Abstract
Type I interferons trigger diverse biological effects by binding a common receptor, composed of IFNAR1 and IFNAR2. Intriguingly, while the activation of an antiviral state is common to all cells, antiproliferative activity and apoptosis affect only part of the population, even when cells are stimulated with saturating interferon concentrations. Manipulating receptor expression by different small interfering RNA (siRNA) concentrations reduced the fraction of responsive cells independent of the interferon used, including a newly generated, extremely tight-binding variant. Reduced receptor numbers increased 50% effective concentrations (EC50s) for alpha interferon 2 (IFN-α2) but not for the tight-binding variant. A correlation between receptor numbers, STAT activation, and gene induction is observed. Our data suggest that for a given cell, the response is binary (+/−) and dependent on the stochastic expression levels of the receptors on an individual cell. A low number of receptors suffices for antiviral response and is thus a robust feature common to all cells. Conversely, a high number of receptors is required for antiproliferative activity, which allows for fine-tuning on a single-cell level.
INTRODUCTION
Type I interferons (IFNs) form a class of cytokines capable of mediating antiviral, growth inhibitory, and immunoregulatory activities (10, 36, 46). Consisting of 18 members in humans (32), all IFNs induce their biological activities through binding to the same receptor complex, composed of the two transmembrane proteins IFNAR1 and IFNAR2 (1). Upon formation of the ternary complex, the interferon signal is transduced through receptor-associated Janus kinases (JAK), which activate the signal transducers and activators of transcription (STAT) proteins. These, in turn, form homo- and heterodimers that translocate to the nucleus to promote the expression of interferon-stimulated genes (ISGs) (45).
Despite their common biological activities and sequence homologies, type I IFNs are not redundant but rather induce their activities differentially (9, 41). These differences take effect in various ways, most notably in the antiviral (AV) and antiproliferative (AP) potencies of interferon subtypes (16, 33) and in their abilities to induce different gene expression patterns (11, 14, 38, 48). The AP activities of IFNs are a result of both apoptosis and cell cycle arrest (17, 20, 40). A profound example for differential activity is the substantially higher AP response induced by beta interferon (IFN-β) than by IFN-α2 (8, 14, 21, 41). However, it should be noted that most of the differences between IFN-α2 and IFN-β are quantitative and not qualitative; thus, higher IFN-α2 concentrations mimic most IFN-β activities.
IFNAR1 and IFNAR2 receptor subunits make distinct contributions to interferon binding, as IFNAR1 binds IFN-α with micromolar affinities, while the IFNAR2 subunit binds at nanomolar affinities (6). Nevertheless, the activation of both receptors is necessary to induce the interferon signal (3, 23). Mutagenesis studies have shown that the binding sites for the two receptor subunits are confined to two surfaces on opposite sides of the interferon molecule (28, 47). Since all type I IFNs signal through the same receptor, the current view is that receptor-ligand interactions play a critical functional role in defining a particular phenotypic readout for a cell. Different potencies of specific IFNs are known to be determined to a large extent by their affinities toward IFNAR1 and IFNAR2 subunits (16). We have previously enhanced the antiproliferative activity of IFN-α2 by increasing its binding affinity to either IFNAR1 or IFNAR2. The H57y-E58N-Q61S triple mutant (YNS mutant), which binds IFNAR1 ∼50-fold tighter than the wild type (WT), exhibited ∼100-fold-higher antiproliferative potency (17), while a mutation on IFN-α2, where the C-terminal tail was replaced with that of IFN-α8, resulted in 20-fold-increased binding affinity to IFNAR2 and 10-fold-increased antiproliferative potency (44). Moreover, we have recently demonstrated that the stability of the ternary interferon-receptor complex rather than the affinity to the individual subunits dictates biological activity (18). In this study, we further extended the repertoire of IFN-α2 variants by generating a mutant possessing both the IFN-α8 tail and YNS.
While there have been numerous efforts to study the effects of alternative ligand binding affinities to the IFNARs and how these relate to differential signaling, it is far less clear whether a simple relationship between surface receptor number and biological responses exists. Moreover, do biological effects change linearly with receptor occupancy, or are these quantized responses? Do all cell types display similar behaviors? It was shown that a decrease in cell surface concentration of one or both of the receptor subunits reduces cell sensitivity and alters signaling (5, 25, 26, 43, 49). In addition, a clear correlation between the efficiency of interferon as an antiviral drug or a cancer drug and surface receptor concentration was suggested (7, 12, 27, 49). Using stably transfected cells, it was shown that an increased number of receptors resulted in an overall elevated sensitivity for IFNs, accompanied by a decrease in interferon differential activity between the low- and high-affinity cytokines IFN-α and -β (29).
Here, we analyzed how antiviral and antiproliferative responsiveness is affected by the number of cell surface receptors and the binding affinity of the interferon used. Surface receptor density was altered using different concentrations of specific small interfering RNA (siRNA), affecting strongly the number of responding cells but less the 50% effective concentrations (EC50s). Single-cell analysis of IFNAR receptor numbers suggests that for each cell the response is binary (+/−), determined only by the number of ligand bound receptors and not by the binding affinity of the interferon added. As fewer receptors are needed to induce an antiviral state than an antiproliferative response, the antiviral state can be considered a robust feature common to all cells, whereas the antiproliferative response is fine-tuned within the population. Finally, we show that the relationship of biological response to receptor number differs among cell lines.
MATERIALS AND METHODS
Cell lines and antibodies.
MDA-MB-231, MDA-MB-468, MCF7, T-47D, BT-549, and HS 578T breast cancer cell lines are part of the NCI60 panel of human tumor cells (the U.S. National Cancer Institute NCI60 human tumor cell line anticancer drug screen). Monoclonal anti-IFNAR1-EC AA3 antibody was a gift from Biogene. Monoclonal anti-IFNAR2-EC 117.7 antibody was a gift from Daniela Novick.
Protein mutagenesis.
Site-directed mutagenesis of IFN-α2 YNS to YNS-α8tail was carried out based on the QuickChange site-directed mutagenesis protocol and was similar to that described elsewhere (50).
Protein expression and purification.
YNS and YNS-α8tail were expressed in the Escherichia coli Rosetta strain and were purified as detailed previously (17). Protein concentrations were determined both by analytical gel filtration chromatography and from the absorbance at 280 nm, with ε280 equal to 19,940 cm−1 M−1. Injection of interferons to an analytical gel filtration column, alone or together with IFNAR2-EC, allows determination of the fraction of active protein that is occluded by the receptor (17). IFN-α2 and IFNAR2-EC were expressed in the E. coli Rosetta strain and purified by ion exchange and size exclusion chromatography (35). IFNAR1-EC was expressed in Sf9 insect cells and purified as described before (19) and was a generous gift from Jacob Piehler.
In vitro binding assays.
Binding affinities of the IFN-α2 wild type and mutants with IFNAR1-EC or IFNAR2-EC were measured using the ProteOn XPR36 protein interaction array system (Bio-Rad), based on surface plasmon resonance technology. A solution of 0.005% Tween 20 in phosphate-buffered saline (PBS), pH 7.4, was used as running buffer at a flow rate of 30 μl/min. For immobilization, an activated ethyl(dimethylaminopropyl)carbodiimide–N-hydroxysuccinimide (EDC/NHS) surface was covered with NeutrAvidin and blocked with ethanolamine. Thereafter, in one of the channels the surface was activated with biotinylated Tris-nitrilotriacetic acid (NTA) for binding the IFNAR1-EC subunit via its His tag. The IFNAR2-EC receptor was biotinylated as described before (44) and later was immobilized to the surface in another channel. Both IFNAR1-EC and IFNAR2-EC were injected in a volume of 180 μl at a concentration of 0.5 μM. The tested interferons were then injected perpendicular to ligands at six different concentrations within a range of 82 to 8,000 nM (IFN-α2) or 2 to 500 nM (YNS and YNS-α8tail) for IFNAR1 binding and 2 to 500 nM for IFNAR2 binding. Data were analyzed using ProteOn Manager 2.1 software, using the standard Langmuir models for fitting kinetic data. Equilibrium dissociation constants (KD) were determined from the rate constants according to the equation
| (1) |
where kd and ka are dissociation and association rate constants, respectively, or from the equilibrium response at six different analyte concentrations, fitted to the mass-action equation (35).
The association rate constants to IFNAR2-EC were measured using an Applied Photophysics fluorescence stopped-flow instrument. Association reactions were measured in PBS, pH 7.4, using the same conditions and data analysis as those previously described (44).
Competitive binding assay.
YNS interferon was labeled with 125I by using the chloramine T iodination method (13). After iodination, the radiolabeled interferon was cleaned using a homemade Sephadex column. For the competition assay, WISH cells were grown on 24-well plates, washed once with PBS plus 0.1% sodium azide, and then incubated for 10 min with the same solution. Cells were then incubated for 1 h at room temperature (25°C) with labeled YNS (500,000 cpm/well) in the presence of unlabeled YNS or YNS-α8tail at different concentrations (100 nM to 0.1 pM) in culture medium plus 0.1% sodium azide. Thereafter, cells were washed three times in PBS. Cells were removed from wells by using 0.1 M NaOH plus 0.1% SDS and transferred into test tubes for measuring of bound, 125I-labeled YNS, using a γ-counter (Packard). Fifty-percent inhibitory concentrations (IC50s) were calculated using Kaleidagraph Synergy Software. For the analysis of the effect of receptor downregulation on interferon binding, WISH cells were transfected for 48 h with various concentrations of siRNA (see below), followed by incubation with 125I-labeled YNS alone or in the presence of 100 nM cold YNS. Thereafter, cells were treated as detailed above. The experiments with 125I-labeled YNS were repeated three times, each time in duplicate.
Antiviral and antiproliferative assays.
