, lanes 11 to 14). From these results, we conclude that the identified regions contribute to determine modality but do not act cooperatively.
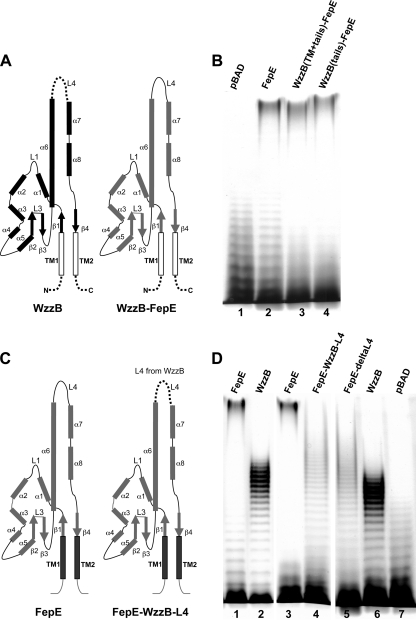
The above-described experiments showed that chimeras of closely related PCPs are functional and mediate intermediate modality compared to the parental molecules. Next, we investigated whether splicing two more divergent PCPs can lead to functional chimeric molecules. We selected two PCPs coexisting in the same E. coli strain, namely, FepE (modal length of >80 repeats) and WzzB from E. coli O157:H7 (WzzBEcO157) (modal length of ∼10 to 19 repeats), which show only 27% identity. WzzBEcO157 is nearly identical (98%) to WzzBSF and closely related (72%) to WzzBST. We selected the same crossover points and constructed six FepE-WzzB and six WzzB-FepE chimeras. However, no phenotype was observed in cells expressing these chimeras, and this was associated with very poor or no expression of the chimeras (data not shown). Since the fusions were correct at the DNA level, we concluded that the resulting proteins were likely not stable. We also investigated whether the lack of functionality of the chimeras could be caused by the mismatch of the transmembrane helices or if the transmembrane helices and cytosolic portions of the protein are involved in determining the specificity toward a very long modal length. We replaced the entire periplasmic domain of WzzBEcO157 with the periplasmic domain of FepE. A second chimera leaving only the N- and C-terminal cytosolic tails of WzzBEcO157 was made by replacing the periplasmic and TM domains of WzzBEcO157 with the corresponding regions of FepE (Fig. 6 A). Both recombinant proteins led to the production of the O-antigen molecules with chain length distribution indistinguishable from that imparted by FepE (Fig. 6B, lanes 3 and 4). This experiment demonstrates that the periplasmic domain of FepE determines the specificity toward the very long modality.
Fig. 6.
Loop 4 of Wzz is important for modality. (A) Diagrams showing the structural components of WzzBEcO157 (WzzB; in black) and its derivative in which the entire periplasmic region was replaced with the corresponding region of FepEEcO157 (WzzB-FepE), indicated in gray. (B) Silver-stained polyacrylamide gel showing the O-antigen LPS profile of E. coli EVV33(pMF19) containing various plasmids. Lane 1, pBAD; lane 2, pSK34 carrying the parental fepEEcO157 (FepE); lane 3, pSK37 encoding WzzB with the FepE periplasmic domain; and lane 4, pSK38 encoding WzzB with FepE periplasmic and TM regions. (C) Diagrams showing the structural components of FepEEcO157 (FepE; in gray) and its derivative in which parental loop 4 (L4) region was replaced with the corresponding region of WzzBEcO157 (FepE-WzzB-L4), as indicated by the dotted line. (D) Silver-stained polyacrylamide gel showing the O-antigen LPS profile of E. coli EVV33(pMF19), with pSK34 carrying parental fepEEcO157 (FepE; lanes 1 and 3), pSK35 carrying parental wzzBEcO157 (WzzB; lanes 2 and 6), pSK36 carrying fepEEcO157 (256–273 > 231–250 wzzBEcO157) (FepE-WzzB-L4; lane 4), pSK40 carrying fepEEcO157 with deletion of residues 256 to 273 (FepE-deltaL4; lane 5), and the pBAD24 vector control (pBAD; lane 7).
