Abstract
The mutant form of lamin A responsible for the premature aging disease Hutchinson-Gilford progeria syndrome (termed progerin) acts as a dominant negative protein that changes the structure of the nuclear lamina. How the perturbation of the nuclear lamina in progeria is transduced into cellular changes is undefined. Using patient fibroblasts and a variety of cell-based assays, we determined that progerin expression in Hutchinson-Gilford progeria syndrome inhibits the nucleocytoplasmic transport of several factors with key roles in nuclear function. We found that progerin reduces the nuclear/cytoplasmic concentration of the Ran GTPase and inhibits the nuclear localization of Ubc9, the sole E2 for SUMOylation, and of TPR, the nucleoporin that forms the basket on the nuclear side of the nuclear pore complex. Forcing the nuclear localization of Ubc9 in progerin-expressing cells rescues the Ran gradient and TPR import, indicating that these pathways are linked. Reducing nuclear SUMOylation decreases the nuclear mobility of the Ran nucleotide exchange factor RCC1 in vivo, and the addition of SUMO E1 and E2 promotes the dissociation of RCC1 and Ran from chromatin in vitro. Our data suggest that the cellular effects of progerin are transduced, at least in part, through reduced function of the Ran GTPase and SUMOylation pathways.
INTRODUCTION
The nuclear lamina provides an architectural framework that defines the size, shape, and physical properties of the nucleus (29). A critical function of the nuclear lamina is to provide a scaffold for chromatin attachment, and there is a growing body of evidence linking the nuclear lamina to the regulation of gene expression and chromosome positioning within interphase cells (49). The nuclear periphery, including the region proximal to the lamina, is rich in heterochromatin and provides a nuclear subcompartment that promotes transcriptional silencing (19). The mechanisms responsible for transcriptional silencing associated with the lamina appear to involve epigenetic regulation and modulation of the higher-order chromatin structure (2). Other functions of the lamina include roles in DNA replication and apoptosis (22, 29). The principal components of the lamina are lamin A/C and lamin B, which are encoded by the LMNA and LMNB genes, respectively (22, 29). More than 300 mutations in LMNA have been described (http://www.umd.be/LMNA/) and have been linked to 12 diseases collectively known as laminopathies. These diseases include dilated cardiomyopathy with conduction defects (DCM-CD), familial partial lipodystrophy (FPLD), atypical Werner's syndrome, Emery-Dreifuss muscular dystrophy (EDMD), and Hutchinson-Gilford progeria syndrome (HGPS) (9, 70, 77).
The nuclear lamina also provides a scaffold for organizing nuclear pore complexes (NPCs) within the nuclear membrane (1). NPCs span the nuclear lamina and both the inner and outer nuclear membranes and serve as conduits for nuclear import and export (73). Nucleoporins that comprise the NPC are organized into subcomplexes that disassemble and reassemble during each round of cell division. The nuclear side of the NPC contains a structure that has the appearance of a basket by electron microscopy (62). The nuclear basket contains ∼16 copies of TPR, a 267-kDa coiled-coil protein described originally in the context of its fusion with the MET oncogene (18, 54). The nuclear basket is a multifunctional scaffold, providing binding sites for import and export receptors, attachment sites for chromatin, a platform for RNP assembly and mRNA quality control, and a location for tethering enzymes that regulate SUMOylation (5, 25, 26, 31, 66, 71, 75, 82).
The interaction of nuclear transport receptors with their respective cargoes is regulated by Ran, a small GTPase which is a member of the Ras superfamily. Conformational changes in Ran induced by GTP binding control the selective disassembly and assembly of import and export complexes, respectively (56). The nucleotide state of Ran is regulated principally by two factors, a GTPase-activating protein (RanGAP) and a nucleotide exchange factor (RCC1). RanGAP stimulates the hydrolysis of GTP by Ran at the NPC and in the cytoplasm (6), and RCC1 promotes GDP-GTP exchange in the nucleus (7, 51, 52). RCC1 binds chromatin, and exchange activity is stimulated by nucleosomes in vitro (51). Ran exits the nucleus bound to transport receptors, while reentry into the nucleus is mediated by NTF2 (61, 69). The compartmentalization and activities of RCC1 (nuclear) and RanGAP (NPC/cytoplasmic), nuclear import mediated by NTF2, and the steady-state distribution of shuttling nuclear transport receptors generate two nuclear/cytoplasmic Ran gradients: a RanGTP gradient estimated to be extremely steep (>500:1) (28) and a Ran protein gradient that at steady state is ∼3:1 (38). The Ran import factor NTF2 is essential in Saccharomyces cerevisiae, indicating that the nuclear concentration of Ran is critical for cell viability (16, 53, 55).
Biochemical analysis of RanGAP led to the discovery that it is covalently attached to the small ubiquitin-like modifier (SUMO) (27, 45, 46). At least three paralogues of SUMO are expressed in mammals, SUMO1, SUMO2, and SUMO3 (4, 63). SUMO1 is ∼50% identical to SUMO2 and SUMO3, while SUMO2 and SUMO3 are 96% identical and may be functionally equivalent. SUMOylation requires an E1-activating enzyme, the SAE1/SAE2 heterodimer, and an E2-conjugating enzyme, Ubc9 (33). SUMO E1 and E2 enzymes are predominantly nuclear, and Ubc9 import is mediated by importin 13 (48). There is also a pool of Ubc9 that is stably associated with the NPC (58, 82). Additionally, the NPC provides binding sites for enzymes that cleave SUMO from modified proteins (SENPs) (14, 50). These observations imply that the NPC plays a central role in organizing the localization and activity of the SUMOylation machinery.
The premature aging disease HGPS is caused predominantly by a de novo C-to-T substitution that activates a cryptic donor splice site within exon 11 of lamin A, resulting in an internal deletion of 50 amino acids (aa) (24). Because the “progerin” form of lamin A (Δ50) lacks the site cleaved by the endoprotease Zmpste24, progerin remains membrane tethered and acts as a dominant negative (DN) protein that disrupts the structure of the lamina (24). While the genetic and protein-processing defects in HGPS are clearly defined, the mechanisms by which progerin mediates defects at the cellular level are largely unknown. Using fibroblasts from progeria patients and transfection approaches in HeLa cells, we show that defects in the nuclear lamina are transduced to the Ran GTPase system. Progerin expression prevents the nuclear localization of Ubc9, disrupts the Ran gradient, inhibits TPR import, and reduces trimethylation on lysine 9 of histone H3 (H3K9me3). These progerin-induced phenotypes are restored by the nuclear localization of Ubc9, suggesting that defects in SUMOylation and Ran-dependent transport might play important roles in HGPS.
MATERIALS AND METHODS
Cell culture.
Primary human fibroblasts from HGPS patients (AGO1972, AG11498, and AGO3199) that express the progerin form of lamin A and primary fibroblasts from a clinically unaffected father (AGO8469) of an HGPS patient were obtained from the Coriell Cell Repository (Camden, NJ). We refer to these fibroblasts as either “HGPS” or “Normal” followed by the last four digits of the Coriell designation. Primary fibroblasts were grown at 37°C in 5% CO2 in minimal essential medium (MEM) (Gibco/Invitrogen, Carlsbad, CA) containing 15% fetal bovine serum, 1× MEM vitamin solution (HyClone, Logan, UT), and 1 mM sodium pyruvate (Gibco/Invitrogen, Carlsbad, CA). The analysis was done on cells at passages 10 to 20. The progerin-induced cellular phenotypes described in this study, and nuclear morphology defects described previously by other groups, become more penetrant during later cell passages. The reason for the variation in penetrance is not clear, but it does not seem to involve senescence, because the fraction of cells expressing senescence-associated β-galactosidase remains low (∼5%) and is similar in HGPS and normal fibroblasts (30; our unpublished data). It is possible that the cellular response to progerin is influenced by a parameter related to growth in culture, given that laminopathies as a group are known to be highly context specific and of variable penetrances in vivo.
HeLa cells were grown at 37°C in 5% CO2 in Dulbecco's modified Eagle's medium (DMEM) (Gibco/Invitrogen, Carlsbad, CA) containing 5% newborn calf serum (Gibco/Invitrogen, Carlsbad, CA), 5% fetal bovine serum (Gibco/Invitrogen, Carlsbad, CA, or Atlanta Biologicals, Lawrenceville, GA), and 1 mM sodium pyruvate (Gibco/Invitrogen, Carlsbad, CA). Cos7 cells were grown in DMEM with 10% fetal bovine serum. Farnesyl transferase inhibitor (FTI) treatments were performed with FTI-277 (Calbiochem/EMD, Gibbstown, NJ) dissolved in dimethyl sulfoxide (DMSO). Cells were treated with 3 μM FTI-277 (41) or 0.1% dimethyl sulfoxide for 4 days and processed for immunofluorescence (IF) microscopy. Lopinavir from the NIH AIDS Research and Reference Reagent Program (Germantown, MD) (catalog number 9481; a gift from D. Rekosh) was dissolved in DMSO. Cells were treated with 20 and 40 μM lopinavir for 4 days and processed for immunoblotting and IF microscopy.
siRNA and plasmids.
Small interfering RNAs (siRNAs) for NTF2 and TPR were obtained from Santa Cruz Biotechnology (Santa Cruz, CA) and were transfected by using Oligofectamine (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. HeLa cells were transfected in 60-mm dishes, split onto glass coverslips at 24 h posttransfection, grown for an additional 48 h, and processed for IF microscopy.
An expression plasmid encoding the progerin form of lamin A was generated by removing the sequence that encodes amino acids 607 to 656. Oligonucleotides matching the relevant regions of exon 11 and exon 12 were used with the QuikChange II site-directed mutagenesis kit (Stratagene/Agilent Technologies, Santa Clara, CA) with a plasmid encoding hemagglutinin (HA)-lamin A as the template (kindly provided by Brian Burke). The cysteine that undergoes farnesylation (Cys611) was changed to serine in progerin by use of the QuikChange II site-directed mutagenesis kit. mCherry-SUMO2 was made by cloning SMO2-GG from pEGFP-SUMO2-GG (kindly provided by Mary Dasso) into pmCherry made by replacing the green fluorescent protein (GFP) from pEGFP with mCherry. pCMV5-FLAG-Ubc9, pCMV5-FLAG-Ubc9-C93S and pCMV5-SENP2-catalytic domain (aa 317 to 590) were all provided by David Wotton. Transport signal fusions to Ubc9 were generated by using the nuclear localization signal (NLS) from the simian virus 40 (SV40) large T antigen and the nuclear export signal (NES) from the protein kinase inhibitor (PKI). We determined that progerin does not inhibit nuclear import mediated by the SV40 NLS (J. B. Kelley and B. M. Paschal, unpublished observations). FLAG-NLS-Ubc9 was made by amplifying Ubc9 from pCMV5-FLAG-Ubc9 to introduce BamHI and XhoI sites and was then cloned into a pcDNA-FLAG-NLS backbone. FLAG-NES-Ubc9 was generated by a similar strategy. The importin β binding (IBB)-β-galactosidase (IBB-β-Gal) construct was provided by Larry Gerace. pEGFP-RCC1 (RCC1-GFP) was a gift from Yixian Zheng. Plasmids were transfected into HeLa cells by using Transfectin (Bio-Rad) according to the manufacturer's protocol and examined at 24 h posttransfection.
