Abstract
Twenty-six antiretroviral drugs (ARVs), targeting five different steps in the life cycle of the human immunodeficiency virus type 1 (HIV-1), have been approved for the treatment of HIV-1 infection. Accordingly, HIV-1 phenotypic assays based on common cloning technology currently employ three, or possibly four, different recombinant viruses. Here, we describe a system to assess HIV-1 resistance to all drugs targeting the three viral enzymes as well as viral assembly using a single patient-derived, chimeric virus. Patient-derived p2-INT (gag-p2/NCp7/p1/p6/pol-PR/RT/IN) products were PCR amplified as a single fragment (3,428 bp) or two overlapping fragments (1,657 bp and 2,002 bp) and then recombined into a vector containing a near-full-length HIV-1 genome with the Saccharomyces cerevisiae uracil biosynthesis gene (URA3) replacing the 3,428 bp p2-INT segment (Dudley et al., Biotechniques 46:458–467, 2009). P2-INT-recombinant viruses were employed in drug susceptibility assays to test the activity of protease (PI), nucleoside/nucleotide reverse transcriptase (NRTI), nonnucleoside reverse transcriptase (NNRTI), and integrase strand-transfer (INSTI) inhibitors. Using a single standardized test (ViralARTS HIV), this new technology permits the rapid and automated quantification of phenotypic resistance for all known and candidate antiretroviral drugs targeting all viral enzymes (PR, RT, including polymerase and RNase H activities, and IN), some of the current and potential assembly inhibitors, and any drug targeting Pol or Gag precursor cleavage sites (relevant for PI and maturation inhibitors) This novel assay may be instrumental (i) in the development and clinical assessment of novel ARV drugs and (ii) to monitor patients failing prior complex treatment regimens.
INTRODUCTION
Treatment of patients infected with the human immunodeficiency virus has evolved considerably since the days of using monotherapy to combat HIV/AIDS in the mid-1980s (87). Introduction of antiretroviral (ARV) regimens based on multidrug combinations in the mid-1990s dramatically changed the approach for treatment of HIV-infected individuals, transforming a usually fatal disease into a more manageable and to some extent chronic illness (29, 87). Unfortunately, broad use of multiple antiretroviral drugs also led to development of drug resistance, a common cause of treatment failure (12, 87), and to the transmission of drug-resistant viruses (69). Measuring viral replication in blood (i.e., plasma viral load) and the levels of CD4+ T cells are the best surrogates to monitor disease progression in HIV-infected individuals and to measure ARV treatment success (48). Treatment failure is typically associated with drug resistance, which cannot be assessed by these more common surrogates. Thus, detecting and quantifying drug resistance have become the standard of care prior to designing new antiretroviral regimens following treatment failure (32, 39, 87).
There are basically two approaches to quantify HIV drug resistance: (i) an indirect method based on detection of specific amino acid substitutions (mutations) previously associated with resistance to specific antiretroviral drugs (i.e., genotyping) or (ii) a more direct method that tests the ability of a patient-derived virus to replicate in the presence of antiretroviral drugs in a cell-based assay (i.e., phenotyping) (23, 39). A third method combines both approaches by taking advantage of a large database to infer a level of HIV drug resistance based on genotyping and its relationship with matched phenotypic data (74). Despite longer turnaround time and higher cost, phenotypic assays involve direct resistance testing of each ARV, including FDA-approved drugs and compounds in preclinical development or under clinical evaluation. More important, phenotypic assays can be performed without any prior knowledge of HIV-1 sequence from the patient. In spite of the complex algorithms currently used in HIV-1 genotypic drug resistance tests, limited sequence length and incomplete assessment of the impact of sequence context frequently restrict the accuracy of drug resistance predictions based on primary or even secondary mutations associated with known drug-resistant viruses. Furthermore, each introduction of a new drug—often to a new target—requires a thorough characterization of mutations associated with drug resistance via phenotypic assays in hundreds to thousands of treated patients. Therefore, an HIV-1 phenotypic platform should involve cloning a viral genomic region encompassing all the drug-targeted genes rather than cloning each gene or coding region in isolation, which would not evaluate possible interactions of different mutations or linkages across the different gene targets (23, 29, 39).
Historically, phenotypic drug susceptibility assays have used HIV-1 isolates (30) or replication-competent recombinant viruses (9, 21, 28, 33, 42, 61, 67, 71) derived from patient samples by cocultivation or PCR amplification, respectively. The use of clinical HIV-1 isolates, in addition to being time-consuming and not amenable for high-throughput process, requires a period of virus propagation that usually alters the original in vivo viral quasispecies distribution, affecting the proportion of viruses which may or may not be harboring drug resistance mutations (34, 44). The ability to construct recombinant viruses carrying patient-derived HIV-1 genomic fragments is more reliable and faster and typically provides a better representation of the patient-derived HIV-1 population for more accurate drug resistance testing (28, 33, 51).
Since the early 1990s multiple approaches have been developed to introduce HIV sequences into a vector with the goal of quantifying virus replication in the presence and absence of antiretroviral drugs. Homologous recombination in mammalian cells of PCR-derived HIV sequences into vectors devoid of the corresponding sequence was one of the first and, thus, far more common, methods used (9, 28, 33, 80). Another frequent technique takes advantage of intrinsic or engineered restriction sites to clone patient-derived PCR products into a vector using restriction digestion and ligation (7, 21, 47, 51, 67). Additional cloning methods to produce recombinant HIV-1 include the use of sequence-specific uracil deglycosylase-mediated cloning (42) or directional cloning by homologous recombination in bacteria (62, 71). The final product of all these methodologies is a replication-competent (28, 42, 62, 67, 71, 80) or pseudotyped (3, 31, 51, 57) virus that is used in multiple- or single-cycle replication assays, respectively. Susceptibility of the recombinant viruses to various HIV inhibitors can be quantified by indirectly monitoring cytopathic effects caused by the replicating virus (28, 33, 61) or by directly measuring full virus production via viral protein levels in the cell-free supernatant, e.g., reverse transcriptase activity (11) or p24 antigen (42, 47). The inclusion or a reporter gene in the viral genome (i.e., firefly luciferase [51, 77], Renilla luciferase [21, 77], and fluorescent proteins [9, 80]) or a virus-induced reporter gene within the target cells (7, 67) provides a measure of virus infection at the step of HIV-1 transcription and is commonly employed with replication-competent or pseudotyped viruses.
There are currently 26 antiretroviral drugs approved for treatment of HIV-infected individuals and at least twice that number in different stages of development (http://aidsinfo.nih.gov/DrugsNew/Default.aspx?MenuItem=Drugs). As a consequence, drug resistance profiles in antiretroviral-experienced patients will become more complex and difficult to interpret. Despite the numerous cloning methods and assays described above, most phenotypic resistance tests require the construction of multiple recombinant viruses carrying different HIV-1 genes or coding sequences (e.g., Gag/protease, protease, reverse transcriptase, 3′ Gag/reverse transcriptase, or integrase) in order to perform drug susceptibility assays with different drug classes. This redundancy in recombinant virus preparation is understandable considering that low virus levels in plasma and labile viral RNA can often limit reverse transcription and PCR to amplification of only subgenomic fragments (23, 29, 39). To optimize cloning of large or multiple subgenomic HIV-1 fragments, we devised a yeast recombination-based cloning system involving both positive and negative selection to ensure insertion of either a single fragment (3,428 nucleotides [nt]) or two overlapping (1,657 nt and 2,002 nt) patient-derived amplicons encompassing the 3′ end of Gag and the entire pol gene (gag-p2/NCp7/p1/p6/pol-PR/RT/IN). Replication-competent recombinant viruses harboring this patient-derived p2-INT fragment are then used to assess resistance to all drugs targeting the three viral enzymes (i.e., protease, reverse transcriptase, and integrase) and virus particle maturation. This HIV-1 phenotypic assay (Viral Antiviral Resistance Test System or ViralARTS HIV) is a more efficient, rapid, accurate, and affordable method to monitor drug resistance in HIV-infected patients under a variety of treatment regimens, including complex combination therapy.
(This research was presented in part at the XVIII International HIV Drug Resistance Workshop, Fort Myers, FL, 9 to 13 June 2009.)
MATERIALS AND METHODS
Cells and viruses.
MT-4 cells were obtained from the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH, from Douglas Richman, and the HEK293T cells were from Stanford University (Stanford, CA). MT-4 cells were maintained in RPMI 1640 medium with 2 mM l-glutamine (Cellgro; Mediatech, Herndon, VA) supplemented with 10% fetal bovine serum (FBS; Cellgro), 10 mM HEPES (Sigma-Aldrich, St. Louis, MO), 100 U of penicillin/ml, and 100 μg of streptomycin/ml (Gibco; Invitrogen, Carlsbad, CA). HEK293T cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with l-glutamine (Gibco), 10% FBS (Cellgro), and penicillin-streptomycin (Gibco). The following viruses were obtained from the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: HIV-1A-92UG029, HIV-1B-HXB2, HIV-1B-92BR003, HIV-1B-93BR019, HIV-1C-96USNG58, HIV-1C-93MW959, HIV-1D-92UG021, HIV-1D-94UG114, HIV-1AE-CMU06, HIV-1AE-92TH021, HIV-1F-93BR20, HIV-1BF-93BR029, HIV-1G-R132, HIV-1AC-92RW009, and HIV-1N-YBF30. the 50% tissue culture infective dose (TCID50) was determined in triplicate for each serially diluted virus using the Reed and Muench method (58), and viral titers are expressed as infectious units per milliliter (IU/ml). Aliquots of DNA and RNA viruses were obtained from Zeptometrix Corporation, Buffalo, NY (BK virus, BKV; Epstein-Barr virus, EBV; hepatitis B virus, HBV; hepatitis C virus, HCV; human herpesvirus 6, HHV-6; human T-lymphotropic viruses type 1 and 2, HTLV-1 and HTLV-2; cytomegalovirus, CMV; herpes simplex virus 1 and 2, HSV-1 and HSV-2; and varicella-zoster virus, VZV) and Advanced Biotechnologies, Inc., Columbia, MD (human herpesvirus 7 [HHV-7] and HIV-2).
Antiretroviral drugs.