The antiproliferative assay (39) was performed on WISH cells and six breast cancer cell lines (see above) by adding interferon (IFN-α2 WT, YNS, YNS-α8tail, or IFN-β) at serial dilutions to the growth medium in flat-bottomed microtiter plates and monitoring cell density after 72 h by staining with crystal violet. The EC50s as well as the sensitivities of cells to increasing amounts of interferon were deduced from an interferon dose-response curve (Kaleidagraph; Synergy Software) using the equation
| (2) |
where y represents the absorbance and reflects the relative number of cells, A0 is the offset, A is the amplitude, c is the interferon concentration, and s is the slope (39). Antiviral activity was assayed as the inhibition of the cytopathic effect of vesicular stomatitis virus on human WISH cells, as described previously (39, 42). In general, interferon was added at serial dilutions to cells grown on flat-bottomed 96-well plates. Four hours later, vesicular stomatitis virus was added to all wells, and after 17 h of incubation, cell density was measured by crystal violet staining. EC50 was calculated as described for the antiproliferative experiment. Both the antiviral and antiproliferative assays were repeated at least three times for each protein. The experimental error (σ) for both assays was 35%. Therefore, a confidence level of 2× the standard error would suggest that differences smaller than 2-fold between interferons are within the experimental error.
siRNA.
WISH cells were transfected for 48 h with either human IFNAR1 or IFNAR2 siGENOME SMARTpool siRNAs, ON-TARGETplus siGENOME nontargeting siRNA number 5 (control siRNA), or a combination of these siRNAs (Dharmacon). IFNAR1 or IFNAR2 siRNAs were used in various concentrations that were completed to the highest siRNA concentration with control siRNA. We adopted this procedure to ensure that the siRNA of interest was transfected with the same efficiency and that the difference in its activity resulted from its lower concentration inside the cell. Transfection was performed using INTERFERin (Polyplus Transfection) according to the manufacturer's recommendations. Importantly, the siRNAs used in this study cannot activate the interferon-induced gene PKR (EIF2AK2) due to their short lengths (∼20 nucleotides [nt]) (22, 24).
FACS analysis of cell surface receptors.
Relative levels of interferon receptor subunits were assessed by indirect fluorescence immunostaining and fluorescence-activated cell sorting (FACS) analysis, as described elsewhere (14, 15). Surface IFNAR levels were quantified by using the median value of their signal histograms and taking the isotypic control levels as background. Quantitative comparisons of allophycocyanin (APC) values were done only between experiments done on the same day using the same instrument setups. In the experiments where siRNA was added, IFNAR levels were relative to control siRNA-transfected cells (100%). Sorting of WISH cells was carried out with a similar fluorescence immunostaining protocol followed by analysis and sorting by FACSAria (BD).
Annexin V/PI assay.
Apoptosis was monitored by the phosphatidylserine content on the outer leaflet of the cell membrane with the annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) assay kit (Bender MedSystems). Cells were detached with 5 mM EDTA in PBS and labeled with annexin V by following the kit manufacturer's instructions.
pSTAT1 and pSTAT3 phosphorylation assays.
WISH cells and the six breast cancer cell lines (see above) were lysed at 4°C with radioimmunoprecipitation assay buffer (150 mM NaCl, 10 mM Tris [pH 7.2], 0.1% SDS, 1% Triton X-100, 1% deoxycholate, 5 mM EDTA). Total protein concentrations were determined using the Bradford assay (Bio-Rad) or bicinchoninic acid (BCA) protein assay (Thermo Scientific), and equal amounts were separated by SDS-PAGE. Levels of phosphorylated STAT1 (pSTAT1) and pSTAT3 were determined by Western blot analysis using polyclonal Tyr(P)701-STAT1- and Tyr(P)705-STAT3-specific antibodies (Santa Cruz Biotechnology, Inc., Santa Cruz, CA). Equal amounts of STAT1 and STAT3 proteins and total protein extracts were verified by reblotting with polyclonal anti-STAT1 and anti-STAT3 antibodies (Delta Biolabs, Gilroy, CA) and monoclonal α-tubulin antibody (Sigma). Quantitative analyses of the Western blots were done using ImageQuant software (GE Healthcare).
Quantitative PCR.
Selected human interferon-stimulated gene expression levels were measured with the ABI Prism 7300 real-time PCR system, using the SYBR green PCR master mix (Applied Biosystems), and cDNA samples were produced with the high-capacity cDNA reverse transcription kit (Applied Biosystems) from 1 μg of total RNA, extracted with the PerfectPure RNA cultured cell kit (5 Prime). Quantitative PCR (qPCR) was performed using 5 ng of cDNA, in a total volume of 20 μl. Relative expression levels were calculated by the ΔΔCT (cycle threshold) relative quantification (RQ) method (ΔΔCT, RQ = 2−ΔΔCT), using the control siRNA-transfected untreated cells as the calibrator sample and hypoxanthine phosphoribosyltransferase 1 (HPRT1) as the endogenous control detector (reference gene levels measured on the same sample).
RESULTS
Fine-tuning of receptor expression using siRNA.
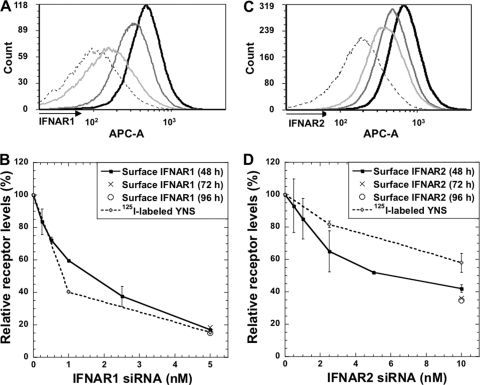
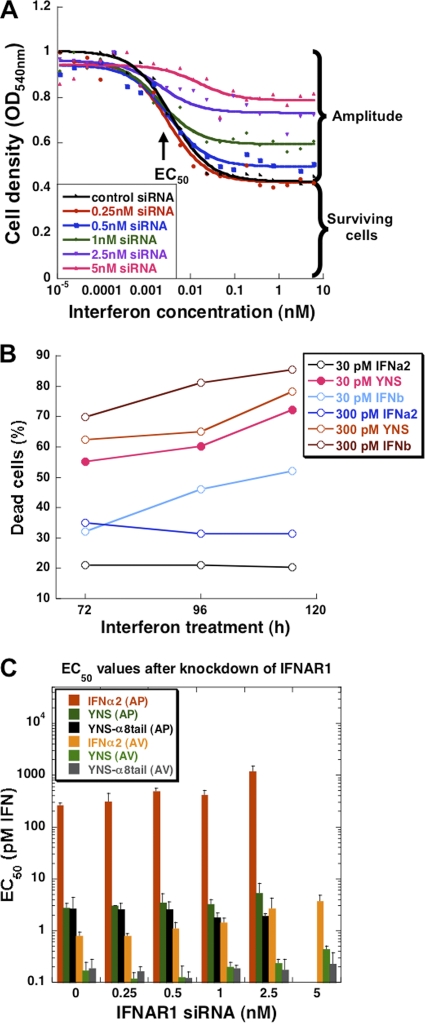
Receptor surface concentration is an important yet mostly neglected variable in dictating signaling. siRNA is a well-established method to decrease gene expression; however, it is usually not used to obtain controlled, partial knockdown, as done here. In order to ensure a reproducible partial knockdown, we kept the total siRNA levels constant, varying only the concentration of the specific siRNA probed (see Materials and Methods). The decrease of IFNAR levels presented on the cell surface (as monitored by fluorescence-activated cell sorting [FACS]) related to the concentration of siRNA applied (Fig. 1). FACS measures the quantity of surface-exposed receptors, which relates to the level of active receptors. As levels of both IFNAR1 and IFNAR2 were measured using the same secondary antibody (see Materials and Methods), their cell surface expression levels could be directly compared, showing 25% more IFNAR2 than IFNAR1 expression (compare Fig. 1A and C). The use of 5 nM siRNA against IFNAR1 and 10 nM siRNA against IFNAR2 resulted in a decrease of ∼85% and 60% of surface receptor, respectively. The reduction in receptor levels was persistent for at least 96 h (Fig. 1B and D, 5 and 10 nM siRNA, respectively). Higher siRNA concentrations did not increase knockdown efficiency. No decrease in IFNAR1 or IFNAR2 levels was observed when cells were transfected with siRNA against the other receptor subunit (data not shown). The reduction in mRNA levels of IFNAR1 after transfection with IFNAR1 siRNA was in line with the FACS results (measured by qPCR [data not shown]).
Fig. 1.
IFNAR1 and IFNAR2 receptor levels upon transfection with specific siRNAs. (A) Histograms plotted from the FACS data upon transfection for 48 h with control siRNA (black line), 1 nM IFNAR1 siRNA (dark-gray line), and 5 nM IFNAR1 siRNA (light-gray line). Dashed line, isotypic control. (B) IFNAR1 surface receptor levels upon increasing amounts of siRNA. Surface levels of IFNAR1 were measured by FACS 48, 72, and 96 h after transfection with different concentrations of IFNAR1 siRNA. Receptor levels were also determined from binding of 125I-labeled YNS upon transfection with control siRNA and 1 or 5 nM siRNA against IFNAR1. (C) Histograms plotted from the FACS data upon transfection for 48 h with control siRNA (black line), 2.5 nM IFNAR2 siRNA (dark-gray line), and 10 nM IFNAR2 siRNA (light-gray line). Dashed line, isotypic control. (D) The same experiment as described for panel B, but siRNA was used against IFNAR2. Note that 125I-labeled YNS binding was measured here upon transfection with control siRNA and 2.5 or 10 nM siRNA against IFNAR2. Error bars represent the standard deviations of results from three independent experiments (each 125I-labeled YNS experiment was performed in duplicates). The error bars (<20% of value) were removed from some of the 5 nM and 10 nM values due to space limitations.