Immunofluorescence microscopy.
Cells were grown on glass coverslips, fixed with 3.7% formaldehyde for 20 min, and permeabilized in 0.2% Triton X-100 for 5 min. For SUMO detection, cells were fixed in 2% formaldehyde for 30 min and extracted with acetone at −20°C for 3 min to release soluble SUMO. Coverslips were incubated in primary antibody diluted in IF microscopy blocking buffer (1× phosphate-buffered saline [PBS], 2% bovine serum albumin [BSA], 2% newborn calf serum) for ≥1 h. Antibodies to the following proteins were used for IF: Ran (monoclonal antibody [MAb], catalog number 610341; BD Biosciences), lamin A (polyclonal antibody [pAb], catalog number PRB-113c; Covance), H3K9me3 (pAb, catalog number ab8898; Abcam), TPR (pAb [25]; kindly provided by Larry Gerace), Nup153 MAb SA1 (8), p62 (catalog number 610498; BD Biosciences), RanBP2 (kindly provided by Frauke Melchior), Ubc9 (pAb, catalog number ab33044; Abcam), SUMO1 (MAb 21C7; Zymed), SUMO2/3 (pAb SUMO-2 [Zymed] and MAb 8A2 [83]), anti-FLAG epitope (MAb M2; Sigma), OctA anti-FLAG (Santa Cruz), RanGAP (MAb 21c7; Zymed), and anti-HA (MAb 16B12 and pAb; both from Santa Cruz). The secondary antibodies for IF microscopy were diluted in blocking buffer and incubated for 1 h. The antibodies used were fluorescein isothiocyanate (FITC)-labeled donkey anti-mouse, Cy3-labeled donkey anti-mouse, FITC-labeled donkey anti-rabbit, and Cy3-labeled donkey anti-rabbit antibodies (all from Jackson Immunoresearch). Wide-field microscopy was performed with a Nikon Eclipse E800 upright microscope (Melville, NY) and recorded with a Hamamatsu C4742-95 charge-coupled device (CCD) (Bridgewater, NJ) using OpenLab software (Improvision, Lexington, MA). Figure 6C contains wide-field images. Confocal imaging was done with Zeiss 510 LSM and a Zeiss 510 Meta LSM microscopes using Zeiss software. Quantitative analysis of IF microscopy images (ratios of the nuclear concentration to the cytoplasmic concentration [N/C ratios]) was performed by using ImageJ (http://rsbweb.nih.gov/ij/) as described previously (38). The “t test: two sample assuming equal variances” function of Excel (Microsoft) was used to calculate P values, the alpha value was 0.05, and the one-tailed P value was used. Spearman's rank correlation was used to quantify the correlation of Ran N/C ratios and H3K9me3 or SUMO2/3 levels in order to account for the nonlinearity of the relationship between the data. Quantification of mCherry-SUMO2 levels was performed by using ImageJ, by outlining individual cells and corresponding nuclei and deriving the ratio of total nuclear SUMO fluorescence intensity to the total cellular SUMO fluorescence. Data were binned prior to plotting as histograms, with bin sizes stated in the figure legends.
Fig. 6.
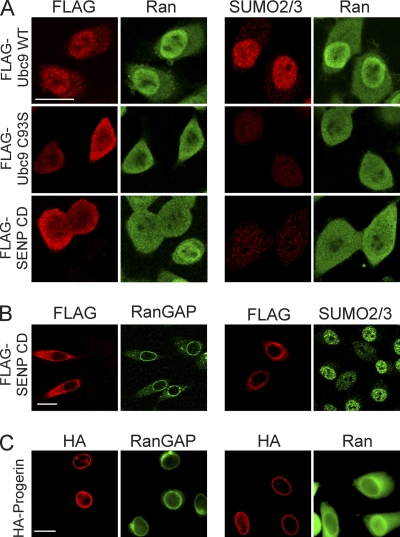
Inhibition of SUMOylation disrupts the Ran protein gradient. (A) HeLa cells were transfected with FLAG-tagged forms of WT Ubc9, a catalytic mutant of Ubc9 (C93S) that acts as a DN protein, and the SENP CD. Double-label IF microscopy for FLAG (red) and endogenous Ran (green) showed that the Ran protein gradient is disrupted by the expression of Ubc9 C93S and the SENP CD (left) under conditions where these factors reduce nuclear SUMO2/3 levels (right). (B and C) The SENP CD (B) and HA-progerin (C) do not disrupt endogenous RanGAP targeting to the NPC. Scale bars, 20 μm.
Live-cell imaging, microinjection, and fluorescence recovery after photobleaching (FRAP).
Live-cell microscopy, photobleaching, and image quantitation were performed by using a Zeiss LSM 510 Meta microscope and a Bioptech heated stage as previously described (38). Injections were performed by using an Eppendorf Femtojet/Injectman NI 2 microinjector. The nonlinear curve fit function of OriginPro (OriginLab Corp.) was used to fit the equation F = A(1−e−Bt), where f is the fluorescence intensity and t is the time after bleaching. The immobile fraction is defined as 1 − A, and the half-life (t1/2) equals ln (0.5)/−B. P values were calculated with Excel (see “Immunofluorescence microscopy”).
Immunoblotting.
SDS-PAGE and immunoblotting were performed by standard methods mostly with the same primary antibodies used for immunofluorescence microscopy, peroxidase-labeled secondary antibodies, and detection by chemiluminescence. Immunoblotting of nucleus-enriched fractions involved the permeabilization of cells with digitonin (0.01%) for 6 min on ice in the presence of 3 mM N-ethylmaleimide and protease inhibitors. The antibody to RCC1 was a mouse monoclonal antibody (KAM-CC225; StressGen). The antibody to prelamin A (sc-6214; Santa Cruz) used in lopinavir experiments is goat polyclonal antibody specific for unprocessed lamin A (57).
Chromatin-RCC1 dissociation assay.
The recombinant proteins His-Ran, glu-tathione S-transferase (GST)-Ran, GST-Ran T24N, His-SUMO2, His- SUMO2-G (nonconjugable), and His-Ubc9 and the His-Aos1/Uba2 heterodimer were expressed in Escherichia coli cells and purified according to standard methods. E1 and E2 activities were confirmed by using the recombinant SUMOylation substrate CtBP. Chromatin was prepared, according to previously established methods (65), from suspension culture HeLa cells grown at the National Cell Culture Center. Buffers were prepared as described previously (65). A frozen pellet from 1 liter of cells (approximately 2 ml) was disrupted in 20 volumes of lysis buffer with 15 strokes in a Dounce homogenizer. The chromatin fraction was collected by centrifugation (2,000 × g) and washed twice by homogenization and centrifugation using the same buffer. The pellet was then washed by homogenization and centrifugation in 20 volumes of buffer B. The pellet (approximately 0.2 ml) was resuspended in 3 volumes of buffer B using a small Dounce homogenizer. The resuspended sample was diluted (dropwise) with an equal volume of buffer B containing 0.6 M KCl and 10% glycerol, mixed end-over-end for 10 min at 4°C, aliquoted (0.2 ml) into 1.5-ml Eppendorf tubes, and centrifuged (20,000 × g). Chromatin pellets were snap-frozen in liquid nitrogen and stored at −80°C.
The chromatin pellet was thawed on ice and homogenized in the Eppendorf tube using a plastic pestle and transport buffer (20 mM HEPES, 110 mM potassium acetate, 2 mM magnesium acetate, 0.5 mM EGTA [pH 7.4]) supplemented with 5 mM MgCl2. The chromatin was centrifuged, and the homogenization was repeated with the same buffer. The chromatin was resuspended to a concentration of 30 mg/ml and incubated with recombinant His- or GST-tagged Ran (1 ng added per 4 μg chromatin) for 30 min at 25°C. The sample was collected by centrifugation, washed twice in the same buffer, and resuspended to a final concentration of 10 μg/μl. Each reaction mixture (20-μl total volume) contained 40 μg chromatin and the combinations of SUMOylation components depicted in the figures. These components included ATP (1 mM), GTP (2 μM), His-SUMO2 (0.25 μM), His-SUMO2-G (0.25 μM), E1 (1 μM), and E2 (1 μM). Reaction mixtures were incubated for 30 min at 25°C and fractionated by centrifugation in a Beckman air-driven ultracentrifuge. Supernatants and pellets were collected, and RCC1 (endogenous) and Ran (recombinant) were analyzed by immunoblotting. The data shown are representative of the results obtained from at least three experiments.
RESULTS
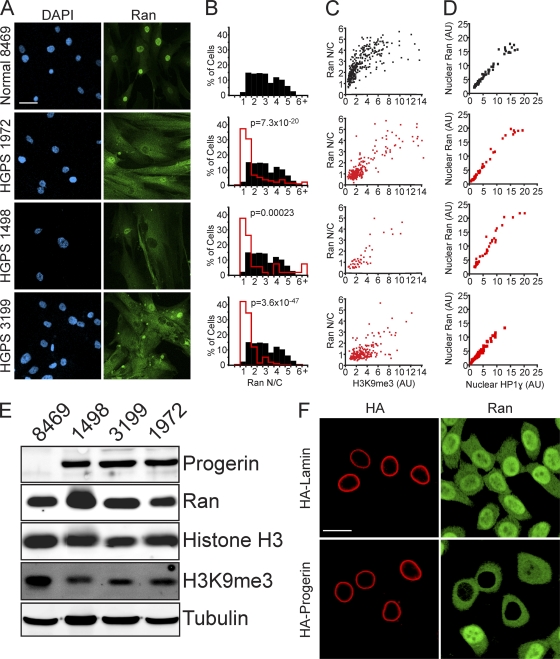
Because the formation of the Ran protein gradient depends on its nucleotide exchange factor RCC1 (72), which is enriched in heterochromatin (chromatin immunoprecipitation [12]), we considered whether the distribution of Ran might be linked to the epigenetic state of chromatin. Primary fibroblasts from progeria patients provide a setting for the testing of this model, as the heterochromatin marks H3K9me3 and H3K27me3 are reduced in HGPS (64, 68). The distribution of Ran was examined in primary fibroblasts from three progeria patients (HGPS 1972, HGPS 1498, and HGPS 3199) and, as a control, in primary fibroblasts of a similar passage number from an unaffected father of a child with progeria (Normal 8469). By IF microscopy, Ran is predominantly nuclear in Normal 8469 fibroblasts, which is the characteristic distribution of Ran, but in HGPS fibroblasts there was a striking reduction in the level of nuclear Ran and an increase in the level of cytoplasmic Ran (Fig. 1 A). In HGPS cells where the Ran distribution was defective, the pattern of Ran localization ranged from cells where the Ran appeared to equilibrate between the nucleus and cytoplasm to cells that displayed a reversal in the N/C Ran gradient.
Fig. 1.