The antiretroviral drugs used in this study were obtained from the following sources: zidovudine (AZT), didanosine (ddI), stavudine (d4T), lamivudine (3TC), abacavir (ABC), tenofovir (TDF), emtricitabine (FTC), nevirapine (NVP), delavirdine, (DLV), efavirenz, (EFV), etravirine, (ETR), saquinavir (SQV), ritonavir (RTV), indinavir (IDV), nelfinavir (NFV), amprenavir (APV), lopinavir (LPV), atazanavir (ATV), tipranavir (TPV), and darunavir (DRV) were from ENZO Life Sciences International, Inc., Plymouth Meeting, PA; raltegravir (RAL) was from Merck & Co., Inc., West Point, PA, and ENZO Life Sciences International, Inc.
Plasma samples.
Plasma samples were obtained during routine patient monitoring from two well-characterized cohorts of HIV-infected individuals at the Hospital Carlos III (Madrid, Spain) and the AIDS Clinical Trials Unit (ACTU) at Case Western Reserve University/University Hospitals of Cleveland (Cleveland, OH) and from ProMedDx (Norton, MA). Plasma samples from Spain were shipped in dry ice and stored at −80°C until analysis. Blood specimens from Cleveland, OH, were collected fresh, and plasma samples were processed and stored at −80°C for further analysis.
Reverse transcription-PCR (RT-PCR) amplification and nucleotide sequence analysis of 3′Gag(p2/p7/p1/p6)/PR/RT/INT-coding sequences.
Plasma viral RNA was purified from pelleted virus particles by centrifuging 1 ml of plasma at 20,000 × g for 60 min at 4°C, removing 860 μl of cell-free supernatant, and resuspending the pellet in the remaining 140 μl to finally extract viral RNA using a QIAamp Viral RNA Minikit (Qiagen, Valencia, CA). Viral RNA was reverse transcribed using AccuScript High Fidelity Reverse Transcriptase (Stratagene Agilent, Santa Clara, CA) and the corresponding antisense external primer in a 20-μl reaction mixture containing 1 mM deoxynucleoside triphosphates (dNTPs), 10 mM dithiothreitol (DTT), and 10 units of RNase inhibitor. Viral cDNA was then PCR amplified using a series of external and nested primers with defined cycling conditions. The HIV-1 genomic region encoding the Gag proteins p2, p7, p1, and p6 and the protease, reverse transcriptase, and integrase enzymes was amplified as a large PCR product (3,428 nt) or two overlapping fragments (1,657 nt and 2,002 nt corresponding to the p2 5′ half RT and 3′ half RT-INT, respectively). External PCRs were carried out in a 50-μl mixture containing 0.2 mM dNTPs, 3 mM MgCl2, and 2.5 units of Pfu Turbo DNA polymerase (Stratagene). Nested PCRs were carried out in a 50-μl mixture containing 0.2 mM dNTPs, 0.3 units of Pfu Turbo DNA polymerase, and 1.9 units of Taq polymerase (Denville Scientific, Metuchen, NJ). PCR products corresponding to the 3′Gag(p2/p7/p1/p6)/PR/RT/INT-coding region of HIV-1 were purified with a QIAquick PCR purification kit (Qiagen) and sequenced using an AP Biotech DYEnamic ET Terminator cycle with Thermosequenase II (Davis Sequencing LCC, Davis, CA). Nucleotide sequences were analyzed using DNASTAR Lasergene Software Suite, version 7.1.0 (Madison, WI).
Virus production.
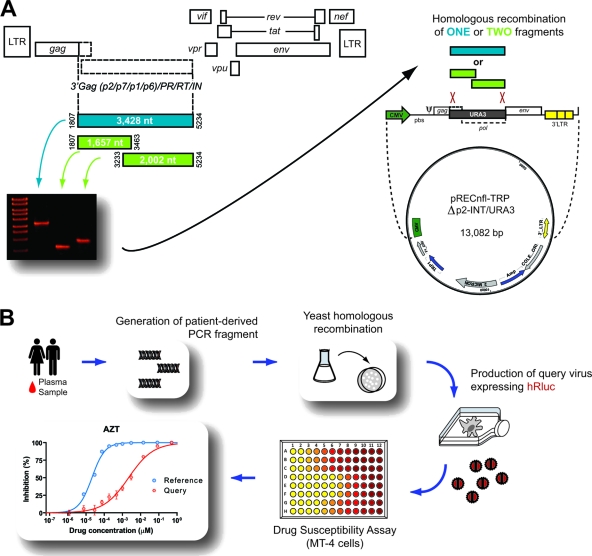
Infectious recombinant viruses were produced using an innovative yeast-based cloning technology (15) with minor modifications. Briefly, PCR products spanning the 3′Gag(p2/p7/p1/p6)/PR/RT/INT-coding region of HIV-1, either as a large fragment (3,428 nt) or as two overlapping fragments (1,657 nt and 2,002 nt), were introduced via yeast homologous recombination into the pRECnfl-TRPΔp2-INT/URA3 vector containing a near-full-length HIV-1 genome with the Saccharomyces cerevisiae uracil biosynthesis (URA3) gene replacing the 3′Gag(p2/p7/p1/p6)/PR/RT/INT HIV-1 coding sequence (Fig. 1A). Following yeast transformation, vector DNA was purified from the entire number of yeast colonies (typically 200 to >1,000 individual colonies) and used to transform Electrocomp TOP10 bacteria (Invitrogen). To guarantee the continuity of the viral population that may have existed in vivo, plasmid DNA from all the bacteria preparations was purified from 10 ml of bacteria culture using a QIAprep Spin Miniprep Kit (Qiagen). Five micrograms was digested with SphI-High-Fidelity (HF) and SalI-HF enzymes to extract a 4,333-bp fragment spanning the viral p24-Vpr coding sequence and purified using E-Gel Clonewell extraction (Invitrogen). It is important to note that intrinsic SphI and SalI restriction sites within the 3′Gag(p2/p7/p1/p6)/PR/RT/INT-coding region occur infrequently in HIV-1 (frequency of <2% according to Los Alamos HIV Database [http://www.hiv.lanl.gov/content/sequence/HIV/mainpage.html]). However, alternative restriction sites could be used to evaluate viruses containing SphI and/or SalI within the region of interest without affecting the sequence of the patient-derived viral fragment. Ten micrograms of the pNL4-3-Δ(SphI-SalI)-hRluc vector expressing the human Renilla luciferase gene (hRluc) (77), where the SphI-SalI fragment was replaced with a short linker, was double digested with SphI-HF and SalI-HF, dephosphorylated with Antarctic phosphatase, and purified (QIAquick PCR purification kit; Qiagen). The SphI-SalI fragment containing the patient-derived 3′Gag(p2/p7/p1/p6)/PR/RT/INT-coding sequence was then subcloned into the recipient vector pNL4-3-Δ(SphI-SalI)-hRluc, and the ligation product was electroporated into Electrocomp Top10 bacteria (Invitrogen). The resulting plasmid DNA was purified (QIAprep Spin Miniprep Kit;Qiagen), and 4 μg was transfected into HEK293T cells using GenDrill (BamaGen Bioscience, Gaithersburg, MD). Cell culture supernatant was harvested at 48 h posttransfection, clarified by centrifugation at 700 × g, filtered through a 0.45-μm-pore-size Steriflip filter (Millipore, Billerica, MA), aliquoted, and stored at −80°C until further use. TCID50 values were determined in triplicate for each serially diluted virus stock as described above.
Fig. 1.
(A) Strategy to introduce patient-derived p2/p7/p1/p6/PR/RT/INT PCR fragments into a proprietary vector via yeast homologous recombination as described previously (15). (B) Overview of the novel HIV-1 phenotyping assay (ViralARTS HIV). Patient-derived viral amplicons were introduced into a vector lacking the corresponding HIV p2-INT sequence. Replication-competent recombinant viruses were produced following transfection of HEK293T cells, and virus replication, in the presence and absence of antiretroviral drugs, was quantified by measuring the expression of the hRluc (Renilla luciferase) gene inserted between the env and nef coding regions (77). LTR, long terminal repeat.
Drug susceptibility based on a multiple cycle replication assay.
Drug susceptibility of 3′Gag(p2/p7/p1/p6)/PR/RT/INT-recombinant viruses was measured by determining the extent to which the antiretroviral drugs inhibited viral replication in MT-4 cells (Fig. 1B). Briefly, serial dilutions spanning empirically determined ranges of each drug were added in triplicate in 96-well plates in RPMI medium with l-glutamine (Cellgro; Mediatech) supplemented with 10% fetal bovine serum, 100 U of penicillin/ml, 100 μg of streptomycin/ml (Mediatech), and 10 mM HEPES (Sigma-Aldrich). MT-4 cells were infected with either the reference virus (HIV-1NL4-3-hRluc) (77) or the corresponding query virus (HIV-1patient-derived-3′Gag(p2/p7/p1/p6)/PR/RT/INT-hRluc) expressing human Renilla luciferase at a multiplicity of infection (MOI) of 0.005 IU/cell for 1 h at 37°C and 5% CO2. HIV-infected MT-4 cells were then resuspended in RPMI medium, and 30,000 cells were added to each well containing preplated antiretroviral drugs. Virus replication was quantified 72 h postinfection by measuring Renilla luciferase activity (relative light units [RLU]) using a Renilla Luciferase Assay System (Promega, Madison, WI) in a multiwell plate reader (Victor V multilabel reader; PerkinElmer, Waltham, MA). Drug concentrations required to inhibit virus replication by 50% (EC50) were calculated by (i) plotting the percent inhibition of luciferase activity versus the log10 drug concentration and (ii) fitting the inhibition curves to the data using nonlinear regression analysis (GraphPad Prism, version 5.01; GraphPad Software, La Jolla, CA). Fold change (FC) resistance values were calculated by dividing the mean EC50 of the query virus (HIV-1patient-derived-3′Gag(p2/p7/p1/p6)/PR/RT/INT-hRluc) by the mean EC50 of the internal control (HIV-1NL4-3-hRluc) in each assay.
HIV-1 replicative fitness determination.
