Abstract
Mechanisms underlying susceptibility to anthrax infection are unknown. Using a phylogenetically diverse panel of inbred mice and spores of Bacillus anthracis Ames, we investigated host susceptibility to pulmonary anthrax. Susceptibility profiles for survival time and organ pathogen load differed across strains, indicating distinct genetic controls. Tissue infection kinetics analysis showed greater systemic dissemination in susceptible DBA/2J (D) mice but a higher terminal bacterial load in resistant BALB/cJ (C) mice. Interestingly, the most resistant strains, C and C57BL/6J (B), demonstrated a sex bias for susceptibility. For example, BALB/cJ females had a significantly higher survival time and required 4-fold more spores for 100% mortality compared to BALB/cJ males. To identify genetic regions associated with differential susceptibility, survival time and extent of organ infection were assessed using mice derived from two susceptibility models: (i) BXD advanced recombinant inbred strains and (ii) F2 offspring generated from polar responding C and D strains. Genome-wide analysis of BXD strain survival identified linkage on chromosomes 5, 6, 9, 11, and 14. Quantitative trait locus (QTL) analysis of the C×DF2 population revealed a significant QTL (designated Rpai1 for resistance to pulmonary anthrax infection, locus 1) for survival time on chromosome 17 and also identified a chromosome 11 locus for lung pathogen burden. The striking difference between genome-wide linkage profiles for these two mouse models of anthrax susceptibility supports our hypothesis that these are multigenic traits. Our data provide the first evidence for a differential sex response to anthrax resistance and further highlight the unlikelihood of a single common genetic contribution for this response across strains.
INTRODUCTION
Anthrax is one of the most ancient and lethal human diseases caused by the virulent (toxigenic and encapsulated) strains of Bacillus anthracis. In humans, anthrax may take three forms depending upon the route of infection, namely, pulmonary (inhalational), cutaneous, and gastrointestinal, with pulmonary being the most lethal and difficult to treat. While the naturally acquired pulmonary anthrax is relatively rare (occurring only ∼5% as often as cutaneous anthrax), this form has a high potential for misuse as a weapon of bioterrorism, considering that environmental dissemination is the most expected mode of release of the agent in mass attacks (26). Pulmonary anthrax often proves fatal, with mortality approaching 100% if not treated early. Anthrax spores have long been considered a potential agent of biological warfare (25, 50, 58). Prior to the 11 cases of pulmonary anthrax that occurred via the U.S. mail delivery system in 2001 (29), fatal cases of pulmonary anthrax in the United States and other countries have been associated with occupational and accidental exposures (37, 60). But the bioterrorism attack in 2001 exposed a new cause for serious concern for future and more widespread attacks on military and civilian populations.
The etiological agent for anthrax, Bacillus anthracis, is a Gram-positive bacterial pathogen that belongs to category A in the CDC list of select agents. The size of its spores (1 to 2 μm) makes them optimal for deposition in the alveolar spaces following inhalation. To be virulent, B. anthracis requires two major components: the capsule and the anthrax toxin. These components are encoded by the plasmids pXO2 and pXO1, respectively, in virulent strains such as Ames. Strains such as Sterne, which lack the capsule (pXO2), are attenuated for human infection and are therefore used for vaccine applications. However, these attenuated strains can cause serious infections in mice. While there is increasing information on the pathogen virulence factors in B. anthracis (3, 7, 12, 34, 71), the mechanisms of host defense and pathogenesis are poorly understood for anthrax.
Although anthrax primarily affects herbivores, virtually all mammals are susceptible to various degrees. Anthrax susceptibility depends on host species (31), strain (69), and the route of infection (35, 46). Interindividual differences in disease onset, survival, and treatment response have been observed in human cases of pulmonary anthrax. In the 2001 anthrax letter attacks, only 5 of the 11 individuals infected by the aerosolized spores succumbed to the disease. The causes underlying these differences in susceptibility are not known but conceivably involve differences in host factors. It is intriguing that some animals (e.g., rats) are very sensitive to B. anthracis toxins yet are difficult to infect by spores, while other animals (e.g., guinea pigs) are more resistant to the toxins but can be killed with relatively few spores (31, 32, 63). Previous investigations on the genetic susceptibility to B. anthracis have mainly focused on the role of anthrax lethal toxin (LT) exposure in vitro (17, 52, 56, 66, 67) and in vivo (36, 40, 45) in laboratory rodent strains. The toxin-based studies indicated that inbred mice vary in their sensitivity to anthrax LT and that host genetic factors underlie the differences in LT susceptibility (4, 36, 40). More importantly, previous observations have revealed a reciprocal trend for murine susceptibility to purified anthrax LT-induced pathology versus spore-induced infection (36, 69). Since spore-induced anthrax better represents a real world infection scenario for bioterrorism, there is an immediate need to fully understand the basis of host defense and susceptibility to B. anthracis spores. Knowledge on host susceptibility to lethal infection during pulmonary anthrax can be a key initiator in the development of new countermeasures.
Lethal anthrax infection caused by B. anthracis spores is a multistep process expected to require spore germination, pathogen multiplication, and systemic dissemination, culminating in death (15, 22, 23, 46). Mechanisms of host resistance could interfere with any of these steps. Hence, there is a need to understand the genetic loci conferring host resistance/susceptibility to anthrax infection. In this context, there is also a continuing need to identify and characterize more appropriate animal models of resistance or susceptibility for dissecting the role of these host factors. Initial investigations in this direction have examined questions of pathogenesis and host resistance in inbred mice by using spores from avirulent anthrax strains, such as Sterne or 34F2 (55, 68–70). While inbred mice offer a valuable tool to understand host susceptibility and response to anthrax infection, little has been done in mice in experiments with the fully virulent strains of B. anthracis. Limited initial studies of mice either used fewer numbers of randomly selected inbred strains (35) or were based on nonpulmonary routes (69) of infection, and they did not identify host genomic regions governing the underlying susceptibility. Whether males and females differ in pulmonary anthrax susceptibility is also not known.
The overall goals of this study were to investigate the role of host genetic background and sex in susceptibility to pulmonary anthrax infection by using the Ames strain and to identify putative genomic loci underlying differential susceptibility by quantitative trait locus (QTL) analysis. The QTL approach, based on associating genetic variation with phenotypic variation, has proven useful for identification of genomic loci involved in many complex diseases, including infectious diseases (16, 54, 57). To begin to examine the role of host genetics, we screened a phylogenetically representative panel of 14 inbred mice strains and F1 progeny from select sensitive-resistant strain breeder pairs. Initial studies established mouse models of host susceptibility and also revealed evidence of a parent-of-origin effect and a differential sex response to pulmonary anthrax. To identify genetic loci responsible for B. anthracis susceptibility, two strategies were used. First, genome-wide single-nucleotide polymorphism (SNP) analysis was performed on a panel of BXD advanced recombinant inbred (ARI) mice. The BXD ARI lines were derived from the female C57BL/6J and male DBA/2J parent strains (48), which represent a progenitor strain pair identified in this study for modeling differential genetic susceptibility. These homozygous, inbred, and genetically distinct ARI lines approximate the genetic diversity in human populations and have been used successfully to map QTLs associated with various clinical conditions and diseases (2, 8, 42, 47). Second, we performed QTL analysis on an F2 population generated from the sensitive DBA/2J and resistant BALB/cJ strains. This work represents the first report of sex bias and a parent-of-origin effect in a murine model of spore-induced pulmonary anthrax infection. These initial genome-wide analyses using two separate mouse models of differential susceptibility, coupled with spore-induced anthrax infection, identified multiple genomic loci associated with pulmonary anthrax outcome. These studies lay the groundwork for follow-on studies to further characterize these susceptibility QTLs to identify the relevant genes.
MATERIALS AND METHODS
Bacterial strain and spore inoculum preparation.
Bacillus anthracis Ames, a fully virulent (toxigenic and encapsulated) strain, was obtained from the Los Alamos National Laboratory. The organism was cultivated, handled, and stored in the biosafety level 3 (BSL-3) Laboratory of the University of Cincinnati College of Medicine, using the protocol and standard operating procedures approved by the university's Institutional Biosafety Committee. Spore inoculum of B. anthracis Ames was prepared using nutrient sporulation medium (13). Briefly, an isolated single colony of B. anthracis Ames was streaked onto nutrient sporulation agar (containing, per liter, 3 g of yeast extract, 3 g of tryptone, 2 g of Bacto agar, 23 g of Lab Lemco agar, and 1 ml of 1% MnCl2), and the plates were incubated at 37°C. Spore formation was monitored microscopically. When the sporulation reached >99% (approximately the fourth day of incubation), the bacterial growth was harvested in 0.01 M phosphate-buffered saline (PBS). The suspension was washed 3 times with PBS by centrifugation (12,000 rpm for 15 min at 4°C) followed by heating at 65°C for 1 h to inactivate the residual vegetative cells. After heating, the spore suspension was washed and resuspended in a suitable suspension medium. An aliquot of the spore suspension was serially diluted and plated out on Trypticase soy agar (TSA) to check for viable spore count. The remaining monodispersed spore stock was frozen in 1-ml aliquots at −80°C in cryovials. On the eve of each infection experiment, a vial of the frozen spore stock was thawed, resuspended in PBS, and quantified to verify the actual viable count in the stock at that point of storage. Before use in mouse inoculations the next day, the suspension medium in the frozen stock (1 ml) was replaced with an equal volume of sterile PBS by centrifugation and resuspension. On the basis of the viable count in the frozen main stock, a working stock of the spore inoculum was prepared in PBS to obtain the desired number of spores per ml. The fresh working inoculum (prequantified as above) was directly used to inoculate mice, as described below. The same batch of inoculum was used for a given treatment group. To generate a stable and consistent spore inoculum for extended use, the effects of two suspension media, PBS versus Trypticase soy broth with 15% glycerol (TSB-G), on the viability of Ames spores during frozen storage (−80°C) were investigated. Due to the significant decrease in spore viability in PBS, occurring as early as 24 h after freezing (data not shown), all inocula were stored frozen (−80°C) in TSB-G broth and resuspended in PBS right before use.