The Ran protein gradient in interphase cells depends on the nuclear lamina and is correlated with markers of heterochromatin. (A) Ran distribution in primary fibroblasts from a control individual (Normal 8469) and three progeria patients (HGPS 1972, HGPS 1498, and HGPS 3199). DAPI, 4′,6-diamidino-2-phenylindole. (B) Histograms (bin size = 0.5) of Ran N/C ratios from control (Normal 8469) (black bars; n = 293) and HGPS patient (HGPS 1972, HGPS 1498, and HGPS 3199) (red lines; n = 264, 53, and 207, respectively) cells. (C) Ran N/C values plotted as a function of nuclear H3K9me3 levels (Spearman's rank correlation coefficient [ρ] values of 0.81 for Normal 8469, 0.74 for HGPS 1972, 0.81 for HGPS 1498, and 0.29 for HGPS 3199). (D) Nuclear Ran values plotted as a function of nuclear HP1γ levels (ρ values of 0.81 for Normal 8469, 0.74 for HGPS 1972, 0.81 for HGPS 1498, and 0.29 for HGPS 3199; n = 64, 50, 40, and 108, respectively). AU, arbitrary units. (E) Immunoblotting of primary fibroblasts. (F) Colocalization of transiently transfected HA-tagged WT lamin A and HA-progerin (red) with endogenous Ran (green) in HeLa cells. Scale bars, 20 μm.
To better assess the Ran distribution changes in HGPS cells, we combined IF microscopy with digital imaging to measure the nuclear and cytoplasmic levels in patient and normal fibroblasts (n > 50 cells per sample). Ran N/C levels were reduced in 3/3 HGPS patient cell lines (P < 0.0005), an effect that we refer to as a disruption of the Ran gradient (Fig. 1B). To our knowledge, this is the first example of a human disease that displays a disruption of the Ran gradient. By double-label IF microscopy, Ran N/C ratios were correlated with nuclear H3K9me3 levels in both patient and normal fibroblasts (Spearman's rank correlation coefficient [ρ] values of 0.81 for Normal 8469, 0.74 for HGPS 1972, 0.81 for HGPS 1498, and 0.29 for HGPS 3199), suggesting that the Ran distribution in these cells is potentially linked to the epigenetic state of chromatin (Fig. 1C). Nuclear Ran levels were also correlated with nuclear levels of heterochromatin protein 1γ (HP1γ), which binds the chromatin mark H3K9me3 (ρ values of 0.98 for Normal 8469, 0.98 for HGPS 1972, 0.96 for HGPS 1498, and 0.97 for HGPS 3199) (Fig. 1D). These correlations could reflect a chromatin regulation of the Ran N/C distribution, the Ran-dependent transport of enzymes that modify chromatin, or a combination of these processes.
By immunoblotting, Ran protein levels were similar in the HGPS (HGPS 1498, HGPS 3199, and HGPS 1972) and normal (Normal 8469) fibroblasts (Fig. 1E). An antibody that recognizes progerin (but not wild-type [WT] lamin A) was generated and used to show that comparable levels of progerin are expressed in the three HGPS fibroblast lines used in this study (Fig. 1E). All three HGPS fibroblast lines displayed a reduction in H3K9me3 levels by immunoblotting, which is consistent with data from previous reports (64, 68) and our IF microscopy.
We tested whether the ectopic expression of progerin is sufficient to disrupt the distribution of endogenous Ran in HeLa cells. By double-label IF microscopy, HA-progerin expression disrupted the Ran gradient, while at comparable expression levels, HA-lamin A did not have this effect (Fig. 1F). Cell-to-cell variation in the extent to which progerin disrupted the Ran distribution was apparent in HeLa cells, consistent with what was observed for HGPS primary fibroblasts. Cell-to-cell variation in the penetrance of progerin was noted previously by others (68), including Scaffidi and Misteli (64), who reported that 60% of HGPS 1972 cells (passages 25 to 30) had reduced levels of HP1α by IF microscopy.
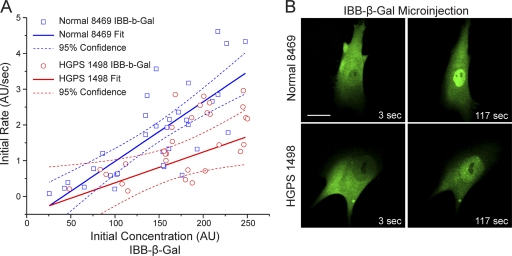
Given the central role played by Ran in nucleocytoplasmic transport (36), we examined whether the disruption of the Ran gradient in HGPS fibroblasts affects nuclear transport. We measured importin β-mediated import into the nucleus by using a microinjection assay with a fluorescent cargo that contains an importin β binding (IBB) domain (from an importin α) fused to β-galactosidase (IBB-β-Gal) labeled with Alexa Fluor 555. Because the molecular mass of the IBB-β-Gal tetramer (∼400 kDa) (44) is well above the diffusion limit of the NPC, the nuclear localization of the IBB-β-Gal reporter is strictly dependent on binding to importin β and the translocation of the importin β/IBB-β-Gal complex through the NPC. Following microinjection into the cytoplasm, IBB-β-Gal underwent rapid nuclear import, both in the Normal 8469 fibroblasts and in the HGPS 1498 fibroblasts (Fig. 2). Plotting of the initial import rate versus the initial concentration (measured in the cytoplasm of the injected cell) revealed a slight reduction in the nuclear import rate in HGPS 1498 fibroblasts (n = 30 cells). The reduced level of import in HGPS cells was observed only at the higher concentrations of IBB-β-Gal used (Fig. 2). Thus, while there is a measureable defect in importin β-dependent transport into the nucleus, the fact that the disruption of the Ran protein gradient in HGPS does not cause a large reduction in transport is unexpected.
Fig. 2.
The importin β-dependent import pathway is functional in HGPS fibroblasts. (A) Rate of importin β-dependent transport measured in living cells. A fluorescent reporter protein (Alexa 544-labeled IBB-β-galactosidase) that undergoes importin β-dependent import was injected into the cytoplasm of control (Normal 8469) and progeria (HGPS 1498) fibroblasts. For each injected cell, the initial rate was plotted versus the initial concentration. The slopes of the solid lines represent the relationship between the import rate and the reporter protein concentration. Dashed lines represent 95% confidence intervals. (B) Images of Normal 8469 and HGPS 1498 fibroblasts 3 s and 117 s after microinjection. Scale bar, 20 μm.
Import of the nucleoporin TPR is defective in HGPS.
We first considered whether the disruption of the Ran gradient in HGPS cells and in HeLa cells expressing progerin (Fig. 1) might be explained by progerin-induced defects in the NPC. Nucleoporins within the central channel of the NPC form a meshwork that acts as a permeability barrier that restricts the free exchange of proteins between the cytoplasm and the nucleus, and age-related changes in nucleoporin composition that disrupt the permeability barrier were described previously (21). It seemed plausible that Ran would leak out of the nucleus if the permeability barrier was defective in HGPS cells. We tested this possibility by cotransfecting progerin with a reporter protein (Myc-tagged pyruvate kinase [PK]) that is normally excluded from the nucleus because it lacks transport signals and is too large to diffuse through the NPC. Myc-pyruvate kinase was completely excluded from the nuclei of cells expressing progerin, which supports the conclusion that the NPC diffusion barrier is functional in this setting (data available for inspection from the authors). Additionally, a disruption of the diffusion barrier would be predicted to result in Ran equilibration between the nucleus and the cytoplasm, whereas in a subset of progerin-expressing cells the cytoplasmic level of Ran exceeded that of the nucleus (Fig. 1F).
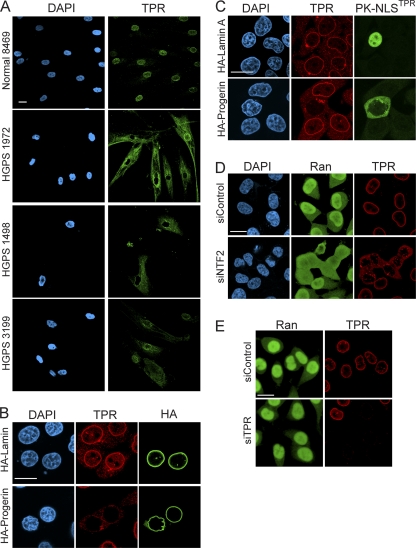
We then considered whether the Ran distribution defects might arise because structural components (i.e., nucleoporins) necessary for Ran import are missing from the NPC in HGPS cells. IF microscopy with antibodies to nucleoporins that comprise the nuclear (Nup153 and TPR), cytoplasmic (Nup358/RanBP2), and central (p62) domains of the NPC was performed. The nucleoporins Nup153, RanBP2, and p62 each localized to the NPC (data not shown). The nucleoporin TPR, however, accumulated in the cytoplasm of HGPS cells and displayed a reduced level of NPC localization (Fig. 3 A). To determine if progerin expression is sufficient to cause TPR localization to the cytoplasm, we transfected HeLa cells with HA-progerin and HA-lamin A and examined the samples by double-label IF microscopy. HA-progerin expression resulted in a clear loss of TPR “rim staining” (Fig. 3B and C). Thus, progerin expression is sufficient to disrupt the subcellular distribution of TPR.
Fig. 3.
Progerin inhibits TPR import. (A) Distribution of endogenous TPR in control (Normal 8469) and progeria (HGPS 1972, HGPS 1498, and HGPS 3199) fibroblasts. (B) HA-lamin A and HA-progerin transfection in HeLa cells costained for endogenous TPR (red) and HA (green). (C) Cotransfection of progerin and a reporter protein that contains the NLS from TPR (pyruvate kinase) (PK-NLSTPR) followed by localization of endogenous TPR (red) and the PK reporter (green). (D) siRNA-mediated knockdown of NTF2 in HeLa cells and localization of endogenous Ran (green) and TPR (red). (E) siRNA-mediated knockdown of TPR in HeLa cells with localization of endogenous Ran (green) and TPR (red). Scale bars, 20 μm.
Although TPR is a component of the NPC, it contains an NLS and undergoes Ran-dependent nuclear import following the postmitotic assembly of the NPC (5, 8, 23). While progerin could induce the cytoplasmic localization of TPR (Fig. 3A and B) by any one of several mechanisms, arguably the simplest explanation was that progerin inhibited the nuclear import of TPR. We tested this by using a PK fusion protein engineered with the 56-amino-acid NLS from TPR (PK-NLSTPR) (17). PK-NLSTPR was localized to the nucleus in cells cotransfected with HA-lamin A (Fig. 3C). In contrast, PK-NLSTPR showed a clear cytoplasmic localization when coexpressed with HA-progerin, and in the same cells there was a nuclear import defect in endogenous TPR (Fig. 3C). Because the progerin-induced TPR transport defect can be recapitulated by using the NLS from TPR, we conclude that cytoplasmic localization of TPR occurs because progerin inhibits the TPR import pathway.
The fact that the Ran N/C gradient is disrupted in HGPS cells (Fig. 1) led us to test whether the TPR import defect is a downstream consequence of the Ran defect. To test this prediction, we disrupted the Ran protein gradient by depleting the Ran import factor NTF2 in HeLa cells and analyzing the distribution of endogenous Ran and TPR by double-label IF microscopy. HeLa cells treated with siRNA to NTF2 showed a marked defect in the Ran N/C distribution, similar to the Ran localization that we observed for HGPS fibroblasts and HeLa cells expressing progerin (Fig. 1). HeLa cells that had a disrupted Ran gradient displayed a predominantly cytoplasmic distribution of TPR (Fig. 3D), again similar to that observed for HGPS cells.