The ability of 20 p2-INT-recombinant viruses, plus the HIV-1NL4-3 wild-type (wt) control, to replicate in the absence of drug pressure was determined by measuring viral growth kinetics as described previously (66, 79). Briefly, 3 × 106 MT-4 cells were infected at an MOI of 0.01 IU/cell in 1 ml of culture medium, incubated for 2 h at 37°C in 5% CO2. HIV-infected cells were then washed two times with 1× phosphate-buffered saline (PBS), then split to be cultured in triplicate wells of a 24-well plate (1 × 106 cells/well). Culture supernatant was assayed using a reverse transcriptase assay on days 0, 3, 4, 5, 6, 7, and 10 postinfection as described previously (56). Viral replication was quantified using the slope of the growth curves and performing linear regression analysis derived from the following equation: log(y) = mt + log(h), where y is virus quantity (cpm), t is time in days, and h is the y-intercept (day 0). All slope values for each virus were used to calculate the mean, standard deviation, and 10th and 90th percentiles. Differences in the mean values were evaluated using a one-way analysis of variance test, and the significant difference from the reference HIV-1NL4-3 virus was calculated using a Bonferroni's multiple comparison test (GraphPad Prism, version 5.01; GraphPad Software, La Jolla, CA).
Statistical analyses.
Descriptive results are expressed as median values and interquartile ranges. A Pearson's correlation coefficient was used to determine the strength of association between categorical variables. All differences with a P value of <0.05 were considered statistically significant. Receiver-operator characteristic (ROC) curves were used to assess the accuracy and concordance between EC50s obtained with ViralARTS HIV and PhenoSense GT for reverse transcriptase and protease inhibitors assay (Monogram Biosciences, South San Francisco, CA). The kappa coefficient, calculated using ComKappa2, version 2.0.4 (60), was used to quantify the concordance between drug susceptibility data obtained with ViralARTS and the current gold standard HIV-1 phenotyping assay. The kappa coefficient calculates a chance-adjusted measure of the agreement between any number of categories, in this case, drug susceptibility determined by two different assays. Finally, as described above, differences in the mean of the slope values for the viral growth kinetics curves were determined using a one-way analysis of variance test, and the significance difference from the reference HIV-1NL4-3 virus was calculated using Bonferroni's multiple comparison test. All statistical analyses were performed using GraphPad Prism, version 5.01 (GraphPad Software, La Jolla, CA), unless otherwise specified.
RESULTS
Characterization of the RT-PCR amplification step.
A subgenomic HIV region spanning the Gag proteins p2, p7, p1, and p6 and the protease, reverse transcriptase, and integrase coding regions was amplified by RT-PCR as a large PCR product (3,428 nt) or two overlapping fragments (1,657 nt and 2,002 nt) from plasma samples to construct p2-INT-recombinant viruses (Fig. 1A). Amplifying these large PCR products can be challenging, particularly using clinical specimens with low viral loads. Thus, sensitivity of the RT-PCR amplification was tested by analyzing 118 plasma samples obtained within a 2-month period after blood extraction from two different clinical sources (i.e., the ACTU, Cleveland, OH, and the Hospital Carlos III, Madrid, Spain). Blood samples from HIV-infected individuals with plasma viral loads ranging from <50 to 10,000 copies of viral RNA/ml were used to PCR amplify the large fragment or two shorter overlapping fragments. RT-PCR products of the correct size were consistently obtained for the large fragment (94%, or 85/90) and two overlapping fragments (98%, or 88/90) in plasma samples with ≥1,000 copies/ml of HIV RNA (Table 1). As expected, a higher success rate in PCR amplification was observed with the two overlapping fragments than with the large 3.4-kbp product, especially when plasma samples with viral loads between 50 and 1,000 copies/ml were used (35% versus 47% for the large fragment versus the two shorter PCR products, respectively) (Table 1).
Table 1.
Sensitivity of RT-PCR amplification of the 3′ end of Gag (p2/p7/p1/p6)- and Pol (PR/RT/INT)-coding sequences as a single large fragment or two overlapping shorter fragments
| Viral load (copies/ml) | % Positive samples by RT-PCR (no. of positive samples/total no. of samples tested)a |
|
|---|---|---|
| Large fragment | Two overlapping fragments | |
| <50b | 0 (0/11) | 0 (0/11) |
| 50–1,000 | 35 (6/17) | 47 (8/17) |
| 1,001–5,000 | 93 (27/29) | 97 (28/29) |
| 5,001–10,000 | 86 (19/22) | 100 (22/22) |
| >10,000 | 100 (39/39) | 97 (38/39) |
RT-PCR amplification of the single large fragment p2/p7/p1/p6/PR/RT/INT (3,428 nt) or the two shorter overlapping fragments p2/p7/p1/p6/PR/5′RT and 3′RT/INT (1,657 nt and 2,002 nt, respectively) performed with plasma samples (n = 118) from HIV-infected individuals with viral loads ranging from <50 to >10,000 copies of viral RNA/ml was performed as described in Materials and Methods.
The plasma viral loads of some of these samples may have been zero.
Highly reproducible success in RT-PCR amplification of the specified products was obtained when 20 plasma samples were tested with different viral loads. Details of this test using two different operators with different lots of critical reagents and over a 7-day period are described in Table S1 of the supplemental material. Finally, the specificity of the different RT-PCR primers and reaction mixtures was analyzed using nucleic acids from a series of RNA and DNA viruses (i.e., HBV, HCV, HIV-2, HTLV-1, HTLV-2, BKV, EBV, HHV-6, HHV-7, CMV, HSV-1, HSV-2, and VZV). As expected, no cross-reactivity was observed with any of these viruses as all RT-PCRs, either for the large fragment or the two shorter overlapping fragments, failed to generate any detectable amplicons (data not shown).
Construction of 3′Gag(p2/p7/p1/p6)/PR/RT/INT-recombinant viruses.
Unlike previous approaches that utilize homologous recombination in mammalian cells (4, 9, 28, 33, 57, 77, 80) or ligation-based cloning techniques (21, 51, 67) to create recombinant viruses, here we used a yeast-based recombination/gap repair method to introduce patient-derived HIV sequences into a vector, with the final goal of producing replication-competent chimeric viruses (15). As described above, a large HIV genomic region from the Gag protein p2 to the integrase coding region was RT-PCR amplified as a large fragment (3,428 nt) or two overlapping fragments (1,657 nt and 2,002 nt) from plasma samples or HIV-1 isolates (Fig. 1A). The p2-INT-recombinant viruses were then constructed by recombining (via gap repair) the large fragment or two overlapping PCR products into a near-full-length HIV-1 genome vector. The URA3 gene was substituted for the p2-INT HIV-1 coding sequence in the nearly full-length NL4-3 vector, i.e., pRECnfl-TRPΔp2-INT/URA3 (Fig. 1A). This vector was engineered to express the Renilla luciferase gene between the env and nef coding regions without affecting the expression of any HIV gene, as previously described (77, 80). Plasmid DNA was isolated from yeast colonies and used to transfect HEK293T cells after a series of intermediate bacterial steps as described above. Recombinant viruses were harvested at 2 days posttransfection and characterized viral stocks used in drug susceptibility assays based on (i) the infection of MT-4 cells in the presence of serial dilutions of antiretroviral drugs and (ii) quantifying virus replication by measuring the activity of Renilla luciferase (Fig. 1B). Finally, global (population) sequencing confirmed that HIV-1 sequences of the original plasma samples and the corresponding p2-INT-recombinant viruses were nearly identical (data not shown).
Performance of the novel drug susceptibility assay.
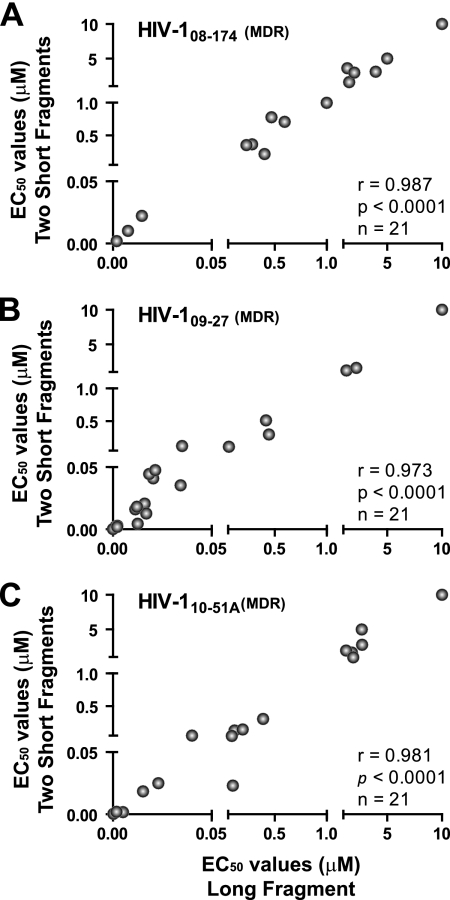
The ability to construct recombinant viruses using one single amplicon or two overlapping amplicons prompted a valid question: would these two viruses contain the same genotype and, as a consequence, the same phenotype? To address this issue, plasma samples from three highly treatment-experienced patients (i.e., 08-174, 09-27, and 10-51A) were used to construct p2-INT-recombinant viruses based on one large or two shorter but overlapping PCR products, as described above. The six p2-INT-recombinant viruses were then used in drug susceptibility assays to compare the intrapatient EC50s for each of the 21 antiretroviral drugs. There was no statistically significant difference in the EC50s between p2-INT-recombinant viruses derived from the single or dual fragments as evidenced by the strong positive correlations shown in Fig. 2 (for sample 08-174, r = 0.987 and P < 0.0001; for 09-27, r = 0.973 and P < 0.0001; and for 10-51A, r = 0.981 and P < 0.0001).
Fig. 2.