Animals.
Adult healthy pathogen-free mice of both sexes (6 to 8 weeks of age) were purchased from the Jackson Laboratory (Bar Harbor, ME). Fourteen inbred mouse strains (129S1/SvImJ, A/J, BALB/cJ [abbreviated as C in crosses], BPL/1J, C3H/HeJ, C57BL/6J [abbreviated as B or B6 in crosses], C57BL/10J, CAST/EiJ, DBA/1J, DBA/2J [abbreviated as D or D2 in crosses], FVB/NJ, NOD/ShiLtJ, NON/ShiLtJ, and SPRET/EiJ) were used, which were representative of the distinct mouse phylogenetic groups recently described (49). The inbred mice were challenged with 500 spores/mouse of B. anthracis Ames.
B6D2F1/J mice (6 weeks old), derived from crossing resistant strain C57BL/6J females and sensitive strain DBA/2J males, were purchased from the Jackson Laboratory (Bar Harbor, ME). Reciprocal F1 crosses from BALB/cJ and DBA/2J strains (i.e., CD2F1 and D2CF1; female strain listed first) were generated by in-house breeding at the Laboratory Animal Medicine Services facility of the University of Cincinnati. Age-matched F1 mice (6 weeks each) and their respective parents were transferred to the BSL-3 facility and inoculated with a lethal dose of B. anthracis Ames spores (500 spores/mouse for B6D2F1 progeny and 2,000 spores/mouse for CD2F1 and D2CF1 progeny).
BXD advanced recombinant inbred (ARI) strains of mice were purchased from the Jackson Laboratory (Bar Harbor, ME) for genome-wide SNP analysis to identify QTLs. The BXD ARI strains were originally made by repeated intercrossing for at least 20 generations of F9 to F14 progeny from the C57BL/6 and DBA/2 parental strains (48). The following 16 genotypically distinct ARI strains were obtained for genetic studies: BXD44, BXD48, BXD50, BXD55, BXD62, BXD66, BXD68, BXD69, BXD86, BXD87, BXD89, BXD90, BXD96, BXD97, BXD98, and BXD100. These 16 BXD ARI strains, along with the C57BL/6J and DBA/2J progenitor strains, were screened for differences in susceptibility to infection by B. anthracis Ames spores. Five mice per BXD ARI strain were challenged with a lethal dose of 500 spores/mouse of B. anthracis Ames. For traditional QTL analysis, a segregant population of 258 F2 mice (146 females, 112 males) was generated in-house from resistant BALB/cJ and sensitive DBA/2J strains. All F2 mice came from intercrossing CD2F1 mice (BALB/cJ females crossed with DBA/2J males).
All mice were housed in autoclaved, individually ventilated static microisolator cages (not more than 4 mice per cage) in the specific-pathogen-free animal facility at the University of Cincinnati. Animals were fed standard pellet diet and autoclaved water ad libitum. The animals were allowed to acclimate for 7 days in the BSL-2 and BSL-3 facility prior to their use in experimental studies. All animal procedures were performed according to the Institutional Animal Care and Use Committee (IACUC)-approved protocols at the University of Cincinnati.
Mouse inoculations.
All inoculations and housing of mice postinfection were carried out in the animal BSL-3 containment area at the University of Cincinnati. For each experiment, mice were age and sex matched, unless noted otherwise. Mice were anesthetized by intraperitoneal injection of Avertin (0.4 mg/g of body weight). An incision was made on the ventral region of the neck, and the trachea was carefully exposed. To deliver the desired spore dose to the lungs, a 50-μl volume of the appropriate working stock was introduced intratracheally followed by 50 μl of air, using a bent needle on a BD ultrafine insulin syringe (Becton Dickinson, San Jose, CA) and placing the animal on an inclined board (∼30° angle). The incision was then closed using 3M Vetbond tissue adhesive.
Determination of survival time and organ bacterial load.
Survival responses of inbred strains (male versus female), F1 progeny, BXD ARI lines, and F2 progeny to pulmonary infection with Ames spores were compared using multiple animals for each treatment group and a constant spore dose, as indicated for the respective experiments. Inoculated mice of each treatment group were frequently monitored for survival to determine the time to death (TTD). The TTD value for a given mouse strain was calculated by averaging the number of hours required for death for the individual animals within that group (i.e., mean survival time, in hours). The BALB/cJ female group was an exception; in this case, the mean TTD was calculated based on the data from the nonsurviving mice only. The median survival times for each strain were also calculated. Lungs, livers, and spleens of the animals who succumbed were immediately harvested and analyzed for total bacterial load, as indicated by CFU. The tissues were homogenized (each in 1 ml of TSB-G) and plated on TSA at 37°C for CFU determination.
Dose-response analysis for BALB/cJ mice.
Dose-response analysis was performed to select the optimal lethal dose of B. anthracis Ames spores for the resistant inbred strain, BALB/cJ (females). Various doses (500 to 2,500 spores/mouse) were used to challenge groups of 10 mice/dose, and the TTD was recorded for each animal. The survival percentage (number of animals surviving divided by the total number of animals infected) was plotted against each spore dose to develop survival curves for the individual doses. The minimum dose that resulted in 100% mortality in the treatment group was confirmed by probit analysis and taken as the optimal lethal dose (LD100) for BALB/cJ mice and their crosses.
Infection kinetics.
A time course experiment was performed to investigate comparative infection kinetics in the most resistant inbred strain, BALB/cJ, and the most sensitive inbred strain, DBA/2J. For either strain, female mice were inoculated with a lethal dose of 2,000 spores/mouse and sampled at increasing time intervals (<1 h, 12 h, 24 h, 48 h, 72 h, and at death). At each time point, mice (n = 3 for each strain) were euthanized using an overdose of Avertin, and tissues (lung, spleen, and liver) were harvested. Tissue homogenates were subjected to CFU analysis as described above. Methods used to determine organ infection loads were the same as those described for screening the 14 inbred strains in the preceding sections.
QTL analysis.
QTL analysis, a statistical assessment of the likelihood of genetic linkage, was performed in two separate cohorts (BXD ARI strains and an F2 population from BALB/CJ and DBA/2J progenitors) to associate phenotype with genotype. The BXD ARI strains were exposed to a pulmonary load of 500 anthrax spores/mouse and screened to identify genetic regions linked with the disparate survival times seen for the C57BL/6J and DBA/2J progenitor inbred strains. Using the online analysis program WebQTL (http://www.genenetwork.org) (65) as the mapping engine and the genome-wide SNP source, survival time and organ CFU data from 16 BXD ARI strains were assessed for putative QTLs. QTL maps were generated using interval regression, and the log of the odds ratio (LOD) score was used in linkage analysis to assess significance. Genome-wide LOD threshold levels for significant and suggestive linkage were determined using 2,000 permutation tests (9). Peak markers from regression analysis were fixed one at a time in composite interval mapping (CIM) to identify potential additional and/or weaker QTLs after variance explained by the first marker was removed from the analysis (30). Bootstrap analysis, which involved 2,000 iterations of random data set reshuffling (with replacement) and remapping of each result, was also performed in WebQTL to assess reliability of QTL peaks and to determine approximate confidence limits.
Genotyping of F2 populations.
DNA was isolated from a tail clip of each F2 and progenitor strain mouse by using either the Wizard genomic DNA isolation kit (Promega, Madison, WI) or the high-throughput DNA extraction service of the Cincinnati Children's Hospital DNA Core Facility. DNA was quantitated using the A260/A280 ratio and diluted to ∼20 ng/μl for PCR genotyping. Primer pairs, chosen based on known polymorphisms between the BALB/cJ and DBA/2J progenitor strains, were purchased from IDT (Coralville, IA). PCR was performed in 15 μl in 96-well plates (Applied Biosystems, Foster City, CA), as described previously (51).
QTL mapping and linkage analyses.
All phenotype data (i.e., survival times or organ CFU) and genotype data were analyzed for main effect QTLs by using the Scan One function of the freely available R/QTL computer package (5). A total of 71 polymorphic markers were genotyped for all F2 mice. Threshold values for significant and suggestive linkages were established using 10,000 permutations of the data set. To identify potential gene-gene interactions, including additivity and epistasis, genome-wide scans for all marker pairs were carried out on the total F2 population using the Scan Two function in R/QTL. Empirical threshold significance values for pairwise interactions were determined using 100 permutations of the data set.
Statistical analyses.