Given that TPR is a key architectural component of the NPC, we tested whether the loss of TPR (independent of progerin expression) affects the Ran N/C distribution. In this experiment we used siRNA to deplete TPR and performed double-label IF microscopy for TPR and Ran. We found that the nuclear distribution of Ran was unaffected by the loss of TPR from the NPC (Fig. 3E). Thus, TPR undergoes Ran-dependent import, TPR cytoplasmic localization in HGPS cells is a consequence of the disruption of the Ran gradient, and nuclear TPR is not required for the Ran gradient.
Progerin reduces nuclear SUMO2/3 levels.
The NPC and the nuclear lamina provide anchoring sites for enzymes that regulate SUMOylation (31, 58, 82). For example, TPR orthologues in S. cerevisiae and Arabidopsis thaliana direct the subnuclear localization and/or activity of the deSUMOylating SENP enzymes (78, 84). Moreover, the SUMO-conjugating enzyme Ubc9 binds to nucleoporins on the cytoplasmic and nuclear sides of the NPC (82). Given these considerations, we postulated that nuclear SUMOylation might be altered by the progerin perturbation of the nuclear lamina structure or by the progerin inhibition of TPR import. To test this idea, we transfected HeLa cells with HA-tagged WT lamin A and HA-tagged progerin and examined the levels of endogenous SUMO1- and SUMO2/3-modified proteins by quantitative IF microscopy. Levels of SUMO1-conjugated proteins in the nucleus detected by IF were unchanged by the expression of HA-WT lamin A and HA-progerin (Fig. 4 A, top). In contrast, HA-progerin expression reduced the levels of SUMO2/3-conjugated proteins detected by IF microscopy (Fig. 4A, bottom). The effect of progerin on SUMO2/3 levels in the nucleus was validated by using HGPS patient cells, which showed a statistically significant reduction in SUMO2/3 levels compared with those of Normal 8469 fibroblasts (Fig. 4B and C). The nuclear signal for SUMO2/3 was correlated with the Ran protein gradient in HGPS cells (ρ values of 0.19 for Normal 8469, 0.82 for HGPS 1972, 0.70 for HGPS 1498, and 0.53 for HGPS 3199) (Fig. 4C and D). As was the case with Ran, there was cell-to-cell variation in HGPS cells, which ranged from normal nuclear levels of SUMO2/3 to significantly reduced levels.
Fig. 4.
SUMO2/3 levels in the nucleus are reduced in response to progerin expression. (A) Overlay of SUMO1 and SUMO2/3 distributions (red) with HA-progerin and HA-WT lamin A (green) in HeLa cells. (B) SUMO2/3 (red) and Ran (green) levels in control (Normal 8469) and progeria (HGPS 1972, HGPS 1498, and HGPS 3199) fibroblasts. (C) Histograms (bin size = 1) of nuclear SUMO2/3 levels from healthy (Normal 8469) (black bars; n = 54) and HGPS patient (HGPS 1972, HGPS 1498, and HGPS 3199) (red lines; n = 56, 65, and 101, respectively) cells. (D) Ran N/C ratio values plotted as a function of nuclear SUMO2/3 levels (ρ values of 0.19 for Normal 8469, 0.82 for HGPS 1972, 0.70 for HGPS 1498, and 0.53 for HGPS 3199). Scale bars, 20 μm.
These data suggest that progerin reduces the nuclear levels of SUMO2/3-modified proteins. Immunoblotting with HGPS cell extracts failed to reveal an obvious reduction in levels of SUMO2/3-modified proteins (n = 6 experiments) (data are available upon request). This finding might be explained by a cell population that is heterogeneous with regard to SUMO2/3 levels. Differences between the cell lines could also be obscured by the loss of the SUMO2/3 modification during handling, despite the inclusion of N-ethylmaleimide to inhibit postlysis deSUMOylation by SENPs.
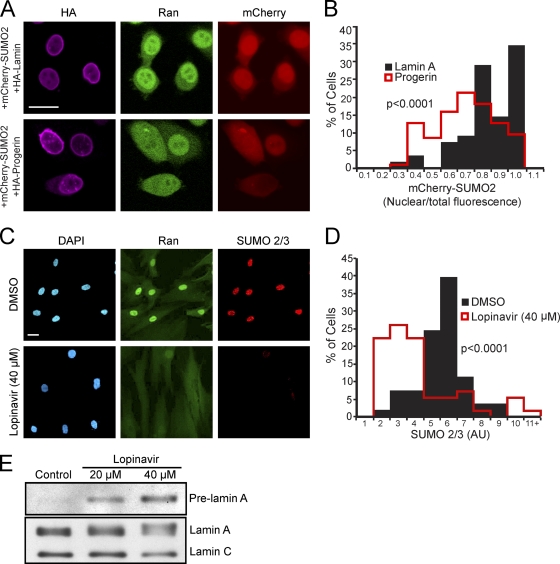
Two approaches were used to corroborate our immunofluorescence data showing that progerin reduces nuclear levels of SUMO2/3. First, we employed a fluorescent protein fusion (mCherry-SUMO2) as a direct readout of progerin effects on the SUMO2 distribution in the cell. mCherry-SUMO2 was cotransfected with HA-progerin and HA-lamin A, and the distributions of mCherry-SUMO2, HA-tagged protein, and endogenous Ran were visualized by triple labeling. We measured both nuclear fluorescence and total cellular fluorescence generated by mCherry-SUMO2 in HA-positive cells and plotted the data as a ratio (Fig. 5 A and B). The mCherry-SUMO2 fluorescence (ratio of nuclear to total fluorescence) was reduced in cells cotransfected with HA-progerin compared with cells cotransfected with HA-lamin A. This result suggests that progerin expression reduces nuclear SUMOylation or reduces the nuclear import of SUMOylated proteins or that nuclear SUMOylation is reduced by a combination of mechanisms.
Fig. 5.
Membrane attachment underlies progerin effects on SUMOylation. (A) Images from HeLa cells triple labeled for HA-progerin and HA-WT lamin A (purple), endogenous Ran (green), and mCherry-SUMO2 (red). (B) Histograms (bin size = 0.1) of Cherry-SUMO2 levels (nuclear/total) in progerin- and lamin A-transfected cells. (C) Endogenous Ran and SUMO2/3 detected in primary human fibroblasts (Normal 8469) treated with lopinavir. (D) Histograms (bin size = 1) of SUMO2/3 levels in cells treated with lopinavir or DMSO. (E) Immunoblotting of primary human fibroblasts treated with lopinavir for 4 days. Total cell extracts were probed with a pan-lamin antibody that recognizes lamin A/C and an antibody that selectively recognizes unprocessed, prelamin A. Scale bars, 20 μm (A and C).
Second, we examined SUMOylation by an approach that does not rely on transfection but instead takes advantage of an aspartyl protease inhibitor shown previously by other groups to inhibit lamin A processing (15). The HIV protease inhibitor lopinavir blocks the Zmpste24 cleavage of prelamin A, and as a consequence, prelamin A remains membrane anchored. Since the lopinavir-induced accumulation of WT prelamin A induces nuclear morphology defects similar to those reported for progerin, we reasoned that lopinavir should mimic the effects of progerin expression with regard to Ran system and SUMOylation defects. Lopinavir treatment induced the appearance of a slower-migrating form of lamin A that was recognized by a prelamin A-specific antibody (Fig. 5E). By double-label IF we found that lopinavir treatment disrupted the Ran gradient and reduced nuclear levels of endogenous SUMO2/3 (Fig. 5C and D). Thus, membrane-tethered WT prelamin A (endogenous) has progerin-like effects on Ran and SUMOylation.
Inhibition of nuclear SUMOylation disrupts the Ran protein gradient.
The correlation between SUMO2/3 levels and the Ran protein gradient in HGPS cells led us to explore whether there is a link between nuclear SUMOylation and the Ran protein gradient. We transfected HeLa cells with a catalytic mutant of Ubc9 (C93S mutant) that reduces SUMOylation by acting as a dominant negative protein (13), and we transfected the catalytic domain (CD) of the SUMO protease SENP2, which cleaves SUMO from target proteins (50). FLAG-tagged WT and C93S forms of Ubc9 and the FLAG-tagged SENP CD were introduced into HeLa cells and examined by double-label IF microscopy for FLAG and Ran or SUMO2/3 and Ran. The expression of FLAG-Ubc9 C93S and the FLAG-SENP CD reduced nuclear levels of Ran (Fig. 6 A). Cells with reduced levels of nuclear SUMO2/3 signal showed a loss of the Ran protein gradient (Fig. 6A). Thus, a reduction of the level of nuclear SUMOylation is sufficient to reduce the level of Ran in the nucleus.
RanGAP is one of the key regulators of the Ran system, and the SUMOylation of RanGAP regulates its anchorage to the NPC (45, 46). We examined whether RanGAP targeting to the NPC is disrupted by progerin and SENP CD expression, settings where SUMOylation is markedly reduced. In cells where the SENP CD and progerin reduced nuclear SUMOylation and disrupted the Ran protein gradient, RanGAP was still targeted to the NPC (Fig. 6B and C). This result together with the fact that RanGAP is localized to the NPC in HGPS patient cells (Kelley and Paschal, unpublished) indicate that the Ran protein gradient disruption in HGPS is not caused by a failure of RanGAP targeting to the NPC.
Defective nuclear localization of Ubc9 in progeria.
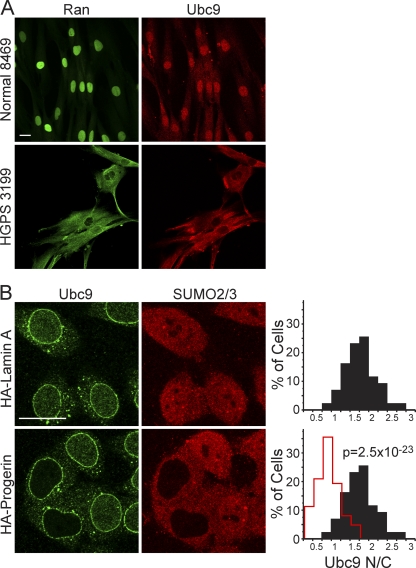
The reduced nuclear levels of SUMO2/3 caused by progerin expression (Fig. 4) led us to hypothesize that progerin inhibits the activity or localization of a component that is critical for SUMOylation. The sole E2 for SUMO conjugation is Ubc9, an enzyme found in both the nucleus and the cytoplasm (40, 82). In normal human fibroblasts, Ubc9 was mostly nuclear, with a small pool detected in the cytoplasm (Fig. 7 A). In progeria fibroblasts, however, Ubc9 was localized predominantly to the cytoplasm. Moreover, in cells where the distribution of Ubc9 was predominantly cytoplasmic, there was a strong disruption of the Ran gradient (HGPS 3199) (Fig. 7A and data not shown). The reduced nuclear localization of Ubc9 was linked to progerin expression by showing that HA-progerin transfection is sufficient to cause a quantitative reduction in the N/C distribution of endogenous Ubc9 in HeLa cells (Fig. 7B and data not shown).