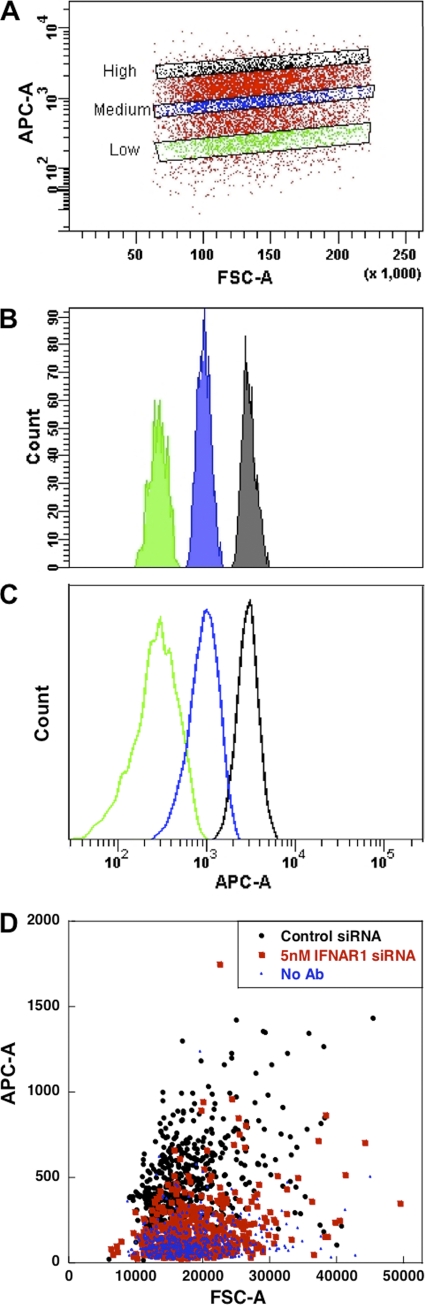
Fig. 5.
Single-cell variation in interferon receptor levels. (A to C) WISH cells from the same population with low, medium, or high intensities of IFNAR2 staining (each of them is ∼10% of the original population [A]) retain their original fluorescence intensities when sorted (B) and remeasured (C), indicating that variation is a reproducible cellular property. (D) Analyzing receptor levels on single cells. FACS measurements of WISH cells transfected with 5 nM control siRNA or IFNAR1 siRNA and the experimental isotypic control (No Ab) were disassembled to single-cell FSC and APC data using MATLAB. The data were randomly sorted, and the first 500 APC values (y axis) were plotted against the corresponding FSC values (x axis).
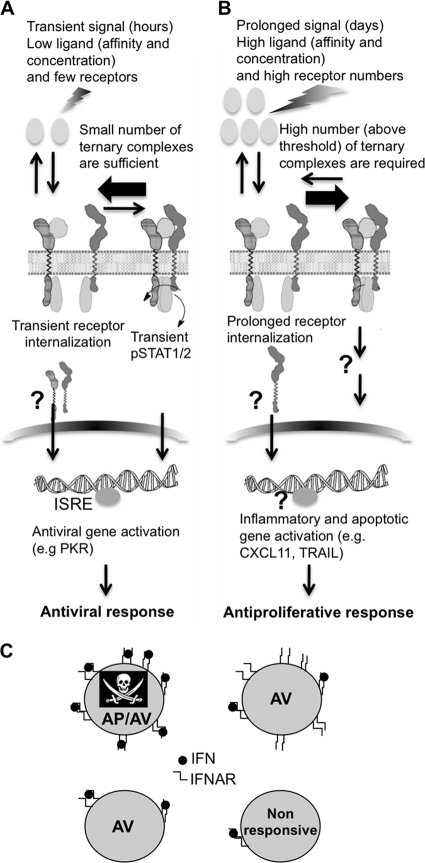
Fig. 10.
Model of differential activation by interferons. (A) The antiviral response is initiated by a transient signal of low concentration of interferon binding few receptors. The activation of a small number of ternary complexes results in STAT phosphorylation and antiviral gene induction. (B) The antiproliferative response in WISH cells is initiated by a prolonged signal of high concentration of interferon binding a minimal threshold of receptors, which is much higher than the threshold needed for the antiviral response. The antiproliferative function cannot be solely explained by STAT activation, and the involvement of additional genes is assumed. (C) Relation between stochastic receptor expression and antiproliferative (AP) or antiviral (AV) activities on single cells. Four cases are portrayed: high receptor, high ligand (AP/AV); high receptor, low ligand (AV); low receptor, high/low ligand (AV); very low receptor (nonresponsive).
Next, we evaluated the effect of IFNAR knockdown on the binding of interferon to the receptors on the cell surface. WISH cells were transfected for 48 h with various siRNAs (control siRNA, 1 or 5 nM IFNAR1 siRNA, 2.5 or 10 nM IFNAR2 siRNA) and then were incubated with 125I-labeled YNS alone or in the presence of 100 nM cold YNS. The reduction in YNS binding was in correlation with the decrease in surface receptor levels as measured by FACS (Fig. 1B and D). Note that relative IFNAR2 levels (upon siRNA) were lower when determined by FACS than when determined from 125I-labeled YNS interferon. This can be attributed to the higher expression of IFNAR2 than of IFNAR1 and the fact that 125I-labeled YNS is washed when not in the ternary complex.
Generating a tighter-binding interferon variant.
The activity of interferon is dictated by its binding affinity. To further extend the range of binding affinities of interferon variants used in this study, we generated the YNS-α8tail, combining the C-terminal tail of IFN-α8 (which increases binding to IFNAR2) and the YNS mutant (which increases binding to IFNAR1) on an IFN-α2 background. The binding affinities of YNS-α8tail toward IFNAR1-EC (extracellular domain only) and IFNAR2-EC (Table 1) correspond to those measured for YNS toward IFNAR1 and IFN-α2-α8tail toward IFNAR2 (44). Using surface plasmon resonance (SPR) measurements, the α8tail increased the affinity toward IFNAR2 5.5-fold (which is only due to a slower kd value), while using stopped-flow data the affinity is increased 38-fold (compared to that of IFN-α2). This difference is due only to a difference in ka values and was previously discussed (35). The higher affinity of YNS-α8tail toward cell surface IFNAR was demonstrated by a binding competition assay on WISH cells. 125I-labeled YNS was mixed with cold interferon (either YNS or YNS-α8tail). YNS-α8tail exhibited a 5-fold-lower IC50 than YNS (Table 1), which is in line with the affinity values determined using SPR but not with the stopped-flow data determined in solution. As IFNAR2 is surface bound on cells, it is not straightforward which of the two methods better represents the difference in binding of interferon to its cell surface receptors.
Table 1.
In vitro binding affinities of the interferon wild type and mutants toward IFNAR1 and IFNAR2 receptor subunitsa
| Interferon | IFNAR1-EC |
IFNAR2-EC |
IC50 on WISH cells |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ka × 105 (M−1 s−1) | kd (s−1) | KD (nM) | Ratioc | ka × 106 (M−1 s−1) | kd × 10−3 (s−1) | KD (nM) | Ratioc | nM | Ratiod | |
| IFN-α2b | 2,000 | 1 | 3 (10) | 6.7 | 2.2 (0.67) | 1 | ||||
| YNS | 2 | 0.009 | 45 | 45 | 3 (10) | 5.6 | 2 (0.5) | 1.1 (1.3) | 0.77 | 1 |
| YNS-α8tail | 2 | 0.008 | 40 | 50 | 3 (68) | 1.2 | 0.4 (0.018) | 5.5 (38) | 0.15 | 5 |
Binding was measured using ProteOn XPR36 and a stopped-flow instrument. KD values were determined from kd/ka over six different concentrations of the analyte. Association rate constants to IFNAR2 were determined both by SPR and by stopped flow. All values in parentheses were obtained from stopped-flow measurements with both proteins being in solution. IC50 values were determined by mixing cold YNS or YNS-α8tail with 125I-labeled YNS (see Materials and Methods).
The affinities for IFN-α2 were determined using the mass-action equation over six different concentrations of the analyte.
Ratios are relative to wild-type IFNα2 (given in the first row).
Ratios are relative to YNS (given in the second row).
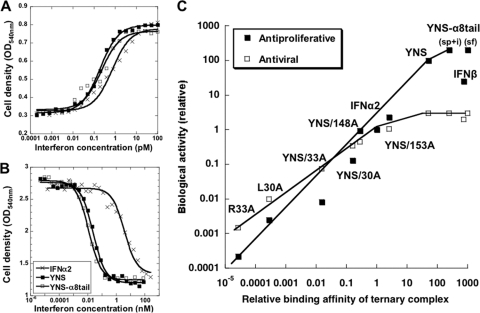
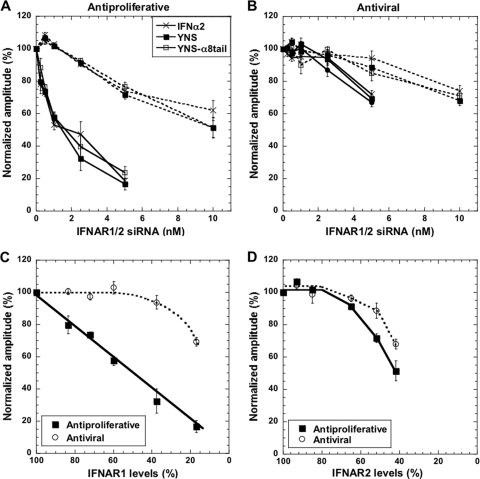
The antiviral potency of YNS-α8tail on WISH cells was similar to that measured for the YNS variant and only ∼3-fold higher than that of the WT (Fig. 2 A; Table 2). The antiproliferative potency of YNS-α8tail only slightly surpassed (2-fold) the potency of the YNS mutant, despite the higher binding affinity of the YNS-α8tail variant (Fig. 2B; Tables 1 and 2). A similar small increase in antiproliferative activity was also observed on MDA-MB-231 human breast cancer cells (Table 2). Plotting the biological potencies of different interferon mutants versus their binding affinities toward both receptors (R1×R2) (Fig. 2C) shows that the increase in antiproliferative activity of YNS-α8tail is somewhat below that expected from the increased affinity, suggesting that we may have reached the limit for antiproliferative activity of IFNs. A similar phenomenon of surpassing the upper limit of affinity required for maximal cellular activity was observed also for growth hormone binding its receptors (31). The different slopes of AV and AP versus binding affinity (over the linear parts of the plots) were discussed previously (18, 34).