We also examined whether the segments previously found to influence chain length determination in WzzBSF and WzzBST may play functional roles in FepE. We selectively targeted several regions of FepE and discovered that various modifications of the flexible region referred to as loop 4 (L4) drastically affected the function. In particular, substitution of FepE region 256 to 273 with the corresponding WzzBEcO157 region 231 to 250 (Fig. 6C) led to a shorter O-antigen modal length of ∼21 repeat units (Fig. 6D, compare lanes 2 and 4). A very similar phenotype also resulted from the deletion of residues 256 to 273 in FepE (Fig. 6D, compare lanes 4 and 5).
Chimeras display differential stability upon cross-linking.
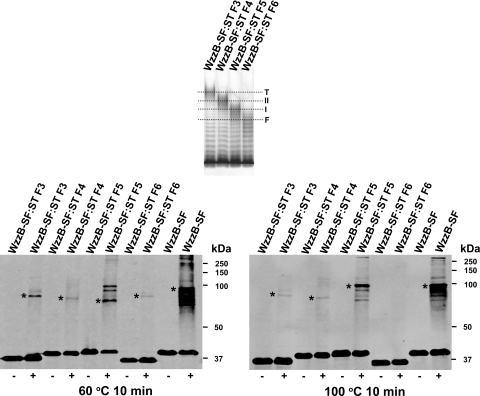
In a previous study, it was shown that WzzBSF stability and oligomerization affect O-antigen modality (26). We investigated whether the chimeras constructed here also exhibit differences in their ability to oligomerize and in their stability after cross-linking. Therefore, WzzBSF-WzzBST and the reciprocal WzzBST-WzzBSF chimeras with swapped regions at the 139, 199, 255, and 274 endpoints were examined for oligomerization and stability. These chimeras were selected because their fusion endpoints defined regions of functional importance for modality (Fig. 7, top). All of these proteins were well expressed and detectable by Western blotting. WzzB oligomers, mainly dimers, were detected by incubation of bacteria with formaldehyde before cell lysis. Using this methodology, different levels of oligomerization were observed, which could not be correlated with a specific swapped region. All of the oligomers were stable at 60°C after cross-linking (Fig. 7, left), but the oligomers of the chimeric 274-endpoint construct were not stable at 100°C (Fig. 7, right). This result suggests that the region corresponding to the C terminus of WzzBSF and spanning helix α4 and TM2 (Fig. 1B and 2B) is likely important for the oligomer stability.
Fig. 7.
Stability of selected WzzB chimeras upon cross-linking. (Top panel) Silver-stained polyacrylamide gel showing the O-antigen LPS profile of E. coli EVV33(pMF19) expressing different recombinant forms of WzzBST containing N-terminal segments of WzzBSF. F3, WzzBSF(Q139)-WzzBST(T140); F4, WzzBSF(Q199)-WzzBST(I200); F5, WzzBSF(Y255)-WzzBST(Q256); and F6, WzzBSF(I274)-WzzBST(H275). Dotted lines indicate the average modalities for the E. coli K-12 (O16) LPS OAg molecules as determined by WzzBST (T), WzzBST-like (II), WzzBSF-like (I), and WzzBSF (F) proteins. (Bottom panels) Immunoblots of total proteins, analyzed by Western blotting with anti-WzzB rabbit polyclonal antiserum. Prior to bacterial cell lysis, cultures were treated with formaldehyde (+), as described in Materials and Methods. Untreated samples (−) were used as controls. Prior to electrophoresis, samples were heated for 10 min at 60°C (left) or at 100°C (right). Asterisks indicate the presence of oligomeric forms of WzzB variants.