Fig. 7.
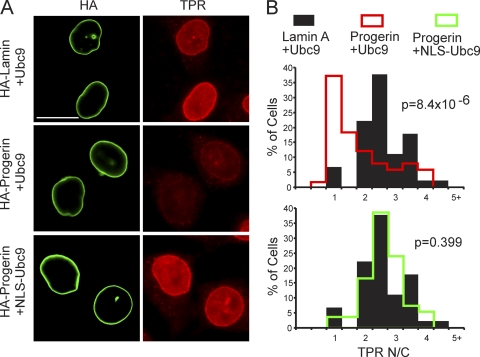
Reduced nuclear localization of Ubc9 in HGPS. (A) Endogenous Ran (green) and Ubc9 (red) in control (Normal 8469) and progeria (HGPS 3199) fibroblasts. (B) Localization of endogenous Ubc9 (green) and SUMO2/3 (red) in HeLa cells cotransfected with HA-WT lamin A and HA-progerin. Histograms (bin size = 0.25) of Ubc9 N/C ratios in HeLa cells expressing HA-lamin A (black bars; n = 43) and HA-progerin (red lines; n = 62). Scale bars, 20 μm.
Constitutive nuclear localization of Ubc9 restores the Ran gradient in cells expressing progerin.
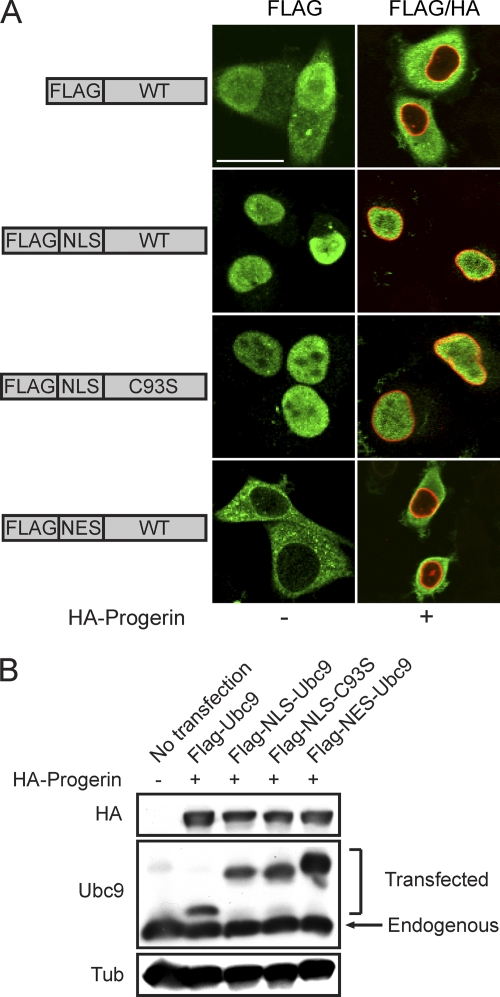
The overexpression of factors that reduce nuclear SUMOylation (SENP CD and dominant negative Ubc9) disrupted the Ran gradient (Fig. 6), suggesting a potential link between SUMOylation and the Ran system. Since progerin induced the cytoplasmic localization of Ubc9, we considered whether the loss of Ubc9 from the nucleus is responsible for the disruption of the Ran gradient in HGPS. We engineered Ubc9 with transport signal fusions (SV40 NLS and PKI NES) to force its subcellular localization to the nucleus and cytoplasm (Fig. 8 A); we then tested whether compartment-specific forms of Ubc9 could rescue the Ran gradient. We also introduced the C93S mutation into the NLS form of Ubc9 to address whether the catalytic function of Ubc9 is necessary for effects on the Ran gradient. Based on the transfection efficiency (∼25%), expression levels of FLAG-tagged forms of Ubc9 were approximately 2-fold that of endogenous Ubc9 (Fig. 8B). As expected, progerin inhibited the nuclear localization of transfected FLAG-tagged WT Ubc9; however, the SV40 NLS fusion rendered both the WT and the catalytically inactive forms of Ubc9 resistant to this effect of progerin (Fig. 8A). The PKI NES fusion with WT Ubc9 was exclusively cytoplasmic in the absence and presence of progerin (Fig. 8A).
Fig. 8.
Transport signal fusions that direct nuclear and cytoplasmic targeting of Ubc9. (A) FLAG-tagged SV40 NLS and PKI NES fusions with Ubc9 (green) were expressed in HeLa cells in the absence and presence of HA-progerin (red). Scale bar, 20 μm. (B) Immunoblotting showing the expression levels of the Ubc9 transport signal fusions relative to those of endogenous Ubc9.
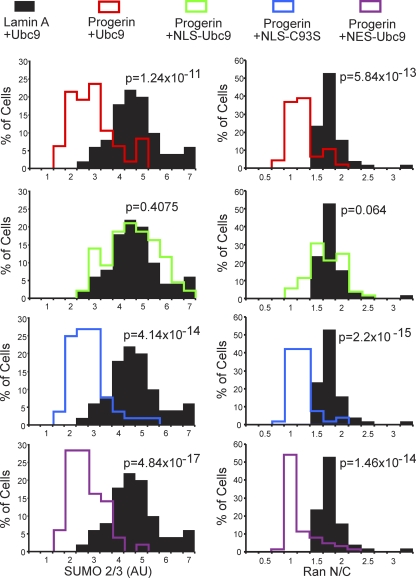
The four engineered forms of Ubc9 (Fig. 8) were each cotransfected with progerin into HeLa cells, and IF microscopy was used to measure Ran N/C levels and the nuclear levels of SUMO2/3. The controls for the experiment were WT Ubc9 cotransfected with WT lamin A (Fig. 9, black histograms) and WT Ubc9 cotransfected with progerin (red histograms). Progerin induced a quantitative reduction in the Ran N/C and SUMO2/3 levels (Fig. 9, red histograms), both of which were rescued by NLS-Ubc9 expression (green histograms). The catalytically inactive Ubc9 (C93S mutant) (blue histograms) targeted to the nucleus and cytoplasmic WT Ubc9 (NES fusion) (plum histograms) targeted to the cytoplasm both failed to restore the Ran N/C and nuclear SUMO2/3 levels in the presence of progerin (Fig. 9). These data show that the SUMO2/3 reduction and Ran gradient disruption by progerin (measured by IF microscopy) can be rescued by forcing Ubc9 into the nucleus and that the restoration of the Ran gradient requires the catalytic activity of Ubc9.
Fig. 9.
Forcing nuclear localization of Ubc9 rescues the Ran gradient in progerin-expressing cells. HeLa cells were cotransfected with HA-WT lamin A and HA-progerin, together with the transport signal fusions of Ubc9. Histograms show the nuclear levels of SUMO2 (bin size = 0.5) and Ran N/C levels (bin size = 0.25) in HA-positive cells expressing ectopic forms of Ubc9.
Constitutive nuclear localization of Ubc9 restores Ran-dependent transport of TPR.
To determine whether the Ubc9-mediated rescue of the Ran gradient in progerin-expressing cells also rescued Ran-dependent nuclear import, we analyzed the distribution of endogenous TPR in HeLa cells cotransfected with progerin, WT Ubc9, and NLS-Ubc9. The reduction in the nuclear import of TPR caused by progerin expression (Fig. 10 A, middle) was restored by the cotransfection of NLS-Ubc9 (Fig. 10A, bottom). By measuring the N/C ratios of TPR in cells expressing WT lamin A and WT Ubc9 (Fig. 10B, black histograms), progerin and WT Ubc9 (red histograms), and progerin and NLS-Ubc9 (green histograms), we determined that NLS-Ubc9 expression results in a quantitative increase in TPR import (Fig. 10B).
Fig. 10.
Nuclear localization of Ubc9 restores TPR import in cells expressing progerin. (A) IF localization of HA-progerin and endogenous TPR in cells cotransfected with Ubc9. Scale bar, 20 μm. (B) Histograms showing TPR N/C levels (bin size = 0.5) in HA-positive cells expressing engineered forms of Ubc9.
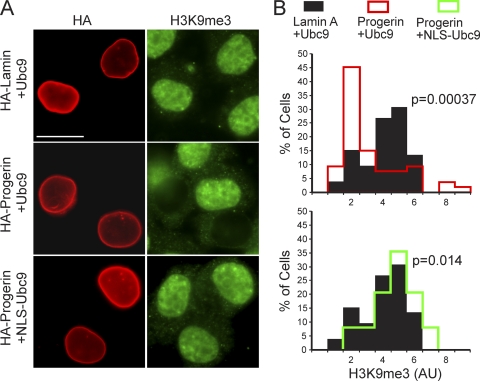
The correlation between the Ran gradient and nuclear levels of the chromatin mark H3K9me3 (Fig. 1) suggested that these pathways might be linked. We tested whether the restoration of the Ran gradient via NLS-Ubc9 could restore H3K9me3 levels. By IF microscopy, HA-progerin transfected into HeLa cells with WT Ubc9 caused a statistically significant reduction of H3K9me3 levels (Fig. 11 A, middle). Significantly, the cotransfection of HA-progerin with NLS-Ubc9 restored nuclear levels of H3K9me3 (Fig. 11A, bottom). Our data suggest that the reduced level of H3K9me3 induced by progerin expression is a consequence of either a Ran gradient disruption, a reduced level of Ubc9 function in the nucleus, or the combined effect of these changes.
Fig. 11.
Nuclear localization of Ubc9 restores H3K9me3 levels in cells expressing progerin. (A) IF localization of HA-progerin and nuclear H3K9me3 in cells cotransfected with Ubc9. Scale bar, 20 μm. (B) Histograms showing H3K9me3 levels (bin size = 1) in HA-positive cells expressing engineered forms of Ubc9.
Ran system defects require progerin attachment to the nuclear membrane.
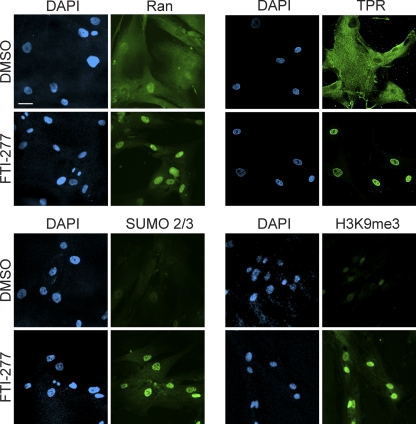
Farnesyl transferase inhibitors (FTIs) block the prenylation of progerin, which prevents its attachment to the inner nuclear membrane (81). Nuclear morphology defects in cultured cells and body weight gain in a mouse model of progeria are both improved by FTIs (11, 79). We tested whether the HGPS phenotypes described in this study are dependent on the constitutive attachment of progerin to the nuclear membrane. We treated HGPS 3199 cells with FTI-277 or vehicle for 3 days and subsequently examined Ran, TPR, SUMO2/3, and H3K9me3 by IF microscopy. The nuclear levels of these proteins and modifications were increased by FTI-277 treatment, indicating that these nuclear phenotypes are induced by the constitutive attachment of progerin to the inner nuclear membrane (Fig. 12). These observations are consistent with the data shown in Fig. 5.