Fig. 2.
Antiviral and antiproliferative potencies of IFN-α2 mutants. Antiviral (A) and antiproliferative (B) dose-response curves of WISH cells treated with the IFN-α2 wild type or the mutants YNS and YNS-α8tail. (C) Relative antiviral and antiproliferative potencies versus relative affinity of IFN-α2 mutants as calculated from the product of IFNAR1 and IFNAR2 affinity differences (R1×R2). The relative affinity of the YNS-α8tail variant was determined by SPR (sp), radiolabeled competition assay (i), and the kd value from SPR and ka from stopped flow (sf). Some of the data are from reference 18. The slope of the linear regression of log(AV)/log(R1×R2) is 0.6 and of log(AP)/log(R1×R2) is 0.9 (over the linear parts of the plots).
Table 2.
Calculated EC50s for wild-type and mutant IFN-α2a
| Interferon | Antiproliferative activity in: |
Antiviral activity in WISH cells |
||||
|---|---|---|---|---|---|---|
| WISH cells |
MDA-MB-231 |
|||||
| EC50 (nM) | Ratio | EC50 (nM) | Ratio | EC50 (pM) | Ratio | |
| IFN-α2 | 3.6 | 1 | 0.29 | 1 | 0.71 | 1 |
| YNS | 0.023 | 150 | 0.0038 | 75 | 0.2 | 3.5 |
| YNS-α8tail | 0.012 | 300 | 0.0014 | 210 | 0.22 | 3.25 |
EC50s were determined from dose-response curves, fitted to equation 2. Ratios are relative to wild-type IFN-α2 (given in the first row).
Reducing the number of receptors results in a significant decrease in the antiproliferative activity but only a minor decrease in antiviral response.
The influence of different receptor levels on antiviral and antiproliferative potencies was examined using IFN-α2 WT, YNS, and YNS-α8tail. WISH cells were transfected with increasing siRNA concentrations of IFNAR1 or IFNAR2 for 48 h prior to the addition of IFNs. The cells were assayed for their antiviral and antiproliferative responses. Importantly, we confirmed by screening hundreds of different siRNAs in combination with interferon treatment that the siRNA transfection by itself has no effect on cell growth or the interferon response (data not shown). Two parameters were examined for each of the treatments: EC50 and the normalized amplitude. The amplitude value represents the proportion of cells undergoing antiproliferative or antiviral response to interferon as measured by crystal violet staining (Fig. 3 A). The determined optical density at 540 nm (OD540) values directly relate to live cell number, as measured by the density of different dilutions and by counting the cells using a cell counter (Bio-Rad; data not shown). The normalized amplitude is the amplitude at a given siRNA concentration divided by the amplitude without siRNA. Figure 3A shows that reducing the receptor numbers by adding siRNA resulted in smaller normalized amplitudes (i.e., the antiproliferative effect was less pronounced). The maximal level of antiproliferative response is reached at a concentration of interferon that saturates the surface receptors (as determined by the binding affinity), suggesting that a certain percentage of cells do not respond, independent of the concentration of interferon. To obtain a better understanding of the nature of interferon-induced cell death in WISH cells, we stained cells grown in a 6-well plate for 72 h after YNS treatment with annexin V, a specific apoptotic marker. FACS measurements showed that ∼60% of the cells were annexin V positive (Fig. 3B). The remaining 40% of the cells were alive and annexin V negative, even in the presence of the highest interferon concentration. This result was confirmed by terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) assay (data not shown).
Fig. 3.
The effect of interferon treatment with or without IFNAR1 knockdown on cell survival and EC50s. (A) Antiproliferative dose-response curve of WISH cells after 48 h of siRNA transfection. Five concentrations (0.25 nM to 5 nM) of siRNA against IFNAR1 were applied, followed by a 72-h treatment with YNS-α8tail. Two parameters were analyzed: EC50 and the amplitude, which is the difference in cell densities between no treatment and the highest interferon concentration for each siRNA. Note that even at the highest interferon concentration, 40% of the cells survive. (B) Cell death following treatment with interferon. WISH cells were incubated with either 30 or 300 pM of IFN-α2, YNS, or IFN-β for 72, 96, or 120 h (x axis). Cells were labeled with annexin V-FITC and PI (propidium iodide) and analyzed by FACS. The percentage of dead cells (y axis) was determined according to the levels of phosphatidylserine in the outer leaflet of the plasma membrane, an apoptotic marker detected by annexin V, and the levels of cell permeability (PI). (C) Tighter interferon-receptor binding compensates for reduced receptor numbers. Five concentrations (0.25 nM to 5 nM) of siRNA against IFNAR1 were applied for 48 h, followed by interferon treatment. The EC50s were determined from the dose-response curves (see panel A) for antiviral (AV) and antiproliferative (AP) activities. Error bars represent the standard deviations of results from three independent experiments.
A second important parameter is the EC50s of antiviral and antiproliferative responses with reduced receptor levels and whether the EC50 depends on the affinity of the given ligand. In Fig. 3C, we show that for IFN-α2, a 5-fold increase in EC50s was determined when IFNAR1 levels were reduced. Conversely, for YNS only, a 2-fold increase in EC50 was observed, and for the YNS-α8tail variant, no change was recorded. This suggests that in terms of EC50, tighter binding IFNs can compensate for reduced receptor numbers.
Figure 4A presents the normalized amplitudes of antiproliferative activities with five siRNA concentrations targeting IFNAR1, using three different IFNs. For example, adding 1 nM siRNA results in 40% fewer surface IFNAR1 receptors (Fig. 1B) and a 45% decrease in the amplitude of the antiproliferative activity, while 5 nM siRNA almost completely diminished the antiproliferative activity of the three IFNs investigated. The change in normalized amplitude was dependent only on the number of surface receptors and not on the type of interferon used. Thus, increased binding affinity of interferon does not compensate for reduced receptor numbers.
Fig. 4.
Relation between surface receptor levels and antiviral and antiproliferative responses. (A) Antiproliferative activity of IFN-α2 and mutants upon transfection with siRNA against IFNAR1 (solid lines) and IFNAR2 (dashed lines). The percentage of responding cells without siRNA is taken as 100% (see also Fig. 3A). (B) The same as described for panel A but for antiviral activity. (C and D) The relation between YNS interferon antiproliferative or antiviral activities and IFNAR1 (C) or IFNAR2 (D) receptor levels on WISH cells. Error bars represent the standard deviations of results from three independent experiments.
The IFNAR2 receptor surface expression is less sensitive to knockdown by siRNA than IFNAR1 knockdown, with 10 nM siRNA resulting in a 60% reduction in the receptor level (Fig. 1D). Reduction in the antiproliferative response of interferon was observed only at siRNA concentrations of >1 nM (Fig. 4A), with 10 nM siRNA reducing the normalized amplitude to 50%. The 25% higher concentration of IFNAR2 receptor than of IFNAR1 receptor (Table 3) may explain why 1 nM siRNA reduces surface expression of IFNAR2 by 15%, with no marked effect on antiproliferative activity. This again confirms that the number of ternary complexes and not the number of individual receptors drives antiproliferative activity.
Table 3.
IFNAR1 and IFNAR2 abundance on WISH cells and six breast cancer cell linesa
| Cell line | MFI |
IFNAR2/IFNAR1 MFI ratio | MFI ratio relative to WISH cells |
||
|---|---|---|---|---|---|
| IFNAR1 | IFNAR2 | IFNAR1 | IFNAR2 | ||
| WISH | 2,280 | 2,855 | 1.25 | 1 | 1 |
| MDA-MB-231 | 2,690 | 1,380 | 0.5 | 1.2 | 0.5 |
| MDA-MB-468 | 3,740 | 2,330 | 0.6 | 1.6 | 0.8 |
| MCF7 | 2,340 | 2,030 | 0.9 | 1.0 | 0.7 |
| T-47D | 2,935 | 980 | 0.35 | 1.3 | 0.3 |
| BT-549 | 1,540 | 2,080 | 1.35 | 0.7 | 0.7 |
| HS 578T | 3,210 | 3,720 | 1.15 | 1.4 | 1.3 |
Surface levels of IFNAR1 and IFNAR2 subunits were measured by FACS analysis. MFI, median fluorescence intensity.
Antiviral potency was much less affected by reducing the number of receptors than antiproliferative activity. Only ≥2.5 nM siRNA against IFNAR1 or ≥5 nM siRNA against IFNAR2 had an effect on the amplitude of the antiviral response (Fig. 4B). Moreover, at 5 nM siRNA, 70% of the cells still showed viral protection against IFNAR1, while the level of receptors decreased to 15% (Fig. 1B), suggesting that a low number of receptors is sufficient to initiate the antiviral response.
Figure 4C and D present a summary of the effects of reducing IFNAR1 or IFNAR2 levels on the biological activity of IFN-YNS. Antiproliferative activity declines linearly with decreasing IFNAR1 receptor levels, whereas antiviral activity does not. On the contrary, IFNAR2 levels required more than 25% knockdown before we observed reduction in antiproliferative activity.