Drug susceptibility of three sets of p2-INT-recombinant viruses constructed using one large fragment (3,428 nt) or two short overlapping (1,657 nt and 2,002 nt) fragments. PCR products were obtained from three treatment-experienced patients, 08-174 (A), 09-27 (B), and 10-51A (C), and cloned as described in Materials and Methods. Pearson correlation coefficient was used to determine the strength of association between the EC50s calculated with recombinant viruses generated with one large and two overlapping PCR products. Extreme reduced susceptibility to NVP (>50 μM), FTC (>100 μM), and 3TC (>300 μM) for the 08-174 virus and FTC (>100 μM) and 3TC (>300 μM) for the 09-27 and 10-51A viruses was converted to 10 μM for graphical purposes. Mutations associated with reduction in drug susceptibility for each virus included the following: 08-174 (in PR, L10I, I47IV, I50IV, I54A, A71V, G73S, I85V, L89V, and L90M; in RT, M41L, E44D, D67N, T69D, V75M, L100I, K103N, V118I, M184V, L210W, T215Y, and K219N; in INT, N155H); 09-27 (in PR, L10V and L90M; in RT, V118I, M184V, and P225H; in INT, N155H); 10-51A (in PR, L10V, V11I, L33F, M46I, I54M, A71V, V82F, and L90M; in RT, M41L, D67G, T69N, K70R, L74I, V75I, M184V, G190A, T215F, and K219Q; in INT, none). MDR, multidrug-resistant virus; r, correlation coefficient; P, two-tailed P value; and n, number of drugs analyzed per set of recombinant viruses. EC50s represent the mean of three independent measurements.
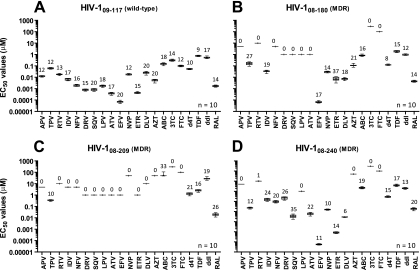
Reproducibility of the drug susceptibility assay was evaluated by testing four different p2-INT-recombinant viruses obtained from one antiretroviral-naïve HIV-infected individual (09-117) and three highly treatment-experienced patients (08-180, 08-209, and 08-240). The mean, standard deviations (SD), 95% confidence intervals (CI), and coefficients of variation (CV) of the EC50s were used to analyze data generated from 10 separate drug susceptibility determinations per virus with 21 antiretroviral drugs. Assay variation, although drug dependent, was similar for all ARVs, ranging from 9% to 20% in the wild-type virus (09-117) and from 1% to 37% in the multidrug-resistant viruses (08-180, 08-209, and 08-240) (Fig. 3). The reproducibility of the entire assay (from RNA extraction to RT-PCR to virus construction and then to the drug susceptibility test) was evaluated by processing three separated aliquots of plasma from an individual infected with a multidrug-resistant virus. The difference between the lowest and highest fold-changes in EC50s among the three replicate assays was less than 2-fold for 16 of the 21 antiretroviral drugs, with three drugs at 2.1-fold and two drugs over 3-fold relative to the reference HIV-1NL4-3 virus (see Fig. S1 in the supplemental material). Population sequencing analysis of the protease, RT, and integrase regions confirmed the concordance among the genotypes and the phenotypes determined for all three viruses (data not shown).
Fig. 3.
Reproducibility of drug susceptibility determination. Four p2-INT-recombinant viruses derived from treatment-naïve (A, 09-117) or treatment-experienced (B, 08-180; C, 08-209; and D, 08-240) patients were used to quantify susceptibility to all 21 antiretroviral drugs in 10 independent determinations (n = 10). The mean EC50, standard deviation, 5th to 95th percentiles, and the coefficient of variation (%) are indicated for each drug. When complete virus inhibition was not achieved using the maximum drug concentration (i.e., virus was completely resistant to a given antiretroviral drug), EC50s were not calculated, and the coefficient of variation was assigned a value of zero (0%). Mutations associated with reduction in drug susceptibility for each virus included the following: 09-117 (in PR, A71T; in RT, none; in INT, none), 08-180 (in PR, L10I, V32I, L33F, K43T, M46I, I47V, I54L, A71V, G73S, I84V, L89V, and L90M; in RT, M41L, E44D, D67N, V75M, F77L, V118I, M184V, L210F, T215Y, K219N, and N348I; in INT, L68V and E92Q); 08-209 (in PR, L10F, V32I, L33F, K43T, M46I, I54L, A71I, G73T, T74P, I84V, I85V, L89V, and L90M; in RT, A62V, D67G, K70E, V75I, F77L, K101E, V106I, Y115F, F116Y, Q151M, Y181C, M184V, and G190S; in INT, E138K, S147G, Q148R, and I203M), and 08-240 (in PR, L10F, V32I, M46I, I47V, I50V, A71I, and V82A; in RT, M41L, E44D, D67N, V118I, M184V, L210W, and T215Y; in INT, N155H). MDR, multidrug-resistant virus.
Finally, we were interested in evaluating the ability of our novel assay to quantify the contribution of minority variants to the overall phenotype of the viral quasispecies. For that, a p2-INT-recombinant virus constructed from a single molecular clone obtained from a multidrug-resistant virus was mixed at different proportions with the wild-type HIV-1NL4-3 reference virus. As expected, the detection of the minority drug-resistant virus depended on the antiretroviral drug tested. Thus, in some instances our novel assay was able to detect resistance in virus mixtures containing as little as 25% of the resistant virus mixed with the wild-type susceptible strain (data not shown).
Natural variation in drug susceptibility of wild-type viruses.
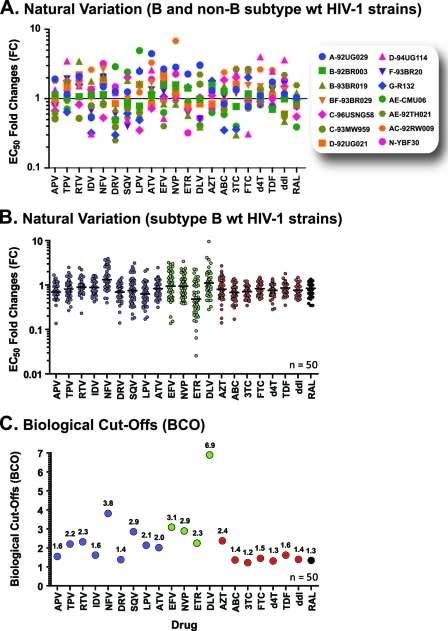
The ViralARTS HIV assay was originally developed using subtype B HIV-1 strains, predominant in North America and Europe (http://www.who.int/hiv/pub/global_report2010/en/index.html); thus, it was important to test the ability of the assay to work with non-B HIV-1 variants that have greater worldwide prevalence. For that, p2-INT-recombinant viruses were generated from 14 diverse HIV-1 isolates, including one subtype A (HIV-1A-92UG029), two subtype B (HIV-1B-92BR003 and HIV-1B-93BR019), two subtype C (HIV-1C-96USNG58 and HIV-1C-93MW959), two subtype D (HIV-1D-92UG021 and HIV-1D-94UG114), one subtype F (HIV-1F-93BR20), one subtype G (HIV-1G-R132), four circulating recombinant forms (HIV-1AE-CMU06, HIV-1AE-92TH021, HIV-1BF-93BR029, and HIV-1AC-92RW009), and a representative of the novel group N virus (HIV-1N-YBF30). Although we were able to amplify the correct fragments by RT-PCR from HIV-1 group O isolates, the respective p2-INT-recombinant viruses were not replication competent (data not shown). Susceptibility to all 21 antiretroviral drugs was evaluated, and the fold changes in EC50s relative to the reference HIV-1NL4-3 virus were calculated (Fig. 4A). As expected, the chimeric viruses derived from diverse HIV-1 isolates displayed variance in drug susceptibility as described by the mean FC values for all 21 drugs (FC, 0.91 to 2; 95% CI, 0.62 to 2.7). However, we observed no evidence of intrinsic resistance to any given antiretroviral drug after comparison with their respective biological cutoffs ([BCOs] see below).
Fig. 4.
Natural variation of wild-type viruses and BCO values. Fourteen wild-type subtype B and non-B HIV-1 isolates (A) (obtained from the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH) and 50 subtype B wild-type HIV-1 strains (B) (obtained from antiretroviral-naïve patients) were used to construct p2-INT-recombinant viruses, and their drug susceptibilities were quantified using the ViralARTS HIV assay. Three independent EC50 replicates for each drug were used to calculate the fold changes (FC) of the different query viruses relative to the HIV-1NL4-3hRluc control, and the mean EC50 fold change is indicated. (C) Upper BCO values for each antiretroviral drug calculated as the 99th percentile of the FC distribution using ViralARTS HIV.
Previous studies have highlighted the importance of analyzing the natural variation in drug susceptibility of viruses obtained from antiretroviral-naïve patients to assess the ability of a given phenotypic assay to reliably measure clinically relevant changes in drug susceptibility (28, 50, 51). Here, we analyzed phenotypic and genotypic drug susceptibility data of 50 wild-type subtype B p2-INT-recombinant viruses derived from antiretroviral-naïve HIV-infected individuals. Fold changes in the EC50s between each virus relative to the reference HIV-1NL4-3 (for each drug) are shown in Fig. 4B. Although the FC values followed a normal distribution, the mean FC was below 1 for several drugs, suggesting that this subset of wt viruses is slightly more susceptible to these particular antiretroviral drugs than the laboratory-adapted HIV-1NL4-3 strain.
This natural variation in drug susceptibility of wt viruses was used to determine the initial biological cutoffs (BCOs) for each one of the 21 antiretroviral drugs used in ViralARTS HIV. Different approaches have been described to calculate BCOs in HIV-1 phenotypic assays (26, 28, 50, 51, 73), which allow the discrimination between viruses with limited, partial, or full susceptibility to any given antiretroviral drug. Here, we tested several definitions for the determination of BCOs, including (i) the mean FC plus 2 standard deviations, (ii) the 97.5th or (iii) the 99th percentile of the FC distribution, and (iv) twice the coefficient of variation of the EC50s plus 1, where a BCO equal to 1 corresponds to zero variability in drug susceptibility among the wt viruses. More important, these different determinations permit cross-comparison of values obtained using other HIV-1 phenotypic assays (28, 50, 51, 73). Slight differences in the BCOs were observed depending on the definition used (see Table S2 in the supplemental material). Based on phenotypic and genotypic correlations, we employed very stringent BCOs for ViralARTS HIV based on the 99th percentile of the FC distribution for all 50 wt subtype B viruses, which accounts for both natural variations (different viruses) and intrinsic assay variability. Figure 4C shows the upper BCO values for each antiretroviral drug. For the PI class, the BCOs ranged from 1.4-fold for darunavir to 3.8-fold for nelfinavir. Delavirdine in the nonnucleoside reverse transcriptase (NNRTI) class had the highest BCO range (6.9-fold) which has been previously described for this drug (51). In the nucleoside reverse transcriptase (NRTI) class, the BCO values were tighter, ranging from 1.2-fold for lamivudine to 1.6-fold for tenofovir, with the exception of zidovudine, which was 2.4-fold. The biological cutoff for raltegravir, the only integrase strand-transfer (INSTI) analyzed in this study, was 1.3-fold (Fig. 4C).