Weighted least-squares analysis (WLS) was performed to analyze the TTD data of 14 inbred strains of 167/169 male and female mice (approximately 99% mortality). Preliminary investigations showed that the distribution of TTDs approximated normality, based on the Shapiro Wilk test. Within-strain variances were found to be heterogeneous, based on Levene's test. The WLS methodology, combined with a normality assumption, has the optimum power to detect differences between means of strains and susceptibility groups, compared to nonparametric methods. A censored distribution of TTD was not analyzed, since the 1% censoring rate (2/169) was believed to be sufficiently low to have a negligible effect on results of hypothesis testing. TTD strain means were sorted in increasing order of magnitude, and two groups (low [most susceptible] and high [resistant]) were identified. The remaining strains comprised a third group whose means were between the endpoints of the two groups. Our initial regression model was a multiway analysis of variance (ANOVA) of TTD that included susceptibility group and strain within susceptibility group as categorical independent variables. Means and standard errors of TTDs by strain were calculated. The linearity of the 14 ordered strain means was tested by linear orthogonal polynomial coefficient analysis within the ANOVA model. In addition, t tests were performed to test differences between all pairs of strain means. Following a significant difference between at least one pair of means, as determined by a global chi-square test evaluating the equality of all strain means (P < 0.001), Fisher's protection level was used to adjust for multiple comparisons of differences between strain means. The same methodology (WLS) was used to analyze organ CFU values, after a loge transformation was applied to approximate normality. Geometric means were calculated to estimate the medians of each strain and of sexes within each strain. Sex-specific analyses of TTDs and loge CFU determinations were also performed using WLS, with ANOVA models to compare strain means by sex. Comparisons of between-strain to within-strain variability were obtained by calculating the ratios of the variance between strains to the residual variance, or the average variance within strains. These were calculated for all TTD data and for each sex.
Probit regression was performed to estimate the LD50 and the minimum effective dose approximating 100% mortality for 5 groups of female BALB/cJ mice (10 mice/group) receiving various spore doses (500, 750, 1,000, 2,000, or 2,500 spores/mouse). Equality of the distribution of TTD values among doses was tested by the Wilcoxon log rank statistic in a survival curve analysis stratified by dose. In addition, mean values of TTDs for 16 strains of ARI-BXD mice were analyzed by WLS, assuming an ANOVA model. The linearity of ordered TTD means was tested by linear orthogonal polynomial coefficients. The ratio of between-strain to within-strain variability was calculated from ANOVA by dividing the model mean square by the residual mean square.
Interstrain and/or sex differences within strains for survival time were also evaluated for reciprocal CD2F1 and D2CF1 progeny and B6D2F1 progeny, and P values were adjusted for multiple comparisons. Means and standard errors for each outcome were used in the bar graphs to facilitate interpretation of results. Statistical analyses were performed using SAS for Windows, version 9.2 (SAS Institute, Cary, NC).
RESULTS
Male and female mice for 14 inbred mouse strains were screened using a lethal dose of 500 spores/mouse of B. anthracis Ames strain. The dose was empirically selected, guided in part by available published information (35), and verified in an initial trial challenge experiment designed to test its potential to cause 100% lethality in selected inbred strains (data not shown).
Survival differences among inbred mouse strains.
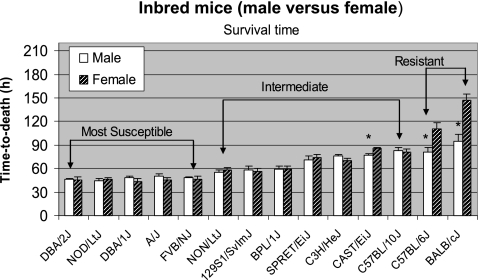
Following exposure to the same lethal dose of B. anthracis spores (500 spores/mouse), significant differences in susceptibility (measured as TTD) were observed among the 14 inbred strains. Using the average TTD of both sexes, three susceptibility groups were identified (Fig. 1): the most susceptible group, with a TTD of <60 h (DBA/2J ∼ NOD/ShiLtJ ∼ DBA/1J ∼ A/J ∼ FVB/NJ); the intermediate susceptible group, with a TTD of 60 to 90 h (NON/ShiLtJ ∼ 129S1/SvImJ ∼ BPL/1J < SPRET/EiJ ∼ C3H/HeJ < CAST/EiJ ∼ C57BL/10J); the least susceptible (or most resistant) group, with a TTD of >90 h (C57BL/6J < BALB/cJ). The susceptibility grouping based on the empirical cutoff limits was verified using pairwise statistical comparisons of the means. TTD means and standard errors of the strains among susceptibility groups obtained from analysis of variance are shown in Table S1A of the supplemental material. The statistical evaluation of paired differences between TTD means of the 14 strains supported the linearly increasing levels of the means and susceptibility group clustering (see Table S1B in the supplemental material). Each susceptibility group differed significantly (P < 0.01) from the other groups. The intermediate group included three subgroups, which are separated by the < signs in the above susceptibility order. Mean TTD values of the 14 strains were significantly different (P < 0.001; degrees of freedom, 13). The ratio of between-strain to within-strain variability for sex-specific and combined sexes TTD was 26.9, implying a greater interstrain than intrastrain variability.
Fig. 1.
Comparative susceptibility of inbred mouse strains of both sexes to pulmonary anthrax infection with spores of B. anthracis Ames. Animals for each inbred strain (n = 5 to 7 mice of each sex; 6 to 8 weeks of age) were intratracheally challenged with 500 spores (per mouse) of B. anthracis Ames and monitored for survival time, as described in Materials and Methods. Survival time was measured as TTD. The TTD values (in hours) were plotted as means and standard errors of the means. All strains showed 100% lethality except BALB/cJ (females); arithmetic means and standard errors of the plotted TTD were therefore calculated based on the nonsurviving mice. The arithmetic mean (146.9 h) and harmonic mean (145.3 h) for BALB/c (females), calculated based on the available TTD values for the mice who succumbed, were comparable. Three susceptibility groups (most sensitive, intermediate, and most resistant) were identified based on survival time, as shown using inverted brackets. Mean values of 14 strains were significantly different for TTD (P < 0.001; degrees of freedom, 13). Significant differences (P < 0.01) in the TTD values between sexes are indicated by an asterisk.
Sex bias in the most resistant group of inbred strains.
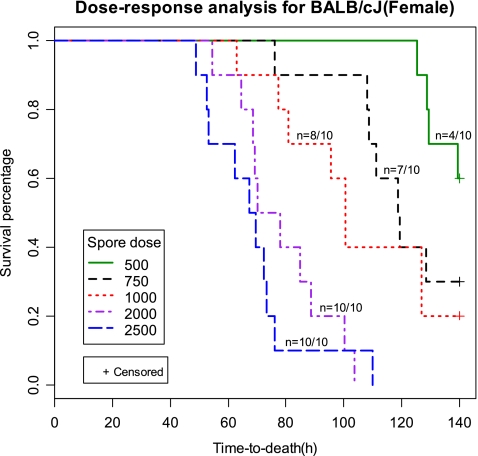
The resistant (least susceptible) group of strains (BALB/cJ and C57BL/6J) showed a significant difference (P < 0.01) in survival times between males and females (Fig. 1). A significant sex difference (P < 0.01) was also observed in CAST/EiJ mice, a member of the intermediate group. The BALB/cJ strain showed a greater sex difference (∼50 h higher mean TTD for female than male mice) compared to the C57BL/6J strain (∼30 h higher mean TTD for females). Furthermore, female BALB/cJ mice groups showed partial mortality (40%) compared to 100% mortality observed for the male counterparts, following the same dose of 500 spores/mouse. Hence, to determine the minimum effective lethal dose causing 100% mortality in female BALB/cJ mice, various doses (500, 750, 1,000, 2,000, and 2,500 spores/mouse) were compared using 10 animals per dose. Dose-response curves showed a decreasing survival percentage with an increasing spore dose. A minimum dose of 2,000 spores per mouse was required (Fig. 2) to achieve 100% mortality (mean TTD of ∼80 h) in female BALB/cJ mice. In contrast, the partial mortality (4 of 10 mice) observed for the 500-spores/mouse dose was characterized by a mean TTD of ∼130 h for the animals who succumbed. Probit regression analysis revealed the following values for the lethal doses: LD50, 584 spores; LD100, 1,978 spores. Based on these observations, a test dose of 2,000 spores was used for further studies on BALB/cJ (female) mice and F1 or F2 progeny derived from using BALB/cJ as a progenitor strain.
Fig. 2.
Dose-response analysis for female BALB/cJ mice for selection of the lethal dose (LD100) of B. anthracis Ames spores. Groups of 10 female mice were intratracheally inoculated with various doses of B. anthracis spores (500 to 2,500 spores/mouse) and monitored for TTD, as described in Materials and Methods. TTD values (in hours) were plotted against the survival percentage for each dose. The mortality ratio (number of dead animals divided by the total number of animals infected) achieved with each dose is represented as n on the respective survival curves. +, censored data (i.e., not all mice died).
Organ colonization and infection kinetics differences among inbred strains.
At the time of death, a marked variation in CFU values was noted in the lungs among the 14 inbred strains challenged with the same lethal dose (500 spores). Mean values of the 14 strains were significantly different (P < 0.001; degrees of freedom, 13). The ratio of between-strain to within-strain variability for sex-specific and total log CFU was 31.5, implying a greater interstrain than intrastrain variability. Based on the overall means, the following ascending order of susceptibility to pathogen accumulation in the lung was obtained: BPL/1J < 129S1/SvImJ < C57BL/6J < NOD/ShiLtJ < FVB/NJ < DBA/1J < NON/ShiLtJ < BALB/cJ < A/J < DBA/2J < SPRET/EiJ < CAST/EiJ < C3H/HeJ < C57BL/10J (data not shown). However, no clear correlation was observed between the susceptibility orders for lung CFU and survival time among these 14 inbred strains.