Analyzing receptor levels at the single-cell level.
FACS measurements of IFNAR1 and IFNAR2 show a Gaussian distribution of expression, suggesting that individual cells might vary in receptor numbers (Fig. 1A and C). We tested experimentally if this apparent distribution of signal indeed correlates with receptor expression rather than a variance in the signal measured by the FACS detector, by gating for cells exhibiting low, medium, and high levels of allophycocyanin (APC) signal corresponding to differing IFNAR1 and IFNAR2 levels within a cell population and reanalyzed these same cells immediately. Figures 5 A to C show that for IFNAR2 the level of APC signal retained its consistency, supporting that a natural distribution of both receptor levels is indeed a physical attribute of WISH cells. The same results were obtained for IFNAR1 (data not shown).
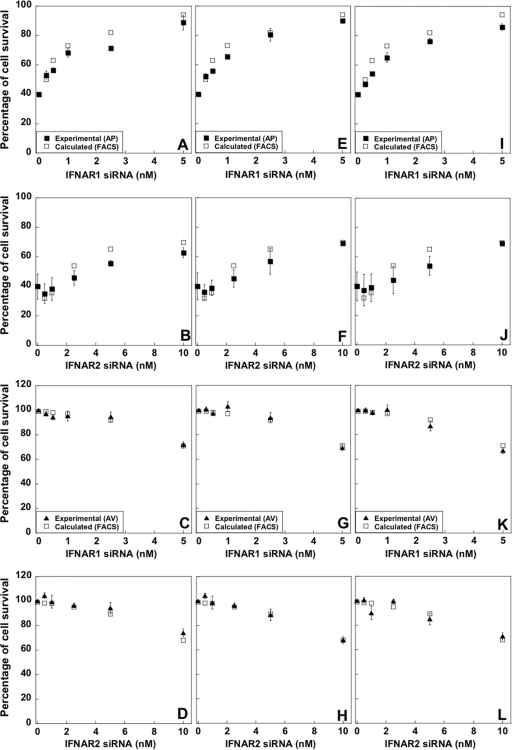
The large differences in surface receptor levels between single cells led us to hypothesize that a critical minimal threshold of receptor numbers may dictate the cells' fate in response to interferon. This would explain why the ability of interferon to induce antiproliferative activity in WISH cells seems to be limited to ∼60% of the population (Fig. 3A and B). Figure 5D shows representative APC values of single cells plotted against FSC (forward scatter characteristics; which are proportional to cell size). Only a weak correlation is observed between the FSC and APC, with the population being homogenous in terms of cell dimension. As expected, most of the control siRNA-transfected cells have higher APC values than IFNAR1 siRNA-transfected cells. Still, there is an overlap between the APC values of control cells and IFNAR1 siRNA-transfected cells, representing specific cells with low levels of IFNAR1 subunit on the surface. As 40% of cells did not undergo apoptosis upon interferon treatment (Fig. 3B), we obtained the APC threshold signal for responding cells, which is the APC value at the same percentile (meaning, 40% of the control siRNA-transfected cells have a weaker and 60% a stronger APC signal). If indeed the APC threshold value corresponds to the minimum number of receptors required to initiate cell death, we should be able to calculate (using the threshold value as a reference) the percentage of surviving cells for each siRNA concentration. Indeed, the calculated percentage of surviving cells reproduced the experimental values using different siRNA concentrations followed by IFN-α2, YNS, and YNS-α8tail treatments with high accuracy (Fig. 6A, E, and I). This suggests that cells undergoing interferon-induced cell death require a minimum number of IFNAR1 receptors for activity, which is equal to 90% of the median receptor number of the population. Next, we performed the same analysis for IFNAR2. From the APC data, we know that the number of IFNAR2 receptors is ∼25% higher than the number of IFNAR1 receptors. Therefore, subtle IFNAR2 knockdown has no effect on cell survival. From 2.5 nM siRNA, the percentage of surviving cells is in line with the predicted number from the APC data of the nontreated cells (Fig. 6B, F, and J).
Fig. 6.
Calculated versus experimental cell survival upon transfection with siRNA and interferon treatment. Experimental percentages of cell survival from the antiproliferative assay (A, B, E, F, I, and J) or antiviral assay (C, D, G, H, K, and L) after treatment with either IFN-α2 (A to D), YNS (E to H), or YNS-α8tail (I to L) are from Fig. 4. Calculated, the percentages of APC values found below (for AP activity) or above (for AV activity) the hypothesized threshold, as obtained from FACS results (i.e., surviving cells; see the text). Error bars of the experimental data represent the standard deviations of results from three independent experiments.
To evaluate the relation between receptor numbers and antiviral cell protection, we chose a different strategy to analyze our data. At the highest siRNA concentrations (5 nM IFNAR1 and 10 nM IFNAR2), the normalized antiviral amplitude was ∼70%. After defining the APC value at 70%, we determined the percentage of cells with higher APC values in the other siRNA treatments. We found that the corresponding APC values for 2.5 and 1 nM siRNA against IFNAR1 were 92.7% and 98.5%, respectively. These values are identical to the experimental data of 93.5% and ∼100% of virus-protected cells when applying 2.5 and 1 nM siRNA, respectively (Fig. 6C, G, and K). Accordingly, to maintain antiviral activity, a cell requires >20% of the median receptor number of the population (without siRNA). Similar results were obtained for the analysis performed for IFNAR2 (Fig. 6D, H, and L). Our data confirm previous suggestions that a much higher number of IFNAR receptors is required to maintain antiproliferative activity than antiviral activity (18).
Relationship between STAT phosphorylation, gene induction, and surface receptor levels.
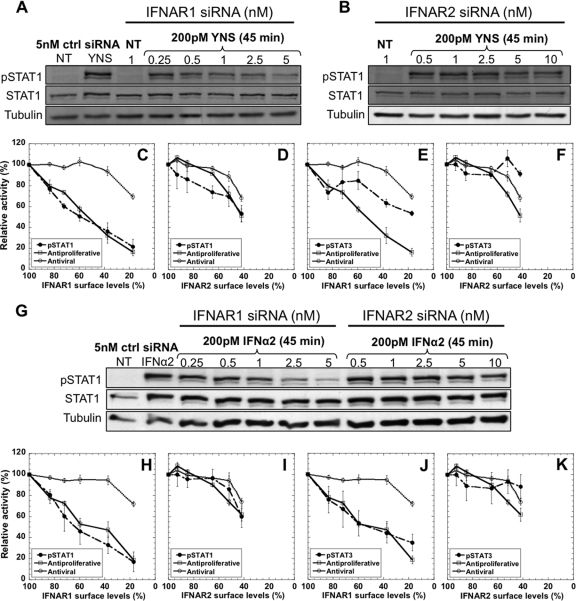
Rapid phosphorylation of STAT1 and -3 is a hallmark of interferon-induced receptor activation. WISH cells were transfected with siRNA for 48 h, followed by 200 pM of IFN-α2 WT or YNS stimulation for 45 min. pSTAT1 and -3 levels were assessed by Western blotting by using specific antibodies and normalized to total protein levels (which are not affected by the knockdown). pSTAT1 decreases in line with the increase in the concentrations of IFNAR1 siRNA (Fig. 7A and G). Although pSTAT1 levels decreased to 30% upon transfection with 5 nM IFNAR1 siRNA, the antiviral activity still remains high (70%) (Fig. 7C and H). Thus, pSTAT1 levels upon interferon stimulation are more than adequate to elicit an antiviral response. Knockdown of IFNAR2 had less of an effect on pSTAT1 levels than knockdown of IFNAR1 (Fig. 7B and G). A decrease in pSTAT1 was observed only after transfection with the three highest IFNAR2 siRNA concentrations, similar to the trend of the antiproliferative and antiviral response levels (Fig. 7D and I). IFNAR1 knockdown also led to less (with YNS treatment) or similar (with IFN-α2 treatment) reduction in pSTAT3 levels compared to the decrease in pSTAT1 (Fig. 7E and J), while IFNAR2 knockdown had only a minor effect on the phosphorylation of STAT3 (Fig. 7F and K).
Fig. 7.
pSTAT1 and pSTAT3 levels upon transfection with siRNA followed by treatment with YNS or IFN-α2. pSTAT1 protein levels as induced by 200 pM of YNS or IFN-α2, after 48 h transfection with either control (ctrl) siRNA or various concentrations of IFNAR1 (A and G) or IFNAR2 (B and G) siRNAs. The same amounts of total protein extracts were loaded per lane (30 μg) and verified by α-tubulin levels. NT, nontreated. Correlations between pSTAT1 levels, antiproliferative activity, and antiviral activity upon IFNAR1 or IFNAR2 downregulation are shown in panels C and D for YNS treatment and panels H and I for IFN-α2 treatment. pSTAT1 levels were normalized to STAT1 total protein. Correlations between pSTAT3 levels, antiproliferative activity, and antiviral activity upon IFNAR1 or IFNAR2 downregulation are shown in panels E and F for YNS treatment and panels J and K for IFN-α2 treatment. pSTAT3 levels were normalized to STAT3 total protein. Error bars represent the standard deviations of results from at least two independent experiments.