DISCUSSION
Chain length distribution of OAgs is imparted by PCPs in a fashion that is independent of the striking differences in the chemical composition of individual O units (4, 8). How each of the chain length regulators imposes a unique pattern of modality, even when heterologously expressed in closely related bacteria, remains unknown. In this work, we used a structure-guided approach to engineer hybrid and variant PCPs and test their ability to support O-antigen chain length determination in a wzz null strain. The two closely related PCPs, WzzBSF and WzzBST, differ by 68 amino acids in their periplasmic domains. These differences are located predominantly on the external surface of the oligomer, with the exception of these within the region ∼60 to 100, which is partially at the protomer interface and partially faces the internal cavity (Fig. 2A).
The analysis of various phenotypes imparted by the chimeras revealed that they are functional and confer modal distributions that are intermediate between those of the parent molecules. From our analysis, we demonstrate that several segments (residues 67 to 95, 200 to 255, and 269 to 274) can influence the specificity toward a given modal length. Generally, the results of these swaps were more pronounced when they caused an increase of the modal length (in WzzBSF background) and were less effective in decreasing the modal length (in WzzBST background). While the reasons for these observations may become apparent when the complete understanding of the molecular mechanism of copolymerase O-antigen chain length control is established, our findings agree well with the previous reports indicating the involvement of more than one region of the protein in determining the specific modality (5, 16, 28).
No functional chimeras resulted from the crossovers between more distantly related PCPs. The sequence differences are scattered throughout the molecules, including those of the interfacial regions, suggesting significant changes in protomers themselves and/or in their association. However, it was possible to transplant the periplasmic domain of FepE onto the transmembrane regions of WzzBEcO157 with the retention of FepE-like very-long-chain modality. Previously, it was reported that a mutation within the cytosolic tail of WzzpHS2 in Shigella flexneri can influence the phenotypes of mutations within the periplasmic domain, implying a direct involvement of this region in chain length control (29). Our results demonstrate that the specificity toward very long modal lengths is determined exclusively by the periplasmic domain and is independent of the cytoplasmic tails.
It was previously speculated that the internal cavity in the center of the oligomer of the crystal structures of FepE and WzzE could play a role in O-antigen polysaccharide length control, but this was not supported by mutagenesis data (35). In contrast, the residues of two of the three functionally important WzzB segments found in this work (residues 201 to 255 and 268 to 274) are predominantly on the external surface of the oligomer, while the third fragment (residues 65 to 96) is partially buried at the protomers interface. These observations lend support for the external surface of the chain length regulators representing the principal site of the modal length control. The observed phenotypic changes resulting from swapping a partially buried segment are perhaps related to the altered arrangement of the protomers within the oligomer. Indeed, Papadopoulos and Morona (28) reported that insertions of short 5-amino-acid sequences within region 65 to 95 of WzzBSF influenced the association of the individual protomers into higher-molecular-weight oligomers, as judged by cross-linking experiments. Here, we show that replacing the region of the WzzBSF C terminus spanning helix α4 and TM2 also affects the stability of the oligomers.
Our findings also correlate well with the previous investigations, which observed that the C-terminal portion of the protein could largely dictate specificity toward a given modality. In particular, a 69-amino-acid swap (D189 to D258) between two closely related WzzB molecules from E. coli (10), which resulted in substantial changes in the O-antigen modal lengths, predominantly consisted of the segment corresponding to the unstructured region of loop 4. Another reported chimeric WzzB with altered modal length specificity contained a 131-amino-acid C-terminal guest fragment containing loop 4 as well as the segment spanning amino acids 269 to 274 (5). Our data also demonstrate the functional importance of loop 4 for distantly related FepE.