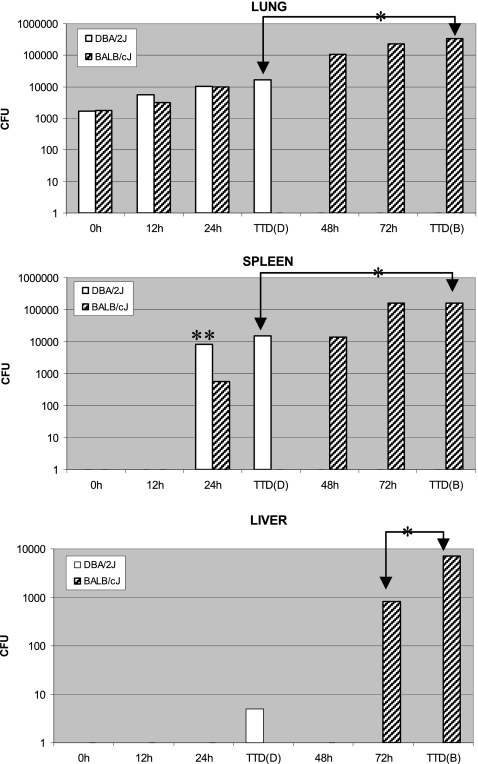
To better understand the differences in organ colonization, we compared predeath kinetics of pathogen accumulation and dissemination in the most resistant strain (BALB/cJ) and most sensitive strain (DBA/2J) (Fig. 3) at the site of spore deposition (lung) and in systemic locations (spleen and liver). Female mice were used for both strains. While the lung bacterial load, measured up to 24 h postinfection, in both strains was comparable (P > 0.05), the final bacterial load at death was ∼20-fold higher (P < 0.01) in resistant BALB/cJ mice. In spleen, live bacilli were first detectable at the same time point (24 h) postinfection for both strains, with the sensitive DBA/2J strain showing significantly higher CFU (P < 0.05). However, the final bacterial load in the spleen at death was significantly (P < 0.01) higher (∼10-fold) in BALB/cJ mice. No CFU were detected in the liver until at the time of death in DBA/2J mice (TTD, ∼30 h), unlike BALB/cJ mice, which did not show bacilli until shortly before death (72 h postinfection) and at the time of death (TTD, ∼80 h). Significantly different (P = 0.01) bacterial loads at 72 h and TTDs implied rapid dissemination and/or multiplication and/or poorer clearance of bacilli at the time of death as possible mechanisms for the BALB/cJ strain. The rate of pathogen dissemination to spleen and liver was greater in the DBA/2J mice, leading to an early and higher systemic bacterial load in this strain at 24 to 30 h postinfection, coinciding with its near-death/death time point. Nevertheless, the absolute systemic load of the pathogen at the time of death was significantly lower (P ≤ 0.01) in DBA/2J mice (∼30 h) than in BALB/cJ mice (∼80 h), implying the former's greater susceptibility to bacterial accumulation during infection as a possible mechanism.
Fig. 3.
Kinetic changes in pathogen dissemination and accumulation during pulmonary infection of the selected resistant (BALB/cJ) and susceptible (DBA/2J) strains of mice with B. anthracis Ames spores. Female mice (6 to 8 weeks of age) for the two inbred strains were intratracheally challenged with 2,000 spores/mouse of B. anthracis Ames and periodically monitored for organ pathogen burden until death, as described in Materials and Methods. Homogenized lung, spleen, and liver from each animal harvested at each time point (n = 3 mice) were subjected to microbiological analyses by agar plating in triplicate. Pathogen load in the infected tissue was expressed as CFU. Values are plotted on a log scale based on the geometric mean and standard error of the mean (the error bars have been plotted but are too small to be visible on the log scale). Abbreviations: TTD(B), time to death for BALB/cJ (∼80 h); TTD(D), time to death for DBA/2J (∼30 h). Time zero corresponds to data measured within the first hour (lag) period, when multiple inoculations and surgeries were performed. Some of the significant differences in organ CFU values between the two strains are indicated by a single (P ≤ 0.01) or double (P < 0.05) asterisk. Other within-strain and between-strain organ CFU differences across different time points were significant at the 1% or 5% level; however, the following were not significant (P > 0.05): lung CFU, DBA/2J (24 h versus TTD), BALB/cJ (72 h versus TTD), and BALB/cJ versus DBA/2J for 0-h, 12-h, and 24-h time points; spleen CFU, DBA/2J (24 h versus TTD), BALB/cJ (72 h versus TTD), BALB/cJ (48 h) versus DBA/2J (24 h), and BALB/cJ (48 h) versus DBA/2J (TTD).
Susceptibility inheritance in the F1 progeny.
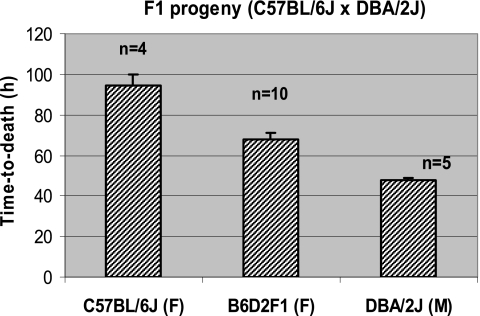
Matings using two strain-pair combinations of the most resistant and most susceptible strains, BALB/cJ × DBA/2J and C57BL/6J × DBA/2J, were performed to investigate the heritability of susceptibility in the F1 generation. F1 hybrid mice were compared with their respective parents for TTD using the same lethal spore dose for the parents and progeny (500 spores/mouse for C57BL/6J crosses and 2,000 spores/mouse for BALB/cJ crosses). The B6D2F1 progeny showed an intermediate TTD (Fig. 4) compared to the C57BL/6J and DBA/2J parental strains. The TTD differences between the B6D2F1 progeny and the parents were statistically significant (P < 0.01), as was the difference between the two parental strains (P < 0.01). The CD2F1 and D2CF1 reciprocal crosses also showed TTD values that were intermediate (Fig. 5A) to the values for their parents (BALB/cJ and DBA/2J), and the differences were significant (P < 0.01). Specifically, the TTD value for the CD2F1 progeny fell between the two parental values and was significantly (P < 0.01) lower (∼53 h) compared to the TTD for the reciprocal cross D2CF1 (∼67 h). The difference between reciprocal F1 mice suggested that one or more genes related to the overall trait may be imprinted. More interesting, the survival times of offspring from these reciprocal F1 crosses correlated more closely with the survival time for the strain of the male breeder, suggesting a possible paternal inheritance pattern. In fact, survival times for D2CF1 males and females (from BALB/cJ sires) did not differ from the mean survival time for BALB/cJ females (P > 0.22 for both). Besides a significant difference (P < 0.01) from its reciprocal F1 group, survival times for CD2F1 males and females differed significantly (P < 0.01) from both parental strains (Fig. 5B) but were more similar to the CD2F1 sire strain (DBA/2J). Survival times of female versus male CD2F1 progeny showed a trend toward a sex bias (P < 0.15), whereas no such trend was found in the reciprocal D2CF1 cross (Fig. 5B).
Fig. 4.
Comparative susceptibilities of the B6D2F1 progeny and the resistant female C57BL/6J (B6) and susceptible male DBA/2J (D2) parental strains. A dose of 500 spores/mouse of B. anthracis Ames was used for all inoculations of age-matched (6 to 8 weeks old) F1 progeny and parental strains. Survival time, expressed as TTD (in hours), is plotted as the mean and standard error of the mean. Abbreviations: M, male; F, female; n, number of animals tested for each strain.
Fig. 5.
Comparative susceptibilities of the reciprocal F1 progeny and the resistant BALB/cJ (C) and susceptible DBA/2J (D2) parental strains. (A) Comparison of the reciprocal F1 crosses D2CF1 and CD2F1 (based on mean values of both sexes) and the parental strains. (B) Comparison of males and females of the crosses (CD2F1 and D2CF1) and the parental strains. Results are consistent with a possible paternal mode of inheritance, with reciprocal F1 offspring having survival times most similar to those for the inbred strain of the sire. A lethal dose of B. anthracis spores (2,000 spores/mouse) optimized for BALB/cJ was used for all inoculations. Survival time, expressed as TTD (in hours) is plotted as the mean and standard error of the mean for the number of animals in a given group. Abbreviations: M, male; F, female; M+F, both sexes averaged; n, total number of animals tested for each mouse strain.
Susceptibility assessments in the BXD ARI strains.
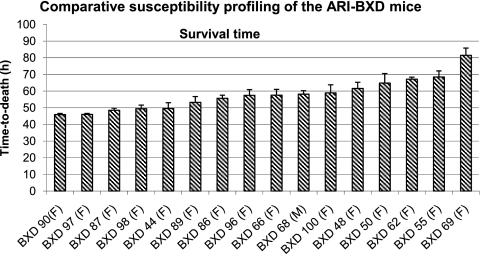
To identify genetic loci responsible for host susceptibility to B. anthracis, we investigated BXD ARI strains, originally generated from female C57BL/6J and male DB/2J mice (48), a parental pair that represents a resistant-susceptible strain pair identified in the current study. Comparison of the individual BXD ARI strains showed an interstrain variability in survival time (Fig. 6). The range of the TTD values for the BXD strains approximated the survival time range for the two progenitor strains (∼40 to ∼80 h) and showed the following descending order of susceptibility: BXD 90 > BXD 97 > BXD 87 > BXD 98 > BXD 44 > BXD 89 > BXD 86 > BXD 96 > BXD 66 > BXD 68 > BXD 100 > BXD 48 > BXD 50 > BXD 62 > BXD 55 > BXD 69. The ratio of between-strain to within-strain variability, calculated from the ANOVA by dividing the model mean square by the residual mean square (406.01/62.03 [6.55]), indicated a statistically significant between-strain variability (P < 0.001).