Fig. 12.
Farnesylation is required for the inhibitory effects of progerin on Ran, TPR, SUMO2/3, and H3K9me3. Progeria fibroblasts (HGPS 3199) were treated with FTI-277 (3 μM) or DMSO (0.1%) for 72 h and examined by IF microscopy for the indicated proteins. Scale bar, 20 μm.
Progerin expression and SUMOylation alter RCC1-chromatin interactions in living cells.
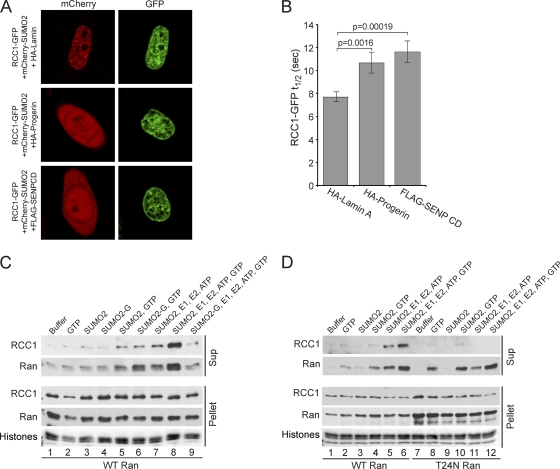
Several properties of RCC1 led us to examine whether progerin transduces its effects to the Ran gradient by modulating RCC1 activity: (i) RCC1 activity is required for the Ran protein gradient (60), (ii) RCC1 binds and dissociates from chromatin during nucleotide exchange on Ran (43), (iii) RCC1 binds preferentially to heterochromatin (12), and (iv) progerin reduces the levels of the heterochromatin marks H3K9me3 and H3K27me3. Additionally, RCC1 remains nuclear in progerin-expressing cells. We used fluorescence recovery after photobleaching (FRAP) to test whether progerin affects RCC1, as the nuclear mobility of RCC1 reflects its binding and dissociation from chromatin (41). We also used FRAP to test whether RCC1 mobility is sensitive to nuclear levels of SUMOylation by transfecting the FLAG-SENP2 CD. HeLa cells expressing HA-progerin and the FLAG-SENP CD were selected for FRAP analysis based on the distribution of cotransfected mCherry-SUMO2, which we found was nuclear in HA-lamin A-transfected cells but additionally localizes to the cytoplasm when cotransfected with HA-progerin and the FLAG-SENP CD (Fig. 13 A). In cells transfected with HA-progerin and the FLAG-SENP CD, the t1/2 values for the recovery of RCC1-GFP were increased by 38% and 50%, respectively (Fig. 13B). Because progerin is mostly restricted to the nuclear membrane but the FRAP measurements reflect RCC1 mobility throughout the nucleus, our data suggest that progerin-induced changes in the nuclear lamina are transduced to RCC1 throughout the nucleoplasm. Finally, the SENP-induced reduction in RCC1 mobility suggests that nuclear SUMOylation can regulate RCC1-chromatin dynamics.
Fig. 13.
SUMOylation regulates RCC1-chromatin interactions. (A) Localizations of mCherry-SUMO2 (red) and RCC1-GFP (green) in HeLa cells cotransfected HA-WT lamin A, HA-progerin, or the FLAG-SENP CD. (B) Nuclear mobility of RCC1-GFP measured by FRAP of cells expressing WT lamin A (n = 31), progerin (n = 24), or the SENP CD (n = 29). Error bars represent standard errors of the means (SEM). (C) SUMOylation-induced dissociation of RCC1 from chromatin. HeLa cell chromatin prebound with His-Ran was incubated with the combinations of recombinant factors indicated. Following a centrifugation step, RCC1 and Ran in supernatant and pellet fractions were analyzed by immunoblotting. Histones in the pellet fraction were visualized by Ponceau S staining. (D) The T24N form of Ran inhibits the SUMOylation-induced dissociation of RCC1 from chromatin. HeLa cell chromatin prebound with GST-Ran (WT and T24N) was treated as described above (C).
SUMOylation promotes RCC1 release from chromatin in vitro.
The current model for the nucleotide exchange cycle includes the formation of a Ran-RCC1-chromatin complex that is dissociated upon GTP binding to Ran (32, 43). The fact that SENP2 CD expression reduced RCC1 mobility (Fig. 13B) suggested that SUMOylation levels in the nucleus influence RCC1-chromatin interactions, which would be predicted to affect nucleotide exchange on Ran. We used a biochemical assay to examine whether SUMOylation affects the interaction between RCC1 and chromatin. Chromatin from HeLa cells was incubated with recombinant Ran (His tagged), which binds to the endogenous RCC1. Following two wash steps, the Ran-RCC1-chromatin complexes were incubated with recombinant factors that mediate SUMOylation (Fig. 13C). A maximal release of RCC1 from chromatin was observed in the presence of SUMO2, E1, E2 (Ubc9), ATP, and GTP (Fig. 13C, lane 8). The substitution of a nonconjugable form of SUMO2 (denoted SUMO2-G) (Fig. 13C, lane 9) or the omission of GTP from the reaction mixture (lane 7) reduced the amount of RCC1 released from chromatin, consistent with the interpretation that, in this assay, RCC1 release is regulated by SUMOylation and GTP binding to Ran. As an additional means of testing whether Ran release from RCC1 is necessary for the SUMOylation-dependent dissociation of RCC1 from chromatin, we performed the experiments using chromatin from HeLa cells prebound with recombinant WT and T24N forms of Ran (both GST tagged). The T24N mutant of Ran was used because it “locks” RCC1 into chromatin (41). The T24N mutant of Ran rendered endogenous RCC1 resistant to dissociation by in vitro SUMOylation (Fig. 13D, compare lanes 6 and 12). This finding is consistent with the possibility that nucleotide binding to Ran and dissociation from RCC1 could be linked to the SUMOylation-stimulated release of these proteins from chromatin. RCC1 itself does not appear to be SUMOylated under the conditions of this assay, leading us to posit that RCC1 dissociation is promoted by a chromatin-associated factor that is SUMOylated.
DISCUSSION
Mutant forms of lamin A are known to alter the structure and properties of the lamina (20), but an understanding of how defects in the lamina generate cellular effects, and particularly how laminopathy-specific phenotypes arise, has been enigmatic. In this study, we found that the mutant form of lamin A expressed in HGPS negatively affects the Ran GTPase system. The founding observation was that the ratio of the nuclear concentration to the cytoplasmic concentration (N/C ratio) of Ran is reduced in fibroblasts from progeria patients, an effect that is caused by progerin (and not simply by the cell passage number), because it can be recapitulated in HeLa cells. Progerin effects on the Ran system can be prevented in HGPS patient cells by FTI treatment, a clear indication that the constitutive attachment of progerin to the nuclear membrane initiates the disruptive mechanism. We also showed that lopinavir, a drug used to inhibit the HIV protease and with known activity against the lamin A-processing enzyme Zmpste24, has progerin-like effects on the Ran system. Thus, the cellular defects described in this study are clearly a consequence of the constitutive membrane attachment of lamin A.
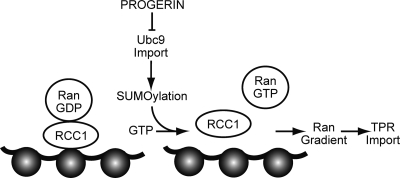
In the course of characterizing how progerin inhibits the Ran system, we made several observations that suggested that the SUMOylation machinery is affected in HGPS. Reduced nuclear levels of SUMO2/3 were observed by IF microscopy of HGPS cells and of HeLa cells expressing progerin, which led us to examine the distribution of the SUMO-conjugating enzyme Ubc9. Nuclear levels of Ubc9 were significantly reduced by progerin expression, suggesting that progerin inhibits Ubc9 import into the nucleus (Fig. 14). The progerin-induced reduction in nuclear Ubc9 levels was correlated with a disruption of the Ran gradient, and forcing Ubc9 into the nucleus rescued the Ran gradient. These observations, together with the fact that the overexpression of proteins that reduce nuclear SUMOylation (SENP CD and DN Ubc9) was sufficient to disrupt the Ran gradient, imply that Ubc9 and SUMOylation play a role in regulating the Ran gradient (Fig. 14). The model depicted could be drawn to include a positive-feedback loop, since Ubc9 is imported by importin 13, which is a Ran-regulated import receptor (46). A positive-feedback loop based on Ubc9 localization would certainly be consistent with the dominant negative effect of progerin on the Ran gradient. Progerin could disrupt the nuclear transport machinery by a mechanism that is not addressed in this study, with the cytoplasmic localization of Ubc9 occurring as a consequence. For example, progerin could alter the expression of a factor that regulates the Ran gradient, and the nuclear localization of Ubc9 might then rescue the Ran gradient via a gene expression mechanism.
Fig. 14.
Working model for how progerin disrupts the Ran gradient in HGPS. The expression of the progerin form of lamin A in HGPS inhibits Ubc9 import, which results in reduced levels of nuclear SUMOylation by SUMO2/3, reduced RCC1 function, and a disruption of the Ran protein gradient. Although classical importin β-dependent import is largely unaffected by progerin, the nuclear import of TPR is defective because of its sensitivity to the Ran protein gradient. TPR modulates multiple nuclear pathways, including those important for posttranscriptional gene regulation (25, 26, 31, 47, 66, 78), raising the possibility that the absence of TPR from the NPC might contribute to some of the gene expression changes associated with HGPS.
Reduced levels of nuclear Ubc9 were correlated with reduced SUMO2/3 levels but without a clear effect on SUMO1 levels. We speculate that this might reflect different turnover rates for SUMO1 and SUMO2/3 modification or progerin effects on SUMO paralog-specific SENPs or that nuclear Ubc9 is dispensable for SUMO1 modification in progerin-expressing cells because Ubc9 on the cytoplasmic side of the NPC performs the SUMO1 modification of proteins, which subsequently undergo nuclear import (59).
How does SUMOylation regulate the Ran gradient? The SUMOylation of RanGAP, which is required for targeting to the NPC (45, 46), is not inhibited by progerin expression. Rather, our data suggest that progerin-induced changes in SUMOylation that affect the Ran gradient could be mediated through RCC1, the chromatin-associated nucleotide exchange factor for Ran (Fig. 13). The data supporting this view include our finding that progerin and SENP CD expressions reduce the nuclear mobility of RCC1, which likely reflects a reduced rate of RCC1 dissociation from chromatin during its nucleotide exchange cycle for Ran. Both the production of RanGTP and the Ran protein gradient are strictly dependent on RCC1 (60, 72). Multiple lines of evidence support the model that RCC1 activity is coupled to a chromatin binding cycle. RCC1 exchange activity can be stimulated by nucleosomes and purified histones (51). By chromatin immunoprecipitation in yeast, RCC1 binding is strongly biased for heterochromatin (12), and by FRAP, RCC1 mobility is reduced in nuclei expressing a histone 2B phosphomimetic (Ser14Asp [76]). RCC1 mobility is also decreased by the Ran GTP binding mutant (T24N) that inhibits the dissociation of RCC1 from chromatin (43). These data suggest a model of exchange whereby RCC1 binds RanGDP, the nucleotide is released, and the Ran-RCC1 complex then binds chromatin. GTP binding to Ran would allow the release of both proteins from chromatin.