STAT phosphorylation leads to gene induction. We examined by qPCR the change in expression of ISGs after 48 h of receptor knockdown followed by 8 or 36 h of treatment with 50 pM of IFN-α2 WT or YNS. This concentration is saturating for both AV and AP activities when using YNS; however, it is saturating for only AV activity when using IFN-α2 and is below the concentration required to initiate the AP response. Therefore, this interferon concentration maximizes differential responsiveness of IFN-α2 WT versus YNS. The expression of PKR (EIF2AK2), CXCL11, interleukin 8 (IL-8), and tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) is highly upregulated by interferon. PKR is a critical component of the cellular antiviral response induced by IFNs, whereas CXCL11, IL-8, and TRAIL are related to the antiproliferative activity of interferon (differential induction by IFN-β, after prolonged time) (4, 38). The decreases in PKR mRNA expression upon transfection with IFNAR1 siRNA were similar whether IFN-α2 WT or YNS was used and were not influenced by the time of interferon treatment (Fig. 8A). The decrease in PKR expression followed the reduction in the antiviral activity (for example, 5 nM siRNA reduced expression and antiviral activity by ∼30%). Conversely, the expression of CXCL11, IL-8, and TRAIL upon IFNAR1 knockdown closely followed the antiproliferative response and the level of IFNAR1 surface receptor (reduction of ∼85% upon treatment with 5 nM IFNAR1 siRNA) (Fig. 8B). Figure 8C demonstrates the direct correlation between the reduction in expression of PKR and IL-8 and the decrease in antiviral and antiproliferative activities, respectively.
Fig. 8.
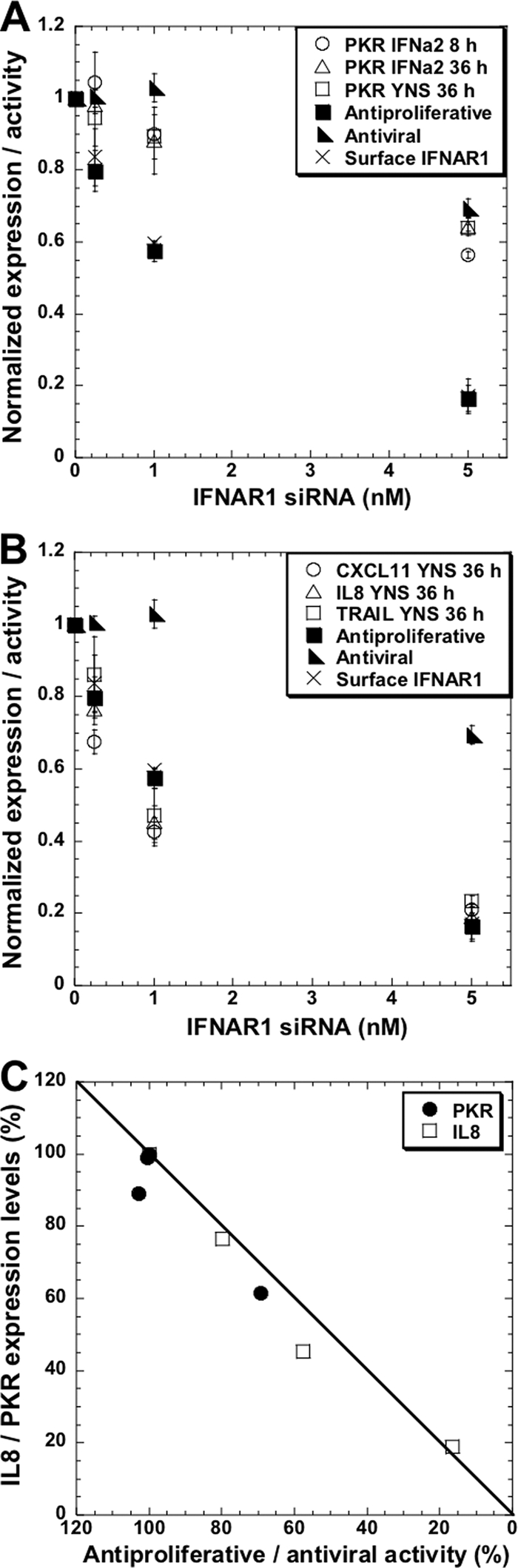
Transcription levels of interferon-stimulated genes upon IFNAR1 knockdown. The expression levels of several interferon-stimulated genes upon 48 h of transfection with IFNAR1 siRNA followed by 8 or 36 h of 50 pM WT IFN-α2 or YNS treatments were monitored by qPCR. (A) A minor decrease in PKR induction upon IFNAR1 knockdown. Notice that the decrease in PKR expression is in line with the reduction in the antiviral activity. (B) The decrease in the induction of several antiproliferative genes (the CXCL11, IL-8, and TRAIL genes) is in line with both the reduction in antiproliferative activity and the reduction in IFNAR1 receptor levels. (C) A linear correlation between the expression of an antiviral gene (the PKR gene) or an antiproliferative gene (the IL-8 gene) with their related biological responses. Error bars represent the standard deviations of results from at least two independent experiments.
Relationship between surface expression and antiproliferative activity in six additional cell lines.
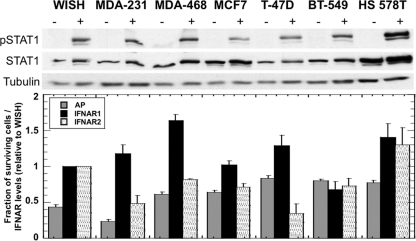
To more generally relate between receptor numbers and antiproliferative response, we determined IFNAR1 and IFNAR2 surface expression levels, measured pSTAT1 activation, and determined the antiproliferative amplitudes of response and EC50s for six breast cancer cell lines from the NCI60 panel of human tumor cells. The cells were treated with serial dilutions of four different IFNs (IFN-α2, IFN-β, YNS, and YNS-α8tail). The EC50s differed ∼20-fold between the cell lines (Table 4), with YNS-α8tail being consistently the most active interferon variant (>100-fold more active than IFN-α2). However, the percentages of responding cells were very different for the different cell lines independent of the type of interferon used. While the percentage of surviving cells was reduced by 75% for MDA-MB-231, for T-47D and BT-549 the reduction was only ∼20% (Fig. 9). Thus, the effect of interferon on growth of these cells is small. We performed FACS to measure IFNAR1 and IFNAR2 levels for these cell lines and also measured their ability to activate STAT1 after interferon stimulation. No direct correlations could be found: even though three out of the six breast cancer cell lines displayed very low levels of interferon-antiproliferative responsiveness (T-47D, BT-549, and HS 578T), this did not correlate directly with receptor expression or pSTAT1 levels (as measured after stimulation with 200 pM of YNS for 45 min) (Fig. 9). These results suggest that factors in addition to receptor numbers determine the scale of responsiveness for interferon-induced cell death.
Table 4.
EC50s obtained for wild-type and mutant IFN-α2 and IFN-β for antiproliferative activity of WISH cells and six breast cancer cell lines
| Interferon | EC50 (nM)a |
||||||
|---|---|---|---|---|---|---|---|
| WISH | MDA-MB-231 | MDA-MB-468 | MCF7 | T-47D | BT-549 | HS 578T | |
| IFN-α2 | 3.6 | 0.29 | 0.69 | 0.73 | 0.4 | 4.2 | 12.9 |
| IFN-β | 0.057 | 0.014 | 0.035 | 0.044 | 0.02 | 0.04 | 0.017 |
| YNS | 0.023 | 0.0038 | 0.0048 | 0.015 | 0.038 | 0.0074 | 0.009 |
| YNS-α8tail | 0.012 | 0.0014 | 0.0031 | 0.014 | 0.0029 | 0.0082 | 0.004 |
EC50s were determined from dose-response curves, fitted to equation 2.
Fig. 9.
Receptor levels, antiproliferative response, and pSTAT1 levels in WISH cells and six breast cancer cell lines. Antiproliferative (AP) activity is displayed as the fraction of surviving cells, which is the cell density with a saturating concentration of YNS interferon divided by the cell density without interferon. Relative cell growth was measured after 72 h of interferon treatment. Surface levels of IFNAR1 and IFNAR2 subunits were taken from Table 3. STAT1 phosphorylation was induced by 200 pM of YNS and detected by Western blot analysis using Tyr(P)701-STAT1-specific antibody. Total STAT1 protein and α-tubulin levels of the same blots were detected by specific general STAT1 and α-tubulin antibodies. The same amounts of total protein extracts were loaded per lane (16 μg). No normalization of these data was done, as STAT1 basal levels differ between cell lines. Error bars represent the standard deviations of results from at least two independent experiments.
DISCUSSION
One of the more intriguing questions in biology is how seemingly similar events result in different biological outcomes. An example for such a paradigm is the interaction of interferon with its two receptors IFNAR1 and IFNAR2. The only mechanism for type I IFNs to generate a biological response is by forming a ternary complex with both receptor subunits. This in turn results in the initiation of a cascade of events that leads to a massive change in transcription regulation, which in WISH cells results in the initiation of antiviral and antiproliferative responses (Fig. 10). In this study, we concentrated our efforts in providing a very detailed analysis of the relationship between ligand and receptor levels for the same cell line. The only variances between experiments were the binding affinities of interferon to each of the receptor subunits and the interferon and receptor concentrations. While the importance of the ligand binding affinity was thoroughly investigated, quantitative studies regarding the significance of receptor numbers in determining activity are scarce. Here, we show that by varying the concentrations of IFNAR1- or IFNAR2-specific siRNAs, we were able to create a well-defined and reproducible gradient of average surface receptor levels with minimal cell disturbances. When using the FACS data to analyze the receptor concentration on individual cells, a significant variation in receptor numbers was observed. Comparing FACS data to other quantification methods showed that the recorded variance in protein levels is real (Fig. 5A to C). Previous work has suggested that the variance is expected to be larger for low-copy-number proteins like IFNAR (2, 30, 37). Our data support that more than 99% of WISH cells harbor a critical threshold of IFNAR to elicit antiviral activity, whereas only ∼60% of the cells have sufficient receptor numbers to elicit an antiproliferative response (Fig. 6). The knockdown experiments clearly show that an antiproliferative response requires a much larger number of ternary complexes than an antiviral response. Notably, decreasing the average IFNAR1 levels by 85% reduces the antiproliferative activity to the same extent, while the effect on the antiviral activity was fairly minor, with a decrease of only 30%. Subtler knockdowns had no effect on the antiviral activities of IFNs, while antiproliferative activities decreased. Taking into account the extremely low concentrations at which antiviral activity is observed (EC50 for IFN-α2 is in the pM range, while EC50 for antiproliferative activity is in the nM range), one may conclude that only a few receptors have to be occupied to initiate an antiviral state, while most receptors have to be bound to initiate an antiproliferative response (see Fig. 10 for a proposed model of action).