Fig. 6.
Relative survival times of different BXD ARI strains intratracheally challenged with B. anthracis Ames spores. A dose of 500 spores/mouse of B. anthracis Ames was used for all strains (5 mice per strain). Survival time was measured as TTD in hours. Values are expressed as means and standard errors of the means. Statistical analysis showed that interstrain variability was significant (P < 0.001) and the intrastrain variability was lower, as indicated by the ratio of the interstrain variability to intrastrain variability (6.55; P < 0.001).
The BXD ARI strains also showed variability for the pathogen burden (CFU) at the site of infection (lung), as well as at two systemic locations (spleen and liver). However, the order of pathogen accumulation (data not shown) did not correlate with the survival susceptibility order (Fig. 6) across the BXD ARI strains. Also, the order of bacterial accumulation in the BXD strains did not correlate among the three organs. As an example, comparison of bacterial buildup showed a significant difference (P < 0.01) between the most resistant (BXD 69) and most susceptible (BXD 90) ARI strains, with a 2-log-higher CFU in the lung but a 1-log-lower CFU in the spleen of the sensitive strain. The pathogen dissemination to the liver was, however, comparable. This indicated a variable rate and extent of both the bacterial multiplication in the lung and systemic dissemination to spleen in these two polar responding BXD strains.
Genome-wide mapping of anthrax susceptibility in BXD ARI mice.
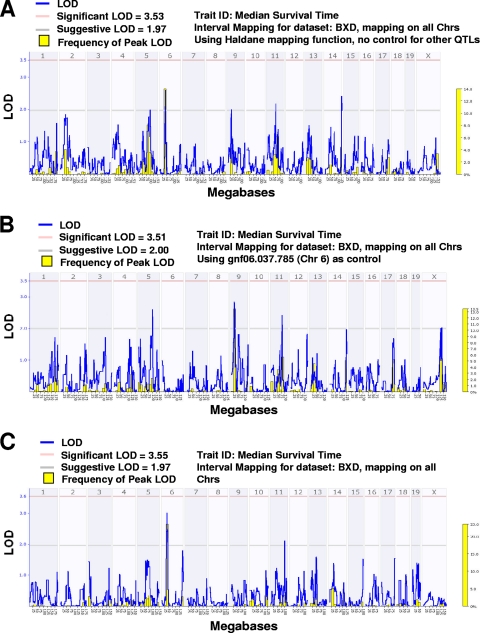
To gain insight into the QTLs underlying the differential susceptibility of C57BL/6 and DBA/2J inbred strains to a pulmonary instillation of B. anthracis Ames spores, median survival time was mapped in 16 BXD ARI strains (n = 76 total mice; 4 to 5 mice per ARI strain) by using WebQTL (65) and the integrated 3,795 polymorphic SNP and microsatellite markers. Significant and suggestive threshold limits of linkage were established at alpha values of 0.05 and 0.1, respectively, and were derived from 2,000 permutations of the data set. A genome-wide QTL map for median survival time in BXD mice is presented in Fig. 7. Five QTLs were identified on chromosomes 5, 6, 9, 11, and 14. The chromosome 6 QTL had the highest peak LOD frequency in a set of 2,000 random genome-wide bootstrap runs. In all cases, the B allele increased the survival time.
Fig. 7.
Genome-wide QTL linkage maps for median survival time of BXD ARI strains after Bacillus anthracis pulmonary infection. Blue plot, LOD ratio; yellow histogram, frequency of peak LOD from 2,000 random bootstrap iterations; pink horizontal line, significant LOD threshold value at genome-wide P level of ≤0.05 (using 2,000 permutations); gray horizontal line, suggestive threshold value at genome-wide P level of ≤0.63 (using 2,000 permutations). (A) A genome-wide LOD plot for median survival time suggested QTLs on chromosomes 5, 6, 9, 11, and 14. CI was then performed after fixing each of the two strongest peaks (chromosomes 6 and 14) to remove their variance and rescanning the genome for linkage. (B) CIM results after removing the chromosome 6 peak marker (gnf06.037.785) variance. Suggestive linkages on chromosomes 5, 9, and 11 were identified. (C) CIM results after removing the chromosome 14 peak marker (rs3691815). Suggestive linkages on chromosomes 6 and 11 were identified.
To further assess these QTLs we performed CIM, in which the variance for each of the best 2 QTLs (chromosomes 6 and 14) was individually removed and the genome rescanned to identify further loci explaining the phenotypic variance (Fig. 7B and C). Interestingly, removing the variance of the chromosome 6 QTL markedly increased the signals of the peaks on chromosomes 5 and 9 and maintained comparable signals on chromosomes 11 and 14 (Fig. 7B). Similarly, chromosome 6 and 11 signals remained when the variance for chromosome 14 was removed from the analysis (Fig. 7C). Unlike survival time, no appreciable QTLs were associated with organ CFU differences across the BXD ARI strains.
QTL analysis of the CD2 F2 population.
We further assessed the genetic linkage of anthrax susceptibility by using traditional QTL analysis on a recombinant F2 population derived from the polar responding BALB/cJ and DBA/2J strain pair. All 258 F2 mice were inoculated with 2,000 spores of Ames strain and phenotyped for survival time and for bacterial burden in the spleen, liver, and lung at the time of death. All F2 mice were also genotyped for 71 microsatellite markers across the genome, and QTL analysis was performed for each of these 4 traits. QTL plots are presented in Fig. 8. Significant linkage was identified on chromosome 17 for survival time, near D17Mit89 (∼64 Mbp), with a peak LOD score of 3.7 (Fig. 8A and C).
Fig. 8.
LOD plots for the CD2F2 population (generated from BALB/cJ and DBA/2J progenitors) infected with Bacillus anthracis Ames spores. A total of 258 F2 mice were exposed to 2,000 Ames spores and monitored for survival time and total lung burden (CFU). (A and C) Results of survival time analysis for genome-wide (A) and chromosome 17 only (C). (B and D) Lung CFU results following infection for genome-wide (B) and chromosome 11 only (D) analysis. Genome-wide threshold values that were significant (solid lines; LOD, 3.37) or suggestive (dashed lines; LOD, 3.00) are shown, as established by 10,000 permutations of the data set. Symbols with numbers represent positions of the listed Mit microsatellite markers genotyped in the full F2 population. The 95% confidence interval for the QTLs (chromosomes 11 and 17) were established by the area marked with a 1.5-LOD drop from the peak.
QTL analyses of organ CFU accumulation did not identify linkage for spleen or liver. However, a chromosome 11 QTL was revealed for lung CFU, with a LOD score of 2.8 (Fig. 8B and D). Because this QTL coincided with the previously identified region on chromosome 11 in BXD SNP analysis (Fig. 7A), there is added confidence for its importance. A separate genome-wide scan of all two-locus pairs did not reveal notable additive or epistatic interactions.
DISCUSSION
Anthrax infection is a complex process that may vary with the specific pathogen strain, as well as host factors. Murine models of anthrax infection have demonstrated an important role for the route of exposure (7, 33, 35, 71), in addition to immune cell function (11, 15). However, little is known about the role of host genetic background in anthrax infection; available studies in this regard are primarily based on the use of avirulent strains of B. anthracis (1, 24, 35, 55, 68, 69). These previous studies of inbred mice did not assess possible sex or parental effects, nor were any genetic loci associated with susceptibility to virulent B. anthracis. Results of this study can begin to fill this information gap.
The concept of same-dose-variable response is the basis of investigating the genetic differences in host susceptibility. However, the delivery of a consistent dose for anthrax infection can be challenging, depending on the route of administration. In anthrax murine models, different routes of pulmonary and extrapulmonary infection have been reported (7, 35, 69, 71). The following rationale was used to select the route for infection in these studies. The pulmonary route of B. anthracis infection is closest to a real world scenario of exposure in bioterrorism situations; thus, a pulmonary route is most reasonable. In addition, a pulmonary challenge requires a more manageable lethal dose of spores (<1,000 to ∼50,000, depending on the intratracheal versus intranasal versus aerosol delivery procedure), compared to the subcutaneous route of infection, which has a very low LD50 and therefore requires only 5 to 50 spores (35, 69). Importantly, slight fluctuations of such a low lethal dose may confound the variability differences due to host genetic background. Given these facts for pulmonary anthrax spore dosing regimens, we chose to use the intratracheal route of infection to deliver a consistent and reliable dose. A dose of 500 spores/mouse was selected to induce a discernible range of survival time and infection responses expected over the entire spectrum of the inbred strain panel tested.
Interstrain variability.
The mean and median survival time differences among the 14 inbred strains supported a possible genetic basis for susceptibility. The most resistant group (i.e., C57BL/6J and BALB/cJ strains) showed an ∼3-fold-higher overall mean survival period (based on TTD of the animals that succumbed to infection) than the most susceptible group, comprised of five equally sensitive strains, DBA/2J, A/J, NOD/ShiLtJ, DBA/1J, and FVB/NJ (Fig. 1). Identification of multiple sensitive strains in this study provides several additional possible combinations to design susceptible-resistant models for future genetic linkage analyses and therapeutics evaluation research. Comparison of the results with the limited previous studies available indicated that the relative extent and/or order of murine interstrain susceptibility varies with the route of infection and virulence status of the B. anthracis strain (virulent versus avirulent) (35, 69). However, this generalization does not seem to be universally true for all inbred strains. For instance, DBA/2J and A/J mice showed higher susceptibility regardless of the bacterial strain virulence or the route of infection.