Comparison of drug susceptibility data obtained with ViralARTS HIV to the current standard HIV-1 phenotypic assay.
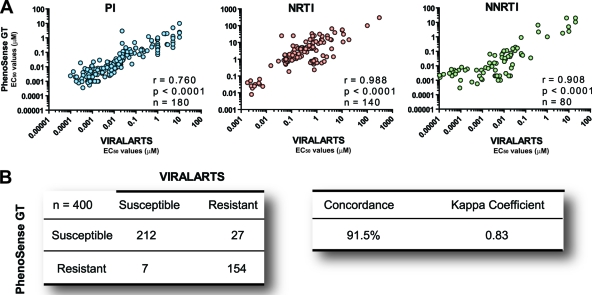
As described above, several HIV-1 drug resistance methods have been developed over the last 10 years; however, only two phenotypic assays, Antivirogram (Virco BVBA, Beerse, Belgium) and PhenoSense (Monogram Biosciences, South San Francisco, CA), have been widely deployed in the clinical setting (28, 51). Twenty plasma samples from highly treatment-experienced or -naïve HIV-infected individuals were tested using two HIV-1 phenotypic assays, i.e., ViralARTS HIV and PhenoSense GT. A strong statistically significant correlation was observed between the 400 EC50s calculated with both HIV-1 phenotyping assays, i.e., r values of 0.760, 0.988, and 0.908 (P < 0.0001, Pearson coefficient correlation) for PIs, NRTIs, and NNRTIs, respectively (Fig. 5A). The accuracy of ViralARTS HIV compared to PhenoSense GT was assessed by comparing the EC50s using ROC analyses. The areas under the ROC values were 0.55 (95% CI, 0.49 to 0.61), 0.74 (95% CI, 0.68 to 0.80), and 0.60 (95% CI, 0.51 to 0.69) for PIs, NRTIs, and NNRTIs, respectively. Finally, the fold changes in EC50s relative to the reference HIV-1NL4-3 virus were calculated and measured up to the corresponding BCO to determine the level of susceptibility for each one of the 21 antiretroviral drugs. The 400 susceptible (EC50 FC less than the BCO) or resistant (EC50 greater than the BCO) determinations obtained with ViralARTS HIV were then compared to the net assessment (i.e., sensitive or resistant) provided by PhenoSense GT. The overall concordance between both assays was 91.5% with a kappa coefficient of 0.83 (P < 0.001) (Fig. 5B).
Fig. 5.
Comparing ViralARTS HIV with the current standard HIV-1 phenotypic assay (PhenoSense GT for reverse transcriptase and protease inhibitors). Twenty plasma samples from highly treatment-experienced or treatment-naïve HIV-infected individuals were used to determine drug susceptibility to 20 antiretroviral drugs. Susceptibility to raltegravir was not included in the analysis since the current standard HIV-1 phenotypic assay requires a separate test to evaluate susceptibility to integrase inhibitors (PhenoSense for integrase inhibitors assay). (A) Pearson correlation coefficient was used to determine the strength of association between the EC50s calculated using the two HIV-1 phenotypic assays viruses. r, correlation coefficient; P, two-tailed P value; and n, number of drugs analyzed per drug class. EC50s represent the mean of three independent measurements. (B) Concordance between ViralARTS HIV and PhenoSense GT for reverse transcriptase and protease inhibitor assay. Four hundred drug susceptibility determinations (i.e., 20 viruses times 20 antiretroviral drugs) were used to calculate the concordance between both assays as follows: number of phenotypic determinations with a concordant result (e.g., susceptible-susceptible or resistant-resistant) with both assays, divided by the total number of determinations (i.e., 400), multiplied by 100. The kappa coefficient was determined as described previously (60). Values of kappa can range from −1.0 to 1.0, with −1.0 indicating perfect disagreement below chance, 0.0 indicating agreement equal to chance, and 1.0 indicating perfect agreement above chance. A rule of thumb is that kappa values that are <0.40 indicate poor agreement, ≥0.40 and <0.75 indicate good agreement, and ≥0.75 and <1.0 indicate excellent agreement.
Replicative fitness of p2-INT-recombinant viruses.
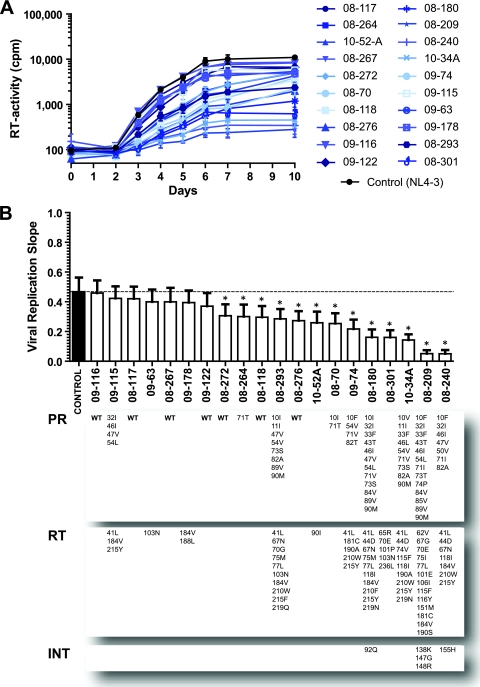
Mutations associated with drug resistance generally reduce viral fitness, i.e., the ability of the virus to replicate in a given environment (54, 55). Moreover, the presence of these replication-impaired viruses, in place of wild-type strains, has been associated with clinical benefits to HIV-infected individuals (53, 55). Our novel HIV-1 phenotypic assay employs replication-competent patient-derived chimeric viruses and supports analyses of replication kinetics during multiple rounds of tissue culture infection. Replicative fitness of 20 p2-INT-recombinant viruses was evaluated using classical viral growth kinetics in MT-4 cells and compared to the reference HIV-1NL4-3 wild-type strains (Fig. 6A). It is important to note that, in order to take into account any detrimental effect in virus replication due to the PCR amplification and cloning by recombination of the p2/p7/p1/p6/PR/RT/INT HIV fragments, the control NL4-3 p2-INT-recombinant virus and the patient-derived chimeric viruses were constructed simultaneously following the same protocol. As expected, a wide range in replicative fitness (based on viral replication slopes) was observed not only among the recombinant viruses carrying p2-INT fragments with mutations associated with drug resistance but also among those containing wild-type p2-INT sequences (Fig. 6B). However, most multidrug-resistant recombinant viruses (containing numerous drug resistance mutations) showed a marked and statistically significant impairment in replicative fitness compared to the HIV-1NL4-3 wild-type control (Fig. 6B).
Fig. 6.
HIV-1 replication fitness determination. (A) Twenty p2-INT-recombinant viruses (i.e., 10 wild-type and 10 multidrug-resistant) were evaluated for their ability to replicate in MT-4 cells in the absence of drug pressure. Virus replication was quantified by measuring reverse transcriptase (RT) activity in the cell-free supernatant. Values and error bars represent the mean and standard deviations of three independent measurements, respectively. (B) Viral replication slopes were calculated using the slopes between RLU values at days 0 and 2, 0 and 3, 0 and 4, 0 and 5, 0 and 6, 0 and 7, and 0 and 10. All seven slope values for each virus were used to calculate the mean and SD of three independent measurements. Differences in the mean values were calculated using a one-way analysis of variance test, and the significance difference from HIV-1NL4-3 was calculated using a Bonferroni's multiple comparison test. The replication kinetics of viruses marked with an asterisk (*) were significantly different from those of the HIV-1NL4-3 control (P < 0.05; 95% confidence interval). Mutations in the protease, reverse transcriptase, and integrase coding regions are indicated for each virus. WT, wild-type virus obtained from treatment-naïve patients.
DISCUSSION
According to the World Health Organization, approximately 6 million HIV-infected individuals worldwide were receiving antiretroviral therapy by the end of 2009, with roughly 685,000 (11.4%) of these patients living in North America and Europe (83). Broader access to antiretroviral drugs has led to considerable reductions in morbidity and mortality (48, 87); however, it has also increased the risk of virologic failure due to selection of drug-resistant viruses. Despite the success of antiretroviral therapy in the developed world, prevalence of antiretroviral resistance among treatment-experienced and treatment-naïve (transmitted resistance) individuals remains elevated, ranging from 37% to 66% and from 8% to 16%, respectively, depending on the cohort analyzed (22, 23, 59, 68). Furthermore, these resistance levels are steadily increasing in developing countries where patients are generally infected with non-subtype B HIV-1 strains.
With the advent of new antiretroviral drugs, there is growing need for universal phenotypic drug resistance assays to monitor patients treated with new and existing antiretroviral drugs spanning multiple HIV-1 targets. Here, we describe the development of a novel HIV-1 drug resistance phenotyping assay based on the generation of 3′Gag(p2/p7/p1/p6)/PR/RT/INT-recombinant viruses using a proprietary yeast-based cloning technology. This yeast-based recombination gap repair technique (15) provides a platform to clone a large DNA fragment (3.43 kb) or two overlapping shorter (1.66 lb and 2.0 kb) HIV-derived fragments into one vector. Unlike previous approaches, this method can use a single chimeric virus containing the entire HIV-1 target for accurate phenotyping of viruses exposed to all protease, reverse transcriptase, and integrase inhibitors, including future RNase H and maturation inhibitors (MIs), in a single assay (ViralARTS HIV).