To determine whether an increase in interferon binding affinity to either one or both receptors will compensate for the reduction of receptor numbers, we repeated the siRNA experiments using IFN-α2 WT, YNS, and YNS-α8tail. Upon receptor knockdown, the antiviral EC50s of YNS and YNS-α8tail were only slightly lower than that of the WT, whereas the antiproliferative EC50 of these tighter-binding IFNs were 100- to 300-fold lower than that of IFN-α2. Reducing the number of receptors (by siRNA) had basically no effect on the EC50s of the tightest-binding YNS-α8tail variant (and had an effect only on the number of responding cells), a small effect for YNS, and a larger effect (5-fold increase) when applying wild-type IFN-α2. This suggests that the differential effect between IFNs is increased at reduced receptor levels. These results are in line with previous measurements on artificial membranes, where kinetic stabilization of the ternary complex (increased half-life) could be attributed to either increased receptor concentration or increased ligand binding affinity, which contribute to increased rebinding (avidity), while increased ligand concentration cannot change the half-life of the receptor complex. Signaling is a function of both the lifetime of the ternary complex and the number of binding events (16, 18). Neither the interferon subtype nor the applied concentration could change the percentage of responding cells, suggesting a threshold of ternary complexes determining whether a specific cell will respond to interferon (with the threshold being higher for AP than for AV activity). These results are complementary to those of Moraga et al., who demonstrated that elevated receptor numbers resulted in increased amplitude of antiproliferative response, a decrease in EC50s, and an elimination of the differential IFN-β versus IFN-α2 activity (29).
The antiviral and antiproliferative responses are driven by the canonical JAK-STAT signaling resulting in ISG activation. Here, we show a very tight relation between receptor knockdown; reduction of STAT1 phosphorylation; a decrease in TRAIL, CXCL11, and IL-8 gene expression; and reduction of the percentage of cells undergoing antiproliferation (the relation is particularly close for IFNAR1 knockdown). Conversely, antiviral potency and PKR gene expression (a gene related to antiviral function) are much less affected by receptor knockdown. From these data, one could assume that pSTAT1 and pSTAT3 activation is tightly linked to the antiproliferative action of interferon, while only minute amounts of these pSTATs are sufficient to induce the antiviral state. Further support for this hypothesis comes from the EC50s for PKR induction, which are 3.5 and 0.2 pM for IFN-α2 and YNS, respectively, while the EC50s for TRAIL induction are 210 and 2.5 pM (similar values were determined for CXCL11). Thus, one may suggest that low pSTAT1 levels are sufficient for PKR but not for TRAIL induction. Still, this hypothesis contradicts the measured EC50 for pSTAT1 activation by IFN-α2 compared to that of YNS (80 pM versus 40 pM, respectively) (18), which is in line with the difference in antiviral EC50s of these two mutants but much below their antiproliferative EC50 difference (which is ∼100-fold). Thus, one has to concede that we do not yet fully understand the roles of pSTAT activation in differential signaling.
No apparent correlation between receptor numbers, STAT1 phosphorylation, and the antiproliferative effect was found when we examined six breast cancer cell lines, including cell lines with low and high levels of responsiveness to interferon (Fig. 9). This finding is in line with a previous study (26), where no clear relation between IFNAR receptor numbers and IFN-α2 and IFN-β signaling potencies was observed among several cell lines. We currently do not have a good explanation for this phenomenon, which may relate to additional cell line-specific parameters involved in interferon signaling. Although we assume that our conclusions regarding the role played by interferon receptors on differential signaling are general, the systematic quantification is valid on a given cell line and cannot be simply transferred to other cell lines. It remains to be seen whether studying additional cell lines with various levels of receptor numbers and different ratios between the two subunits will reveal a different profile of reduction in activity upon their downregulation. In this context, studies in hepatocellular carcinoma and CD34-positive cells suggested that also in those systems, surface receptor concentration is important to the efficiency of interferon as a cancer drug (7, 12).
It has been previously shown that surface IFNAR1 and IFNAR2 levels decrease by ∼60% upon interferon induction without siRNA (14). To investigate how siRNA will affect interferon-induced downregulation (which may relate to signaling), we repeated the FACS experiment after 48 h of transfection with 1 nM IFNAR1-specific siRNA and 1 h of treatment with 200 pM YNS. Interferon treatment reduced the IFNAR1 levels to ∼40% of that of the pre-interferon-treated cells, independent of whether this level was already reduced by siRNA or not (data not shown). In other words, interferon-induced downregulation works on top of the siRNA knockdown. These results suggest that the analysis done in this work is valid, despite the interferon-induced downregulation of receptors.
An as-yet-unresolved issue is how to correctly subtract the APC-nonspecific signal (isotypic control in the FACS experiments) from the specific signal on a single cell, as the noise is also stochastic (thus, subtraction of an average value is meaningless). However, because the difference in signal intensity is relatively large (Fig. 5D), this issue should have a minor effect on the analysis performed here. A second issue is whether there is a correlation on the same cell between expression levels of IFNAR1 and IFNAR2. The siRNA data suggest that the answer is no, as reducing expression of one receptor has no effect on the expression of the other receptor. Still, while on average there are 25% more IFNAR2 receptors than IFNAR1 receptors (in WISH cells), on a single cell the ratio may vary.
In this work, we went from the characterization of an IFN-α2 superagonist toward a quantitative analysis of the role of interferon receptors on biological functions. The transient formation of a small number of ternary interferon receptor complexes is sufficient to promote antiviral activity through STAT signaling (Fig. 10A). Conversely, a high number of complexes, formed over a prolonged period of time, is required for antiproliferative activity. This function cannot be solely explained by STAT activation. Other, as-yet-unknown factors are expected to be involved (Fig. 10B). By single-cell analysis, we observed an intriguing correlation between stochastic receptor levels and the interferon biological activities (Fig. 10C). This is particularly conspicuous in the relation of IFNAR1 surface expression and the antiproliferative and antiviral activities of IFNs. We found that individual cells respond in a binary manner upon interferon induction; hence, the interferon response operates in a digital and not an analog mode. Although related as homogenous, the population is composed of cells with diverse surface IFNAR1 and IFNAR2 levels (according to FACS measurements, up to 50% standard deviation of the mean). Thus, according to its receptor levels, the individual cell may enter a state of ceasing growth or apoptosis upon the addition of sufficient interferon or it may keep replicating. It should be noted that the variability of surface expression levels is not inherited and that for a population the numbers are actually quite constant. Our results demonstrate that the antiviral response, which is the primary action of interferon on all cells (clearing viral infections in their initial stages), is much less sensitive to receptor number than the antiproliferative response and thus is robust and stable. In contrast, more complex activities, such as antiproliferative activity, which often are linked with apoptosis and tissue damage, need to be under tighter control. Thus, antiproliferative activity has to be more tunable and may differ between single cells to protect the organ from excessive damage. The biochemical mechanism presented here may provide a novel explanation for disengagement of tumors from interferon responsiveness, leading to cellular resistance. A similar mechanism of tuned responsiveness may also occur for other cellular receptors, particularly for low-expressed receptors, for which surface density is expected to vary significantly between individual cells.
ACKNOWLEDGMENTS
This research has received funding from the European Community's Seventh Framework Program (FP7/2007-2013) under grant agreement no. 223608.
We thank Jacob Piehler, Daniela Novick, and Darren Baker for providing us with IFNAR1-EC and the AA3 and 117.7 antibodies and Jerome Langer for critical reading of the manuscript.
We declare that we have no conflicts of interest.
Footnotes
Published ahead of print on 20 June 2011.