Previous murine infection studies using the Sterne strain (24, 68) correlated anthrax susceptibility to natural complement C5 (encoded by the Hc gene; also called hemolytic complement) deficiency genotype, as in other microbial infections (19, 27). The studies using the Sterne strain were based either on C5 replenishment (68) in a C5-deficient strain (A/J) or C5 depletion with cobra venom factor (24) in a C5-sufficient strain (C57BL/6 strain). Our study using the virulent Ames strain, however, showed that correlation between hereditary C5 deficiency and anthrax susceptibility is not universal among different inbred mouse strains. For instance, the C5-sufficient DBA/1J strain (72) demonstrated equally high susceptibility (i.e., decreased survival time) to pulmonary anthrax as the C5-deficient strains DBA/2J, A/J, and NOD/ShiLtJ (Fig. 1). Similarly, an earlier study (35) reported comparable susceptibility of a complement C5-deficient inbred strain (DBA/2) and a C5-sufficient inbred strain (C3HeB/FeJ) to the virulent strain of B. anthracis. These observations collectively highlight the genetic complexities of the overall host response to anthrax infection.
Sex bias and parent-of-origin effect.
The significant (P < 0.01) sex difference we observed for survival time (Fig. 1) of BALB/cJ and C57BL/6J strains contrasts with earlier findings from murine studies of anthrax susceptibility (35, 69). For BALB/cJ, a sex bias was observed for the lethal dose (Fig. 2) required to cause 100% mortality, an observation that may or may not extrapolate as a general phenomenon for the other resistant strains. Although the resistant group strains did not show a common sex bias for lung bacterial burden (data not shown), C57BL/6J resistant female mice (individual animals that succumbed to infection) had lower bacterial burdens than their male counterparts, implying divergent genetic determinants for the two phenotypes (survival and pathogen multiplication in the host lung). The sex difference could be the result of hormonal differences or other sex-related expression changes. Although anthrax spore infection and anthrax LT showed opposing susceptibilities (69), a role for a perturbed balance of endocrine function in a purified toxin-induced murine anthrax model has been indicated (41). However, no such hormonal effect information has yet been identified for anthrax infection. Importantly, the physiological basis for, and therapeutic implications of, this sex disparity could be further investigated using the sex bias models optimized in this study.
Comparing total progeny from the reciprocal CD2F1 and D2CF1 crosses revealed a significant difference in TTD (Fig. 5A); however, males and females within each F1 cross did not differ (Fig. 5B). This differential TTD in reciprocal F1 populations supports a possible parent-of-origin effect, such as imprinting in response to anthrax infection. A parent-of-origin effect was also demonstrated for susceptibility differences in parasitic infections, including toxoplasmosis (28), Plasmodium falciparum malaria (18), and trypanosomiasis (10). Of interest, the parent-of-origin effect described for a mouse model of trypanosomiasis was also associated with overall survival time (10), with resistance attributed to the paternal allele.
Interestingly, the mean survival times of offspring from the two reciprocal F1 crosses were consistent with the susceptibility of the male strain in each cross. Specifically, offspring derived from the F1 cross using the sensitive DBA/2J sire (CD2F1) were sensitive, and offspring from the F1 cross with the resistant BALB/cJ sire (D2CF1) were resistant. Although relatively uncommon in the literature until recently, such epigenetic inheritance through the paternal lineage was elegantly demonstrated over multiple generations in mice evaluated for obesity resistance of Obrq2a and for reduced food intake (73). Data for survival time to hyperoxic acute lung injury was also found to be consistent with imprinting and transgenerational inheritance (64). But, epigenetic inheritance can also result from a heritable epigenetic effect that is independent of inheritance through the paternal lineage (43, 73), so additional testing of cross populations with paternal strain variation and strict environmental controls is needed to better understand the exact mode of inheritance for anthrax susceptibility.
Pathogen fate in a resistant versus susceptible genetic background.
Lung pathogen burden, when measured at death, showed interstrain variability but did not correlate with the survival pattern for inbred strains or BXD ARI strains (data not shown). Our comparative infection kinetics data for the most resistant (BALB/cJ) versus most susceptible (DBA/2J) strains showed that the initial steps in bacterial establishment in the lung (germination of spores, initial multiplication) are independent of the genetic background, but the rate of systemic dissemination was greater in the sensitive host background. However, once the pathogen-host interaction underlying the disease pathology comes into play, the resistant host genetic background seems to confer greater tolerance to bacterial buildup, possibly leading to the longer survival. These observations for inbred mice contrast with those of previous studies (31) in other animal species, which reported that naturally resistant (e.g., rats) or actively immunized animals had a lower systemic load of anthrax bacilli at death than naturally susceptible (e.g., guinea pigs) animals. Our observed infection kinetics pattern may be a net outcome of innate immune defense (pathogen clearance) and pathogen growth or dissemination rate (31), one or both of which may vary in the two genetic backgrounds. The identified resistant and susceptible strains of inbred mice thus offer potential infection models for evaluations of vaccines and therapeutics.
Multigenic nature of host susceptibility.
Genetic matings (F1 crosses) using two progenitor pairs of resistant and sensitive parents, e.g., C57BL/6J × DBA/2J and BALB/cJ × DBA/2J, suggested multigenic control of susceptibility to Ames infection. F1 progeny from either pair showed an intermediate survival phenotype, consistent with the involvement of multiple genes in host susceptibility to anthrax infection process. This observation based on the fully virulent Ames strain of B. anthracis is in contrast to previous observations with the avirulent B. anthracis strains Sterne or 34F2 (55, 68, 69). The previous Sterne studies did not observe an intermediate phenotype in the F1 progeny, which implied that host resistance is genetically linked to a dominant autosomal locus or gene complex. Although it is also possible that the intermediate F1 response is due to partial dominance, decreased penetrance, or variable expressivity of a single gene, our subsequent genetic analyses lend strong support to a role for multiple genes.
This study showed a reverse survival time phenotype for pulmonary anthrax spores in inbred strains compared to that reported for anthrax LT (32, 69). In addition, comparison of our data with those from subcutaneous challenge studies (35) demonstrated that a different route of administration of the same anthrax strain spores can lead to opposing responses in the same inbred mouse strain. For example, data here showed that C57BL/6J mice are resistant to a pulmonary instillation of B. anthracis spores, but this same strain died quickly when these spores were given via a subcutaneous route (35). Therefore, data suggest that a response to pulmonary infection with B. anthracis spores is likely controlled by a different set of genes than that controlling the response to anthrax LT or to a subcutaneous challenge. It is reasonable to expect, however, that one or more of the genes involved in each of these responses may overlap.
Genome-wide QTLs underlying host susceptibility to pulmonary anthrax.
In an initial effort to identify a genetic link to susceptibility after pulmonary instillation of anthrax spores, we performed genome-wide SNP analysis for the survival time of 16 BXD ARI strains. Several QTLs linked to median survival time were identified, with loci mapping to chromosomes 6, 11, and 14. Additional loci were identified on chromosomes 5 and 9 when CIM was used to remove the variance of the QTL on chromosome 6. As a second strategy to identify genetic regions linked to anthrax susceptibility, QTL analysis was performed on an F2 population derived from the BALB/cJ-DBA/2J resistant-sensitive mouse pair. A significant QTL was identified on distal chromosome 17, with support for a second locus on chromosome 11, based on its concurrence with the results of the BXD analysis.
An earlier report (55) proposed that resistance of inbred mice to a subcutaneous injection of spores of the avirulent strain 34F2 (Sterne) of B. anthracis was controlled by a single dominant gene in the C57BL/6 (sensitive) and C3H/He (resistant) mouse model. If a single gene was relevant in our infection model (with the fully virulent strain), a small ARI panel generated from C57BL/6 (resistant) and DBA/2 (sensitive) mice would have been sufficient to detect a significant linkage. In fact, if under the control of a single gene, the BXD strains would be expected to show a biphasic (i.e., sensitive or resistant) survival time. The continuous distribution of survival times for the panel of 16 BXD strains strongly supports a multigenic trait. A set of 16 RI strains has the power to detect linkage with 95% probability if the QTL accounts for about 60% of the between-strain variance (44). Given that ARI lines were used in this study, the power to detect a major locus was further improved (44). But, because several loci of apparent similar strength were detected in this set of 16 ARI strains, it is not surprising that a significant linkage was not identified using this strategy. Thus, unlike the subcutaneous route, a single major effect locus controlling survival to a pulmonary instillation of B. anthracis is not supported by our results for BXD strains and for a separate CD2 F2 population.