Multiple commercial (28, 51, 57, 71) or in-house (3, 7, 9, 21, 30, 31, 33, 42, 47, 61, 62, 67, 77) phenotypic assays are currently available to quantify recombinant virus susceptibility to different drug classes; however, none has been able to simultaneously evaluate resistance to antiretroviral drugs targeting gag, protease, reverse transcriptase, and integrase coding regions. One of the main advantages of the ViralARTS HIV system is the ability to construct and test recombinant viruses carrying larger HIV-derived fragments. The yeast-based recombination/gap cloning system from HIV-1 is capable of accommodating large DNA fragments as well as combinations of two and even three overlapping DNA cassettes (15). Cloning of the entire HIV-1 genome as three overlapping DNA products amplified by RT-PCR from plasma samples and construction of several full-length infectious clones have been successful using this methodology (E. J. Arts, unpublished data). Moreover, yeast-based cloning is approximately 100-fold more efficient than bacteria-based restriction enzyme cloning or mammal-based recombination. As such, a two- or three-fragment recombination into our DNA vector still provides more unique clones (better representation of the HIV-1 intrapatient population) than other cloning methodologies (15). Altogether, the ability (i) to clone large patient-derived HIV fragments and (ii) to provide a better representation of the in vivo HIV quasispecies has led to the development of a complementary HIV phenotypic assay to be used with antiretroviral drugs targeting the env gene, i.e., viral binding, fusion, and entry inhibitors (J. Weber and M. E. Quinones-Mateu, unpublished data).
The ability to use two smaller and overlapping PCR products is particularly relevant for resistance testing on patients with low plasma HIV RNA loads. However, another potential issue relates to a possible loss of in vivo genetic linkage found in some clones within the intrapatient HIV-1 population when two as opposed to a single viral fragment are recombined. Although more definitive evidence will be provided once we complete ongoing studies based on next-generation sequencing (Weber and Quinones-Mateu, unpublished), in this study we clearly demonstrated that the drug resistance genotype and phenotype of p2-INT-recombinant viruses constructed using one single or two overlapping HIV fragments were indistinguishable. It is important to note that potential loss of linkage via yeast recombination of two products may be somewhat irrelevant considering the impact of RT or PCR recombination between HIV-1 clones of an intrapatient population during the amplification step, necessary for all recombinant virus methods. Even though our multiple-cycle assay may have enhanced sensitivity for lower frequency drug resistance polymorphisms, the greatest impact on drug resistance is likely related to the dominant and linked drug resistance mutations across the entire Gag protein p2 to the integrase coding region. Thus, all potential mutations associated with resistance to MIs, PIs, NRTIs, NNRTIs, and INSTIs can be analyzed using a single recombinant virus in this HIV-1 phenotypic assay. Numerous studies have shown that mutations outside the protease and the polymerase domain of the RT coding region have an effect on susceptibility to PIs and RTIs, respectively. Mutations downstream of the Gag protease cleavage site p24(CA)/p2 have been associated with reduced susceptibility to PIs (10, 35–37, 40, 61) while amino acid substitutions in the connection (5, 18, 24, 27, 43, 46, 65, 84) and RNase H (5, 19, 63) domains of the reverse transcriptase have been shown to have an effect on NRTI and NNRTI resistance. Recombinant viruses used in the ViralARTS HIV system contain not only patient-derived active sites/domains of relevant HIV-1 enzymes but also the majority of the HIV-1 substrates, providing a future assay for maturation (38) and RNase H inhibitors (41) still in preclinical development (78).
The new HIV-1 phenotypic assay provides accurate and reproducible drug susceptibility data to all currently available MIs, PIs, NRTIs, NNRTIs, and INSTIs. The overall amplification success of the p2-INT fragment from plasma samples with ≥1,000 copies/ml of HIV RNA was 96%, with even higher success rates obtained with the two shorter fragments (98%). The use of proprietary universal primers ensured not only amplification success with samples of diverse HIV-1 subtypes but also the absence of nonspecific products from any endogenous or related virus. Moreover, the subtype B backbone used to construct the recombinant viruses (HIV-1NL4-3) was compatible not only with p2-INT fragments from subtype B wild-type and multidrug-resistant strains but also with that from all non-B HIV-1 group M subtypes tested. The assay is efficient and reproducible, as evidenced by the repeated testing of the entire process. Finally, the ViralARTS HIV system was able to detect a drug-resistant virus present at a level as low as 25% in a mixture with wild-type virus, similar to what has been previously reported for other HIV phenotypic assays (45, 51). Of course, detection of low-frequency drug resistance is applicable only if (i) the cloning technique can adequately sample from the intrapatient HIV-1 population (e.g., a minimal of 200 clones, a feat easily achieved by yeast-based cloning) and (ii) the relative replicative fitness of the drug-resistant variant compared to the susceptible viral strain allows for its quantification.
The fold changes in EC50s determined with wild-type recombinant viruses constructed from 50 patient-derived samples were used to calculate preliminary biological cutoffs for each antiretroviral drug. Several approaches have been used to calculate BCOs in HIV-1 phenotypic assays (26, 28, 50, 51, 73), which then sets the standard for characterizing a patient-derived virus as susceptible or resistant to any given drug. Here, the BCOs for ViralARTS HIV were established based on the 99th percentile of the FC distribution, as described by Parkin et al (50), for the PhenoSense assay. Although the BCOs calculated for our new HIV-1 phenotyping assay are comparable to those determined for the two most utilized HIV-1 phenotyping assays (28, 50, 51, 73), these BCOs are still a work in progress and will be periodically updated as additional wild-type viruses are continually analyzed and added to our database. On the other hand, clinical cutoffs (CCOs) may have greater relevance since in vitro data (i.e., fold changes) are compared to clinical response information from treatment-experienced patients before and after a defined period of antiretroviral therapy (81, 82). Therefore, future studies will be designed to determine CCOs for each antiretroviral drug using this novel HIV-1 phenotyping assay.
Even though the ability to detect and quantify HIV-1 drug resistance can vary among laboratories (29), there is usually high concordance between drug resistance methodologies. Multiple studies have compared different genotypic assays (8, 20, 25, 64, 85), genotype versus phenotype (8, 16, 49, 72), and different phenotypic (52, 64, 76, 86) drug resistance assays. In the case of phenotypic assays, agreement among the tests varies with drug classes, usually showing better a correlation for PIs and lower correlation for NRTIs (64, 86). Discrepancies in identifying drug resistance often arise when FC values are too close to the assay's BCOs or CCOs (low resistance determination) for specific antiretroviral drugs (52, 64, 76). Comparative analyses of two of the most used commercial HIV-1 phenotypic assays, PhenoSense and Antivirogram, have shown variable concordance depending on the study, i.e., 71.4% (76), 86.9% (64), and 91.5% (52). As described here, drug resistance phenotypes as determined by the ViralARTS HIV assay resulted in a 91.5% concordance (kappa coefficient of 0.83) with the established PhenoSense GT assay. Percent concordance between these two assays decreased slightly (87.5%) when the net drug resistance assessment from the PhenoSense GT assay not only was based on phenotypic data but also used genotypic interpretation (e.g., those cases where the identification of certain drug resistance mutations overshadowed the susceptible call of the phenotypic part of the test, and the final call was based solely on the genotype). Regardless, the concordance between the drug susceptibility determinations based on ViralARTS HIV and PhenoSense was not different from that calculated between PhenoSense and Antivirogram (52, 64, 76).
HIV, as any other RNA virus, is constantly probing the mutation landscape in order to maintain and improve its ability to replicate in any given environment (i.e., viral fitness) (14, 55). Drug resistance mutations, although providing a selective advantage in the presence of drug pressure, usually reduce viral fitness in the absence of antiretroviral drugs (reviewed in references 53–55). Multiple methodologies have been used to measure HIV replicative fitness in vitro (55); however, a single-cycle infection assay is commonly used to estimate the replication capacity of patient-derived pseudotyped viruses (1, 2, 13). Although this replication capacity assay remains an interesting research tool, it has not found a definitive role in patient management (39). Multiple cycle replication assays tend to amplify small differences in viral replication (17, 80), which are often indistinguishable in single-cycle infection, especially where virus infection is monitored only until HIV-1 mRNA transcription in trans (i.e., luciferase expression) and does not account for viral protein translation, assembly, and virion maturation. In our system, replicative fitness can be measured using replication-competent p2-INT viruses in both monoinfections using viral growth kinetics assays as well as in growth competition experiments for a more accurate measure of fitness relative to the control HIV-1 strain. It is important to note that replicative fitness of this chimeric p2-INT virus may or may not reflect the replicative fitness of the primary HIV-1 isolate. Nonetheless, current approaches based on subdividing the HIV-1 target genes into multiple chimeric viruses are likely less representative of replicative fitness than cloning one-third of the HIV-1 genome and avoiding intragenic cloning within the pol gene. For example, the connection subdomain and RNase H domain of the RT have been shown to have an impact on both RTI resistance and replicative fitness of the virus (75).
Drug resistance testing has increased 3-fold in the United States since 1999, which includes testing of treatment-experienced patients as well as increasing numbers of genotypic and phenotypic resistance assays performed on antiretroviral-naïve patients (6). In newly infected or treatment-naïve individuals, drug resistance genotyping is preferred, given its low cost and rapid turnaround time; however, phenotypic assays continue to provide a real measure of virus replication in the presence of any antiretroviral drug (23, 39, 70). As HIV therapy continues to progress toward more diverse and complicated regimens (i.e., combinations of drugs targeting different viral genes), drug resistance assays must evolve to optimally quantify virus susceptibility against all available and future antiretroviral drugs. Our unique HIV-1 phenotyping assay may provide the platform for more efficient and affordable monitoring of HIV-infected individuals treated with antiretroviral therapy.
Supplementary Material
ACKNOWLEDGMENTS
We thank Guido Vanham and Katrien Fransen (Institute of Tropical Medicine, Antwerp, Belgium) for providing clinical samples for the validation studies. We also thank Michael D. Miller (Gilead Sciences, Inc., Foster City, CA) for providing access to critical plasma samples from the GS-US-183-0105 study for the characterization and verification studies. We are grateful to Michael D. Miller (Merck & Co., Inc., West Point, PA) for a generous supply of raltegravir.
Diagnostic Hybrids, Inc., acknowledges the contribution of the State of Ohio, Department of Development and Third Frontier Commission, which provided funding in support of the Platform for Antiviral Resistance Testing and Vaccine Development project. This publication was prepared with financial support from the State of Ohio.
The content of this paper reflects the views of Diagnostic Hybrids, Inc., and does not purport to reflect the views of the State of Ohio.
Footnotes
Supplemental material for this article may be found at http://aac.asm.org/.
Published ahead of print on 31 May 2011.