We showed that in vitro SUMOylation promotes the release of RCC1 and Ran from chromatin. As the released fractions of RCC1 and Ran failed to display a gel shift indicative of SUMOylation, we speculate that the target of SUMOylation that promotes the RCC1 dissociation is probably a chromatin-associated factor. It remains formally possible that RCC1 is directly SUMOylated but undergoes rapid deSUMOylation by chromatin-associated SENP activity. It also deserves mention that histones, known to bind to RCC1 and stimulate its exchange activity, are SUMOylated (64). We found that the SUMOylation-stimulated RCC1 release from chromatin in vitro was reduced by the addition of recombinant T24N Ran, a mutant defective for nucleotide binding. This result, along with the fact that in reaction mixtures containing WT Ran and SUMOylation machinery, RCC1 release was more efficient in the presence of GTP, leads us to suggest that SUMOylation might contribute to a step in the nucleotide exchange reaction.
The reduced nuclear transport of Ubc9 might explain some of the epigenetic changes associated with HGPS. SUMOylation by Ubc9 is thought to modulate the chromatin structure via the modification of histones and subunits of chromatin-remodeling enzymes in pathways linked to gene repression (67, 74, 80). The reduced levels of the repressive chromatin mark H3K9me3 in HGPS and in HeLa cells expressing progerin, and the restoration of H3K9me3 levels upon the forcing of Ubc9 into the nucleus, suggest that nuclear Ubc9 levels in HGPS might be insufficient for the maintenance of heterochromatin.
Mislocalization of TPR in progeria.
One of the changes in protein distribution in HGPS fibroblasts was the mislocalization of TPR to the cytoplasm. The TPR import pathway is clearly sensitive to the disruption of the Ran protein gradient because it can be recapitulated by progerin expression in HeLa cells and by reducing levels of the Ran import factor NTF2. The TPR import defect was also observed by using a reporter protein that contains the 56-amino-acid NLS. This result provides clear evidence that the DN effect of progerin on TPR localization reflects the inhibition of its import, possibly because the TPR import pathway is highly sensitive to nuclear Ran levels. TPR import is mediated by the importin α/β heterodimer (5). As importin β activity is only slightly reduced in HGPS cells, the TPR import defect might be caused by a reduced function of the TPR NLS or importin α, or some other step in the pathway. The fact that TPR import is dependent on a steep Ran protein gradient could be physiologically important and contribute to the temporal control of NPC assembly. TPR is the last nucleoporin imported into the nucleus during the reassembly of the NPC after mitosis, occurring after its NPC-anchoring protein Nup153 has already been incorporated into the nuclear side of the pore (8, 10). TPR import is delayed until the NPC diffusion barrier is formed, at which point an N/C gradient of Ran can be established. The notion that the progerin-mediated disruption of the Ran system affects the import of a subset of proteins is interesting in light of a previous study by Hernandez and colleagues, which showed that a mutant form of lamin A that lacks part of the Ig fold and fails to undergo proper processing induces progerin-like phenotypes in mice (35). This included a reduced nuclear localization of the transcription factor Lef1, which those authors showed is the basis of defective Wnt signaling in the model.
TPR and its orthologues have been linked to multiple nuclear processes, including protein export in vertebrates, RNA processing and export in yeast, dosage compensation in flies, and SUMOylation in diverse organisms (25, 26, 31, 47, 66, 78). The disruption of the nuclear basket has the potential to alter gene expression through posttranscriptional mechanisms that would not be detected by microarray analysis. Additionally, the nuclear basket helps exclude heterochromatin from the vicinity of the NPC, implying that the reduction or absence of TPR in HGPS cells would have an impact on chromatin organization at the nuclear periphery (39).
Phylogenetic considerations.
Yeasts lack a nuclear lamina and do not encode a protein with discernible homology to the intermediate filament superfamily. Our findings linking the nuclear lamina to SUMOylation and NPC assembly in mammalian cells may nonetheless parallel a pathway that has been described for S. cerevisiae. Heterochromatin in S. cerevisiae is tethered at the nuclear periphery by Sir proteins, which bind to the membrane-associated protein Esc1. It was shown previously that Esc1 is required for the proper formation of the nuclear basket of the NPC and for the regulation of the SUMO protease Ulp1 (42). In light of the fact that premature aging is caused by progerin expression, it is interesting that Sir proteins are involved in the regulation of the replicative life span in S. cerevisiae (37). In mice, Sirt1, which is regulated by SUMOylation (80), protects the cardiovascular system from stress-induced apoptosis (3). Notably, progeria patients die of cardiovascular complications at an average age of 13 years (34). These data together with the loss of heterochromatin in HGPS suggest that highly conserved pathways link architectural elements of the nuclear periphery (including the NPC) with the chromatin structure. By extension, nuclear transport and posttranslational modifications, including SUMOylation, might be important in the aging process in organisms ranging from yeasts to humans.
ACKNOWLEDGMENTS
We thank B. Burke, V. Cordes, M. Dasso, L. Gerace, D. Gilbert, I. Macara, D. Rekosh, D. Wotton, and Y. Zheng for their generous gifts of reagents. We also thank V. Cordes, T. Misteli, and especially D. Wotton for helpful discussions.
These studies were supported by the Progeria Research Foundation, the NIH, and the NSF.
Footnotes
Published ahead of print on 13 June 2011.
REFERENCES
- 1. Aebi U., Cohn J., Buhle L., Gerace L. 1986. The nuclear lamina is a meshwork of intermediate-type filaments. Nature 323:560–564 [DOI] [PubMed] [Google Scholar]
- 2. Akhtar A., Gasser S. M. 2007. The nuclear envelope and transcriptional control. Nat. Rev. Genet. 8:507–517 [DOI] [PubMed] [Google Scholar]
- 3. Alcendor R., et al. 2007. Sirt1 regulates aging and resistance to oxidative stress in the heart. Circ. Res. 100:1512–1521 [DOI] [PubMed] [Google Scholar]
- 4. Ayaydin F., Dasso M. 2004. Distinct in vivo dynamics of vertebrate SUMO paralogues. Mol. Biol. Cell 15:5208–5218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ben-Efraim I., Frosst P. D., Gerace L. 2009. Karyopherin binding interactions and nuclear import mechanism of nuclear pore complex protein Tpr. BMC Cell Biol. 10:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bischoff F. R., Klebe C., Kretschmer J., Wittinghofer A., Ponstingl H. 1994. RanGAP1 induces GTPase activity of nuclear Ras-related Ran. Proc. Natl. Acad. Sci. U. S. A. 91:2587–2591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bischoff F. R., Ponstingl H. 1991. Catalysis of guanine nucleotide exchange on Ran by the mitotic regulator RCC1. Nature 354:80–82 [DOI] [PubMed] [Google Scholar]
- 8. Bodoor K., et al. 1999. Sequential recruitment of NPC proteins to the nuclear periphery at the end of mitosis. J. Cell Sci. 112:2253–2264 [DOI] [PubMed] [Google Scholar]
- 9. Broers J., Ramaekers F., Bonne G., Yaou R., Hutchison C. 2006. Nuclear lamins: laminopathies and their role in premature ageing. Physiol. Rev. 86:967. [DOI] [PubMed] [Google Scholar]
- 10. Burke B., Ellenberg J. 2002. Remodelling the walls of the nucleus. Nat. Rev. Mol. Cell Biol. 3:487–497 [DOI] [PubMed] [Google Scholar]
- 11. Capell B. C., et al. 2005. Inhibiting farnesylation of progerin prevents the characteristic nuclear blebbing of Hutchinson-Gilford progeria syndrome. Proc. Natl. Acad. Sci. U. S. A. 102:12879–12884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Casolari J., et al. 2004. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell 117:427–439 [DOI] [PubMed] [Google Scholar]
- 13. Chakrabarti S. R., Nucifora G. 1999. The leukemia-associated gene TEL encodes a transcription repressor which associates with SMRT and mSin3A. Biochem. Biophys. Res. Commun. 264:871–877 [DOI] [PubMed] [Google Scholar]
- 14. Cheng J., Bawa T., Lee P., Gong L., Yeh E. T. 2006. Role of desumoylation in the development of prostate cancer. Neoplasia 8:667–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Coffinier C., et al. 2007. HIV protease inhibitors block the zinc metalloproteinase ZMPSTE24 and lead to an accumulation of prelamin A in cells. Proc. Natl. Acad. Sci. U. S. A. 104:13432–13437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Corbett A. H., Silver P. A. 1996. The NTF2 gene encodes an essential, highly conserved protein that functions in nuclear transport in vivo. J. Biol. Chem. 271:18477–18484 [DOI] [PubMed] [Google Scholar]
- 17. Cordes V. C., Hase M. E., Muller L. 1998. Molecular segments of protein Tpr that confer nuclear targeting and association with the nuclear pore complex. Exp. Cell Res. 245:43–56 [DOI] [PubMed] [Google Scholar]
- 18. Cordes V. C., Reidenbach S., Rackwitz H. R., Franke W. W. 1997. Identification of protein p270/Tpr as a constitutive component of the nuclear pore complex-attached intranuclear filaments. J. Cell Biol. 136:515–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cremer T., Cremer C. 2001. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat. Rev. Genet. 2:292–301 [DOI] [PubMed] [Google Scholar]
- 20. Dahl K. N., et al. 2006. Distinct structural and mechanical properties of the nuclear lamina in Hutchinson-Gilford progeria syndrome. Proc. Natl. Acad. Sci. U. S. A. 103:10271–10276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. D'Angelo M. A., Raices M., Panowski S. H., Hetzer M. W. 2009. Age-dependent deterioration of nuclear pore complexes causes a loss of nuclear integrity in postmitotic cells. Cell 136:284–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dechat T., et al. 2008. Nuclear lamins: major factors in the structural organization and function of the nucleus and chromatin. Genes Dev. 22:832–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dultz E., et al. 2008. Systematic kinetic analysis of mitotic dis- and reassembly of the nuclear pore in living cells. J. Cell Biol. 180:857–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Eriksson M., et al. 2003. Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome. Nature 423:293–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Frosst P., Guan T., Subauste C., Hahn K., Gerace L. 2002. Tpr is localized within the nuclear basket of the pore complex and has a role in nuclear protein export. J. Cell Biol. 156:617–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Galy V., et al. 2004. Nuclear retention of unspliced mRNAs in yeast is mediated by perinuclear Mlp1. Cell 116:63–73 [DOI] [PubMed] [Google Scholar]
- 27. Geiss-Friedlander R., Melchior F. 2007. Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell Biol. 8:947–956 [DOI] [PubMed] [Google Scholar]