FepE normally operates within the same cell as WzzB but imparts a very long modality (>80 repeat units) (25). We show that like in WzzBSF/WzzBST, replacement and deletion of short segments within loop 4 are important for the specificity of the modal distribution and play a critical role in the ability of FepE to affect synthesis of very long polysaccharide chains. This agrees with a recent study showing that insertion of linker peptides in this region leads to marked changes of modality by WzzBSF (28). The involvement of loop 4 in modal length control in all PCPs examined to date is significant, considering that it is positioned approximately 100 Å away from the inner membrane. This segment is disordered in all the available crystal structures of the chain length regulators (35). A recent reconstitution of the O-antigen polymerization in vitro demonstrates no need for participation of any proteins other than WzzB and Wzy to obtain O-antigen polymers with medium modal lengths (39). Wzy has a predicted large periplasmic loop (15, 36). Therefore, we speculate that the flexible loop 4 region allows for additional interactions with other protomers or could mediate a conformational change necessary for function. Indeed, our structural studies of the periplasmic domains of chimeric WzzB indicate that the loop 4 region is largely devoid of secondary structure elements and is at least partially involved in interprotomer contacts (our unpublished data).
Swapping multiple segments did not completely change the specificity of a given PCP toward a different modality, an observation that emphasizes the critical contribution of the flanking sequences for chain length control. Furthermore, PCPs among many E. coli serotypes differ only in several amino acids, many of which represent conservative substitutions, located mostly on the external face of the protein. These subtle changes might confer sufficiently different properties to the molecular surface of the PCP, which in turn could influence the extent of the nonspecific weak polysaccharide binding. The absence of a single, well-defined binding site is not surprising, given the sheer size of the synthesized O-antigen polymer. An extensive surface area may be necessary to form numerous noncovalent interactions along the exposed parts of the chain length regulator oligomers. The binding would have to be relatively weak to allow for a polysaccharide transfer to an LPS core ligation step without much energy expenditure in the ATP-depleted environment of a periplasm. In support of this hypothesis, conformational changes of the chain length regulators have been shown to occur in response to the O-antigen addition, as revealed both by circular dichroism and by small-angle small scattering studies (12, 34).
The current models used to explain the molecular mechanism of modal length control suggest interplay between the polymerase and the PCP (23), although evidence of a physical association between the two proteins is still lacking. The short (268-to-274-residue) segment imparting the greater alteration of the modality by the two closely related PCPs is located ∼30 Å from the inner membrane, sufficiently close to provide an interaction interface with Wzy. Thus, the observed changes in modal length upon swapping the two segments could in fact be caused by the different affinities of these regions toward Wzy. Since the interaction between the two proteins has not been directly shown, this hypothesis remains to be experimentally tested. Alternatively, this surface-exposed patch could be part of a carbohydrate interaction interface. In addition, the oligomerization state of Wzz protomers is critical for function, as recently demonstrated by Papadopoulos and Morona (28). Our cross-linking studies also confirm these observations.
In summary, through extensive structure-guided mutagenesis of class I PCPs, we identified several segments within the protein structure that contribute to defining the modality of the O antigen.
Supplementary Material
ACKNOWLEDGMENTS
We thank Cristina L. Marolda for initial input in this study.
This work was supported by grant MOP-89787 from the Canadian Institutes of Health Research to M.C. and M.A.V. M.A.V. holds a Canada Research Chair in Infectious Diseases and Microbial Pathogenesis.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 3 June 2011.