REFERENCES
- 1. Barbour J. D., et al. 2004. Higher CD4+ T cell counts associated with low viral pol replication capacity among treatment-naive adults in early HIV-1 infection. J. Infect. Dis. 190:251–256 [DOI] [PubMed] [Google Scholar]
- 2. Bates M., Wrin T., Huang W., Petropoulos C., Hellmann N. 2003. Practical applications of viral fitness in clinical practice. Curr. Opin. Infect. Dis. 16:11–18 [DOI] [PubMed] [Google Scholar]
- 3. Bona R., et al. 2006. Development of a human immunodeficiency virus vector-based, single-cycle assay for evaluation of anti-integrase compounds. Antimicrob. Agents Chemother. 50:3407–3417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boucher C. A., et al. 1996. Human immunodeficiency virus type 1 drug susceptibility determination by using recombinant viruses generated from patient sera tested in a cell-killing assay. Antimicrob. Agents Chemother. 40:2404–2409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brehm J. H., et al. 2007. Selection of mutations in the connection and RNase H domains of human immunodeficiency virus type 1 reverse transcriptase that increase resistance to 3′-azido-3′-dideoxythymidine. J. Virol. 81:7852–7859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Buchacz K., Baker R. K., Young B., Brooks J. T. 2010. Changes in the use of HIV antiretroviral resistance testing in a large cohort of U.S. patients, 1999 to 2006. J. Acquir. Immune. Defic. Syndr. 53:625–632 [DOI] [PubMed] [Google Scholar]
- 7. Choi J. Y., Kwon O. K., Choi S. Y., Park Y. K., Kim S. S. 2011. Drug susceptibility of human immunodeficiency virus type 1-derived pseudoviruses from treatment-experienced patients to protease inhibitors and reverse transcriptase inhibitors, using a modified single-round assay. J. Clin. Virol. 50:19–25 [DOI] [PubMed] [Google Scholar]
- 8. Church J. D., et al. 2009. Comparison of laboratory methods for analysis of non-nucleoside reverse transcriptase inhibitor resistance in Ugandan infants. AIDS Res. Hum. Retroviruses 25:657–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Covens K., et al. 2009. Novel recombinant virus assay for measuring susceptibility of human immunodeficiency virus type 1 group M subtypes to clinically approved drugs. J. Clin. Microbiol. 47:2232–2242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dam E., et al. 2009. Gag mutations strongly contribute to HIV-1 resistance to protease inhibitors in highly drug-experienced patients besides compensating for fitness loss. PLoS Pathog. 5:e1000345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Debyser Z., et al. 1991. An antiviral target on reverse transcriptase of human immunodeficiency virus type 1 revealed by tetrahydroimidazo-[4,5,1-jk] [1,4]benzodiazepin-2 (1H)-one and -thione derivatives. Proc. Natl. Acad. Sci. U. S. A. 88:1451–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Deeks S. G. 2003. Treatment of antiretroviral-drug-resistant HIV-1 infection. Lancet 362:2002–2011 [DOI] [PubMed] [Google Scholar]
- 13. Deeks S. G., et al. 2003. Persistence of drug-resistant HIV-1 after a structured treatment interruption and its impact on treatment response. AIDS 17:361–370 [DOI] [PubMed] [Google Scholar]
- 14. Domingo E., Holland J. J. 1997. RNA virus mutations and fitness for survival. Annu. Rev. Microbiol. 51:151–178 [DOI] [PubMed] [Google Scholar]
- 15. Dudley D. M., et al. 2009. A novel yeast-based recombination method to clone and propagate diverse HIV-1 isolates. Biotechniques 46:458–467 [DOI] [PubMed] [Google Scholar]
- 16. Dunne A. L., et al. 2001. Comparison of genotyping and phenotyping methods for determining susceptibility of HIV-1 to antiretroviral drugs. AIDS 15:1471–1475 [DOI] [PubMed] [Google Scholar]
- 17. Dykes C., Wu H., Sims M., Holden-Wiltse J., Demeter L. M. 2010. Human immunodeficiency virus type 1 protease inhibitor drug-resistant mutants give discordant results when compared in single-cycle and multiple-cycle fitness assays. J. Clin. Microbiol. 48:4035–4043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ehteshami M., et al. 2008. Connection domain mutations N348I and A360V in HIV-1 reverse transcriptase enhance resistance to 3′-azido-3′-deoxythymidine through both RNase H-dependent and -independent mechanisms. J. Biol. Chem. 283:22222–22232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ehteshami M., Gotte M. 2008. Effects of mutations in the connection and RNase H domains of HIV-1 reverse transcriptase on drug susceptibility. AIDS Rev. 10:224–235 [PubMed] [Google Scholar]
- 20. Gallego O., Martin-Carbonero L., Aguero J., de M. C., Corral A., Soriano V. 2004. Correlation between rules-based interpretation and virtual phenotype interpretation of HIV-1 genotypes for predicting drug resistance in HIV-infected individuals. J. Virol. Methods 121:115–118 [DOI] [PubMed] [Google Scholar]
- 21. Garcia-Perez J., Sanchez-Palomino S., Perez-Olmeda M., Fernandez B., Alcami J. 2007. A new strategy based on recombinant viruses as a tool for assessing drug susceptibility of human immunodeficiency virus type 1. J. Med. Virol. 79:127–137 [DOI] [PubMed] [Google Scholar]
- 22. Geretti A. M. 2007. Epidemiology of antiretroviral drug resistance in drug-naive persons. Curr. Opin. Infect. Dis. 20:22–32 [DOI] [PubMed] [Google Scholar]
- 23. Grant P. M., Zolopa A. R. 2009. The use of resistance testing in the management of HIV-1-infected patients. Curr. Opin. HIV AIDS 4:474–480 [DOI] [PubMed] [Google Scholar]
- 24. Gupta S., et al. 2010. Combinations of mutations in the connection domain of human immunodeficiency virus type 1 reverse transcriptase: assessing the impact on nucleoside and nonnucleoside reverse transcriptase inhibitor resistance. Antimicrob. Agents Chemother. 54:1973–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hales G., et al. 2006. A randomised trial comparing genotypic and virtual phenotypic interpretation of HIV drug resistance: the CREST study. PLoS Clin. Trials 1:e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Harrigan P. R., et al. 2001. World-wide variation in HIV-1 phenotypic susceptibility in untreated individuals: biologically relevant values for resistance testing. AIDS 15:1671–1677 [DOI] [PubMed] [Google Scholar]
- 27. Harrigan P. R., et al. 2002. A mutation in the 3′ region of the human immunodeficiency virus type 1 reverse transcriptase (Y318F) associated with nonnucleoside reverse transcriptase inhibitor resistance. J. Virol. 76:6836–6840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hertogs K., et al. 1998. A rapid method for simultaneous detection of phenotypic resistance to inhibitors of protease and reverse transcriptase in recombinant human immunodeficiency virus type 1 isolates from patients treated with antiretroviral drugs. Antimicrob. Agents Chemother. 42:269–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hirsch M. S., et al. 2008. Antiretroviral drug resistance testing in adult HIV-1 infection: 2008 recommendations of an International AIDS Society-USA panel. Clin. Infect. Dis. 47:266–285 [DOI] [PubMed] [Google Scholar]
- 30. Japour A. J., et al. 1993. Standardized peripheral blood mononuclear cell culture assay for determination of drug susceptibilities of clinical human immunodeficiency virus type 1 isolates. Antimicrob. Agents Chemother. 37:1095–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jarmy G., Heinkelein M., Weissbrich B., Jassoy C., Rethwilm A. 2001. Phenotypic analysis of the sensitivity of HIV-1 to inhibitors of the reverse transcriptase, protease, and integrase using a self-inactivating virus vector system. J. Med. Virol. 64:223–231 [DOI] [PubMed] [Google Scholar]
- 32. Johnson V. A., et al. 2010. Update of the drug resistance mutations in HIV-1: December 2010. Top. HIV Med. 18:156–163 [PubMed] [Google Scholar]
- 33. Kellam P., Larder B. A. 1994. Recombinant virus assay: a rapid, phenotypic assay for assessment of drug susceptibility of human immunodeficiency virus type 1 isolates. Antimicrob. Agents Chemother. 38:23–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kusumi K., et al. 1992. Human immunodeficiency virus type 1 envelope gene structure and diversity in vivo and after cocultivation in vitro. J. Virol. 66:875–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lambert-Niclot S., et al. 2008. Impact of gag mutations on selection of darunavir resistance mutations in HIV-1 protease. J. Antimicrob. Chemother. 62:905–908 [DOI] [PubMed] [Google Scholar]
- 36. Larrouy L., et al. 2010. Gag mutations can impact virological response to dual-boosted protease inhibitor combinations in antiretroviral-naive HIV-infected patients. Antimicrob. Agents Chemother. 54:2910–2919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Larrouy L., et al. 2011. Positive impact of HIV-1 gag cleavage site mutations on virological response to darunavir boosted with ritonavir. Antimicrob. Agents Chemother. 55:1754–1757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li F., et al. 2003. PA-457: a potent HIV inhibitor that disrupts core condensation by targeting a late step in Gag processing. Proc. Natl. Acad. Sci. U. S. A. 100:13555–13560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. MacArthur R. D. 2009. Understanding HIV phenotypic resistance testing: usefulness in managing treatment-experienced patients. AIDS Rev. 11:223–230 [PubMed] [Google Scholar]
- 40. Maguire M. F., et al. 2002. Changes in human immunodeficiency virus type 1 Gag at positions L449 and P453 are linked to I50V protease mutants in vivo and cause reduction of sensitivity to amprenavir and improved viral fitness in vitro. J. Virol. 76:7398–7406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Marchand C., et al. 2008. Madurahydroxylactone derivatives as dual inhibitors of human immunodeficiency virus type 1 integrase and RNase H. Antimicrob. Agents Chemother. 52:361–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Martinez-Picado J., Sutton L., De Pasquale M. P., Savara A. V., D'Aquila R. T. 1999. Human immunodeficiency virus type 1 cloning vectors for antiretroviral resistance testing. J. Clin. Microbiol. 37:2943–2951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. McCormick A. L., et al. 2011. The impact of the N348I mutation in HIV-1 reverse transcriptase, on non-nucleoside reverse transcriptase inhibitor resistance in non subtype B HIV-1. Antimicrob. Agents Chemother. 55:1806–1809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Meyerhans A., et al. 1989. Temporal fluctuations in HIV quasispecies in vivo are not reflected by sequential HIV isolations. Cell 58:901–910 [DOI] [PubMed] [Google Scholar]