Several preliminary observations can be made for the identified QTLs. A similar chromosome 11 locus was identified for survival time in the BXD ARI strains and for lung burden at death (CFU) in the CD2 F2 QTL analysis. This chromosome 11 linkage maps near Ltxs1, a major QTL identified for macrophage susceptibility to anthrax LT in an in vitro model using C57BL/6 (sensitive) and C3H/He (resistant) inbred strains (52). The gene for Ltxs1 was recently identified as Nalp1b (an NLR family pyrin domain containing 1B) (4). The effects of allelic variation of this gene on the outcome of LT challenge and infection have been recently investigated (39, 61). However, the four SNPs that define the chromosome 11 QTL peak in our study map about 7 Mbp proximal to Nalp1b, suggesting one or more possible new candidate genes. The QTL on chromosome 6 coincides with the confidence interval of the recently reported Rsvs1 locus (59). This QTL controls the differential susceptibility to respiratory syncytial virus infection, as identified in C57BL/6J and AKR/J mice by measuring lung viral titers (CFU) at 4 days postinoculation.
The significant QTL on distal chromosome 17 has been tentatively designated Rpai1, for resistance to pulmonary anthrax infection, locus 1. This QTL is distinct from the plague resistance locus, Prl1, which maps to the major histocompatibility complex (MHC) region on proximal chromosome 17 (62). This distinction was expected, given that C57BL/6J mice are resistant to B. anthracis but sensitive to Yersinia pestis. Many other QTLs for differential host defense map to the MHC region of chromosome 17, including susceptibility/resistance loci for retroviruses (6), malaria (20), tuberculosis severity (54), chlamydial pneumonia (38), leishmaniasis (53), and mouse cytomegalovirus (14). Based on mapping location, these loci must differ from Rpai1, identified in this study. Survival time of mice to Trypanosoma congolense infection maps just distal to the MHC region at ∼30 Mbp (21) but is still considerably proximal to Rpai1. Therefore, the mapped location for Rpai1 identifies a heretofore-unrecognized region of chromosome 17 for host susceptibility and thus represents a distinct gene or set of closely linked genes affecting the differential response to pulmonary anthrax.
In conclusion, in a survey of 14 inbred mouse stains, this study identified several potential mouse models of spore-induced pulmonary anthrax susceptibility and also revealed a possible role of sex and the parent of origin in anthrax susceptibility. These mouse models will allow us to further investigate sex-specific factors underlying anthrax disease susceptibility and progression, as well as the responses to treatment therapeutics, vaccines, and other countermeasures. Results of genetic analysis of the BXD ARI strains and a separate CD2 F2 population supported multiple genes in the host susceptibility to pulmonary anthrax, with at least four genetic loci identified on chromosomes 6, 11, 14, and 17. The chromosome 11 locus was identified for both survival time and lung burden post-anthrax spore infection and was also in the vicinity (7 Mbp) of the previously identified Ltxs1 locus for anthrax LT susceptibility. The significant novel QTL on chromosome 17 appears to be distinct from all other mapped QTLs for host defense. The striking differences between the genome-wide linkage profiles from the two susceptibility mouse models support our hypothesis that the anthrax phenotypes are multigenic. The data further highlight that the genetic basis for this differential susceptibility likely differs for the different inbred strains and suggest that there is not a single common genetic contribution for resistance to pulmonary anthrax across strains. Future research will focus on efforts to characterize these genetic loci and to identify positional candidate genes affecting host susceptibility to pulmonary anthrax.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by NIH grant AI070865 (J.S.Y.) from the National Institute of Allergy and Infectious Diseases.
We thank Manish Gupta for occasional backup technical assistance and Shu Zheng for assistance in statistical analysis. We also thank William Gibbons, Jr., for training and technical support with the genotyping of microsatellite markers. We gratefully acknowledge Babetta Marrone of the Los Alamos National Laboratory for providing the Bacillus anthracis Ames strain.
Footnotes
Supplemental material for this article may be found at http://iai.asm.org/.
Published ahead of print on 31 May 2011.
REFERENCES
- 1. Abalakin B. A., Cherkasskii B. L. 1978. The use of inbred mice as models for the indication and differentiation of Bacillus anthracis strains. Zh. Mikrobiol. Epidemiol. Immunobiol. 55:146–147 [PubMed] [Google Scholar]
- 2. Aziz R. K., et al. 2007. Susceptibility to severe Streptococcal sepsis: use of a large set of isogenic mouse lines to study genetic and environmental factors. Genes Immun. 8:404–415 [DOI] [PubMed] [Google Scholar]
- 3. Barua S., et al. 2009. The mechanism of Bacillus anthracis intracellular germination requires multiple and highly diverse genetic loci. Infect. Immun. 77:23–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boyden E. D., Dietrich W. F. 2006. Nalp1b controls mouse macrophage susceptibility to anthrax lethal toxin. Nat. Genet. 38:240–244 [DOI] [PubMed] [Google Scholar]
- 5. Broman K. W., Wu H., Sen S., Churchill G. A. 2003. R/qtl: QTL mapping in experimental crosses. Bioinformatics 19:889–890 [DOI] [PubMed] [Google Scholar]
- 6. Case L. K., et al. 2008. Replication of beta- and gammaretroviruses is restricted in I/LnJ mice via the same genetic mechanism. J. Virol. 82:1438–1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chand H. S., et al. 2009. Discriminating virulence mechanisms among Bacillus anthracis strains by using a murine subcutaneous infection model. Infect. Immun. 77:429–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chesler E. J., et al. 2005. Complex trait analysis of gene expression uncovers polygenic and pleiotropic networks that modulate nervous system function. Nat. Genet. 37:233–242 [DOI] [PubMed] [Google Scholar]
- 9. Churchill G. A., Doerge R. W. 1994. Empirical threshold values for quantitative trait mapping. Genetics 138:963–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Clapcott S. J., Teale A. J., Kemp S. J. 2000. Evidence for genomic imprinting of the major QTL controlling susceptibility to trypanosomiasis in mice. Parasite Immunol. 22:259–263 [DOI] [PubMed] [Google Scholar]
- 11. Cote C. K., Rea K. M., Norris S. L., van Rooijen N., Welkos S. L. 2004. The use of a model of in vivo macrophage depletion to study the role of macrophages during infection with Bacillus anthracis spores. Microb. Pathog. 37:169–175 [DOI] [PubMed] [Google Scholar]
- 12. Cybulski R. J., Jr., et al. 2009. Four superoxide dismutases contribute to Bacillus anthracis virulence and provide spores with redundant protection from oxidative stress. Infect. Immun. 77:274–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dang J. L., et al. 2001. Bacillus spore inactivation methods affect detection assays. Appl. Environ. Microbiol. 67:3665–3670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Desrosiers M. P., et al. 2005. Epistasis between mouse Klra and major histocompatibility complex class I loci is associated with a new mechanism of natural killer cell-mediated innate resistance to cytomegalovirus infection. Nat. Genet. 37:593–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dixon T. C., Fadl A. A., Koehler T. M., Swanson J. A., Hanna P. C. 2000. Early Bacillus anthracis-macrophage interactions: intracellular survival and escape. Cell Microbiol. 2:453–463 [DOI] [PubMed] [Google Scholar]
- 16. Fortin A., et al. 2001. Identification of a new malaria susceptibility locus (Char4) in recombinant congenic strains of mice. Proc. Natl. Acad. Sci. U. S. A. 98:10793–10798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Friedlander A. M., Bhatnagar R., Leppla S. H., Johnson L., Singh Y. 1993. Characterization of macrophage sensitivity and resistance to anthrax lethal toxin. Infect. Immun. 61:245–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fry A. E., et al. 2008. Common variation in the ABO glycosyltransferase is associated with susceptibility to severe Plasmodium falciparum malaria. Hum. Mol. Genet. 17:567–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gervais F., Desforges C., Skamene E. 1989. The C5-sufficient A/J. congenic mouse strain. Inflammatory response and resistance to Listeria monocytogenes. J. Immunol. 142:2057–2060 [PubMed] [Google Scholar]
- 20. Goncalves L. A., Almeida P., Mota M. M., Penha-Goncalves C. 2008. Malaria liver stage susceptibility locus identified on mouse chromosome 17 by congenic mapping. PLoS One 3:e1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Goodhead I., et al. 2010. A comprehensive genetic analysis of candidate genes regulating response to Trypanosoma congolense infection in mice. PLoS Negl. Trop. Dis. 4:e880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Guidi-Rontani C. 2002. The alveolar macrophage: the Trojan horse of Bacillus anthracis. Trends Microbiol. 10:405–409 [DOI] [PubMed] [Google Scholar]
- 23. Hanna P. C., Ireland J. A. 1999. Understanding Bacillus anthracis pathogenesis. Trends Microbiol. 7:180–182 [DOI] [PubMed] [Google Scholar]
- 24. Harvill E. T., Lee G., Grippe V. K., Merkel T. J. 2005. Complement depletion renders C57BL/6 mice sensitive to the Bacillus anthracis Sterne strain. Infect. Immun. 73:4420–4422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Inglesby T. V., et al. 1999. Anthrax as a biological weapon: medical and public health management. Working Group on Civilian Biodefense JAMA 281:1735–1745 [DOI] [PubMed] [Google Scholar]