- 28. Gorlich D., Seewald M. J., Ribbeck K. 2003. Characterization of Ran-driven cargo transport and the RanGTPase system by kinetic measurements and computer simulation. EMBO J. 22:1088–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gruenbaum Y., Margalit A., Goldman R. D., Shumaker D. K., Wilson K. L. 2005. The nuclear lamina comes of age. Nat. Rev. Mol. Cell Biol. 6:21–31 [DOI] [PubMed] [Google Scholar]
- 30. Han X., et al. 2008. Tethering by lamin A stabilizes and targets the ING1 tumour suppressor. Nat. Cell Biol. 10:1333–1340 [DOI] [PubMed] [Google Scholar]
- 31. Hang J., Dasso M. 2002. Association of the human SUMO-1 protease SENP2 with the nuclear pore. J. Biol. Chem. 277:19961–19966 [DOI] [PubMed] [Google Scholar]
- 32. Hao Y., Macara I. G. 2008. Regulation of chromatin binding by a conformational switch in the tail of the Ran exchange factor RCC1. J. Cell Biol. 182:827–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hay R. T. 2005. SUMO: a history of modification. Mol. Cell 18:1–12 [DOI] [PubMed] [Google Scholar]
- 34. Hennekam R. C. 2006. Hutchinson-Gilford progeria syndrome: review of the phenotype. Am. J. Med. Genet. A 140:2603–2624 [DOI] [PubMed] [Google Scholar]
- 35. Hernandez L., et al. 2010. Functional coupling between the extracellular matrix and nuclear lamina by Wnt signaling in progeria. Dev. Cell 19:413–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Izaurralde E., Kutay U., von Kobbe C., Mattaj I. W., Gorlich D. 1997. The asymmetric distribution of the constituents of the Ran system is essential for transport into and out of the nucleus. EMBO J. 16:6535–6547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kaeberlein M., McVey M., Guarente L. 1999. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 13:2570–2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kelley J. B., Paschal B. M. 2007. Hyperosmotic stress signaling to the nucleus disrupts the Ran gradient and the production of RanGTP. Mol. Biol. Cell 18:4365–4376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Krull S., et al. 2010. Protein Tpr is required for establishing nuclear pore-associated zones of heterochromatin exclusion. EMBO J. 29:1659–1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lee G. W., et al. 1998. Modification of Ran GTPase-activating protein by the small ubiquitin-related modifier SUMO-1 requires Ubc9, an E2-type ubiquitin-conjugating enzyme homologue. J. Biol. Chem. 273:6503–6507 [DOI] [PubMed] [Google Scholar]
- 41. Lerner E., et al. 1995. Ras CAAX peptidomimetic FTI-277 selectively blocks oncogenic Ras signaling by inducing cytoplasmic accumulation of inactive Ras-Raf complexes. J. Biol. Chem. 270:26802–26806 [DOI] [PubMed] [Google Scholar]
- 42. Lewis A., Felberbaum R., Hochstrasser M. 2007. A nuclear envelope protein linking nuclear pore basket assembly, SUMO protease regulation, and mRNA surveillance. J. Cell Biol. 178:813–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li H. Y., Wirtz D., Zheng Y. 2003. A mechanism of coupling RCC1 mobility to RanGTP production on the chromatin in vivo. J. Cell Biol. 160:635–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lyman S. K., Guan T., Bednenko J., Wodrich H., Gerace L. 2002. Influence of cargo size on Ran and energy requirements for nuclear protein import. J. Cell Biol. 159:55–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mahajan R., Delphin C., Guan T., Gerace L., Melchior F. 1997. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell 88:97–107 [DOI] [PubMed] [Google Scholar]
- 46. Matunis M. J., Coutavas E., Blobel G. 1996. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J. Cell Biol. 135:1457–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mendjan S., et al. 2006. Nuclear pore components are involved in the transcriptional regulation of dosage compensation in Drosophila. Mol. Cell 21:811–823 [DOI] [PubMed] [Google Scholar]
- 48. Mingot J., Kostka S., Kraft R., Hartmann E., Görlich D. 2001. Importin 13: a novel mediator of nuclear import and export. EMBO J. 20:3685–3694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Misteli T. 2007. Beyond the sequence: cellular organization of genome function. Cell 128:787–800 [DOI] [PubMed] [Google Scholar]
- 50. Mukhopadhyay D., Dasso M. 2007. Modification in reverse: the SUMO proteases. Trends Biochem. Sci. 32:286–295 [DOI] [PubMed] [Google Scholar]
- 51. Nemergut M. E., Mizzen C. A., Stukenberg T., Allis C. D., Macara I. G. 2001. Chromatin docking and exchange activity enhancement of RCC1 by histones H2A and H2B. Science 292:1540–1543 [DOI] [PubMed] [Google Scholar]
- 52. Ohtsubo M., Okazaki H., Nishimoto T. 1989. The RCC1 protein, a regulator for the onset of chromosome condensation locates in the nucleus and binds to DNA. J. Cell Biol. 109:1389–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Oki M., Nishimoto T. 1998. A protein required for nuclear-protein import, Mog1p, directly interacts with GTP-Gsp1p, the Saccharomyces cerevisiae ran homologue. Proc. Natl. Acad. Sci. U. S. A. 95:15388–15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Park M., et al. 1986. Mechanism of met oncogene activation. Cell 45:895–904 [DOI] [PubMed] [Google Scholar]
- 55. Paschal B. M., Fritze C., Guan T., Gerace L. 1997. High levels of the GTPase Ran/TC4 relieve the requirement for nuclear protein transport factor 2. J. Biol. Chem. 272:21534–21539 [DOI] [PubMed] [Google Scholar]
- 56. Pemberton L. F., Paschal B. M. 2005. Mechanisms of receptor-mediated nuclear import and nuclear export. Traffic 6:187–198 [DOI] [PubMed] [Google Scholar]
- 57. Pendas A. M., et al. 2002. Defective prelamin A processing and muscular and adipocyte alterations in Zmpste24 metalloproteinase-deficient mice. Nat. Genet. 31:94–99 [DOI] [PubMed] [Google Scholar]
- 58. Pichler A., Gast A., Seeler J. S., Dejean A., Melchior F. 2002. The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell 108:109–120 [DOI] [PubMed] [Google Scholar]
- 59. Pichler A., Melchior F. 2002. Ubiquitin-related modifier SUMO1 and nucleocytoplasmic transport. Traffic 3:381–387 [DOI] [PubMed] [Google Scholar]
- 60. Ren M., Drivas G., D'Eustachio P., Rush M. 1993. Ran/TC4: a small nuclear GTP-binding protein that regulates DNA synthesis. J. Cell Biol. 120:313–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ribbeck K., Lipowsky G., Kent H., Stewart M., Görlich D. 1998. NTF2 mediates nuclear import of Ran. EMBO J. 17:6587–6598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ris H. 1989. Three-dimensional imaging of cell ultrastructure with high resolution low voltage SEM. Inst. Phys. Conf. Ser. 98:657–662 [Google Scholar]
- 63. Saitoh H., Hinchey J. 2000. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J. Biol. Chem. 275:6252–6258 [DOI] [PubMed] [Google Scholar]
- 64. Scaffidi P., Misteli T. 2005. Reversal of the cellular phenotype in the premature aging disease Hutchinson-Gilford progeria syndrome. Nat. Med. 11:440–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Schnitzler G. R. 2000. Isolation of histones and nucleosome cores from mammalian cells. Curr. Protoc. Mol. Biol. 21:1–215. [DOI] [PubMed] [Google Scholar]
- 66. Shah S., Tugendreich S., Forbes D. 1998. Major binding sites for the nuclear import receptor are the internal nucleoporin Nup153 and the adjacent nuclear filament protein Tpr. J. Cell Biol. 141:31–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Shiio Y., Eisenman R. N. 2003. Histone sumoylation is associated with transcriptional repression. Proc. Natl. Acad. Sci. U. S. A. 100:13225–13230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Shumaker D. K., et al. 2006. Mutant nuclear lamin A leads to progressive alterations of epigenetic control in premature aging. Proc. Natl. Acad. Sci. U. S. A. 103:8703–8708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Smith A., Brownawell A., Macara I. G. 1998. Nuclear import of Ran is mediated by the transport factor NTF2. Curr. Biol. 8:1403–1406 [DOI] [PubMed] [Google Scholar]
- 70. Smith E. D., Kudlow B. A., Frock R. L., Kennedy B. K. 2005. A-type nuclear lamins, progerias and other degenerative disorders. Mech. Ageing Dev. 126:447–460 [DOI] [PubMed] [Google Scholar]
- 71. Soop T., et al. 2005. Nup153 affects entry of messenger and ribosomal ribonucleoproteins into the nuclear basket during export. Mol. Biol. Cell 16:5610–5620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Tachibana T., Imamoto N., Seino H., Nishimoto T., Yoneda Y. 1994. Loss of RCC1 leads to suppression of nuclear protein import in living cells. J. Biol. Chem. 269:24542–24545 [PubMed] [Google Scholar]
- 73. Tran E. J., Wente S. R. 2006. Dynamic nuclear pore complexes: life on the edge. Cell 125:1041–1053 [DOI] [PubMed] [Google Scholar]
- 74. Uchimura Y., et al. 2006. Involvement of SUMO modification in MBD1- and MCAF1-mediated heterochromatin formation. J. Biol. Chem. 281:23180–23190 [DOI] [PubMed] [Google Scholar]
- 75. Walther T. C., et al. 2001. The nucleoporin Nup153 is required for nuclear pore basket formation, nuclear pore complex anchoring and import of a subset of nuclear proteins. EMBO J. 20:5703–5714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Wong C. H., et al. 2009. Apoptotic histone modification inhibits nuclear transport by regulating RCC1. Nat. Cell Biol. 11:36–45 [DOI] [PubMed] [Google Scholar]
- 77. Worman H., Bonne G. 2007. “Laminopathies”: a wide spectrum of human diseases. Exp. Cell Res. 313:2121–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Xu X. M., et al. 2007. Nuclear pore anchor, the Arabidopsis homolog of Tpr/Mlp1/Mlp2/megator, is involved in mRNA export and SUMO homeostasis and affects diverse aspects of plant development. Plant Cell 19:1537–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Yang S. H., Qiao X., Fong L. G., Young S. G. 2008. Treatment with a farnesyltransferase inhibitor improves survival in mice with a Hutchinson-Gilford progeria syndrome mutation. Biochim. Biophys. Acta 1781:36–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Yang Y., et al. 2007. SIRT1 sumoylation regulates its deacetylase activity and cellular response to genotoxic stress. Nat. Cell Biol. 9:1253–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Young S. G., Fong L. G., Michaelis S. 2005. Prelamin A, Zmpste24, misshapen cell nuclei, and progeria—new evidence suggesting that protein farnesylation could be important for disease pathogenesis. J. Lipid Res. 46:2531–2558 [DOI] [PubMed] [Google Scholar]
- 82. Zhang H., Saitoh H., Matunis M. J. 2002. Enzymes of the SUMO modification pathway localize to filaments of the nuclear pore complex. Mol. Cell. Biol. 22:6498–6508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Zhang X. D., et al. 2008. SUMO-2/3 modification and binding regulate the association of CENP-E with kinetochores and progression through mitosis. Mol. Cell 29:729–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Zhao X., Wu C. Y., Blobel G. 2004. Mlp-dependent anchorage and stabilization of a desumoylating enzyme is required to prevent clonal lethality. J. Cell Biol. 167:605–611 [DOI] [PMC free article] [PubMed] [Google Scholar]