- 45. Mo H., Lu L., Pithawalla R., Kempf D. J., Molla A. 2004. Complementation in cells cotransfected with a mixture of wild-type and mutant human immunodeficiency virus (HIV) influences the replication capacities and phenotypes of mutant variants in a single-cycle HIV resistance assay. J. Clin. Microbiol. 42:4169–4174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nikolenko G. N., et al. 2007. Mutations in the connection domain of HIV-1 reverse transcriptase increase 3′-azido-3′-deoxythymidine resistance. Proc. Natl. Acad. Sci. U. S. A. 104:317–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Paolucci S., Baldanti F., Zavattoni M., Gerna G. 2004. Novel recombinant phenotypic assay for clonal analysis of reverse transcriptase mutations conferring drug resistance to HIV-1 variants. J. Antimicrob. Chemother. 53:766–771 [DOI] [PubMed] [Google Scholar]
- 48. Paredes R., Clotet B. 2010. Clinical management of HIV-1 resistance. Antiviral Res. 85:245–265 [DOI] [PubMed] [Google Scholar]
- 49. Parkin N., et al. 2002. Phenotypic and genotypic HIV-1 drug resistance assays provide complementary information. J. Acquir. Immune. Defic. Syndr. 31:128–136 [DOI] [PubMed] [Google Scholar]
- 50. Parkin N. T., et al. 2004. Natural variation of drug susceptibility in wild-type human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 48:437–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Petropoulos C. J., et al. 2000. A novel phenotypic drug susceptibility assay for human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 44:920–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Qari S. H., et al. 2002. Comparative analysis of two commercial phenotypic assays for drug susceptibility testing of human immunodeficiency virus type 1. J. Clin. Microbiol. 40:31–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Quinones-Mateu M. E., Arts E. J. 2001. HIV-1 fitness: implications for drug resistance, disease progression, and global epidemic evolution, p. 134–170 In Kuiken C., et al. (ed.), HIV sequence compendium 2001. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, NM [Google Scholar]
- 54. Quinones-Mateu M. E., Arts E. J. 2002. Fitness of drug resistant HIV-1: methodology and clinical implications. Drug Resist. Updat. 5:224–233 [DOI] [PubMed] [Google Scholar]
- 55. Quinones-Mateu M. E., Arts E. J. 2006. Virus fitness: concept, quantification, and application to HIV population dynamics. Curr. Top. Microb. Immunol. 299:83–140 [DOI] [PubMed] [Google Scholar]
- 56. Quinones-Mateu M. E., et al. 2000. A dual infection/competition assay shows a correlation between ex vivo human immunodeficiency virus type 1 fitness and disease progression. J. Virol. 74:9222–9233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Race E., Dam E., Obry V., Paulous S., Clavel F. 1999. Analysis of HIV cross-resistance to protease inhibitors using a rapid single-cycle recombinant virus assay for patients failing on combination therapies. AIDS 13:2061–2068 [DOI] [PubMed] [Google Scholar]
- 58. Reed L. J., Muench H. 1938. A simple method of estimating fifty percent endpoints. Am. J. Hyg. 27:493–497 [Google Scholar]
- 59. Richman D. D., et al. 2004. The prevalence of antiretroviral drug resistance in the United States. AIDS 18:1393–1401 [DOI] [PubMed] [Google Scholar]
- 60. Robinson B. F., Bakeman R. 1998. ComKappa: a Windows 95 program for calculating kappa and related statistics. Behav. Res. Methods Instruments Computers 30:731–732 [Google Scholar]
- 61. Robinson L. H., Gale C. V., Kleim J. P. 2002. Inclusion of full-length human immunodeficiency virus type 1 (HIV-1) gag sequences in viral recombinants applied to drug susceptibility phenotyping. J. Virol. Methods 104:147–160 [DOI] [PubMed] [Google Scholar]
- 62. Roman F., et al. 2006. A new recombinant virus system for the study of HIV-1 entry and inhibition. J. Virol. Methods 131:99–104 [DOI] [PubMed] [Google Scholar]
- 63. Roquebert B., Marcelin A. G. 2008. The involvement of HIV-1 RNAse H in resistance to nucleoside analogues. J. Antimicrob. Chemother. 61:973–975 [DOI] [PubMed] [Google Scholar]
- 64. Ross L., et al. 2005. A direct comparison of drug susceptibility to HIV type 1 from antiretroviral experienced subjects as assessed by the antivirogram and PhenoSense assays and by seven resistance algorithms. AIDS Res. Hum. Retroviruses 21:933–939 [DOI] [PubMed] [Google Scholar]
- 65. Schuckmann M. M., et al. 2010. The N348I mutation at the connection subdomain of HIV-1 reverse transcriptase decreases binding to nevirapine. J. Biol. Chem. 285:38700–38709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Selhorst P., et al. 2011. Human immunodeficiency virus type 1 resistance or cross-resistance to nonnucleoside reverse transcriptase inhibitors currently under development as microbicides. Antimicrob. Agents Chemother. 55:1403–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Shi C., Mellors J. W. 1997. A recombinant retroviral system for rapid in vivo analysis of human immunodeficiency virus type 1 susceptibility to reverse transcriptase inhibitors. Antimicrob. Agents Chemother. 41:2781–2785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Sista P., et al. 2009. Nine-year trends in clinically relevant reduced susceptibility of HIV-1 to antiretrovirals. J. Clin. Virol. 44:190–194 [DOI] [PubMed] [Google Scholar]
- 69. Tang J. W., Pillay D. 2004. Transmission of HIV-1 drug resistance. J. Clin. Virol. 30:1–10 [DOI] [PubMed] [Google Scholar]
- 70. Thompson M. A., et al. 2010. Antiretroviral treatment of adult HIV infection: 2010 recommendations of the International AIDS Society-USA panel. JAMA 304:321–333 [DOI] [PubMed] [Google Scholar]
- 71. Van Baelen K., et al. 2009. A combined genotypic and phenotypic human immunodeficiency virus type 1 recombinant virus assay for the reverse transcriptase and integrase genes. J. Virol. Methods 161:231–239 [DOI] [PubMed] [Google Scholar]
- 72. Van Houtte M., et al. 2009. A comparison of HIV-1 drug susceptibility as provided by conventional phenotyping and by a phenotype prediction tool based on viral genotype. J. Med. Virol. 81:1702–1709 [DOI] [PubMed] [Google Scholar]
- 73. Verlinden Y., et al. 2005. Assessment of the antivirogram performance over time including a revised definition of biological test cut-off values. Antivir. Ther. 10:S51 [Google Scholar]
- 74. Vermeiren H., et al. 2007. Prediction of HIV-1 drug susceptibility phenotype from the viral genotype using linear regression modeling. J. Virol. Methods 145:47–55 [DOI] [PubMed] [Google Scholar]
- 75. Wang J., Bambara R. A., Demeter L. M., Dykes C. 2010. Reduced fitness in cell culture of HIV-1 with nonnucleoside reverse transcriptase inhibitor-resistant mutations correlates with relative levels of reverse transcriptase content and RNase H activity in virions. J. Virol. 84:9377–9389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Wang K., Samudrala R., Mittler J. E. 2004. Antivirogram or PhenoSense: a comparison of their reproducibility and an analysis of their correlation. Antivir. Ther. 9:703–712 [PubMed] [Google Scholar]
- 77. Weber J., Quinones-Mateu M. E. 2007. Novel anti-HIV-1 screening system based on intact recombinant viruses expressing synthetic firefly and Renilla luminescent proteins. Antivir. Ther. 12:S155 [Google Scholar]
- 78. Weber J., et al. 2010. Characterization of mutations associated with resistance to MPC-4326 and their effect on HIV-1 replicative fitness. Antivir. Ther. 15:A56 [Google Scholar]
- 79. Weber J., et al. 2010. Resistance mutations in protease, reverse transcriptase, and integrase genes: do they have an epistatic effect on drug susceptibility and/or HIV-1 replicative fitness? Antivir. Ther. 15:A94 [Google Scholar]
- 80. Weber J., et al. 2006. Use of a novel assay based on intact recombinant viruses expressing green (EGFP) or red (DsRed2) fluorescent proteins to examine the contribution of pol and env genes to overall HIV-1 replicative fitness. J. Virol. Methods 136:102–117 [DOI] [PubMed] [Google Scholar]
- 81. Winters B., et al. 2008. Determination of clinically relevant cutoffs for HIV-1 phenotypic resistance estimates through a combined analysis of clinical trial and cohort data. J. Acquir. Immune. Defic. Syndr. 48:26–34 [DOI] [PubMed] [Google Scholar]
- 82. Winters B., et al. 2009. Clinical cut-offs for HIV-1 phenotypic resistance estimates: update based on recent pivotal clinical trial data and a revised approach to viral mixtures. J. Virol. Methods 162:101–108 [DOI] [PubMed] [Google Scholar]
- 83. World Health Organization 2010. Towards universal access: scaling up priority HIV/AIDS interventions in the health sector. Progress report 2010. World Health Organization, Geneva, Switzerland: http://www.who.int/hiv/pub/2010progressreport/en/ [Google Scholar]
- 84. Yap S. H., et al. 2007. N348I in the connection domain of HIV-1 reverse transcriptase confers zidovudine and nevirapine resistance. PLoS Med. 4:e335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Zazzi M., et al. 2004. Comparative evaluation of three computerized algorithms for prediction of antiretroviral susceptibility from HIV type 1 genotype. J. Antimicrob. Chemother. 53:356–360 [DOI] [PubMed] [Google Scholar]
- 86. Zhang J., Rhee S. Y., Taylor J., Shafer R. W. 2005. Comparison of the precision and sensitivity of the Antivirogram and PhenoSense HIV drug susceptibility assays. J. Acquir. Immune. Defic. Syndr. 38:439–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Zolopa A. R. 2010. The evolution of HIV treatment guidelines: current state-of-the-art of ART. Antiviral Res. 85:241–244 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.