REFERENCES
- 1. Brierley M. M., Fish E. N. 2002. Review: IFN-alpha/beta receptor interactions to biologic outcomes: understanding the circuitry. J. Interferon Cytokine Res. 22:835–845 [DOI] [PubMed] [Google Scholar]
- 2. Cai L., Friedman N., Xie X. S. 2006. Stochastic protein expression in individual cells at the single molecule level. Nature 440:358–362 [DOI] [PubMed] [Google Scholar]
- 3. Cajean-Feroldi C., et al. 2004. Identification of residues of the IFNAR1 chain of the type I human interferon receptor critical for ligand binding and biological activity. Biochemistry 43:12498–12512 [DOI] [PubMed] [Google Scholar]
- 4. Chawla-Sarkar M., Leaman D. W., Borden E. C. 2001. Preferential induction of apoptosis by interferon (IFN)-beta compared with IFN-alpha2: correlation with TRAIL/Apo2L induction in melanoma cell lines. Clin. Cancer Res. 7:1821–1831 [PubMed] [Google Scholar]
- 5. Claudinon J., Monier M.-N., Lamaze C. 2007. Interfering with interferon receptor sorting and trafficking: impact on signaling. Biochimie 89:735–743 [DOI] [PubMed] [Google Scholar]
- 6. Cohen B., Novick D., Barak S., Rubinstein M. 1995. Ligand-induced association of the type I interferon receptor components. Mol. Cell. Biol. 15:4208–4214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Damdinsuren B., et al. 2007. Interferon alpha receptors are important for antiproliferative effect of interferon-alpha against human hepatocellular carcinoma cells. Hepatol. Res. 37:77–83 [DOI] [PubMed] [Google Scholar]
- 8. da Silva A. J., et al. 2002. Comparison of gene expression patterns induced by treatment of human umbilical vein endothelial cells with IFN-alpha 2b versus IFN-beta 1a: understanding the functional relationship between distinct type I interferons that act through a common receptor. J. Interferon Cytokine Res. 22:173–188 [DOI] [PubMed] [Google Scholar]
- 9. Grumbach I. M., et al. 1999. Activation of the Jak-Stat pathway in cells that exhibit selective sensitivity to the antiviral effects of IFN-beta compared with IFN-alpha. J. Interferon Cytokine Res. 19:797–801 [DOI] [PubMed] [Google Scholar]
- 10. Haller O., Kochs G., Weber F. 2006. The interferon response circuit: induction and suppression by pathogenic viruses. Virology 344:119–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hilkens C. M. U., Schlaak J. F., Kerr I. M. 2003. Differential responses to IFN-alpha subtypes in human T cells and dendritic cells. J. Immunol. 171:5255–5263 [DOI] [PubMed] [Google Scholar]
- 12. Ito K., et al. 2004. Initial expression of interferon alpha receptor 2 (IFNAR2) on CD34-positive cells and its down-regulation correlate with clinical response to interferon therapy in chronic myelogenous leukemia. Eur. J. Haematol. 73:191–205 [DOI] [PubMed] [Google Scholar]
- 13. Jacobs S., Hazum E., Shechter Y., Cuatrecasas P. 1979. Insulin receptor: covalent labeling and identification of subunits. Proc. Natl. Acad. Sci. U. S. A. 76:4918–4921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jaitin D. A., et al. 2006. Inquiring into the differential action of interferons (IFNs): an IFN-alpha2 mutant with enhanced affinity to IFNAR1 is functionally similar to IFN-beta. Mol. Cell. Biol. 26:1888–1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jaitin D. A., Schreiber G. 2007. Upregulation of a small subset of genes drives type I interferon-induced antiviral memory. J. Interferon Cytokine Res. 27:653–664 [DOI] [PubMed] [Google Scholar]
- 16. Jaks E., Gavutis M., Uzé G., Martal J., Piehler J. 2007. Differential receptor subunit affinities of type I interferons govern differential signal activation. J. Mol. Biol. 366:525–539 [DOI] [PubMed] [Google Scholar]
- 17. Kalie E., Jaitin D. A., Abramovich R., Schreiber G. 2007. An interferon alpha2 mutant optimized by phage display for IFNAR1 binding confers specifically enhanced antitumor activities. J. Biol. Chem. 282:11602–11611 [DOI] [PubMed] [Google Scholar]
- 18. Kalie E., Jaitin D. A., Podoplelova Y., Piehler J., Schreiber G. 2008. The stability of the ternary interferon-receptor complex rather than the affinity to the individual subunits dictates differential biological activities. J. Biol. Chem. 283:32925–32936 [DOI] [PubMed] [Google Scholar]
- 19. Lamken P., Lata S., Gavutis M., Piehler J. 2004. Ligand-induced assembling of the type I interferon receptor on supported lipid bilayers. J. Mol. Biol. 341:303–318 [DOI] [PubMed] [Google Scholar]
- 20. Laude H., La Bonnardiere C. 1984. Cytocidal effect of interferons on porcine renal cells. J. Interferon Res. 4:101–110 [DOI] [PubMed] [Google Scholar]
- 21. Leaman D. W., et al. 2003. Novel growth and death related interferon-stimulated genes (ISGs) in melanoma: greater potency of IFN-beta compared with IFN-alpha2. J. Interferon Cytokine Res. 23:745–756 [DOI] [PubMed] [Google Scholar]
- 22. Lemaire P. A., Anderson E., Lary J., Cole J. L. 2008. Mechanism of PKR activation by dsRNA. J. Mol. Biol. 381:351–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lutfalla G., et al. 1995. Mutant U5A cells are complemented by an interferon-alpha beta receptor subunit generated by alternative processing of a new member of a cytokine receptor gene cluster. EMBO J. 14:5100–5108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Manche L., Green S. R., Schmedt C., Mathews M. B. 1992. Interactions between double-stranded RNA regulators and the protein kinase DAI. Mol. Cell. Biol. 12:5238–5248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Marchetti M., et al. 2006. Stat-mediated signaling induced by type I and type II interferons (IFNs) is differentially controlled through lipid microdomain association and clathrin-dependent endocytosis of IFN receptors. Mol. Biol. Cell 17:2896–2909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Marijanovic Z., Ragimbeau J., van der Heyden J., Uzé G., Pellegrini S. 2007. Comparable potency of IFNalpha2 and IFNbeta on immediate JAK/STAT activation but differential down-regulation of IFNAR2. Biochem. J. 407:141–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Meng F., et al. 2011. Alteration of interferon-α/β receptors in chronic hepatitis B patients. J. Clin. Immunol. [Epub ahead of print.] doi:10.1007/s10875-011-9518-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mitsui Y., Senda T. 1997. Elucidation of the basic three-dimensional structure of type I interferons and its functional and evolutionary implications. J. Interferon Cytokine Res. 17:319–326 [DOI] [PubMed] [Google Scholar]
- 29. Moraga I., Harari D., Schreiber G., Uzé G., Pellegrini S. 2009. Receptor density is key to the alpha2/beta interferon differential activities. Mol. Cell. Biol. 29:4778–4787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Newman J. R. S., et al. 2006. Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature 441:840–846 [DOI] [PubMed] [Google Scholar]
- 31. Pearce K. H., Jr., Cunningham B. C., Fuh G., Teeri T., Wells J. A. 1999. Growth hormone binding affinity for its receptor surpasses the requirements for cellular activity. Biochemistry 38:81–89 [DOI] [PubMed] [Google Scholar]
- 32. Pestka S., Krause C. D., Walter M. R. 2004. Interferons, interferon-like cytokines, and their receptors. Immunol. Rev. 202:8–32 [DOI] [PubMed] [Google Scholar]
- 33. Pfeffer L. M. 1997. Biologic activities of natural and synthetic type I interferons. Semin. Oncol. 24:S9–63-S9-69 [PubMed] [Google Scholar]
- 34. Piehler J., Roisman L. C., Schreiber G. 2000. New structural and functional aspects of the type I interferon-receptor interaction revealed by comprehensive mutational analysis of the binding interface. J. Biol. Chem. 275:40425–40433 [DOI] [PubMed] [Google Scholar]
- 35. Piehler J., Schreiber G. 1999. Biophysical analysis of the interaction of human ifnar2 expressed in E. coli with IFNalpha2. J. Mol. Biol. 289:57–67 [DOI] [PubMed] [Google Scholar]
- 36. Platanias L. C. 2005. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 5:375–386 [DOI] [PubMed] [Google Scholar]
- 37. Raj A., van Oudenaarden A. 2008. Nature, nurture, or chance: stochastic gene expression and its consequences. Cell 135:216–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rani M. R., et al. 1996. Characterization of beta-R1, a gene that is selectively induced by interferon beta (IFN-beta) compared with IFN-alpha. J. Biol. Chem. 271:22878–22884 [DOI] [PubMed] [Google Scholar]
- 39. Roisman L. C., Piehler J., Trosset J. Y., Scheraga H. A., Schreiber G. 2001. Structure of the interferon-receptor complex determined by distance constraints from double-mutant cycles and flexible docking. Proc. Natl. Acad. Sci. U. S. A. 98:13231–13236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Roos G., Leanderson T., Lundgren E. 1984. Interferon-induced cell cycle changes in human hematopoietic cell lines and fresh leukemic cells. Cancer Res. 44:2358–2362 [PubMed] [Google Scholar]
- 41. Rosenblum M. G., et al. 1990. Growth inhibitory effects of interferon-beta but not interferon-alpha on human glioma cells: correlation of receptor binding, 2′,5′-oligoadenylate synthetase and protein kinase activity. J. Interferon Res. 10:141–151 [DOI] [PubMed] [Google Scholar]
- 42. Rubinstein M., Pestka S. 1981. Purification and characterization of human leukocyte interferons by high-performance liquid chromatography. Methods Enzymol. 78:464–472 [DOI] [PubMed] [Google Scholar]
- 43. Severa M., et al. 2006. Differential responsiveness to IFN-alpha and IFN-beta of human mature DC through modulation of IFNAR expression. J. Leukoc. Biol. 79:1286–1294 [DOI] [PubMed] [Google Scholar]
- 44. Slutzki M., Jaitin D. A., Yehezkel T. B., Schreiber G. 2006. Variations in the unstructured C-terminal tail of interferons contribute to differential receptor binding and biological activity. J. Mol. Biol. 360:1019–1030 [DOI] [PubMed] [Google Scholar]
- 45. Stark G. R., Kerr I. M., Williams B. R., Silverman R. H., Schreiber R. D. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227–264 [DOI] [PubMed] [Google Scholar]
- 46. Theofilopoulos A. N., Baccala R., Beutler B., Kono D. H. 2005. Type I interferons (alpha/beta) in immunity and autoimmunity. Annu. Rev. Immunol. 23:307–336 [DOI] [PubMed] [Google Scholar]
- 47. Uzé G., et al. 1994. Domains of interaction between alpha interferon and its receptor components. J. Mol. Biol. 243:245–257 [DOI] [PubMed] [Google Scholar]
- 48. Velazquez L., Fellous M., Stark G. R., Pellegrini S. 1992. A protein tyrosine kinase in the interferon alpha/beta signaling pathway. Cell 70:313–322 [DOI] [PubMed] [Google Scholar]
- 49. Wagner T. C., et al. 2004. Interferon receptor expression regulates the antiproliferative effects of interferons on cancer cells and solid tumors. Int. J. Cancer 111:32–42 [DOI] [PubMed] [Google Scholar]
- 50. Zheng L., Baumann U., Reymond J.-L. 2004. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res. 32:e115. [DOI] [PMC free article] [PubMed] [Google Scholar]