- 26. Inglesby T. V., et al. 2002. Anthrax as a biological weapon, 2002: updated recommendations for management. JAMA 287:2236–2252 [DOI] [PubMed] [Google Scholar]
- 27. Jagannath C., et al. 2000. Hypersusceptibility of A/J. mice to tuberculosis is in part due to a deficiency of the fifth complement component (C5). Scand. J. Immunol. 52:369–379 [DOI] [PubMed] [Google Scholar]
- 28. Jamieson S. E., et al. 2008. Genetic and epigenetic factors at COL2A1 and ABCA4 influence clinical outcome in congenital toxoplasmosis. PLoS One 3:e2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jernigan J. A., et al. 2001. Bioterrorism-related inhalational anthrax: the first 10 cases reported in the United States. Emerg. Infect. Dis. 7:933–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jiang C., Zeng Z. B. 1995. Multiple trait analysis of genetic mapping for quantitative trait loci. Genetics 140:1111–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jones W. I., Jr., et al. 1967. In vivo growth and distribution of anthrax bacilli in resistant, susceptible, and immunized hosts. J. Bacteriol. 94:600–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lincoln R. E., Walker J. S., Klein F., Rosenwald A. J., Jones W. I., Jr. 1967. Value of field data for extrapolation in anthrax. Fed. Proc. 26:1558–1562 [PubMed] [Google Scholar]
- 33. Loving C. L., Kennett M., Lee G. M., Grippe V. K., Merkel T. J. 2007. Murine aerosol challenge model of anthrax. Infect. Immun. 75:2689–2698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Loving C. L., et al. 2009. Role of anthrax toxins in dissemination, disease progression, and induction of protective adaptive immunity in the mouse aerosol challenge model. Infect. Immun. 77:255–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lyons C. R., et al. 2004. Murine model of pulmonary anthrax: kinetics of dissemination, histopathology, and mouse strain susceptibility. Infect. Immun. 72:4801–4809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. McAllister R. D., et al. 2003. Susceptibility to anthrax lethal toxin is controlled by three linked quantitative trait loci. Am. J. Pathol. 163:1735–1741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Meselson M., et al. 1994. The Sverdlovsk anthrax outbreak of 1979. Science 266:1202–1208 [DOI] [PubMed] [Google Scholar]
- 38. Min-Oo G., et al. 2008. Genetic control of susceptibility to pulmonary infection with Chlamydia pneumoniae in the mouse. Genes Immun. 9:383–388 [DOI] [PubMed] [Google Scholar]
- 39. Moayeri M., et al. 2010. Inflammasome sensor Nlrp1b-dependent resistance to anthrax is mediated by caspase-1, IL-1 signaling and neutrophil recruitment. PLoS Pathog. 6:e1001222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Moayeri M., Martinez N. W., Wiggins J., Young H. A., Leppla S. H. 2004. Mouse susceptibility to anthrax lethal toxin is influenced by genetic factors in addition to those controlling macrophage sensitivity. Infect. Immun. 72:4439–4447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Moayeri M., Webster J. I., Wiggins J. F., Leppla S. H., Sternberg E. M. 2005. Endocrine perturbation increases susceptibility of mice to anthrax lethal toxin. Infect. Immun. 73:4238–4244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mountz J. D., et al. 2005. Genetic segregation of spontaneous erosive arthritis and generalized autoimmune disease in the BXD2 recombinant inbred strain of mice. Scand. J. Immunol. 61:128–138 [DOI] [PubMed] [Google Scholar]
- 43. Nadeau J. H. 2009. Transgenerational genetic effects on phenotypic variation and disease risk. Hum. Mol. Genet. 18:R202–R210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Neumann P. E. 1992. Inference in linkage analysis of multifactorial traits using recombinant inbred strains of mice. Behav. Genet. 22:665–676 [DOI] [PubMed] [Google Scholar]
- 45. Nye S. H., et al. 2008. Rat survival to anthrax lethal toxin is likely controlled by a single gene. Pharmacogenomics J. 8:16–22 [DOI] [PubMed] [Google Scholar]
- 46. Passalacqua K. D., Bergman N. H. 2006. Bacillus anthracis: interactions with the host and establishment of inhalational anthrax. Future Microbiol. 1:397–415 [DOI] [PubMed] [Google Scholar]
- 47. Peirce J. L., Chesler E. J., Williams R. W., Lu L. 2003. Genetic architecture of the mouse hippocampus: identification of gene loci with selective regional effects. Genes Brain Behav. 2:238–252 [DOI] [PubMed] [Google Scholar]
- 48. Peirce J. L., Lu L., Gu J., Silver L. M., Williams R. W. 2004. A new set of BXD recombinant inbred lines from advanced intercross populations in mice. BMC Genet. 5:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Petkov P. M., et al. 2004. An efficient SNP system for mouse genome scanning and elucidating strain relationships. Genome Res. 14:1806–1811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pile J. C., Malone J. D., Eitzen E. M., Friedlander A. M. 1998. Anthrax as a potential biological warfare agent. Arch. Intern. Med. 158:429–434 [DOI] [PubMed] [Google Scholar]
- 51. Prows D. R., et al. 2007. Genetic analysis of hyperoxic acute lung injury survival in reciprocal intercross mice. Physiol. Genomics 30:271–281 [DOI] [PubMed] [Google Scholar]
- 52. Roberts J. E., Watters J. W., Ballard J. D., Dietrich W. F. 1998. Ltx1, a mouse locus that influences the susceptibility of macrophages to cytolysis caused by intoxication with Bacillus anthracis lethal factor, maps to chromosome 11. Mol. Microbiol. 29:581–591 [DOI] [PubMed] [Google Scholar]
- 53. Roberts L. J., Baldwin T. M., Curtis J. M., Handman E., Foote S. J. 1997. Resistance to Leishmania major is linked to the H2 region on chromosome 17 and to chromosome 9. J. Exp. Med. 185:1705–1710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sanchez F., et al. 2003. Multigenic control of disease severity after virulent Mycobacterium tuberculosis infection in mice. Infect. Immun. 71:126–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Shibaya M., Kubomichi M., Watanabe T. 1991. The genetic basis of host resistance to Bacillus anthracis in inbred mice. Vet. Microbiol. 26:309–312 [DOI] [PubMed] [Google Scholar]
- 56. Singh Y., Leppla S. H., Bhatnagar R., Friedlander A. M. 1989. Internalization and processing of Bacillus anthracis lethal toxin by toxin-sensitive and -resistant cells. J. Biol. Chem. 264:11099–11102 [PubMed] [Google Scholar]
- 57. Sissons J., et al. 2009. Multigenic control of tuberculosis resistance: analysis of a QTL on mouse chromosome 7 and its synergism with SstI. Genes Immun. 10:37–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Spencer R., Wilcox M. 1993. Agents of biological warfare. Rev. Med. Microbiol. 4:138–143 [Google Scholar]
- 59. Stark J. M., et al. 2010. Genomewide association analysis of respiratory syncytial virus infection in mice. J. Virol. 84:2257–2269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Suffin S. C., Carnes W. H., Kaufmann A. F. 1978. Inhalation anthrax in a home craftsman. Hum. Pathol. 9:594–597 [DOI] [PubMed] [Google Scholar]
- 61. Terra J. K., et al. 2010. Cutting edge: resistance to Bacillus anthracis infection mediated by a lethal toxin sensitive allele of Nalp1b/Nlrp1b. J. Immunol. 184:17–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Turner J. K., McAllister M. M., Xu J. L., Tapping R. I. 2008. The resistance of BALB/cJ mice to Yersinia pestis maps to the major histocompatibility complex of chromosome 17. Infect. Immun. 76:4092–4099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Vilas-Boas G. T., Peruca A. P., Arantes O. M. 2007. Biology and taxonomy of Bacillus cereus, Bacillus anthracis, and Bacillus thuringiensis. Can. J. Microbiol. 53:673–687 [DOI] [PubMed] [Google Scholar]
- 64. Wang C., et al. 2010. A model for transgenerational imprinting variation in complex traits. PLoS One 5:e11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wang J., Williams R. W., Manly K. F. 2003. WebQTL: web-based complex trait analysis. Neuroinformatics 1:299–308 [DOI] [PubMed] [Google Scholar]
- 66. Watters J. W., Dewar K., Lehoczky J., Boyartchuk V., Dietrich W. F. 2001. Kif1C, a kinesin-like motor protein, mediates mouse macrophage resistance to anthrax lethal factor. Curr. Biol. 11:1503–1511 [DOI] [PubMed] [Google Scholar]
- 67. Watters J. W., Dietrich W. F. 2001. Genetic, physical, and transcript map of the Ltxs1 region of mouse chromosome 11. Genomics 73:223–231 [DOI] [PubMed] [Google Scholar]
- 68. Welkos S. L., Friedlander A. M. 1988. Pathogenesis and genetic control of resistance to the Sterne strain of Bacillus anthracis. Microb. Pathog. 4:53–69 [DOI] [PubMed] [Google Scholar]
- 69. Welkos S. L., Keener T. J., Gibbs P. H. 1986. Differences in susceptibility of inbred mice to Bacillus anthracis. Infect. Immun. 51:795–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Welkos S. L., Trotter R. W., Becker D. M., Nelson G. O. 1989. Resistance to the Sterne strain of B. anthracis: phagocytic cell responses of resistant and susceptible mice. Microb. Pathog. 7:15–35 [DOI] [PubMed] [Google Scholar]
- 71. Welkos S. L., Vietri N. J., Gibbs P. H. 1993. Non-toxigenic derivatives of the Ames strain of Bacillus anthracis are fully virulent for mice: role of plasmid pX02 and chromosome in strain-dependent virulence. Microb. Pathog. 14:381–388 [DOI] [PubMed] [Google Scholar]
- 72. Wortis H. H. 1965. A gene locus concerned with an antigenic serum substance in Mus musculus. Genetics 52:267–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Yazbek S. N., Spiezio S. H., Nadeau J. H., Buchner D. A. 2010. Ancestral paternal genotype controls body weight and food intake for multiple generations. Hum. Mol. Genet. 19:4134–4144 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.