REFERENCES
- 1. Batchelor R. A., Haraguchi G. E., Hull R. A., Hull S. I. 1991. Regulation by a novel protein of the bimodal distribution of lipopolysaccharide in the outer membrane of Escherichia coli. J. Bacteriol. 173:5699–5704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chenna R., et al. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31:3497–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Clay C. D., Soni S., Gunn J. S., Schlesinger L. S. 2008. Evasion of complement-mediated lysis and complement C3 deposition are regulated by Francisella tularensis lipopolysaccharide O antigen. J. Immunol. 181:5568–5578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cuthbertson L., Kos V., Whitfield C. 2010. ABC transporters involved in export of cell surface glycoconjugates. Microbiol. Mol. Biol. Rev. 74:341–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Daniels C., Morona R. 1999. Analysis of Shigella flexneri wzz (Rol) function by mutagenesis and cross-linking: wzz is able to oligomerize. Mol. Microbiol. 34:181–194 [DOI] [PubMed] [Google Scholar]
- 6. Daniels C., Vindurampulle C., Morona R. 1998. Overexpression and topology of the Shigella flexneri O-antigen polymerase (Rfc/Wzy). Mol. Microbiol. 28:1211–1222 [DOI] [PubMed] [Google Scholar]
- 7. Duerr C. U., et al. 2009. O-antigen delays lipopolysaccharide recognition and impairs antibacterial host defense in murine intestinal epithelial cells. PLoS Pathog. 5:e1000567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Feldman M. F., et al. 1999. The activity of a putative polyisoprenol-linked sugar translocase (Wzx) involved in Escherichia coli O antigen assembly is independent of the chemical structure of the O repeat. J. Biol. Chem. 274:35129–35138 [DOI] [PubMed] [Google Scholar]
- 9. Franco A. V., Liu D., Reeves P. R. 1996. A Wzz (Cld) protein determines the chain length of K lipopolysaccharide in Escherichia coli O8 and O9 strains. J. Bacteriol. 178:1903–1907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Franco A. V., Liu D., Reeves P. R. 1998. The wzz (cld) protein in Escherichia coli: amino acid sequence variation determines O-antigen chain length specificity. J. Bacteriol. 180:2670–2675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gouet P., Robert X., Courcelle E. 2003. ESPript/ENDscript: extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 31:3320–3323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Guo H., et al. 2006. Overexpression and characterization of Wzz of Escherichia coli O86:H2. Protein Expr. Purif. 48:49–55 [DOI] [PubMed] [Google Scholar]
- 13. Guzman L. M., Belin D., Carson M. J., Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hug I., et al. 2010. Helicobacter pylori lipopolysaccharide is synthesized via a novel pathway with an evolutionary connection to protein N-glycosylation. PLoS Pathog. 6:e1000819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kim T. H., et al. 2010. Characterization of the O-antigen polymerase (Wzy) of Francisella tularensis. J. Biol. Chem. 285:27839–27849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Klee S. R., Tzschaschel B. D., Timmis K. N., Guzman C. A. 1997. Influence of different rol gene products on the chain length of Shigella dysenteriae type 1 lipopolysaccharide O antigen expressed by Shigella flexneri carrier strains. J. Bacteriol. 179:2421–2425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Larue K., Kimber M. S., Ford R., Whitfield C. 2009. Biochemical and structural analysis of bacterial O-antigen chain length regulator proteins reveals a conserved quaternary structure. J. Biol. Chem. 284:7395–7403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Marolda C. L., Haggerty E. R., Lung M., Valvano M. A. 2008. Functional analysis of predicted coiled coil regions in the Escherichia coli K-12 O antigen polysaccharide chain length determinant Wzz. J. Bacteriol. 196:2128–2137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Marolda C. L., Lahiry P., Vines E., Saldias S., Valvano M. A. 2006. Micromethods for the characterization of lipid A-core and O-antigen lipopolysaccharide. Methods Mol. Biol. 347:237–252 [DOI] [PubMed] [Google Scholar]
- 20. Marolda C. L., et al. 2010. Membrane topology and identification of critical amino acid residues in the Wzx O-antigen translocase from Escherichia coli O157:H4. J. Bacteriol. 192:6160–6171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Marolda C. L., Tatar L. D., Alaimo C., Aebi M., Valvano M. A. 2006. Interplay of the Wzx translocase and the corresponding polymerase and chain length regulator proteins in the translocation and periplasmic assembly of lipopolysaccharide O antigen. J. Bacteriol. 188:5124–5135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Marolda C. L., Vicarioli J., Valvano M. A. 2004. Wzx proteins involved in biosynthesis of O antigen function in association with the first sugar of the O-specific lipopolysaccharide subunit. Microbiology 150:4095–4105 [DOI] [PubMed] [Google Scholar]
- 23. Morona R., Purins L., Tocilj A., Matte A., Cygler M. 2009. Sequence-structure relationships in polysaccharide co-polymerase (PCP) proteins. Trends Biochem. Sci. 34:78–84 [DOI] [PubMed] [Google Scholar]
- 24. Morona R., Van Den Bosch L., Daniels C. 2000. Evaluation of Wzz/MPA1/MPA2 proteins based on the presence of coiled-coil regions. Microbiology 146:1–4 [DOI] [PubMed] [Google Scholar]
- 25. Murray G. L., Attridge S. R., Morona R. 2003. Regulation of Salmonella Typhimurium lipopolysaccharide O antigen chain length is required for virulence; identification of FepE as a second Wzz. Mol. Microbiol. 47:1395–1406 [DOI] [PubMed] [Google Scholar]
- 26. Murray G. L., Attridge S. R., Morona R. 2006. Altering the length of the lipopolysaccharide O antigen has an impact on the interaction of Salmonella enterica serovar Typhimurium with macrophages and complement. J. Bacteriol. 188:2735–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Paixao T. A., et al. 2009. Establishment of systemic Brucella melitensis infection through the digestive tract requires urease, the type IV secretion system, and lipopolysaccharide O antigen. Infect. Immun. 77:4197–4208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Papadopoulos M., Morona R. 2010. Mutagenesis and chemical cross-linking suggest that Wzz dimer stability and oligomerization affect lipopolysaccharide O-antigen modal chain length control. J. Bacteriol. 192:3385–3393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Purins L., Van Den Bosch L., Richardson V., Morona R. 2008. Coiled-coil regions play a role in the function of the Shigella flexneri O-antigen chain length regulator WzzpHS2. Microbiology 154:1104–1116 [DOI] [PubMed] [Google Scholar]
- 30. Raetz C. R., Whitfield C. 2002. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71:635–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ruiz N., Kahne D., Silhavy T. J. 2009. Transport of lipopolysaccharide across the cell envelope: the long road of discovery. Nat. Rev. Microbiol. 7:677–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Saldias M. S., Ortega X., Valvano M. A. 2009. Burkholderia cenocepacia O antigen lipopolysaccharide prevents phagocytosis by macrophages and adhesion to epithelial cells. J. Med. Microbiol. 58:1542–1548 [DOI] [PubMed] [Google Scholar]
- 33. Sperandeo P., Dehò G., Polissi A. 2009. The lipopolysaccharide transport system of Gram-negative bacteria. Biochim. Biophys. Acta 1791:594–602 [DOI] [PubMed] [Google Scholar]
- 34. Tang K. H., Guo H., Yi W., Tsai M. D., Wang P. G. 2007. Investigation of the conformational states of Wzz and the Wzz. O-antigen complex under near-physiological conditions. Biochemistry 46:11744–11752 [DOI] [PubMed] [Google Scholar]
- 35. Tocilj A., et al. 2008. Bacterial polysaccharide co-polymerases share a common framework for control of polymer length. Nat. Struct. Mol. Biol. 15:130–138 [DOI] [PubMed] [Google Scholar]
- 36. Valvano M. A. 2003. Export of O-specific lipopolysaccharide. Front. Biosci. 8:s452–s471 [DOI] [PubMed] [Google Scholar]
- 37. Vines E. D., Marolda C. L., Balachandran A., Valvano M. A. 2005. Defective O-antigen polymerization in tolA and pal mutants of Escherichia coli in response to extracytoplasmic stress. J. Bacteriol. 187:3359–3368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. West N. P., et al. 2005. Optimization of virulence functions through glucosylation of Shigella LPS. Science 307:1313–1317 [DOI] [PubMed] [Google Scholar]
- 39. Woodward R., et al. 2010. In vitro bacterial polysaccharide biosynthesis: defining the functions of Wzy and Wzz. Nat. Chem. Biol. 6:418–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.